Abstract
Chronic kidney disease (CKD) ultimately causes renal fibrosis and end-stage renal disease, thus seriously threatens human health. However, current medications for CKD and fibrosis are inefficient, which is often due to poor targeting capability to renal tubule. In this study, we discover that biomimetic high-density lipoprotein (bHDL) lipid nanoparticles possess excellent targeting ability to injured tubular epithelial cells by kidney injury molecule-1(KIM-1) mediated internalization. Thus, we co-load anti-inflammatory drug triptolide (TP) and anti-fibrotic drug nintedanib (BIBF) on bHDL nanoparticles to treat CKD. Based on the targeted delivery and mutual enhancement of the efficacy of co-delivered drugs, the bHDL-based system effectively reduces kidney injury and alleviates renal fibrosis in different CKD mouse models. The mechanistic study shows that BIBF and TP synergistically remodel the fibrotic niches by decreasing inflammatory cytokines, limiting immune cell infiltration and inhibiting the activation of myofibroblasts. The bHDL vehicle also possesses high manufacturability, good safety and adequately reduces the toxicity of TP. Thus, this system is promising for the treatment of CKD and bHDL has good potential for delivering agents to damaged renal tubular epithelial cells.
Similar content being viewed by others
Introduction
Chronic kidney disease (CKD) is a common syndrome characterized as chronic structural and functional dysfunction of the kidney1,2,3, which can be triggered by various factors, including hypertension, diabetes, injury and complex genetic factors. CKD often persists for months leading to permanent renal damage, which finally develop to renal fibrosis and end-stage renal disease that requires dialysis or even kidney transplantation for the patients2,4. As CKD advances, a range of fatal complications may occur, such as cardiovascular disorders, metabolic acidosis and mineral and bone disorder5,6,7. CKD’s rising incidence has rendered it a crucial public health issue, posing a significant threat to human well-being, as well as a substantial financial burden8,9. Unfortunately, no effective treatments are currently available for patients with CKD, especially for renal fibrosis3. Hence, there are unmet clinical needs for new interventions that can effectively halt the progression of CKD and alleviate renal fibrosis.
Renal tubular epithelial cells (RTECs) play a vital role in the progression of CKD and renal interstitial fibrosis10. During kidney injury, these cells are not merely the victim of injury, but they also play a vital role in aggravating it11, indicating their potential as promising target cells for CKD targeted therapy. However, efficient and safe delivery of therapeutic drugs to RTECs is challenging due to the unique physiological and structural characteristics of the kidney, including the size barrier and charge barrier of the glomerular filtration apparatus comprising of fenestrated endothelium, glomerular basement membrane and podocytes12,13. Various strategies have been tested to achieve drug delivery to RTECs such as controlling the charge and particle size of nanoparticles14 and applying targeting ligands15,16. Even though these strategies effectively enhance drug accumulation in kidney, the efficiency of these systems is usually compromised by the hepatic distribution partly resulting from the formation of protein corona17.
Kidney injury molecule-1 (KIM-1), a type I transmembrane glycoprotein which is highly expressed on the injured proximal TEC, is being investigated as a potential renal-targeted delivery target18,19. Initially, KIM-1 was thought to be an early marker of renal tubular injury in the proximal area. In recent years, research has shown that it can serve as a receptor and mediates the uptake of a wide array of substances20,21,22. KIM-1 is able to serve as a scavenger receptor to clear apoptotic cells to protect against acute kidney injury(AKI)21,23. Therefore, the selective expression and functional specificity render KIM-1 an exemplary target for renal tubule-specific drug delivery.
Biomimetic high-density lipoprotein (bHDL) nanoparticles are simple delivery system comprised of apolipoproteins and phospholipids24. bHDL nanoparticles exhibit ultra-small particle sizes, a key characteristic enabling them to traverse the glomerular filtration barrier effectively, enhancing their chances to arrive at RTECs. Furthermore, bHDL nanoparticles retain the inherent structure of natural HDL, granting them prolonged circulation time with slow clearance by reticuloendothelial system. Besides, bHDL nanoparticles possess the capability of naturally targeting to scavenger receptors25,26. As previously mentioned, KIM-1 operates as a scavenger receptor, binding to phosphatidylserine (PS), oxidized LDL (ox-LDL), as well as natural LDL and fatty acids. Ox-LDL is a typical scavenger receptor ligand and binds to a variety of scavenger receptors, including LOX-1, scavenger receptor class A, CD36 and scavenger receptor class B type I (SR-BI)27. Notably, natural LDL does not bind to class A, C, and D scavengers and is a specific ligand for SR-BI. SR-BI can also effectively bind PS and fatty acids, indicating that KIM-1 possesses more functional similarities with SR-BI than others scavenger receptor. HDL is a type of classical receptor of SR-BI, implying a possible link between KIM-1 and HDL. However, the potential interaction between KIM-1 and HDL remains uncertain. Given the facts, it is crucial to explore whether KIM-1 can augment the uptake of b-HDL nanoparticles by damaged RTECs, which may imply the promising application of bHDL nano-delivery system for precise drug delivery within the renal milieu.
Current treatments for CKD and fibrosis primarily aim to suppress typical symptoms and usually target single signaling pathway. However, therapeutic outcomes of these treatments are unsatisfactory, which is likely due to the intricate nature of CKD and the presence of fibrotic niches where inflammation and fibrosis invariably coexist, as well as a complex interplay between multiple cells28. Simultaneous regulation of inflammation and fibrosis may be an effective therapeutic approach. For this purpose, we proposed that triptolide (TP) and nintedanib (BIBF) may be a good combination that could remodel the fibrotic niche. Here, TP is the main active ingredient obtained from the root bark of Tripterygium wilfordii., which is widely used in the treatment of renal diseases in China, showing impressive anti-inflammatory and immunosuppressive activities29. However, its clinical use is severely restricted by its low solubility/permeability, poor bioavailability, and potential off-target toxicity30. BIBF is a small-molecule tyrosine kinase inhibitor that has been approved for the treatment of idiopathic pulmonary fibrosis31. It impedes the proliferation, migration, and transformation of fibroblasts, thereby exhibiting a potent antifibrotic effect32. Despite its efficacy, the absolute bioavailability of its oral formulation is only 4.69%33. Therefore, we aimed to develop a targeted approach based on bHDL nanoparticles that can improve their bioavailability and co-deliver TP and BIBF preferentially to the disease site to modify the renal malfunctions, while mitigating unintended drug distribution and reducing systemic toxicity.
In this study, we constructed bHDL lipid nanoparticles co-loaded with TP and BIBF for the treatment of CKD and explored the renal targeting ability of bHDL nanoparticles as well as the targeting mechanism. As expected, bHDL exhibited pronounced renal targeting, particularly accumulating in impaired RTECs. Further mechanistic studies highlighted the pivotal role of KIM-1 in the endocytic uptake of bHDL nanoparticles. Meanwhile, the simultaneous delivery of anti-inflammatory and antifibrotic drugs to the kidneys via bHDL nanoparticles can effectively reduce renal injury and delay fibrosis in multiple CKD mouse models.
Results
KIM-1 is markedly upregulated after kidney injury
Several studies have shown that KIM-1 is not expressed in normal kidney tissues, and the expression level rise significantly after kidney damage in AKI and diabetic kidney disease (DKD) mouse models, which is positively correlated with the degree of kidney injury20,34,35. To explore the KIM-1 expression level in CKD, three CKD mouse models with different etiologies, unilateral ureteral obstruction (UUO), folic acid-induced and adenine-induced mouse model, were constructed and the KIM-1 expression was determined. KIM-1 was highly expressed in all kinds of model mice as compared to the control mice (Fig. 1a–c). Co-staining KIM-1 with lotus tetragonolobus lectin (LTL), a marker of proximal tubule, demonstrated that KIM-1 is mainly expressed in the proximal tubules (Fig. 1a). Experimental results based on western blotting as well as qPCR also proved that KIM-1 is significantly raised after injury from the protein and transcriptional level (Fig. 1b–d). Additionally, KIM-1 expression level on cisplatin-induced RTECs was also assessed and the protein level of KIM-1 was dramatically elevated in the HK-2 cells after treating with cisplatin for 24 h (Supplementary Fig. 6c), which is in consistent with previous study36. In conclusion, KIM-1 is highly expressed on injured RTECs and could be a specific delivery target for injured RTEC-targeted renal therapy.
a Representative immunofluorescence images(left) and the Pearson correlation coefficient(right) of colocalization of KIM-1(red) and LTL (green) in the kidney of control, UUO model, folic acid model and adenine model mice. Scale bars, 100 µm. Data are mean ± SD (n = 3 independent samples). b Western blot analysis of KIM-1 protein expression in the kidney of control, UUO model, folic acid model and adenine model mice. The value of 60 and 55 kDa are referred to the predicted protein weight of KIM-1 and α-tubulin. α-tubulin was used as internal control. n = 3 biologically independent samples. c Semi-quantification of KIM-1 protein expression by Image J based the western blots shown in b and it was normalized with corresponding α-tubulin signal. Data are mean ± SD (n = 3 independent samples, two-tailed unpaired t-test). d Real-time qPCR analysis of Havcr1 mRNA expression level in kidney of control, UUO model, folic acid model and adenine model mice. Data are displayed as normalized fold expressions relative to control group, and Acta mRNA was used as internal control. Data are mean ± SD (n = 4 independent samples, two-tailed unpaired t-test). Control: untreated healthy mice, UUO model: unilateral ureteral obstruction model mice, folic acid model: mice treated with folic acid to induce kidney injury, adenine model: mice treated with adenine to induce kidney injury, LTL lotus tetragonolobus lectin. Source data are provided as a Source Data file.
