Abstract
Bacteremia, a leading cause of death, generally arises after bacteria establish infection in a particular tissue and transit to secondary sites. Studying dissemination from primary sites by solely measuring bacterial burdens does not capture the movement of individual clones. By barcoding Klebsiella pneumoniae, a leading cause of bacteremia, we track pathogen dissemination following pneumonia. Variability in organ bacterial burdens is attributable to two distinct dissemination patterns distinguished by the degree of similarity between the lung and systemic sites. In metastatic dissemination, lung bacterial clones undergo heterogeneous expansion and the dominant clones spread to secondary organs, leading to greater similarity between sites. In direct dissemination, bacterial clones exit the lungs without clonal expansion, leading to lower burdens in systemic sites and more dissimilarity from the lung. We uncover bacterial and host factors that influence the dynamics of clonal sharing and expansion. Here, our data reveal unexpected heterogeneity in Klebsiella bacteremia dynamics and define a framework for understanding within-host bacterial dissemination.
Similar content being viewed by others
Introduction
Sepsis is a systemic inflammatory syndrome with high mortality rates caused by a dysregulated immune response to infection1,2. Sepsis is highly associated with bloodstream infections, ~90% of which are caused by bacteria3,4,5. Gram-negative species are leading causes of bacteremia and exhibit high levels of antimicrobial resistance (AMR)6. Enterobacterales are pathogens of urgent concern7,8 and are the most common cause of Gram-negative bacteremia. Klebsiella pneumoniae is a leading pathogen in both Gram-negative bacteremia and deaths associated with AMR3,6,9.
Gram-negative bacteremia pathogenesis involves three phases10. First, pathogens infect initial sites. Second, bacteria cross host barriers and gain bloodstream access. Third, pathogens sustain bacteremia by avoiding immune clearance mechanisms in filtering organs like the spleen and liver11. Pneumonia and bloodstream infections are the two most common conditions associated with death due to AMR organisms9, and the lung is a common site of infection leading to secondary bacteremia. To prevent the progression of pneumonia to bacteremia, dissemination mechanisms must be further investigated.
Experimentally defining host-pathogen interactions during dissemination is difficult since this process often occurs simultaneously with the phases of initial site infection and bloodstream survival. Murine pneumonia models can identify bacterial or host factors required for lung fitness and can assess bloodstream fitness by measuring bacterial abundance in tissues like the spleen and liver. However, these data must be carefully interpreted, as decreases in pathogen abundance at secondary sites can either be due to lower fitness at that site or differential dissemination from the lungs. Models that do not involve primary site infections, such as a tail vein injection, define bacteremia fitness defects in a dissemination-independent manner. For example, we previously identified GmhB, involved in ADP-heptose biosynthesis for the lipopolysaccharide (LPS) inner core and a ligand for the pro-inflammatory sensor alpha-kinase 1 (ALPK1)12,13, as a conserved Gram-negative bacteremia fitness factor14 in both pneumonia and tail-vein models. However, the defect of the K. pneumoniae gmhB mutant in the spleen was larger in the pneumonia model despite the absence of a defect in the lung, suggesting that GmhB affects both dissemination and sustaining bacteremia. Thus, bacterial tissue burden can measure defects at the first and last bacteremia phases (initial site infection and replication at systemic sites), but metrics to characterize dissemination are lacking10.
Isogenic barcoding of bacteria has been used to measure infection dynamics in various models15,16,17,18,19,20,21,22,23,24,25,26,27, and can be applied to study dissemination. This approach enables the quantitative tracking of individual bacterial clones during different phases of infection, including the measurement of infection bottlenecks and clonal replication within tissues. One approach for analyzing barcoded bacteria is STAMPR (Sequence-Tagged Analysis of Microbial Populations in R), which employs population genetics and resampling approaches to quantify bacterial expansion, infection bottlenecks, and dissemination patterns23,28. By comparing the presence and abundance of barcodes across sites relative to the inoculum, STAMPR can trace clones between tissues, enabling inferences regarding in vivo dissemination routes.
Here, we applied bacterial barcoding and STAMPR to a murine model of bacteremia originating from pneumonia. Initially, wild-type K. pneumoniae bacteremia dynamics were measured. Two dissemination patterns termed metastatic and direct were identified by the degree of similarity between the lung and secondary sites. These patterns are generally correlated with clonal expansion in the lung or absence thereof, respectively. Metastatic dissemination led to a higher bacterial burden in systemic organs. STAMPR analyses using bacterial and host mutants were used to define how dynamics change after disruption of bacterial ADP-heptose biosynthesis (GmhB) or protein translocation (TatC), and host Nox2 NADPH oxidase (CybB) or monocyte chemokine receptor (Ccr2). Disruption of gmhB increased bacterial founding populations in the lung and influenced replication within secondary sites. TatC was required for fitness across sites, yet did not influence bottlenecks between sites. Disruption of Nox2 yielded only direct dissemination but led to increased bacterial burden within tissues. Ccr2 did not influence dissemination patterns but may influence the degree of clonal expansion in the lung. This study uncovers informative yet previously hidden patterns that define bacterial spread from the lung to the bloodstream and correlate with bacterial burdens at systemic sites. Furthermore, the framework for analysis of pathogen dissemination presented here should be broadly useful for studying bacteremia-associated infection dynamics.
Results
Bacterial barcoding reveals the dynamics of K. pneumoniae dissemination from pneumonia
To characterize K. pneumoniae infection dynamics, we barcoded the hypervirulent strain KPPR129. This library, KPPR1-STAMPR, contained ~40,000 unique barcodes inserted at the Tn7 site. The 25-nucleotide barcodes did not influence bacterial fitness, and KPPR1-STAMPR had an even abundance of barcodes across the library (Supplementary Fig. 1A, B). Deep sequencing and resampling of the barcodes detected in a sample can enable estimation of the founding population (Ns), the number of individual clones from the inoculum that give rise to the population at the site of infection. The founding population size is a measure of the bottleneck, the combination of barriers that prevent cells in the inoculum from colonizing a given site. Smaller founding populations indicate tighter bottlenecks. To confirm that the library enabled accurate calculations of founding population, we created a standard curve using in vitro bottlenecks through serial dilutions, and verified the library is appropriate for estimating founding populations up to ~8 x 105 (Supplementary Fig. 1C).
To measure baseline dynamics of K. pneumoniae infection in a model of pneumonia that progresses to bacteremia30, WT C57BL/6 J mice were retropharyngeally inoculated with 1 × 106 CFU KPPR1-STAMPR and then observed at 24- or 48- hours. Compared to the inoculum, the K. pneumoniae population expanded in the lung 10-100x at 24-hours and ~1000x by 48-hours (Fig. 1A). At 24-hours, systemic translocation and bacteremia can be consistently detected with minimal animal morbidity. Bacterial CFU recovery from the spleen and liver were typically lower than the inoculum size, indicating replication restriction within tissues or tight bottlenecks between primary and secondary sites. At 48-hours, animal morbidity was common and CFU recovery in the spleen and liver was generally higher than the inoculum size but lower than the lung, indicating that continued dissemination or bacterial replication may eventually overcome host barriers at secondary tissues. Bacterial abundance in the liver, spleen, and blood were variable but largely correlated with lung burden (Supplementary Fig. 2A). The lung contained a large founding population (Fig. 1B) that produced the observed CFU, with a median Ns of 6397 at 24-hours and 1629 at 48-hours (Supplementary Data 1). Secondary organs had lower Ns values. At 24-hours, the median founding population was 34, 78, and 16 clones in the spleen, liver, and blood, representing 0.5, 1.2, and 0.25% the size of the lung population, respectively, although the range of Ns was wide. Sex differences were not detected in either CFU or Ns values (Supplementary Fig. 2B–D).
