Abstract
Cyclic nucleotide-gated channel 5 (CNGC5), CNGC6, and CNGC9 (CNGC5/6/9 for simplicity) control Arabidopsis root hair (RH) growth by mediating the influx of external Ca2+ to establish and maintain a sharp cytosolic Ca2+ gradient at RH tips. However, the underlying mechanisms for the regulation of CNGCs remain unknown. We report here that calcium dependent protein kinase 1 (CPK1) directly activates CNGC5/6/9 to promote Arabidopsis RH growth. The loss-of-function mutants cpk1-1, cpk1-2, cngc5-1 cngc6-2 cngc9-1 (shrh1/short root hair 1), and cpk1 shrh1 show similar RH phenotypes, including shorter RHs, more RH branching, and dramatically attenuated cytosolic Ca2+ gradients at RH tips. The main CPK1-target sites are identified as Ser20, Ser27, and Ser26 for CNGC5/6/9, respectively, and the corresponding alanine substitution mutants fail to rescue RH growth in shrh1 and cpk1-1, while phospho-mimic versions restore the cytosolic Ca2+ gradient at RH apex and rescue the RH phenotypes in the same Arabidopsis mutants. Thus we discover the CPK1-CNGC modules essential for the Ca2+ signaling regulation and RH growth in Arabidopsis.
Similar content being viewed by others
Introduction
The plant root system is essential for anchorage and the acquisition of nutrient ions and water, and forms an important interface with soil microbes1,2,3. Root hairs (RHs), the tubular-shaped structures derived from the outgrowth of root epidermal cells, play essential roles in these processes1,2,3. RHs adhere to the surface of soil particles and release adhensive exudates that aid root penetration into the soil4,5,6,7,8. It has been also reported that the roots of English ivy (Hedera helix) exude a yellowish mucilage that promotes the capacity of the plant to climb vertical surfaces, and RHs play an important role in the plant climbing9. RHs greatly increase the area of the root-soil interface and so play a major role in the uptake of nutrients and water, and, along with root system architecture, largely determine the volume of the root-soil interface that plants can exploit. An early study reported that the RH interface of a two-week old winter rye (Secale cereale) plant is approximately 4321 square feet (401 square meter), which is almost twice that of the remaining root surface3,10. The diameter of the RH cylinder around the root is approximately ten times larger than that of the root, and thus the volume of the RH cylinder is about 100 times larger than that of the root3,10.
The essential role that RHs play in the uptake of water and nutrient ions from the soil, including the macronutrients nitrate, potassium, phosphate and calcium, and micronutrients, is indisputable4,6,11,12. This is reflected by the strong effect of ion nutrient availability (deficiency or sufficiency) on RH density, RH length, and plant biomass13,14,15,16,17,18. Moreover, impairment of RH growth results in the decreased uptake of nutrient ions and water, and consequently reduces the tolerance of plants to abiotic stresses and results in the reduction of plant biomass4,6,11,12,19. Thus, the ability of the root system to take up water and important nutrient ions, especially immobile ions such as phosphate, can be significantly increased by promoting RH growth. This is extremely important for the breeding of new crop cultivars with enhanced performance in less fertile soils3.
RHs are distributed in the differentiation zone of the root. In Arabidopsis, the diameter of RHs is approximate 10 µm, and RHs can grow to approximate 1 mm in length at rate of more than 1 µm per min at a fixed angle of about 85° relative to the root surface20. The development of RHs can be roughly categorized into three steps: cell-fate determination of the root epidermal cells, the initiation of RHs, and subsequent elongation through tip growth. The epidermal cells of roots are determined to be either hair (H) cells (trichoblasts) or non-hair (N) cells (atrichoblasts), and this process follows a strict cell position-dependent pattern. H cells are present over the intercellular space between two underlying cortex cells, and N cells are present over a single cortex cell20. The cell-position-dependent pattern for cell-fate determination of root epidermis is mainly controlled genetically by an intrinsic regulating network21,22, and GLABRA2 (GL2), a homeodomain transcription factor protein, functions as a central component of the regulating network21,22. GL2 is mainly expressed in the epidermal cells adopting an N cell-fate21, and loss of GL2 results in the production of excessive RHs21,22. Thus GL2 is a positive regulator of N cell-fate differentiation and a negative regulator of H cell-fate differentiation in the epidermis. The expression of GL2 is positively regulated by a number of upstream transcription factors, including TRANSPARENT TESTA GLABRA (TTG)23,24, the basic helix-loop-helix (bHLH) transcription factors GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3)25,26, and the R2R3-MYB transcription factors WEREWOLF (WER) and MYB2327,28. WER, GL3, EGL3, and TTG form a complex with WER as the central component. By contrast, CAPRICE (CPC) and its redundant partners positively regulate H cell-fate differentiation by negatively regulating the WER-GL3-EGL3-TTG pathway. CPC binds to WER in a competitive manner to prevent the binding of GL3/EGL3 to WER29,30,31,32. Moreover, CPC is capable of traveling from one epidermal cell to another to repress the N cell-fate32,33. In addition to this network governing epidermal cell-fate determination, a number of other genes are also involved in this process, acting either independently or through interactions with the network components20.
Once H cell fate is determined, RH growth is initiated. The initiation of RHs is a process of cell-wall loosening and bulge/swelling formation34,35,36. The acidification of the apoplastic space and the production of reactive oxygen species (ROS) facilitate cell wall loosening for the RH initiation34,35,36. ROS production during RH initiation is mainly catalyzed by the RESPIRATORY BURST OXIDASE HOMOLOG C/ROOT HAIR DEFECTIVE2 (RBOHC/RHD2)37. The ROS then triggers external Ca2+ influx by activating the Ca2+-permeable channel Annexin 1 to elevate the cytosolic Ca2+ concentration in the bulges38. The accumulation of ROS also facilitates the softening of the cell wall in the bulges to allow the turgor pressure-driven expansion of the bulge for RH tip growth39,40. Thus ROS and Ca2+ form a positive regulatory loop for the RH initiation and tip growth41.
RHs grow uni-directionally by depositing cell wall and plasma membrane (PM) materials at their apex via exocytosis, and, similar to pollen tubes, RHs use turgor pressure as the driving force to expand their volume and surface at RH tips42,43. This process is referred to as tip growth. The tip growth of RHs is regulated by phytohormones, including auxin, ethylene, jasmonate (JA), abscisic acid (ABA), gibberellin (GA), strigolactones (SL), cytokinin (CK), and brassinosteroids (BR)44. RH growth is also affected by changes in environmental conditions, such as the availability of nutrients and water, and interactions with soil microbes3. These factors regulate RH tip growth through intrinsic regulatory machinery through changes in Ca2+, ROS, pH, the cytoskeleton, ROPs etc.44
External Ca2+ influx occurs during RH initiation37, and the influx of Ca2+ at the bulges of trichoblasts forms a Ca2+ gradient at their apex, where the cytosolic Ca2+ concentration can reach about 1 µM and decreases to about 0.1 µM at the base of RHs45,46. The Ca2+ gradient at the RH apex oscillates with a frequency of 2-4 peaks per min, as does the tip growth of RHs, but the Ca2+ oscillation lags the RH growth oscillation by a few seconds47,48. The cytosolic Ca2+ gradient at the RH apex functions as a key regulator of both RH tip growth and orientation and eventually vanishes when tip growth ceases45,48,49. During RH growth, the cytosolic Ca2+ oscillation is highly coordinated with and interacts to other regulating factors, including ROS, pH, ROPs, and cytoskeleton, suggesting a central role for Ca2+ as a signal in RH growth regulation. Several ROPs function as PM-localized switches for the tip growth of RHs and pollen tubes50,51,52,53. The activated ROPs are involved in RH growth through their regulation of exocytosis and endocytosis, changes in the cytoskeleton, ROS production, and Ca2+ signaling39,54,55,56,57,58. ROS are known as signaling molecules, and polarized ROS production is required for RH tip growth37. The knockout mutant rhd2/rbohc failed to accumulate ROS, and developed only very short RHs37. Further analysis revealed that the ROP2-RHD2-ROS cascade is involved in RH tip growth regulation by activating PM Ca2+ channels and external Ca2+ influx37,39. The elevation of cytosolic Ca2+ can also stimulate ROS production through the activation of RBOHC/RHD241. Thus, ROS and Ca2+ form a positive regulatory loop in RHs. F-actin and microtubules are involved in the movement of organelles and the nucleus along the cytoskeleton for their proper positioning to support RH growth59,60,61,62. Ca2+ signaling regulates cytoskeletal dynamics via Ca2+-binding proteins, and the elevation of cytosolic Ca2+ facilitates the de-polymerization of F-actin filaments and microtubules63.
Despite the central role of the cytosolic Ca2+ in RH growth, the underlying mechanism by which the Ca2+ gradient is retained and regulated for RH growth is still largely unknown. It has been well established that external Ca2+ influx is mainly focused at RH tips rather than the shank area, and consequently the cytosolic Ca2+ gradient is established and retained at RH apex for RH growth45,64,65,66. Thus, the Ca2+ channels responsible for the external Ca2+ influx into RHs were thought to be localized mainly at RH tips. Later on, electrophysiological analysis provided evidence for the presence of the PM Ca2+-permeable channels at RH tips37,67. Ca2+-permeable channels belong to a few families, including cyclic nucleotide-gated channels (CNGCs), glutamate receptor-like channels (GLRs), and Annexins, all of which are candidates to mediate RH tip growth. However, the PM Ca2+ channels involved in RH growth remained undiscovered for decades. Recently, three CNGCs, namely CNGC5, CNGC6, and CNGC9 (CNGC5/6/9 for simplicity), were revealed to be the PM Ca2+ channels essential for the tip growth of Arabidopsis RHs, and the triple mutant cngc5-1 cngc6-2 cngc9-1 designated as short root hair1 (shrh1) and the quadruple mutant cngc5-1 cngc6-2 cngc9-1 cngc14-1 designated as short root hair 2 (shrh2) showed similar defects in RH growth50,51. The subcellular localization of the CNGC5/6/9 at RH tips has been clearly demonstrated50,51. These studies demonstrate that the CNGCs are the main Ca2+ channels that mediate RH growth48,49. Another study revealed that CNGC14 is essential for the touch sensing of RHs because the knockout mutant cngc14 showed RH defects only in a condition that RHs grow in solid medium68. However, it is still unknown how these CNGCs are regulated to control the tip-focused Ca2+ gradient and RH tip growth.
The Arabidopsis genome contains 20 CNGC members. Each CNGC has six transmembrane domains (S1-S6) with a pore-forming loop between S5 and S6, a short N terminus, and a long C terminus with a cyclic nucleotide-binding domain (CNBD) and a calmodulin (CaM)-binding domain (CaMBD), similar to their mammalian orthologs cyclic nucleotide-gated (CNG) channels69,70. Both mammalian CNGs and plant CNGCs can form PM Ca2+ channels as either homo- or hetero-tetramers70,71,72. Mammalian CNGs are activated by the binding of cyclic nucleotides (cAMP and cGMP)70, but no activation of plant CNGCs by cyclic nucleotides was observed in Xenopus laevis oocytes72. Thus, it appears that the cyclic nucleotides may not be upstream regulators of CNGCs. On the other hand, protein phosphorylation/dephosphorylation is a general regulatory mechanism for regulating the activity of diverse ion channels. For instance, the Ca2+-permeable channels Annexin1 and CNGCs can be activated by OPEN STOMATA 1(OST1)-mediated phosphorylation in response to cold and drought stress/abscisic acid (ABA), respectively73,74. The anion channels slow anion channel associated 1 (SLAC1), SLAC1 homologues (SLAHs), and K+ channels from Shaker family can be phosphorylated by OST1 and calcium-dependent protein kinase (CPK/CDPK) in guard cells for stomatal movement regulation75,76,77,78. Thus, the CNGCs are very likely regulated by kinase- and phosphatase-mediated protein phosphorylation and dephosphorylation, and the kinases from diverse protein kinase families, including CPK/CDPK, calcineurin B-like (CBL) protein-interacting protein kinase (CIPK), SNF1-related protein kinase 2 (SnRK2), and mitogen-activated protein kinase (MAPK) cascade, are attractive candidates.