Fabrication and characterization of the kidney targeted nanoparticles
KIM-1, markedly expressed on impaired RTECs, presents a potential target for renal-specific drug delivery37,38. Considering the features of bHDL nanocarrier, we postulated that it holds promise as a means of targeting KIM-1. To explore the KIM-1 targeting of bHDL, we firstly fabricated blank bHDL lipid nanoparticles, with the phospholipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and D-4F, a widely used apoA1 mimic peptide. Meanwhile, we co-loaded the anti-inflammatory drug TP and the antifibrotic drug BIBF into bHDL to obtain co-loaded nanoparticles TP/BIBF-bHDL for CKD treatment. Morphological analysis using transmission electron microscopy (TEM) and dynamic light scattering (DLS) revealed that blank bHDL possessed a predominantly discoidal structure with an average diameter of 10.9 nm (Fig. 2a, Supplementary Fig. 1). Upon drug loading, TP/BIBF-bHDL became less discoidal with a slight increase in diameter. Cryo-EM images showed that TP/BIBF-bHDL had higher contrast than blank bHDL, and displayed particles with both spherical and discoidal shapes (Fig. 2b). Zeta potential results showed that both blank and drug-loaded bHDL nanoparticles exhibited positive charge (Fig. 2c), which could facilitate their renal accumulation39. The positive charge of the nanoparticles may be attributed to the D-4F peptide, which contains multiple positively charged lysine residues. Upon intravenous injection, the dense positive charge on the nanoparticles may damage red blood cells, leading to potential toxicity. Therefore, the hemolytic activity of TP/BIBF-bHDL was investigated to assess its biocompatibility. Result showed that TP/BIBF-bHDL did not cause significant hemolysis with a hemolysis rate lower than 5% (Supplementary Fig. 2), confirming its safety.
a TEM images of bHDL and TP/BIBF-bHDL. Scale bars, 50 nm. b Cryo-EM images of bHDL and TP/BIBF-bHDL. Scale bars, 25 nm. c Size and zeta potential of bHDL, TP/BIBF-bHDL, TP-bHDL and BIBF-bHDL measured by DLS. Data are mean ± SD (n = 3 independent samples). d Changes in particle size of TP/BIBF-bHDL under 37 °C and 4 °C. Data are mean ± SD (n = 3 independent samples). e TEM images of TP/BIBF-bHDL and TP/BIBF-bHDL after incubation with 10%FBS or 10% lipoprotein-depleted FBS at 37 °C for 2 hours. Scale bars, 50 nm. n = 3 independent experiments. f Size of TP/BIBF-bHDL displayed in e and measured by image J. Data are mean ± SD (n = 20). g Quantification of serum protein absorbed on the bHDL nanoparticles and liposomes by BCA assay. Data are mean ± SD (n = 3 independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). h In vitro release profiles of TP/BIBF-bHDL in the presence of PBS and FBS. Data are mean ± SD (n = 3 independent samples). bHDL blank biomimetic high-density lipoprotein (HDL) nanoparticles, TP/BIBF-bHDL triptolide and nintedanib co-loaded biomimetic HDL nanoparticles, FBS fetal bovine serum. Source data are provided as a Source Data file.
Furthermore, we investigated the stability of the drug-laden bHDL nanoparticles, which exhibited no significant alterations in the particle size when they were suspended in phosphate buffer saline (PBS) at 37 °C for 12 h and 4 °C for 5 days (Fig. 2d). The stability of the bHDL nanoparticles in serum was also performed by monitoring changes in particle size and serum protein absorption. Given the presence of endogenous lipoproteins in serum, both normal fetal bovine serum (FBS) and lipoprotein-deproteinized FBS were used. After a 2-hour incubation at 37 °C with 10% FBS and 10% lipoprotein-deproteinized FBS, the TP/BIBF-bHDL exhibited no aggregation with only a slight increase in particle size of approximately 4 nm (Fig. 2e, f), demonstrating excellent stability in serum. The serum protein level absorbed on bHDL nanoparticles was estimated by Bicinchoninic acid (BCA) assay, with liposomes included as a control. The bHDL nanoparticles absorbed only ~25% proteins compared to liposomes (Fig. 2g). Interestingly, bHDL nanoparticle absorbed ~60% less protein when incubated in the lipoprotein-deproteinized FBS, indicating that lipoproteins are the primary proteins to be absorbed. Drug release profiles of TP/BIBF-bHDL in both PBS and FBS exhibited comparable trends, affirming the robust stability of TP/BIBF-bHDL in FBS (Fig. 2h). Additionally, we quantified the encapsulation efficiency (EE%) and drug loading capacity (LC%), with values of 97.7% and 6.01% for BIBF and 84.78% and 1.3% for TP, respectively (Table S1).
bHDL nanoparticles present excellent capability of targeting injured RTECs
To investigate the renal targeting efficacy of bHDL nanoparticles, we prepared DiD-labelled bHDL (DiD-bHDL) to assess the biodistribution in UUO mice, a classical mouse model of renal fibrosis. DiD-liposomes with a particle size of 80 nm were prepared (Supplementary Fig. 3), with the same lipid composition as bHDL but lacking D-4F. Following intravenous injection of DiD, DiD-liposomes and DiD-bHDL, vital organs were harvested at predetermined time points and placed in an in vivo imager to visualize the distribution. As shown in Fig. 3a-b, DiD-bHDL were rapidly taken up by the kidney, liver and lung after injection, and the renal localization of DiD-bHDL is higer than free DiD and DiD-liposomes. However, it should be noted that free DiD dye may form micelles, which could hinder its ability to pass through the kidney filter, thereby reducing its distribution in the kidney. The accumulation of bHDL in the kidney displayed a time-dependent increase, peaking at 2 hours post-injection. After 24 hours, substantial fluorescence was still visible in the injured kidney in the bHDL group. The fluorescence intensity was also semi-quantitatively analyzed (Fig. 3c–e, Supplementary Fig. 4a–c). The liver serves as the primary site for the uptake and elimination of nanoparticles, with both bHDL and liposomes distributing to this organ. It’s worth noting that the distribution of bHDL in the liver is lower than that of normal liposomes, which are composed of the same phospholipid DMPC, cholesterol and lack the D-4F component. Furthermore, we compared the biodistribution of bHDL in UUO model mice and healthy mice. The bHDL nanoparticles showed greater accumulation in the ligation kidneys, suggesting that bHDL can specifically target the damaged kidneys (Fig. 3b and d, Supplementary 4g). The localization of bHDL within the injured kidneys was further elucidated through immunofluorescence, revealing a preferential accumulation of bHDL in the damaged RTECs (Fig. 3f).
a Bioluminescence images of major organs in UUO mice after intravenous administration of DiD, DiD-liposome or DiD-bHDL at 0.5, 2, 8 and 24 hours. b Bioluminescence images of kidneys in UUO and healthy mice after DiD-bHDL administration at various time points. c Fluorescence intensity in organs of UUO mice treated with DiD, DiD-liposome or DiD-bHDL at 2 hours. d Fluorescence intensity in kidneys of healthy and UUO mice treated with DiD-bHDL. e Fluorescence intensity in the injured kidney, liver and lung of UUO mice treated with DiD-bHDL. Data in b–d are mean ± SD (n = 3 biologically independent samples for a–d, two-way ANOVA with Sidak’s multiple comparisons test and adjustment applied). f Immunofluorescence images of DiD-bHDL localization in kidneys of UUO mice at 2- and 24-hour post-treatment. LTL (green) marks the renal tubules. Scale bars, 50 µm. n = 3 independent samples. g BIBF concentration in major organs of UUO mice 2 hours after intravenous administration of TP/BIBF or TP/BIBF-bHDL. h BIBF concentration in normal and ligated kidneys of UUO mice treated with TP/BIBF-bHDL at different time points. i Mean concentration-time curves of BIBF in kidneys of UUO mice after intravenous administration of TP/BIBF or TP/BIBF -bHDL. Data in g–i are mean ± SD (n = 5 biologically independent samples, two-way ANOVA with Sidak’s multiple comparisons test). j Uptake of DiD-bHDL in normal and cisplatin-injured HK-2 cells for 2 hours. k Uptake of DiD-bHDL in normal and TGF-β1-stimulated NRK-49F cells for 2 hours. Data in j–k are mean ± SD (n = 3 independent samples, two-tailed unpaired t-test). l Mean concentration-time curves of BIBF in the plasma of rats treated with TP/BIBF or TP/BIBF-bHDL. Data are mean ± SD (n = 6 biologically independent samples, two-way ANOVA with Sidak’s multiple comparisons test and adjustment applied). DiD free DiD, DiD-liposome DiD-laden liposomes, DiD-bHDL DiD-loaded biomimetic high-density lipoprotein nanoparticles, UUO mice unilateral ureteral obstruction model mice, TP/BIBF free triptolide and nintedanib solution, TP/BIBF-bHDL triptolide and nintedanib co-loaded biomimetic high-density lipoprotein nanoparticles. Source data are provided as a Source Data file.
To more precisely delineate the systemic distribution of the therapeutic agents, we further investigated the targeting ability of bHDL to the damaged kidney by quantifying the concentration of BIBF in tissues. The distribution of the drug in each organ after tail vein injection of TP/BIBF or TP/BIBF-bHDL is shown in Fig. 3g and Supplementary Fig. 4d–f. These findings align with the results obtained from fluorescence imaging. Encapsulation of the drug in bHDL markedly increased its distribution in the kidneys, especially in the damaged kidneys (Fig. 3h), while concurrently reducing its distribution in the spleen. Analysis of the time profile of drug concentration in the kidney showed that the renal drug concentration in the TP/BIBF-bHDL group was significantly higher than that in the free drug group at all time points after drug administration (Fig. 3i), and the area under curve (AUC0-∞) in the kidney of the TP/BIBF-bHDL group was 72.85 ± 5.70 µg ml−1h, approximately double that of the free drug group (Table S3).
Additionally, we conducted in vitro cellular uptake experiments using HK-2 cells, with and without cisplatin pretreatment, to elucidate the capacity of bHDL to target injured RTECs. Our findings revealed that damaged HK-2 cells exhibited a higher uptake of bHDL compared to their normal counterparts (Fig. 3j). Concurrently, we assessed the uptake of bHDL by NRK-49F cells, a type of renal fibroblast cell, which revealed notably lower uptake relative to HK-2 cells, as shown in Supplementary Fig. 4h. Moreover, stimulation of fibroblast cells with TGF-β1 did not result in an increased uptake of bHDL (Fig. 3k). TGF-β1 could induce the fibroblast cells transition to myofibroblast cells which have decreased endocytosis ability due to the increased contractility and membrane tension40. Collectively, these results provide compelling evidence supporting the enhanced targeting capability of bHDL toward injured RETCs.
In addition, we performed pharmacokinetic studies to elucidate the in vivo behaviors of bHDL nanoparticles. After the administration of TP/BIBF-bHDL and TP/BIBF through tail vein injection and gavage administration of TP/BIBF, the concentration of BIBF in plasma at various time points was measured by LC-MS/MS. The key pharmacokinetic parameters were calculated using DAS statistical software and displayed in Table S2. As shown in Fig. 3l, BIBF exhibited low oral bioavailability, due to its low solubility and poor absorption in the intestinal environment. At all sampling time points, the blood drug concentration in the TP/BIBF-bHDL group was significantly higher than that in the TP/BIBF group, with 80% increase in AUC. Meanwhile, the half-life time (t1/2) of TP/BIBF-bHDL is also 30% higher. These results indicate that encapsulation of drugs in bHDL significantly improved drug bioavailability, reduced drug clearance and prolonged its half-life.
