Abstract
Physiological adaptations to fasting enable humans to survive for prolonged periods without food and involve molecular pathways that may drive life-prolonging effects of dietary restriction in model organisms. Mobilization of fatty acids and glycerol from adipocyte lipid stores by canonical neutral lipases, including the rate limiting adipose triglyceride lipase (Pnpla2/ATGL), is critical to the adaptive fasting response. Here we discovered an alternative mechanism of lipolysis in adipocytes involving a lysosomal program. We functionally tested lysosomal lipolysis with pharmacological and genetic approaches in mice and in murine and human adipocyte and adipose tissue explant culture, establishing dependency on lysosomal acid lipase (LIPA/LAL) and the microphthalmia/transcription factor E (MiT/TFE) family. Our study establishes a model whereby the canonical pathway is critical for rapid lipolytic responses to adrenergic stimuli operative in the acute stage of fasting, while the alternative lysosomal pathway dominates with prolonged fasting.
Similar content being viewed by others
Introduction
The metabolic adaptation to fasting enables humans to survive for months without exogenous energy intake1,2. However, the biology of fasting may be relevant beyond starvation. Caloric restriction, including when achieved by intermittent fasting, prolongs life in model organisms ranging from yeast to mammals3. Yet, dysregulation of pathways crucial to survival with starvation are maladaptive in other contexts; indeed, whereas lipid stored in adipocytes is a critical energy source during fasting, inappropriate lipolysis in obesity drives lipotoxicity and insulin resistance in distant tissues such as the liver4. In short, the tuning of adipocyte lipid turnover to systemic energy balance is an important determinant of metabolic homeostasis.
The canonical view of how adipocytes mobilize lipid stores invokes the coordinated action of lipases, which release three fatty acid chains and one glycerol backbone from each triglyceride molecule5. Adipocyte triglyceride lipase (ATGL) cleaves the first fatty acid chain and is considered rate limiting, followed by hormone sensitive lipase (HSL) and monoacylglycerol lipase (MGL)6,7. The action of lipases at the surface of adipocyte lipid droplets is regulated by cAMP-dependent transduction of hormonal signaling and genetic targeting of canonical lipases attenuates lipolytic release of fatty acids in response to adrenergic stimuli6,8,9,10,11. Consistent with the broader importance of lipolysis to metabolic homeostasis, human mutations in genes encoding members of this canonical pathway, including LIPE (HSL) and PNLPA2 (ATGL), are associated with phenotypic manifestations that can include abnormal adiposity, systemic lipid derangements, and insulin resistance12,13.
The centrality of the canonical lipolysis pathway in adipocytes is called into question by several observations. First, residual adipose tissue lipolytic activity is detectable even when canonical lipases are genetically targeted6,9,14,15. Second, efflux of triglyceride-containing exosomes from adipocytes bypasses canonical lipases and can be scavenged and metabolized by resident macrophages16. Third, across fasting studies, genes encoding canonical lipases are not consistently upregulated in adipose tissue within a time-scale that can support the adaptive fasting response17,18,19,20. We previously conducted a longitudinal transcriptomics analysis of human subcutaneous adipose tissue during a rigorously controlled prolonged inpatient fast and found that lipolysis gene transcripts were unchanged or even declined with fasting, whereas fasting augmented transcripts related to lysosomes and lysosomal acid lipase (LIPA), the enzyme capable of digesting lipid at the low pH in lysosomes18. Such transcriptional data may not correlate with either the levels or the functionality of lipases known to be regulated by post-translational modifications21,22,23, and therefore provide rationale to revisit fasting lipolytic mechanisms and functionally test the hypothesis that an alternative lysosomal pathway is operative in adipocytes during fasting.
