Abstract
S-palmitoylation is a reversible and widespread post-translational modification, but its role in the regulation of ferroptosis has been poorly understood. Here, we elucidate that GPX4, an essential regulator of ferroptosis, is reversibly palmitoylated on cysteine 66. The acyltransferase ZDHHC20 palmitoylates GPX4 and increases its protein stability. ZDHHC20 depletion or inhibition of protein palmitoylation by 2-BP sensitizes cancer cells to ferroptosis. Moreover, we identify APT2 as the depalmitoylase of GPX4. Genetic silencing or pharmacological inhibition of APT2 with ML349 increases GPX4 palmitoylation, thereby stabilizing the protein and conferring resistance to ferroptosis. Notably, disrupting GPX4 palmitoylation markedly potentiates ferroptosis in xenografted and orthotopically implanted tumor models, and inhibits tumor metastasis through blood vessels. In the chemically induced colorectal cancer model, knockout of APT2 significantly aggravates cancer progression. Furthermore, pharmacologically modulating GPX4 palmitoylation impacts liver ischemia–reperfusion injury. Overall, our findings uncover the intricate network regulating GPX4 palmitoylation, highlighting its pivotal role in modulating ferroptosis sensitivity.
Similar content being viewed by others
Introduction
Ferroptosis is an iron-dependent form of cell death characterized by excessive lipid peroxidation, distinct from apoptosis and necrosis1,2,3. Ferroptosis is tightly regulated by glutathione peroxidase 4 (GPX4), ferroptosis-suppressor protein 1 (FSP1), dihydro-orotate dehydrogenase (DHODH), and a GTP cyclohydrolase 1 (GCH1)/tetrahydrobiopterin (BH4) system and membrane-bound O-acyltransferase domain containing 1/2 (MBOAT1/2)4,5,6,7,8,9. The antioxidant enzyme GPX4 can directly reduce phospholipid hydroperoxide to hydroxyphospholipid, thus acting as a central repressor of ferroptosis4. FSP1, DHODH and GCH1 produce metabolites such as CoQ10 and BH4 with radical-trapping antioxidant activity, thus terminating lipid peroxidation5,6,7,8. MBOAT1/2 inhibit ferroptosis by increasing the cellular content of monounsaturated fatty acids (PE-MUFA) and correspondingly decreasing the cellular content of polyunsaturated fatty acids (PE-PUFA), which are the preferred substrates for phospholipid peroxidation, thereby remodeling the cellular phospholipid profile and suppressing ferroptosis9. Accumulating evidence indicates the strong correlation of ferroptosis with pathological conditions such as neurodegenerative diseases, stroke, and multiple organ damage. Moreover, ferroptosis plays a profound role in tumor growth, metastasis, and response to cancer therapies, highlighting the therapeutic potential of drug-induced ferroptosis in cancer treatment10,11.
GPX4 is the sole known mammalian selenoprotein that utilizes glutathione (GSH) as a co-factor to convert toxic lipid hydroperoxides (L-OOH) on the cytoplasmic membrane to non-toxic lipid alcohols (L-OH), thus protecting cells from ferroptotic cell death12,13,14. The functional significance of GPX4 has been implicated in tumor development, drug resistance of cancer cells, and the pathology of several diseases like inflammatory bowel disease (IBD) and Sedaghatian-type spondylometaphyseal dysplasia (SSMD)15,16,17,18,19. In addition, homozygous GPX4 knockout leads to embryonic lethality, and inducible inactivation of GPX4 causes acute renal failure in mice, underscoring the physiological functions of GPX420,21. GPX4 has been reported to undergo several post-translational modifications, including ubiquitination, phosphorylation, and succinylation, which affect its activity and stability22,23,24. Nevertheless, whether other mechanisms regulate GPX4 function and, more importantly, whether the regulation can be exploited to manage ferroptosis-associated diseases requires further investigation.
Palmitoylation is a reversible post-translational modification involving the attachment of palmitic acid to proteins25. It is catalyzed by 23 mammalian palmitoyltransferases (DHHCs), characterized by the Asp-His-His-Cys sequence motif, and is reversed by acyl protein thioesterases, including APT1/2 and alpha/beta hydrolase domain-containing (ABHD) protein family members26. Palmitoylation plays a pivotal role in regulating protein function, localization, stability, and signaling pathways27. Aberrant protein palmitoylation has been implicated in tumorigenesis, immune evasion, and the pathogenesis of various diseases such as inflammatory bowel disease and Alzheimer’s disease28,29,30,31,32. However, whether palmitoylation regulates GPX4 activity, which is instrumental for ferroptosis sensitivity, remains unknown.
In this study, we demonstrate that GPX4 is reversibly palmitoylated on cysteine 66, catalyzed by ZDHHC20 and APT2, respectively. The palmitoylation of GPX4 affects the stability of the GPX4 protein, thereby regulating cellular sensitivity to ferroptosis in vitro and in vivo. Specifically, inhibiting palmitoylation of GPX4 through knockdown of ZDHHC20 or general palmitoylation inhibitor 2-BP sensitizes cells to ferroptosis, whereas maintaining palmitoylation via the knockdown of APT2 or APT2 inhibitor ML349 renders cells resistant to ferroptosis. Our study suggests that targeting the palmitoylation-depalmitoylation cycle of GPX4 to influence the ferroptosis response may be a therapeutic target for ferroptosis-related diseases.
Results
Identification of 2-BP as a potent contributor to ferroptosis
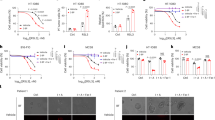
Our study initially aimed to identify critical metabolic regulators that affect ferroptosis sensitivity. We screened a customized library of ~3200 metabolism-focused small molecules in an RSL3-induced ferroptosis assay using A375 human melanoma cells (Fig. 1a). This screening revealed several known ferroptosis inducers, such as iFSP1 (an FSP1 inhibitor), buthionine-sulfoximine (BSO), and indoleamine 2,3-dioxygenase-1 (IDO1) inhibitors, as well as numerous reported inhibitors of ferroptosis, including Ferrostatin-1 (Fer-1), (±)-α-Tocopherol, Vitamin K1 and K2, Vitamin E, and Trolox, thereby validating the robustness of our screening (Fig. 1a). Particularly, 2-bromopalmitate (2-BP), a stable analog of palmitate known for inhibiting various lipid metabolism-related enzymes and protein palmitoylation33,34,35, emerged as a potent enhancer of ferroptosis susceptibility (Fig. 1a). We confirmed that 2-BP effectively promoted ferroptosis induced by GPX4 inhibitors (including RSL3, ML162 and ML210) and the cystine/glutamate antiporter (xCT) inhibitor erastin in a dose-dependent manner in A375 and A549 human lung cancer cells (Fig. 1b and Supplementary Fig. 1a). It was similarly noted under conditions of cystine starvation (Fig. 1c and Supplementary Fig. 1b). 2-BP substantially increased RSL3-induced cell death, lipid peroxidation, and PTGS2 mRNA levels, which was blocked by the ferroptosis inhibitor Fer-1 (Fig. 1d-g and Supplementary Fig. 1c, d). In 2-BP-stimulated cancer cells that were treated with RSL3 alone or alongside various cell death inhibitors, RSL3-induced cell death could be fully reversed by Fer-1 but not the apoptosis inhibitor z-VDA-fmk, the necrosis inhibitor Necrostatin-1, and the autophagy inhibitor Bafilomycin A1 (BafA1) (Fig. 1h). Furthermore, 2-BP consistently aggravated RSL3-induced ferroptotic cell death in different types of tumors (Supplementary Fig. 1e). We also observed that while 2-BP alone did not alter cellular reactive oxygen species (ROS) levels, it significantly amplified the ROS increase triggered by RSL3 (Fig. 1i). Additionally, 2-BP treatment did not exhibit detectable cytotoxicity in A375 and A549 cells (Supplementary Fig. 1f). Taken together, these results demonstrate that 2-BP effectively sensitizes cancer cells to ferroptosis in vitro.
a Volcano plot showing ferroptotic cell death changes in A375 cells treated with ~3200 metabolism-focused small molecules for 24 h, followed by 24 h RSL3 treatment. b Cell viability analysis in A375 cells pre-treated with 2-BP for 24 h, followed by 48 h treatment with GPX4 inhibitors (RSL3, ML162, ML210) and SLC7A11 inhibitor (erastin). c Cell viability analysis of A375 cells pre-treated with 2-BP for 24 h, followed by 48 h cystine starvation-induced ferroptosis, with or without Fer-1 (2 μM). Phase contrast and fluorescence images (d) and quantitative analysis of SYTOX Green-stained dead cells (e) in A375 cells pre-treated with 2-BP for 24 h, followed by 6 h RSL3 (4 μM) treatment, with or without Fer-1 (2 μM). Scale bar, 100 μm. f Lipid peroxidation analysis in A375 cells pre-treated with 2-BP for 24 h, followed by RSL3 (4 μM) treatment for 6 h, with or without Fer-1 (2 μM). g PTGS2 mRNA level detection in A375 cells pre-treated with 2-BP for 24 h, then treated with RSL3 (4 μM) for 6 h, with or without Fer-1 (2 μM). h Heatmap of cell viability in A375 cells pretreated with 2-BP for 24 h, followed by 24 h RSL3 (1 μM) treatment with various inhibitors, including z-VAD-fmk (20 μM), necrostatin-1 (2 μM), Bafilomycin A1 (10 nM), and Fer-1 (1 μM). i ROS quantification in A375 cells pretreated with 2-BP for 24 h, followed by 6 h RSL3 treatment using CM-H2DCFDA probe. Xenograft experiment with subcutaneously inoculated A375 cells in nude mice treated with IKE (20 mg/kg), 2-BP (20 mg/kg), a combination of IKE (20 mg/kg) and 2-BP (20 mg/kg), and a combination of IKE (20 mg/kg), 2-BP (20 mg/kg), and Lip-1 (10 mg/kg) (i.p., once every two days) for two weeks. Measurements include dissected tumors (j), growth curve (k), and tumor weight (l). n = 6. m, n Orthotopic xenograft tumors from RKO cells transplanted into the cecum of nude mice, treated with 2-BP (20 mg/kg) and a combination of 2-BP and Lip-1 (i.p., once every three days). Tumor volume indicated by fluorescence intensity. n = 8. Scale bar, 2 cm. Data are presented as mean ± SD. Statistical analyses: two-way ANOVA with Tukey’s multiple comparisons test for (c, e–g, i); one-way ANOVA with Tukey’s multiple comparisons test for (a, b, k, l, n). Sample sizes: (a–c, e–i) n = 3 independent experiments; (j–l) n = 6 individual mice; (m, n) n = 8 individual mice. Image in d shows representative results from three independent experiments. ns not significant.