In this research, we screened an Arabidopsis protein kinase mutant library for RH mutants. This led to the identification of the loss-of-function mutants cpk1-1 and cpk1-2 as having shrh1-like RH defects. We found that CPK1 positively regulates RH growth by activating CNGC5/6/9 through protein phosphorylation at a conserved serine within the N terminal region of the three CNGCs. We found that the expression of phospho-mimic CNGC5/6/9 with an S-to-D point mutation at the main CPK1-target sites is sufficient to rescue the RH phenotypes of cpk1 and shrh1, revealing a CPK1-CNGC signaling module that is essential for RH growth, and shedding light on the molecular mechanism underlying the regulation of cytosolic Ca2+ in this process.
Results
CPK1 is required for RH growth in Arabidopsis
Cyclic nucleotides are unlikely to be upstream regulators of CNGCs because they are unable to activate CNGCs in Xenopus leavis oocytes72. We then focused on protein kinases as potential regulators of CNGCs in RHs.
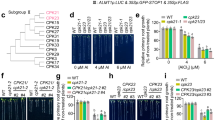
To determine whether any kinases are essential for RH growth, we screened a kinase mutant library that contains mutants from most protein kinase families, including CPKs, SnRK2s, MAP kinases, and CBL-CIPKs, by analyzing RH phenotypes (Supplementary Table 1). Columbia (Col-0) wild-type plants and the triple mutant shrh1, the latter which shows significant defects in RH growth48, were used as controls. Four-day old seedlings grown in plates containing solid half-strength Murashige and Skoog (1/2 MS) medium were analyzed. We performed optical sectioning under bright field, and each set of sectioning images were merged into a single 2-D image. Most RHs growing at different angles and layers can be observed clearly in the merged 2-D images, so we used them for RH phenotype analysis. We found that the T-DNA insertion mutants cpk1-1 and cpk1-2 showed significant defects in RH growth, including shorter RHs (Fig. 1a–c) and more RH branching (Fig. 1b, d), but their RH density was normal (Fig. 1a, e), compared to the wild type. Interestingly the RH-related phenotypes in cpk1-1 and cpk1-2 were similar to that of shrh1 (Fig. 1a–e).
4-5 day-old seedlings growing in Petri dishes were used for RH phenotype analysis. A set of optical section images of the RHZ of the seedlings with a 22.5 µm sectioning step and a sectioning speed of 5 steps/sec were captured under a stereo microscope at room temperature (25 ± 1 °C), and the set of images were automatically and immediately merged into a 2-D picture for each Petri dish. The 2-D pictures were used for RH phenotype analysis. For the time-lapse analysis of RH elongation rate and growth time, the optical sectioning was performed every min for 300 min for each dish, and the merged 2-D photos were used to analyze the RH elongation rate and time. a, b Typical 2-D photos derived from optical sectioning images of the RHZ of seedlings (a) and individual RHs showing the typical RH shapes (b). c–g Statistical analyses of RH length (c), RH branching rates (d), RH density (e), RH elongation rate (f), and RH elongation time (g). Scale bars, 0.2 mm in (a), and 10 μm in (b). c–e The numbers of biologically independent roots with approximate 50 RHs per root examined in this study are 11, 14, 10, 15, 11, 11, 14, 12, 17, and 12 for RH length (c), 12, 14, 11, 10, 10, 11, 10, 13, 15, and 12 for RH branching (d), and 10, 10, 11, 13, 10, 12, 13, 12, 13, and 9 for RH density (e), for the 10 Arabidopsis lines as shown from left to right in each panel. f, g n = 20 roots for both WT and cpk1-1. Samples with different letters are significantly different with P < 0.05 (one-way ANOVA) in (c–e), and P < 0.01 (two-tailed Student’s t-test) in (f, g). Data are presented as means ± SEM. Source data are provided as a Source Data file.
To monitor RH growth, we performed a time-lapse analysis for no less than 300 min. The data showed that the RHs of cpk1-1 grew at a significantly lower speed (Supplementary Fig. 1a, b; Fig. 1f), and also ceased growth earlier (Supplementary Fig. 1a, b; Fig. 1g), compared to that of wild type. These phenotypes resemble the phenotypes of shrh1 as previously reported48.
We conducted RT-qPCR experiments, and the results showed that CPK1 was knocked out in the mutants cpk1-1 and cpk1-2 (Supplementary Fig. 2a). We generated complemented (COM) lines by expressing wild type CPK1 under its native promoter in the mutant cpk1-2 background, and two COM lines pCPK1-COM1 (CPK1 COMPLEMENTATION 1 under CPK1 native promoter in cpk1-2) and pCPK1-COM5 were selected for further analysis. The RT-qPCR data showed that the CPK1 was expressed in pCPK1-COM1 and pCPK1-COM5 at a level similar to that of wild type plants (Supplementary Fig. 2a). RH phenotype analysis showed that the defects in RH growth were successfully rescued in pCPK1-COM1 and pCPK1-COM5 compared to cpk1-2 and wild type (Fig. 1a, c, d). We further generated overexpression (OE) lines by overexpressing wild type CPK1 under a Ubiquitin 10 (UBQ10) promoter in the background of the wild type plants, and two OE lines CPK1-OE6 (CPK1 OVEREXPRESSION 6 in wild type background) and CPK1-OE13 were selected for RH phenotype analysis. Results from RT-qPCR showed that the expression of CPK1 in the two OE lines was increased by approximately 7-10 fold compared to that of wild type plants (Supplementary Fig. 2a). We analyzed the RH phenotypes, and observed significant longer RHs in the two OE lines compared to that in the wild type (Fig. 1a, c). We rarely observed branching RHs (Fig. 1d), and the RH density was not obviously altered (Fig. 1a, e), compared to that of wild type plants.
To further analyze the functions of CPK1 in RHs, we generated transgenic Arabidopsis lines by expressing wild type CPK1 under a RH-specific promoter EXPANSIN A7 (EXPA7)79 in the background of cpk1-1. Two transgenic lines pEXPA7-COM13 (CPK1 COMPLEMENTATION 13 under the promoter pEXPA7 in the background of cpk1-1) and pEXPA7-COM14 were selected for further experiments. RT-qPCR analysis was performed. The data showed that the CPK1 was expressed in pEXPA7-COM13 and pEXPA7-COM14 in a level similar to that in the wild type plants (Supplementary Fig. 2a). We analyzed the RH phenotypes in the transgenic lines, and found that the RH length was restored to levels similar to that of wild type plants (Fig. 1a, c), the RH branching rates were dramatically reduced from more than 20% in cpk1-1 to less than 0.5% in the two transgenic lines (Fig. 1d), and the RH density was not obviously altered in the pEXPA7-COM13 and pEXPA7-COM14 lines compared to cpk1-1 and the wild type (Fig. 1a, e). These results together suggest that CPK1 plays an important role in RH growth.
CPK1 is highly expressed in RHs, and CPK1 is localized in the periphery of RHs
To investigate the expression profile of CPK1 in Arabidopsis, we generated transgenic Arabidopsis lines containing the GUS gene under the control of the native promoter of CPK1 in the wild type background. GUS staining results showed that CPK1 is highly expressed in RHs and the xylem of primary roots (Supplementary Fig. 3a).
A transgenic line, pEXPA7-COM13 with eGFP fused to the C terminus of CPK1, was generated to study the subcellular localization of CPK1 in RHs. Strong eGFP fluorescence in the apexes of most RHs was observed (Supplementary Fig. 3b). We further monitored the subcellular localization of CPK1 in RHs, and the optical sections of individual RHs with 2 µm sectioning steps showed that CPK1 was mainly distributed in the periphery of RH cells, especially at the RH apex (Supplementary Fig. 3c), consistent with a potential role in RH growth.
CPK1 functions upstream of CNGC5/6/9 for RH growth
We hypothesized that CPK1 may function as a direct upstream regulator of the CNGCs in RHs. To test this hypothesis, we generated cpk1-1 cngc5-1, cpk1-1 cngc6-2, and cpk1-1 cngc9-1 double mutants and analyzed their RH phenotype. We observed strong defects of RH growth in the cpk1-1 mutant (Supplementary Fig. 4a–c), while the single mutants cngc5-1, cngc6-2, and cngc9-1 had wild type-like RHs (Supplementary Fig. 4a–d). The wild type-like RH growth observed in the three single cngc mutants is consistent with a previous report48. However, we observed cpk1-1-like defects of RH growth in the cpk1-1 cngc5-1, cpk1-1 cngc6-2, and cpk1-1 cngc9-1 double mutants, including shorter RHs and higher RH branching frequencies (Supplementary Fig. 4a–c). The RH density was not obviously altered in the double mutants compared to the wild type (Supplementary Fig. 4a, d).
We next generated a cpk1 shrh1 quadruple mutant by crossing the single mutant cpk1-1 to the triple mutant shrh1 (cngc5-1 cngc6-2 cngc9-1)48. We analyzed the RH phenotypes, and found that cpk1-1, shrh1, and cpk1 shrh1 showed similarly strong RHs phenotypes with shorter RHs and more branches than wild type (Fig. 2a–c). Like the cpk1-1 and shrh1 mutants, the RH density was not obviously altered in the quadruple mutant cpk1 shrh1 (Fig. 2a, d). This data demonstrates that the effects of the mutations in CPK1 and CNGC5/6/9 on RH growth are not additive, suggesting that CPK1 and CNGC5/6/9 function in the same signaling pathway in Arabidopsis RHs.
A set of optical sectioning images of the RHZ of the 4-5 day-old seedlings with a 22.5 µm sectioning step and a sectioning speed of 5 steps per sec were captured under a stereo microscope at room temperature (25 ± 1 °C), and the images were automatically and immediately merged into a 2-D picture for each Petri dish. The merged 2-D pictures were used for RH phenotype analysis. a Typical merged 2-D photos of RHZs of individual seedlings. Scale bars, 0.2 mm in (a). b–d Statistical analyses of RH length (b), branching rates (c), and RH density (d). b–d Numbers of biologically independent roots with approximate 50 RHs per root examined are 11, 13, 13, 13, 10, and 12 for RH length (b), 12, 12, 12, 16, 10, and 11 for RH branching (c), and 16, 14, 9, 13, 11, and 13 for RH density (d), for the lines WT, shrh1, cpk1-1, cpk1 shrh1, CPK1-OE3(shrh1), and CNGC9-OE1(cpk1-1), respectively. Samples with different letters are significantly different with P < 0.05 (one-way ANOVA), and data are presented as means ± SEM in (b–d). Source data are provided as a Source Data file.
It was previously shown that the Arabidopsis CNGC9-OE42 line overexpressing CNGC9 under the UBQ10 promoter in the wild type Arabidopsis background conferred a longer RH phenotype48. As described above, we found that overexpression of CPK1 under the UBQ10 promoter conferred a similar phenotype (Fig. 1a, c)48. To study whether CPK1 functions upstream of CNGC5/6/9 in RH growth, we generated transgenic lines expressing CPK1 under the UBQ10 promoter in the background of shrh1, and one transgenic line designated as CPK1-OE3 (shrh1) (CPK1 OEVEREXPRESSION 3 in the background of shrh1) was selected for further study. We also generated transgenic lines by overexpressing CNGC9 under the UBQ10 promoter in the background of cpk1-1, and one transgenic line was selected and designated as CNGC9-OE1 (cpk1-1) (CNGC9 OVEREXPRESSION 1 in the background of cpk1-1). RT-qPCR data verified the over-expression of CPK1 in CPK1-OE3 (shrh1) (Supplementary Fig. 2a) and CNGC9 in CNGC9-OE1 (cpk1-1) (Supplementary Fig. 2d). Analysis of the RH phenotypes of the two overexpression lines revealed shorter RHs in both CPK1-OE3 (shrh1) and CNGC9-OE1 (cpk1-1), similar to the cpk1-1 and shrh1 mutants (Fig. 2a, b). In addition, a strong RH branching phenotype was seen in CPK1-OE3(shrh1) and CNGC9-OE1(cpk1-1) compared to the wild type, and the RH branching phenotypes in the two transgenic lines were weaker than that of shrh1, cpk1-1, and cpk1 shrh1 (Fig. 2c). The RH density was not obviously altered in the two overexpression lines compared to the wild type (Fig. 2d). The failure to rescue cpk1-1 RH phenotypes and to induce longer RHs in the same mutant by the overexpression of CNGC9 is consistent with CPK1 being an essential upstream activator of CNGC9. Similarly, the failure to rescue RH phenotypes and to induce longer RHs by the overexpression of CPK1 in shrh1 is consistent with CNGC5/6/9 being the downstream targets of CPK1. Thus, together the results strongly support our hypothesis that CPK1 functions upstream of CNGC5/6/9 in the regulation of RH growth in Arabidopsis.