Mice were infected with KPPR1-STAMPR, a wild-type K. pneumoniae barcoded library in a model of pneumonia that progresses to bacteremia. Tissues were harvested 24- or 48-hours post-infection (black circles or blue squares, respectively) and analyzed with quantitative culture and the STAMPR pipeline. A The bacterial burden in each tissue at the time of harvest is displayed as log10 CFU/site. The dotted line indicates the infection inoculum, 1 x 106 CFU/mouse. B The founding population size in each tissue was estimated by the STAMPR pipeline and displayed as log10 Ns. The dotted line indicates the resolution limit of the library, ~8 × 105 founders, the maximum complexity for any site. C The extent of clonal replication is estimated by log10(CFU/Ns). D The extent of replication evenness is displayed as log10(Ns/Nb). The dotted line indicates a theoretical value 0, representing even replication. E The genetic distance (GD) between the lung and secondary organs, or (F) the spleen and other secondary organs was modeled by the STAMPR pipeline at 24- or 48-hours. In all, data is represented as a box plot with points representing individual animals, whisker end points indicating the minimum and maximum observed values, box boundaries representing the 25th and 75th percentile, and the middle line representing the median value. In (A–F), differences between tissues at 24- or 48- hours were assessed with two-tailed unpaired t-tests and corrected for multiple comparisons; p-values are displayed above each comparison. In (A–F), comparisons between the lung and secondary organs (A–E) or the spleen and other organs F within each timepoint was assessed by ordinary one-way ANOVA and corrected for multiple comparisons: #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001 for the 24- hour group and +p < 0.05, ++p < 0.01, +++p < 0.001, ++++p < 0.0001 for the 48-hour group. Specifically, in (A) the p-value between the lung and spleen, liver, or blood at 24-hours is <0.0001, <0.0001, and 0.0003; the p-value between the lung and spleen, liver, or blood at 48-hours is 0.0002, 0.0022, and 0.6254, respectively. In (B) the p-value between the lung and spleen, liver, or blood at 24-hours is <0.0001 for each comparison; the p-value between the lung and spleen, liver, or blood at 48-hours is <0.0001, 0.0012, and <0.0001, respectively. In (C) the p-value between the lung and spleen, liver, or blood at 24-hours is 0.2747, 0.0738, and 0.9801; the p-value between the lung and spleen, liver, or blood at 48-hours is 0.4235, 0.5367, and 0.2186, respectively. In (D) the p-value between the lung and spleen, liver, or blood at 24-hours is 0.0004, 0.0172, and 0.0012; the p-value between the lung and spleen, liver, or blood at 48-hours is 0.0384, 0.1405, and 0.0042, respectively. In (E) the p-value between the spleen and liver is 0.9775 and between the spleen and blood is 0.9994 at 24-hours; the p-value between the spleen and liver is 0.9932 and between the spleen and blood is 0.6551 at 48-hours. In (F) the p-value between the lung and liver or the lung and blood is <0.0001 for both comparisons at 24-hours; the p-value between the lung and liver is 0.0223 and between the liver and blood is 0.9897 at 48-hours. For the 24-hour group, n = 17 mice in 5 independent trials. For the 48-hour group, n = 9 mice in a single trial. STAMPR analysis was excluded for animals with no detectable tissue CFU or any sample with low sequencing quality. Source data are provided within the Source Data file.
The significant population expansion at secondary sites over time could be explained by the expansion of clones present early in infection or the accumulation of new clones over time. At 24- and 48- hours, founding populations in secondary sites were similar, indicating that a narrow bottleneck limits K. pneumoniae exit from the lung and this bottleneck is not significantly modulated by time or disease stage. Instead, the expansion of founding clones, measured by CFU/Ns, was greater in all organs at 48-hours (Fig. 1C), indicating either high replication or reseeding of the same clones. In particular, there was a very large expansion of clones in the blood at 48-hours. Expansion can be even throughout the clones in the population or uneven, measured by Ns/Nb (a metric for evenness) where Nb is a STAMPR metric that calculates founding population sizes based on the frequency of barcodes, rather than the number of unique barcodes (Ns)23. When barcode abundances are relatively even, Ns and Nb ratios are close to one. During uneven bacterial replication with no reduction in the number of unique clones, Nb will decrease but Ns will remain constant, and the Ns/Nb ratio will increase. At 24- and 48- hours, replication was uneven in the lungs compared to other organs, indicating expansion of a subset of founding clones (Fig. 1D, even replication indicated by the dotted line).
We next evaluated the extent of barcode similarity between sites using genetic distance (GD). Higher GD values indicate two sites have dissimilar populations. Lower GD values indicate similarity, due to either sharing many barcodes or sharing a few highly abundant barcodes. At 24-hours, the GD between the lung and secondary sites was high, indicating that the lung was dissimilar from the spleen, liver, and blood (Fig. 1E), which could be explained by the sharing of the subset of clones that are predominant in the lung. At 48-hours, systemic sites became more similar to the lungs, likely due to expansion of shared clones. GDs between the spleen and other secondary sites were generally low, demonstrating that systemic sites were more similar to each other than the lung throughout the course of infection (Fig. 1F). The combination of increased expansion of a similar number of founders over time, increased relatedness between the lung and secondary sites, and the uneven nature of replication in the lung suggests that dominant clones from the lung seed secondary organs and expand over time. Accordingly, bacteremia observed at 48-hours, when mice have substantial morbidity, is attributable to the large expansion of a small number of clones.
Two distinct modes of K. pneumoniae dissemination from the lungs
In the experiments above, there was a marked heterogeneity of CFU in secondary sites at 24-hours (for example, a 6-log variability in the liver; Fig. 1A). Analysis of lung and spleen barcode frequency plots revealed two distinct patterns of barcode sharing between these sites (Fig. 2A–F). In one subset of mice there were clearly dominant (highly abundant) clones in the lung (Fig. 2A, red circles) that were also dominant in the spleen (Fig. 2B) and liver (Fig. 2C). We termed this pattern as metastatic dissemination, since it appeared that clones which had disproportionally expanded at the initial site translocated to the secondary site. In a second subset of animals, there was little variation in clonal abundance (more evenly distributed barcodes) across the bacterial population in the lung; the most abundant lung clones (Fig. 2D, red circles) were not typically abundant in the spleen (Fig. 2E) and liver (Fig. 2F). We termed this pattern as direct dissemination since clones in the lung translocated to the secondary site without substantial expansion in the lung.
Mice were infected with a wild-type K. pneumoniae barcoded library in a model of pneumonia that progresses to bacteremia. Tissues were harvested 24-hours post-infection and analyzed with the STAMPR pipeline. A–F Frequency plots are displayed for a representative mouse experiencing either metastatic (A–C) or direct (D–F) dissemination. Unique 25-nt barcodes were assigned a random tag and plotted on the x-axis, the log10 frequency of each barcode within the indicated tissue is plotted on the y-axis. The 30 most abundant barcodes within the lung for each mouse are highlighted in red and indicated in the spleen or liver if the barcode was also found at that site. During metastatic dissemination, the most abundant barcodes in the lung (A) replicated more than other clones and were often found in the corresponding spleen (B) or liver (C). During direct dissemination, the most abundant barcodes in the lung did not have extensive replication beyond other barcodes (D) and were not often found in the corresponding spleen (E) or liver (F). Representative heat maps for two mice, one experiencing metastatic (G, H) and one experiencing direct (I, J) dissemination are displayed for genetic distance (GD; G, I) and fractional genetic distance (FRD; H, J). For FRDx-y heatmaps, x is the row and y is the column. The boxed comparisons are mentioned in the text and highlighted for clarity. Source data for Fig. 2G-J are provided within the Source Data file.