To determine the role of 2-BP in enhancing ferroptosis in vivo, we subcutaneously implanted A375 xenograft tumors into nude mice, and treated with 2-BP and imidazole ketone erastin (IKE) individually or in combination. We observed that although 2-BP alone did not significantly impact the growth of A375 xenograft tumors, it significantly further enhanced the efficacy of IKE to reduce the size and weight of the tumors, and this effect could be reversed by the ferroptosis inhibitor liproxstatin-1 (Lip-1), providing evidence that the observed effects are caused by ferroptosis (Fig. 1j–l). Meanwhile, none of these treatments had effects on mouse body weight, suggesting low in vivo toxicity (Supplementary Fig. 2a). Immunohistochemical (IHC) staining revealed that neither 2-BP nor IKE, individually or in combination, altered the expression of cleaved caspase-3 (Supplementary Fig. 2b, c), suggesting that the inhibition of tumor growth was not mediated through apoptosis. Significantly, IKE treatment increased levels of 4-hydroxynonenal (4-HNE), a biomarker of lipid peroxidation36. This increase was further amplified by the presence of 2-BP, underscoring the role of 2-BP in enhancing IKE-induced ferroptosis in vivo (Supplementary Fig. 2d, e). Additionally, the elevated expression of PTGS2 mRNA in tumor tissues of the combined treatment group further supported the stimulatory effects of 2-BP on ferroptosis (Supplementary Fig. 2f). To further investigate whether 2-BP alone can stimulate ferroptosis in physiological contexts, we developed an orthotopic tumor model of colorectal cancer (CRC) using human RKO cells. Unlike the observations in the subcutaneous tumor model, 2-BP significantly reduced the growth of the orthotopically implanted RKO tumors, and treatment with Lip-1 markedly reversed the tumor growth inhibition induced by 2-BP (Fig. 1m, n). 2-BP treatment also significantly increased the levels of 4-HNE staining and PTGS2 mRNA expression, which were substantially mitigated by co-treatment with Lip-1 (Supplementary Fig. 2g–i). In summary, these findings indicate that 2-BP could significantly enhance the sensitivity of tumor cells to ferroptosis in vivo.
GPX4 is palmitoylated at Cys 66
We next investigated the molecular mechanism by which 2-BP sensitizes tumor cells to ferroptosis. 2-BP inhibits a variety of proteins and enzymes involved in lipid metabolism37. Since iron-dependent lipid peroxidation is a hallmark of ferroptosis, we performed a lipidomic analysis to evaluate whether 2-BP modulates the formation of various lipid species, thereby rendering cancer cells susceptible to ferroptosis. Interestingly, our observations indicate that treatment with 2-BP did not substantially affect the overall lipid composition, including the levels of individual free fatty acids or specific lipid species (Supplementary Fig. 3a–c). Considering previous research has indicated that phosphatidylcholine or phosphatidylethanolamine polyunsaturated fatty acids (PC/PE-PUFAs), such as PC/PE-arachidonoyl (AA)/adrenoyl (AdA), are the primary targets for oxidation in ferroptosis38, we specifically investigated changes in these lipid species. We found that treatment with 2-BP did not change the levels of these lipid species (Supplementary Fig. 3d, e). This suggests that contribution of 2-BP to enhancing ferroptosis susceptibility is unlikely to be through a broad regulation of lipid composition.
To elucidate whether 2-BP regulates ferroptosis at the transcriptional level, we performed RNA sequencing analysis on A375 cells with 2-BP treatment or not. We compared gene expression levels between the control group and the 2-BP-treated group. We identified that 109 genes were significantly downregulated, and 99 genes were significantly upregulated with a fold change greater than 2 (Supplementary Fig. 4a). According to the KEGG enrichment pathway analysis, the impact of 2-BP treatment on ferroptosis was not substantial, indicating 2-BP alone was not sufficient to induce ferroptosis in vitro (Supplementary Fig. 4b). We further analyzed the expression of genes associated with the ferroptosis signaling pathway, including glutathione synthesis, oxidative reduction regulation, lipid metabolism, and iron homeostasis. Our findings showed that 2-BP treatment did not significantly impact the mRNA expression levels of these genes (Supplementary Fig. 4c). Additionally, we carried out quantitative PCR (qPCR) analysis on key ferroptosis regulators. The results showed that 2-BP only slightly upregulated the mRNA expression of SLC7A11 and FSP1, but not GPX4, ACSL4, ACSL1 and PTGS2 (Supplementary Fig. 4d). However, the modest upregulation of SLC7A11 and FSP1 did not align with the stimulatory effects of 2-BP on ferroptosis. Therefore, 2-BP does not alter the ferroptosis sensitivity at the transcriptional level.
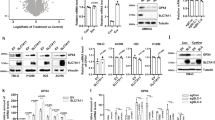
As 2-BP is widely used as a general protein palmitoylation inhibitor, we hypothesized that 2-BP enhances cellular vulnerability to ferroptosis by altering the palmitoylation of ferroptosis-relevant proteins. To explore this, we performed the proteomic analysis of palmitoylated proteins in hydroxylamine-treated or untreated A375 melanoma cells using the alkyne-tagged palmitic acid analogue Alk14 as a metabolic label, particularly focusing on proteins associated with ferroptosis (Fig. 2a). We observed that GPX4, a crucial inhibitor of ferroptotic cell death, undergoes palmitoylation. Notably, the counts of palmitoylated GPX4 peptides were significantly reduced following 2-BP treatment (Fig. 2b, c). Palmitoylation of both endogenous GPX4 and exogenously expressed Flag-GPX4 in A375 and 293 T cells was confirmed using streptavidin to detect GPX4 protein labeled using the acyl-biotin exchange (ABE) assay (Fig. 2d, e and Supplementary Fig. 5a). Supporting our hypothesis, 2-BP treatment substantially inhibited GPX4 palmitoylation (Fig. 2f, g). Intriguingly, 2-BP treatment also significantly reduced GPX4 protein levels (Fig. 2f, h). To further explore the connection between 2-BP and GPX4 in ferroptosis sensitivity, we created GPX4-knockout A375 cells using CRISPR/Cas9-mediated genomic deletion. These GPX4-deficient cells did not exhibit increased sensitivity to ferroptosis upon 2-BP treatment (Supplementary Fig. 5b, c). In addition, 2-BP still induced ferroptosis in FSP1-KO A375 cells, indicating that 2-BP primarily targets GPX4 rather than FSP1 (Supplementary Fig. 5d, e). Furthermore, we induced ferroptosis with different concentrations of RSL3 and observed that 2-BP significantly increased the occurrence of ferroptosis induced by low concentrations of RSL3, but did not further enhance the ferroptosis induced by high concentrations of RSL3 (Supplementary Fig. 5f). Subsequent overexpression of GPX4 in A375 and HT1080 cells significantly mitigated the effects of 2-BP on RSL3-induced ferroptosis (Supplementary Fig. 5g–j), suggesting that 2-BP intensifies cellular susceptibility to ferroptosis primarily through modulating GPX4.
a Schematic of the workflow for identifying palmitoylated proteins using the click chemistry assay and LC-MS/MS. b Mass spectrometry results from three independent samples were used to identify palmitoylated proteins. The numbers in the figure represent the palmitoylated proteins identified in each sample (with proteins identified in the HAM group excluded). c Peptide spectral counts for GPX4 in control and 2-BP treated groups, with and without HAM. Palmitoylation analysis of Flag-GPX4 in A375 (d) and 293T (e) cells using the ABE assay, with and without HAM. f–h Palmitoylation of Flag-GPX4 in A375 cells pre-treated with 2-BP and incubated with Alk14, analyzed using the click chemistry assay with and without HAM (f). Quantitative analysis of GPX4 palmitoylation (g) and total GPX4 levels (h). i, j Palmitoylation analysis of exogenous GPX4 and its mutants in A375 cells using the ABE assay with and without HAM (i). Quantitative analysis of GPX4 palmitoylation (j). Immunoblot detection (k) and quantification (l) of GPX4 in A375 cells infected with Flag-GPX4-WT or mutant constructs. m Cell viability analysis in A375-GPX4-KO cells infected with WT or mutant Flag-GPX4 constructs, pre-treated with 2-BP, and then treated with RSL3. Data are presented as mean ± SD. Statistical analysis: two-way ANOVA with Tukey’s test for (c, j); one-way ANOVA with Tukey’s test for (g, h, l). Sample sizes: (c, g, h, j, l, m) n = 3 independent experiments. Image in d, e, f, i, k shows representative results from three independent experiments.
To pinpoint the palmitoylation sites on GPX4, we individually mutated each of the seven conserved cysteine residues in Flag-tagged GPX4 to serine and assessed the palmitoylation status of these mutants (Supplementary Fig. 6a). The palmitoylation level of GPX4 was nearly abolished when C66 was mutated, and mildly reduced with a C75 mutation (Fig. 2i, j and Supplementary Fig. 6b, c). To control for potential variations in plasmid transcription, we measured the mRNA levels of each mutant, confirming that transcriptional levels were consistent across all variants (Supplementary Fig. 6d). Remarkably, the mutation of GPX4 at C66 to serine (GPX4-C66S), but not the C75S mutation, led to a significant reduction in GPX4 protein levels (Fig. 2k, l and Supplementary Fig. 6e–g). To further confirm whether both C66 and C75 residues of GPX4 are palmitoylated, we conducted the acyl-PEG exchange (APE) assays and observed that only a single palmitate was incorporated into GPX4 (Supplementary Fig. 6h). Given that the C66S mutation nearly completely abolished GPX4 palmitoylation, while the C75S mutation led to a moderate reduction, we conclude that C66 is the primary palmitoylation site on GPX4, with C75 likely playing a facilitative role in the palmitoylation process. In GPX4-knockout A375 cells, re-expression of GPX4-WT, GPX4-C66S, and GPX4-C75S mutants maintained cell viability in the absence of Fer-1. However, the dose-dependent stimulatory effects of 2-BP on RSL3-induced ferroptosis were fully abrogated by the C66S mutation (Fig. 2m). These findings strongly indicate that C66 is a critical palmitoylation site on GPX4, with a significant role in maintaining its protein stability and sensitivity to ferroptosis.
GPX4 palmitoylation regulates protein stability
To elucidate how 2-BP sensitizes tumor cells to ferroptosis by affecting GPX4 protein levels, we treated A375 and A549 cancer cells with varying concentrations and durations of 2-BP. We consistently observed a dose-dependent and time-dependent reduction in GPX4 protein levels, while other ferroptosis-related proteins such as FSP1, SLC7A11, and ACSL4 remained unaffected (Fig. 3a, b). This specificity was further confirmed across multiple cancer cell lines, including WM1361A and SK-MEL-30 melanoma, RKO and HT29 colon cancer, and HT1080 fibrosarcoma cells (Supplementary Fig. 7a). Immunofluorescence staining corroborated these findings, showing a reduction in endogenous and ectopically overexpressed GPX4 protein (Fig. 3c and Supplementary Fig. 7b). To investigate the mechanism by which 2-BP diminishes GPX4 protein levels, we used cycloheximide (CHX) to inhibit mRNA translation and observed a shortened half-life of GPX4 in the presence of 2-BP, suggesting post-transcriptional regulation (Fig. 3d, e). The proteasome inhibitor MG132, but not the lysosomal inhibitor BafA1, hindered the 2-BP-induced GPX4 degradation, indicating a proteasome-dependent process (Fig. 3f, g). The ubiquitination assay further revealed that 2-BP enhances the ubiquitination of GPX4 (Fig. 3h). Taken together, these results suggest that 2-BP decreases GPX4 protein levels via proteasome-dependent degradation.