Here, we used genetic and pharmacological approaches in cultured adipocytes, adipose tissue explants, and murine models to test for a lysosomal role in adipose tissue lipolysis with fasting. In murine adipose tissue and adipocytes we discovered induction of a lysosomal program with fasting. Pharmacological inhibition of lysosomal lipolysis attenuated the fasting surge of lipolytic products into the circulation of fasting mice as did the inducible adipocyte-specific genetic targeting of the lysosomal lipase (Lipa/LAL), which was further dependent on a microphthalmia/transcription factor E (MiT/TFE)-LIPA regulatory axis. We also performed experiments in which we compared the dynamics of the canonical ATGL-dependent lipolysis during fasting to the lysosomal mechanism, finding distinct temporal roles with dependency on the canonical pathway for rapid lipolytic responses to adrenergic stimuli operative in the acute stage of fasting, while the alternative lysosomal pathway dominated with prolonged fasting.
Results
Prolonged fasting drives a lysosomal program in murine adipose tissue
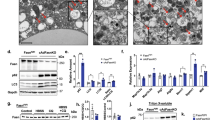
We first tested whether fasting activates a lysosomal program in murine adipose tissue. We used a 24-hour fasting protocol, which enabled timing the start and end of the fast in the morning just after the nocturnal period of maximal caloric intake in mice. This resulted in a reproducible increase in circulating lipolytic products in fasted 8-week-old mice, including non-esterified free fatty acids (NEFA) and glycerol (Fig. 1a). We performed immunoblotting of adipose tissue samples from fasted mice, and we detected augmentation of the lysosomal lipase (LAL) and members of the MiT/TFE transcription factor family, which are regulators of lysosomal biogenesis and function: TFEB, TFE3, and MITF (Fig. 1b). By contrast, the canonical rate limiting lipase, ATGL, was reduced in inguinal adipose (iWAT). The analyses of gonadal adipose tissue (gWAT) were generally less dramatic (Fig. 1b). These protein-level data demonstrating a preferential augmentation of lysosomal programs with prolonged fasting in murine adipose tissue are directionally consistent with the transcriptional signal previously observed in human adipose tissue18.
a Murine model of fasting-induced lipolysis. Mice were fasted for 24 h and refed for 6 h (C57Bl6 male n = 5; female mice n = 6 cont, 6 fast, 5 refed, 8-weeks-old). Statistical significance was assessed by one-way ANOVA/Tukey’s test. Left: male serum glycerol: *p = 0.03; male serum non-esterified fatty acids (NEFA): ****p < 0.0001. Right: female glycerol: ***p = 0.0001, **p = 0.007; NEFA: ****p < 0.0001. When present error bars indicate s.d.m. (a). b Inguinal (iWAT) and gonadal (gWAT) adipose tissue immunoblots for canonical lipase (ATGL), lysosomal transcriptional regulators (TFEB, TFE3, MITF), and the lysosomal lipase (LAL). The control, fasting, refeeding protocol was the same as in ‘a’. Note: tubulin controls run on different gels. c qPCR of adipocytes isolated from inguinal adipose tissue, including canonical lipolytic genes and lysosomal genes. Male n = 6 con, 5 fast, 6 refed; Female n = 6 con, 6 fast, 5 refed: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, two-way ANOVA/Dunnett’s test. The control, fasting, refeeding protocol was the same as in ‘a’. d Heat-map comparing isolated adipocytes (from C) to nutrient-restricted adipocyte cultures derived from primary adipocyte progenitor (AP) cells (n = 3 biological replicates) or 3T3L1 cells (n = 3 biological replicates). e. Whole mount immunofluorescence staining for marker of lysosomes (LAMP1) in adipose tissue. Arrow=putative LAMP1+ puncta indicating lysosomes. LD=lipid droplet. N = nucleus. Scale = 10 micron. N = 5 control, 7 fasted stained in this manner and used for blinded counting shown in ‘f’. f Blinded observer counted perinuclear LAMP1+ puncta in perilipin+ adipocytes: n = 5 control, 7 fasted; two-sided T-test; error bars S.D.M. g Perilipin levels were measured by ELISA in lysosome preparations from isolated adipocytes. Perilipin was undetectable in the lysosome preparations from fed mice; therefore the lower-limit of the assay (0.055 ng/mL) was used for the purpose of the graph. Each dot represents one mouse, n = 8, significance assessed by two-tailed t-test; error bars S.D.M.