RH phenotypes of cpk1 mutants can be partially rescued by high external Ca2+
To test the hypothesis that CPK1 regulates RH growth by triggering Ca2+ influx through the direct regulation of CNGC5/6/9, we analyzed the effects of external Ca2+ on the RH phenotypes of cpk1-1 and cpk1-2. The elongating growth of RHs was strongly promoted by Ca2+ in a dose-dependent manner in all the lines, including the wild type, cpk1-1, cpk1-2, and CPK1-OE13 (Supplementary Fig. 5a, b). RH elongation was promoted to a lesser extent in the cpk1-1 and cpk1-2 mutants compared to that of wild type and CPK1-OE13, especially under normal Ca2+ level (1.5 mM; Supplementary Fig. 5a, b). The strong RH branching phenotype in cpk1-1 and cpk1-2 was strongly suppressed in both low (0 and 0.1 mM) and high (10 mM) Ca2+ medium compared to normal levels, and the branching RH was very rarely observed in the wild type and CPK1-OE13 plants regardless of the Ca2+ level (Supplementary Fig. 5c). RH density was not obviously altered by changes in the Ca2+ concentration in any of the Arabidopsis lines tested (Supplementary Fig. 5a, d). These data together suggest that CPK1 functions in RH growth in a Ca2+-dependent manner, supporting our hypothesis that CPK1 is the direct upstream regulator of CNGCs and external Ca2+ influx in RHs.
CPK1 physically interacts with CNGC5/6/9
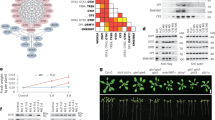
It is well-established that CNGC5/6/9 function as the main components of the plasma membrane (PM) Ca2+ channels essential for the regulation of cytosolic Ca2+ signaling and RH growth in Arabidopsis48,49. To test the possibility that CPK1 functions as the direct upstream regulator of CNGC5/6/9, we analyzed the protein-protein interactions between CPK1 and CNGC5/6/9 using the yeast 2-hybrid (Y2-H) technique based on a mating split-ubiquitin system (mbSUS)80. We found that CPK1 interacts strongly with full-length CNGC5/6/9 (Fig. 3a). We then analyzed the interactions of CPK1 with CNGC5/6/9 using split luciferase system in N. Benthamiana leaves, and WRKY72 was used as a negative control. We observed clear luciferase luminescent signal in the leaves co-expressing CPK1-nLUC and cLUC-CNGCs, but not in the leaves co-expressing WRKY72-nLUC and cLUC-CNGCs (Fig. 3b). We conducted immuno-blot assays in N. benthamiana leaves after the luciferase luminescence detection, and the expression of full length CPK1, WRKY72, and CNGC5/6/9 was detected (Fig. 3c), supporting our luciferase data. We also conducted a bimolecular fluorescence complimentary (BiFC) assay using split YFC in Xenopus laevis oocytes as previously described73,78. The C terminus (YFPC) and N terminus (YFPN) of YFP were fused to the C termini of CPK1 and CNGC5/6/9, respectively. The fused proteins were designated as CPK1-YFPC, CNGC5-YFPN, CNGC6-YFPN, and CNGC9-YFPN, respectively. cRNA was prepared in vitro, and was micro-injected into the Xenopus oocytes. After a 2-3-day incubation at 16 °C to allow transient gene expression, we observed clear YFP signal in the periphery of the Xenopus oocytes co-injected with the cRNA of CPK1-YFPC and one of the CNGC5-YFPN, CNGC6-YFPN, and CNGC9-YFPN, but failed to observe any YFP signal in the Xenopus oocytes co-injected with the cRNA of CPK1-YFPC and the free YFPN (Fig. 3d), supporting the interaction of CPK1 with CNGC5/6/9, and suggesting that this occurs in the PM. These data together demonstrate clearly that CPK1 physically interacts with CNGC5/6/9.
a Y-2H results show the interactions of CPK1 with CNGC5/6/9. b Luciferase complementation analysis in N. benthamiana leaves show the protein interactions of CPK1 with CNGC5/6/9, but no interaction of WRKY72 with CNGC5/6/9. c Immuno-blot assay data show the expression of CPK1, WRKY72, and CNGC5/6/9 in N. benthamiana leaves, supporting the luciferase complementation results. d BiFC assays in Xenopus laevis oocytes show the interactions of CPK1 with CNGC5/6/9 and the subcellular localization of CNGC5/6/9 in the plasma membrane of oocytes. The Y-2H assay was performed using the mbSUS system, and the CDS of CPK1 and CNGC5/6/9 were cloned into the vectors pNXgate32-3HA (Nub) and pMetYCgate (Cub), respectively. For luciferase complementation assay in N. benthamiana leaves, the CDS of CPK1 was cloned into the modified vector pCambia1300-nLUC, the CDS of CNGC5/6/9 were cloned into the modified vector pCambia1300-cLUC. Agrobacterium (strain GV3101) carrying the nLUC and cLUC vectors were co-injected into N. benthamiana leaves. The leaves were sprayed with 1 mM/L luciferin in the dark 48 hours after the injection, and photos were taken 7 min later. For the BiFC assay in Xenopus laevis oocytes, the CDS of CPK1 and CNGC5/6/9 were cloned into the vectors pGEMKN-YFPC and pGEMKN-YFPN, respectively. cRNA was transcribed in vitro. A total of volume of 50 nL of the cRNA mixture containing 25 ng of each vector was micro-injected into the oocytes. The oocytes were incubated in the ND96 solution for 2-3 days at 16 ± 0.5 °C, and images of the oocytes were captured under a confocal microscope. Three biological replicates were performed for Y2-H, Luciferase complementation, and immune-blot assays. 5 oocytes were analyzed in BiFC assay for each experiment in (d). Scale bars, 0.1 mm (d). W72 denotes WRKY72 in (b) and (c). Source data are provided as a Source Data file.
We further analyzed the interactions of CPK1 to the fragments of CNGC5/6/9 through classical GAL4 Y2-H assay81, and observed strong interactions between CPK1 and the N termini of CNGC5/6/9 (CNGC5-N, CNGC6-N, and CNGC9-N) (Fig. 4a). We failed to observe obvious interaction of CPK1 with the C termini and transmembrane (TM) domains of the three CNGCs (Fig. 4a). It has been reported that CNGC14 is involved in RH growth for touch sensing68, but the triple mutant shrh1 and quadruple mutant shrh2 showed similar RH phenotypes48, suggesting that CNGC14 is not essential for RH growth. We thus conducted an Y2-H assay to analyze the potential interactions of CPK1 with fragments of CNGC14. No interaction was detected between CPK1 and the N- or C-terminus or TM domain of CNGC14 (Fig. 4a), implying that CNGC14 isn’t a downstream target of CPK1.
a, b The interactions of CPK1 with the N termini of CNGC5/6/9 were observed using the Y2-H assay (a) and firefly luciferase complementation assay in N. benthamiana leaves (b). The expression of CPK1 and the N termini of CNGC5/6/9 in N. benthamiana leaves was verified by immuno-blot assay (c). The interactions of CPK1 to the N termini of CNGC5/6/9 were further verified by in vitro Co-IP: CNGC5-N (d), CNGC6-N (e), and CNGC9-N (f). The Y2-H assay was performed using the classical GAL4 Y2-H system. For the in vitro Co-IP assay, the CDS of CPK1 and CNGC5-N, CNGC6-N, CNGC9-N were cloned into the vectors 1305-UBQ10: Myc and 1305-UBQ10-Venus-Flag, respectively, downstream the UBQ10 promoter. The vectors were co-transformed into N. benthamiana leaves, and the combined expression of mCherry-Myc and CNGC9-N-Venus-Flag was used as control. The fused proteins were extracted, and the antibodies against the Myc and Flag tags were used for immuno-blot analysis. Three biological replicates were conducted for all experiments. Abbreviations: C5 (CNGC5), C6 (CNGC6), C9 (CNGC9), C14 (CNGC14), and TM (transmembrane) in (a), W72 (WRKY72), 5 N (CNGC5-N), 6 N (CNGC6-N), and 9 N (CNGC9-N) in (b), C5-N-Ve (CNGC5-N-Venus) in (d), C6-N-Ve (CNGC6-N-Venus) in (e), and C9-N-Ve (CNGC9-N-Venus) in (f). Source data are provided as a Source Data file.
We then conducted a luciferase complementation assay in N. benthamiana leaves to test for potential interactions between CPK1 and the N termini of CNGC5/6/9, and WRKY72 was used as a negative control. Strong protein interactions of CPK1 with the N termini of CNGC5/6/9 were observed (Fig. 4b), and no obvious protein interaction of WRKY72 with the N termini of CNGC5/6/9 was detected (Fig. 4b). We conducted immuno-blot assays after the luciferase luminescence detection in N. benthamiana leaves, which confirmed the successful expression of CPK1, WRKY72, and the N termini of CNGC5/6/9 (Fig. 4c), supporting our luciferase complementation results.
We also performed a co-immunoprecipitation (Co-IP) assay in N. benthamiana leaves, which further confirmed the interaction of CPK1 with CNGC5-N (Fig. 4d), CNGC6-N (Fig. 4e), and CNGC9-N (Fig. 4f). All those data together demonstrate that CPK1 physically interacts with the N termini of CNGC5/6/9, supporting our hypothesis that CPK1 is a direct upstream regulator of CNGC5/6/9 for RH growth.
CPK1 strongly activates CNGC5/6/9
To test whether CPK1 is capable of directly activating CNGC5/6/9, we transiently expressed CNGC9 and CPK1 in Xenopus oocytes, and measured the whole-oocyte currents using the two-electrode voltage-clamping (TEVC) technique in 30 mM Ba2+-based bath solution as described previously73. The Ba2+-based bath solution is more suitable for divalent cation channel current recordings than a Ca2+-based bath solution because Ca2+-influx-triggered endogenous anion channel currents can be almost completely abolished in the Ba2+-based bath solution in Xenopus oocytes73. We observed small background whole-oocyte currents in the oocytes injected with H2O or expressing CPK1 alone, a modest activation of whole-oocyte currents in oocytes expressing CNGC9 alone, and a much stronger activation of whole-oocyte currents in the oocytes co-expressing CNGC9 and CPK1 (Supplementary Fig. 6a, b). We also tested the effects of inactivated CPK1 with a K179R point mutation (designated as CPK1K179R). We observed modest whole-oocyte currents in the oocytes coexpressing CPK1K179R and CNGC9, and the activation of CNGC9 was similar to that in the oocytes expressing CNGC9 alone, but significantly smaller than that in the oocytes co-expressing wild type CPK1 and CNGC9 (Supplementary Fig. 6a, b). The CNGC9-mediated whole-oocyte currents were strongly inhibited by the Ca2+ channel blocker Gd3+ (100 µM) (Supplementary Fig. 6a, b). These data together indicate that CPK1 can activate CNGC9.
We next analyzed the activation of CNGC5/6/9 by CPK1 in human embryonic kidney 293 T (HEK293T) cells which lack the Ca2+-influx-activated endogenous anion channel currents. We first performed cytosolic Ca2+ imaging experiments by monitoring the FRET/CFP ratio of the Ca2+ indicator Cameleon version 3.6 (YC3.6) in HEK293T cells as described82. We observed a small cytosolic Ca2+ increase in control HEK293T cells, a modest cytosolic Ca2+ increase in HEK293T cells expressing wild-type CNGC5 alone, and a larger cytosolic Ca2+ increase in the HEK293T cells co-expressing CPK1 and CNGC5, upon the application of 10 mM external Ca2+ (Fig. 5a), demonstrating the activation of CNGC5 by CPK1. Similarly we also observed the activation of CNGC6 (Fig. 5b) and CNGC9 (Fig. 5c) by CPK1 by monitoring the cytosolic Ca2+ elevation in HEK293T cells. We tested the activation of CNGC9 by CPK1D274A which is an inactivated version of CPK1 with a D274A point mutation and observed a modest cytosolic Ca2+ increase in the HEK293T cells co-expressing CNGC9 and CPK1D274A, similar to that seen in the HEK239T cells expressing CNGC9 alone (Fig. 5c), suggesting that CNGC9 was not activated by CPK1D274A. These data together demonstrate the activation of CNGC5/6/9 by CPK1.
