Abstract
While all native tRNAs undergo extensive post-transcriptional modifications as a mechanism to regulate gene expression, mapping these modifications remains challenging. The critical barrier is the difficulty of readthrough of modifications by reverse transcriptases (RTs). Here we use Induro—a new group-II intron-encoded RT—to map and quantify genome-wide tRNA modifications in Induro-tRNAseq. We show that Induro progressively increases readthrough over time by selectively overcoming RT stops without altering the misincorporation frequency. In a parallel analysis of Induro vs. a related RT, we provide comparative datasets to facilitate the prediction of each modification. We assess tRNA modifications across five human cell lines and three mouse tissues and show that, while the landscape of modifications is highly variable throughout the tRNA sequence framework, it is stabilized for modifications that are required for reading of the genetic code. The coordinated changes have fundamental importance for development of tRNA modifications in protein homeostasis.
Similar content being viewed by others
Introduction
All native tRNA molecules contain post-transcriptional modifications at 10-20% of the nucleobases across the three domains of life1. Many of these modifications occur in the anticodon stem-loop (ASL) with a role to modulate the quality of anticodon-codon base-pairings, while others are in the body with a role to stabilize the tRNA L-shaped tertiary structure. Whether these two groups of modifications coordinate with each other in the development of a tRNA is unknown. Importantly, tRNA modifications can be dynamic, changing abundance in response to stress and disease2,3,4,5,6,7,8, thus fine-tuning the process of protein synthesis and providing a mechanism to regulate gene expression. However, mapping and quantifying changes in tRNA modifications remains difficult, as many modifications interfere with the end-to-end cDNA synthesis of an RT for Illumina sequencing, whose high-throughput and high-capacity platform is a strength over other platforms9,10. Indeed, due to the difficulty to read through modifications that cluster in high density in the compact tRNA structure, most published genome-wide tRNAseq datasets provide little information on tRNA modifications11,12,13,14,15,16,17,18,19,20,21.
The central component of a successful tRNAseq using the Illumina platform is a processive and efficient RT that can read through as many modifications as possible. The traditional RTs are represented by SuperScript III and SuperScript IV, which are recombinant variants of the native RT of Moloney Murine Leukemia virus with improved stability and processivity22. A newer generation of RTs are encoded by group-II introns that express ribozymes, which are self-spliced from precursor RNAs as a mobile element to invade new genomes. As intron-encoded RTs must efficiently copy the intron-released ribozymes, which are large and highly structured, they are typically highly processive, with a stronger strand displacement activity23 than viral RTs24. However, only two group-II intron RTs are in use for tRNAseq (i.e., TGIRT25,26 in mim-tRNAseq18,27 and Marathon28,29 in tRNA structure-seq20 and in ALL-tRNAseq19), emphasizing the need for new discoveries.
Each RT has an intrinsic “signature” of nucleotide incorporation in response to a modification—to stop, to mis-incorporate, or to jump over. This signature can vary depending on the chemical nature and the structural context of the modification. For example, while N1-methylation of guanosine (m1G) interferes with Watson-Crick (W-C) pairing, it can occur at position 9, where the tRNA acceptor stem turns from the D stem to stack on the T stem, or at position 37 on the 3’-side of the anticodon, or at both. Similarly, while the m1A methylation interferes with W-C pairing, it predominantly occurs at both positions 9 and 58 of tRNA, each with a distinct local structural environment that can confer a distinct RT signature. Thus, the mechanism that determines the RT signature at each modification can change with the local structure and with the reaction condition. Even if the reaction condition of an RT is provided by a commercial source, this does not mean that it is the best condition to read through tRNA modifications. However, the RT signature at each modification is poorly defined in current tRNAseq’s. Addressing this unmet need will permit a more reliable prediction of modifications, annotated or non-annotated, and a stronger basis to evaluate changes in modifications across tissues and cell types.
Here we address this unmet need by establishing Induro-tRNAseq, using the recently developed and commercially available Induro (NEB M0681) as the RT. Induro is a new member of group-II intron RTs and has shown higher processivity than other RTs30. We show that the readthrough of Induro is both temperature- and time-dependent, and that the driver to maximum readthrough is selective overcoming of RT stops, without altering the misincorporation frequency. This mechanistic insight, not yet available for other RTs, enables users to choose and modify the condition at hand. We establish the readthrough profiles of Induro at the tRNA modifications that occur in the W-C face, which can change RT signatures and are thus “RT-readable”. Comparison of the Induro’s signature at each RT-readable modification to that of the related group-II-intron RT TGIRT18 provides two datasets for the same modification, which will strengthen the power of prediction of each modification. Using Induro, we have compared profiles of tRNA modifications across 5 human cell lines and across 3 mouse tissues. We observed large variations throughout the L-shaped sequence framework among isodecoders (tRNAs sharing the same anticodon and thus decoding the same codon) and isoacceptors (tRNAs sharing the same amino-acid identity while reading different codons), but a stabilized landscape of modifications for those that are responsible for decoding of the genetic code. This observation, previously unrecognized, suggests a coordinated mechanism across tissues and cell lines to develop tRNA modifications to ensure protein homeostasis.
Results
A multiplexing Induro-tRNAseq workflow
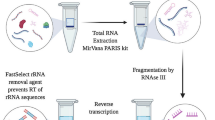
In our development of Induro-tRNAseq, to reduce the number of experimental steps, we used total RNA from cultured human cells as input, rather than gel- or beads-purified tRNA. To determine the tRNA charging level of each input, we performed a periodate oxidation of the terminal 2’,3’-hydroxyls of uncharged tRNAs and removed the terminal A76 nucleotide by an alkaline β-elimination reaction, generating a CC-end21. In contrast, charged aminoacyl-tRNA (aa)-tRNAs were resistant to the oxidation and retained the normal CCA-end after deacylation in alkaline (Fig. 1a, step 1). The CC- and CCA-ending tRNAs, after repair of the 3’-end by T4 PNK, were each joined with a barcoded 3’-adapter via a splint ligation (step 2, Supplementary Tables 1, 2). The ligation efficiency was high, typically reaching >75% (quantification in Methods) and generating ligation products in the predicted size of ~120 nucleotides (Fig. 1b). Several pools of tRNA, each with a specific barcode, were combined at this step to start a multiplexing workflow. The Induro RT reaction was then initiated with a primer (step 3) common to the barcoded adapters, generating a range of cDNA products (Fig. 1c), indicating the presence of modifications that inhibited end-to-end cDNA synthesis. To remove primer molecules that were extended without tRNA, cDNA products of 90-180 nucleotides were gel-isolated (step 4, Fig. 1c) and circularized (step 5). We next used the high-fidelity Q5 DNA polymerase for PCR amplification of each circular cDNA to add Illumina adapters (step 6, Supplementary Table 3). The extended cDNA libraries, after 7-9 cycles of amplification (Fig. 1d), were isolated from a native PAGE and sequenced on an Illumina NextSeq 500. A side-by-side comparison of the workflow, using total RNA vs. purified tRNA as input, showed similar performance throughout, most notably in the final step of generating PCR-amplified cDNA libraries (Fig. 1d). Thus, as starting with total RNA was simpler, it was used in the Induro workflow.
a Preparing tRNAseq library. Step 1, total RNA including charged and uncharged tRNAs, where a black ball depicts the amino acid, is extracted from cells or tissues and used directly unless specified. The A76 of uncharged tRNA is removed by β-elimination, while aa-tRNAs are unaffected and are deacylated in a buffer, pH 9.5. Step 2, a 3’-barcoded adapter is ligated to the 3’-end of deacylated tRNAs by T4 Rnl2 via a DNA splint (turquoise). Several tRNA libraries are combined to start a multiplex workflow. Step 3, an RT primer is hybridized to the 3’-adapter to initiate cDNA synthesis by Induro. Step 4, cDNA products are gel purified and circularized by Circligase. Step 5, PCR amplification of the circularized cDNA by Q5 DNA polymerase with a barcode 2-containing primer and a universal primer (purple). Step 6, the final gel-purified barcoded library contains tRNA sequence (black), barcode 1 (red), and barcode 2 (blue). b A denaturing gel of tRNA ligated with the 3’-adapter after step 2. L: 62-mer and 121-mer DNA ladders. c A denaturing gel of cDNA products after reverse transcription by Induro in step 3. cDNA products of 90–180 nts were excised and extracted from the gel. L: small range RNA ladder (NEB); none: a control RT reaction without input RNA; FL: cDNA of full-length tRNA. d A non-denaturing gel image of the DNA library after PCR amplification in step 5. Double-stranded DNAs (dsDNAs) of 150–240 base pairs (bps) were excised and extracted. L: O’RangeRuler 10 bp DNA ladder + GeneRuler 50 bp DNA ladder (Thermo Fisher). All gels were stained with SYBR gold. b–d Each reaction step was analyzed by gels at least 5 times, showing a representative gel, while the uncropped gel scan is in Source Data 1.
We performed Induro-tRNAseq on 5 human cell lines (K562, HEK293T, HeLa, SH-SY5Y, and HAP1), generating 2-7 million total raw reads of each. The trimmed and processed reads were aligned to the high-confidence set of human tRNA sequences annotated in gtRNAdb of the hg38 reference genome31, where we allowed mismatches up to 10% of each sequence. In all cell lines, >75% of total reads were uniquely mapped to gtRNAdb sequences, similar between samples starting with total RNA or with purified tRNA (Supplementary Fig. 1a). This high frequency of mapping indicates high quality of the workflow and lack of contaminating sequences or inappropriate alignment. Histograms of mapping quality (MAPQ) scores using total RNA as input showed high confidence reads ranging in 36–40 (Supplementary Fig. 1b). Reproducibility between biological replicates was high, with the coefficient of determination r2 for each replicate = 0.9999.
Validating Induro-tRNAseq
We evaluated the quality of the Induro workflow by assessing the integrity of the conserved tRNA 3’-end CCA sequence, where the terminal A76 is the universal site for aminoacylation. This sequence is protruded from the tRNA structure and is most sensitive to degradation during the workflow. We showed that the fraction of incomplete CCA in each isoacceptor family, including 3’-N, 3’-NC, and 3’-NCC, where N is the discriminator base at position 73, among all 3’-end sequences, was low (6.2% for total RNA of K562 and 6.8% for total RNA of HEK293T cells, Supplementary Fig. 2a), indicating an overall high quality of tRNA throughout the workflow.