Next, we performed qPCR analyses of lipolysis genes in adipocytes isolated from fed and fasted mice. Genes involved in lipogenesis and those encoding canonical lipases trended down with fasting in the adipocyte fraction, with Lipe reaching statistical significance. By contrast, fasting and refeeding resulted in the dynamic augmentation of MiT/TFE family members and genes involved in lysosomal function, including the lysosomal lipase (Lipa) (Fig. 1c, S1). For these analyses, we applied the standard approach to isolating adipocytes with sequential enzymatic digestion of adipose tissue, centrifugation, and collection of floating, buoyant adipocytes24. One consideration with this approach is whether transcriptional changes are attributable to contaminating cells from the stromal-vascular fraction, which include macrophages that also have the capacity to catabolize lipid16. Importantly, the changes we observed in the adipocyte fractions were not robustly seen in corresponding stromal-vascular fractions (Fig. S1). As an orthogonal approach, however, we also studied in vitro models. We generated adipocytes from 3T3L1 murine preadipocytes and by differentiation of primary adipocyte progenitor (AP) cells25. We then restricted nutrients and insulin in the standard adipocyte media, finding that the adipocyte cultures remained viable and released fatty acids and glycerol into the media consistent with lipolysis (Fig. S1). Shifts in the expression of canonical lipolytic genes and lysosomal genes were directionally consistent with primary adipocytes isolated from fasted mice relative to fed controls: there was a relative down-regulation of canonical lipolysis and lipogenesis genes and upregulation of lysosomal genes including lysosomal lipase, Lipa (Fig. 1d). The surge in lysosomal genes, including Tfeb, was not reflected in the stromal-vascular fraction with fasting although both Tfeb and Lipa increased with refeeding (Fig. S1A, C). The stromal-vascular fraction of adipose tissue is comprised of heterogenous cell-types including macrophages, which are known to be involved in adipose tissue lipid metabolism and may play a complementary role in responding to the dramatic lipid fluxes occurring with fasting/feeding transitions including those potentially mediated by adipocyte-derived exosomes16. These results therefore suggest activation of a lysosomal gene expression program in adipocytes with fasting, with the caveat that the in vitro nutrient restriction model does not fully reflect the complex physiology of organismal fasting.
To test whether the lysosomal gene program with fasting translates into augmented lysosomal activity in adipose tissue, we performed immunofluorescence microscopy of whole mount adipose tissue from fed and 24-hour fasted mice. A blinded observer counted LAMP1-positive puncta in the peri-nuclear region of adipocytes, the cytosolic compartment that forms a triangle-like shape, bordered by the plasma membrane, the nucleus, and the perilipin-positive margin of the dominant lipid droplet. Adipocytes from fasted mice displayed an increase in the average number of putative LAMP1-positive lysosomes (Fig. 1e, f, S1). Given that it can be challenging to definitively distinguish adipocyte nuclei from closely approximated stromal vascular cells by light microscopy, we sought to assess for an association between lysosomes and lipid droplets with an orthogonal method. We performed an ELISA for the lipid droplet protein perilipin in lysosomal preparations isolated from the adipocytes of fed and fasting mice. Whereas perilipin was generally below the limit of detectability in lysosome preparations from fed mice, we detected a surge in lysosomal-associated perilipin in adipocytes collected from fasting mice (Fig. 1g). Collectively, these data demonstrate augmentation of a lysosomal program in adipose tissue and adipocytes with prolonged fasting in mice.
Systemic pharmacological inhibition of lysosomal function attenuates fasting lipolysis
Having demonstrated evidence of increased lysosomal markers in adipose tissue with fasting, we first examined potential functional significance with pharmacologic lysosomal inhibition. We used two lysosomal inhibitors: bafilomycin, an inhibitor of lysosomal acidification, or lalistat2, an inhibitor of LAL. We included an ATGL inhibitor (atglistatin) group in parallel. We treated mice for three days with twice daily I.P. administration, timing a 24 h fast during the final day. Both lysosomal inhibitors attenuated the fasting surge in NEFA, whereas we detected no difference in mice treated with the ATGL inhibitor (Fig. S2). By contrast, when we administered inhibitors prior to isoproterenol—a classical lipolytic stimulus—atglistatin neutralized the NEFA surge (Fig. S2). Bafilomycin also partially attenuated isoproterenol-stimulated NEFA release, albeit not to the same degree as atglistatin (Fig. S2). It was also notable that the effect of lysosomal inhibitors was most evident in the NEFA measurements, with no significant effect noted in glycerol measurements.