A deeper analysis of the relatedness of barcodes between the lung and spleen further distinguished the two patterns of K. pneumoniae lung dissemination. While the lung was largely genetically distinct from secondary sites (Fig. 1E), individual frequency plots indicated some degree of barcode sharing between the lung and these tissues. To examine the contribution of individual clones to genetic similarity, a fractional genetic distance metric (FRDx-y) was used to quantify the fraction of clones shared between site x and y relative to the total number of clones in y. For example, a high FRDlung-spleen indicates that their shared barcodes are a high proportion of the total spleen barcodes. A corresponding lower FRDspleen-lung would suggest the lung is a large reservoir of clones, only some of which are shared with the spleen. Measuring FRDlung-spleen, two patterns of clonal sharing were evident (Fig. 2G–J). In mice experiencing metastatic dissemination, many clones in the spleen were present in the lung (high FRDlung-spleen, Fig. 2G, H: lung:spleen comparisons boxed), even though the lungs and spleens were dissimilar overall (high GD). In mice experiencing direct dissemination, clones in the lung and spleen were dissimilar (high GD) and largely distinct (low FRDlung-spleen, Fig. 2I, J: lung:spleen comparisons boxed).
Using mice in the 24-hour cohort to design cutoffs, we classified metastatic dissemination as any animal experiencing a higher fraction of shared clones between the lung and a secondary site, defined by a FRDlung-spleen or FRDlung-liver > 0.336 (Fig. 3A–C, Supplementary Fig. 3A, B). This cut off was set to classify all subsequent groups within the study and identified metastatic dissemination in 8/17 and direct dissemination in 9/17 mice at 24- hours. Dissimilar lungs and spleens with many shared clones can be explained by uneven expansion of clones in the lung that are present in the spleen, but at distinct frequencies. Thus, to quantify uneven replication, we plotted Ns/Nb ratios (Fig. 3A). Mice with metastatic dissemination had higher Ns/Nb ratios, indicating increased uneven replication. The metastatic dissemination group had significantly elevated bacterial burdens in the lungs, spleens, and blood with trends toward increased burden in the liver (Fig. 3D–G, replicated in 4A–D). Thus, metastatic dissemination in K. pneumoniae occurs when uneven clonal expansion (Ns/Nb) in one organ is associated with translocation (low GD, high FRD) to another and tends to predict higher bacterial burdens across sites (high CFU).
Mice were infected with KPPR1-STAMPR, in a model of pneumonia that progresses to bacteremia. Tissues were harvested 24-hours (A–G) or 48-hours (H–N) post-infection and analyzed with quantitative culture and the STAMPR pipeline. A, H The extent of replication evenness within the lung is estimated by log10(Ns/Nb) for each mode of dissemination, metastatic (dark purple closed circles) and direct (pink open circles). B, C, I, J Genetic distance (GD) and number of barcodes shared (FRD) between the lung:spleen (B, I) or the lung:liver (C, J) are displayed for each mouse, with indications for defining dissemination patterns as metastatic or direct, further defined in Supplementary Fig. 3. Lines connect values from the same mouse. Bacterial burden, displayed as log10 CFU, is compared across the lung (D, K), spleen (E, L), liver (F, M), or blood (G, N). For (A, D–G, H, K–N), data is represented as a box plot with points representing individual animals, whisker end points indicating the minimum and maximum observed values, box boundaries representing the 25th and 75th percentile, and the middle line representing the median value. For (A, D–G), comparisons between metastatic and direct groups were assessed by two-tailed unpaired t-tests and corrected for multiple comparisons; p-values are displayed above each comparison. In (A–G), n = 17 mice infected across five independent trials; in (H–N), n = 9 mice in a single trial. STAMPR analysis was excluded for one mouse with no detectable blood CFU, and any sample with low sequencing quality. Source data are provided within the Source Data file.
In the subset experiencing direct dissemination, secondary site clones were not abundant in the lung (FRDlung-spleen or FRDlung-liver < 0.336; Fig. 3A–C, Supplementary Fig. 3A, B). Ns/Nb ratios confirmed that these animals had more even expansion in the lung (Fig. 3A). In direct dissemination, clones from the inoculum translocated to the spleen without substantial expansion in the lungs and was associated with lower bacterial burdens in secondary sites (Fig. 3D–G; low lung Ns/Nb, high GD, low FRD, low CFU). These differences in CFU at secondary sites did not correlate with differences in the number of founders (Fig. 4E–H, Supplementary Fig. 2E). Instead, in mice with metastatic dissemination there was greater expansion of barcodes (CFU/Ns) in the lungs, spleen and blood (Fig. 4I–L). Therefore, metastatic dissemination does not result in a greater number of disseminating clones, but rather results in an increased abundance of the clones that disseminate. Together, these data reveal two dissemination patterns that were not apparent from CFU alone, termed metastatic and direct dissemination. Importantly, male and female mice exhibited similar patterns of dissemination (Supplementary Fig. 2F, G).
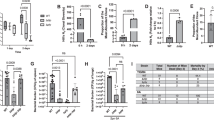
Wild-type mice were infected with a KPPR1-STAMPR (purple circles or pink hexagons), gmhB-STAMPR (blue triangles), or tatC-STAMPR (green triangles) K. pneumoniae barcoded library. Nox2-/- (gold diamonds) or Ccr2-/- (yellow squares) mice were infected with the KPPR1-STAMPR barcoded library. For all, an inoculum of 1 × 106 CFU/mouse was administered in a model of pneumonia that progresses to bacteremia and tissues were harvested 24-hours post infection. Wild-type mice infected with the KPPR1 library were divided into groups demonstrating metastatic (KPPR1-met, purple circles) or direct (KPPR1-dir, pink hexagons) dissemination as defined in Fig. 3. The bacterial burden at the time of harvest in the (A) lung, (B) spleen, (C) liver, or (D) blood for each infection group is displayed as log10 CFU. The founding population in each tissue was estimated by the STAMPR pipeline for the (E) lung, (F) spleen, (G) liver, or (H) blood for each group and displayed as log10 Ns. A measure of total CFU per founder is represented by log10(CFU/Ns) for the (I) lung, (J) spleen, (K) liver, or (L) blood for each group. In (A–L), bars represent the mean and points represent individual animals. For all, n = 5–9 mice in at least two independent trials; specifically, KPPR1-met n = 8, KPPR1-dir n = 9, gmhB n = 9, tatC n = 5, Nox2-/- n = 5, Ccr2-/- n = 8. Comparisons between the KPPR1-met group and each other group were assessed using an ordinary one-way ANOVA with Dunnet’s multiple comparisons correction and indicated by *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Comparisons between the KPPR1-dir group and each other group were also assessed using an ordinary one-way ANOVA with Dunnet’s multiple comparisons correction and indicated by #p < 0.05, ###p < 0.001, ####p < 0.0001. For clarity, only p-values < 0.1 are displayed on in A–L. Source data for each panel are provided within the Source Data file, including the every p-value measured for the one-way ANOVA comparisons.