Immunoblots of GPX4, FSP1, SLC7A11, and ACSL4 in A375 and A549 cells treated with varying concentrations of 2-BP (a) and at different time points (b). c Immunofluorescence staining of GPX4 in A375 cells treated with 2-BP for 48 h. Scale bar, 10 μm. d, e Immunoblots of GPX4 in A375 cells pre-treated with 2-BP for 24 h, followed by cycloheximide (CHX) treatment for indicated times (d). Quantification of GPX4 protein levels (e). f GPX4 detected by immunoblots in A375 cells pre-treated with 2-BP for 24 h, followed by 12 h proteasome inhibitor MG132 treatment. g GPX4 detected by immunoblots in A375 cells pre-treated with 2-BP for 24 h, followed by 12 h lysosome inhibitor BafA1 treatment. h Immunoblots were performed on A375 cells that were pre-treated with 2-BP for 12 h, followed by the addition of 10 μM MG132 for an additional 12 h, after which GPX4 was pulled down using an anti-GPX4 antibody. i, j Immunoblots of GPX4 in A375 cells infected with Flag-GPX4-WT or Flag-GPX4-C66S, followed by CHX treatment for indicated times (i). Quantification of GPX4 protein levels (j). k, l Immunoblots of GPX4 in A375 cells infected with Flag-GPX4-WT or Flag-GPX4-C75S, followed by CHX treatment for indicated times (k). Quantification of GPX4 protein levels (l). GPX4 protein levels detected by immunoblots (m) and relative mRNA levels by RT-qPCR (n) in A375 cells infected with various GPX4 constructs. o GPX4 detected by immunoblots in A375 cells infected with Flag-GPX4-WT or Flag-GPX4-C66S, treated with or without MG132 for 12 h. p, q Immunoblots of GPX4 in A375 cells infected with various GPX4 constructs, followed by 48 h 2-BP treatment (p). Quantification of GPX4 protein levels (q). Data are presented as mean ± SD. Statistical analysis: two-way ANOVA with Tukey’s test for (q); one-way ANOVA with Tukey’s test for (e, j, l, n). Sample sizes: (e, j, l, n, q) n = 3 independent experiments. Image in a–d, f–i, k, m, o, p shows representative results from three independent experiments.
We next examined the impact of GPX4 palmitoylation on protein stability using GPX4-C66S and GPX4-C75S mutants. Cycloheximide chase assays revealed that the C66S mutation, but not C75S, significantly reduced GPX4 half-life (Fig. 3i–l) and introducing C75S into GPX4-C66S did not further decrease GPX4 levels (Fig. 3m, n). We also observed that the degradation of GPX4-C66S could be inhibited by the proteasome inhibitor MG132 (Fig. 3o). We overexpressed wild-type GPX4 (GPX4-WT), GPX4-C66S, and GPX4-C75S mutants in A375 cells and observed that 2-BP reduced levels of endogenous GPX4, GPX4-WT, and GPX4-C75S, but not GPX4-C66S (Fig. 3p, q), highlighting C66 as a crucial palmitoylation site for GPX4 stability.
Previous studies identified cysteine 66 as an allosteric binding site for GPX4 inhibitors including RSL3 and LOC188639. These inhibitors not only block the enzymatic activity of GPX4 but also trigger GPX4 protein degradation through an as-yet-unclear mechanism. Since palmitoylation at the GPX4 C66 residue enhances the stability of GPX4 protein, we propose that GPX4 inhibitors binding to the C66 allosteric site may decrease the palmitoylation of GPX4. Supporting the hypothesis, both RSL3 and LOC1886 significantly reduced the palmitoylation levels of GPX4 as shown in click chemistry assays with metabolic label Alk14 (Supplementary Fig. 8a, b). Notably, LOC1886 decreased protein levels of both endogenous and exogenous GPX4-WT, but not the GPX4-C66S mutant (Supplementary Fig. 8c), and inhibited the 2-BP enhancement of RSL3-induced ferroptosis sensitivity (Supplementary Fig. 8d). We also observed a synergistic decrease in GPX4 protein levels beyond RSL3 treatment alone (Supplementary Fig. 8e). Taken together, these results indicate that palmitoylation at C66 is pivotal for GPX4 stability and modulates ferroptosis sensitivity.
ZDHHC20 is the palmitoyl acyltransferase of GPX4
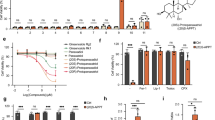
To identify the specific protein S-acyl transferases responsible for GPX4 palmitoylation, we overexpressed 23 HA-tagged ZDHHC palmitoyl acyltransferases along with Flag-tagged GPX4 in 293T cells and performed the click chemistry-based assay to detect palmitoylation. Among these, only ZDHHC20 overexpression significantly enhanced GPX4 palmitoylation (Fig. 4a). This interaction between ZDHHC20 and GPX4 was further confirmed through co-immunoprecipitation experiments (Fig. 4b). We subsequently verified the role of ZDHHC20 in regulating GPX4 palmitoylation. Our observations revealed that ZDHHC20 knockdown did not impact GPX4 mRNA levels (Fig. 4c, d), yet markedly reduced its palmitoylation and overall protein expression (Fig. 4e). Moreover, the overexpression of wild-type (WT) ZDHHC20 increased GPX4 palmitoylation and protein levels, but not the catalytically inactive C156S mutant of ZDHHC20 (Fig. 4f). We further explored how ZDHHC20-mediated palmitoylation of GPX4 affects the susceptibility of cancer cells to ferroptosis. ZDHHC20-deficient cells were more prone to ferroptosis (Fig. 4g), whereas overexpression of ZDHHC20 conferred increased ferroptosis resistance (Fig. 4h, i). We found that overexpression of ZDHHC20 to increase GPX4 palmitoylation enhanced the protein levels of GPX4 but did not affect its localization on the cell membrane (Fig. 4h, j). We also utilized the Cancer Treatment Response Portal (CTRP) database (https://portals.broadinstitute.org/ctrp.v2.1/) to analyze the relationship between ZDHHC20 expression and ferroptosis. In consistence, high expression levels of ZDHHC20 were associated with increased tolerance to ferroptosis (Fig. 4k). Collectively, these results indicate that ZDHHC20 plays a crucial role in the palmitoylation of GPX4, significantly influencing the ferroptosis sensitivity of cancer cells.
a Palmitoylation of GPX4 in 293T cells expressing Flag-GPX4 and transfected with either empty vector or HA–DHHC plasmids, incubated with 50 μM Alk14 for 12 h, determined using the click chemistry assay. b Co-immunoprecipitation in 293T cells transiently transfected with Flag-GPX4 and HA-ZDHHC20 plasmids to investigate ZDHHC20-GPX4 interaction, with GPX4 and ZDHHC20 detection by immunoblots. RT-qPCR analysis of ZDHHC20 (c) and GPX4 (d) mRNA levels in A375 cells infected with lentiviruses encoding control shRNA or ZDHHC20 shRNA. e GPX4 palmitoylation in A375 cells infected with lentiviruses encoding control shRNA or ZDHHC20 shRNA, and incubated with 50 μM Alk14 for 12 h, determined using the click chemistry assay. f GPX4 palmitoylation in A375 cells infected with lentiviruses encoding HA-empty vector, HA-ZDHHC20 or HA-ZDHHC20-C156S and incubated with 50 μM Alk14 for 12 h, determined using the click chemistry assay. g Cell viability analysis in A375 cells infected with lentiviruses encoding control shRNA or ZDHHC20 shRNA, followed by 48 h RSL3 treatment. h Immunoblots of GPX4 and ZDHHC20 in A375 cells infected with lentiviruses encoding empty vector or ZDHHC20. i Cell viability analysis in A375 cells infected with lentiviruses encoding empty vector or ZDHHC20, followed by 24 h RSL3 treatment. j Immunofluorescence staining for GPX4 and ZDHHC20 was performed in A375 cells infected with lentiviruses encoding a control plasmid or an HA-ZDHHC20 plasmid. Scale bar, 4 μm. k Analysis of the correlation between ZDHHC20 gene expression and ferroptosis sensitivity to RSL3, ML162 and ML210 using the CTRP database (https://portals.broadinstitute.org/ctrp.v2.1/). ZDHHC20 was correlated with resistance to RSL3 (P = 0.139), ML162 (P = 0.135) and ML210 (P = 0.154) according to their Pearson correlation score. “In Fig. 4k, the box plot is presented to illustrate the correlation between the sensitivity of [860 cancer cell lines to 481 compounds] and [different levels of ZDHHC20 gene expression]. The central box extends from the lower quartile (-1, Q1, 25th percentile) to the upper quartile (1, Q3, 75th percentile), indicating the interquartile range (2, IQR), which contains the middle 50% of the data. The median (0, Q2, 50th percentile) is represented by a line inside the box, dividing the data into two halves. The whiskers extend from the edges of the box to the minimum and maximum data points within a certain range, typically 1.5 times the IQR, unless there are outliers. Outliers are data points that fall outside the range of -3 to 3 and are plotted as individual points marked by circles. Data are presented as mean ± SD. Statistical analysis: one-way ANOVA with Tukey’s test for (c, d). Sample sizes: (c, d, g, i) n = 3 independent experiments. k n = 860 cancer cell lines. Image in a, b, e, f, h, j shows representative results from three independent experiments.
APT2 mediates the depalmitoylation of GPX4
To determine the enzyme responsible for the depalmitoylation of GPX4, we overexpressed Flag-tagged GPX4 protein in 293T and A375 cell lines. Using Flag-affinity magnetic beads, we isolated GPX4-interacting proteins and conducted mass spectrometry-based proteomic analysis. This analysis revealed 37 proteins interacting with GPX4 in both cell lines (Fig. 5a). APT2 (encoded by LYPLA2), a known protein acyltransferase (PAT) involved in depalmitoylation, was identified as an interacting partner of GPX4 (Fig. 5a). We further verified this interaction via co-immunoprecipitation and immunostaining assays (Fig. 5b, c). Next, we investigated the role of APT2 in GPX4 depalmitoylation. Our experiments demonstrated an increase in GPX4 palmitoylation following APT2 deletion (Fig. 5d). Moreover, APT2 knockdown led to an elevation in GPX4 protein levels, while the levels of other ferroptosis-related proteins like FSP1, SLC7A11, and ACSL4 remained unchanged (Fig. 5e). Additionally, GPX4 transcript levels were unaffected by APT2 knockdown (Fig. 5f). The use of ML349, a specific APT2 inhibitor, corroborated these observations as treatment with ML349 raised GPX4 protein levels without altering its mRNA levels (Fig. 5g, h). We also examined GPX4 protein stability in the absence of APT2. Our findings showed that knocking down APT2 resulted in increased stability of the GPX4 protein (Fig. 5i, j). Collectively, these results establish APT2 as the regulatory enzyme of GPX4 depalmitoylation.
a Analysis of proteins interacting with GPX4 in A375 and 293T cells infected with lentiviruses encoding empty vector or Flag-GPX4, using anti-Flag magnetic beads enrichment and LC-MS/MS. b Co-immunoprecipitation in 293T cells transiently transfected with Flag-GPX4 and HA-APT2 plasmids to investigate GPX4-APT2 interaction detected by immunoblots. c Immunofluorescence staining for GPX4 and APT2 was performed in A375 cells infected with lentiviruses encoding a control plasmid or an HA-APT2 plasmid. Scale bar, 4 μm. d Palmitoylation analysis of exogenous GPX4 in A375 cells infected with lentiviruses encoding control or APT2 shRNA, using the ABE assay with or without HAM. e Immunoblots of APT2, GPX4, SLC7A11, and ACSL4 in A375 and HT1080 cells infected with lentiviruses encoding control or APT2 shRNA. f RT-qPCR detection of GPX4 mRNA levels in A375 and HT1080 cells infected with lentiviruses encoding control or APT2 shRNA. g Immunoblots of GPX4, FSP1, SLC7A11, and ACSL4 in A375, HT1080, and A549 cells treated with ML349 at indicated concentrations. h RT-qPCR analysis of GPX4 mRNA levels in A375 cells treated with ML349 at indicated concentrations. i, j Immunoblots of GPX4 and APT2 in A375 cells infected with lentiviruses encoding control or APT2 shRNA, treated with CHX for indicated times (i). Quantification of GPX4 protein levels (j). Data are presented as mean ± SD. Statistical analysis: two-way ANOVA with Tukey’s test for (j); one-way ANOVA with Tukey’s test for (f, h). Sample sizes: (f, h, j) n = 3 independent experiments. Image in b, c, d, e, g, i shows representative results from three independent experiments.