Cytosolic Ca2+ imaging and patch clamping analysis were performed in HEK293T cells, and the activation of CNGC5/6/9 by CPK1, but not by CPK1D274A, was observed. CPK1, CPK1D274A, and CNGC5/6/9 were cloned into the vector pCI-neo-IRES-YC3.6, and the vectors were then transformed into HEK293T cells using the Lipofectamine 2000 Transfection Reagent Kit individually or in a combination as indicated. HEK293T cells were cultured for 2-3 days after transformation, and the HEK293T cells with bright YC3.6 fluorescent signal were used for cytosolic Ca2+ imaging and patch clamping analysis. a–c Cytosolic Ca2+ imaging data show the activation of CNGC5 (a), CNGC6 (b), and CNGC9 (c) by CPK1, but not by CPK1D274A (c). 10 mM external Ca2+ was added as indicated by the arrows. d–i Typical whole-cell recordings (d, f, and h) and average current-voltage curves (e, g, and i) of patch clamping results show the activation of CNGC5 (d and e), CNGC6 (f and g), and CNGC9 (h and i) by CPK1. C5, C6, and C9 represent CNGC5, CNGC6, and CNGC9, respectively. The numbers of HEK293T cells tested are indicated in brackets, and data are presented as means ± SEM in (a–c), (e), (g), and (i). Source data are provided as a Source Data file.
We next analyzed the activation of CNGC5/6/9 by CPK1 using patch clamping technique by measuring whole-cell Ca2+ channel currents in HEK293T cells. We observed small background currents in the mock control HEK293T cells, modest whole cell currents in HEK293T cells expressing CNGC5 alone, and a stronger activation of whole-cell currents in HEK293T cells co-expressing CNGC5 and CPK1 (Fig. 5d, e), demonstrating the activation of CNGC5 by CPK1. Similar activation of CNGC6 (Fig. 5f, g) and CNGC9 (Fig. 5h, i) by CPK1 was observed in the patch clamping analysis in HEK293T cells. We also found that the inactivated CPK1D274A did not activate CNGC9 in HEK293T cells (Fig. 5h, i). These patch clamping data are consistent with the cytosolic Ca2+ imaging data and verify the activation of CNGC5/6/9 by CPK1.
CPK1 phosphorylates CNGC5/6/9 at a conserved serine site within their N termini
Our data show that CPK1 can interact with and activate CNGC5/6/9 (Figs. 4, 5). We thus hypothesized that CPK1 phosphorylation of these CNGCs may regulate their Ca2+ channel activity. To identify the CPK1-target sites in CNGC9, we analyzed its N-terminal region using mass spectrometry, and 11 candidate sites were identified (Fig. 6a, b; Supplementary Fig. 7a). It has been established that the sites targeted by protein kinases from the CPK and SnRK2 families mainly reside in RxxS motifs83,84. Of the 11 candidate sites identified, only Ser26 and Ser73 reside in an RxxS motif (Supplementary Fig. 7a), and Ser26 was previously identified as the main SnRK2.6/OST1-target site73. We then generated point-mutated versions of CNGC9-N by substituting the candidate serine residues with Ala (A) to prevent the phosphorylation of these sites (CNGC9-NS26A and CNGC9-NS73A). We expressed CPK1, CNGC9-NS26A and CNGC9-NS73A in the E. coli strain Rosetta-gami(DE3)pLysS, and purified the proteins as previously described85. We then performed in vitro protein phosphorylation assays in the presence of 0.5 mM CaCl2, and found that both wild-type CNGC9-N and CNGC9-NS73A were strongly phosphorylated by CPK1, but the phosphorylation of CNGC9-NS26A by CPK1 was dramatically attenuated (Fig. 6c). These results suggest that the Ser26 is the main CPK1-target site on CNGC9-N, and Ser73 may be a secondary CPK1-target site.
a, b LC-MS/MS spectrum show that the peptides LDSMDSR and KGFRKGSK contain the phospho-serines Ser26 (a) and Ser73 (b). c In vitro phosphorylation assay shows that the CPK1-mediated phosphorylation of CNGC9-NS26A was dramatically reduced, whereas the phosphorylation of CNGC9-NS73A was not obviously attenuated, compared to wild-type CNGC9-N. For the in vitro phosphorylation assays, the wild type and point-mutated CNGC9-N were cloned into the vector pCold-TF with a cleavable 6×His tag, and CPK1 was cloned into the vector pMAL-c5X with a MBP tag. The recombinants were expressed in E. coli and purified, and the recombinants with the tags retained were used for the in vitro protein phosphorylation assays. Three biological replicates were conducted for both the LC-MS/MS and the in vitro phosphorylation assays. Source data are provided as a Source Data file.
We then analyzed the Ca2+ dependence of CPK1-mediated protein phosphorylation. Autophosphorylation of CPK1 was observed in the presence of 0.5 mM CaCl2, but was strongly inhibited in Ca2+-free reaction buffer supplemented with 5 mM EGTA (Fig. 6c). Moreover, we found that CPK1 phosphorylation of CNGC9-N was slightly weaker in the absence of Ca2+ (Fig. 6c). This indicates that the autophosphorylation of CPK1 is strongly dependent on Ca2+, but the phosphorylation of CNGC9-N by CPK1 is largely independent of the Ca2+-dependent autophosphorylation of CPK1.
We conducted an alignment of the amino sequences among the N termini of CNGC5/6/9, which showed that Ser26 in CNGC9 corresponds to Ser20 in CNGC5 and Ser27 in CNGC6, which in both cases similarly reside in an RxxS motif (Supplementary Fig. 7b). The other CPK1-target site, S73 in CNGC9-N, was found to correspond to Ser58 in CNGC5 and Ser73 in CNGC6 (Supplementary Fig. 7b). Note, Ser73-CNGC6 and Ser73-CNGC9 reside within an RxxS motif, and Ser58-CNGC5 does not reside within an RxxS motif (Supplementary Fig. 7b), suggesting that these three serines are secondary CPK1-target sites that may not be essential for the activation of CNGC5/6/9. Thus, the results from our protein phosphorylation assays (Fig. 6c) suggest that the main CPK1-target site in RH-expressed CNGCs is conserved, corresponding to Ser20 in CNGC5, Ser27 in CNGC6, and Ser26 in CNGC9.
Phosphorylation of CNGC5/6/9 at the main CPK1-target sites is required for the activation of CNGC5/6/9
To study whether the phosphorylation CNGC5/6/9 at their main CPK1-target sites is required for their activity as Ca2+ channels, we substituted their main CPK1-target Ser (S) sites with Ala (A) to create phospho-dead versions. The point-mutated versions of CNGC5/6/9 were designated as CNGC5S20A, CNGC6S27A, and CNGC9S26A. We transiently co-expressed CPK1 with either wild-type or phospho-dead CNGC5/6/9, or each one individually, in HEK293T cells, and performed cytosolic Ca2+ imaging experiments. We observed a small cytosolic Ca2+ increase in the mock control HEK293T cells and in the HEK293T cells expressing either CPK1 or CNGC5S20A alone, a modest cytosolic Ca2+ increase in the HEK293T cells expressing wild-type CNGC5 alone, and a much larger cytosolic Ca2+ increase in the HEK293T cells co-expressing CPK1 and CNGC5, upon the application of 10 mM external Ca2+ (Fig. 7a). The cytosolic Ca2+ increase in the HEK293T cells expressing CNGC5S20A alone was similar to that in the HEK293T cells co-expressing CPK1 and CNGC5S20A (Fig. 7a), demonstrating that the S20A point-mutation dramatically impaired the activation of CNGC5 by CPK1. These data together established that the phosphorylation of CNGC5 by CPK1 at Ser20 is necessary for the activation of CNGC5 by CPK1. In other words, Ser20 is the main CPK1-target site for the activation of CNGC5. Similar cytosolic Ca2+ imaging results were also obtained for CNGC6 (Fig. 7b) and CNGC9 (Fig. 7c) in HEK293T cells. These results together establish that S20, S27, and S26 are the main CPK1-target sites for the respective activation of CNGC5/6/9.
CPK1, the wild type CNGC5/6/9, and iCNGCs with a S-to-A point mutation were cloned into the vector pCI-neo-IRES-YC3.6, and were then expressed in HEK293T cells individually or in a combination as indicated. HEK293T cells with bright YC3.6 fluorescent signal were used for cytosolic Ca2+ imaging and patch clamping analysis. a–c Cytosolic Ca2+ imaging data show the strong inhibition of CNGC5 (a), CNGC6 (b), and CNGC9 (c) by the S-to-A point mutations at the main CPK1-target sites compared to the strong activation of wild type CNGCs by CPK1. d–i Patch clamping data show the inhibition of CNGC5 (d, e), CNGC6 (f, g), and CNGC9 (h, i) by the S-to-A point mutations at the main CPK1-target sites compared to the activation of wild type CNGC5/6/9 by CPK1. C5S20A, C6S27A, and C9S26A represent CNGC5S20A, CNGC6S27A, and CNGC9S26A, respectively. The numbers of HEK293T cells tested are indicated in brackets, and data are presented as means ± SEM in (a–c), (e), (g), and (i). Source data are provided as a Source Data file.
We next performed patch clamping experiments to analyze the activation of CNGC5/6/9 by CPK1 by measuring whole-cell Ca2+ channel currents in HEK293T cells in a 10 mM Ca2+-based bath solution as previously described73. We observed small background conductance in the mock control HEK293T cells and the HEK293T cells expressing CPK1 alone. Similarly, a small activation of whole-cell currents was seen in cells expressing CNGC5S20A alone or coexpressing CNGC5S20A and CPK1. By contrast, a modest activation of whole-cell Ca2+ channel currents was seen in cells expressing wild-type CNGC5, and much larger whole-cell Ca2+ channel currents were seen in cells co-expressing wild-type CPK1 and CNGC5 (Fig. 7d, e). The whole-cell currents in HEK293T cells expressing CNGC5S20A alone were similar to those in HEK293T cells coexpressing CNGC5S20A and CPK1, but were significantly smaller than those of cells either expressing wild-type CNGC5 or co-expressing CNGC5 and CPK1 (Fig. 7d, e). Similar patch clamping results were also observed in HEK293T cells for the CPK1-activation of CNGC6 (Fig. 7f, g) and CNGC9 (Fig. 7h, i). These data are consistent with our cytosolic Ca2+ imaging results in HEK293T cells, and further demonstrate that the phosphorylation of CNGC5/6/9 by CPK1 is necessary for the activation of the three CNGCs.
The phosphorylation of CNGC5/6/9 at the main CPK1-target sites is necessary and sufficient to rescue the RH phenotypes of the shrh1 triple mutant and cpk1-1
It has been reported that OST1 phosphorylation of CNGC5/6/9/12 at the main OST1-target sites is required for ABA-induced stomatal closure73, and the previously identified OST1-target sites in CNGC5/6/9 are identical to the CPK1-target sites identified in this study. Phospho-dead versions of CNGC5/6/9/12, including CNGC5S20A, CNGC6S27A, CNGC9S26A, and CNGC12S13A, largely lack Ca2+ channel activity, were designated as iCNGCs (inactivated CNGCs)73. Conversely, the phospho-mimic versions of CNGC5/6/9/12, including CNGC5S20D, CNGC6S27D, CNGC9S26D, and CNGC12S13D, showed a strong activity that was similar to their activation by OST173, so these were designated as aCNGCs (activated CNGCs)73. Hereafter we will use the same iCNGC and aCNGC designations to refer to these mutants.
It has been well-established that CNGCs are capable of forming homo- and hetero-tetramers71,72, and that CNGC5/6/9 are important for RH growth in Arabidopsis48. We thus hypothesized that RH phenotypes of shrh1 may be caused by the lack of CPK1-activated CNGCs. To investigate this, we generated transgenic Arabidopsis lines expressing the iCNGCs and aCNGCs under their respective native promoters in the background of shrh1, and designated these lines as CNGC5S20A(shrh1) (expressing CNGC5S20A under CNGC5 native promoter in the background of shrh1), CNGC5S20D(shrh1), CNGC6S27A(shrh1), CNGC6S27D(shrh1), CNGC9S26A(shrh1), and CNGC9S26D(shrh1). The transgenic expression of these CNGC variants was verified by RT-qPCR (Supplementary Fig. 2b–d), and the RH phenotypes were evaluated. We found that the short RH phenotype of shrh1 was successfully rescued by the expression of aCNGCs in the transgenic lines CNGC5S20D(shrh1), CNGC6S27D(shrh1), and CNGC9S26D(shrh1), and the average RH length of CNGC9S26D(shrh1) was even longer than that in the wild type (Fig. 8a, b). Moreover, the RH branching phenotype was also successfully rescued by the expression of aCNGCs (Fig. 8a, c). By contrast, we found that the short RHs and RH branching phenotypes were not rescued in the CNGC5S20A(shrh1), CNGC6S27A(shrh1), and CNGC9S26A(shrh1) transgenic lines (Fig. 8a–c). Notably, the RH density was not affected in any of the transgenic lines (Fig. 8a, d). This demonstrates that the CPK1 phosphorylation of CNGC5/6/9 is necessary for RH growth in Arabidopsis.