Because metastatic dissemination is characterized by clonal expansion in the lung, we hypothesized that this pattern may be more dominant in later infection after bacteria have experienced multiple rounds of replication. Indeed at 48-hours, metastatic dissemination was the dominant pattern (Fig. 3H–J, 8/9 mice) with high FRDlung-spleen, FRDlung-liver. Notably, the animal displaying direct dissemination at 48-hours had lower CFU across sites compared to the corresponding metastatic group (Fig. 3K–N).
K. pneumoniae fitness factors GmhB and TatC make distinct contributions to metastatic dissemination and population expansion at secondary sites
We next sought to determine how K. pneumoniae fitness factors that differ in their importance for primary site infection and bloodstream survival affect patterns of K. pneumoniae dissemination from the lung. GmhB is an enzyme involved in the production of ADP-heptose, which is incorporated into the inner core of LPS and is detected by the pro-inflammatory host receptor ALPK112,31,32. Our previous study showed that K. pneumoniae GmhB is dispensable for lung fitness but enhances spleen and liver fitness14. Another bacterial factor, TatC, is part of the twin-arginine transporter required for moving multiple folded protein substrates across the cytoplasmic membrane33. Unlike GmhB, TatC is required for both primary and secondary site fitness during bacteremia in multiple species34,35. Based on bacterial burden, it is unclear if either GmhB or TatC is involved in dissemination. Thus, barcoded libraries were generated in gmhB and tatC mutants (Supplementary Fig. 1D, E), and dissemination patterns were measured after 24-hours (Figs. 4 and 5).
Wild-type mice were infected with a (A, B) gmhB-STAMPR (blue circles) or (C, D) tatC-STAMPR (green circles) K. pneumoniae barcoded libraries, while (E, F) Nox2-/- (gold circles) or (G, H) Ccr2-/- (yellow circles) mice were infected with the wild-type KPPR1-STAMPR library. In all experiments, an inoculum of 1×106 CFU/mouse was administered in a model of pneumonia that progresses to bacteremia and tissues were harvested 24-hours post-infection. In (A–H), genetic distance (GD) and number of barcodes shared (FRD) between the lung:spleen (A, C, E, G) or lung:liver (B, D, F, H) are displayed with indications for whether dissemination is classified as metastatic (closed symbols) or direct (open symbols), further defined in Supplementary Fig. 3. I The lung log10(Ns/Nb) is displayed for all groups. In (J–N), wild-type mice infected with the KPPR1 library were divided into groups demonstrating metastatic (KPPR1-met) or direct (KPPR1-dir) dissemination. The GD between the lung:spleen (J), FRD between the lung:spleen (K), GD between the lung:liver (L), FRD between the lung:liver (M), and the GD between the spleen:liver (N) is displayed for all infected groups. In (I–N), data is represented as a box plot with points representing individual animals, whisker end points indicating the minimum and maximum observed values, box boundaries representing the 25th and 75th percentile, and the middle line representing the median value. KPPR1-met is displayed as purple circles, KPPR1-dir is displayed as pink open circles, gmhB-STAMPR is displayed as blue triangles, tatC-STAMPR is displayed as green triangles, Nox2-/- mice are displayed as gold triangles, and Ccr2-/- mice are displayed as yellow squares. For all, n = 5–9 mice in at least two independent trials; specifically, KPPR1-met n = 8, KPPR1-dir n = 9, gmhB n = 9, tatC n = 5, Nox2-/- n = 5, Ccr2-/- n = 8. Comparisons between the KPPR1-met group and each other group were assessed using an ordinary one-way ANOVA with Dunnet’s multiple comparisons correction, p-values for each comparison are indicated in (I–N). Source data for each panel are provided within the Source Data file.
As previously described, the gmhB mutant had high CFU in the lung, similar to KPPR1-met (metastatic; Fig. 4A)14. However, there was a larger gmhB founding population in the lung than observed with either KPPR1-met or KPPR1-dir (direct; Fig. 4E), suggesting that the absence of GmhB may enable better survival of K. pneumoniae at this site. Interestingly, the higher number of founders did not translate to higher lung CFU, perhaps due to less expansion of the population (Fig. 4I, CFU/Ns, p = 0.10). The expansion that occurred was uneven, with Ns/Nb similar to KPPR1-met (Fig. 5I). Despite high CFU and uneven replication, the systemic spread of gmhB resembled direct dissemination, with significantly higher GDlung-spleen and GDlung-liver (dissimilar populations) and lower FRDlung-spleen and FRDlung-liver (fewer shared clones) than KPPR1-met (Fig. 5A, B, J–M, Supplementary Fig. 3C). In the spleen, gmhB CFU was significantly lower than KPPR1-met, although the number of founders was not (Fig. 4B, F). Instead, there was lower expansion (CFU/Ns) in the gmhB mutant (Fig. 4J), consistent with published findings that gmhB is defective in the spleen ex vivo and in vivo14. Similarly, in the blood gmhB CFU was lower than that of KPPR1-met (Fig. 4D); lower CFU is attributed to reduced expansion (Fig. 4L) rather than tighter infection bottlenecks (Fig. 4H, similar Ns). In contrast, gmhB experienced a wide bottleneck in the liver (Fig. 4G, higher Ns) but had similar CFU (Fig. 4C), with significantly less expansion than KPPR1-met (Fig. 4K, lower CFU/Ns). These combined observations indicate that GmhB contributes to bacterial expansion in the lungs associated with metastatic dissemination. The reduced capacity of the gmhB mutant to replicate in the spleen and blood is interestingly associated with higher survival (wider bottlenecks), yet poor replication ability, in the lung and liver.
In contrast to GmhB, TatC was required for both survival and expansion in the lung as evidenced by lower CFU, lower founders (Ns) and less expansion (CFU/Ns) compared to KPPR1-met (Fig. 4A, E, I). In the lung, the median Ns/Nb was low, with even expansion resembling KPPR1-dir (Fig. 5I). Dissemination to the spleen also resembled KPPR1-dir, with significantly higher GD and lower FRD than KPPR1-met (Fig. 5J, K; Supplementary Fig. 3D); relatedness of clones between the lung and liver was variable (Fig. 5L, M). CFU in the spleen and blood, but not liver, were significantly lower than KPPR1-met (Fig. 4B–D). There were no differences in founding population sizes, although liver Ns trended higher than wild-type. Instead, the CFU defects were attributable to a lack of expansion at all sites (Fig. 4I–L). While the gmhB mutant is defective for replication in the lungs, the tatC mutant is defective for both survival and replication, which does not appear to influence the number of clones that reach systemic sites, yet both mutants are associated with reduced capacity to replicate in secondary sites.