KIM-1 mediates bHDL nanoparticles uptake by injured RTECs
We further explored the cellular uptake mechanism of bHDL nanoparticles. To clarify the mechanism, we first investigated the endocytic pathways involved in the uptake of bHDL nanoparticles using various endocytosis inhibitors. As shown in Fig. 4a, the uptake of bHDL was significantly impeded by low temperature and cytochalasin D (an actin-disrupting agent), as well as O-Phospho-L-serine and BLT-1 (an inhibitor of SR-BI). The cubilin antibody slightly reduces the uptake of bHDL, but this reduction is not statistically significant, suggesting that the cubilin receptor is not the primary endocytic pathway for bHDL nanoparticles. Conversely, inhibitors like chlorpromazine (clathrin-mediated endocytosis inhibitor), amiloride (macropinocytosis inhibitor), cyclodextrins (lipid raft inhibitor), and gentamicin (megalin-mediated endocytosis inhibitor) did not affect bHDL uptake. These findings indicate that the uptake of bHDL is mainly via phagocytosis, phosphatidylserine receptor and SR-BI-mediated endocytosis. Moreover, pre-incubation of cells with D-4F significantly reduced the uptake of bHDL, highlighting the critical role of D-4F in RTECs uptake processes (Fig. 4b).
a Uptake of bHDL under 4°C and with different endocytic inhibitors. Data are mean ± SD (n = 3 independent samples, two-tailed unpaired t-test). b Effect of D-4F on bHDL uptake in HK-2 cell. Data are mean ± SD (n = 3 independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). c Uptake of DiD-bHDL in normal and cisplatin-injured HK-2 cells for 2 hours, with or without pretreatment using KIM-1 antibody (30 µg/ml, 0.5 hours). Data are mean ± SD (n = 3 independent samples, two-tailed unpaired t-test). The control refers to untreated cells; KIM-1 antibody pretreated refers to cells with antibody pretreatment. d Representative confocal images of cisplatin-injured HK-2 cells after 2 hours of exposure. The nuclei were stained DAPI (blue), KIM-1 was labeled in green and DiD-bHDL was visualized in red. Scale bars, 20 µm. e Representative immunofluorescence images of colocalization of KIM-1(green) with DiD-bHDL or DiD-liposome (red) in the kidney. Nuclei were stained DAPI (blue). Scale bars, 50 µm. f Quantification of the colocalization of KIM-1 and DiD-bHDL or DiD-liposome by Pearson’s correlation coefficient. The Pearson’s correlation coefficient was measured by Colocalization Finder with image J. Data are mean ± SD (n = 3 biologically independent samples, two-tailed unpaired t-test). g, h Uptake of DiD-bHDL in HK-2 cells with KIM-1 protein knockdown(g) or overexpression(h) for 2 hours after 24-hour cisplatin treatment(2 µg/ml). Data are mean ± SD (n = 3 biologically independent samples, two-tailed unpaired t-test). i Western blot analysis of SR-BI expression in normal and cisplatin-injured HK-2 cells treated with cisplatin(2 µg/ml) for 24 hours. n = 3 independent samples. j Semi-quantification of SR-BI protein level (i) using image J, with β-actin as the internal control. Predicted protein weights: β-actin (43 kDa) and SR-BI (74 kDa). Data are mean ± SD (n = 3 biologically independent samples, two-tailed unpaired t-test). DiD-liposome DiD-laden liposomes, DiD-bHDL DiD-loaded biomimetic high-density lipoprotein nanoparticles, control untreated cells, CDDP-induced cisplatin-induced injured cells, KIM-1-/- HK-2 HK-2 cells with HAVCR1 knocked down, OE-KIM-1 HK-2 HK-2 cells with HAVCR1 overexpression. Source data are listed in the Source Data file.
Injury resulted in a noteworthy upregulation of KIM-1 protein on RTECs (Supplementary Fig. 6c), accompanied by a substantial augmentation in their uptake of bHDL (Fig. 3j). The enhanced cellular uptake of bHDL in injured HK-2 cells may be attributed to the interaction between bHDL and KIM-1. To substantiate this hypothesis, a competitive inhibition experiment was firstly performed to examine the effect of KIM-1 antibody on bHDL uptake. HK-2 cells and cisplatin-induced injured HK-2 cells were incubated with DiD-bHDL for 2 hours, with or without pretreatment using the KIM-1 antibody. As shown in the Fig. 4c, the KIM-1 antibody effectively inhibited the uptake of bHDL by 30% in the damaged cells, while paradoxically augmenting its uptake in normal HK-2 cells. The reason for this is that normal HK-2 cells express less KIM-1, and the addition of antibody stimulated the damaged cells to express KIM-1 to a certain extent and increase their uptake. Whereas in cisplatin-induced injured HK-2 cells with high KIM-1 expression, bHDL could enter the injured HK-2 cells through KIM-1-mediated endocytosis, which was inhibited by the addition of antibody. Then confocal laser scanning microscopy was applied to explore whether the uptake of bHDL was mediated by KIM-1, and the colocalization of KIM-1 and bHDL in cisplatin-induced injured HK-2 cells were observed with a Pearson’s correlation coefficient of 0.9, indicating there is an interaction between KIM-1 and bHDL (Fig. 4d). The colocalization between KIM-1 and bHDL was also evaluated in vivo. Kidneys from UUO mice were harvested 2 hours following the intravenous administration of DiD-bHDL. Immunofluorescence staining was subsequently performed to observe the co-localization of DiD-bHDL with KIM-1. A DiD-liposome group was added as a comparison. As shown in Fig. 4e and f, DiD-bHDL co-localized well with KIM-1 in kidney tissue with a Pearson’s correlation coefficient of 0.94, indicating the direct association of bHDL and KIM-1. In contrast, a modest co-localization of KIM-1 with liposomes was observed, as evidenced by a Pearson’s correlation coefficient of 0.64. The reason for this may be that expression of KIM-1 confers a phagocytic phenotype on RETCs, leading to their uptake by liposomes. All these results reveal that bHDL enters cells through endocytosis mediated by KIM-1.
To further characterize the role of KIM-1 in the endocytosis of bHDL in HK-2 cells, we performed a cellular uptake experiment in HK-2 cells overexpressing KIM-1. KIM-1 was overexpressed in HK-2 cells using lentivirus tool ( ~ 54% increase, supplementary Fig. 5). The uptake results showed that the overexpression of KIM-1 in HK-2 cells resulted in a significant increase ( ~ 60%) of bHDL uptake after injury (Fig. 4h). This represents a notable increase compared to HK-2 cells without KIM-1 overexpression, which exhibited only around 20% uptake. We also attenuated the expression of KIM-1 in HK-2 cells by transfecting them with KIM-1-specific small interfering RNA. The downregulation of KIM-1 was confirmed using fluorescence microscopy (Supplementary Fig. 6a), qPCR (Supplementary Fig. 6b), and western blot analysis (Supplementary Fig. 6c-d). Flow cytometry analyses revealed a significant decrease in the uptake of bHDL in the KIM-1 knockdown HK-2 cells compared to the normal HK-2 cells after injury (Fig. 4g).
To verify the increased uptake of bHDL by damaged cells compared with normal cells was caused by increased expression of KIM-1, not the SR-BI receptor, a widely known HDL receptor41, we assessed the expression of SR-BI on cisplatin-induced injured HK-2 cells. Western blot results showed that cisplatin injury did not increase the expression of SR-BI on HK-2 cells but rather slightly decreased it (Fig. 4i and j). In addition, we also detected the expression of SR-BI in the injured kidney of UUO mice by immunofluorescence. The data obtained from the fluorescence semi-quantitative analysis demonstrated that the injury did not significantly enhance the expression of SR-BI in the damaged kidneys (Supplementary Fig. 7a, b). It is noteworthy that injured kidneys are often accompanied by macrophage infiltration, which is highly expressed SR-BI. To gain a more accurate understanding of the impact of injury on SR-BI expression in RTECs, we conducted a detailed analysis of published single-cell sequencing data from UUO-injured kidneys42. No notable elevation in SR-BI expression could be found in RETCs (Supplementary Fig. 7c). Conversely, in macrophages, SR-BI expression demonstrated a substantial increase and diminished with the prolongation of UUO modelling time. Thus, all those evidences showed that more bHDL uptake in injured HK-2 cells was not associated with SR-BI.
TP/BIBF-bHDL relieves renal injury and fibrosis in UUO model, folic acid model and adenine model mice
Then we evaluated the efficacy of TP/BIBF-bHDL in alleviating kidney injury and fibrosis in UUO mouse model. As shown in Fig. 5a, following ureteral ligation, the mice received various treatments through intravenous administration via the tail vein, repeated bi-daily over a 14-day period. On the fifteenth day after ligation, mice were sacrificed and samples were collected to assess renal damage and fibrosis levels. Protein-to-creatinine ratio (PCR) is used to assess the severity of total proteinuria. Compared to the control group, UUO mice showed higher urinary protein levels in urine and the protein level decreased after drug treatments, with the TP/BIBF-bHDL group showing the greatest decrease, approaching the levels observed in the control group (Supplementary Fig. 8a). Upon UUO, the contralateral kidney undergoes adaptive changes to compensate for the loss of function in the obstructed kidney. These changes initially provide benefit but eventually causes damages to the contralateral kidney, leading to increased urinary protein. The anti-proteinuric effect of TP/BIBF-bHDL is likely exerted on the contralateral side, as the obstructed kidney cannot contribute to urine output. We evaluated the level of creatinine (Cre) and urea in serum of mice, which are common indicators of renal function, and ureteral ligation causes increased urea levels that gradually returned to normal after treatment (Supplementary Fig. 8b-c). And the differences in blood creatinine levels among the groups were not significant (Supplementary Fig. 8b), which may be caused by the strong compensatory ability of the kidneys. Hematoxylin and eosin staining (HE) staining revealed severe glomerular atrophy, notable tubular dilatation, and proteinaceous tubular casts in UUO model mice, indicative of pronounced kidney injury. Post-treatment, these renal tissue alterations were ameliorated, with the TP/BIBF-bHDL group showing the highest degree of injury relief (Fig. 5b–e, Supplementary Fig. 8d). In addition, apoptosis studies also demonstrated that TP/BIBF-bHDL treatment effectively alleviate renal injury (Fig. 5d, Supplementary Fig. 8f). Masson staining identified collagen deposition in the renal interstitium of the model mice, which was significantly reduced in the group treated with the co-loaded preparation (Fig. 5c, f, Supplementary Fig. 8e). Fibrosis is characterized by the deposition of extracellular matrix, which is produced and secreted by activated myofibroblasts. To assess the anti-fibrotic effects of TP/BIBF-bHDL, we tested its impact on level of α-SMA expression. Consistent with the Masson staining results, TP/BIBF-bHDL effectively decreased α-SMA expression to the greatest extent (Fig. 5 g, o). In conclusion, our findings collectively demonstrate that TP/BIBF-bHDL significantly alleviates renal injury and fibrosis in UUO model mice.
a Scheme of UUO model and administration regimen. b–d HE (b), Masson staining (c) and TUNEL assay (d) of kidney tissues of UUO mice after different treatments. Scale bars, 50 μm. n = 3 biologically independent samples. e, f The injury score (e) and fibrosis area (f) of kidney of UUO mice after different treatments. Data are mean ± SD (n = 3 biologically independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). g Western blot analysis of α-SMA expression in kidney of UUO mice after different treatments. n = 3 biologically independent samples. h Description of folic acid model and administration regimen. i–k HE (i), Masson staining (j) and TUNEL assay (k) of kidney tissues from folic acid-induced mice after different treatments. Scale bars, 50 μm. n = 3 biologically independent samples. l, m The injury score (l) and fibrosis area (m) of kidney of folic acid model mice after treatment. Data are mean ± SD (n = 3 biologically independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). n Western blot analysis of α-SMA expression in kidney of folic acid-induced mice after different treatments. n = 3 biologically independent samples. o, p Semi-quantification of α-SMA protein expression in UUO model mice and folic acid model mice with different treatments by Image J based the western blots shown in g (o) and n (p). It was normalized with corresponding α-tubulin signal. α-tubulin was used as internal control. The value of 42 and 55 kDa are referred to the predicted protein weight of α-SMA and α-tubulin. Data are mean ± SD (n = 3 biologically independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). a, h created in BioRender. He, S. (2024) https://BioRender.com/k11e324. UUO model unilateral ureteral obstruction model mice, control healthy mice without any treatment, PBS Phosphate buffered saline, TP/BIBF mixed solution of triptolide and nintedanib, TP/BIBF-bHDL triptolide and nintedanib co-loaded biomimetic high-density lipoprotein nanoparticles, HE hematoxylin-eosin staining, TUNEL Terminal Deoxynucleotidyl Transferase-mediated dUTP Nick-End Labeling. Source data are listed in the Source Data file.