Inhibition of APT2 suppresses ferroptosis
To solidify the evidence that APT2 is a critical regulator of GPX4 depalmitoylation, thereby impacting cellular susceptibility to ferroptosis, we individually silenced APT2 and other depalmitoylases, including protein acyltransferase APT1 and the abhydrolase domain-containing proteins known to mediate protein depalmitoylation, including ABHD17A, ABHD17B, ABHD17C, and ABHD1040 (Supplementary Fig. 9a). Of these proteins, only the silencing of APT2 led to resistance against RSL3-induced ferroptosis (Fig. 6a, b and Supplementary Fig. 9b, c). SYTOX green staining assays further substantiated that APT2 knockdown inhibited RSL3-mediated ferroptotic cell death (Fig. 6c, d). Furthermore, APT2-deficient cells demonstrated resistance to other ferroptosis inducers, including ML162, ML210, and erastin (Supplementary Fig. 9d).
a Cell viability analysis in A375 cells infected with lentiviruses encoding control, APT1, APT2, ABHD17A, ABHD17B, ABHD17C, and ABHD10 shRNA, followed by 48 h RSL3 treatment. b Cell death analysis in A375 cells infected with lentiviruses encoding control or depalmitoylase shRNAs, treated with RSL3 (4 μM) for 6 h, with or without Fer-1 (2 μM). Phase contrast and fluorescence images (c) and quantitative analysis of dead cells stained with SYTOX Green (d) in A375 cells infected with control or APT2 shRNA and treated with RSL3 (4 μM) for 6 h. Scale bar, 20 μm. e Cell viability analysis in A375 cells pre-treated with ML348, ML349, Palm B, and ABHD957 for 24 h, followed by 48 h RSL3 treatment. f Cell death analysis in A375 cells pre-treated with 2-BP and various depalmitoylase inhibitors for 24 h, followed by RSL3 treatment for 48 h, with or without Fer-1. g Immunoblot confirmation of APT2 and GPX4 expression in A375 cells with APT2 knockdown, infected with HA-APT2 lentivirus. h Cell viability in A375 cells with APT2 knockdown, infected with HA-APT2 lentivirus, followed by 48 h RSL3 treatment. i Immunoblot confirmation of APT2 and GPX4 expression in A375 cells with APT2 knockdown, infected with lentiviruses encoding empty vector, HA-APT2, HA-APT2-C2S, or HA-APT2-S122A. j Cell viability analysis in A375 cells with APT2 knockdown, infected with lentiviruses encoding various HA-APT2 constructs, followed by 48 h RSL3 treatment. k Cell viability analysis in A375 cells infected with control or APT2 shRNA, pre-treated with 2-BP for 24 h, then treated with RSL3 for 48 h. l Cell viability analysis in A375 and HT1080 cells pre-treated with 2-BP and ML349 for 24 h, followed by 48 h RSL3 treatment. Data are presented as mean ± SD. Statistical analysis: two-way ANOVA with Tukey’s test for (b, f, k, l). Sample sizes: (a, b, d–f, h, j–l) n = 3 independent experiments. Image in c, g, i shows representative results from three independent experiments. ns not significant.
These observations were also supported by experiments using specific depalmitoylase inhibitors: ML348 for APT1, ML349 for APT2, Palm B for both APT1 and APT2, and ABD957 targeting ABHD17A, ABHD17B, and ABHD17C. Remarkably, only the APT2 inhibitor ML349 consistently conferred resistance to ferroptosis in the treated cells (Fig. 6e, f and Supplementary Fig. 9e, f). To confirm APT2’s regulatory impact on GPX4, we reintroduced a shRNA-resistant form of APT2 into APT2-knockdown cells. This re-expression of APT2 reversed the increase in GPX4 protein levels observed following APT2 deletion, and re-sensitized the APT2-depleted cells to ferroptosis (Fig. 6g, h). To further validate the crucial role of APT2’s depalmitoylase activity in modulating ferroptosis, we overexpressed either the wild-type APT2 or its enzymatically inactive mutants (APT2-C2S and S122A) in APT2-deficient cells. The overexpression of wild-type APT2 reduced GPX4 protein levels and increased cellular susceptibility to ferroptosis, in contrast to the inactive mutant forms which had no effect (Fig. 6i, j). Moreover, we demonstrated that both APT2 knockdown and pharmacological inhibition with ML349 mitigated the sensitizing effect of 2-BP on ferroptosis (Fig. 6k, l). In summary, our data suggest that targeting APT2 can significantly enhance the resistance of tumor cells to ferroptosis.
APT2 ablation aggravates colon cancer development
Substantial evidence from previous studies has established ferroptosis as a critical factor in colorectal tumor development41. In line with this, our recent findings have revealed that inhibiting ferroptosis through IDA can promote CRC tumor growth42. Given that APT2 modulation preserves GPX4 palmitoylation and strengthens cellular resistance to ferroptosis, we sought to further investigate the role of APT2 in CRC progression. We engineered mice with a targeted APT2 knockout in their intestinal epithelial cells and utilized the azoxymethane (AOM)/dextran sodium sulfate (DSS)-induced CRC model to evaluate the impact of APT2 deletion on colon cancer development (Fig. 7a and Supplementary Fig. 10a, b). Compared to WT mice, APT2 heterozygous or homozygous knockout mice exhibited a marked increase in tumor numbers in both colorectal and small intestinal regions (Fig. 7b, c and Supplementary Fig. 10c, d), suggesting that APT2 knockout significantly exacerbated CRC development in the AOM/DSS-induced model. Hematoxylin and eosin staining of Swiss-rolled colons further confirmed that APT2-knockout colorectums displaying a higher incidence of colonic tumors (Fig. 7d). Additionally, immunohistochemical analysis revealed that compared to those in tumors from APT2 wild-type mice, GPX4 levels were higher in APT2-knockout colorectal tumors (Fig. 7e). Further examination revealed a marked reduction in 4-HNE levels across APT2-knockout colorectal tumors, indicating that APT2 knockout suppressed ferroptosis in vivo (Fig. 7e, f). These findings suggest that APT2 depletion leads to increased GPX4 protein levels and aggravates CRC progression.
a Timeline of the AOM/DSS-induced colon cancer model in C57BL/6J transgenic mice. b Representative images of large intestine tumorigenesis in Vil1-Cre; APT2+/+, Vil1-Cre; APT2flox/+, and Vil1-Cre; APT2flox/flox mice. Scale bar, 10 μm. c Quantification of tumor numbers in the large intestine of Vil1-Cre; APT2+/+, Vil1-Cre; APT2flox/+, and Vil1-Cre; APT2flox/flox mice. d H&E staining of colorectal lesions induced by AOM/DSS in Vil1-Cre; APT2+/+, Vil1-Cre; APT2flox/+, and Vil1-Cre; APT2flox/flox mice. Scale bar, 1 mm and 100 μm. e IHC staining for APT2, GPX4, and 4-HNE in colorectal lesions induced by AOM/DSS in Vil1-Cre; APT2+/+, Vil1-Cre; APT2flox/+, and Vil1-Cre; APT2flox/flox mice. Scale bar, 10 μm. f Intensity scoring of 4-HNE staining in colorectal lesions induced by AOM/DSS in Vil1-Cre; APT2+/+, Vil1-Cre; APT2flox/+, and Vil1-Cre; APT2flox/flox mice. Data are presented as mean ± SD. Statistical analysis: one-way ANOVA with Tukey’s test for (c, f). Sample sizes: (c, f) n = 6 independent tumors. Image in d, e shows representative results from three independent experiments.
GPX4 palmitoylation and depalmitoylation regulate ferroptosis
Previous studies have indicated that ferroptosis affects the metastatic spread of cancer cells43. We then investigated the impact of palmitoylation-mediated ferroptosis sensitivity on tumor metastasis. In the colorectal cancer transplantation model, we found that the administration of 2-BP effectively hindered the metastasis of colorectal cancer, and this inhibition was reversed by the treatment with ferroptosis inhibitor Lip-1 (Fig. 8a, b). Further, to determine if 2-BP could prevent tumor cell metastasis through the vascular system, we conducted a tail vein-to-lung metastasis assay. Our observations revealed that 2-BP significantly inhibited the vascular-to-lung metastasis of A375 cells (Fig. 8c, d).
a, b Orthotopic xenograft tumors by transplanting RKO cells into the cecum of nude mice, treated with 2-BP (20 mg/kg, ip, every three days) and a combination of 2-BP and Lip-1 (20 mg/kg, ip, every three days). Representative image of distal metastatic tumors (a) and quantitative analysis of metastatic tumor numbers (b). n = 8 individual mice. Scale bar, 500 μm. c, d Nude mice pre-treated with 2-BP (20 mg/kg, ip, daily) and a combination of 2-BP and Lip-1 (20 mg/kg, ip, daily) for 3 days, followed by tail vein injection of 2 million A375-GFP cells and continued treatment for one week. Observation for lung metastatic tumor development three weeks later. Representative image of lung metastatic tumors (c) and quantitative analysis of metastatic tumor numbers (d). n = 6 individual mice. Scale bar, 500 μm. e C57BL/6J mice were intraperitoneally injected with ML349 (5 mg/kg) and Lip-1 (10 mg/kg) one hour before the induction of liver injury by ischemia-reperfusion. Representative images of the livers under different treatments, as well as the corresponding hematoxylin and eosin (HE) staining of the livers and immunohistochemical staining for MDA from liver tissues, are presented. n = 6 individual mice. Scale bar, 50 μm. f, g The analysis of serum alanine transaminase (ALT) and aspartate aminotransferase (AST) levels in C57BL/6J mice with liver injury induced by ischemia-reperfusion. h, i RT-qPCR analysis of the relative mRNA levels of Ptgs2 and Chac1 in liver tissue from C57BL/6J mice with liver injury induced by ischemia-reperfusion. j Quantitative analysis of MDA in liver tissue from C57BL/6J mice with liver injury induced by ischemia-reperfusion. Data are presented as mean ± SD. Statistical analysis: one-way ANOVA with Tukey’s test for (b, d, f–j). Sample sizes: (a, b) n = 8; (c, d) n = 6; (e–j) n = 6 individual mice. Image in a, c, e shows representative results from three independent experiments. ns not significant.
We also explored the physiological roles of palmitoylation-mediated ferroptosis resistance through ischemia-reperfusion experiments. The results demonstrated that the APT2 inhibitor ML349 substantially reduced liver injury caused by ischemia-reperfusion, paralleling the effects of Lip-1 (Fig. 8e–g). This finding was supported by the notable decrease in mRNA levels of ferroptosis markers Ptgs2 and Chac1, and further confirmed by malondialdehyde (MDA) IHC staining, indicating that ML349 inhibits ferroptosis (Fig. 8h–j). In contrast, the palmitoylation inhibitor 2-BP markedly exacerbated liver injury in the ischemia-reperfusion model by promoting ferroptosis, although 2-BP alone did not induce liver damage (Supplementary Fig. 11a–f).