Host oxidative burst determines the lung bottleneck, mode of lung expansion, and the type of dissemination
To determine how host defenses influence K. pneumoniae population dynamics, we performed STAMPR analysis of wild-type K. pneumoniae in mice defective in NADPH oxidase Nox2 (Nox2-/- a.k.a. Cybb-/-), and the monocyte chemokine receptor Ccr2 (Ccr2-/-), which are required for the phagocyte oxidative burst and monocyte recruitment, respectively. Nox2 restricts K. pneumoniae replication in the lung but has more subtle influences in the spleen and liver34. After lung infection with KPPR1-STAMPR, mice lacking Nox2 displayed significantly elevated lung CFU compared to wild-type mice (Fig. 4A). The increase in CFU was equal in magnitude to the increase in founders, indicating that the increase in CFU is explained by wider bottlenecks rather than increased replication (Fig. 4E). However, the expansion observed in Nox2-/- mice was significantly more even than wild-type metastatic group, with consistently low Ns/Nb (Fig. 5I). Correspondingly, all Nox2-/- mice exhibited direct dissemination to both the liver and spleen, with high GD and low FRD relative to the lungs (Fig. 5E, F, Supplementary Fig. 3E). However, bacterial burdens in the liver, spleen, and blood of Nox2-/- mice were similar to KPPR1-met mice (Fig. 4B–D), with similar founding population sizes (Fig. 5F–H) and expansion of founders (Fig. 4J–L). Nox2-/- mice had high morbidity at this timepoint and dose, indicative of severe infection and precluding more replicates.
Since Nox2 is involved in oxidative bursts across neutrophils and monocytes that are highly recruited during K. pneumoniae lung infection14, we sought to resolve if monocytes influence K. pneumoniae dissemination by infecting Ccr2-/- mice with KPPR1-STAMPR. In contrast to the Nox2-/- mice, mice lacking Ccr2 had similar bacterial burden, founding populations, and expansion across sites (Fig. 4A–L) as KPPR1-met mice. However, Ccr2-/- mice had more even replication in the lung (Fig. 5I). Based on GD and FRD, both metastatic and direct dissemination were observed in Ccr2-/- mice (Fig. 5G, H, Supplementary Fig. 3F). Thus, monocytes may contribute to uneven replication of K. pneumoniae expansion in the lung. In contrast, Nox2 tightens infection bottlenecks in the lung, drives heterogeneity in K. pneumoniae replication in the lung, and appears to be required for metastatic dissemination. Thus, oxidative bursts from innate immune cells, partially from Ccr2+ subsets, may impose stresses that result in heterogenous K. pneumoniae replication in the lungs that influence downstream translocation patterns.
Patterns of dissemination from the lung influence the similarity of populations in the liver and spleen
While the lung was largely distinct from secondary organs, clonal sharing between secondary sites varied with the host or bacterial genotype and mode of dissemination. Spleen and liver populations were more similar in KPPR1-infected wild-type mice with metastatic dissemination (Fig. 5N). Clonal sharing between the spleen and liver in animals with direct dissemination varied, with some animals having similar clones and others having distinct clones (Fig. 5N). Across perturbations of bacterial and host factors, there were also examples of mixed dissemination types, with metastatic dissemination (high FRD) to one organ and direct dissemination (low FRD) to the other in a single mouse. For example, in gmhB-STAMPR infected mice, the three animals where metastatic dissemination to the spleen was observed had direct dissemination to the liver, evidenced by lower FRD values, and similar patters were observed in tatC-STAMPR infections (Fig. 5A–D, Supplementary Fig. 4). These observations may indicate that bacterial mutants with tissue-specific fitness defects may consequently undergo distinct modes of dissemination between different host tissues. KPPR1-STAMPR infections in Ccr2-/- mice also had mixed dissemination patterns. In 2/6 mice, metastatic dissemination to the spleen and direct dissemination to the liver were observed. These data reveal complex tissue-specific interactions in which the extent of replication in the lungs, which in turn influences metastatic or direct dissemination, can influence the genetic similarity of bacteria between systemic sites.
Discussion
Here, using a murine model of bacteremic pneumonia, we leveraged barcoded K. pneumoniae to investigate bacterial dissemination from the lung. This second phase of bacteremia pathogenesis has been impossible to study independently from other phases using tissue CFU alone. Using high-complexity libraries of barcoded bacteria with varied bacterial and mouse genotypes, we defined two modes of lung dissemination and identified bacterial and host factors that influence infection dynamics. Metastatic dissemination, broadly defined as dissemination that requires replication in an upstream site, was associated with uneven clonal expansion of K. pneumoniae in the lung and translocation of dominant lung clones to secondary sites. Metastatic dissemination was also associated with high bacterial burdens. Direct dissemination, defined as clones in the lung translocating to the secondary site without substantial expansion in the lung, was associated with lower tissue CFU in wild-type infections (Fig. 6). Both patterns occurred in early infections, whereas metastatic dissemination was the dominant pattern in late disease. The bacterial factors GmhB and TatC had distinct contributions to metastatic dissemination, infection bottlenecks, and expansion at primary and secondary sites. The host defense effector Nox2 was required for both metastatic dissemination and control of the initial infecting population, leading to severe infection with high bacterial burdens in Nox2-/- mice. These findings uncover a critical role for replication patterns at the primary site of infection in modulating dissemination. We propose that uncovering the hidden variables that influence dissemination and bacteremia dynamics will reveal principles of infection that underlie disease severity during pneumonia and progression to bacteremia.
Bacterial barcoding and STAMPR analysis revealed infection dynamics that are unresolved with quantitative culture and CFU values alone. In direct dissemination, replication is not necessary prior to translocation from the initial site. This pattern may arise due to defects in the host barrier integrity during infection. In metastatic dissemination, clonal replication in the lung leads to the presence of dominant clones in secondary sites. It is likely that bacterial factors influencing replication ability at the initial site also influence the ability for metastatic dissemination to occur. Metastatic dissemination is influenced by time, where clones which have more time to replicate are more likely to translocate and be identified at a secondary site. While two patterns of dissemination are described here, these modes likely represent a spectrum of translocation dynamics.
The primary distinguishing feature between metastatic and direct dissemination is the extent to which bacterial replication promotes the transit of clones from primary to secondary compartments (Fig. 6). Metastatic dissemination, detectable with high FRD values between the lung and secondary tissues, involves clones prominent in the lung population disseminating to the blood, liver, and spleen. This metastatic pattern suggests that clones surviving initial host defenses replicate in the lung and eventually disseminate. Similarity between the lung and other sites could be explained by a single dissemination event with expansion in the second site, or repeated dissemination of dominant lung clones. In contrast, direct dissemination occurs when bacteria do not undergo high clonal expansion in the initial site. Several biological mechanisms may explain how direct dissemination occurs. One is that a small degree of transit occurs early following infection, or even during the inoculation process, where organs are seeded with different subpopulations of clones that progress on different trajectories of survival and replication. Alternatively, perhaps low-abundance clones occupy privileged niches in the primary site that enable more efficient dissemination.
Identifying which organ reservoirs drive persistent bacteremia is important to understand pathogenesis and potentially direct patient care, as the reservoir likely differs by pathogen. In Streptococcus pneumoniae, lung infection seeds the spleen which in turn is the reservoir for bacteremia36. Treatment with low doses of azithromycin that concentrated in the spleen without reaching inhibitory levels in the blood did not affect lung CFU, but lowered spleen CFU and cleared bacteremia. This provided a mechanism to support the observed clinical efficacy of azithromycin combined with a beta-lactam. These dynamics resemble the direct dissemination of K. pneumoniae, where lung and spleen populations diverge. However, the lung appears to be an important reservoir in mice with metastatic dissemination with predominant clones driving bacteremia. This is further supported by the pattern in 48-hour infections, with higher systemic burdens without an increase in founding populations, and closer relatedness between the lung and systemic sites. Guiding antimicrobial therapy to short-circuit the dynamics driving bacteremia could be a focus for future treatment strategies.