Encouraged by the results, we further investigated the efficacy of TP/BIBF-bHDL in folic acid model and adenine model mice. In comparison to the control mice, the administration of both folic acid and adenine led to severe kidney injury and fibrosis. As expected, TP/BIBF-bHDL was most effective in improving kidney function, relieving renal injury and reducing collagen deposition caused by folic acid (Fig. 5i–n, p, Supplementary Fig. 8g–k) and adenine (Supplementary Fig. 9). The combined administration of TP and BIBF demonstrated superior antifibrotic effects compare to single treatment, indicating that the simultaneous anti-inflammatory and antifibrotic effects were more effective for disease remission (Fig. 5i and n, Supplementary Fig. 9e and i).
TP/BIBF-bHDL reverses renal fibrosis by remodeling the fibrosis niches
To elucidate the underlying therapeutic mechanisms, we further employed RNA sequencing for comprehensive analysis. Total RNA was extracted from the kidneys of normal mice and UUO model mice treated with PBS or TP/BIBF-bHDL, and then subjected to transcriptome sequencing for analysis. Comparative gene expression studies among the different mouse groups identified 6044 and 2196 differentially expressed genes (DEGs) in the kidneys of control versus UUO-PBS mice, and UUO-PBS versus UUO-TP/BIBF-bHDL mice, respectively, with 2018 DEGs common to both comparisons. A heatmap of these DEGs indicated a significant upregulation of genes related to inflammation, immune cell infiltration, and extracellular matrix (ECM) generation post-model induction, whereas TP/BIBF-bHDL treatment markedly downregulated these genes (Fig. 6c). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that the significantly up-regulated differential genes in UUO mice were mainly enriched for viral protein-cytokine receptor interaction, ECM-receptor interaction, cell adhesion molecules, cytokines-cytokine receptor interaction, complement and coagulation cascades and PI3K-Akt signaling pathways (Fig. 6d). In contrast, the significantly downregulated differential genes in TP/BIBF-bHDL-treated UUO mice were mainly enriched in ECM-receptor interaction, cytokines-cytokine receptor interaction and PI3K-Akt signaling pathways (Fig. 6e). Gene Ontology (GO) enrichment analysis of DGEs indicated a predominant enrichment in inflammatory response, immune response, extracellular matrix degradation, and activation of cytokines and chemokines (Fig. 6f, g). These findings reveal that TP/BIBF-bHDL treatment effectively lessen immune cell infiltration, dampen inflammatory factor production, and reduce ECM synthesis. Consequently, this implies the potential of TP/BIBF-bHDL in remodeling the fibrotic microenvironment and impeding the progression of fibrosis.
a, b Volcano of DEGs in kidneys among healthy mice and UUO mice treated with PBS and TP/BIBF-bHDL. The genes associated with fibrosis were marked. c Heatmap of genes related to ECM production, ECM degradation, inflammation and immune infiltration in different groups. d The top 20 KEGG pathways enrichment up in the UUO mice compared to the control mice. e The top 20 KEGG pathways enrichment down in the TP/BIBF-bHDL-treated UUO mice compared to PBS-treated UUO mice. f The top 30 GO terms up in the UUO mice compared to the control mice. g The top 30 GO terms down in the TP/BIBF-bHDL-treated UUO mice compared to PBS-treated UUO mice. UUO model unilateral ureteral obstruction model mice, control untreated healthy mice, PBS phosphate buffered saline, TP/BIBF-bHDL triptolide and nintedanib co-loaded biomimetic high-density lipoprotein nanoparticles.
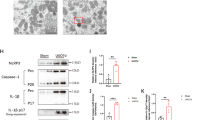
To explore the capability of TP/BIBF-bHDL in modulating the fibrotic niches, we conducted a detailed analysis of its impact on myofibroblast activation, ECM deposition, immune cell infiltration, and inflammatory factors secretion in the kidney, employing immunohistochemical techniques. In line with our transcriptomic findings, significant elevations were observed in the kidneys of UUO mice in pivotal markers associated with myofibroblast activation (α-smooth muscle actin, α-SMA), ECM deposition (collagen type I alpha 1 chain, COL1A1), immune cell infiltration (macrophages identified by F4/80, and T-cells by CD3), as well as inflammatory factors (tumor necrosis factor-alpha, TNF-α, and interleukin-1 beta, IL-1β). These elevations were notably reduced in TP/BIBF-bHDL-treated UUO mice (Fig. 7a–g). qPCR experiments further substantiated the effective amelioration of the fibrotic microenvironment and mitigation of kidney injury by TP/BIBF-bHDL (Fig. 7h). We further investigated the ability of TP/BIBF-bHDL to regulate the fibrotic microenvironment in folic acid model and adenine model mice. Results indicated that TP/BIBF-bHDL was also effective in remodeling the fibrotic microenvironment and attenuating renal fibrosis in the folic acid model (Supplementary Fig. 10) and adenine model mice (Supplementary Fig. 11). TP/BIBF-bHDL treatment significantly reduced fibroblast activation and ECM production in the kidneys of folic acid-induced and adenine-induced mice. The levels of inflammatory factors and chemokines were lower in the kidneys of folic acid model and adenine model mice treated with TP/BIBF-bHDL compared to the PBS group. Collectively, these results suggest that TP/BIBF-bHDL, which simultaneously delivers anti-inflammatory and antifibrotic drugs to RTECs, mitigates renal injury and attenuates fibrosis by improving the fibrotic niches from an inflammatory, immune and fibrotic perspective, a mechanism that distinguishes it from those antifibrotic strategies that target a single pathogenetic process or pathway.
a Representative immunohistochemical images for COL1A1、α-SMA、F4/80、CD3、TNF-α and IL-1β in the kidney of UUO mice with different treatments. Scale bars, 40 µm. b–g Semi-quantitation of the COL1A1、α-SMA、F4/80、CD3、TNF-α and IL-1β expression level in the kidney of UUO mice with different treatments. Data are mean ± SD (n = 5 biologically independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). h mRNA expression of Acta2、Col1a1、Fn1、Tgfb1、Tnf、Il1b and Ccl2 in the kidney of UUO mice with different treatments. Data are mean ± SD (n = 4 biologically independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). UUO model unilateral ureteral obstruction model mice, control untreated healthy mice, PBS phosphate buffered saline, TP triptolide solution, BIBF nintedanib solution, TP/BIBF a mixed solution of free triptolide and nintedanib, TP-bHDL, BIBF-bHDL and TP/BIBF-bHDL triptolide-loaded, nintedanib-loaded and triptolide and nintedanib co-loaded biomimetic high-density lipoprotein nanoparticles. Source data are listed in the Source Data file.
bHDL possess a good safety profile and ameliorate the toxicity of triptolide
Then the safety profiles of bHDL and TP/BIBF-bHDL were investigated in vitro and in vivo. First, we assessed the cytotoxicity of blank bHDL nanoparticles in HK-2 and NRK-49F cells. Blank bHDL nanoparticles has good biocompatibility and does not show obvious toxicity even when the concentration of bHDL reaches 32 times the therapeutic concentration, indicating its excellent biocompatibility (Supplementary Fig. 12a, b). Next, we conducted the safety evaluation in vivo and the safety of bHDL and TP/BIBF-bHDL after intravenous administration for 7 days, 14 days and 28 days were explored separately. As shown in Supplementary Fig. 12c, there was no difference in body weight change among control, bHDL, TP/BIBF, and TP/BIBF-bHDL groups. Even after the long-term administration (injection once every two days for 28 days), no obvious injury was found in major organs showed by HE staining (Fig. 8a). Moreover, blood routine examination demonstrated that there were no significant differences in all groups (Fig. 8c). It’s worth noting that long-term administration of TP/BIBF resulted in liver damage, with an increase in the liver function index ALT after 14 and 28 days of administration (Fig. 8b, Supplementary Fig. 12d and e). Neither the bHDL group nor the TP/BIBF-bHDL group exhibited significant differences in the liver function markers ALT and AST at 14 and 28 days compare to the control group. These results showed that bHDL nanoparticles have good biocompatibility and do not bring toxic side effects even when administered for a long period of time and reduce the toxic side effects of drugs to some extent.
a HE staining of heart, liver, spleen, kidney, brain, testis in mice treated with different formulations for 28 days. Scale bars, 50 µm. b, c Hematological tests and Blood routine examination of healthy mice on day 28 after different treatments. Data are mean ± SD (n = 5 biologically independent samples, one-way ANOVA with Dunnett’s multiple comparisons test and adjustment applied). d HE staining of heart, liver, spleen, kidney, brain in mice treated with TP, TP/BIBF, TP-bHDL and TP/BIBF-bHDL for 3 times at the concentration of 0.5 mg/mL TP, respectively. Scale bars, 100 µm. Control healthy mice, bHDL blank biomimetic high-density lipoprotein (HDL) nanoparticles, TP free triptolide, TP/BIBF mixed solution of free triptolide and nintedanib, TP-bHDL triptolide-loaded biomimetic HDL nanoparticles, TP/BIBF-bHDL triptolide and nintedanib co-loaded biomimetic HDL nanoparticles, ALT alanine aminotransferase, AST aspartate aminotransferase, Cre Creatinine, UREA urea nitrogen, CKMB creatine kinase MB, LDHI lactate dehydrogenase I, WBC white blood cell, RBC red blood cell, LYM lymphocyte, RDW red cell distribution width, PLT platelet, HGB hemoglobin, HE hematoxylin-eosin staining. Source data are listed in the Source Data file.