As Induro is a new RT for tRNAseq, we evaluated its quality and robustness vs. TGIRT in mim-tRNAseq18, using cDNA libraries of K562 and HEK293T cells prepared in the respective workflow. As both RTs are expressed from group-II introns, which are generally thermostable, we assayed Induro at 42 °C, the same temperature used for TGIRT18. Based on similar numbers of detectable tRNA species, we showed that the two enzymes produced similar abundance profiles of cytosolic (cyto)-tRNAs and mitochondrial (mt)-tRNAs (Fig. 2a). The only exception was that the TGIRT workflow under-detected mt-Ser(GCT) by 423-fold in K562 cells and by 11-fold in HEK293T cells (Fig. 2a, Supplementary Fig. 2b). Notably, mt-Ser(GCT) is unique among all human tRNAs as having the shortest sequence (62 nucleotides), lacking the entire D stem-loop32 (Fig. 2a). The low detection of this tRNA in the TGIRT workflow was likely due to the input, which was a gel-purified tRNA pool in the range of 60-100 nucleotides18. Indeed, using the Induro workflow, analysis of total RNA vs. purified tRNA as input showed similar abundance profiles between the two, but under-detected mt-Ser(GCT) in the purified pool by 13-fold in K562 (Fig. 2a) and by 38-fold in HEK293T (Supplementary Fig. 2c). Thus, the Induro workflow starting with total RNA can detect a wider range of tRNA sizes. Indeed, as mt-tRNAs vary more broadly in size relative to cyto-tRNAs, the Induro workflow detected more of these species relative to the TGIRT workflow in both HEK293T and K562 cells (Supplementary Fig. 2d).
a Differential abundance of isodecoders of cyto-tRNAs and mt-tRNAs of K562 cells as identified by Induro-tRNAseq (n = 2, biological replicates) and by mim-tRNAseq (n = 2). The Induro workflow detected 266 cyto-tRNAs and the TGIRT workflow detected 265 cyto-tRNAs, while both detected all 22 isodecoders of mt-tRNAs. Shown in each comparison are log-transformed read counts normalized by DESeq2, showing the Pearson correlation coefficient r. b Differential abundance of isodecoders of cyto-tRNAs and mt-tRNAs starting with total RNA vs. total tRNA from K562 cells by Induro-tRNAseq (n = 2, biological replicates). The dataset consisted of cyto-tRNAs (266, 281 in two duplicates) and mt-tRNAs (22 in each sample). c Differential changes of abundance of isoacceptors of cyto-tRNAs and mt-tRNAs of HEK293T cells in sodium arsenite (SA)-induced oxidative stress as detected by the Induro workflow. The abundance of each isoacceptor in the presence of 100 µM or 300 µM of SA relative to the absence of SA was calculated (n = 4, two biological replicates and two technical replicates). d Bar graphs represent the abundance of mt-Met(CAT) and iMet(CAT) (n = 4) of data in (c). Individual data points are indicated. Error bars, mean ± 95% confidence interval. e Differential changes of tRNA charging levels of HEK293T cells in SA-induced oxidative stress as detected by the Induro workflow. The charging level of each isoacceptor was the percentage of the charged reads with 3’-CCA end relative to the total reads of the isoacceptor. X-axis and Y-axis indicate the difference in charging at 100 µM and 300 µM of SA (n = 4, two biological replicates and two technical replicates). f Bar graphs showing the percentage of charged reads of mt-Met(CAT) and iMet(CAT) (n = 4) of data in (e). Individual data points are indicated. Error bars, mean ± 95% confidence interval.
We compared the Induro workflow with the MSR-seq workflow21, which uses Superscript IV for multiplexing of small RNAs to assess stress-induced tRNA response. We analyzed HEK293T cells, treated with sodium arsenite (SA) – an inducer of oxidative stress33 – at three concentrations (0, 100, and 300 μM) with 0 μM as the untreated control. As both treated and untreated samples produced uniquely mapped reads at >70% (Supplementary Fig. 2e), we compared tRNA in each stress condition with the unstressed control at the isoacceptor level by analyzing pooled reads that shared the same anticodon. Analysis of tRNA abundance showed no obvious difference between isoacceptors and between stress conditions (Fig. 2c, d), consistent with data of MSR-seq21. We then calculated the tRNA charging level for each anticodon family, by taking the sum of charged isodecoder reads (CCA-ending) over the sum of both charged and uncharged reads (NCC-, NC-, and N-ending). This analysis identified two notable changes, one of which was a decrease of 1.12-fold in charging of iMet(CAT) for initiation of cytosolic protein synthesis (Fig.2e, f), also consistent with data of MSR-seq21, thus validating the quality of the Induro workflow. The other change was a more pronounced decrease of 1.24-fold in charging of mt-Met(CAT) for initiation of mitochondrial protein synthesis (Fig. 2e, f), indicating a higher sensitivity of the mitochondrial machinery to the arsenite stress than its cytosolic counterpart. This insight, previously unknown, would be an avenue for future studies.
Profiling Induro for readthrough of tRNA modifications
Having validated the Induro workflow for reproducing results of established tRNAseq’s, we profiled the enzyme for its readthrough of RT-readable modifications. In the canonical tRNA sequence framework, these modifications are m1A9/58, m1G9/37, acp3U20 (acp3U = 3-amino-3-carboxy propyl-uridine at position 20), m22G26/27 (m22G = N2, N2 - dimethyl guanosine at position 26 or 27), m3C32 (m3C = N3-methyl cytidine at position 32), I34 (I = inosine at position 34), m1I37, yW37 (yW = wybutosine at position 37), ms2i6A37 and ms2t6A37 (ms2i6A = 2-methylthio-N6-isopentenyl-adenosine and ms2t6A = 2-methylthio-N6-threonyl-carbamoyl-adenosine, at position 37) (Fig. 3a). For each isodecoder, we defined the readthrough frequency as the end-to-end reads of both correct and incorrect incorporation over the total mapped reads. While modifications that occur outside of the W-C face (e.g., ψ (pseudouridine), D (dihydrouridine), m5C, τm5U (5-taurine-methyl-uridine), and T (ribo-thymine)) are not RT-readable in assays with Mg2+, we explored their readability in assays with Mn2+ (see below). We did not consider chemical treatment of these latter modifications, due to the potential of chemical-induced biases.
a The L-shaped tRNA structure showing RT-readable modifications. b Yield of readthrough (%) in end-to-end cDNA synthesis at 25, 37, 42, and 55 °C after the Induro RT reaction for 1, 2, or 16 h. Data were collected from total RNA of K562 cells as the input (n = 2; technical replicates). Center line: median; box limits: upper and lower quartiles; whiskers: 1.5X interquartile range; points: outliers. N.D.; Not detectable. **p < 0.01; two-tailed unpaired t-test. c Heatmaps of the frequency (%) of the RT misincorporations and RT stops at the annotated modifications. d Left: A scatter plot showing the frequency (%) of the RT misincorporation as a function of the G9:m1G9 ratio of the transcript of human mt-Leu(TAA) (n = 2; technical replicates). Right: The frequency of misincorporation of the native mt-Leu(TAA) in sequencing analysis of a total RNA sample. Data were collected from K562 cells with total RNA as the input under the RT reaction at 42 °C for 16 h (n = 2; technical replicates). Source data containing p values and n values are provided in Source Data 2.
We began by testing the temperature and time of the Induro reaction to identify the condition that maximized readthrough, using K526 as a model. While Induro is recommended to perform at 55 °C (NEB M0681), we also tested 42, 37, and 25 °C to determine whether lowering the temperature would reduce enzyme dynamics at each modification to promote readthrough. Additionally, while Induro is capable of readthrough of a 12 kb RNA in less than 10 min at 55 °C (NEB M0681), we tested its readthrough of tRNA by monitoring at 1, 2, and 16 h. Unexpectedly, at 55 °C, the frequency of readthrough was low (10%) and did not improve over time. In contrast, the frequency of readthrough increased over 16 h to 60% at 42 °C, to 90% at 37 °C, and to 40% at 25 °C, but was not detectable at 25 or 37 °C in 1–2 h (Fig. 3b). Thus, the Induro readthrough is both temperature- and time-dependent, showing the highest readthrough at 37 °C overnight. Notably, the lack of Induro readthrough at non-optimal conditions was not due to reduced read counts, as all conditions produced uniquely mapped reads at >70% of total reads (Supplementary Fig. 3a).
The temperature- and time-dependent readthrough of tRNA at the global level was also shown for cyto-tRNAs and mt-tRNAs when each was analyzed separately (Supplementary Fig. 3b), indicating that it is an intrinsic feature of Induro. The specific increase in readthrough at 42, 37, and 25 °C, but not at 55 °C, supports the notion of reduced dynamics of the RT at lower temperatures that prevented the enzyme from falling off the tRNA template. Notably, while readthrough increased over time at 42, 37, and 25 °C, the average misincorporation rate through all tRNA sequences remained constant at ~3% (Supplementary Fig. 3c), indicating that the increase in readthrough was driven by decreases of RT stops.
We determined whether the decreases of RT stops were uniform at all modifications or were site-specific. We developed a comprehensive map of the interplay between RT stop and RT misincorporation at each readable modification (Fig. 3c). At each of these modifications, defined as position 0, we only found RT stops or RT misincorporation from –1 to +1, despite an exhaustive search from positions –2 to +2, indicating no RT jump-over (Supplementary Fig. 3d). We also confirmed that, relative to each position 0, there was no adjacent modification that would have caused RT stops, allowing precise assignment of RT stops. The map showed a diverse interplay between RT stops and RT misincorporations across all readable modifications, indicating the distinct response of Induro to each modification. At most RT-readable modifications, Induro responded primarily by misincorporation at position 0, with a low level of RT stop (Supplementary Fig. 3d). Notably, at I34, the enzyme responded exclusively by misincorporation without RT stop, whereas at two other modifications—acp3U20a and ms2i6A37/ms2t6A37—the enzyme responded primarily by RT stops, although both at the +1 position after the site of the modification. Despite these, the overall preference of the enzyme for misincorporations relative to RT stops across most modifications indicates its high propensity of readthrough. Notably, we showed that Induro read through m3C32 (Fig. 3c), whereas many tRNAseq methods require a chemical treatment of the modified base for detection34. Additionally, at 42 and 55 °C, where time-dependent data were available, we observed specifically at 42 °C that the frequency of RT stops indeed decreased over time at all modifications, while that of misincorporation remained constant (Fig. 3c). These results provide insight into global analysis of tRNA (Fig. 3b), showing that the time-dependent decreases of RT stops across all modifications were the basis for increases of the RT readthrough.
While maximum readthrough of Induro occurred at 37 °C overnight (Fig. 3b), we chose 42 °C overnight for the workflow. One reason was that the information at 37 °C was limited to a single timepoint at 16 h, whereas a complete dataset was available at 42 °C across three timepoints for RT misincorporations vs. RT stops (Fig. 3c), which would facilitate users to decide the condition for a specific reaction. Additionally, analysis of all potential modifications that were identified by RT misincorporations or RT stops, annotated or non-annotated, showed the detection of approximately 30 more modifications at 42 than at 37 °C (Supplementary Fig. 3e), indicating a broader scope of detection of modifications at 42 °C.