Given the discrepancy in patterning between serum glycerol and NEFA lipolytic products and the possibility that systemic levels could reflect complex metabolism of lipids and/or lipolytic products in non-adipose tissues, we tested the inhibitors, in vitro, with adipose tissue explants (Fig. S2). The two lysosomal inhibitors neutralized NEFA and glycerol release in explants exposed to nutrient-restricted conditions, whereas atglistatin had no detectable inhibitory effect (Fig. S2). We also assessed the three drugs in the context of catecholamine exposure. Atglistatin inhibited catecholamine-stimulated lipolysis, whereas neither lysosomal inhibitor consistently modified the catecholamine effect (Fig. S2). The in vitro nutrient restriction model does not fully reflect the physiology of in vivo fasting and there is also concurrent dynamic modulation of autophagy pathways that may be distinct from the physiological regulation of lipolysis and therefore ex vivo lipolysis experiments must be interpreted with caution. These collective data, however, reaffirm the role of the canonical lipolytic pathway in rapid lipolysis induced by an adrenergic stimulus and identify a potential role for lysosome-dependent lipolysis with nutrient-restricted conditions, in vitro, or with prolonged fasting, in vivo.
A limited role for ATGL in the mobilization of lipid from adipocytes with prolonged fasting in mice
Given that systemic pharmacological inhibition of ATGL had no detectable effect on lipolysis metrics with prolonged fasting, we re-examined its functional role at the adipocyte level through genetic targeting of the ATGL gene, Pnlpa2. We used a previously published model, crossing Adiponectin-Cre (Adipoq-Cre) and Pnlpa2-floxed mice to achieve adipocyte-specific ATGL loss of function (KO=AdipoqCre+/+;Pnpla2 f/f)(Fig. 2a)14. We also revised our phenotyping approach to maximize statistical power, performing repeated blood sampling to assess within group changes in circulating lipolytic products in response first to isoproterenol and then a week later to a 24-hour fast (Fig. 2a). As with pharmacological inhibition (Fig. S2), adipocyte-specific targeting of ATGL neutralized isoproterenol-stimulated release of lipolytic products into circulation (Fig. 2b, c). However, we did not detect an inhibitory effect of ATGL loss of function on NEFA or glycerol with a 24-hour fast (Fig. 2c). We observed modest attenuation of fasting weight loss in male mice, but not in females (Fig. 2d).
a Schematic of adipocyte specific ATGL/Pnpla2 loss of function (KO) relative to control (FLOX), using mice that were 8–9 weeks old at study start. Paired blood draws were used to assess the lipolytic response to adrenergic stimulus (isoproterenol) and fasting. For panels d–h, data shown as mean, S.D.M. b Change in plasma NEFA with isoproterenol (ISO) or fast. Significance for genotype effect assessed by two-way ANOVA. FLOX male n = 5; KO male n = 7; FLOX female n = 5 with iso and n = 6 with fasting; KO female n = 7. c Change in plasma glycerol with isoproterenol (ISO) or fast. Significance for genotype effect assessed by two-way ANOVA. FLOX male n = 5; FLOX female n = 5; KO male n = 7; KO female n = 6. d Change in body weight assessed by two-tailed t-test: FLOX male n = 6; FLOX female n = 5; KO male n = 7; KO female n = 7. Significance assessed by two-tailed t-test. e Terminal plasma ketone levels. FLOX male n = 6; FLOX female n = 5; KO male n = 7; KO female n = 6. Significance assessed by two-tailed t-test. f Terminal plasma FGF21 levels. FLOX male n = 5; FLOX female n = 5; KO male n = 7; KO female n = 7. Significance assessed by two-tailed t-test. g Terminal plasma glucose levels. FLOX male n = 5; FLOX female n = 5; KO male n = 7; KO female n = 6. Significance assessed by two-tailed t-test. h qPCR of isolated inguinal (iWAT) and gonadal (gWAT) adipocytes for lipolysis and lysosome genes. FLOX n = 10 (male n = 5, female n = 5; KO n = 13 (male n = 7; female n = 6). Two-way ANOVA, **p = 0.008; ***p = 0.0002; ****p < 0.0001. i Immunoblots for iWAT and gWAT adipose tissue collected at the 24-hour fasting timepoint. Note: tubulin controls run on different gels.