To further validate that 2-BP and ML349 modulate ferroptosis sensitivity by targeting GPX4, we generated GPX4-knockout A375 cells and then injected these cells subcutaneously into nude mice for xenograft assays. The A375 GPX4-KO xenograft tumors were then treated with IKE alone, or in combination with either 2-BP or ML349, over four weeks. Consistent with previous studies, GPX4-KO A375 tumors exhibited robust growth15. IKE treatment significantly reduced tumor size and weight, yet the addition of 2-BP or ML349 neither enhanced nor diminished the effectiveness of IKE (Supplementary Fig. 12a–c). These results confirm that 2-BP and ML349 specifically alter ferroptosis occurrence by modulating GPX4 palmitoylation in vivo.
It has been reported that GPX4-R152H mutation is associated with SSMD due to a partial loss of GPX4 function, but this variant is less prone to degradation44. We hypothesized that this variant might affect palmitoylation levels as well. Indeed, the GPX4-R152H variant showed enhanced palmitoylation (Supplementary Fig. 13a, b). Consistent with previous reports, our data showed that the GPX4-R152H variant, although losing most of its enzymatic activity, has not been completely inactivated (Supplementary Fig. 13c–e). We then observed that 2-BP reduced the GPX4-R152H protein level and activity against ferroptosis (Supplementary Fig. 13f, g), and pharmacological inhibition of APT2 with ML349 to maintain GPX4 palmitoylation enhances its activity (Supplementary Fig. 13h). Considering ferroptosis’s link to various diseases, targeting GPX4 palmitoylation may represent a potential strategy to inhibit ferroptosis in these contexts.
Discussion
Palmitoylation is a dynamic post-translational modification that plays a crucial role in modulating protein transport, localization, stability, and protein-protein interaction, thereby influencing many essential biological processes25. Despite established knowledge on the broad impact of protein palmitoylation, its specific involvement in ferroptosis regulation has remained largely unexplored. Our report revealed that GPX4 is subject to palmitoylation, which enhances its protein stability and decreases cellular ferroptosis sensitivity. This study began with a screening of metabolism-related small molecules to assess their impact on ferroptosis sensitivity, and identified 2-BP, a general inhibitor of palmitoylation, as a significant enhancer of ferroptosis susceptibility. Through palmitoylation mass spectrometry analysis and targeted mutation of GPX4’s conserved cysteine residues, we established that GPX4 undergoes palmitoylation at Cys 66. Two recent pieces of research revealed that SLC7A11 undergoes palmitoylation modification to regulate ferroptosis, although the identified palmitoylation sites of SLC7A11 reported in the two studies were inconsistent45,46. However, we found that the enhanced ferroptosis sensitivity induced by 2-BP was negated in cells expressing palmitoylation-defective GPX4 C66S mutant. Given the pivotal role of GPX4 in mitigating ferroptosis as downstream of SLC7A11 with the unique function of reducing toxic lipid peroxides, the palmitoylation-dependent modulation of GPX4 is likely to have a more pronounced impact on ferroptosis protection.
GPX4 is the only known selenoprotein in mammals capable of reducing lipid hydroperoxides to their corresponding lipid alcohols within cell membranes to prevent ferroptotic cell death4,14. It has been observed that cancer cells transitioning into therapy-resistant states rely on GPX4, identifying it as a potential target in cancer therapy strategies15,16. To this end, numerous small molecule inhibitors, notably RSL3 and ML162, have been developed to inhibit GPX4 activity. Recent discoveries have highlighted that RSL3 and ML162 can interact with GPX4 beyond the selenocysteine 46 (U46) active site, specifically binding to cysteine 66 (C66), suggesting an alternative inhibitory pathway. These inhibitors, along with LOC1886 which also binds covalently to C66, induce a conformational alteration in GPX4, leading to its degradation via a mechanism that remains to be elucidated. In our investigation, we demonstrated that RSL3 and LOC1886 impeded GPX4 palmitoylation at the C66 site, which is crucial for its protein stability. RSL3 was reported to have poor in vivo efficacy4,47. Our findings indicated that inhibiting GPX4 palmitoylation amplified the effects of RSL3 to trigger ferroptosis and markedly suppressed the growth of xenografted tumors. This suggests that targeting GPX4 palmitoylation could potentiate the effectiveness of ferroptosis inducers in cancer treatment.
A substantial body of research has established a robust link between ferroptosis and a wide spectrum of diseases, including cancer, immunological disorders, neurodegenerative conditions, stroke, and damage to multiple organs11. Suppressing ferroptosis has been shown to exacerbate tumor progression in colorectal cancer41,42. Our study further demonstrated that the deletion of APT2 in the colitis-induced colorectal cancer model strongly promoted tumor development by preserving GPX4 palmitoylation and inhibiting ferroptosis. Tumor cells are susceptible to ferroptosis within the bloodstream, indicating that the hematogenous spread of tumors depends on GPX443. Our results showed that 2-BP significantly inhibited the metastasis of both melanoma and colon cancer cells by suppressing GPX4 palmitoylation. The induction of ferroptosis in various organ damages and the specific role of GPX4 inactivation leading to acute kidney injury in mice were reported21,48. Through ischemia-reperfusion experiments, we elucidated the protective role of GPX4 palmitoylation-mediated resistance to ferroptosis, with the APT2 inhibitor ML349 significantly reducing liver injury caused by ischemia-reperfusion. Furthermore, the GPX4 R152H mutation’s association with SSMD suggests that targeting APT2 to maintain GPX4 palmitoylation may enhance GPX4-R152H activity, proposing APT2 as a viable target for therapeutic intervention44. In summary, our research underscores the potential of modulating the GPX4 palmitoylation-depalmitoylation cycle as a strategy for treating ferroptosis-related diseases.
Methods
Ethical statement
This research complies with all relevant ethical regulations. All animal experiments were approved by the Animal Research Ethics Committee of Shenzhen Bay Laboratory (Approved Protocol ID: AEYCQ202101), and all relevant ethical regulations were followed.
Cell lines and cell culture
Human melanoma cell lines A375, SK-MEL-28, and A2058, human colorectal cancer cell lines SW480, SW620, RKO, HT-29, and HCT116, human fibrosarcoma cell line HT1080, human non-small cell lung cancer cell line A549, human breast cancer cell line MCF7, human pancreatic cancer cell line PANC-1, human cervical cancer cell line HeLa, human embryonic kidney cell line 293T, and mouse melanoma cell line B16-F0 were obtained from the American Type Culture Collection (ATCC). Human melanoma cell lines WM1361A, SK-MEL-30, A875, WM115, and WM2664 were provided by the Cell Bank, Chinese Academy of Sciences. The human glioblastoma cell line U251-MG was provided by Professor Tengfei Guo of the Institute of Biomedical Engineering at Shenzhen Bay Laboratory (Shenzhen, China). A875 and WM115 cells were cultured in Minimum Essential Medium (MEM) (Gibco, USA). SK-MEL-28, SW480, SW620, HT-29, and B16-F0 cells were cultured in RPMI 1640 medium (Gibco, USA). All other cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA). All culture media were supplemented with 10% fetal bovine serum (FBS) (ExCell, China) and 1% penicillin-streptomycin antibiotics (Gibco, USA). All cells were cultured at 37 °C in a humidified incubator with 5% CO2 and were routinely tested and confirmed to be negative for mycoplasma.
High-throughput screening of metabolism-focused small molecule library
A375 cells were seeded at a density of 5000 cells per well in 96-well plates. The Biomek i7 Automated Workstation (Beckman Coulter) was used to add a metabolism-focused library containing ~3200 small molecule compounds to the 96-well plates for pretreatment of cells for 24 h. Subsequently, these cells were treated with RSL3 or DMSO for an additional 24 h. After that, cell viability was measured. The cell viability of the compound alone treatment group was used to exclude the toxicity of the compound library to the cells. The data were calculated using the following formula: (OD value of small molecules plus RSL3-treated cells - OD value of blank well)/(OD value of RSL3-treated cells - OD value of blank well). This screening was repeated three times, and the average data were taken. Log2(fold change) >1 or <–1, indicating a fold change >2 or <–2, and -log10(P value) >1.3, indicating P < 0.05, were used for analysis.
Cell viability assay
Cell viability was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions. Briefly, cells were seeded at a density of 5000 cells per well in 96-well plates and treated as indicated in each individual experiment. Subsequently, the CellTiter-Glo reagent was added to the cells, and the luminescence was read within 10 min.
Cell death assay
Cells were seeded at an appropriate density and cultured for 24 h. Subsequently, the cells were treated according to the descriptions in individual experiments. Then, RSL3 or cysteine deprivation was used to induce ferroptosis, and SYTOX Green (5 nM) was used to monitor cell death. Pictures of the cultured cells were taken at the indicated time points. The percentage of dead cells was determined by dividing the number of SYTOX Green-positive cells by the total number of cells.
Measurement of lipid peroxidation
A375 cells were seeded at a density of 1 × 105 cells per well in 6-well plates. The next day, the cells were treated with 2-BP for 24 h, followed by RSL3 to induce ferroptosis. Before trypsinization, the cells were incubated with C11-BODIPY (581/591) (1 μM) for 30 min at 37 °C. Subsequently, the cells were suspended in 500 μl of fresh PBS (Gibco), strained through a 35 μM cell strainer (Falcon tube with cell strainer CAP). The analysis was performed using a flow cytometer (FACS Canto II, BD Biosciences) with a 488-nm laser for excitation. Data were collected from the FL1 detector (C11-BODIPY) using a 502LP and 530/30 BP filter. At least 10,000 events were analyzed for each sample. The data were analyzed using FlowJo software.
Intracellular ROS measurement
A375 cells are cultured for 24 h in a 60-mm cell culture dish, followed by treatment with 2-BP for an additional 24 h, and then treated with RSL3 for another 6 h. The culture medium is replaced with serum-free Fluoro-brite medium (A1896701, Gibco, USA) containing 10 μM of the CM-H2DCFDA probe (C6827, Thermo Fisher Scientific, USA), and the cells are incubated for 30 min at 37 °C. Images are taken using fluorescence microscopy photography and analyzed with ImageJ software.
Real-time quantitative PCR (qPCR) assays
Total RNA was extracted using the GeneJET RNA Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions, and first-strand cDNA synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Real-time PCR was conducted in triplicate using the Dream TaqTM Green PCR Master Mix (Thermo Fisher Scientific) on the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad). GAPDH served as the internal control. Relative quantification was performed using the ΔΔCT method. The primers used are listed in Supplementary Table 1.
Western blotting
Cells were washed three times with ice-cold PBS and lysed in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and a cocktail of proteinase inhibitors) for 30 min at 4 °C. After sonication and centrifugation, the total protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific, USA). Equal amounts of protein from each group were separated by SDS-PAGE and electro-transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) at 100 V for 2 h. The PVDF membrane was then blocked with 5% non-fat powdered milk solution (Bio-Rad) for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C, followed by incubation with the HRP-conjugated secondary antibody. Immunoreactivity was detected using chemiluminescent substrate (Clarity Western ECL Substrate; Bio-Rad). The following primary antibodies were used: GPX4 (1:1,000, A21440, Abclonal), FSP1 (1:1000, 20886-1-AP, Proteintech), SLC7A11 (1:1,000, A2413, Abclonal), ACSL4 (1:1000, 22401-1-AP, Proteintech), Flag (1:1000, F1804, Merck), HA (1:1000, 3724S, CST), APT2 (1:1000, ab151578, Abcam), ZDHHC20 (1:1000, SAB4501054, Sigma-Aldrich), and β-actin (1:3,000, 66009-1-IG, Proteintech).