In the realm of targeted drug delivery systems, a key objective, alongside enhancing drug efficacy, is to minimize drug toxicity. To evaluate the potential of bHDL in reducing toxicity, we performed acute toxicity experiment. Given TP’s high toxicity and narrow therapeutic window, we selected it as model drug to measure the median lethal dose (LD50) of TP and TP-bHDL. As the results shown in Table S4, the LD50 of TP-bHDL was 0.88 mg/kg, significantly greater than TP (0.48 mg/kg), underscoring bHDL’s substantial capability in mitigating TP’s toxicity. For a more comprehensive assessment of its toxicity reduction, various formulations, including TP solution, TP/BIBF solution, TP-bHDL, and TP/BIBF-bHDL, were administered at a dose equivalent to 0.5 mg/kg of TP (every other day). On the 7th day, all mice in the TP group had succumbed, whereas 20% survival was noted in the TP/BIBF group, 80% in the TP-bHDL group, and complete survival in the TP/BIBF-bHDL group. The major organs of the mice in each group were taken for pathological examination, revealing that TP group mice showed severe liver damage and cardiac damage (Fig. 8d). Additionally, significant cardiomyocyte atrophy, cardiac myofibrillar lysis, and hepatocellular atrophy could be observed. In the TP/BIBF co-administration group, significant cardiac injury was seen, but to a lesser extent than in the TP group, suggesting that co-administration could alleviate the toxicity of TP to some extent. In contrast, in the TP-bHDL as well as TP/BIBF-bHDL groups, no significant cardiac injury and liver injury were found. These findings collectively demonstrated that bHDL represents an efficient and safe drug delivery system, markedly reducing drug toxicity and enhancing overall safety.
Discussion
As mentioned, developing drug delivery systems for RTECs is challenging due to multiple hard-to-pass barriers14, and for clinical translation there are even more practical issues. For example, lipid-based liposomes and micelles are often too big to pass through the glomerulus filter14,43; polymeric solid nanoparticles usually suffer from fast protein corona formation and liver accumulation44,45; and a large number of more complicated designs are hard to manufacture in large quantity and have safety concerns. These all limited the potential of current RTEC targeted delivery systems.
In this study, we revealed that the simple and biocompatible bHDL nanoparticles have strong natural targeting properties to the injured RTECs and KIM-1 mediated the phagocytosis of injured RETCs. The bHDL nanoparticles possess several advantages. First, the mediator KIM-1 is only expressed on damaged RTECs but not normal RTECs, which grant the bHDL NPs high delivery selectivity35. Though KIM-1, also named T-cell immunoglobulin and mucin domain 1 (TIM-1), is extensively expressed in activated T and B cells too18,19, this seems to not decisively affect the RTEC targeting capability of bHDL. Second, we found that KIM-1 likely initiated an endocytosis process that engulf the full bHDL. This is distinct to the endocytosis mediated by conventional bHDL binding partner SR-BI, where the endocytosis selectively uptake HDL cholesteryl esters and not whole nanoparticles25,46. Third, bHDL, like other HDLs, are small biocompatible particles that are not primary objects for blood clearance nor prone to protein corona attachment47. These characteristics all help reduce its liver accumulation, enhance glomerulus passage, and elongate its circulation time, which ultimately lead to better RTEC delivery efficiency. Fourth, these vehicles are built from simple phospholipids and apoA-I mimicking peptide D-4F, with a straightforward method. The phospholipid DMPC has been applied in the Visudyne, which is a marketed product. Additionally, the D-4F has been tested in clinical trials for the treatment of cardiovascular diseases48. The simple structure and ingredient improve its manufacturability and make it easier to pass safety inspections. Nonetheless, several unresolved issues remain that warrant attention in future studies. For example, further investigation is needed into the distribution of surface charges after the D-4F peptide binds with phospholipids. Notably, the L-4F peptide, which exhibits similar properties and behavior to the D-4F peptide. Therefore, our future research should explore the potential of the L-4F peptide as an alternative for constructing a RTECs drug delivery system.
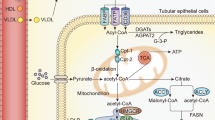
As for the therapeutic strategy, consensus remains elusive regarding an effective treatment modality for fibrosis49,50. It is known that inflammation is a pivotal driver in the pathogenesis of fibrosis49,51, however, simple anti-inflammatory therapies, such as corticosteroids, has proven inadequate in mitigating fibrosis50,52. Many attempts have been made to develop drugs targeting specific cells or pathways in the fibrosis process53. Presently, Nintedanib and Pirfenidone are the only anti-fibrotic agents with clinical approval, yet their therapeutic responses exhibit notable variability among patients54. As understanding of the pathology of fibrosis grows, it is increasingly recognized as a complex process involving multiple factors, with the fibrotic niches playing a crucial role in its progression55,56. This suggests that multifaceted interventions in the fibrotic process might yield improved therapeutic outcomes. In this study, we delivered both anti-inflammatory and anti-fibrotic drugs to RTECs, with the aim of improving the treatment effect. TP not only inhibited the release of pro-inflammatory cytokines such as TNF-α and MCP-1 from RETCs, thereby reducing the infiltration of inflammatory cells in the renal interstitium57; but also suppressed the antigen-presenting capabilities of RETCs, thereby reducing T-cell activation and exerting immunosuppressive effects58. Nintedanib mitigates fibroblast proliferation, migration, and transformation by targeting PDGFR, VEGFR, and FGFR, thus plays a significant role in antifibrotic therapy59,60. In addition, nintedanib could also suppress the inflammation of lung epithelial cell and inhibit the fibroblast-to myofibroblast transition by suppressing the TGF-β/Smad signaling pathways61,62. The co-delivery system effectively targets both inflammation and fibroblast activation, thereby improving its capability to combat fibrosis by remodeling the fibrotic niches. The fibrotic niche not only provides an ideal environment for the activation and proliferation of fibroblasts but also negatively impacts the regenerative capacity and plasticity of surviving parenchymal cells, thereby hindering tissue repair and regeneration. Targeting and disrupting the fibrotic niches might be an effective therapeutic strategy to halt the progression of fibrosis. Our research indicates that combination therapies addressing both anti-inflammatory and anti-fibrotic pathways may offer a more effective approach to modulating the fibrotic niches than strategies focusing solely on a single aspect.
In summary, we introduced a new renal tubule-targeted approach using bHDL nanoparticles and loaded both anti-inflammatory agent triptolide and antifibrotic compound nintedanib for CKD management (Fig. 9). We found that biocompatible bHDL is an excellent targeted drug delivery system for injured RETCs and revealed the underlying targeting mechanism that KIM-1 facilitates the endocytosis of these nanoparticles. This insight lays a groundwork for the practical application of bHDL in renal therapeutics. Our findings also indicate that co-administration of anti-inflammatory and anti-fibrotic drugs offers a more effective means of impeding the progression of fibrosis in CKD. Our study thus contributes several insights into the development of renal tubule-targeted drug delivery systems and the therapeutic approach for kidney diseases.
DMPC 1,2-dimyristoyl-sn-glycero-3-phosphocholine, KIM-1 kidney injury molecule 1, D-4F D-4F peptide, Injured RTECs injured renal tubular epithelial cells. Created in BioRender. He, S. (2024) https//BioRender.com/u96q289.
Methods
All the experiments in this study were conducted in strict accordance with the ethical standards and guidelines established by the Animal Ethics Committee of Sichuan University. The experimental protocols were reviewed and approved by the Ethics committee of Sichuan University to ensure compliance with relevant ethical regulations (KS2020022).
Materials
1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) were purchased from Shochem (Shanghai) Co., Ltd. D-4F (Ac-DWFKAFYDKVAEKFKEAF-NH2) was synthesized by GL Biochem (Shanghai) Ltd. Lotus Tetragonolobus Lectin (LTL), fluorescein (FITC)(L32480), KIM-1 Polyclonal Antibody (PA5-79345,Lot35FAFA04), and Protein Ladder (26619) were bought from Thermo Fisher Scientific Inc. Triptolide was purchased from chengdu Bio-purify phytochemicals Ltd. MTT assay kit, Nystatin, Dextran Sulfate(98%,CAS: 9011-18-1), Puromycin (99%, CAS: 58-58-2), Chlorpromazine Hydrochloride(98%,CAS: 69-69-0) and Amiloride HCl dihydrate (98%,CAS: 17440-83-4) was bought from Beijing Solarbio Science & Technology Co., Ltd. α-Smooth Muscle Actin Rabbit mAb (19245 T), COL1A1 (E8F4L,72026S) Rabbit mAb and F4/80 Rabbit mAb(D2S9R, 70076S) were purchased from Cell Signaling Technology, Inc. Beta-actin antibody(sc81178, Lot#J1116),Alpha Tubulin antibody(sc-5286, Lot#L1522), SR-BI antibody(sc-518140, Lot#12322), mouse TIM-1 antibody(A-12)(sc-518008, Lot#J2617), TNF alpha antibody(sc-52746) and CD3 antibody(sc-20047, Lot#G2821) were bought from Santa Cruz Biotechnology, Inc. SR-BI antibody(ab217318) for immunofluorescence was bought from Abcam. Rabbit IL-1β antibody (bioss, BS-0812R) was bought from Bioss CO., LTD. Lentivirus-RNAi human HAVCR1 (also referred as KIM-1) and HAVCR1 overexpression Lentivirus vector were produced by GeneChem.
Cell lines and Animals
HK-2 cells were purchased from National Intra-structure of Cell Line Resource of China and NRK-49F were obtained from the Cell Resource Center, Peking Union Medical College. HK-2 cells were cultured in the DMEM/F12 medium containing 10% FBS, 1%ITS-X, and 1% streptomycin/penicillin at 37°C under 5% CO2 humidified environment. NRK-49F cells were cultured in DMEM medium containing 10% FBS, 1%ITS-X, and 1% streptomycin/penicillin at 37°C under 5% CO2 humidified environment.
Male Sprague Dawley rats (6-8weeks), male BALB/c mice (6 to 8 weeks) and male C57BL/6 mice (4 to 6 weeks) were purchased from Beijing HFK bioscience co., LTD. All animals used in the study were fed with a standard laboratory chow diet, providing a balanced nutrient profile for the animals. The animals were housed in a controlled environment with a 12-hour light/dark cycle, maintained at a temperature of 22 ± 2 °C and humidity of 50 ± 10%. Food and water were provided freely. All the animal experiments were completed under the guidance of the Animal Ethics Committee of Sichuan University and all experiments received ethical approval (KS2020022).
HK-2 cell lentiviral transduction
Lentivirus-RNAi human HAVCR1 (also named KIM-1) and HAVCR1 overexpression Lentivirus vector were produced by GeneChem. HK-2 cells were seeded onto 6-well plates and when they grew to 50% confluence, transfected them with the lentivirus vectors for 48 hours. Puromycin was used to select stably transduce KIM-1 knockdown or KIM-1 overexpression HK-2 cells and the validation was confirmed by qPCR.