Barcoding provided insights on bacteremia fitness factors and host defenses that could not be elucidated from CFU alone14. Mutations of the bacteria or host led to infection dynamics that were distinct from both metastatic and direct dissemination observed in wild-type infections. Mutation of gmhB led to a wider bottleneck in the lung but with similar CFU and uneven expansion seen in KPPR1-met mice, direct dissemination to the liver and spleen, and decreased expansion at these sites. GmhB is involved in the biosynthesis cascade of ADP-heptose, which can be integrated into the LPS core or sensed as a soluble mediator by pro-inflammatory cytosolic ALPK113,14,32. These data suggest a fitness tradeoff between survival of founders in the lung, and perhaps the liver, versus the consistent ability for the population to expand in diverse tissue. Since LPS protects bacteria from multiple host threats, it is likely that a modified LPS molecule in the absence of ADP-heptose leaves bacteria vulnerable to specific environmental stressors but may mask the bacteria from host defenses elicited by ALPK1. Interestingly, our previous work did not detect a difference in overall immune cell infiltration or cytokine production in lung homogenate of mice infected with KPPR1 vs. gmhB14. The exact interactions between tissues, ADP-heptose, and ALPK1 need to be fully investigated. In contrast, TatC was important for survival in the lung and population expansion across sites, with mixed patterns of dissemination. Perhaps TatC is dispensable for exiting the lung via direct dissemination and reaching systemic sites but is required for expansion amidst host threats (lower CFU/Ns). Alternatively, reductions in CFU/Ns values (Fig. 4I–L) could reflect a reduction in reseeding of the same clones from the lung to the spleen.
Nox2-/- mice had unique bacteremia dynamics, consistent with defective killing of initial founders and loss of a barrier for dissemination, leading to higher lung founding populations, lung burdens, and populations in secondary organs that develop independently of growth in the lung. It is also interesting that the initial inoculum was highly diverse (~40,000 barcodes) but that lungs of wild-type mice were less complex (25th to 75th percentile Ns ranging from 2500 to 10,000 barcodes). We hypothesize that innate immune cells effectively clear most of the inoculum but cannot control the entire infection, leading to the reduction in initial barcodes. This hypothesis is supported by the finding that Nox2-/- animals have a higher number of founding clones in the lungs (Fig. 4E). These data indicate that Nox2 is required for metastatic dissemination. Considering that Ccr2 was dispensable, metastatic dissemination may be independent of inflammatory monocytes and instead may depend on other immune subsets, like neutrophils or alveolar macrophages. Our study did not specifically deplete neutrophils during K. pneumoniae infection so we cannot make specific conclusions about this subset. However, given that most immune cells in the lung during K. pneumoniae infection are neutrophils and Ccr2+ monocytes14,37, both of which express Nox2, we believe that the dispensability of Ccr2+ cells in determining patterns of dissemination supports a role for neutrophils in mediating these modes. Since metastatic dissemination is marked by high bacterial replication and lung CFU, it is curious that this pattern requires oxidative bursts from immune cells, which is typically associated with bacterial clearance. Neutrophil killing of K. pneumoniae can be inhibited by soluble mediators secreted by myeloid-derived suppressor cells38. Potentially, the lung inflammatory environment inhibits effective containment of K. pneumoniae. Nox2-dependent oxidative stress may impose heterogeneity on bacterial populations, leading to uneven replication and metastatic dissemination.
The dynamics of bacteremia likely depend on the initial site of infection. Here, we used pneumonia as the primary infection due to its relevance in K. pneumoniae disease and AMR infections9. The microenvironment of the lung during K. pneumoniae infection can change bacterial metabolism and may influence fitness at secondary sites39. DsbA, involved in disulfide bond formation, is required for splenic fitness after tail vein injection but is dispensable when infection originates in the lung34. In metastatic dissemination, perhaps bacteria that replicate in the lung are primed for increased splenic survival in comparison to those that translocate before replication (as seen in direct dissemination). The large variation in blood CFU detected in our data further imply that dissemination likely occurs through multiple rounds of shedding, rather than a continuous event. This characteristic may be initial site-dependent and requires investigation. Deeper understanding of dissemination during Gram-negative bacteremia will integrate knowledge of primary infections across other relevant sites.
Limitations of this study include the use of a single K. pneumoniae strain and lack of clinical outcome data. K. pneumoniae is a highly diverse species with remarkable strain variation. We used KPPR1, a well characterized hypervirulent strain that has been used across laboratories for decades. This strain demonstrates the utility of bacterial barcoding to provide insight into the dynamics underlying previously established paradigms and provides insight into broad mechanisms of K. pneumoniae dissemination. Other K. pneumoniae pathotypes may display different dynamics. Here, we measured bacterial abundance in secondary sites during systemic disease but not specific outcomes like body weight, temperature, or serum markers of tissue damage. However, no wild-type animals were noted to appear moribund at the 24-hour timepoint, at which both dissemination patterns were similarly present. Nox2-/- animals, which all experienced direct dissemination, and wild-type mice analyzed at 48-hours, which mostly experienced metastatic dissemination, were all noted to have severe morbidity. Notably, dissemination patterns did not seem to correlate with mouse age or sex across trials.
In summary, bacterial barcoding unveiled infection dynamics hidden when measuring only bacterial tissue burden. We discovered that patterns of heterogenous clonal expansion at primary sites underlie variability in dissemination and bacterial burden in tissue. Future work defining mechanisms of metastatic or direct dissemination will substantially deepen our insights into how pathogens traverse throughout the host and contextualize how host-pathogen interactions control infection. Our work demonstrates how high-complexity libraries of barcoded bacteria can be leveraged to reveal frameworks of infection and better understand host-pathogen interactions during bacteremia.
Methods
Bacterial strains and reagents
All reagents were sourced from Sigma Aldrich unless noted otherwise. K. pneumoniae cultures were grown shaking overnight at 37 °C in LB broth (Fisher Bioreagent, Ottowa, ON), and bacteria grown on LB plates were incubated at 30 °C. Growth media was supplemented with 40 µg/mL kanamycin (Sigma Aldrich, St. Louis, MO) to select for bacteria containing barcodes and/or with 50 µg/mL hygromycin (Sigma-Aldrich) to select for knockout-strains generated by Lambda red mutagenesis. The bacterial strains used in this study are described in Supplementary Table 1, and primers used in the study are in Supplementary Table 2.
Lambda Red mutagenesis was used to generate gmhB (VK055_2352) and tatC (VK055_3142) isogenic knockouts containing a hygromycin resistance cassette14,30,40. To generate electrocompetent KPPR1 harboring the pKD46 plasmid, an overnight culture was first grown at 30 °C. The culture was then diluted into LB broth containing 50 µg/mL spectinomycin, 50mM L-arabinose, 0.5 mM EDTA (Promega, Madison, WI), and 10 µM salicyclic acid and cultured until reaching exponential phase. The culture was placed on ice for 30 minutes, pelleted at 8000xg for 15 minutes at 4 °C, and serially washed with 50 mL 1 mM HEPES (pH 7.4; Gibco, Grand Island, NY), 50 mL diH2O, and 20 mL 10% glycerol. A hygromycin resistance cassette from the pSIM18 plasmid was amplified with primers containing 65 base pair homology at the 5’ end of the chromosomal site flanking the open reading frame of gmhB14 or tatC (Supplementary Table 2). The purified fragment was electroporated into competent KPPR1-pKD46. Transformants were allowed to recover overnight, shaking at 30 °C and then selected on agar with hygromycin. Each knockout was confirmed using primers flanking either gmhB or tatC.