Immunofluorescence staining
Cells grown on sterile coverslips in a 6-well plate were washed once with 1× PBS, fixed with 4% PFA for 20 min, and then permeabilized with 0.2% Triton X-100 for 20 min. After blocking with 3% bovine serum albumin (BSA) for 1 h, the cells were incubated with primary antibody overnight at 4 °C, followed by incubation with secondary antibody for 1 h at room temperature. Nuclei were labeled with DAPI staining solution (C1006, Beyotime, China). Finally, the coverslips were mounted onto a clean glass microscope slide using Mowiol® 4–88 (81381, Sigma-Aldrich) mounting medium. Fluorescence images were obtained on a confocal laser-scanning microscope (Carl Zeiss Confocal LSM980).
LC-MS analysis of phospholipids
For each sample, 200 μl water was added to the sample. After 30 s vortex, the samples were freezed and thawed with liquid nitrogen for 3 times. Then the samples were sonicated for 10 min in ice-water bath. Add 480 μl MTBE: MEOH = 5:1 (containing internal standard) to the EP tube. After 30 s vortex, then the samples were sonicated for 10 min in ice-water bath. Then the samples were incubated at –40 °C for 1 h and centrifuged at 900 × g for 15 min at 4 °C. 300 μl of supernatant was transferred to a fresh tube and dried in a vacuum concentrator at 37 °C. Then, the dried samples were reconstituted in 100 μl of 50% methanol in dichloromethane by sonication on ice for 10 min. The constitution was then centrifuged at 16,200 × g for 15 min at 4 °C, and 85 μl of supernatant was transferred to a fresh glass vial for LC/MS analysis. LC-MS/MS analyses were performed using an UHPLC system (1290, Agilent Technologies), equipped with a Kinetex C18 column (2.1 × 100 mm, 1.7 μm, Phenomen). The mobile phase A consisted of 40% water, 60% acetonitrile, and 10 mmol/L ammonium formate. The mobile phase B consisted of 10% acetonitrile and 90% isopropanol, which was added with 50 mL 10 mmol/L ammonium formate for every 1000 mL mixed solvent. The analysis was carried with elution gradient as follows: 0~1.0 min,40% B;1.0~12.0 min,40%~100% B; 12.0~13.5 min, 100% B; 13.5~13.7 min, 100%~40% B; 13.7~18.0 min, 40% B. The column temperature was 55 °C. The auto-sampler temperature was 4 °C, and the injection volume was 2 μl (pos) or 2 μl (neg), respectively. The QE mass spectrometer was used for its ability to acquire MS/MS spectra on data-dependent acquisition (DDA) mode in the control of the acquisition software (Xcalibur 4.0.27, Thermo). In this mode, the acquisition software continuously evaluates the full scan MS spectrum. The ESI source conditions were set as following: sheath gas flow rate as 30 Arb, Aux gas flow rate as 10 Arb, capillary temperature 320 °C (positive), 300 °C (negative), full MS resolution as 70,000, MS/MS resolution as 17,500, collision energy as 15/30/45 in NCE mode, spray Voltage as 5 kV (positive) or –4.5 kV (negative), respectively.
RNA-sequencing analysis
A375 cells treated with 20 μM 2-BP or vehicle control DMSO for 48 h were collected, and the samples were sent to Novogene Company (China) for RNA extraction (RNA integrity number >9), library preparation, sequencing, and data analysis. Total RNA was used as input material for the RNA sample preparations. Briefly, mRNA was purified from total RNA by using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in First Strand Synthesis Reaction Buffer(5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase, then use RNaseH to degrade the RNA. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and dNTP. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, Adaptor with hairpin loop structure were ligated to prepare for hybridization. To select cDNA fragments of preferentially 370~420 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then PCR amplification, the PCR product was purified by AMPure XP beads, and the library was finally obtained.
After the library is qualified, the different libraries are pooling according to the effective concentration and the target amount of data off the machine, then being sequenced by the Illumina NovaSeq 6000. The end reading of 150 bp pairing is generated. The basic principle of sequencing is to synthesize and sequence at the same time (Sequencing by Synthesis). Four fluorescent labeled dNTP, DNA polymerase and splice primers were added to the sequenced flow cell and amplified. When the sequence cluster extends the complementary chain, each dNTP labeled by fluorescence can release the corresponding fluorescence. The sequencer captures the fluorescence signal and converts the optical signal into the sequencing peak by computer software, to obtain the sequence information of the fragment to be tested.
Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using the DESeq2 R package (1.20.0). DESeq2 provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Padj <0.05 and |log2(foldchange)| >1 were set as the threshold for significantly differential expression.
LC-MS analysis of proteomics
Cells treated with different conditions were incubated with 50 μM Alk14 (13266, Cayman Chemical) for 12 h and then collected and lysed in lysis buffer (100 mM Tris-HCl pH 7.2, 5 mM EDTA, 150 mM NaCl, 2.5% SDS, inhibitor cocktail). After centrifugation at 16,000 × g for 20 min at 4 °C, the supernatant was collected. Protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific, USA). The protein was precipitated with chloroform/methanol/water (v/v 1:4:3), briefly air-dried, and dissolved in 50 μl of lysis buffer. Click chemistry reagents were added to the samples in the following order: 1 μl of 4 mM TAMRA azide (47130, Lumiprobe), 1.2 μl of 10 mM tris[(1-benzyl-1H-1,2,3-triazol-4-yl) methyl]amine (TBTA) (678937, Sigma-Aldrich), 1 μl of 40 mM CuSO4 (451657, Sigma-Aldrich), and 1 μl of 40 mM tris(2-carboxyethyl)phosphine HCl (TCEP hydrochloride) (C4706, Sigma-Aldrich). The reaction mixtures were thoroughly mixed and incubated for 30 min in the dark at room temperature. The protein was precipitated with chloroform/methanol/water (v/v 1:4:3), briefly air-dried, and dissolved in 200 μl of lysis buffer. Two additional rounds of methanol-chloroform-H2O precipitations were performed on the samples as described above, and then dissolved in 100 μl of lysis buffer. The samples were then equally divided into two parts and incubated with 50 μl of 1 M hydroxylamine or a negative control (1 M NaCl) respectively at room temperature for 2 h. The protein was precipitated with chloroform/methanol/water (v/v 1:4:3), briefly air-dried, and dissolved in 200 μl of lysis buffer. Two additional rounds of methanol-chloroform-H2O precipitations were performed on the samples as described above, and then dissolved in 50 μl of lysis buffer. The sample was diluted 1:10 with PBS and incubated with 20 μl of streptavidin beads at 4 °C overnight with shaking. The beads were washed three times with PBS containing 0.2% SDS.
The magnetic beads were resuspended in 400 μl of 25 mM ammonium bicarbonate and then washed five times to remove residual contaminants. Subsequently, the pelleted beads were resuspended in 200 μl of ammonium bicarbonate buffer containing 10 mM dithiothreitol (DTT) and incubated at 55 °C with rotation for 40 min to reduce disulfide bonds. After reaching room temperature, 40 mM iodoacetamide (IAA) was added to the suspension, which was then rotated in the dark for 30 min at room temperature to block free cysteine residues. Following this, 250 ng of sequencing-grade trypsin was added to the supernatant, and the mixture was incubated overnight (16 h) at 37 °C with rotation in the dark to perform digestion. Pierce C18 Spin Columns were employed for peptide desalting, with 200 μl of 50% acetonitrile (ACN) used to rinse the walls of the spin column and wet the resin. Centrifugation at 1500 × g for 1 min was performed to ensure complete washing, followed by discarding the flow-through and repeating the washing process once to fully activate the C18 resin bed. Subsequently, 200 μl of 0.5% trifluoroacetic acid (TFA) in 5% ACN was added, centrifuged at 1500 × g for 1 min, and the flow-through discarded and repeated once to further clean the sample. For every 3 μl of sample, 1 μl of 2% TFA in 20% ACN was added, followed by loading the sample on top of the resin bed, placing the column into a receiver tube, and centrifuging at 1500 × g for 1 min. The flow-through was recovered to ensure complete binding and the steps were repeated twice. Then, 200 μl of 0.5% TFA in 5% ACN was added to the column, centrifuged at 1500 × g for 1 min, and the flow-through discarded and repeated three times. The column was placed in a new receiver tube, and 20 μl of 70% ACN was added to the top of the resin bed. Centrifugation at 1500 × g for 1 min was performed, followed by repeating the step three times to ensure thorough drying of the sample. The combined sample was then re-dissolved in 20 μl of ultrapure water plus 0.1% formic acid, and the peptide concentration was measured using a nanodrop at 205 nm. The sample concentration was adjusted to 250 ng/μl, and after centrifugation at 14,000 × g for 5 min, the supernatant was transferred to a sample vial. The nLC-MS/MS analysis was conducted on an Easy nLC 1200 (Thermo Fisher Scientific, Bremen, Germany) coupled to an Orbitrap Fusion Lumos equipped with a nanospray flex ion source (Thermo Fisher Scientific, Bremen, Germany). The MS raw data for each sample were combined and searched using the Proteome Discoverer 2.5 software (Thermo Fisher Scientific, Bremen, Germany) for identification and quantitation analysis. The detailed mass spectrometry data is shown in Supplementary Data 1.
Acyl-biotin exchange assay
The acyl-biotin exchange (ABE) assays were conducted in accordance with previously established protocols. Cell pellets were homogenized on ice with a lysis buffer consisting of 100 mM Tris-HCl (pH 7.2), 5 mM EDTA, 150 mM NaCl, 2.5% SDS, and an inhibitor cocktail for a duration of 30 min. Following lysis, the samples were centrifuged at 16,000 × g for 20 min to pellet the cellular debris. The protein concentration in the supernatant was quantified using the BCA Protein Assay Kit (Thermo Fisher Scientific, USA). For each sample, 1 mg of protein was mixed with 500 μl of lysis buffer supplemented with 50 mM N-ethylmaleimide (NEM) (E3876, Sigma) and incubated at room temperature for 2 h with gentle agitation. Subsequently, the protein was precipitated with chloroform/methanol/water (1:4:3, v/v), air-dried briefly, and resuspended in 200 μl of lysis buffer. Two additional rounds of methanol-chloroform-H2O precipitations were performed on the samples as previously described, and the precipitated proteins were then solubilized in 1 ml of lysis buffer containing 5 mM biotin-BMCC by gentle mixing at room temperature. The samples were divided equally into two parts and incubated with 0.5 ml of 1 M hydroxylamine or a negative control (1 M NaCl) at room temperature for 3 h. After precipitation, the samples were resuspended in 200 μl of lysis buffer. Each sample was loaded with 20 μl as a loading control, and the remaining 180 μl was diluted 1:10 with PBS and incubated with 20 μl of streptavidin beads at 4 °C overnight with shaking. The beads were washed three times with PBS containing 0.2% SDS. The beads and loading controls were mixed with SDS loading buffer and incubated at 95 °C for 10 min. The samples were then separated by SDS-PAGE and subjected to western blot analysis. The detailed mass spectrometry data is shown in Supplementary Data 2.