Mouse models
UUO mice model
Male BALB/C mice (4-6 weeks old) were anesthetized by intraperitoneal injection with tribromoethanol, restrained on the operating table, and incised the left abdomen of the mice, exposing the left ureter. Two ligatures were created at the opening of the renal pelvis and the upper 1/3 of the ureter, and the left ureter was then cut at the midpoint of the two ligature points. After ligation, the kidney was repositioned and the muscle and skin were sutured layer by layer. In the control group, after opening the abdomen, the ureter was isolated without ligation and sutured directly to the abdomen. On the day after the ligation, UUO mice were treated with PBS, TP, BIBF, TP/BIBF, TP-bHDL, BIBF-bHDL and TP/BIBF-bHDL by tail vein injection at an equal dose (0.1 mg/kg TP and 2 mg/kg BIBF), every two days for fourteen days. The control group were treated with PBS. After the completion of treatment, blood samples were obtained for the analysis of blood-related indices. The mice were euthanized, and their kidneys were harvested and subsequently fixed for further experimentation.
Folic acid mice model
Male C57BL/6 mice were injected intraperitoneally on day 1 and 14 with folic acid solution prepared by dissolving folic acid in 0.3 M sodium bicarbonate solution at a dose of 250 mg/kg. From the 15th day of modelling, PBS, TP, BIBF, TP/BIBF, TP-bHDL, BIBF-bHDL and TP/BIBF-bHDL were administered to the mice via tail vein injection at a dose of 0.1 mg/kg TP and 2 mg/kg BIBF, every other day for a period of 14 days. Healthy male C57BL/6 mice treated with PBS were used as a control group. After the administration, blood was collected for blood-related indices. Mice were executed and kidneys were removed and fixed for subsequent experiments.
Adenine mice model
Male BALB/C mice (4-6 weeks) were gavaged with adenine at a dose of 70 mg/ (kg.d) for 21 consecutive days for modelling. Healthy male BALB/C mice treated with PBS were used as a control group. The treatment for adenine mice was started from the 15th day of modeling and treatment protocols were consistent with the folic acid model.
Preparation of bHDL nanoparticles
bHDL and drug-laden bHDL nanoparticles were prepared as previously reported63, and the detailed steps were as follows:
Blank bHDL nanoparticles
20 mg of DMPC were dissolved in a 3 mL mixture of chloroform and methanol solvent (9:1,v/v), then evaporated under vacuum at 37 °C for 15 min to remove the solvent and hydrated the formed lipid film through sonication with 2 mL of PBS solution. Following this, 10 mg of the D-4F mimetic peptide dissolved in 2 mL of water was added to the hydrated lipid solution and sonicated for 10 minutes by a probe ultrasonic instrument in ice water bath. After completion of ultrasonication, the preparation solution underwent 3 cycles of hot and cold immersion, with each cycle involving a 60 °C and ice water bath respectively.
Drug-laden bHDL nanoparticles
Briefly, DMPC (20 mg), TP (0.2 mg) and BIBF (2 mg) were dissolved with 3 mL mixture solvent (chloroform: methanol = 9:1,v/v) in a 25 ml round-bottom flask. Then, the solvent was removed by evaporation and 2 mL PBS solution was used to hydrate the lipid film. Then, 2 mL of D-4F(10 mg) peptide solution was added to the lipid-drug solution and sonicated them by probe ultrasound instrument (200 W,15 min). After that, the mixture was subjected to three thermal (60°C) and cold cycles (10 min for each cycle) to obtain TP/BIBF-bHDL nanoparticles. The preparation method of TP-bHDL and BIBF-bHDL were similar to TP/BIBF-bHDL, except that they were only encapsulated with TP or BIBF.
DiD-bHDL nanoparticles
DiD-bHDL nanoparticles are prepared in a similar way to drug-laden nanoparticles, except that the drug is replaced with DiD(120 µg).
Characterization of bHDL nanoparticles
The particle size distribution and zeta potential were evaluated by a dynamic light scatter (Zetasizer Nano ZS90, Malvern Instrument, UK). Transmission electron microscopy and cryo-electron microscopy were used to examine the morphology of bHDL nanoparticles.
A combination of ultrafiltration centrifugation and HPLC methods were employed to measure the EE% and LC% of TP/BIBF-bHDL. Initially, free drugs and nanoparticles were separated through ultrafiltration centrifugation method. 2 mL of TP/BIBF-bHDL nanoparticle solution was added to an Amicon centrifugal filter tube with a 3 kDa molecular weight cut-off (MWCO) and centrifuged for 10 minutes (9280 x g) to separate free drugs and drug-laden nanoparticles. Subsequently, the concentrations of TP and BIBF in the nanoparticles were analyzed using HPLC after breaking the emulsion with methanol at three times the volume. EE% and LC% were calculated using the following equations:
and
where We means the amount of TP or BIBF in the nanoparticles, Wt means the total amount of the drugs and Wd means the total weight of the nanoparticles.
The HPLC analysis was conducted on a high-performance liquid chromatography (Agilent 1260, USA) equipped with a C18 column (250 mm× 4.6 mm, 5 µm). The mobile phase consisted of acetonitrile and 0.05% formic acid in water, with a gradient elution at a flow rate of 1 ml/min. Detection was performed on a UV detector, set at wavelengths of 210 nm for TP and 385 nm for BIBF. And the injection volume of each sample was 20 µL and the column temperature was set at 35 °C. All samples were prepared via protein precipitation using methanol prior to analysis.
Preparation of DiD-liposomes
DiD-liposomes were prepared by thin-film hydration method. Briefly, DMPC (20 mg), cholesterol (1 mg) and DiD (120 µg) were dissolved in 3 mL of chloroform and evaporated the solvent to form a lipid film. Then, hydrated the lipid film with 4 ml water and sonicated it for 20 min to form the DiD-Liposomes.
The stability study of bHDL nanoparticles
The stability of bHDL nanoparticles was estimated by observing the particle size alterations in different conditions, as well as comparing the in vitro release behavior of bHDL nanoparticles in PBS and 10% FBS. First, TP/BIBF-bHDL nanoparticles were placed at 4 °C and 37 °C for various durations, and the changes in particle size were detected by DLS. Then, to explore the stability of TP/BIBF-bHDL in serum, TEM was used to observe the size and shape of bHDL nanoparticles after incubating with 10% FBS for 2 hours at 37 °C. Given the endogenous lipoproteins in FBS may affect the observation, we observed the particle size of nanoparticles in both normal FBS as well as lipoprotein-removed FBS.
The in vitro drug release of bHDL nanoparticles was evaluated by dialysis method. Briefly, 0.5 ml of TP/BIBF-bHDL was added into the dialysis bag (MWCO = 3 kDa) and incubated in 30 mL of PBS or 10%FBS with gentle stirring. At designated time points, the buffer was extracted and replaced with an equal volume of fresh medium. The concentration of the drugs in the buffer was determined by HPLC.
Analysis of serum protein absorbed on bHDL nanoparticles
The protein level absorbed on the bHDL nanoparticles in serum was then estimated and liposome group was added as a control. Briefly, 0.5 mL of bHDL nanoparticles or liposomes were incubated with 0.5 mL of FBS or lipoprotein-removed FBS for 2 hours at room temperature. Subsequently, the bHDL-protein complexes were separated by chemical precipitation with a mixture of dextran sulfate and magnesium chloride, while the liposome-protein complexes were isolated by ultracentrifugation at 180,000 × g for four hours. Then, the complexes were lysed with 200 µl of RIPA, and the amount of adsorbed protein was quantified using the BCA assay.
Hemolysis Assay
Mice blood was collected and centrifuged at 3341 × g for 5 minutes to obtain red blood cells(RBCs). The collected RBCs were washed three times with PBS and then prepared into RBCs suspension with a concentration of 8%(v/v). The RBCs suspension was mixed with TP/BIBF-bHDL in equal volume and incubated for 1 hour at 37°C. 1%Triton X-100 was used as a positive control and PBS solution was used as the negative control. After incubation, the supernatant was centrifuged (3341 x g, 5 min) and the absorbance was tested at 570 nm via microplate Reader. The hemolysis rate was calculated according to the equation:
In vivo biodistribution assay
UUO model mice were applied to conduct the in vivo biodistribution experiment. Forty-eight UUO model mice were randomly divided into 3 groups and injected with DiD solution, DiD-liposome and DiD-bHDL (6 μg DiD each mouse) via tail vein. At 0.5, 2, 8 and 12 hours after administration, the mice were euthanized, and their organs were isolated and analyzed for fluorescence intensity using in vivo imaging. To investigate the localization of bHDL in the kidney, isolated kidney tissues were promptly fixed and subjected to analysis through immunofluorescence staining. To compare the distribution of bHDL nanoparticles in healthy and injured kidney, healthy male mice were also applied to conduct the biodistribution study.
To achieve a more precise characterization of the in vivo distribution of bHDL, we utilized the Liquid Chromatography Mass Spectrometry (LC-MS/MS) method for distribution experiment. Fifty UUO model mice were injected with TP/BIBF solution or TP/BIBF-bHDL into the tail vein at a dose of 2 mg/kg BIBF. At each predetermined time point, and the mice were euthanized, their organs were isolated, weighed, and homogenized with twice the tissue weight in saline to prepare a homogenate. Subsequently, the quantity of BIBF in each tissue was determined through LC-MS/MS. Pharmacokinetics parameters of TP/BIBF or TP/BIBF-bHDL in kidney after intravenous administration were calculated by Data and Statistics Software (DAS3.0; Shanghai, China). Relative uptake efficiency (Re kidney) and concentration efficiency (Ce kidney) were calculated using the following equations:
Pharmacokinetics experiment
To investigate the in vivo behavior of TP/BIBF-bHDL, we also performed pharmacokinetic study in vivo. Eighteen male SD rats were randomly divided into three groups and received equal doses (2 mg/kg BIBF) of TP/BIBF or TP/BIBF-bHDL by tail vein injection or administration of TP/BIBF (2 mg/kg BIBF) by gavage, and blood was collected from the orbital region of the rats in each group at scheduled time points after administration. Then the plasma was obtained through centrifugation (3341 × g, 10 min) and the drug content in plasma was estimated via the LC-MS/MS method.
In vitro cellular uptake assay
HK-2 cells were seeded onto 6-well plates, cultured at 37 °C, 5% CO2 for 24 hours, then treated with cisplatin for another 24 hours at the concentration of 2 µg/mL to induce injury. Then the injured HK-2 cells and normal HK-2 cells were treated with DiD-bHDL at a dose of 200 ng/ml at 37°C for 2 hours. After incubation, the cells were washed with PBS, digested with trypsin and the fluorescence intensity in the cells was evaluated by Flow Cytometry. The uptake experiment of bHDL was also performed in KIM-1 knockdown and KIM-1 overexpression HK-2 cells with or without cisplatin-induced injury.