To determine if the Induro workflow was quantitative in detecting a readable modification, we noted the 100% frequency of RT misincorporation at I34 and the near 100% frequency at m1I37 and yW37 (Supplementary Fig. 3d), indicating that each is homogeneously modified. Thus, a fractional reduction of RT misincorporation at each of these would represent a fractional loss of the modification. For other modifications, as the stoichiometry cannot be determined solely by the frequency of RT misincorporation, we performed a calibration analysis using m1G9 in the native mt-Leu(TAA)32 as an example, where TAA is the gene sequence for the anticodon. We prepared a transcript of the tRNA, lacking any modification, and a separate transcript containing m1G9 at 100% as the single modification. Sequencing analysis of the two transcripts, mixed at different ratios, revealed a linear correlation between the frequency of misincorporation and the stoichiometry of m1G9, with the Pearson r of 0.99 (Fig. 3d), thus demonstrating the quantitative precision of the workflow. However, we found that the RT misincorporation rate at saturating m1G9 reached 60% with the transcript tRNA but reached 80% with the native tRNA. Notably, the native mt-Leu(TAA) has m2G10 following m1G9 in an LC-MS (liquid chromatography-mass spectrometry) analysis32, which is absent from the transcript tRNA, suggesting that Induro was sensitive to the local environment of each modification.
Induro sensitive to chemical and structural environments of a modification
To test if Induro was sensitive to the environment of a modification, we developed a high-resolution map for m1A and m1G, each occurring in at least two positions of the tRNA tertiary structure. While other modifications also occur at multiple positions, their frequency was much lower (e.g., m22G26/27). Analysis of m1A9 vs. m1A58, both important for tRNA biology4,35, showed that while readthrough at m1A9 decreased with temperature, that of m1A58 was steady throughout the duration of time (Fig. 4a). The decrease of readthrough of m1A9 with increasing temperature, but not of m1A58, suggests that m1A9 is in a stronger structural barrier relative to m1A58, such that the RT would fall off the tRNA template with increasing dynamics at higher temperature. Indeed, position 9 is in a structure that is stabilized by the conserved U8-A14 base pair within a network of many tertiary interactions, whereas position 58 is in the T loop with fewer tertiary interactions (Fig. 3a). In contrast, analysis of m1G9 vs. m1G37, both also important for tRNA biology36,37,38,39,40,41,42, showed that while readthrough at m1G9 was mildly decreased with increasing temperature and time, that at m1G37 increased robustly, except for the lack of a time effect at 55 °C (Fig. 4b). The temperature-dependent increase of readthrough at m1G37, localized on the 3’-side of the anticodon, indicates a decrease of RT stops, possibly due to the temperature-assisted unstacking of the modified base from the anticodon to facilitate RT readthrough. These diverse responses to both temperature and time demonstrate the sensitivity of Induro to the chemical and structural environments of each modification.
a The frequency (%) of readthrough at m1A9 and m1A58. Data were collected from total RNA of K562 cells (n = 2; technical replicates for individual RT conditions). b The frequency (%) of readthrough at m1G9 and m1G37. Data were collected from total RNA of K562 cells (n = 2; technical replicates for individual RT conditions). c Top chart: the frequency (%) of RT misincorporation. Middle chart: the frequence (%) of RT stop in the presence of Mg2+ or Mn2+ as the only divalent metal ion. Data were collected from total RNA of K562 cells (n = 2; technical replicates for individual RT conditions). Unmodified natural nucleotides at tRNA positions 70, 74, and 75 were used as the control. A modification is identified if misincorporation is detected over 10%, which is marked by the red lines. Bottom chart: the number of detectable modifications in the presence of Mg2+ or Mn2+. a–c Center line: median; box limits: upper and lower quartiles; whiskers: 1.5X interquartile range; points: outliers. *p < 0.05, **p < 0.01, ***p < 0.001; two-tailed unpaired t-test. All p values and n values used for statistical analysis are available in Source Data 4–6. d, e The frequency (%) of RT misincorporation in the presence of Mg2+ vs. Mn2+ at m7G (d) and at Ψ (e), showing the Pearson correlation coefficient r.
We determined whether Induro responded to the divalent metal ion Mg2+ vs. Mn2+ in the RT reaction. Nucleic-acid polymerases generally display decreased fidelity with Mn2+ relative to Mg2+ as the divalent metal ion43,44, indicating the possibility to use Mn2+ to increase the misincorporation frequency to increase readthrough. Indeed, the related group-II intron RT Marathon was recently shown45 to detect modifications in rRNAs with Mn2+ at sites that were not detectable with Mg2+. In a titration analysis of Induro with Mn2+ as the only divalent metal ion in the RT reaction, we showed that the highest activity was achieved with 1 mM (Supplementary Figs. 4a–c). Additionally, we showed that the workflow with Mn2+ as the only divalent metal ion generated >80% uniquely mapped reads, producing quantification of the abundance of each tRNA similar to that with Mg2+ (Supplementary Figs. 4d, e).
Using unmodified natural nucleotides at tRNA positions 70, 74, and 75 as the control, and using RT misincorporations or RT stops at >10% as the proxy for a modification, we showed that the major improvement of detection with Mn2+ relative to Mg2+ was at ms2i6A37/ms2t6A37, specifically by increasing the frequency of misincorporation while decreasing RT stops (Fig. 4c). Nonetheless, we were unable to detect non-W-C face modifications, such as ψ or m7G. However, with a cut-off of misincorporation at a frequency of 4–12%, we detected m7G, but not ψ (Fig. 4d, e), although the recent study with Marathon was able to detect both at a 0.1–1% frequency of misincorporation45. Overall, with Mn2+ as the divalent metal ion, the readthrough of the Induro workflow improved specifically at ms2i6A37/ms2t6A37, the most dominant barrier to the RT, and additionally the sensitivity of detection improved for m7G using a lower threshold of misincorporation. Unless otherwise stated, we maintained Mg2+ in the workflow, due to the substantial data that we have collected with this divalent metal ion.
Two datasets of tRNA modifications using Induro and TGIRT readouts
While tRNA modifications are present across the tree of life, systematic analysis of RT-readable modifications has been done by limited studies on limited cell types, each using a specific RT and generating a specific dataset17,18. Due to variations in each RT and in each workflow, whether one dataset with one RT can serve as the reference for analysis with a different RT or a different cell type is unknown. Specifically, while modifications of known chemical moieties and locations in the tRNA structure are well defined (Fig. 3a), there remain modifications that are ambiguous to predict due to low readout signals, difficult sequence contexts, or unknown structures in less-studied or disease-associated cell types16. In this case, having two datasets of tRNA modifications, each with a different RT in a different workflow, would help resolve ambiguity, strengthen the prediction, and provide the basis for cross-reference of each dataset. We thus built a comparison of two datasets, each generated from the respective Induro or TGIRT workflow.
Quantitative analysis of RT-readable modifications showed that the two RTs had essentially identical misincorporation rates across the tRNA sequence (Fig. 5a). However, one notable difference was at the two modifications that caused mainly RT stops for Induro—acp3U20 and ms2i6A37/ms2t6A37. While both RTs stalled at these two modifications, Induro stopped at acp3U20 with a higher frequency relative to TGIRT, but at ms2i6A37/ms2t6A37 with a lower frequency (Fig. 5b), providing a distinct signature of each. Nonetheless, the two RTs detected the same number of annotated modifications in the respective workflows (Fig. 5c).
a Cumulative bar graphs show the frequency (%) of RT stops (pink) and RT misincorporations (blue) detected by Induro and by TGIRT at each readable modification as averaged from all tRNA isodecoders of K562 cells, in relation to the frequency (%) of correct reads (white) and of lost coverage (gray). The % of RT stops at a given position was the number of stops relative to the number of total reads. b The frequency (%) of RT stops at each readable and annotated modification detected by Induro and by TGIRT. c The number of reads at each annotated modification site among all isodecoders detected by Induro and by TGIRT. d The distribution of read identity (in %) of each readable modification as detected from total tRNA of K562 cells by Induro and by TGIRT in response to the upstream nucleotide. e, f The read identity (%) of m1A9 vs. m1A58 (e), and of m1G9 vs. m1G37 (f) generated by Induro and by TGIRT. All data were collected from K562 cells. For all of the plots, the analysis of Induro was based on this work (n = 4, biological replicates), while that of TGIRT was extracted from the published dataset18. Center line: median; box limits: upper and lower quartiles; whiskers: 1.5X interquartile range; points: outliers. Student’s t-test was performed by a two-sided analysis (*p < 0.05, **p < 0.01). Source data are provided in Source Data 8-10, where p values and n values used for statistical analysis are available.
We then developed a detailed map for each modification to determine if the two RTs would differ in the signature profile to produce a different “read identity” that represents their intrinsic difference. We considered the sequence context of each modification, based on the notion that Induro was sensitive to environmental changes of each modification (Fig. 4a, b). We defined the read identity as the direct sequencing read of the RT misincorporation at a modification. For example, a misincorporation of C in response to m1A would produce a read identity of G as the complement to C, indicating that G was the nucleotide that was produced in sequencing.
We found a subset of modifications whose read identity was independent of the sequence context, such as acp3U20a, I34, m1I34, and yW37 (Supplementary Fig. 5a). Of these, yW37 was the only one that elicited a difference between the two RTs, where Induro produced primarily G as the read identity while TGIRT produced primarily T. Separately, we found a subset of modifications whose read identity was context-dependent, such as m1G9/37, m3C32, m22G26/27, and ms2i6A37/ms2t6A37. These also included m1A modifications at both the major (positions 9 and 58) and minor (positions 14 and 16) sites. Of these, the two RTs showed more differences in response to the upstream than the downstream nucleotide (Fig. 5d, Supplementary Fig. 5b). Data for each modification was presented by combining reads at all positions where the modification occurred in a tRNA, which would be dominated by those at the most prominent positions (e.g., m1A9 and m1A58 relative to m1A14 and m1A16). The most notable difference between the two RTs was in response to the upstream nucleotide C of the combined m1A9/14/16/58 modifications, where Induro produced A as the read identity, while TGIRT produced T (Fig. 5d). Additionally, in response to the upstream nucleotide G of the combined m22G26/27 modifications, Induro produced a mixture of A/T as the read identity, whereas TGIRT produced largely A. In response to the upstream nucleotide U of the combined ms2i6A37/m2t6A37 modifications, Induro produced A as the read identity, whereas TGIRT produced largely T. In contrast, only one clear difference existed in response to the downstream nucleotide. In response to the downstream nucleotide A of the combined m22G26/27 modifications, Induro produced a mixture of A/T as the read identity, whereas TGIRT produced A (Supplementary Fig. 5b). While the differences between the two RTs could be due to different coverages in the library preparation, they are likely driven by the intrinsic differences between them.
At m1A and m1G, both Induro and TGIRT showed a similar but position-specific response (Fig. 5e, f). Both produced T as the read identity of m1A9 but a mixture of T/G for m1A58. Similarly, both produced C as the read identity of m1G9 but a mixture of G/C/T for m1G37. This parallel display of read identity demonstrates that, like Induro, TGIRT is also sensitive to chemical and structural environments of each modification, which was not reported previously. Combined, the similarities and differences between the two RTs constitute a layer of information that can be further explored to enhance the confidence in profiling of tRNA modifications.