A set of optical sectioning images of the RHZ of the 4-5 day-old seedlings with a 22.5-µm sectioning step and a sectioning speed of 5 steps per sec were captured under a stereo microscope at room temperature (25 ± 1 °C), and each set of images were automatically and immediately merged into a 2-D picture for each Petri dish. The merged 2-D pictures were used for RH phenotype analysis. a–d Typical merged 2-D photos of the RHZ of the seedlings (a), and the statistical analyses of RH length (b), RH branching rates (c), and RH density (d), showing that the RH phenotypes of shrh1 were rescued by the aCNGCs, but not by the iCNGCs. b–d Numbers of biologically independent roots with approximately 50 RHs per root examined are 7, 14, 13, 9, 15, 10, 25, and 12 in (b), 12, 14, 14, 12, 19, 12, 25, and 14 in (c), and 9, 12, 13, 8, 16, 11, 11, and 13 in (d), for the Arabidopsis lines as shown from left to right in each panel. e–h Typical merged 2-D photos of the RHZ of the seedlings (a) and statistical analyses of RH length (f), RH branching rates (g), RH density (h), showing that the RH phenotypes of cpk1-1 were rescued by the aCNGCs, but not by the iCNGCs. e–h Numbers of biologically independent roots with approximate 50 RHs per root examined are 11, 10, 10, 10, 10, 10, 20, and 12 in (f), 12, 8, 11, 10, 10, 10, 13, and 14 in (g), and 8, 10, 10, 10, 10, 10, 12, and 13 in (h), for the Arabidopsis lines as shown from left to right in each panel. Samples with different letters were found to be significantly different with a P < 0.05 (one-way ANOVA), and the data are presented as means ± SEM in (b–d) and (f–h). Source data are provided as a Source Data file.
As described above, we hypothesized that the RH phenotypes of cpk1 mutants are caused by the failure of CPK1 activation of CNGC5/6/9. We then tested whether aCNGCs can rescue the RH growth defects in the cpk1 mutant. We generated transgenic lines expressing the iCNGCs and aCNGCs under their respective native promoters in the cpk1-1 background, and designated these lines as CNGC5S20A(cpk1-1) (expressing CNGC5S20A under CNGC5 native promoter in the background of cpk1-1), CNGC5S20D(cpk1-1), CNGC6S27A(cpk1-1), CNGC6S27D(cpk1-1), CNGC9S26A(cpk1-1), and CNGC9S26D(cpk1-1). The expression of the iCNGCs and aCNGCs in those transgenic lines was verified by RT-qPCR (Supplementary Fig. 2b–d), and the RH phenotypes were analyzed. This revealed that the expression of the aCNGCs, but not the iCNGCs, could fully rescue the RH length and branching phenotypes of cpk1-1 (Fig. 8e–g). Similar to the shrh1 mutant, the RH density of cpk1-1 was not significantly altered by the expression of either iCNGCs or aCNGCs (Fig. 8e, h). These results indicate that CPK1 phosphorylates CNGC5/6/9 to promote RH growth in Arabidopsis.
We also generated transgenic Arabidopsis lines by overexpressing the iCNGCs and aCNGCs under the UBQ10 promoter in the wild type background. The overexpression lines were selected and designated as CNGC5S20A-OE(WT) (OVEREXPRESSING CNGC5S20A under UBQ10 promoter in the wild type background), CNGC5S20D-OE(WT), CNGC6S27A-OE(WT), CNGC6S27D-OE(WT), CNGC9S26A-OE(WT), and CNGC9S26D-OE(WT). The overexpression of aCNGCs and iCNGCs in the transgenic lines was confirmed by RT-qPCR analysis (Supplementary Fig. 2b–d). We analyzed the RH phenotypes, and observed cpk1- and shrh1-like defects of RH growth, including shorter RHs and more RH branching, in the lines overexpressing the iCNGCs, compared to the wild type (Supplementary Fig. 8a–c). This is consistent with a previous study that showed the overexpression of iCNGCs strongly suppresses the endogenous wild type CNGCs dominant-negatively73. We did not observe any obvious RH length and branching phenotypes in the transgenic lines overexpressing the aCNGCs compared to the wild type (Supplementary Fig. 8a–c). RH density was not obviously altered across the transgenic lines expressing either iCNGCs or aCNGCs (Supplementary Fig. 8a, d). These results support the requirement of CPK1-mediated phosphorylation of CNGCs for RH growth in Arabidopsis.
We then studied the effects of CPK1-mediated phosphorylation at the secondary CPK1-target sites in vivo. We generated point-mutated versions CNGC5S58A, CNGC5S58D, CNGC6S73A, CNGC6S73D, CNGC9S73A, and CNGC9S73D. We performed cytosolic Ca2+ imaging analysis in HEK293T cells and found that the Ca2+ channel activity of CNGC9S73A was similar to that of wild type CNGC9 (Supplementary Fig. 7c), suggesting that the phosphorylation of that site is not essential for the activation of CNGC9. We then fused an eGFP to the C termini of the mutated versions of CNGC5/6/9 and generated transgenic Arabidopsis lines by expressing these mutated CNGC5/6/9 under their respective native promoters in cpk1-1. The lines were designated as CNGC5S58A(cpk1-1) (expressing CNGC5S58A under CNGC5 native promoter in the background of cpk1-1), CNGC5S58D(cpk1-1), CNGC6S73A(cpk1-1), CNGC6S73D(cpk1-1), CNGC9S73A(cpk1-1), and CNGC9S73D(cpk1-1), respectively. RT-qPCR was then used to verify the expression of these transgenes in these lines (Supplementary Fig. 2b–d), and their RH phenotypes were evaluated. We observed cpk1-1-like short RHs (Supplementary Fig. 9a, b) and RH branching (Supplementary Fig. 9a, c) phenotypes, and saw no changes in RH density in these transgenic lines (Supplementary Fig. 9a, d), compared to wild type. Those data establish that the secondary CPK1-target sites at the N termini of CNGC5/6/9 are not essential for RH growth in Arabidopsis.
The phosphorylation of CNGC5/6/9 by CPK1 is required to retain the cytosolic Ca2+ gradient at RH tips
To study whether CPK1 regulates RH growth by regulating the cytosolic Ca2+ signaling at RH tips via its regulation of CNGC5/6/9, we introduced the gene encoding the calcium indicator YC3.6 under the UBQ10 promoter in the mutants and transgenic lines described above. To be sure that the results across the lines were comparable, we analyzed their YFP signal fluorescence at RH apices. This revealed that the lines had similar YFP signal intensities (Supplementary Fig. 10), suggesting that the YC3.6 reporter is expressed at similar levels across the lines. We then analyzed the Ca2+ gradient at the apex of RHs in these lines. A sharp Ca2+ gradient was observed at the RH apex in the wild type plants, but the Ca2+ gradients were severely attenuated in the cpk1-1, shrh1, and cpk1 shrh1 mutants (Fig. 9a, b). Further analysis revealed that the RH cytosolic Ca2+ gradients in the cpk1-1 and shrh1 mutants were successfully restored by the expression of the aCNGCs, but not by the iCNGCs (Fig. 9a, b).
The cytosolic Ca2+ gradient at the tips of growing RH was monitored by measuring the FRET/CFP ratio of YC3.6 in the Arabidopsis lines expressing the calcium sensor YC3.6. A sharp cytosolic Ca2+ gradient was observed in wild-type RHs, but this was dramatically attenuated in the cpk1-1, shrh1, and cpk1 shrh1 mutants. The sharp cytosolic Ca2+ gradient was successfully restored to wild-type-like levels in the RHs of cpk1-1 and shrh1 by the expression of aCNGCs, but not by the expression of the iCNGCs, under their respective native promoters. a Pseudo images of typical YC3.6 FRET/CFP ratios in RHs of wild-type, cpk1-1, shrh1, and cpk1 shrh1 mutants, and the CNGC5S20A(shrh1), CNGC5S20D(shrh1), CNGC6S27A(shrh1), CNGC6S27D(shrh1), CNGC9S26A(shrh1), CNGC9S26D(shrh1), CNGC5S20A(cpk1-1), CNGC5S20D(cpk1-1), CNGC6S27A(cpk1-1), CNGC6S27D(cpk1-1), CNGC9S26D(cpk1-1), and CNGC9S26A(cpk1-1) transgenic lines. b YC3.6 FRET/CFP ratios reflecting cytosolic Ca2+ gradients at RH apices. Scale bars, 10 μm in (a). n = 24, 20, 25, 25, 16, 25, 11, 23, 24, 24, 12, 24, 18, 18, 19, and 23 biologically independent RHs for the lines as shown from left to right in (b). The data are presented as means ± SEM, and samples with different letters are significantly different with P < 0.05 (one-way ANOVA) in (b). Source data are provided as a Source Data file.
Overall, our findings indicate that CPK1-mediated phosphorylation of CNGC5/6/9 is necessary and sufficient to maintain the sharp cytosolic Ca2+ gradient at RH tips which is required to maintain their growth.
Discussion
It has been well-established that the cytosolic Ca2+ gradient at the apex of RHs plays a central role in RH growth45,46, and that PM Ca2+ channels are essential for RH elongation86. Multiple CNGCs have been identified as being essential for RH elongation by our group and others48,49,68. In this research, we reveal the requirement of CPK1-mediated phosphorylation of CNGC5/6/9 to establish and maintain the sharp cytosolic Ca2+ gradient at the RH apex that drives RH growth. In a word, CPK1 is the main upstream activator of CNGC5/6/9 in RHs. However, we noticed that iCNGCs exhibited a low level of Ca2+ channel activity in HEK293T cells in the absence of CPK1 compared with that of wild-type CNGCs (Figs. 5, 7), suggesting that wild-type CNGCs were modestly activated. It is very likely that the three wild-type CNGCs were slightly phosphorylated at the main CPK1-target sites or other yet-to-be-identified sites by endogenous protein kinases in HEK293T cells, and the impairment of CPK1-activation of the three iCNGCs (Fig. 7) supports this explanation.
It has been reported that CNGC14 was expressed in RHs, but the cngc14 mutant showed a short RH phenotype only when the roots and RHs were embedded in solid medium68. Thus, CNGC14 is important for touch sensing for RHs. Our previous study found that the triple mutant shrh1 (cngc5/6/9) and quadruple mutant shrh2 (cngc5/6/9/14) showed quite similar RH phenotypes48. In this research we found that CPK1 interacts with CNGC5/6/9, and did not observe obvious protein interaction of CPK1 to CNGC14 in Y2-H assays (Fig. 4a). The negative Y2-H results can’t completely exclude the possible protein interaction between CPK1 and CNGC14. Taken together, it is reasonable to conclude that CPK1 is involved in RH growth mainly through activating CNGC5/6/9, the three main CPK1-targets, and CNGC14 may function as a secondary CPK1-target in RHs.
Our data indicate that CPK1-CNGC modules control RH growth by mediating and regulating external Ca2+ influx at RH tips. However, the RH phenotypes of the single mutants cpk1-1 and cpk1-2 were strongly diminished under both low Ca2+ (0 mM and 0.1 mM) and high Ca2+ (10 mM) conditions relative to that under the standard (1.5 mM) Ca2+ condition (Supplementary Fig. 5a-c). RH elongation was inhibited to a similar extent in wild type, cpk1 mutants and OE lines, suggesting that the amount of Ca2+ absorbed by the RHs of wild type and the CPK1-OE13 transgenic line was reduced to an extent similar to that of cpk1 under low Ca2+. Conversely, the RH growth defects in the cpk1 mutants were partially rescued by high Ca2+, suggesting that over-loading of Ca2+ occurred to a similar extent in these lines.
The autophosphorylation of CPK1 showed a strong Ca2+-dependence, while the CPK1 phosphorylation of CNGC9-N was less so (Fig. 6c). It is unknown whether CPK1 autophosphorylation is important for RH growth, but it is reasonable to speculate that CPK1 mediates and activates CNGC5/6/9 for cytosolic Ca2+ elevation which consequently enhances the activity of CPK1 via autophosphorylation to further elevate cytosolic Ca2+ levels. Thus, CPK1 and CNGC5/6/9 may operate as a positive feedback loop to maintain the sharp cytosolic Ca2+ gradient at RH tips to facilitate RH growth.