NRK-49F cells were also applied to evaluate the uptake of bHDL. NRK-49 cells were inoculated in 6-well plates and cultured for 24 hours with or without 10 ng/mL TGF-β1, which could induce transformation of fibroblasts into myofibroblasts. Then DiD-bHDL nanoparticles were added to the cells for 2 hours incubation and the fluorescence intensity were measured with the flow cytometry.
In vitro competitive inhibition assay
HK-2 cells were inoculated in 6-well plates and cultured overnight at 37 °C, 5% CO2. HK-2 cells were pretreated with different endocytosis inhibitors (Chlorpromazine at a concentration of 25 µM, Nystatin at a concentration of 50 µM, Amiloride at a concentration of 50 µM, β-CD and Dextran sulfate sodium at a concentration of 50 µg/mL, Gentamicin and O-Phospho-L-serine at a dose of 20 µg/ml, Cytochalasin D at a concentration of 1.25 µg/mL, cubilin antibody at a concentration of 30 µg/mL, BLT-1 at a concentration of 25 µM and D-4F with different dose) for 0.5 hours, and then administrated with DiD-bHDL at a concentration of 200 µg/mL for 2 hours. After incubation, the cells were washed with PBS and tested by Flow cytometry. Moreover, HK-2 cells were treated with DiD-bHDL at a dose of 200 µg/mL for 2 hours at 4°C and the uptake of DiD-bHDL was estimated.
To explore whether KIM-1 mediated the uptake of bHDL by the injured HK-2 cells, the competitive inhibition assay was also conducted with the KIM-1 antibody. HK-2 cells were incubated with 2ug/mL cisplatin for 24 hours to induce injury and then treated the cells with or without KIM-1 antibody(30 µg/mL) for 0.5 hours. Then DiD-bHDL nanoparticles were added to the cells for another 2 hours and flow cytometry was applied to evaluate the uptake of bHDL. Normal HK-2 cells were also added as control.
KIM-1 targeting assay
To further examine whether KIM-1 mediates bHDL’s uptake, HK-2 cells were inoculated in laser confocal petri dishes and cultured overnight. HK-2 cells were treated with cisplatin at a concentration of 2 µg/mL for 24 hours and then administrated with DiD-bHDL for 2 hours. After incubation, the cells were washed with PBS, stained with KIM-1 antibody and FITC-labeled secondary antibody. DAPI was used to stain the nuclei. After staining, CLSM was applied to examine the co-localization of KIM-1 and DiD-bHDL.
Moreover, co-localization of KIM-1 and DiD-bHDL were estimated in vivo. UUO mice were administered with DiD-bHDL or DiD-liposome at a dose of 6 µg by tail vein injection on the seventh day after ureteral ligation. Two hours after administration, the mice were euthanized and kidneys were extracted, fixed and subjected to immunofluorescence staining, whereby KIM-1 protein was labelled using KIM-1 antibody and FITC-labeled secondary antibody. The fluorescent sections were subjected to CLSM to observe co-localization. Normal mice were added as control.
Renal function assessment
The levels of Cre and urea in the serum were assessed to evaluate the renal function of mice after different treatment. After the completion of treatments, the blood was collected, centrifuged to take the upper serum layer, and the levels of Cre and urea were measured using a Roche blood biochemistry test.
Histopathology and Immunohistochemistry staining
Renal tissues were fixed in 4% paraformaldehyde, dehydrated, hyalinized and embedded in paraffin. Then the HE staining and Masson staining were conducted with the paraffin-embedded kidney tissues. HE-stained sections were examined under the microscope to observe specific lesions and kidney damage was scored using a four-point scale: no damage (0 points), slight (1 point), mild (2 points), moderate (3 points) and severe (4 points), respectively. Masson sections were examined using a microscope and images were taken. The fibrous tissue area (Area) in the images was determined using an Image-Pro Plus 6.0 image analysis system (Media Cybernetics, USA). The fibrous tissue expression area percentage was calculated by the following formula:
For immunohistochemistry staining, paraffin-embedded sections were obtained, enclosed in serum and then incubated sequentially with primary antibodies against α-SMA, COL1A1, F4/80, CD3, IL-1β and TNF-α, and secondary antibodies. Finally, staining was conducted using 3,3’-diaminobenzidine (DAB) and re-stained with hematoxylin. Photographs of the sections were taken and the percentage of positive area per image (% DAB Positive Tissue) was calculated using the Halo data analysis system.
Immunofluorescence staining
Renal tissues were embedded in the Optimal Cutting Temperature (OCT) compound and sectioned. The sections were stained with FITC-labeled LTL, and sequentially incubated with primary antibodies against KIM-1 and fluorescently labeled secondary antibodies. DAPI was applied to stain the nuclei. To estimate the expression of SR-BI in kidney, sections were stained with primary antibodies against SR-BI and subsequently with fluorescently labeled secondary antibodies. The nuclei were stained using DAPI.
For TUNEL staining, Paraffin-embedded kidney sections were stained with fluorescent TUNEL incubation solution according to the operation of the TUNEL kit (Roche, Swiss). The nuclei were stained with DAPI.
Western blot assay
The protein expression of KIM-1, SR-BI and α-SMA in kidney and HK-2 cells were evaluated by Western blot. Renal tissues and HK-2 cells were lysed by RIPA Lysis buffer containing 1% protease inhibitor for 30 minutes. After lysis, the protein concentration of each sample was estimated by BCA assay kit. Loading buffer was added to the samples and samples were boiled for 10 minutes. Then samples were uploaded into the PAGE gel to separate the proteins and transferred them with PVDF films. Then these films were closed with 5% milk solution for 1 hour, incubated with primary antibodies at 4°C overnight and followed by HRP-conjugated secondary antibodies. Proteins were detected using a ECL detection kit.
Quantitative PCR assay
Gene expression in both kidney tissue and HK-2 cells was estimated by qPCR. Total RNA was extracted using FastPure cell/Tissue Total RNA Isolation Kit provided by Nanjing Vazyme Biotech Co., Ltd. Then total RNA was reverse transcribed to obtain cDNA using 5ˣ HiScript III qRT SuperMix from Vazyme Biotech Co., Ltd. ChamQ SYBR qPCR Master Mix was used in the PCR study and the experimental procedures was conducted according to the protocol provided by Vazyme. The primers for quantitative real-time PCR were provided by Sangon Biotech and listed in Table S5. Relative expression of the target genes was normalized to β-actin levels in kidney tissues and GAPDH levels in HK-2 cells.
RNA-sequencing
Total RNA was extracted from kidneys of control mice or UUO model mice treated with PBS or TP/BIBF-bHDL (0.1 mg/kg TP, 2 mg/kg BIBF). Then the RNA sequencing was conducted by Shanghai OE Biotech Co., Ltd and the transcriptome sequencing data was collected on an Ilumina Novaseq 6000 platform. DESeq2 software was used to analyze differentially expressed genes.
Safety evaluation assay
HK-2 cells and NRK-49Fcells were seeded onto 96-well plates and cultured overnight. Blank bHDL was diluted with culture medium into its multiplicative concentration solution based on the normal administered concentration, which ranges from 0.5 to 32 times. The cells were treated with the diluted bHDL solutions for 24 hours and then cell counting kit-8 (CCK-8) was applied to test the cell viability of the cells.
Sixty Male BALB/C mice aged 4-6 weeks were randomly divided into four groups and administrated with PBS, bHDL, TP/BIBF and TP/BIBF-bHDL via tail vein injection at a concentration of 0.1 mg/kg TP and 2 mg/kg BIBF. The drug was administered every two days for 28 consecutive days, and the body weights of mice were recorded. On the 7th, 14th and 28th days of administration, blood samples were collected for Routine blood tests and blood biochemistry examination, and tissue samples were taken for pathological examination by H&E staining.
To further investigate the toxicity-reducing effect of bHDL, we performed acute toxicity experiments to measure the LD50 of both TP/BIBF and TP/BIBF-bHDL. Due to the high toxicity of TP, we selected to calculate the LD50 based on the concentration of TP. Fifty male BALB/C mice (4-6 weeks) were randomly divided into ten groups and were injected via the tail vein with varying concentrations of TP (0.3, 0.375, 0.47, 0.58, and 0.74 µg/mL TP) and TP-bHDL (0.18, 0.3, 0.5, 0.83, and 1.38 µg/mL TP). After administration, the condition of mice in each group was monitored and the death of mice at varying doses was recorded over 7 days. Moreover, twenty male BALB/C mice (4-6 weeks) were randomly assigned to four groups and injected by tail vein with TP, TP-bHDL, TP/BIBF, and TP/BIBF-bHDL at a concentration of 0.5 mg/kg TP for three times. The condition of the mice was recorded under close observation. On the seventh day after administration, mice were executed and tissues of the heart, liver, spleen, lungs and kidneys were taken for histopathological examination.
Statistical analysis
All data analyses were performed using GraphPad software (Vision 8). All experimental data are presented as mean ± standard deviation. For two-group comparison, unpaired T test with two tailed test was used for comparison. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test and two-way ANOVA with Sidak’s multiple comparisons test were conducted for multiple comparisons. Statistical significance was defined as a p value of less than 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the article and its supplementary information files contain the main data supporting the results of this study. And a reporting summary for this article is also provided. The transcriptome sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive under accession code PRJNA1064395. Source data are provided with this paper.
References
Bikbov, B. et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 395, 709–733 (2020).
Romagnani, P. et al. Chronic kidney disease. Nat. Rev. Dis. Prim. 3, 17088 (2017).
Chen, T. K., Knicely, D. H. & Grams, M. E. Chronic kidney disease diagnosis and management: a review. Jama 322, 1294–1304 (2019).
DeFronzo, R. A., Reeves, W. B. & Awad, A. S. Pathophysiology of diabetic kidney disease: impact of SGLT2 inhibitors. Nat. Rev. Nephrol. 17, 319–334 (2021).
Go, A. S., Chertow, G. M., Fan, D., McCulloch, C. E. & Hsu, C. Y. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N. Engl. J. Med 351, 1296–1305 (2004).
Jankowski, J., Floege, J., Fliser, D., Böhm, M. & Marx, N. Cardiovascular disease in chronic kidney disease: pathophysiological insights and therapeutic options. Circulation 143, 1157–1172 (2021).
Reiss, A. B. et al. CKD, arterial calcification, atherosclerosis and bone health: Inter-relationships and controversies. Atherosclerosis 278, 49–59 (2018).
Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 395, 709–733 (2020).
Wang, V., Vilme, H., Maciejewski, M. L. & Boulware, L. E. The economic burden of chronic kidney disease and end-stage renal disease. Semin Nephrol. 36, 319–330 (2016).
Doke, T. et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat. Immunol. 23, 947–959 (2022).
Carney, E. F. Acute kidney injury: Proximal tubule cells modulate inflammation after renal injury. Nat. Rev. Nephrol. 11, 254 (2015).
Du, B. et al. Glomerular barrier behaves as an atomically precise bandpass filter in a sub-nanometre regime. Nat. Nanotechnol. 12, 1096–1102 (2017).