Changes of tRNA modifications across tissues
We used the Induro workflow to probe potential changes in tRNA modifications across tissues. While tissue-specific changes in tRNA abundance were reported17,46, showing a correlation with tissue-specific gene expression, little is known about tissue-specific changes in tRNA modifications17, limiting a broader understanding of the changes. We used mouse as the model, due to its accessibility to genetic manipulation for further studies. We validated the compatibility of the Induro workflow for analysis of tissues, showing mapping rates of tRNA of the brain, kidney, and spleen ≥ 70% (Supplementary Fig. 6a). We then monitored changes of the brain-specific mouse Arg(TCT)-4 isodecoder (Fig. 6a), whose C50U mutation in the T stem decreases the abundance of the mature tRNA, leading to neurodegeneration47. We analyzed the cerebellum tissue of the B6N wild-type (WT) mouse and the C50U-expressing B6J mutant mouse, each in triplicate. A comparative analysis of tRNA abundance readily detected the loss of Arg(TCT)-4 in the mutant relative to the WT brain among cyto-tRNAs by 27-fold, consistent with the published work47, but no difference among mt-tRNAs (Fig. 6b). Additionally, while the workflow detected no difference between the WT and mutant brains in RT-readable modifications in the Arg(TCT)-4 tRNA or in total tRNA, it displayed the sensitivity to detect the C50U mutation in the mutant brain (Supplementary Figs. 6b, c). Thus, while the abundance of Arg(TCT)-4 in the mutant brain is reduced, there is no change in the tRNA modification profile in the isodecoder or global tRNA, indicating that the genetic phenotype of the mutant mouse is primarily driven by the loss of the tRNA. To further validate the compatibility of the workflow with tissues, we analyzed three tissues of the WT mouse (cerebellum, kidney, and spleen), each in triplicate. Analysis of tRNA abundance of all isodecoders readily identified the brain-specific enrichment of Arg(TCT)-4 and Ala(TGC)-6 relative to kidney and to spleen (Supplementary Figs. 6d, e), consistent with a previous report17.
a Sequence and cloverleaf structure of mouse Arg(TCT)−4, showing the C50U mutation. b A differential abundance analysis of isodecoders of cyto- and mt-tRNAs from 12-week-old WT (B6N) and mutant (B6J) mice (n = 6, 3 biological replicates, 2 technical replicates of each). The number of cyto-tRNAs detected was 205 and 213 in WT and mutant, respectively, while all 22 mt-tRNAs were detected in both. Each comparison is shown with log-transformed read counts normalized by DESeq2 with the Pearson correlation coefficient r. c The frequency (%) of RT misincorporation at major modification sites in all tRNAs of cerebellum, kidney, or spleen of B6N mice (n = 6, from 3 biological replicates and 2 technical replicates). Center line: median; box limits: upper and lower quartiles; whiskers: 1.5X interquartile range; points: outliers. Student’s t-test was performed by a two-sided analysis. d, e The relative level of acp3U detected by LC-MS/MS in three tissues of B6N mouse (d, n = 9; 3 biological replicates, each 3 technical replicates) and B6J mouse (e, n = 6; 2 biological replicates, each 3 technical replicates) normalized by total nucleosides of each tissue sample. The acp3U level in the spleen was used as the reference. Error bars, mean ± 95% confidence interval. Student’s t-test was performed by a two-sided analysis. Source data are provided for ion counts in Source data 11. f, g Differential misincorporation frequencies (%) between the cerebellum and the kidney of B6N (f) and between the cerebellum and the spleen of B6N (g), showing isodecoders that exhibit lower frequencies in the cerebellum at position 20.
We then compared the profile of tRNA modifications across the three mouse tissues, using the WT B6N mouse as the model. Analysis of all RT-readable modifications based on the frequency of misincorporation of each showed that, while the three tissues shared an overall similar profile in most of these modifications, the cerebellum had a 25% loss of the modification at position 20 relative to the other two tissues (Fig. 6c), which is most likely the loss of acp3U20 (Fig. 3a). Indeed, an LC-MS/MS analysis of nucleosides generated from total tRNA of each tissue confirmed that the level of acp3U in the cerebellum was reduced by ~25% relative to kidney and spleen (Fig. 6d). Importantly, this loss was not specific to the B6N mouse but was also observed in the B6J mouse by the LC-MS/MS analysis (Fig. 6e), indicating that it is inherent to the cerebellum. The loss is also consistent with a reduced level of the modification enzyme DTWD1 for biosynthesis of acp3U in the cerebellum of B6N relative to the kidney in a proteomic study48 (Supplementary Fig. 6f), indicating a brain-specific differential regulation of the modification. This was most strongly represented by the loss of acp3U20 in the isodecoder Cys(GCA)-9 (Fig. 6f, g). Additionally, the cerebellum had other unique features, where the isodecoder Ala(TGC)-6 had a notable reduction of modifications at positions 15, 48, and 60, but an increase in the modification at position 58 (Fig. 6f, g). Some of these changes were predicted earlier17.
Coordinated changes of tRNA modifications
It was shown recently that, while isodecoders vary broadly in tRNA abundance across tissues and cell types, isoacceptors vary little17,27, indicating a mechanism that buffers the level of each anticodon family. We confirmed this buffering effect in our dataset of 5 human cell lines and 3 mouse tissues. Using human neuroblastoma SH-SY5Y cells and mouse brain as the respective reference, we observed considerable variations in the abundance of isodecoders, but minimal variation in the abundance of isoacceptors (Supplementary Figs. 7a, b). The largest variations in abundance among isodecoders were a 236-fold change for Lys(TTT)-5 between SH-SY5Y and HeLa and a 902-fold change for Arg(TCT)-4 between mouse brain and kidney. In contrast, the largest variations among isoacceptors were a 10-fold for Ile(GAT) between SH-SY5Y and HAP1 and a 3-fold for Gln(TTG) between mouse brain and kidney. These contrasts were much larger than those reported recently27.
We then determined whether tRNA modifications would change across cell types and tissues. We used the frequency of RT misincorporation (%) as the readout of each modification and calculated the level of each modification by multiplying the frequency of misincorporation (%) with the expression level of the associated tRNA normalized by the DESeq2 method49. The weighted misincorporation frequency of each modification in brain cells was then subtracted from the corresponding frequency in non-brain cells to calculate the value for the cell-type- or tissue-specific change. In each tRNA, we focused on misincorporations at positions that are known to harbor modifications in the sequence framework (i.e., the 8 sites in Fig. 3a), thus including modifications of different but related chemical structures at each position (e.g., ms2i6A, ms2iO6A, and ms2t6A at position 37). Among these positions, we chose the one with the most variability across cell types and tissues for genome-wide analysis of all tRNAs. We analyzed the frequency of each weighted misincorporation among unique isodecoders (139 in human cells, 103 in mouse tissues), and the average of weighted misincorporations of each modification among unique anticodons (46 in human cells, 45 in mouse tissues). In contrast to the reduced variation of tRNA abundance from isodecoders to isoacceptors (Supplementary Figs. 7a, b), for modifications, we observed a similar level of variability between the two groups (Supplementary Figs. 7c, d).
We next determined whether changes of tRNA modifications across cell types and tissues were function dependent. We analyzed separately the tRNA modifications that are within the ASL, responsible for decoding, and those that are outside of the ALS (i.e., non-ASL), responsible for stabilizing the tRNA tertiary structure. For each group, we focused on the position of modification that showed the highest variability in the misincorporation rate. We found that the two groups segregated from each other across cell types and tissues, with the ASL group less variable, while the non-ASL group significantly more variable (Fig. 7a, b). Thus, there is a coordinated emphasis to stably express modifications in the ASL across tissues and cell types, but less so on modifications outside of the ASL.
a Violin plots showing differences of the weighted RT misincorporation frequencies (%) at the most variable site of ASL or non-ASL modifications among isodecoders. SH-SY5Y & HeLa & K562, 2 biological replicates, each 2 technical replicates; HEK293T & HAP1, 2 biological replicates; mouse tissues, each 3 biological replicates. b Dataset of a in bar graphs. c Dataset of b in bar graphs at the major sites of ASL and non-ASL modifications in mouse tissues. d Datasets of c in bar graphs at position 32 (left) and 20 (right). e Dataset of (b) in bar graphs at the major sites of ASL and non-ASL modifications in human cells. f Datasets of e in bar graphs at position 32 (left) and 20 (right). Error bars, mean ± 95% standard error, by a two-sided Wilcoxon test (*p < 0.05, **p < 0.01, ***p < 0.001). g Violin plots showing the fraction of modifications (%) in the consistency scale across human cell lines and mouse tissues. A modification of consistency of 100% means that it is present in all 5 human cell lines, while a consistency of 80% means that it is present in 4 of the 5 cell lines. Student’s t-test by a two-sided analysis. h A landscape of coordinated changes of tRNA modifications across tissues and cell types. The charged aa-tRNA is shown with the amino acid as a star; the ribosome is drawn as a cartoon with the large and small subunits, while the mRNA is drawn with abundant codons (triplets without a margin) and rare codons (triplets with a margin), where the distance between each marked codon indicates the presence of unmarked codons. a–f Individual data points, p values and n values for statistical analysis are available in Source Data 12−19.
We found that the position of modifications that drove the stability in the ASL group or the position of modifications that drove the variability in the non-ASL group was each distinct and was associated with a specific isoacceptor family. Among mouse tissues, position 32 in the ASL group is the least variable, driven by the stability of the modifications in Ser(GCT) (e.g., isodecoders Ser(GCT)-3/2/4), whereas position 20 in the non-ASL group is the most variable, driven by the variability of the modifications in Ala(TGC) (e.g., isodecoder Ala(TGC)-2) (Figs. 7c, d, Supplementary Fig. 7e, f). Human cell lines showed a similar pattern; position 32 in the ASL has the most stable modifications, driven by the stability of the modifications in Ser(GCT) and Ser(AGA) (e.g., isodecoders Ser(GCT)-3/1/4 and Ser(AGA)-1/2), whereas position 20 in the non-ASL group has the most variability, driven by the variability of the modifications in Ala(TGC) (e.g., isodecoder Ala(TGC)-1) (Figs. 7e, f, Supplementary Fig. 7e, f). The close parallel between mouse tissues and human cell lines emphasizes a conserved landscape of coordinated changes of tRNA modifications across tissues and cell types. The most likely modification at position 32 is m3C32, whose stability is consistent with a role in translation of genes with biased codons required for regulation of cell cycle and DNA replication pathways50. The most likely modification at position 20 is acp3U20, whose variability is consistent with the variability between neuronal cells and other cell types (Fig. 6c, d).
Given the coordinated changes of tRNA modifications, we sought to identify modifications that were constant across tissues and cell types. These constant modifications would be important to stabilize the tRNA structure or to improve decoding. We determined how these constant modifications were associated with major and minor tRNAs for translation of abundant and rare codons in each genome51. Among isodecoders of one anticodon family, we defined those representing ≥ 50% of the cumulative abundance as major, while those representing < 50% as minor. We found that 96% of the modifications in the major and 93% of the modifications in the minor are at the 100% consistency across the 5 cell lines, while 99% of the modifications in the major and 96% of the modifications in the minor are at the 100% consistency across the 3 tissues (Fig. 7g, left). Conversely, among isoacceptors of one amino-acid family, we defined those representing ≥ 50% of the cumulative abundance as major, while those representing < 50% as minor. We found that 93% of the modifications in the major and 97% of the modifications in the minor are at the 100% consistency across the 5 cell lines, while 94% of the modifications in the major and 100% of the modifications in the minor are at the 100% consistency across the 3 tissues (Fig. 7g, right). Thus, across diverse cell types and tissues, both major and minor tRNAs are associated with constant modifications, with an average of 3 per tRNA.