It has been recently made clear that CNGC5/6/9/12 are phosphorylated and activated by OST1/SnRK2.6 for ABA-induced stomatal closure73, and we show here that the same residues in CNGC5/6/9 are phosphorylated by CPK1 to promote RH growth. Interestingly the Arabidopsis mutant ost1/snrk2.6 showed normal RHs. Moreover, OsCNGC9, a close homolog of Arabidopsis CNGC6 in rice, is phosphorylated and activated by OsSAPK8, a close homolog of Arabidopsis OST1, for chilling response in rice, and the site targeted by OsSAPK8 was found to be Ser645, which is localized at the C terminus rather than the N terminus87. Clearly, diverse protein kinases can phosphorylate and activate specific CNGCs using the same or different target sites for different biological processes. Furthermore, it seems that the mechanisms for Ca2+ signaling regulation vary across biological processes, cell/tissue types, and plant species.
It is well established that ROS-triggered Ca2+ uptake plays an important role in RH growth37. RHD2, a NADPH oxidase, is responsible for ROS formation in RHs, and the rhd2 mutant showed a short RH phenotype37. Annexin 1 was identified as a ROS-activated Ca2+- and K+-permeable channel in the plasma membrane of root epidermal cells, but the mutant ann1 showed normal RHs38. It seems that Annexin 1 is not the main ROS-activated Ca2+ channel for RH growth. In this research, we reveal the role of CPK1-CNGC modules in establishing and maintaining the Ca2+ gradient and RH growth in Arabidopsis. However, H2O2 failed to activate CNGC6 in Xenopus laevis oocytes72, suggesting that CNGCs are not the ROS-activated Ca2+ channels in both RHs and guard cells. The ROS-activated Ca2+ channels essential for RH growth thus remain to be identified. Nevertheless it is reasonable to speculate that both yet-to-be-identified ROS-activated Ca2+ channels and CNGCs (ROS-insensitive Ca2+ channels) are important for RH growth, and that loss-of-function mutations in both types of Ca2+ channels will confer stronger RH phenotypes than that reported here.
RHs elongate mainly via tip growth. Interestingly it has been recently reported that shank-localized cell wall growth also contributes to RH elongation in Arabidopsis88. Considering that CPK1 is mainly distributed at the apex of RHs rather than in the shank area, it is very likely that the CPK1-CNGC modules are not involved in the shank-localized cell wall growth in RHs.
Methods
Plant materials and growth conditions
Arabidopsis thaliana plants (Columbia-0 ecotype) were grown in an environment-controlled growth room under a 16-h light/8-h dark daily cycle in the light of cold daylight lamps with a photon fluence rate of approximate 70–80 µmol·m−2·s−1 during the daytime at 21 ± 1 °C, and the relative ambient humidity was ≥ 75% controlled by an ultrasonic humidifier as described48,72. The Arabidopsis T-DNA insertion line SALK_080155 (cpk1-1) and SALK_010530C (cpk1-2) were obtained from ABRC. The triple mutant shrh1 (short root hair1/cngc5-1 cngc6-2 cngc9-1) was generated as described previously48. The quadruple mutant cpk1 shrh1 was generated by crossing the single mutant cpk1-1 and triple mutant shrh1.
Generation of transgenic Arabidopsis lines
The promoters (2 kb genomic DNA upstream of the coding regions) of CNGC5/6/9 and CPK1 were PCR amplified from Arabidopsis genomic DNA. The full length CDS (coding DNA sequences) of CPK1 and the three CNGCs were PCR amplified from Arabidopsis cDNA. The promoters and CDS were cloned into vector pCambia1305 as described48. For the generation of iCNGCs (CNGC5S20A, CNGC6S27A, and CNGC9S26A) and aCNGCs (CNGC5S20D, CNGC6S27D, and CNGC9S26D), the point-mutations were introduced into the wild type CNGC5/6/9 by a PCR reaction. The CNGC5/6/9 point mutants were verified by sequencing. See Supplementary Data 1 for primers for the vector construction. The vectors were transformed into Agrobacterium tumefaciens strain GV3101, and the mutants cpk1-1 and shrh1 expressing the calcium indicator YC3.648 were transformed using floral dip method89. T1 seedlings were screened on ½ MS plates containing 50 mg/L hygromycin (Catalog # CH6361, Coolaber).
RT-PCR and RT-qPCR
RT-PCR and RT-qPCR were conducted as described72. 7-day-old seedlings grown on ½ MS medium in a plate were ground, and total RNA was extracted from the samples using TRIzol reagent (Invitrogen, USA). RT-PCR was performed using a Hifair® V one-step RT-gDNA digestion SuperMix (Catalog # 11142ES10; Yeasen, China). RT-qPCR analysis was performed using Hieff® qPCR SYBR Green Master Mix (Catalog # 11204ES08; Yeasen, China) on a Bio-Rad CFX Connect Real-Time PCR System (Bio-Rad; USA) according to the manufacture’s protocol with the primers listed in Supplementary Data 1.
Y2-H assay
To detect the protein interactions of CPK1 with the full length CNGC5/6/9, Y2-H assays using the mbSUS system80 were conducted. The full length CDS of CNGC5/6/9 were cloned into the pMetYCgate (Cub) vector with Pst I and Hind III as described73, and the CDS of CPK1 was cloned into the pNXgate32-3HA (Nub) vector with EcoR I and Sma I. The vectors were transformed into the THY.AP4. yeast (Saccharomyces cerevisiae) strain using the lithium acetate transformation method80. The yeast clones were selected on SD/–Leu–Trp medium (Catalog # PM2220, Coolaber), and protein interactions were confirmed on selective SD/–Leu–Trp–His–Ade medium (Catalog # PM2110, Coolaber) as described73. Photographs were taken using a single-lens reflex (SLR) camera (EOS 7D, Canon). See Supplementary Data 1 for primer sequences.
To analyze the protein interactions of CPK1 with the fragments of CNGC5/6/9/14 (The N and C termini), Y2-H assays using the classical GAL4 Y2-H system81 were conducted. The coding sequences of the N termini (aa1-102 for CNGC5-N, aa1-116 for CNGC6-N, aa1-117 for CNGC9-N, and aa1-101 for CNGC14-N) and the C termini (aa413-717 for CNGC5-C, aa429-747 for CNGC6-C, aa428-733 for CNGC9-C, and aa411-726 for CNGC14) of CNGC5/6/9/14 were cloned into the vector pGBK-T7, and the CDS of CPK1 was cloned into the vector pGAD-T7, with EcoR I and BamH I, as described73. The vectors were co-transformed into the yeast strain AH109 using the lithium acetate method80. The protein interactions of CPK1 with the transmembrane (TM) domains of CNGC5/6/9/14 were analyzed using mbSUS system. The coding sequences of the TM domains (aa103-412 for CNGC5-M, aa117-428 for CNGC6-M, aa118-427 for CNGC9-M, and aa102-410 for CNGC14-M) of CNGC5/6/9/14 were cloned into the pMetYCgate (Cub) vector with Pst I and Hind III as described73. Those vectors and Nub-CPK1 were co-transformed into the THY.AP4. yeast (Saccharomyces cerevisiae) strain. The yeast clones of AH109 and THY.AP4 were selected on SD/-Leu-Trp medium (Catalog # PM2220, Coolaber), and the protein interactions were confirmed on selective SD/–Leu–Trp–His–Ade medium (Catalog # PM2110, Coolaber). Pictures were taken using the single-lens reflex camera (EOS 7D, Canon). Please see Supplementary Data 1 for primer sequences.
Co-IP assay
Co-IP assays in N. benthamiana leaves were conducted as described73. The CDS of CNGC5-N, CNGC6-N, and CNGC9-N were cloned into the vector 1305-UBQ10-Venus-Flag upstream the venus-Flag tag and downstream the UBQ10 promoter, and the CPK1 CDS was cloned into the vector 1305-UBQ10: Myc upstream the Myc tag and downstream the UBQ10 promoter. The vectors CNGC5-N-Venus-Flag, CNGC6-N-Venus-Flag, and CNGC9-N-Venus-Flag were respectively co-transformed with either CPK1-Myc or mCherry-Myc into N. benthamiana leaves for co-expression. Total proteins were extracted from 3 g (fresh weight) of plant samples with lysis buffer (150 mM NaCl, 1 mM EDTA, 0.05% (v/v) NP40, 1 mM PMSF, 5 mg/L Dnase, Protease Inhibitor Cocktail (Catalog # GRF101, Epizyme, China), 20 mM Tris-HCl, and pH 7.5). The extracts were incubated at 4 °C for 2 h and centrifuged at 12,000 g for 3 min at 4 °C. Anti-Myc agarose (Catalog # M20012M, Abmart) was incubated with the supernatant for 6 h at 4 °C. The Co-IP products were washed 5 times using washing buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5). CPK1 and either mCherry or the N termini of CNGC5/6/9 were detected with anti-Myc antibodies (Catalog # Ab62928, Abcam) (1:5,000) and anti-Flag antibodies (Catalog # M20008, Abmart) (1:5,000), respectively. The chemi-luminescence signal was detected using autoradiography.
Luciferase complementation and immuno-blot experiments in N. benthamiana leaves
For luciferase complementation assays in N. benthamiana leaves, the vectors pCambia1300-nLUC and pCambia1300-cLUC were modified. In brief, 4 x Myc was fused upstream the nLUC in the vector pCambia1300-nLUC, and 3 x Flag followed by a stop codon was fused downstream the cLUC in the vector pCambia1300-cLUC. The CDS of CPK1 and WRKY72 were cloned into the modified vector pCambia1300-nLUC upstream the Myc tag, while the CDS of the CNGC5, CNGC6, CNGC9, CNGC5-N, CNGC6-N, and CNGC9-N were cloned into the modified vector pCambia1300-cLUC downstream the cLUC and upstream the 3 x Flag. Agrobacterium (strain GV3101) carrying nLUC and cLUC vectors were co-injected into the N. benthamiana leaves. 48 hours after the injection, the N. benthamiana leaves were sprayed with 1 mM luciferin solution in darkness, and the luciferase luminescence was detected by taking photos with a biosystem (Model C280, Azure, USA) 7 mins after the luciferin spraying.
For immuno-blot assay in N. benthamiana leaves after luciferase luminescence detection, 0.2 g N. benthamiana leaves (fresh weight) was ground in 300 µL extraction buffer (150 mM NaCl, 1 mM EDTA, 0.05%(v/v) NP40, 1 mM PMSF, 5 mg/L Dnase, Protease Inhibitor Cocktail (Catalog # GRF101, Epizyme, China), 20 mM Tris-HCl, and pH 7.5). The total protein was extracted from the sample in the extraction buffer as described90. The homogenate was centrifuged at 13,000 g for 20 min at 4 °C, and the supernatant was used for immuno-blot assay. The fused protein with nLUC was detected using anti-Myc antibody (Catalog # Ab62928, Abcam) (1:5,000), while the fused protein with cLUC was detected using anti-Flag antibody (Catalog # M20008, Abmart) (1:5,000).
RH phenotype analysis
RH phenotypes were analyzed in Arabidopsis seedlings as described48. Arabidopsis seeds were surface sterilized with 0.5% sodium hypochlorite for 10 min, washed 3 times with sterile H2O, and then sowed in Petri dishes containing solid medium modified from a previous method91. The basic solid medium contained 5 mM KNO3, 1 mM MgSO4, 1 mM KH2PO4, 0.1 mM NaFeEDTA, 5 mM Mes, 1% (w/v) sucrose, 0.68% (w/v) agarose, MS microelements at full strength, and pH 6.0 adjusted with Tris-HCl. 5 mM EGTA was added to the basic medium for 0 mM Ca2+ medium. CaCl2 was added to the basic medium at a concentration of 0.1 mM, 1.5 mM, and 10 mM as indicated for 0.1 mM, standard Ca2+ (1.5 mM), and high Ca2+ (10 mM) medium, respectively. The Petri dishes with Arabidopsis seeds were vertically placed in a growth chamber with a controlled growth condition same as that in the Arabidopsis plant growth room as described in this research (See Plant Growth in the Methods and Materials). RHs located within the 2 mm root hair zone (RHZ) of 4-5 day-old seedlings were used for RH phenotype analysis under a stereo microscope (Model M205 FCA, Leica, Germany) as described48. A Petri dish with 4-5 day seedlings was placed horizontally on the stage of the stereo microscope, and a set of optical sectioning images along Z axis with a 22.5 µm sectioning step and a sectioning speed of 5 steps per second were captured at room temperature (25 ± 1 °C). 20-40 optical sectioning images were captured within 4-8 sec for each seedling. The set of optical sectioning images of the bright-field were automatically and immediately merged into a 2-D photo. By that method, most RHs growing at different angles and distributed at different optical sectioning layers around the primary roots were able to be seen with consistent spatial resolution in the merged 2-D bright-field photo. The 2-D fluorescent photos merged from a set of fluorescence optical sectioning images were obtained by the same method. The length, shapes, and branching phenotypes of RHs in the merged 2-D photos were analyzed after the picture capture. No less than 10 roots with approximate 50 RHs each root were analyzed for each experimental condition each Arabidopsis line. Software GraphPad Prism 9.0 was used to plot the data.