Du, B., Yu, M. & Zheng, J. Transport and interactions of nanoparticles in the kidneys. Nat. Rev. Mater. 3, 358–374 (2018).
Huang, Y., Wang, J., Jiang, K. & Chung, E. J. Improving kidney targeting: The influence of nanoparticle physicochemical properties on kidney interactions. J. Controlled Release 334, 127–137 (2021).
Wang, J., Masehi-Lano, J. J. & Chung, E. J. Peptide and antibody ligands for renal targeting: nanomedicine strategies for kidney disease. Biomater. Sci. 5, 1450–1459 (2017).
Wang, J., Tripathy, N. & Chung, E. J. Targeting and therapeutic peptide-based strategies for polycystic kidney disease. Adv. Drug Deliv. Rev. 161-162, 176–189 (2020).
Xiao, W. et al. The protein corona hampers the transcytosis of transferrin-modified nanoparticles through blood-brain barrier and attenuates their targeting ability to brain tumor. Biomaterials 274, 120888 (2021).
Karmakova, Т. et al. Kidney Injury Molecule 1 (KIM-1): a multifunctional glycoprotein and biological marker (Review). Sovrem. Tekhnologii Med 13, 64–78 (2021).
Song, J. et al. Understanding kidney injury molecule 1: a novel immune factor in kidney pathophysiology. Am. J. Transl. Res 11, 1219–1229 (2019).
Wajda, J. et al. The Marker of Tubular Injury, Kidney Injury Molecule-1 (KIM-1), in Acute Kidney Injury Complicating Acute Pancreatitis: A Preliminary Study. J. Clin. Med 9, 1463 (2020).
Ichimura, T. et al. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J. Clin. Invest 118, 1657–1668 (2008).
Yamanishi, Y. et al. TIM1 is an endogenous ligand for LMIR5/CD300b: LMIR5 deficiency ameliorates mouse kidney ischemia/reperfusion injury. J. Exp. Med 207, 1501–1511 (2010).
Ichimura, T., Brooks, C. R. & Bonventre, J. V. Kim-1/Tim-1 and immune cells: shifting sands. Kidney Int 81, 809–811 (2012).
Mo, Z.-C., Ren, K., Liu, X., Tang, Z.-L. & Yi, G.-H. A high-density lipoprotein-mediated drug delivery system. Adv. Drug Deliv. Rev. 106, 132–147 (2016).
Yu, H. HDL and Scavenger Receptor Class B Type I (SRBI). Adv. Exp. Med Biol. 1377, 79–93 (2022).
Pedersbæk, D. & Simonsen, J. B. A systematic review of the biodistribution of biomimetic high-density lipoproteins in mice. J. Controlled Release 328, 792–804 (2020).
Khan, M. A. et al. Oxidized LDL receptors: a recent update. Curr. Opin. Lipido. 34, 147–155 (2023).
Nastase, M. V., Zeng-Brouwers, J., Wygrecka, M. & Schaefer, L. Targeting renal fibrosis: Mechanisms and drug delivery systems. Adv. Drug Deliv. Rev. 129, 295–307 (2018).
Liu, Q. Triptolide and its expanding multiple pharmacological functions. Int Immunopharmacol. 11, 377–383 (2011).
Xi, C., Peng, S., Wu, Z., Zhou, Q. & Zhou, J. Toxicity of triptolide and the molecular mechanisms involved. Biomedicine Pharmacother. 90, 531–541 (2017).
Richeldi, L. et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N. Engl. J. Med 370, 2071–2082 (2014).
Lamb, Y. N. Nintedanib: A Review in Fibrotic Interstitial Lung Diseases. Drugs 81, 575–586 (2021).
Velagacherla, V., Suresh, A., Mehta, C. H. & Nayak, U. Y. Advances and challenges in nintedanib drug delivery. Expert Opin. Drug Deliv. 18, 1687–1706 (2021).
Carney, E. F. Proximal tubule cells modulate inflammation after renal injury. Nat. Rev. Nephrol. 11, 254–254 (2015).
Mori, Y. et al. KIM-1 mediates fatty acid uptake by renal tubular cells to promote progressive diabetic kidney disease. Cell Metab. 33, 1042–1061.e1047 (2021).
Yang, C. et al. A renal YY1-KIM1-DR5 axis regulates the progression of acute kidney injury. Nat. Commun. 14, 4261 (2023).
Tang, T. T. et al. Kim-1 Targeted Extracellular Vesicles: A New Therapeutic Platform for RNAi to Treat AKI. J. Am. Soc. Nephrol. 32, 2467–2483 (2021).
Liu, D. et al. ROS-responsive chitosan-SS31 prodrug for AKI therapy via rapid distribution in the kidney and long-term retention in the renal tubule. Sci. Adv. 6, eabb7422 (2020).
Wu, Q. et al. Targeted delivery of celastrol to glomerular endothelium and podocytes for chronic kidney disease treatment. Nano Res 15, 3556–3568 (2022).
Zhang, D. et al. Enhancing CRISPR/Cas gene editing through modulating cellular mechanical properties for cancer therapy. Nat. Nanotechnol. 17, 777–787 (2022).
Damiano, M. G., Mutharasan, R. K., Tripathy, S., McMahon, K. M. & Thaxton, C. S. Templated high density lipoprotein nanoparticles as potential therapies and for molecular delivery. Adv. Drug Deliv. Rev. 65, 649–662 (2013).
Conway, B. R. et al. Kidney single-cell atlas reveals myeloid heterogeneity in progression and regression of kidney disease. J. Am. Soc. Nephrol. 31, 2833–2854 (2020).
Ma, Y. et al. A review of the application of nanoparticles in the diagnosis and treatment of chronic kidney disease. Bioact. Mater. 5, 732–743 (2020).
Xiao, Q. et al. The effects of protein corona on in vivo fate of nanocarriers. Adv. Drug Deliv. Rev. 186, 114356 (2022).
Cao, M. et al. Molybdenum derived from nanomaterials incorporates into molybdenum enzymes and affects their activities in vivo. Nat. Nanotechnol. 16, 708–716 (2021).
Linton, M. F., Tao, H., Linton, E. F. & Yancey, P. G. SR-BI: A Multifunctional Receptor in Cholesterol Homeostasis and Atherosclerosis. Trends Endocrinol. Metab. 28, 461–472 (2017).
Li, M. et al. Nanoparticle elasticity affects systemic circulation lifetime by modulating adsorption of apolipoprotein A-I in corona formation. Nat. Commun. 13, 4137 (2022).
Wolska, A., Reimund, M., Sviridov, D. O., Amar, M. J. & Remaley, A. T. Apolipoprotein Mimetic Peptides: Potential New Therapies for Cardiovascular Diseases. Cells 10, 597 (2021).
Henderson, N. C., Rieder, F. & Wynn, T. A. Fibrosis: from mechanisms to medicines. Nature 587, 555–566 (2020).
Senior, M. Fighting fibrosis. Nat. Biotechnol. 40, 1169–1173 (2022).
Rockey, D., Bell, P. & Hill, J. Fibrosis — a common pathway to organ injury and failure. N. Engl. J. Med 372, 1138–1149 (2015).
Setten, E. et al. Understanding fibrosis pathogenesis via modeling macrophage-fibroblast interplay in immune-metabolic context. Nat. Commun. 13, 6499 (2022).
Wynn, T. A. & Ramalingam, T. R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med 18, 1028–1040 (2012).
Finnerty, J. P., Ponnuswamy, A., Dutta, P., Abdelaziz, A. & Kamil, H. Efficacy of antifibrotic drugs, nintedanib and pirfenidone, in treatment of progressive pulmonary fibrosis in both idiopathic pulmonary fibrosis (IPF) and non-IPF: a systematic review and meta-analysis. BMC Pulm. Med 21, 411 (2021).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019).
Joshi, N. et al. A spatially restricted fibrotic niche in pulmonary fibrosis is sustained by M-CSF/M-CSFR signalling in monocyte-derived alveolar macrophages. Eur. Respir. J. 55, 1900646 (2020).
Jiang, S. et al. Friend or foe? The dual role of triptolide in the liver, kidney, and heart. Biomedicine Pharmacother. 161, 114470 (2023).
Cheng, Y., Zhao, Y. & Zheng, Y. Therapeutic potential of triptolide in autoimmune diseases and strategies to reduce its toxicity. Chin. Med 16, 114 (2021).
Jamadar, A. et al. The tyrosine-kinase inhibitor Nintedanib ameliorates autosomal-dominant polycystic kidney disease. Cell Death Dis. 12, 947 (2021).
Roth, G. J. et al. Nintedanib: from discovery to the clinic. J. Med Chem. 58, 1053–1063 (2015).
Tu, J. et al. Nintedanib mitigates radiation-induced pulmonary fibrosis by suppressing epithelial cell inflammatory response and inhibiting fibroblast-to-myofibroblast transition. Int. J. Biol. Sci. 20, 3353–3371 (2024).
Ihara, H. et al. Nintedanib inhibits epithelial-mesenchymal transition in A549 alveolar epithelial cells through regulation of the TGF-β/Smad pathway. Respir. Investig. 58, 275–284 (2020).
Yu, X. et al. Melittin-lipid nanoparticles target to lymph nodes and elicit a systemic anti-tumor immune response. Nat. Commun. 11, 1110 (2020).
Acknowledgements
This research was financially supported by the National Science Fund for the Regional Innovation and Development Joint Fund (No. U20A20411, ZRZ and U23A20495, LZ), the National Key R&D Program of China (2023YFC2509302, LZ), Natural Science Foundation of Sichuan Province (23NSFC3777, QL) and Science and Technology Project of Chengdu City (2022-YF05-01398-SN, QL). We would like to thank Dr.Yang Liu (College of Polymer Science and Engineering, Sichuan University) for helping us with Molecular dynamics simulation analysis. We sincerely thank Dr. Zhenhua Shao and Xiaowen Tian (State Key Laboratory of Biotherapy, Sichuan University) for helping us with cryo-EM measurement. We would like to thank Dr.Meiju Xie(Analytical and Testing Center, Sichuan University) for her help with TEM measurements.
Author information
Authors and Affiliations
Contributions
The original idea was conceived by L.Z., S.S.H. and Z.R.Z. S.S.H., X.Y.L., Y.Y.H., Y.Z.D., L.Y., S.Y.H., J.L.F. and L.L.W. participated in the experiments and collected the data. Data analysis was performed by S.S.H., X.Y.L., Y.Y.H., L.G., L.L. and Q.L. The manuscript was drafted by S.S.H., and Z.L. The whole work was supervised by L.Z. and Z.R.Z. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mohammad M. Al-bataineh, Joseph Vincent Bonventre, Jens Simonsen, Chengguo Wei and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, S., Li, X., He, Y. et al. High-density lipoprotein nanoparticles spontaneously target to damaged renal tubules and alleviate renal fibrosis by remodeling the fibrotic niches. Nat Commun 16, 1061 (2025). https://doi.org/10.1038/s41467-025-56223-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56223-z