Click chemistry assay
Cells with different treatments were incubated with 50 μM Alk14 (13266, Cayman Chemical) for 12 h and then collected and lysed in lysis buffer (100 mM Tris-HCl pH 7.2, 5 mM EDTA, 150 mM NaCl, 2.5% SDS, inhibitor cocktail). The supernatant was collected after centrifugation at 16,000 × g for 20 min at 4 °C. The protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific, USA). The target protein was purified using anti-Flag magnetic beads, and the beads were suspended in 50 μl of IP washing buffer. Click chemistry reagents were added to the beads in the following order: 1 μl of 4 mM TAMRA azide (47130, Lumiprobe), 1.2 μl of 10 mM tris[(1-benzyl-1H-1,2,3-triazol-4-yl) methyl]amine (TBTA) (678937, Sigma-Aldrich), 1 μl of 40 mM CuSO4 (451657, Sigma-Aldrich), and 1 μl of 40 mM tris(2-carboxyethyl)phosphine HCl (TCEP hydrochloride) (C4706, Sigma-Aldrich). The reaction mixtures were thoroughly mixed and incubated for 30 min in the dark at room temperature. Next, 20 μl of 4×SDS loading buffer was added, and the resulting mixture was heated at 95 °C for 10 min. Half of the mixture was also treated with hydroxylamine (438227, Sigma) (pH 7.4, final concentration 500 μM) and heated for another 5 min at 95 °C to remove S-palmitoylation. The samples were then separated by SDS-PAGE, the gel was briefly washed in water, and then scanned to record the rhodamine fluorescence signal using a ChemiDoc MP Imaging system (Bio-Rad). After that, the gel was incubated with destaining buffer (50% CH3OH, 40% water, and 10% acetic acid) by shaking for 2–8 h and then incubated in water to help lower the background. After scanning, the gel was stained with Coomassie Brilliant Blue (B7920, Sigma) to check for protein loading. The detailed mass spectrometry data is shown in Supplementary Data 2.
Acyl-PEG exchange assay
Acyl-PEG exchange assays were performed essentially as previously described. Samples were suspended in lysis buffer (100 mM Tris-HCl pH 7.2, 5 mM EDTA, 150 mM NaCl, 2.5% SDS, inhibitor cocktail). The protein concentration in the supernatant was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific, USA). For each sample, 1 mg of total protein in 92.5 µL of lysis buffer was treated with 5 µL of 200 mM neutralized TCEP (Thermo Fisher Scientific, USA) for final concentration of 10 mM TCEP for 30 min with nutation. NEM (Sigma), 2.5 µL from freshly made 1 M stock in ethanol, was added for a final concentration of 25 mM and incubated for 2 h at room temperature. Protein was precipitated with chloroform/methanol/water (v/v 1:4:3), briefly air-dried, and then dissolved in 100 µL of lysis buffer. To ensure complete removal of NEM from the protein pellets, the samples were subjected to two additional rounds of methanol-chloroform-H2O precipitations, as described above. For hydroxylamine (NH2OH) cleavage and mPEG-maleimide alkylation, the protein pellet was resuspended in 30 µL lysis buffer and treated with 90 µL of 1 M neutralized NH2OH in lysis buffer to obtain a final concentration of 0.75 M NH2OH. Control samples not treated with NH2OH were diluted in 90 µL lysis buffer. Samples were incubated at room temperature for 1 h with nutation. The samples were then subjected to methanol-chloroform-H2O precipitation, as described above, and resuspended in 30 µL lysis buffer, warmed to 37 °C for 10 min, and briefly (∼5 s) sonicated and treated with 90 µL lysis buffer with 0.2% Triton X-100 and 1.33 mM mPEG-Mal (5 kDa; Sigma) for a final concentration of 1 mM mPEG-Mal. Samples were incubated for 2 h at room temperature with nutation before a final methanol-chloroform-H2O precipitation. Dried protein pellets were resuspended in 50 µL 1× Laemmli buffer (Bio-Rad) and then heated for 5 min at 95 °C. The samples were then separated by SDS-PAGE and subjected to western blot analysis.
Lentivirus-delivered shRNA knockdown and gene overexpression
All shRNA and overexpression vectors were purchased from IGE Biotechnology Company (Guangzhou, China). These selected shRNA sequences were cloned into the pLKO.1-puro lentiviral vector. Recombinant lentiviral particles were produced by transfecting the plasmids into 293T cells. In brief, 3.25 μg of psPAX3 and 1.75 μg of pMD2.G plasmids, along with 5 μg of target gene plasmids, were co-transfected into 293T cells using polyethylenimine linear (PEI) from Polysciences (23966, Polysciences, USA). The lentivirus supernatant was collected 48 h after transfection and filtered. After infecting the target cells with 8 μg/ml of polybrene for 24 h, infected cells were selected with 2 μg/ml of puromycin (P8833, Sigma-Aldrich). The target sequences of the genes in this study are listed in Supplementary Table 2.
CRISPR-Cas9-mediated gene knockout
Single guide RNAs (sgRNAs) were designed to target the critical exons of the genes of interest for inactivation. The sgRNAs were cloned into the lentiCRISPR v2-puro vector (Addgene, #98290). Recombinant lentiviral particles were produced by transfecting the plasmids into 293T cells. In brief, 3.25 μg of psPAX2 and 1.75 μg of pMD2.G plasmids were co-transfected with 5 μg of the target gene plasmids into 293T cells using linear polyethyleneimine (PEI) from Polysciences (23966, Polysciences, USA). The lentivirus supernatant was collected 48 h post transfection and filtered. The target cells were infected with 8 μg/ml of polybrene for 24 h, followed by selection of infected cells with 2 μg/ml of puromycin (P8833, Sigma-Aldrich). The target sequences of the genes in this study are listed in Supplementary Table 3.
Immunohistochemistry
IHC staining was conducted using the IHC Detection Kit (ab64264, Abcam, UK) according to the manufacturer’s instructions. In brief, fresh tissue samples were fixed with 4% paraformaldehyde, dehydrated, and then embedded in paraffin. The embedded tissue was sectioned into 5 μm thick slices. The tissue sections were then deparaffinized, rehydrated, boiled in citrate buffer for 20 min to retrieve antigens, permeabilized, and blocked for endogenous peroxidase activity and non-specific antibody binding. After that, the sections were incubated with primary antibodies at 4 °C overnight. After 10 min of incubation with biotinylated secondary antibodies, the positive protein signal was visualized using the DAB Histochemistry Kit. Nuclei were counterstained with hematoxylin (H3136, Merck, Germany). Images were captured at a magnification of 20× using the VS200 Research Slide Scanner (Olympus). For hematoxylin and eosin (H&E) staining, mouse intestinal tissues were dehydrated and embedded in paraffin in the shape of a Swiss roll, with the distal end of the colon located at the innermost segment of the roll. Paraffin sections were cut as described above and stained with hematoxylin and eosin using an automated H&E staining workstation (HistoCore SPECTRA, Leica).
Procedures for chemical synthesis of ABD957
The synthesis of 2-((1R,5S)-8-(2-(((S)-4-(3-(N, N-dimethylsulfamoyl)-1H-pyrazole-1-carbonyl)-3-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)-3,8-diazabicyclo[3.2.1]octan-3-yl) acetic acid (ABD957) was performed as previously described40. The detailed chemical synthesis steps are presented in Supplementary Fig. 14.
Co-immunoprecipitation (Co-IP)
In brief, cells were washed three times with ice-cold phosphate-buffered saline (PBS) and lysed in NP40 lysis buffer containing protease and phosphatase inhibitors (Sigma) on ice for 30 min. The cell lysates were centrifuged at 15,000 × g for 15 min at 4 °C, and 250 μg of the supernatant was incubated with anti-Flag/HA magnetic beads (Thermo Fisher Scientific) for 1 h at room temperature. Then, the protein-bound magnetic beads were collected using a magnetic stand and gently washed with lysis buffer for three times. The bound proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Ubiquitination assays
For the ubiquitination analysis of GPX4 in the indicated cells, the cells were treated with 2-BP for 12 h followed by the addition of 10 μM MG132 (MCE, HY-13259) for an additional 12 h. Subsequently, the cells were harvested in 500 μL of SDS lysis buffer (1% SDS, 50 mM Tris-Cl) and heated at 100 °C for 20 min. Then, 100 μL of the cell lysate was taken as the input sample, and the remaining lysate was diluted tenfold with NP40 lysis buffer. The cell lysates were immunoprecipitated with anti-rabbit IgG (AC005, Abclonal) or anti-GPX4 (14432-1-AP, Proteintech) antibody overnight at 4 °C. On the second day, Protein A/G magnetic beads (MCE, HY-K0202) were added to the cell lysate and incubated at 4 °C for 4 h, followed by immunoblotting analysis with an anti-ubiquitin (CST, #3936T) antibody to detect the ubiquitination of GPX4.
Animal studies
All animals were housed at a suitable temperature (22–24 °C) and humidity (40–70%) under a 12/12-h light/dark cycle with unrestricted access to food and water for the duration of the experiment. The xenograft experiment follows the humane endpoint. Indicators such as huddled posture, immobility, ruffled fur, failure to eat, hypothermia (colonic temperature of <34 °C), or weight loss (>20%) may be useful objective criteria for early euthanasia. The animals will be euthanized immediately if they are unable to stand or if they display agonal breathing, severe muscular atrophy, severe ulceration, or uncontrolled bleeding. The subcutaneous tumor maximum volume was 2000 mm3 and authorized by the Committees on Animal Research and Ethics and was not exceeded at any time during the experiments.
Subcutaneous tumor model
Six- to eight-week-old athymic nude mice were subcutaneously (s.c.) injected into the flank with 2 × 106 A375 cells or 2 × 106 A375-GPX4-KO cells. The tumors were allowed to grow until their volume reached 80 mm³. Mice bearing tumors derived from A375 cells were randomly assigned to receive treatment with IKE (20 mg/kg), 2-BP (20 mg/kg), a combination of IKE (20 mg/kg) and 2-BP (20 mg/kg), and a combination of IKE (20 mg/kg), 2-BP (20 mg/kg), and Lip-1 (10 mg/kg), via intraperitoneal injection once every three days for two weeks. Mice-bearing tumors derived from A375-GPX4-KO cells were randomly assigned to receive treatment with IKE (20 mg/kg), IKE plus 2-BP (20 mg/kg), or IKE plus ML349 (5 mg/kg), respectively, via intraperitoneal injection once every two days for four weeks. Body weight and tumor volume were monitored throughout the entire process. Tumor volume (mm³) was calculated using the formula: π/6 × length (mm) × [width (mm)]2.
Orthotopic colorectal xenograft tumor model
RKO cancer cells were resuspended in PBS at a density of one million per 25 µL and mixed with 25 µL of Matrigel matrix (354234, Corning, USA). Six- to eight-week-old athymic nude mice were anesthetized with a 1.25% tribromoethanol solution (T4840-2, Sigma-Aldrich), and surgery was performed to open the mouse’s abdominal cavity and expose the cecum. A total of 50 µL of the RKO cancer cell mixture was slowly injected into the cecal wall using an insulin needle, and the injection site was inspected to ensure that the mixture did not leak into the cecal lumen or around the cecum. The cecum was then returned to the abdominal cavity, and the peritoneum, abdominal muscle layers, and skin layer of the mouse were sutured closed. Three weeks after the in-situ inoculation, different groups of mice were intraperitoneally injected with 2-BP (20 mg/kg) or a combination of 2-BP and Lip-1 (10 mg/kg) once every three days for another three weeks. Then, the mice were sacrificed, and the large intestine tissues were imaged using an Olympus fluorescence microscope (MVX10, Japan).