For time-lapse analysis of RH growth, optical sectioning images were captured each 1 min for no less than 300 min at 21 ± 1 °C, and the merged 2-D photos generated immediately after optical sectioning by the software Leica Application Suite X (Leica, Germany) were used for the time-lapse analysis.
Protein expression, purification, and in vitro phosphorylation assay
The encoding sequences of wild type CNGC9-N (aa1-117) and the mutated versions of CNGC9-N, including CNGC9-NS26A and CNGC9-NS73A, were cloned into the vector pCold-TF downstream the TF, and this vector contained a 6×His tag upstream of TF92. A MBP tag was fused to the N terminus of CPK1, and the recombinant was cloned into the vector pMAL-c5X. The recombinants were expressed in E. coli strain Rosetta-gami(DE3)pLysS and purified using the standard protocols as described85. The recombinants CPK1 and CNGC9-N with the tags retained were incubated with 1 µM ATP plus 1 µCi of [γ-32P] ATP (Catalog # NEG502Z500UC, PerkinElmer) in a reaction buffer (20 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, and pH 7.4) at 30 °C for 30 min. The reactions were terminated by boiling in SDS sample buffer and separated by 10% SDS-PAGE.
Liquid chromatography tandem mass spectrometry (LC-MS/MS)
The target sites of CPK1 in CNGC9-N were identified by LC-MS/MS as described93. The recombinant GST-CPK1 and CNGC9-N were incubated with 1 mM ATP in the reaction buffer (20 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, and pH 7.4) at 30 °C for 1.5 h. The reaction was diluted by adding 4 volumes of 50 mM triethylammonium bicarbonate, digested with trypsin with a 1:100 (w/w) ratio, and was incubated at 37 °C overnight. After desalting, the tryptic peptides were enriched with IMAC and analyzed by LC-MS/MS. Experiments were performed on a Q Exactive HF-X mass spectrometer coupled to Easy nLC (Thermo Fisher Scientific). The peptide mixture was loaded onto the C18-reversed phase column (25 cm long, 75 μm inner diameter) packed in-house with RP-C18 1.9 μm resin in buffer A (0.1% Formic acid in HPLC-grade water) and separated with a linear gradient of buffer B (80% acetonitrile, 0.1% Formic acid in 20% HPLC-grade water) at a flow rate of 300 nL/min. MS data was acquired using a data-dependent top-20 method by dynamically choosing the most abundant precursor ions from the survey scan (350–1800 m/z) for higher energy collision-induced dissociation (HCD) fragmentation. Determination of the target value is based on predictive Automatic Gain Control (pAGC). Dynamic exclusion duration was 45 s. Survey scans were acquired at a resolution of 60,000 at m/z 200, and resolution for HCD spectra was set to 15,000 at m/z 200. Normalized collision energy was 28 eV.
BiFC and TEVC analysis in Xenopus laevis oocytes
For BiFC assays, the CDS of CPK1 was cloned into the vector pGEMKN-YFPC, and the CDS of CNGC9 was cloned into the pGEMKN-YFPN vector as described78. The cRNA were transcribed in vitro using T7 RiboMAXTM Large-scale RNA Pro-duction System (Catalog # p1712, Promega, USA). cRNA mixture was micro-injected into oocytes with 25 ng per vector in a total volume of 50 nL. The oocytes were incubated in ND96 solution at 16 ± 0.5 °C for 2-3 days after the micro-injection to allow the genes to be expressed. Images were captured using a 20× objective (NA 0.75, water) with an excitation of 487 nm and an emission of 500 to 550 nm after the incubation under a confocal microscope equipped with a detector (HyD S) and a white light laser (Stellaris 5, Leica, Germany) as described73.
For TEVC experiments in Xenopus laevis oocytes, the K179R point mutation was introduced into CPK1 by PCR, the CDS sequences of CNGC9, CPK1, and CPK1K179R were cloned into the vector pGEMHE with the primers listed in Supplementary Data 1, and the cRNA of CNGC9 and the kinases were prepared in vitro using the T7 RiboMAXTM Large-scale RNA Pro-duction System (Catalog # p1712, Promega, USA). Frogs (Xenopus laevis) were from Shen’s lab at Hangzhou Normal University (Hangzhou City, China). The oocytes were isolated from Xenopus laevis and microinjected with the cRNA of a single gene or cRNA mixture with an amount of 25 ng per gene in a total volume of 50 nL for each oocyte, and the oocytes microinjected with 50 nL water were used as control. The oocytes were incubated at 16 ± 0.5 °C for 2-3 days after the microinjection to allow the expression of the proteins. Channel currents were recorded using Axopatch-900A setup (Axon Instruments, USA) after the incubation, and the membrane voltage with a 2-sec duration for each voltage was stepped from -180 mV to 0 mV with a + 20 mV increment as described previously73. The pipette solution contained 3 M KCl. The bath solution contained 30 mM BaCl2, 2 mM NaCl, 1 mM KCl, 130 mM mannitol, and 5 mM Mes-Tris (pH 5.5). Software pClampex 10.2 (Axon Instruments, USA) was used for data acquisition and analyses. Software Sigmaplot 14.0 was used to plot the data.
Cytosolic Ca2+ imaging and patch clamping experiments in HEK293T cells
For cytosolic Ca2+ imaging and patch clamping experiments in HEK293T cells (Catalog # FH0242, FuHeng Biology, China), the CDS of CPK1, CPK1D274A, wild type CNGC5/6/9, iCNGCs, and aCNGCs were cloned into the vector pCI-neo-IRES-YC3.6.
HEK293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 IU/mL), and streptomycin (100 µg/mL) in a water-injected incubator (Model 3111; Thermo-Fisher Scientific, USA) with 5% CO2 at 37 °C in a moist atmosphere as described82.
Vectors were transformed into HEK293T cells individually or in a combination as indicated using a Lipofectamine 2000 Transfection Reagent Kit (Invitrogen, USA), and the cells were cultured in the medium for 2-3 days to allow the gene expression. The HEK293T cells were digested with 0.25% trypsin-EDTA for no more than 1 min, collected by centrifugation at 1000 g for 1 min, washed twice by centrifugation at 1000 g for 1 min, and resuspended in a Ca2+-free CIB solution (130 mM NaCl, 3 mM KCl, 0.6 mM MgCl2, 1.2 mM NaHCO3, 10 mM glucose, and 10 mM Hepes, and the pH adjusted to 7.2 with NaOH). The HEK293T cells with bright YC3.6 signal were used for Ca2+ imaging and patch clamping experiments.
Cytosolic Ca2+ imaging experiments in HEK293T cells were performed by monitoring the ratio of FRET/CFP of YC3.6 with a 50 ms exposure to excitation light and a 3-sec-inter-pulse period in the Ca2+-free CIB solution under an inverted microscope (Carl Zeiss, Model D1, Germany) at room temperature (25 ± 1 °C), and 10 mM external Ca2+ was applied as indicated. The pictures and the ratio values of interested cells/regions were automatically saved in the hard drive of a personal computer. The software MetaFluor (Version 7.8.0.0; Molecular Devises, USA) was used for data acquisition and analyses.
Patch clamping experiments in HEK293T cells were performed using the Axopatch-200B patch-clamp setup (Axon Instruments, USA) with a Digidata 1440 A digitizer (Axon Instruments, USA) combined with an inverted microscope (Model A1; Carl Zeiss, Germany) as described82. The bath solution contained 120 mM NMDG-Cl, 10 mM CsCl, 1 mM MgCl2, 10 mM CaCl2, and 10 mM Hepes, and the pH was adjusted to 7.2 with HCl. The pipette solution contained 120 mM Cs-Glutamate, 8 mM NaCl, 6.7 mM EGTA, 3.35 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, and pH was adjusted to 7.2 with CsOH. The osmolarity was adjusted to 310 mmol kg-1 using D-glucose for both bath and pipette solutions. Whole-cell currents were monitored and recorded by applying a 2-sec-duration ramp voltage from -180 mV to +20 mM every 12 sec for 40 times for each cell after the accession to whole cell configuration with a seal resistance of no less than 10 GΩ as described82. Software pClampex 10.2 (Axon Instruments, USA) was used for data acquisition and analysis. Software Sigmaplot 14.0 was used to plot the data.
Cytosolic Ca2+ imaging analysis in RHs
Triple mutant shrh1 expressing YC3.6 was generated as described48. YC3.6 was introduced into the mutant cpk1-1 by crossing the mutant to the wild type plants expressing YC3.6, and the homozygous cpk1-1 mutant with bright YC3.6 signal in RHs were isolated from the progeny plants. The Arabidopsis transgenic lines were generated based on the wild type and mutants expressing YC3.6 in RHs. FRET/CFP-based cytosolic Ca2+ imaging analysis were performed in RHs of 4-day-old seedlings adhered to a cover glass with the medical adhesive glue (Hollister, USA) under the inverted microscope (Model Dl; Carl Zeiss, Germany). The cover glass with roots and RHs was immersed in the working solution (5 mM KCl, 100 µM CaCl2, 10 mM Mes-Tris, pH 5.7) in a chamber. The ratio of FRET/CFP of YC3.6 was monitored with a 50 ms exposure to excitation light and a 3-sec-inter-pulse period, and the data were automatically saved in the hard drive of a personal computer. In situ calibration was performed as described94.
Statistical analysis
Student’s t-test and one-way ANOVA were used to determine significant differences between samples. No data were excluded from all analyses. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are incorporated into the article and its online supplementary materials. The LC-MS/MS data generated in this study have also been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with an accession code PXD055717. Gene sequence data in this article can be found in GenBank database using the accession numbers AT5G04870 (CPK1), AT5G57940 (CNGC5), AT2G23980 (CNGC6), AT4G30560 (CNGC9), and AT2G24610 (CNGC14). Source data are provided with this paper.
References
Cui, S., Suzaki, T., Tominaga-Wada, R. & Yoshida, S. Regulation and functional diversification of root hairs. Semi Cell Dev. Biol. 83, 115–122 (2017).
Ibáñez, F., Wall, L. & Fabra, A. Starting points in plant-bacteria nitrogen-fixing symbioses: intercellular invasion of the roots. J. Exp. Bot. 68, 1905–1918 (2017).
Rongsawat, T., Peltier, J. B., Boyer, J. C., Very, A. A. & Sentenac, H. Looking for root hairs to overcome poor soils. Trends Plant Sci. 26, 83–94 (2021).
Tanaka, N. et al. Characteristics of a root hair-less line of Arabidopsis thaliana under physiological stresses. J. Exp. Bot. 65, 1497–1512 (2014).
Bengough, A. G., Loades, K. & McKenzie, B. M. Root hairs aid soil penetration by anchoring the root surface to pore walls. J. Exp. Bot. 67, 1071–1078 (2016).
Haling, R. E. et al. Root hairs improve root penetration, root-soil contact, and phosphorus acquisition in soils of different strength. J. Exp. Bot. 64, 3711–3721 (2013).
Choi, H.-S. & Cho, H.-T. Root hairs enhance Arabidopsis seedling survival upon soil disruption. Sci. Rep. 9, 11181–11181 (2019).
Galloway, A. F. et al. Cereal root exudates contain highly structurally complex polysaccharides with soil-binding properties. Plant J. 103, 1666–1678 (2020).
Huang, Y. et al. Nanospherical arabinogalactan proteins are a key component of the high-strength adhesive secreted by English ivy. Proc. Natl Acad. Sci. USA 113, E3193–E3202 (2016).
Dittmer, H. J. A quantitative study of the roots and root hairs of a winter rye plant (Secale cereale). Am. J. Bot. 24, 417–420 (1937).
Ahn, S. J., Shin, R. & Schachtman, D. P. Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134, 1135–1145 (2004).