While most constant modifications are shared between human and mouse, we searched for those that were specific to one or the other, indicating an organism-specific role. Using the RT misincorporation rate as the proxy for a modification, we found 2 high-confidence constant modifications at the isoacceptor level that were specific to human or to mouse (Supplementary Fig. 8a). One was acp3U20a of Asn(GTT) and the other was m22G26 of Tyr(GTA)). As both modifications are specific to human, we found correlation with codon usage in the human genome, but not the mouse genome. Specifically, in a codon-usage analysis that was normalized by the abundance of each mRNA in the RNAseq data of HEK293T cells52 and of mouse kidney53, we found that the human genome has a higher usage of the optimal codon for the tRNA with the constant modification. For example, AAC is the optimal codon for Asn(GTT), while UAC is the optimal codon for Tyr(GTA), and both are in a higher usage in the human genome, but not in the mouse genome (Supplementary Fig. 8b). Thus, the association of the constant modification in each case may help the tRNA with better reading of the optimal codon during protein synthesis.
Finally, our dataset of 5 human cell lines and 3 mouse tissues clarified an open question in the field – whether m1G37 is stably associated with Pro(AGG) in human and mouse. Previous work was unable to detect m1G37 in human Pro(AGG)18, but was able to detect it in the other two isoacceptors. As m1G37 is essential for cell survival and is conserved in evolution39,54,55, its potential absence from human Pro(AGG) was difficult to understand. We now showed that, by analysis of all raw reads for Pro(AGG) across a wide range of eukaryotic cell types, m1G37 was consistently read as a mixture of G/C/T, while the associated I34 was consistently read as a G (Supplementary Fig. 8c), which are respectively the read identity of each (Supplementary Figs. 5a, Fig. 5f). Thus, m1G37 is stably associated with Pro(AGG) and is conserved from yeast to human (Supplementary Fig. 8d).
Discussion
Here we provide a genome-wide tRNAseq dataset that defines the RT signature at each readable modification across the tRNA sequence framework, thus addressing an unmet need in the current field. We use the recently developed group-II Induro RT and provide both practical and mechanistic insight into how to maximize its readthrough and how to strengthen the prediction of each modification after readthrough. Importantly, we produce information on how modifications can undergo a coordinated shift across tissues and cell types, providing a conceptual framework that will open the door to innovative research with impact on the vastly expanding fields of tRNA modifications.
Maximizing RT readthrough
The key to profile tRNA modifications is to maximize the RT readthrough. Yet, most published tRNAseq datasets used one condition, without an exploration, raising the question of whether the chosen condition was the best for the cells of interest (e.g., cells in cold stress or in thermophiles). The lack of a well-defined condition that maximizes readthrough could have contributed to the incomplete profiling of tRNA modifications. With Induro, we have explored a range of reaction conditions, showing that the condition that maximizes readthrough and detection of modifications—at 42 °C and overnight—is distinct from the provider’s condition. Additionally, we provide a dataset to show the progress of Induro to achieve the maximum readthrough over time and temperature, facilitating users to make an informed decision for their experiments.
We show that Induro increases its readthrough with time to 60% at 42 °C and to 90% at 37 °C, but that its readthrough is limited at 10% at 55 °C without a time-dependent increase. The increase in readthrough overcomes all but two modifications (acp3U20 and ms2i6A37/ms2t6A37), thus laying out a clear landscape to read tRNA sequences. Mechanistically, the increase in readthrough is achieved by overcoming RT stops without altering RT misincorporation frequencies, consistent with the notion that the latter is an intrinsic feature of Induro. In contrast, overcoming RT stops is thermally controlled and is dependent on the position and structure of each modification. A positional effect is observed at m1A and m1G, showing the sensitivity of the RT to the local environment of each modification. The RT also responds to Mn2+ by overcoming the most dominant barrier – at ms2i6A37/ms2t6A37, while showing a higher sensitivity to detect the non-W-C modification m7G. Notably, we did not detect other non-W-C modifications (e.g., ψ), due to low-confidence reads. Nonetheless, the comprehensive dataset, and the accompanying mechanistic insight, establishes the utility of the Induro workflow to probe modifications in tRNA and in other RNAs that are structured and are extensively modified (e.g., rRNAs).
Strengthen the power of prediction of tRNA modifications
The detection of a tRNA modification in the Illumina platform is by the misincorporation signature of the RT, which provides the basis for predicting the modification at annotated and non-annotated sites. However, ambiguity arises if the readout is naturally low or is reduced by stress. While RNA mass spectrometry is a direct approach to determine the site and chemical nature of the modification16, it is low throughput and requires more materials. Here we compare the fidelity mechanism of Induro and TGIRT to generate two datasets for each RT-readable modification, providing a basis to use the comparison to strengthen the power of prediction.
As both Induro and TGIRT are derived from a group-II intron family, they likely share a similar structure and mechanism for the nucleotide selectivity at each modification. Indeed, we show a largely similar read-identity profile of the two at most modifications, providing confidence for each prediction. Notably, at acp3U20, which naturally has a low readout of misincorporation, the use of RT stops would be a more reliable readout (Fig. 3c). In this case, Induro has a stronger readout of RT stops than TGIRT (Fig. 5b), indicating that it is a better choice for studies of acp3U20 and the associated modification enzyme. Additionally, while Induro is used here, TGIRT has been used in multiple studies and in a broader range of cell types (different strains of bacteria, yeast, fly, and cultured human cells, including iPSCs and differentiated neuronal and cardiac cells16,18,27). The close similarity of the two enzymes will permit the use of TGIRT-generated datasets as the basis to extend the application of Induro to strains and cell types that were covered by TGIRT. Thus, Induro can be added to the growing list of group-II intron RT enzymes that are applicable for genome-wide tRNAseq. Although the sequence information of Induro is not available, this does not hinder its ability to produce high-quality and reproducible data.
Neuron-specific tRNA modifications
We have analyzed the profile of each RT-readable tRNA modification across 5 human cell lines and 3 mouse tissues, generating a large and diverse dataset. From a comparative analysis of the 3 mouse tissues, we find that neuron cells have a distinct profile relative to the others (Fig. 6c), indicating a special need of neurons for tRNA modifications and for protein synthesis. Indeed, synapses during learning and memory require rapid re-programming of neuronal proteins, which in turn requires rapid responses of neuronal tRNAs and the associated modifications. We show that the most notable difference associated with neuronal tRNA modifications is the substantial loss of acp3U20 (Fig. 6c–e), which is proposed to stabilize the local structure of the D loop through Mg2+ coordination56. The substantial loss of acp3U20 suggests that a more flexible D loop is important for neuronal tRNAs. Indeed, the flexibility of the D loop is necessary to rapidly change its conformation to interact with different protein factors in each elongation cycle of protein synthesis. One such protein factor is the maturation enzyme for CCA addition to the tRNA 3’-end52, and another is the elongation factor that brings the charged aa-tRNAs to the ribosome53. As each enzyme uses a different structure to interact with the D loop, the increased flexibility of the loop, via loss of acp3U20 in neuronal tRNAs, would fulfill this multi-partner requirement.
Coordinated changes of tRNA modifications in protein homeostasis
We have discovered a landscape of coordinated changes of tRNA modifications across tissues and cell types that has fundamental importance for understanding the development and maintenance of protein homeostasis. We show that, while the tRNA modifications required for stabilizing the L-shaped tertiary structure are variable in different tissues and cell types, those required for decoding the genetic code are stabilized. This landscape of coordinated changes is summarized in a model (Fig. 7h) that supports both isodecoders and isoacceptors.
First, at the level of isodecoders, each isodecoder of an anticodon family has a unique sequence and requires a unique set of tRNA modifications to stabilize its folding into the L-shaped structure, which is uniform across all tRNAs. In this L-shaped structure, every tRNA will have an equal opportunity to dock into the ribosome decoding site to allow for screening for the ones that have the matching anticodons for the codon at hand. The high variability of tRNA modifications is to match with the high variability of tRNA sequences across tissues and cell types to endow every sequence with the necessary modifications to be sampled by the decoding site with equal affinity57,58. Second, at the level of isoacceptors, while the ones that have the matching anticodons are selected by the decoding site, the high variability of tRNA modifications is again to stably fold members of distinct sequences in the family for docking into the decoding site. However, to ensure cognate docking, and to discriminate against near-cognate docking, the modifications that support the best quality of the anticodon-codon interaction are stabilized. These modifications can help to shape the structure of the ASL59,60 (e.g., those at positions 32 and 37) and/or to improve the base-pairing quality and capacity40 (e.g., those at positions 34 and 37). An example of the latter is I34, which in Pro(AGG) not only expands the base-pairing capacity to A/C/U at the wobble position61, but also compensates for the lack of Pro(GGG) in eukaryotes62 (Supplementary Fig. 8d) to cover the CCC codon for decoding.
Third, at the global level of protein synthesis, we show that every tRNA, whether major or minor, has a set of constant modifications that are stably expressed across tissues and cell types. These constant modifications are to stabilize each tRNA structure and/or to improve the quality of decoding. As major tRNAs in a cellular pool are to satisfy the demand of abundant codons, while minor ones to satisfy the demand of rare codons, these constant modifications are required to maintain a supply-demand balance that is necessary for protein homeostasis51. Thus, based on the need of each genome, its GC content, its tRNA repertoire63, and the modifications available, the ribosome will then determine the speed and quality of translation one codon at a time. The lack of a corresponding tRNA for a codon, due to the lack of modifications, has been linked to many pathologies2.
Overall, we describe the development and validation of Induro-tRNAseq to provide a comprehensive dataset for analysis of RT-readable tRNA modifications, which will facilitate a better understanding of tRNA biology. While non-RT-readable modifications (e.g., ψ) were not addressed here, we will explore different chemical reactions for each in future studies. For example, we will determine if ψ should be pre-treated with CMC (cyclohexyl-(2-morpholino-ethyl)-carbodiimide) or bisulfite64,65,66,67,68,69 for the most robust readout. Importantly, this work unveils a landscape of coordinated changes of tRNA modifications that is necessary to maintain protein homeostasis across tissues and cell types.
Methods
Unless otherwise noted, we followed the manufacturer’s instructions of each kit. Additionally, we extracted the TGIRT data from mim-tRNAseq18 for comparison with the Induro data.
Cell culture
Human embryonic kidney HEK293T cells (ATCC, #CRL-3216), chronic myeloid leukemia K562 cells (ATCC, #CL-243), cervical cancer HeLa cells (ATCC, #CCL-2), neuroblastoma SH-SY5Y (ATCC, #CRL-2266), and near-haploid HAP1 cells (Horizon, #C631)70 were cultured at 37 °C in Dulbecco’s Modified Eagle’s Medium (DMEM) (Corning, #10-013-CV) with 5% CO2 and 10% fetal bovine serum (Corning, #35-010-CV). To induce oxidative stress, HEK293T cells were treated with 100 µM and 300 µM sodium arsenite (RICCA, 7140-16) for 2 h prior to harvest.