AOM/DSS induced spontaneous inflammatory carcinogenesis model
Eight- to twelve-week-old homozygous (Vil1-Cre; APT2flox/flox), heterozygous (Vil1-Cre; APT2flox/+), and wild-type (Vil1-Cre; APT2+/+) transgenic C57BL/6J mice with APT2 gene knockout (KO) were intraperitoneally injected with a dose of 10 mg/kg azoxymethane (AOM) (#A5486, Sigma-Aldrich, USA). One week after the injection of AOM, the normal drinking water in the cages was replaced with 2% dextran sodium sulfate (DSS) (MP biomedical) for one week, followed by two weeks of normal drinking water, which was referred to as one DSS cycle. This process was repeated twice as shown in the timeline in Fig. 7a. The body weight of all mice was monitored regularly throughout the entire process, and the mice were sacrificed at the end of the experiments on day 71. Both the large and small intestines were harvested for imaging, and the number of tumors in the intestines was quantified.
Cancer cell metastasis assay
Six- to eight-week-old athymic nude mice were intraperitoneally injected with 2-BP (20 mg/kg) or a combination of 2-BP (20 mg/kg) and Lip-1 (10 mg/kg) once a day for three consecutive days. Following this, two million A375-EGFP cancer cells were injected through the tail vein. Then, the intraperitoneal injection of 2-BP or 2-BP plus Lip-1 was continued for one week. Three weeks after the tail vein injection, lung tissues were harvested and imaged using an Olympus fluorescence microscope (MVX10, Japan). The green lung colonies formed by metastatic A375-EGFP cells in the left lungs of the different groups of mice were quantified to evaluate the lung metastatic ability of the cancer cells.
Liver ischemia/reperfusion injury (IRI)
Six- to eight-week-old male C57BL/6J wild-type mice were anesthetized with a 1.25% solution of tribromoethanol (T4840-2, Sigma-Aldrich) via intraperitoneal injection. The mice were placed on the surgical table with their abdomens facing up. Following that, the abdomen was cleaned by wiping the skin with a 0.5% iodine solution, and the abdomen was then opened along the midline of the mouse, starting from the median abdominal membrane, and ending at the xiphoid process. The quadrate lobe was separated from its attachment with the left lateral lobe to reveal the portal triad. Next, non-invasive clamps were placed through the portal vein, hepatic artery, and bile duct, above the branches of the right lobe, for a duration of 45 min. When the middle lobe and the left lobe appeared whitened, the abdomen was covered with moistened (sterile 0.9% saline) gauze, and the mouse was then transferred to a heating pad to maintain a body temperature of 37 °C. The clamps were removed, and the abdomen was closed by suturing the muscle layer and skin. Blood samples and liver lobes were collected after a 24 h reperfusion period. According to the grouping requirements, mice were intraperitoneally injected with 2-BP (20 mg/kg), ML-349 (5 mg/kg), or Lip-1 (10 mg/kg) 1 h before ischemia.
Statistics and reproducibility
All statistical data with error bars are presented as the mean ± standard deviation (SD), and all experiments were independently repeated at least three times or more than six mice. Statistical significance between groups was determined by Student’s t-test, one-way analysis of variance (ANOVA), or two-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, and **** p < 0.0001 were considered statistically significant, while ns was considered not statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are included within the article, Supplementary Information, the Source Data file, and the protein mass spectrometry raw data are available through the ProteomeXchange Consortium via the PRIDE partner repository with dataset identifiers PXD051426 and PXD051448. The RNA-Seq data generated in this study have been deposited in the Genome Sequence Archive (GSA) database under accession code HRA007165. The correlation between ZDHHC20 gene expression and ferroptosis sensitivity were derived from the CTRP database (https://portals.broadinstitute.org/ctrp.v2.1/). Source data are provided with this paper.
References
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Yang, W. S. & Stockwell, B. R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 26, 165–176 (2016).
Conrad, M. & Pratt, D. A. The chemical basis of ferroptosis. Nat. Chem. Biol. 15, 1137–1147 (2019).
Yang, W. S. et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014).
Doll, S. et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019).
Bersuker, K. et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019).
Kraft, V. A. et al. GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 6, 41–53 (2019).
Mao, C. et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 593, 586–590 (2021).
Liang, D. et al. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell 186, 2748–2764 (2023).
Jiang, X., Stockwell, B. R. & Conrad, M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282 (2021).
Stockwell, B. R. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell 185, 2401–2421 (2022).
Ursini, F., Maiorino, M., Valente, M., Ferri, L. & Gregolin, C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim. Biophys Acta (BBA) 710, 197–211 (1982).
Ursini, F., Maiorino, M. & Gregolin, C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim. Biophys. Acta 839, 62–70 (1985).
Kühn, H. & Borchert, A. Regulation of enzymatic lipid peroxidation: the interplay of peroxidizing and peroxide reducing enzymes. Free Radic. Biol. Med. 33, 154–172 (2002).
Hangauer, M. J. et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017).
Viswanathan, V. S. et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017).
Mayr, L. et al. Dietary lipids fuel GPX4-restricted enteritis resembling Crohn’s disease. Nat. Commun. 11, 1775 (2020).
Smith, A. C. et al. Mutations in the enzyme glutathione peroxidase 4 cause Sedaghatian-type spondylometaphyseal dysplasia. J. Med. Genet. 51, 470–474 (2014).
Fedida, A. et al. Sedaghatian-type spondylometaphyseal dysplasia: whole exome sequencing in neonatal dry blood spots enabled identification of a novel variant in GPX4. Eur. J. Med. Genet. 63, 104020 (2020).
Seiler, A. et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent-and AIF-mediated cell death. Cell Metab. 8, 237–248 (2008).
Friedmann Angeli, J. P. et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014).
Dong, K. et al. HOIP modulates the stability of GPx4 by linear ubiquitination. Proc. Natl Acad. Sci. 119, e2214227119 (2022).
Wu, K. et al. Creatine kinase B suppresses ferroptosis by phosphorylating GPX4 through a moonlighting function. Nat. Cell Biol. 25, 714–725 (2023).
Kerins, M. J., Milligan, J., Wohlschlegel, J. A. & Ooi, A. Fumarate hydratase inactivation in hereditary leiomyomatosis and renal cell cancer is synthetic lethal with ferroptosis induction. Cancer Sci. 109, 2757–2766 (2018).
Linder, M. E. & Deschenes, R. J. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 (2007).
Blaskovic, S., Adibekian, A., Blanc, M. & van der Goot, G. F. Mechanistic effects of protein palmitoylation and the cellular consequences thereof. Chem. Phys. Lipids 180, 44–52 (2014).
Ko, P. J. & Dixon, S. J. Protein palmitoylation and cancer. EMBO Rep. 19, e46666 (2018).
Rocks, O. et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 (2005).
Chen, S. et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature 549, 399–403 (2017).
Chen, S. et al. Targeting MC1R depalmitoylation to prevent melanomagenesis in redheads. Nat. Commun. 10, 877 (2019).
Sun, Y. et al. AMPK phosphorylates ZDHHC13 to increase MC1R activity and suppress melanomagenesis. Cancer Res. 83, 1062–1073 (2023).
Zhang, M. et al. A STAT3 palmitoylation cycle promotes TH17 differentiation and colitis. Nature 586, 434–439 (2020).
Coleman, R. A., Rao, P., Fogelsong, R. J. & Bardes, E. S. 2-Bromopalmitoyl-CoA and 2-bromopalmitate: promiscuous inhibitors of membrane-bound enzymes. Biochim. Biophys Acta 1125, 203–209 (1992).
Webb, Y., Hermida-Matsumoto, L. & Resh, M. D. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 275, 261–270 (2000).
Fukata, M., Fukata, Y., Adesnik, H., Nicoll, R. A. & Bredt, D. S. Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 (2004).
Zhong, H. & Yin, H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 4, 193–199 (2015).
Chase, J. & Tubbs, P. Specific inhibition of mitochondrial fatty acid oxidation by 2-bromopalmitate and its co-enzyme A and carnitine esters. Biochem. J. 129, 55–65 (1972).
Kagan, V. E. et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90 (2017).
Liu, H. et al. Small-molecule allosteric inhibitors of GPX4. Cell Chem. Biol. 29, 1680–1693.e1689 (2022).
Remsberg, J. R. et al. ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth. Nat. Chem. Biol. 17, 856–864 (2021).
Chaudhary, N. et al. Lipocalin 2 expression promotes tumor progression and therapy resistance by inhibiting ferroptosis in colorectal cancer. Int. J. Cancer 149, 1495–1511 (2021).
Cui, W. et al. Gut microbial metabolite facilitates colorectal cancer development via ferroptosis inhibition. Nat. Cell Biol. 26, 124–137 (2024).
Ubellacker, J. M. et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature 585, 113–118 (2020).
Liu, H. et al. Characterization of a patient-derived variant of GPX4 for precision therapy. Nat. Chem. Biol. 18, 91–100 (2022).
Shi, Z. et al. Loss of LncRNA DUXAP8 synergistically enhanced sorafenib induced ferroptosis in hepatocellular carcinoma via SLC7A11 de‐palmitoylation. Clin. Transl. Med. 13, e1300 (2023).
Wang, Z. et al. AMPKα1-mediated ZDHHC8 phosphorylation promotes the palmitoylation of SLC7A11 to facilitate ferroptosis resistance in glioblastoma. Cancer Lett. 584, 216619 (2024).
Feng, H. & Stockwell, B. R. Unsolved mysteries: how does lipid peroxidation cause ferroptosis? PLoS Biol. 16, e2006203 (2018).
Stockwell, B. R. et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 (2017).
Acknowledgements
This work was supported by National Key R&D Program of China (2022YFA0912600), Shenzhen Medical Research Fund (B2302018), National Natural Science Foundation of China (32100579 and 82341011), Major Program (S201101004) and Open Fund Program (SZBL2021080601004) of Shenzhen Bay Laboratory. The authors are grateful to Biochemical Analysis Core, Multi-omics Mass Spectrometry Core, Bio-Imaging Core, and Animal Laboratory Center of Shenzhen Bay Laboratory for help with technical assistance.
Author information
Authors and Affiliations
Contributions
B.H. and C.Y. provided conceptualization and wrote the original draft. B.H., H.W., S.L., M.H., D.L., Y.Z., Y.H., and Y.N. performed the experiments and analyzed the data. L.Z., C.B., and C.Y. reviewed and edited the paper. C.Y. provided supervision and resources and performed funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Eun-Woo Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, B., Wang, H., Liu, S. et al. Palmitoylation-dependent regulation of GPX4 suppresses ferroptosis. Nat Commun 16, 867 (2025). https://doi.org/10.1038/s41467-025-56344-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56344-5
This article is cited by
-
LRP8 Promotes colorectal cancer progression by suppressing ferroptosis through the SLC3A2/GPX4 signalling axis
European Journal of Medical Research (2026)
-
Rottlerin triggers dual degradation of SLC7A11 and GPX4 to drive ferroptosis and chemosensitization in hepatocellular carcinoma
Cell Death Discovery (2026)
-
Hyperoside Inhibits Doxorubicin-Induced Ferroptosis in Cardiomyocytes via the Nrf2/GPX4 Pathway
Journal of Cardiovascular Translational Research (2026)
-
Molecular mechanisms and therapeutic strategies of GPX4 regulation in acute kidney injury
Pharmacological Reports (2026)
-
Regulatory Landscapes of Protein Acylations in Neuroinflammation: From Molecular Mechanisms to Therapeutic Targets
Neurochemical Research (2026)