Canales, J., Contreras-Lopez, O., Alvarez, J. M. & Gutierrez, R. A. Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J. 92, 305–316 (2017).
Bates, T. R. & Lynch, J. P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 19, 529–538 (1996).
Janes, G. et al. Cellular patterning of Arabidopsis roots under low phosphate conditions. Front Plant Sci. 9, 735 (2018).
Xue, C., Li, W., Shen, R. & Lan, P. Impacts of iron on phosphate starvation-induced root hair growth in Arabidopsis. Plant Cell Environ. 46, 215–238 (2023).
Huang, L. et al. Zinc finger protein 5 (ZFP5) associates with ethylene signaling to regulate the phosphate and potassium deficiency-induced root hair development in Arabidopsis. Plant Mol. Biol. 102, 143–158 (2020).
Shibata, M. et al. Trihelix transcription factors GTL1 and DF1 prevent aberrant root hair formation in an excess nutrient condition. N. Phytol. 235, 1426–1441 (2022).
De Pessemier, J. et al. Root system size and root hair length are key phenes for nitrate acquisition and biomass production across natural variation in Arabidopsis. J. Exp. Bot. 73, 3569–3583 (2022).
Saengwilai, P. et al. Root hair phenotypes influence nitrogen acquisition in maize. Ann. Bot. 128, 849–858 (2021).
Grierson C., Nielsen E., Ketelaarc T. & Schiefelbein J. Root hairs. The Arabidopsis Book 12, e0172 (2014).
Masucci, J. D. et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260 (1996).
Lin, Y. & Schiefelbein, J. Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128, 3697–3705 (2001).
Hung, C. Y. et al. A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117, 73–84 (1998).
Walker, A. R. et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350 (1999).
Bernhardt, C. et al. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439 (2003).
Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A. & Schiefelbein, J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132, 291–298 (2005).
Lee, M. M. & Schiefelbein, J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483 (1999).
Kang, Y. H. et al. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21, 1080–1094 (2009).
Lee, M. M. & Schiefelbein, J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14, 611–618 (2002).
Tominaga, R., Iwata, M., Okada, K. & Wada, T. Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19, 2264–2277 (2007).
Song, S. K. et al. Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 157, 1196–1208 (2011).
Kang, Y. H., Song, S. K., Schiefelbein, J. & Lee, M. M. Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol. 163, 193–204 (2013).
Kurata, T. et al. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132, 5387–5398 (2005).
Bibikova, T. N., Jacob, T., Dahse, I. & Gilroy, S. Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125, 2925–2934 (1998).
Liszkay, A., van der Zalm, E. & Schopfer, P. Production of reactive oxygen intermediates O2.-, H2O2, and.OH by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 136, 3114–3123 (2004).
Monshausen, G. B., Bibikova, T. N., Messerli, M. A., Shi, C. & Gilroy, S. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl Acad. Sci. USA 104, 20996–21001 (2007).
Foreman, J. et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446 (2003).
Laohavisit, A. et al. Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca2+- and K+-permeable conductance in root cells. Plant Cell 24, 1522–1533 (2012).
Jones, M. A., Raymond, M. J., Yang, Z. & Smirnoff, N. NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58, 1261–1270 (2007).
Macpherson, N. et al. NADPH oxidase involvement in cellular integrity. Planta 227, 1415–1418 (2008).
Takeda, S. et al. Local positive feedback regulation determines cell shape in root hair cells. Science 319, 1241–1244 (2008).
Ketelaar, T., Galway, M. E., Mulder, B. M. & Emons, A. M. Rates of exocytosis and endocytosis in Arabidopsis root hairs and pollen tubes. J. Microsc 231, 265–273 (2008).
Rounds, C. M. & Bezanilla, M. Growth mechanisms in tip-growing plant cells. Annu Rev. Plant Biol. 64, 243–265 (2013).
Li, M. et al. Regulation of phytohormones on the growth and development of plant root hair. Front Plant Sci. 13, 865302 (2022).
Wymer, C. L., Bibikova, T. N. & Gilroy, S. Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12, 427–439 (1997).
Bibikova, T. N., Zhigilei, A. & Gilroy, S. Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta 203, 495–505 (1997).
Monshausen, G. B., Messerli, M. A. & Gilroy, S. Imaging of the yellow cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 147, 1690–1698 (2008).
Tan, Y.-Q. et al. Three CNGC family members, CNGC5, CNGC6, and CNGC9, are required for constitutive growth of Arabidopsis root hairs as Ca2+-permeable channels. Plant Commun. 1, 100001 (2020).
Brost, C. et al. Multiple cyclic nucleotide-gated channels coordinate calcium oscillations and polar growth of root hairs. Plant J. 99, 910–923 (2019).
Feiguelman, G., Fu, Y. & Yalovsky, S. ROP GTPases structure-function and signaling pathways. Plant Physiol. 176, 57–79 (2017).
Yalovsky, S., Bloch, D., Sorek, N. & Kost, B. Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 147, 1527–1543 (2008).
Yang, Z. Cell polarity signaling in Arabidopsis. Ann. Rev. Cell Dev. Biol. 24, 551–575 (2008).
Ou, H. & Yi, P. ROP GTPase-dependent polarity establishment during tip growth in plants. N. Phytol. 236, 49–57 (2022).
Feiguelman, G. et al. Microtubule-associated ROP interactors affect microtubule dynamics and modulate cell wall patterning and root hair growth. Development 149, dev200811 (2022).
Fu, Y., Wu, G. & Yang, Z. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152, 1019–1032 (2001).
Jones, M. A. et al. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14, 763–776 (2002).
Bloch, D. et al. Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol. Biol. Cell 16, 1913–1927 (2005).
Molendijk, A. J. et al. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20, 2779–2788 (2001).
Ketelaar, T., de Ruijter, N. C. A. & Emons, A. M. C. Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell 15, 285–292 (2003).
Brueggeman, J. M., Windham, I. A. & Nebenführ, A. Nuclear movement in growing Arabidopsis root hairs involves both actin filaments and microtubules. J. Exp. Bot. 73, 5388–5399 (2022).
Wang, Y. et al. Disruption of actin filaments induces mitochondrial Ca2+ release to the cytoplasm and [Ca2+]c changes in Arabidopsis root hairs. BMC Plant Biol. 10, 53 (2010).
Bibikova, T. N., Blancaflor, E. B. & Gilroy, S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 17, 657–665 (1999).
Pei, W., Du, F., Zhang, Y., He, T. & Ren, H. Control of the actin cytoskeleton in root hair development. Plant Sci. 187, 10–18 (2012).
Schiefelbein, J. W., Shipley, A. & Rowse, P. Calcium influx at the tip of growing root-hair cells of Arabidopsis thaliana. Planta 187, 455–459 (1992).
Herrmann, A. & Felle, H. H. Tip growth in root hair cells of Sinapis alba L.: significance of internal and external Ca2+ and pH. N. Phytol. 129, 523–533 (1995).
Felle, H. H. & Hepler, P. K. The cytosolic Ca2+ concentration gradient of Sinapis alba root hairs as revealed by Ca2+-selective microelectrode tests and fura-dextran ratio imaging. Plant Physiol. 114, 39–45 (1997).
Véry, A. A. & Davies, J. M. Hyperpolarization-activated calcium channels at the tip of Arabidopsis root hairs. Proc. Natl Acad. Sci. USA 97, 9801–9806 (2000).
Zhang, S. et al. Arabidopsis CNGC14 mediates calcium influx required for tip growth in root hairs. Mol. Plant 10, 1004–1006 (2017).
Kaplan, B., Sherman, T. & Fromm, H. Cyclic nucleotide-gated channels in plants. FEBS Lett. 581, 2237–2246 (2007).
Kaupp, U. B. & Seifert, R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824 (2002).
Chiasson, D. M. et al. A quantitative hypermorphic CNGC allele confers ectopic calcium flux and impairs cellular development. eLife 6, e25012 (2017).
Tan, Y.-Q. et al. Multiple cyclic nucleotide-gated channels function as ABA-activated Ca2+ channels required for ABA-induced stomatal closure in Arabidopsis. Plant Cell 35, 239–259 (2023).
Yang, Y. et al. OPEN STOMATA 1 phosphorylates CYCLIC NUCLEOTIDE-GATED CHANNELs to trigger Ca2+ signaling for ABA-induced stomatal closure in Arabidopsis. Plant Cell 36, 2328–2358 (2024).
Liu, Q. et al. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 40, e104559 (2021).
Lee, S. C., Lan, W., Buchanan, B. B. & Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl Acad. Sci. USA 106, 21419–21424 (2009).
Geiger, D. et al. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal 4, ra32 (2011).
Geiger, D. et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl Acad. Sci. USA 107, 8023–8028 (2010).
Geiger, D. et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl Acad. Sci. USA 106, 21425–21430 (2009).
Lin, C., Choi, H.-S. & Cho, H.-T. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Mol. Cells 31, 393–397 (2011).
Obrdlik, P. et al. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl Acad. Sci. USA 101, 12242–12247 (2004).
Fields, S. & Song, O. A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 (1989).
Gao, Q.-F. et al. Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl Acad. Sci. USA 113, 3096–3101 (2016).
Wang, P. et al. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl Acad. Sci. USA 110, 11205–11210 (2013).
Umezawa, T. et al. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal 6, rs8 (2013).
Lin, Z. et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 11, 613 (2020).
Kiegle, E., Gilliham, M., Haseloff, J. & Tester, M. Hyperpolarisation-activated calcium currents found only in cells from the elongation zone of Arabidopsis thaliana roots. Plant J. 21, 225–229 (2000).
Wang, J. et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 14, 315–329 (2021).
Herburger, K., Schoenaers, S., Vissenberg, K. & Mravec, J. Shank-localized cell wall growth contributes to Arabidopsis root hair elongation. Nat. Plants 8, 1222–1232 (2022).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Liu, H. et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 (2008).
Zhao, S., Zhang, M. L., Ma, T. L. & Wang, Y. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28, 3005–3019 (2016).
Ma, D. et al. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl Acad. Sci. USA 113, 224–229 (2016).
Wang, P. et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69, 100–112.e106 (2018).
Swanson S. J. & Gilroy S. Imaging changes in cytoplasmic calcium using the yellow cameleon 3.6 biosensor and confocal microscopy. In: Plant Lipid Signaling Protocols (eds Munnik T., Heilmann I.). Humana Press (2013).
Acknowledgements
We thank Dr. Shanshan Wang and Dr. Wenjuan Cai (CAS Center for Excellence in Molecular Plant Sciences, CAS, Shanghai 200032, China) for assistance in LC-MS/MS and BiFC experiments, respectively, and Dr. Jeremy Murray (CAS Center for Excellence in Molecular Plant Sciences, CAS, Shanghai 200032, China) for a critical reading of this article. Funding This research was mainly supported by the National Natural Science Foundation of China (grant numbers 32470332 and 32270279 to W.Y.-F., 32300253 to T.Y.-Q., and 32400214 to Y.Y.) and the Key Laboratory of Plant Carbon Capture of CAS (CNXT002 to W.Y.-F.), and was partially supported by the Postdoctoral Fellowship Program of CPSF (GZB20240744 to Y.Y.), the China Postdoctoral Science Foundation (2024M753231 to Y.Y.), and Shanghai Post-doctoral Excellence Program (2024652 to Y.Y.).
Author information
Authors and Affiliations
Contributions
Z.M. screened the Arabidopsis mutant library, constructed most vectors, generated quadruple mutants cpk1 shrh1 and transgenic Arabidopsis lines, and performed most experiments, including Y2-H, luciferase complementation, Co-IP, and cytosolic Ca2+ imaging experiments in both HEK293T cells and RHs. D.B.-Y. performed RH time-lapse, BiFC, TEVC, and patch clamping experiments. T.Y-Q. and Z.M. performed LC-MS/MS experiments and identified the CPK1-target sites. Y.Y. and Z.Y. were involved in the vector construction. W.Y.-F. proposed the project, designed the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Xu Na Wu, Wang Tian and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, M., Du, BY., Tan, YQ. et al. CPK1 activates CNGCs through phosphorylation for Ca2+ signaling to promote root hair growth in Arabidopsis. Nat Commun 16, 676 (2025). https://doi.org/10.1038/s41467-025-56008-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56008-4
This article is cited by
-
A new phenotyping method for root growth studies in compacted soil validated by GWAS in barley
Plant Methods (2025)
-
Unearthing the hidden organ: vital role of the root in drought tolerance of plants
Plant Molecular Biology (2025)