Construction of STAMPR Libraries
The MFDλpir-pSM1 plasmid donor library, containing random 30 nucleotide barcodes, was cultured in LB broth with kanamycin and 600 µM DAP15. To construct a K. pneumoniae library, a 1:1:1 ratio of the MFDλpir-pSM1 plasmid donor library, a helper donor (MFDλpir pLMP1039), and either wild-type KPPR1 (for KPPR1-STAMPR), ΔgmhBhygro (for gmhB-STAMPR), or ΔtatChygro (for tatC-STAMPR) were spotted on a total of ten 0.45 µm filters and incubated on plates containing 300 µM DAP for 20-24 hours. After incubation, filters were combined and vigorously vortexed with PBS to release the transconjugants. The concentration of the recovered bacteria was determined by quantitative plating, and remaining transconjugants were plated onto multiple LB agar plates containing kanamycin and grown overnight at 30 °C. All plates were pooled together by scraping recovered lawns into LB + 25% glycerol and mixing thoroughly. The pools were split into multiple aliquots, labeled as the appropriate barcoded library, and stored at −80 °C until further use.
Murine Bacteremia
This study included male and female mice used between the ages of 8-12 weeks from the C57BL/6J lineage and included wild-type animals purchased directly (Jackson Laboratory, Bar Harbor, ME), Nox2-/- (Cybb-/-, B6129S-Cybbtm1Din/J41), and Ccr2-/- genotypes (B6129F2-Cmkbr2tm1Kuz42). Animals were housed in accordance with to humane guidelines for animal handling43, including a 12-hour light and dark cycle, ambient temperatures between 20–26 °C, and humidity of roughly 30–35%. Prior to each infection, K. pneumoniae overnight cultures for each strain were pelleted at 5000 x g for 15 minutes. The bacterial pellets were resuspended in PBS and the OD600 measured to adjust cultures to the desired concentration. For the pneumonia model, mice were anesthetized with isoflurane and 1×106 K. pneumoniae CFU in 50 µL PBS was administered retropharyngeally. This dose is above the seven-day LD50 for KPPR1 infected mice, but at 24-hours post infection robust pneumonia and bacteremia is observed with no signs of clinical distress37. Mice were sacrificed at 24- or 48-hours post-infection and lung, spleen, liver, and blood were collected. Blood samples were collected by cardiac punctures and dispensed into heparin-coated tubes (BD, Franklin Lakes, NJ) to prevent clotting. Spleens were homogenized in 1 mL PBS and livers were homogenized in 2 mL PBS, and a 100 µL sample was removed for quantitative plating to calculate the bacterial burden at each site. The remaining organ homogenate was plated onto LB dishes with kanamycin to recover the total number of barcodes within each site and plates were incubated overnight at 30 °C. The following day, bacterial lawns were scraped into 10 mL PBS + 25% glycerol, keeping each mouse and tissue separate, and mixed thoroughly. ~1 x 109 CFU was removed from each sample and boiled to extract bacterial DNA. The remaining bacteria collected from lawn scraping were stored at −80 °C.
DNA sequencing and STAMPR analysis
The barcode-containing region was amplified from the bacterial DNA using the boiled bacterial cells in a PCR with OneTaq HS Quick-Load Master Mix (New England Biolabs) and custom primers (Supplementary Table 2). Primers contained TruSeq indexes and adapters for Illumina sequencing. PCR products were pooled, purified with a column kit (GeneJet PCR Purification Kit), and sequenced on either an Illumina MiSeq or Illumina NextSeq 1000.
Illumina sequencing reads were demultiplexed with a custom R script, then trimmed and mapped to the appropriate reference library using either CLC Genomics Workbench (Qiagen) or custom R scripts. Mapped reads were exported as a CSV table. Any sample deemed to have low-quality sequencing results, defined as <10,000 reads, was removed from the study and not included in further STAMPR pipeline analysis. Noise correction was performed to adjust for increased index hopping in samples sequenced on the Illumina NextSeq. Founding population estimates were determined using STAMPR scripts28 and genetic distance was estimated with Cavalli-Sforza cord distance27.
Ns values were calculated as follows. After removing noise due to index hopping according to the STAMPR algorithm28, the number of unique nonzero barcodes was determined. Then, for each output sample, the reference library was resampled according to a multinomial distribution with a sampling size equal to the total read count of the output sample. This resampled input vector was again resampled according to a multinomial distribution with varying sampling sizes. At each sampling size, the number of unique barcodes was calculated and a reference resample curve was created by plotting the sampling depth as the X-axis and the number of unique barcodes as the Y-axis. The number of unique barcodes in the output sample was used for inverse linear interpolation from the reference resample curve to calculate Ns.
GD was calculated as the Cavalli-Sforza chord distance27. FRD was calculated as follows. For two samples A and B, GD was first calculated (iteration 0). The most abundant shared barcode (by geometric mean) in Samples A and B was removed, and GD was calculated again (iteration 1). Then, the first and second most abundant barcode were removed, and GD was calculated again (iteration 2). This procedure was repeated for 1000 iterations or until one sample had all barcodes >0 reads removed. The number of iterations that yield GD < 0.8, the threshold established for genetic relatedness, is defined as RD. Note that 0.8 is a relatively strict threshold, as samples with no overlapping barcodes have GD = (2√2)/π = 0.9. For pairs of samples where all 1000 iterations yielded GD < 0.8, RD was set to equal the number of unique barcodes with >0 reads in each sample. FRD(A-B) is defined as ln(RD(A-B) + 1)/ln(number of nonzero barcodes in sample B + 1).
Statistical analysis
At least two independent infections were performed for each mouse and K. pneumoniae mutant group. Statistical significance was defined as a p-value < 0.05 (GraphPad Prism) as determined using: ordinary one-way ANOVA with Dunnett’s multiple comparison to assess differences among multiple groups with one reference mean, ordinary one-way ANOVA with Tukey’s multiple comparison to assess differences among any groups, or unpaired t-tests to assess differences between two groups. All box plots included in this study display individual data points, with the center line representing the median, the box displaying the 25th and 75th percentiles, and whiskers extending to the minimum and maximum values.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The STAMPR pipeline output underlying all data generated in this study have been deposited in GitHub at https://github.com/hullahalli/stampr_rtisan/tree/main/Holmes2024. All underlying data for Figs. 1, 2G–J, Figs. 3–5 and Supplementary Fig. 1A, Supplementary Figs. 2, and 3 are derived from the STAMPR pipeline output provided in Supplementary Data 1 and grouped as individual figure panels in the Source Data file. The data from Fig. 2A–F and Supplementary Fig. 4 are publicly available at the GitHub link provided above.
Code availability
Barcode counts and original scripts have been deposited on Github (https://github.com/hullahalli/stampr_rtisan). Individual heat maps for GD and FRD for each mouse in the study can be found in Supplementary Data 2.
References
Hajj, J., Blaine, N., Salavaci, J. & Jacoby, D. The “Centrality of Sepsis”: A review on incidence, mortality, and cost of care. Healthcare 6, 90 (2018).
Singer, M. et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 315, 801–810 (2016).
Verway, M. et al. Prevalence and mortality associated with bloodstream organisms: a population-wide retrospective cohort study. J. Clin. Microbiol. 60, e0242921 (2022).
Wisplinghoff, H. et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–317 (2004).
Marra, A. R. et al. Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J. Clin. Microbiol. 49, 1866–1871 (2011).
Diekema, D. J. et al. The Microbiology of Bloodstream Infection: 20-Year Trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 63, e00355-19 (2019).