RNA extraction and tRNA isolation
Total RNA of cultured human cells was extracted with TRIzol (Invitrogen, #10296010). To determine tRNA charging levels, extracted total RNA was dissolved in 25 mM sodium acetate, pH 5.0. With mouse tissues, 12-week-old C57BL/6 J (B6J) WT mice were extracted for total RNA of cerebellum, kidney, and spleen, while the age-matched congenic B6J.B6NnTr20 mice were extracted for total RNA of cerebellum59. The mouse brain, kidney, and spleen used in this study were obtained from animals that were maintained under the institutional IACUC guidelines, compliant with those of the American Veterinary Medical Association for husbandry and euthanasia. The tRNA pool was isolated from total RNA by 12% denaturing PAGE/7 M urea in TBE (90 mM Tris-borate pH 8.3 and 2 mM EDTA) as gel slices, which were crushed with a disposable pestle in a tRNA extraction buffer (0.3 M Na Acetate (NaOAc), pH 5.0, 0.25% SDS, and 1 mM EDTA) as described18,71. The tRNA pool was filtered through a Costar Spin-X 8163 centrifuge tube filter (Corning #8163), precipitated with isopropanol in the presence of 20 µg glycogen (RNA grade; Thermo Scientific #R0551), and dissolved in RNase-free water. Approximately 12% of the input total RNA was recovered as tRNA. To compare with the TGIRT-based mim-tRNAseq dataset18, Induro-tRNAseq was performed with 4 biological replicates of total RNA from K562 cells, 2 biological replicates of total RNA from HEK293T cells, and 2 biological replicates of gel-purified tRNA pool of K562 and HEK293T cells. Two technical replicates were performed for each biological replicate.
Induro-tRNAseq library preparation
Prior to starting the workflow, the integrity of each sample (500 ng of total RNA or 125 ng of gel-purified tRNA pool) was verified by 12% denaturing PAGE/7 M urea. For Induro time-course experiments, the workflow was carried out twice to provide two technical replicates using one of the 4 biological replicates of total RNA of K562 cells.
Standard tRNA deacylation and T4 PNK (polynucleotide kinase) 3´-end repair
The tRNA in each sample was first deacylated by incubation for 45 min at 37 oC in 10 µL 75 mM glycine, pH 9.5, with 1 U/µL Superase-In (Thermo Fisher #AM2694). The reaction was diluted to 100 µL with 1X T4 PNK buffer and incubated for 30 min with 1 µL T4 PNK (10 U/µL; NEB M0201) to remove 3’ phosphoryl groups. The deacylated RNA was ethanol precipitated with 20 µg of glycogen and the pellet was dissolved in 7.0 µL water.
Oxidation, β-elimination, and deacylation of tRNA and T4 PNK 3´-end repair
To determine tRNA charging levels, 4 µL of 90 mM sodium acetate buffer, pH 4.8-5.0, was added to 500 ng of total tRNA (as part of ~2.5 µg total RNA) in 28 µL of water. Next, 4 µL of a freshly prepared 150 mM NaIO4 solution (Thermo Fisher #419610050) was added to give final concentrations of 16 mM NaIO4, 10 mM NaOAc, pH 4.8–5.0. The oxidation reaction was incubated 30 min at room temperature (r.t.) and was then quenched with the addition of 4 µL of 1 M glucose to 0.1 M final concentration and incubated 5 min at r.t. Next, 20 µL of 100 mM sodium borate, pH 9.5, was added for a final concentration of 33 mM. This mixture was incubated for 30 min at 45 °C for β-elimination and deacylation. The reaction was neutralized, and 3´-end repair was initiated by addition of 20 µL of 200 mM Tris-HCl, pH 6.8, 40 mM MgCl2, and 1 µL T4 PNK, and incubated at 37 °C for 30 min. The T4 PNK was heat-inactivated at 65 °C for 10 min. The deacylated RNA was ethanol precipitated with 20 µg of glycogen and the pellet was dissolved in 7.0 µL water.
Ligation of 3´-RNA adapter
T4 Rnl2 (NEB M0239L) was used for splint-mediated 3’-ligation of each tRNA library to a barcoded 35-mer RNA/DNA adapter bearing a 5’-phosphate and a 3’-amino group (oligonucleotides listed in Tables S1, S2). Typically, 4 ligation reactions were carried out in parallel with each in 20 µL and containing a separate tRNA library, a uniquely barcoded 3’-adapter (0.25 µM), two paired DNA splints (0.32 µM) with both sharing the same barcode as the 3’-adapter, T4 Rnl2 (0.5 U/µL), and Superase-In RNase inhibitor (1 U/µL) in a buffer containing 400 µM ATP, 10 mM MgCl2, 1 mM DTT, 10% PEG8000, and 50 mM Tris-HCl, pH 7.5. The sequences of the two corresponding splint DNAs are listed in Table S2− one for the 3’-NCCA end derived from charged tRNAs and the other for the 3’-NCC end derived from uncharged tRNAs. Reactions were briefly heat-cooled at 60 °C for 2.5 min prior to adding T4 Rnl2 and kept at 16 °C for 45 min and then at 25 °C for 45 min. Ligation was terminated by combining all 4 reactions into 20 µL 0.1 M EDTA, followed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1), pH 5.2, and purified through a Zymo Oligo Clean & Concentrator cartridge (Zymo Research #D4061). The nucleic acid in the multiplexed library was eluted in 11 µL of RNase-free water. Analysis of the tRNA ligated with the 3’-adapter was performed on a denaturing gel (Fig. 1b). Total RNA isolated from cells or a gel-purified tRNA pool was used as the input. The migration position of the starting tRNA was marked by a bracket (indicating a range of sizes), while the ligated tRNA was shown by an arrow. The intensity of each band was corrected by the molecular mass of each species and by removing the contaminating 5S rRNA band from the ligated band. The yield of ligation was measured by dividing the band intensity of the ligated tRNA (corrected for adapter addition and 5S RNA contamination) over the sum of the starting tRNA and the ligated tRNA, showing typically ≥ 75%.
Reverse transcription
Each library was converted to cDNA with Induro RT (NEB M0681S) using a 57-mer ssDNA as the primer: 5’pRNAGATCGGAAGAGCGTCGTGTAGGGAAAGAG/iSp18/GTGACTGGAGTTCAGACGTGTGCTC-3’, where p is a phosphate, R is purine, N is any nucleotide, and iSP is an 18-carbon chain spacer. Each RT reaction was performed in 20 µL with 0.5 µM RT primer, 0.75 mM dNTPs, 10 mM DTT, 1 U/µL Superase-In, and 10 U/µL Induro RT in 1X Induro buffer. The reaction was briefly heat-cooled at 60 °C prior to adding the RT and was incubated overnight (12-16 h) at 42 °C unless otherwise specified. After cDNA synthesis, the RNA was hydrolyzed by addition of 1 µL of 5 M KOH and heated at 90-95 °C for 3 min. The solution was neutralized with 0.25 M NaOAc, pH 5.0, extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and the cDNA precipitated with 3 volumes of ethanol. The pellet was dissolved in 8 µL of 7 M urea-TBE with 0.01% xylene cyanol and bromophenol blue. After heat-cool at 80-85 °C for 2 min, the cDNA was loaded onto a denaturing 10% PAGE/7 M urea gel (8 × 7 × 0.1 cm) and electrophoresed at 200 V in hot TBE for 20 min. Included in the gel as controls were an RT reaction conducted in the absence of tRNA and a Small Range RNA Ladder (NEB N0364S). The gel was stained with SYBR gold and visualized by a ChemiDoc imager (BioRad). The gel was excised in the range of 90–180 nucleotides (nts) to include full-length and truncated cDNA and was extracted into 500 µL TE by incubation in a MultiTherm Shaker (Benchmark Scientific) at 70 °C/1500 rpm for 1-2 h. After gel removal, the cDNA was precipitated with isopropanol in the presence of 20 µg glycogen carrier and the pellet dissolved in 5.5 µL water.
cDNA circularization and PCR
The cDNA was circularized by incubation with Circligase (Lucigen CL4111K) for 3 h at 60 °C in a 10 µL reaction54, which was terminated by heat inactivation (at 80 °C for 10 min) and stored at –20 °C until PCR amplification. The circularized cDNA (1 µL) was amplified by PCR using Q5 Hot Start High-Fidelity 2X Master Mix (NEB M0494L) in a 25 µL reaction containing 12.5 pmoles of forward and reverse primers (primers listed in Table S3). A unique barcoded reverse primer was used for each group of 4 multiplexed cDNA pools. The thermocycler was programmed for 30 sec at 98 °C, followed by at least 5 cycles of 10 sec at 98 °C, 10 sec at 62 °C, and 10 sec at 72 °C. To determine the optimal cycle number, separate reactions were terminated after 5, 6, 7, and 8 cycles. After clean-up by ethanol precipitation, these reactions were electrophoresed on a non-denaturing 8% PAGE gel alongside O’RangeRuler 10 bp DNA (Thermo Fisher #SM1313) and GeneRuler 50 bp DNA ladders (Thermo Fisher #SM0371), followed by SYBR gold staining. The dsDNA product (150-240 bp in length) with the highest yield but without contaminating higher MW DNA was excised from the gel and eluted into 500 µL TE by continuous mixing at r.t. overnight. After clarification of the suspension through a Costar Spin-X cartridge, the cDNA was ethanol precipitated with 20 µg glycogen and dissolved in 20 µL TE. The average yield of gel-purified cDNA from each multiplexed sample of 4 was approximately 50 ng as determined by Qubit (Invitrogen) and Bioanalyzer (Agilent) analysis. At least 20 ng of each multiplexed PCR product should be provided for sequencing. If the yield of PCR product was insufficient, the gel purification step could be eliminated, resulting in a 2-4-fold increase in the amount of recovered cDNA. In this case a Zymo Oligo Clean & Concentrator cartridge should be used to remove the PCR primers.
Sequencing
Equimolar amounts of the PCR-amplified cDNA libraries were pooled and loaded onto the Illumina NextSeq 500 platform and a 2×75 paired-end sequencing run was performed.
Induro activity with MnCl2 as the single divalent metal ion
We replaced the Induro buffer with the buffer 50 mM Tris-HCl, pH 8.3, containing 1 mM MnCl2. Reverse transcription of a DNA-RNA hairpin with a single-stranded 5’-overhang (5’-P-rGrGrCrGrGrArArUrCrATCTCACTCTACATCATGGCCGGCCGGGTAATCGGCCGGC-3’, Supplementary Fig. 4a) was used to titrate the optimal concentration of MnCl2 for Induro. The reaction was 5 min at r.t. followed by 15 min at 55 °C. We found that MnCl2 at greater than 1 mM was inhibitory to the enzyme.
Synthesis of mt-Leu(TAA) for calibration analysis
The 5’-RNA fragment of mt-Leu(TAA) was chemically synthesized encoding the sequence from nucleotides 1 to 17, containing either G9 or m1G9. The 3’-RNA fragment of the tRNA was transcribed by T7 RNA polymerase and gel purified. The 5’- and 3’-RNA fragments were mixed in equal molar concentration, heat-cooled, and covalently joined by T4 Rnl2. The reconstituted full-length mt-Leu(TAA) was separated from the component RNAs by a denaturing PAGE/7 M urea gel and extracted from the gel. For calibration analysis, the G9-containing and the m1G9-containing full-length tRNAs were mixed in fractions of 1:0, 3:1, 1:3, and 0:1, where the methylated tRNA represented 0, 25, 75, and 100% of the input.
LC-MS/MS analysis of acp3U in mouse tRNAs
Total RNA isolation. E. coli yfiP-KO strain of the Keio collection, where the gene for acp3U47 biosynthesis was replaced by a Kan marker, was obtained from Mark Goulian (U Pennsylvania). P1 lysate was made from this strain and transduced to MG1655 to generate the MG1655 yfiP-KO strain72. E. coli yfiP-WT and yfiP-KO in MG1655 were grown overnight at 37 °C in LB. Cells were harvested and total tRNA was isolated from each strain73. Total RNA (up to 650 μg) of each mouse tissue was extracted with TRIzol59 and was run in a denaturing 12% PAGE/7 M in 1x TBE (90 mM Tris-borate pH 8.3 and 2 mM EDTA) at 200 V, 30 min, to isolate the tRNA fraction in the gel extraction buffer (0.3 M NaOAc pH 5.0, 0.25% SDS, 1 mM EDTA)18. The eluted tRNA was filtered through a Costar Spin-X 8163 centrifuge tube filter (Corning, #8163), precipitated with one volume of isopropanol and 20 µg of RNA grade glycogen (Thermo Scientific, #R0551), and dissolved in RNase-free water. Approximately 10% of the input total RNA was recovered as tRNA.
Digestion of tRNAs to nucleosides. Total tRNA (0.3–2 µg) was incubated at 95 °C, 5 min, cooled on ice, 3 min, and digested to ribonucleotide monophosphates in 23 µL with nuclease P1 (0.3 U, Wako #M0660S) in 10 mM ammonium acetate pH 5.0 at 45 °C, 2 h, followed by dephosphorylation with 0.025U phosphodiesterase (Worthington Biochemical #LS003926) and 1U recombinant shrimp alkaline phosphatase (NEB #M0371S) in 100 mM ammonium bicarbonate pH 8.0, 2 h, 37 °C in a 28 μL reaction74. The mixture of nucleosides after digestion was separated from proteins by centrifugation through a 10,000-Da MWCO spin filter (Sartorius #VN01H02).
LC-MS/MS analysis of tRNA nucleosides. Nucleosides dissolved in LC/MS grade water was analyzed on an Agilent 1290 Infinity II HPLC coupled to an Agilent 6470 QQQ-MS in the positive ion mode, using the multiple reaction monitoring method75. Samples were run through a Hypersil GOLD aQ C18 Polar Endcapped HPLC column (Thermo Fisher, 25303-152130; 3-µm particle size, 175-Å pore size, 2.1 × 150 mm) at a flow rate of 0.35 mL/min with a gradient of Buffer A (5 mM ammonium acetate pH 5.0) and Buffer B (100% acetonitrile) at 37 °C. The gradient was 0% Buffer B from 0-5 min, 0-20% Buffer B from 5-7 min, and 30% Buffer B from 7-9 min, followed by 5 min of re-equilibration to 0% Buffer B. The parameters used for the mass spectrometer were as follows: gas temperature at 175 °C, gas flow at 12 L/min, nebulizer pressure at 20 psi, sheath gas temperature at 325 °C, sheath gas flow at 12 L/min, and capillary voltage at 2500 V. The Multiple Reaction Monitoring (MRM) transitions for detection of each nucleoside were: m/z 268 → 136 for A, m/z 244 → 112 for C, m/z 284 → 152 for G, m/z 346 → 214 for acp3U, m/z 245 → 113 for U. Fragmentor, collision energy, and retention time for each nucleoside are described in Supplementary Table 4.
Nucleosides digested from total tRNA (2 µg) of yfiP-WT and yfiP-KO cells were used for LC-MS/MS analysis. Digested total tRNA from WT cells was used to determine the optimal LC-MS/MS parameters for detection of acp3U, which was confirmed by the absence of the peak from the digested total tRNA of the KO cells. Three biological replicates of each mouse tissue were digested and analyzed by LC-MS/MS. Each tissue sample was injected three times as technical replicates. Ion counts for each nucleoside were determined using Agilent Qualitative Analysis. Briefly, the peaks of each nucleoside were integrated, and the area of each peak was determined, corresponding to the ion counts for each nucleoside. Quantification of the relative acp3U level in each sample was performed by comparing ion counts of acp3U over the sum of ion counts for A,C,G,U in each sample.
Data analysis
Read processing was performed as described34. Paired-end reads were merged using PEAR v 0.9.6 (ref76), which were then demultiplexed using cutadapt v2.5 as described77 and a fasta file of the first 10 nts for the different 3’-barcoded adapters was created. Indels in the alignment to the adapter were removed with --no-indels. The two 5’-RN nucleotides introduced by reverse transcription (see Library Preparation above) were trimmed from reads with -u 2. Reads shorter than 10 nts were discarded by cutadapt77 according to the parameters in -m 10. Processed reads of human samples were mapped to tRNA reference transcripts derived from human genome hg38 and those of mouse samples mapped to the reference derived from the mouse genome mm39 using mimseq v1.2. Mapping was as described18 (github.com/nedialkova-lab/mim-tRNAseq), where matured and processed tRNA sequences were mapped to MODOMICS entries using BLAST and clustered with the --cluster parameter using a user-defined sequence identity threshold. After clustering, reads were aligned using GSNAP to the representative cluster sequences of mature tRNAs. The following mapping parameters were used:
H. sapiens: --species Hsap --cluster --cluster-id 0.95 --min-cov 2000 --max-mismatches 0.1 --remap --remap-mismatches 0.075
M. musculus: --species Mmus --remap --remap-mismatches 0.075
Mapping rates were calculated by the number of uniquely mapped reads relative to the sum of the number of unmapped reads, multiple-mapped reads, and uniquely mapped reads. Additional quantification and statistical analysis were also based on methods described34. Fractions of incomplete 3’-ends (3’-N, 3’-NC and 3’-NCC, where N = the discriminator nucleotide at position 73) relative to all 3’-ends (including 3’-NCCA) were calculated per unique tRNA sequence. For differential tRNA abundance analysis, read counts of cyto-tRNAs and mt-tRNAs were normalized with DESeq2 (ref49) separately and the Pearson correlation coefficient (r) was calculated. Rates of misincorporation were calculated by summing up counts of mismatches for all four nucleotides relative to total read counts at the position of interest. Frequency of stops was determined by dividing the number of stops at a given position by the total number of reads at that position. Readthrough at a position was obtained by subtracting the ratio of RT stops at that position from 1.0. For a given modification, RT stops were reported as the maximum value observed in a 3-nt window (–1, 0, and +1) centering on the modification to reduce the likelihood of over-estimation. Modifications in human tRNAs at known positions (9, 20, 26, 32, 34, 37, and 58), as well as non-annotated positions, were identified by RT stop or RT misincorporation of >10%. Annotated modifications were as defined34,56. All data analysis was performed with in-house python written codes using Python v3.7.0. Analysis of the mim-tRNAseq dataset was performed using the following parameters:
H. sapiens: --species Hsap --cluster --cluster-id 0.95 --min-cov 2000 --max-mismatches 0.1 --remap --remap-mismatches 0.075
S. cerevisiae: --species Scer --cluster --cluster-id 0.90 --min-cov 2000 --max-mismatches 0.1 --remap --remap-mismatches 0.075
S. pombe: --species Spom --cluster --cluster-id 0.95 --min-cov 2000 --max-mismatches 0.1 --remap --remap-mismatches 0.075
D. melanogaster: --species Dmel --cluster --cluster-id 0.95 --min-cov 2000 --max-mismatches 0.1 --remap --remap-mismatches 0.075
In human cells, the weighted misincorporation difference of each site was calculated by subtracting the frequency in SH-SY5Y, whereas in mouse tissues, it was calculated by subtracting the frequency in cerebellum from that of kidney or spleen.
Calculation of variability of misincorporation
Misincorporation difference was obtained by subtracting the data of each sample group from that of the SH-SY5Y cell line for human cells, and cerebellum tissue for mouse tissues, respectively. The most variable sites were selected by ordering all positions with a misincorporation difference and taking the highest varying site(s) for individual tRNAs. Two-sided Wilcoxon test was used to assess statistical significance, where a p value < 0.05 is considered statistically significant.
Codon usage analysis
The number of each codon was counted based on each transcript sequence of protein-coding gene retrieved from Ensembl (human; GRCh38, mouse; GRCm39) and weighted by RPKM (reads per kilobase per million mapped reads) of each transcript determined previously to calculate the codon usage in human HEK293T cell line52 and in mouse kidney53.
Data sources
The raw FASTQ files of mim-tRNAseq18 of wild-type human cells (HEK293T and K562), yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe), and fly (Drosophila melanogaster) were retrieved from Gene Expression Omnibus (GEO): GSE152621.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. The tRNA sequencing data generated in this study have been deposited in the sequence read archive (SRA) database of NCBI under accession code PRJNA996215 [http://www.ncbi.nlm.nih.gov/bioproject/996215]. Source data are provided with this paper.
Code availability
Scripts for data processing and figure creation are available at https://github.com/YaMing-Hou-lab/Induro-tRNAseq (https://doi.org/10.5281/zenodo.14419721).
References
Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res 28, 395–404 (2018).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Dedon, P. C. & Begley, T. J. Dysfunctional tRNA reprogramming and codon-biased translation in cancer. Trends Mol. Med 28, 964–978 (2022).
Liu, F. et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 167, 816–828 e816 (2016).
Chan, C. T. et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6, e1001247 (2010).
Chan, C. T. et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3, 937 (2012).
Chionh, Y. H. et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7, 13302 (2016).
Patil, A. et al. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 11, 3656–3665 (2012).
Lucas, M. C. et al. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. 42, 72–86 (2023).
Thomas, N. K. et al. Direct Nanopore Sequencing of Individual Full Length tRNA Strands. ACS Nano 15, 16642–16653 (2021).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Pang, Y. L., Abo, R., Levine, S. S. & Dedon, P. C. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res 42, e170 (2014).
Erber, L. et al. LOTTE-seq (Long hairpin oligonucleotide based tRNA high-throughput sequencing): specific selection of tRNAs with 3’-CCA end for high-throughput sequencing. RNA Biol. 17, 23–32 (2020).
Shigematsu, M. et al. YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucleic Acids Res 45, e70 (2017).
Kimura, S., Dedon, P. C. & Waldor, M. K. Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat. Chem. Biol. 16, 964–972 (2020).
Pinkard, O., McFarland, S., Sweet, T. & Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 11, 4104 (2020).