CDC. Antibiotic Resistance Threats in the United State, 2013. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; (2013).
CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; (2019).
Collaborators, A. R. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Holmes, C. L., Anderson, M. T., Mobley, H. L. T. & Bachman, M. A. Pathogenesis of gram-negative Bacteremia. Clin. Microbiol. Rev. 34, e00234–20 (2021).
Broadley, S. P. et al. Dual-track clearance of circulating bacteria balances rapid restoration of blood sterility with induction of adaptive immunity. Cell Host Microbe 20, 36–48 (2016).
Milivojevic, M. et al. ALPK1 controls TIFA/TRAF6-dependent innate immunity against heptose-1,7-bisphosphate of gram-negative bacteria. PLoS Pathog. 13, e1006224 (2017).
Zhou, P. et al. Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose. Nature 561, 122–126 (2018).
Holmes, C. L. et al. The ADP-Heptose Biosynthesis Enzyme GmhB is a conserved gram-negative bacteremia fitness factor. Infect. Immun. 90, e0022422 (2022).
Fakoya, B. et al. Nontoxigenic Vibrio cholerae challenge strains for evaluating vaccine efficacy and inferring mechanisms of protection. mBio 13, e0053922 (2022).
Hullahalli, K., Dailey, K. G. & Waldor, M. K. Innate immune responses yield tissue-specific bottlenecks that scale with pathogen dose. Proc. Natl Acad. Sci. USA 120, e2309151120 (2023).
Liu, X. et al. Exploration of bacterial bottlenecks and Streptococcus pneumoniae Pathogenesis by CRISPRi-Seq. Cell Host Microbe 29, 107–20.e6 (2021).
Wincott, C. J. et al. Cellular barcoding of protozoan pathogens reveals the within-host population dynamics of Toxoplasma gondii host colonization. Cell Rep. Methods 2, 100274 (2022).
Aggarwal, S. D. et al. BlpC-mediated selfish program leads to rapid loss of Streptococcus pneumoniae clonal diversity during infection. Cell Host Microbe 31, 124–34.e5 (2023).
Woodward, S. E. et al. Gastric acid and escape to systemic circulation represent major bottlenecks to host infection by Citrobacter rodentium. ISME J. 17, 36–46 (2023).
Campbell, I. W., Hullahalli, K., Turner, J. R. & Waldor, M. K. Quantitative dose-response analysis untangles host bottlenecks to enteric infection. Nat. Commun. 14, 456 (2023).
Zhang, T., Sasabe, J., Hullahalli, K., Sit, B. & Waldor, M. K. Increased Listeria monocytogenes dissemination and altered population dynamics in Muc2-deficient mice. Infect Immun. 89, e00667–20 (2021).
Hullahalli, K. & Waldor, M. K. Pathogen clonal expansion underlies multiorgan dissemination and organ-specific outcomes during murine systemic infection. Elife. 10, e70910 (2021).
Mahmutovic, A. et al. RESTAMP - Rate estimates by sequence-tag analysis of microbial populations. Comput Struct. Biotechnol. J. 19, 1035–1051 (2021).
Bachta, K. E. R., Allen, J. P., Cheung, B. H., Chiu, C. H. & Hauser, A. R. Systemic infection facilitates transmission of Pseudomonas aeruginosa in mice. Nat. Commun. 11, 543 (2020).
Zhang, T. et al. Deciphering the landscape of host barriers to Listeria monocytogenes infection. Proc. Natl Acad. Sci. USA 114, 6334–6339 (2017).
Abel, S. et al. Sequence tag-based analysis of microbial population dynamics. Nat. Methods 12, 223–226 (2015).
Hullahalli, K., Pritchard, J. R. & Waldor, M. K. Refined quantification of infection bottlenecks and pathogen dissemination with STAMPR. mSystems 6, e0088721 (2021).
Broberg, C. A., Wu, W., Cavalcoli, J. D., Miller, V. L. & Bachman, M. A. Complete Genome Sequence of Klebsiella pneumoniae Strain ATCC 43816 KPPR1, a Rifampin-Resistant Mutant Commonly Used in Animal, Genetic, and Molecular Biology Studies. Genome Announc. 2, (2014).
Bachman, M. A. et al. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6, e00775 (2015).
García-Weber, D. et al. ADP-heptose is a newly identified pathogen-associated molecular pattern of Shigella flexerni. EMBO Rep. 19, e46943 (2018).
Garcia-Weber, D. & Arrieumerlou, C. ADP-heptose: a bacterial PAMP detected by the host sensor ALPK1. Cell Mol. Life Sci. 78, 17–29 (2021).
Anderson, M. T., Mitchell, L. A., Zhao, L. & Mobley, H. L. T. Citrobacter freundii fitness during bloodstream infection. Sci. Rep. 8, 11792 (2018).
Holmes, C. L. et al. Klebsiella pneumoniae causes bacteremia using factors that mediate tissue-specific fitness and resistance to oxidative stress. PLoS Pathog. 19, e1011233 (2023).
Mobley, H. L. T. et al. Fitness factor genes conserved within the multi-species core genome of gram-negative enterobacterales species contribute to Bacteremia Pathogenesis. PLoS Pathog. 20, e1012495 (2024).
Carreno, D. et al. Splenic macrophages as the source of bacteraemia during pneumococcal pneumonia. EBioMedicine 72, 103601 (2021).
Xiong, H. et al. Distinct contributions of neutrophils and CCR2+ monocytes to pulmonary clearance of different Klebsiella pneumoniae Strains. Infect. Immun. 83, 3418–3427 (2015).
Ahn, D. et al. Acquired resistance to innate immune clearance promotes Klebsiella pneumoniae ST258 pulmonary infection. JCI Insight 1, e89704 (2016).
Wong Fok Lung, T. et al. Klebsiella pneumoniae induces host metabolic stress that promotes tolerance to pulmonary infection. Cell Metab. 34, 761–74.e9 (2022).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Werner, J. L. et al. Induction of Pulmonary Granuloma formation by Propionibacterium acnes is regulated by MyD88 and Nox2. Am. J. Respir. Cell Mol. Biol. 56, 121–130 (2017).
Moore, B. B. et al. Protection from pulmonary fibrosis in the absence of CCR2 signaling. J. Immunol. 167, 4368–4377 (2001).
(US) NRC. Guide for the care and use of laboratory animals. Washington, D.C.: National Academies Press; (2011).
Acknowledgements
The authors were supported by the following funding sources: K99A1175481 – CLH; HHMI and R01 AI042347 – MKW.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.L.H., K.G.D., K.H., M.K.W., M.A.B. Methodology: C.L.H., K.G.D., K.H., A.E.W., S.M., B.S.M., L.V.U. Investigation: C.L.H., K.G.D., K.H., G.B., M.K.W., M.A.B. Visualization: C.L.H., K.G.D., K.H. Funding acquisition: M.K.W., M.A.B. Writing: C.L.H., K.G.D., K.H., M.K.W., M.A.B. Manuscript Review and Editing: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
The study was approved by the University of Michigan Institutional Animal Care and Use Committee (protocol: PRO00011097), and performed with careful adherence to humane guidelines for animal handling43.
Peer review
Peer review information
Nature Communications thanks Kelly Bachta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Holmes, C.L., Dailey, K.G., Hullahalli, K. et al. Patterns of Klebsiella pneumoniae bacteremic dissemination from the lung. Nat Commun 16, 785 (2025). https://doi.org/10.1038/s41467-025-56095-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56095-3