Behrens, A., Rodschinka, G. & Nedialkova, D. D. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 81, 1802–1815.e1807 (2021).
Scheepbouwer, C. et al. ALL-tRNAseq enables robust tRNA profiling in tissue samples. Genes Dev. 37, 243–257 (2023).
Yamagami, R., Sieg, J. P., Assmann, S. M. & Bevilacqua, P. C. Genome-wide analysis of the in vivo tRNA structurome reveals RNA structural and modification dynamics under heat stress. Proc. Natl Acad. Sci. USA 119, e2201237119 (2022).
Watkins, C. P., Zhang, W., Wylder, A. C., Katanski, C. D. & Pan, T. A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat. Commun. 13, 2491 (2022).
Werner, S. et al. Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes. Nucleic Acids Res 48, 3734–3746 (2020).
Stamos, J. L., Lentzsch, A. M. & Lambowitz, A. M. Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications. Mol. Cell 68, 926–939 e924 (2017).
Belfort, M. & Lambowitz, A. M. Group II Intron RNPs and reverse transcriptases: from retroelements to research tools. Cold Spring Harb Perspect Biol. 11, a032375 (2019).
Mohr, S. et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19, 958–970 (2013).
Lentzsch, A. M., Yao, J., Russell, R. & Lambowitz, A. M. Template-switching mechanism of a group II intron-encoded reverse transcriptase and its implications for biological function and RNA-Seq. J. Biol. Chem. 294, 19764–19784 (2019).
Gao, L. et al. Selective gene expression maintains human tRNA anticodon pools during differentiation. Nat. Cell Biol. 26, 100–112 (2024).
Zhao, C., Liu, F. & Pyle, A. M. An ultraprocessive, accurate reverse transcriptase encoded by a metazoan group II intron. RNA 24, 183–195 (2018).
Chung, K. et al. Structures of a mobile intron retroelement poised to attack its structured DNA target. Science 378, 627–634 (2022).
Maguire, S. & Guan, S. Rolling circle reverse transcription enables high fidelity nanopore sequencing of small RNA. PLoS One 17, e0275471 (2022).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44, D184–D189 (2016).
Suzuki, T. et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11, 4269 (2020).
Hei, T. K., Liu, S. X. & Waldren, C. Mutagenicity of arsenic in mammalian cells: role of reactive oxygen species. Proc. Natl Acad. Sci. USA 95, 8103–8107 (1998).
Cui, J., Liu, Q., Sendinc, E., Shi, Y. & Gregory, R. I. Nucleotide resolution profiling of m3C RNA modification by HAC-seq. Nucleic Acids Res 49, e27 (2021).
Oerum, S., Degut, C., Barraud, P. & Tisne, C. m1A post-transcriptional modification in tRNAs. Biomolecules 7, 20 (2017).
Bowles, I. E. & Jackman, J. E. A tRNA-specific function for tRNA methyltransferase Trm10 is associated with a new tRNA quality control mechanism in Saccharomyces cerevisiae. RNA 30, 171–187 (2024).
Gamper, H. B., Masuda, I., Frenkel-Morgenstern, M. & Hou, Y. M. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat. Commun. 6, 7226 (2015).
Gamper, H. et al. Insights into genome recoding from the mechanism of a classic +1-frameshifting tRNA. Nat. Commun. 12, 328 (2021).
Masuda, I. et al. tRNA methylation is a global determinant of bacterial multi-drug resistance. Cell Syst. 8, 302–314.e308 (2019).
Masuda, I. et al. tRNA methylation resolves codon usage bias at the limit of cell viability. Cell Rep. 41, 111539 (2022).
Gamper, H. et al. Twice exploration of tRNA +1 frameshifting in an elongation cycle of protein synthesis. Nucleic Acids Res 49, 10046–10060 (2021).
Demo, G. et al. Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation. Nat. Commun. 12, 4644 (2021).
Vashishtha, A. K., Wang, J. & Konigsberg, W. H. Different divalent cations alter the kinetics and fidelity of DNA polymerases. J. Biol. Chem. 291, 20869–20875 (2016).
Hou, Y. M., Gu, S. Q., Zhou, H. & Ingerman, L. Metal-ion-dependent catalysis and specificity of CCA-adding enzymes: a comparison of two classes. Biochemistry 44, 12849–12859 (2005).
Araujo Tavares, R. C., Mahadeshwar, G., Wan, H. & Pyle, A. M. MRT-ModSeq - rapid detection of RNA modifications with marathonRT. J. Mol. Biol. 435, 168299 (2023).
Dittmar, K. A., Goodenbour, J. M. & Pan, T. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2, e221 (2006).
Ishimura, R. et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 (2014).
Giansanti, P. et al. Mass spectrometry-based draft of the mouse proteome. Nat. Methods 19, 803–811 (2022).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Cui, J. et al. m(3)C32 tRNA modification controls serine codon-biased mRNA translation, cell cycle, and DNA-damage response. Nat. Commun. 15, 5775 (2024).
Rak, R., Dahan, O. & Pilpel, Y. Repertoires of tRNAs: the couplers of genomics and proteomics. Annu Rev. Cell Dev. Biol. 34, 239–264 (2018).
Lyabin, D. N. et al. YB-3 substitutes YB-1 in global mRNA binding. RNA Biol. 17, 487–499 (2020).
Anisimova, A. S. et al. Multifaceted deregulation of gene expression and protein synthesis with age. Proc. Natl Acad. Sci. USA 117, 15581–15590 (2020).
Christian, T., Evilia, C., Williams, S. & Hou, Y. M. Distinct origins of tRNA(m1G37) methyltransferase. J. Mol. Biol. 339, 707–719 (2004).
Bjork, G. R. et al. A primordial tRNA modification required for the evolution of life? EMBO J. 20, 231–239 (2001).
Takakura, M., Ishiguro, K., Akichika, S., Miyauchi, K. & Suzuki, T. Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat. Commun. 10, 5542 (2019).
Gromadski, K. B., Daviter, T. & Rodnina, M. V. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol. Cell 21, 369–377 (2006).
Gromadski, K. B. & Rodnina, M. V. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell 13, 191–200 (2004).
Maehigashi, T., Dunkle, J. A., Miles, S. J. & Dunham, C. M. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc. Natl Acad. Sci. USA 111, 12740–12745 (2014).
Ledoux, S., Olejniczak, M. & Uhlenbeck, O. C. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat. Struct. Mol. Biol. 16, 359–364 (2009).
Lyu, X. et al. Adaptation of codon usage to tRNA I34 modification controls translation kinetics and proteome landscape. PLoS Genet 16, e1008836 (2020).
Pernod, K. et al. The nature of the purine at position 34 in tRNAs of 4-codon boxes is correlated with nucleotides at positions 32 and 38 to maintain decoding fidelity. Nucleic Acids Res 48, 6170–6183 (2020).
Diwan, G. D. & Agashe, D. Wobbling Forth and drifting back: the evolutionary history and impact of bacterial tRNA modifications. Mol. Biol. Evol. 35, 2046–2059 (2018).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Lovejoy, A. F., Riordan, D. P. & Brown, P. O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014).
Zhang, W. & Pan, T. Pseudouridine RNA modification detection and quantification by RT-PCR. Methods 203, 1–4 (2022).
Khoddami, V. et al. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl Acad. Sci. USA 116, 6784–6789 (2019).
Dai, Q. et al. Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat. Biotechnol. 41, 344–354 (2023).
Carette, J. E. et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011).
Behrens, A. & Nedialkova, D. D. Experimental and computational workflow for the analysis of tRNA pools from eukaryotic cells by mim-tRNAseq. STAR Protoc. 3, 101579 (2022).
Masuda, I. & Hou, Y. M. Protocol to identify the core gene supported by an essential gene in E. coli bacteria using a genome-wide suppressor screen. STAR Protoc. 4, 102196 (2023).
Avcilar-Kucukgoze, I., Gamper, H., Hou, Y. M. & Kashina, A. Purification and use of tRNA for enzymatic post-translational addition of amino acids to proteins. STAR Protoc. 1, 100207 (2020).
Ammann, G., Berg, M., Dalwigk, J. F. & Kaiser, S. M. Pitfalls in RNA modification quantification using nucleoside mass spectrometry. Acc. Chem. Res 56, 3121–3131 (2023).
Dai, W. et al. Activity-based RNA-modifying enzyme probing reveals DUS3L-mediated dihydrouridylation. Nat. Chem. Biol. 17, 1178–1187 (2021).
Zhang, J., Kobert, K., Flouri, T. & Stamatakis, A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Acknowledgements
We thank Danny Nedialkova and Andrew Behrens for assistance with the computational tool kit associated with mim-tRNAseq. We thank Susan Ackerman and Mridu Kapur for providing tissues of mouse B6N and B6J, Lorraine Iacovitti and Andrew Gray for tissues of B6J, and Tao Pan, Satoshi Kimura, Zoya Ignatova, and Wendy Gilbert for discussion. This work was supported by NIH R35-GM149336 to J.P., NIH R01-GM132189 to R.K., MCB-2427729 to B.D.G., and NIH R35-GM134931 to Y.M.H.
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.N., H.G., and Y.M.H.; Methodology, Y.N., H.G., H.M., J.L., Z.S., E.Y., S.G., K.K., N.S.L.; J.A.P., R.K., B.D.G., and N.N.; Investigation, Y.N., H.G., H.M., S.M., J.L., Z.S., E.Y., S.G., K.K.; Software, Y.N., and Z.S.; Writing – original draft, Y.N., H.G., and Y.M.H.; Writing – review and editing, Y.N., H.G., E.Y., Z.S., and Y.M.H.; Supervision and funding acquisition, J.P., R.K., B.D.G., and Y.M.H.
Corresponding author
Ethics declarations
Competing interests
Y.N., H.G., H.M, S.M, J.L., N.S.L., J.A.P., R.K, B.D.G., and Y.M.H. declare no competing interests. Z.S., K.K., E.Y., S.G., and N.N. are employees of New England Biolabs, Inc., where Z.S. performed data analysis, K.K. and S.G. performed Illumina sequencing of tRNAseq libraries, and E.Y. and N.N. provided discussion. New England Biolabs is a manufacturer and vendor of molecular biology reagents, including Induro, Rnl2, T4 PNK, restriction enzymes, and several buffers used in this study. These authors declare no competing interests. The affiliation of Z.S., K.K., E.Y., S.G., and N.N. with New England Biolabs does not affect these authors’ impartiality, adherence to journal standards and policies, or objective data presentation, analysis, and interpretation.
Peer review
Peer review information
Nature Communications thanks Qi Chen, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nakano, Y., Gamper, H., McGuigan, H. et al. Genome-wide profiling of tRNA modifications by Induro-tRNAseq reveals coordinated changes. Nat Commun 16, 1047 (2025). https://doi.org/10.1038/s41467-025-56348-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56348-1
This article is cited by
-
Hidden in plain sight: illuminating the tRNA landscape by sequencing
Genome Biology (2026)
-
Prime editing-installed suppressor tRNAs for disease-agnostic genome editing
Nature (2025)