We next examined additional metrics of the critical transition to lipid metabolism, including systemic ketones and the prototypical fasting hormone, FGF21. We observed no difference in ketones at the 24-hour time-point and FGF21 was modestly attenuated in male mice (Fig. 2e, f). Fasting glucose was similar between the two groups (Fig. 2g). With qPCR analyses of the adipocyte fractions, we detected increased transcription of Mitf and a trend towards increased Tfeb (Fig. 2h). Augmentation of lysosome-related factors was more evident at the protein level in adipose tissue immunoblots, including TFEB and LAL, raising the question of whether there was compensatory activation of a lysosomal program with targeting of the rate limiting canonical lipase (Fig. 2i). It is possible that this compensatory lysosomal program was more rapid in female mice given no apparent effect of ATGL loss of function on either weight loss or FGF21 production in female mice.
Recognizing potential non-adipose tissue sources of systemic NEFA levels, we next focused our testing of lipolytic responses to ex vivo adipose tissue explant culture (Fig. S3). We studied tissues from adipocyte-specific ATGL loss of function (KO=AdipoqCre+/+;Pnpla2 f/f) and floxed controls. The response to 24 h of nutrient restriction was statistically indistinguishable between control tissue and ATGL loss of function tissue (Fig. S3). With short-term catecholamine stimulation, however, adipocyte specific genetic targeting of ATGL neutralized NEFA and glycerol release, consistent with inhibition of stimulated lipolysis (Fig. S3). One question is whether the in vitro nutrient restriction protocol was associated with induction of autophagy. While we observed some evidence of dynamic regulation of autophagy genes with nutrient restriction (Fig. S3), upregulation tended to be associated with restoration of nutrients rather than the removal of nutrients and the degree of change was generally not dissimilar from that observed in adipocytes collected from fasting mice (Fig. S3). Nonetheless, these data demonstrating release of lipolytic products from adipose tissue despite genetic neutralization of ATGL/Pnpla2 in adipocytes suggests possible functional involvement of non-canonical pathways.
Overall, the results from adipocyte-specific genetic targeting of ATGL were consistent with the non-targeted, systemic pharmacologic experiments in demonstrating no detectable modulation of direct lipolytic products with prolonged fasting, even though some indirect metrics such as FGF21 release and weight loss were modestly attenuated with ATGL loss of function in male mice. In the absence of an obvious technical explanation (e.g. sex differences in Pnpla2 recombination), it is possible that female mice exhibited more rapid fasting activation of lysosomal mechanisms, obscuring effects of ATGL loss of function at prolonged fasting timepoints. Therefore, our results reinforced the role of the canonical lipolytic mechanism in response to a short-term adrenergic stimulus, while raising the question of whether ATGL is operative during earlier phases of fasting.
Targeting transcriptional regulators of lysosomal function attenuates fasting lipolysis
Given the limitations of targeting lipolysis pathways with pharmacological methods, which are not cell-type specific and have potential off-target effects, we next sought to manipulate lysosomal pathways in adipocytes with genetic methods, first focusing on candidate upstream transcriptional regulators of lysosomal lipolysis. To facilitate loss of function testing of a panel of candidate genes, we first performed experiments in vitro in 3T3L1 cells, which are amenable to both viral transgenesis and efficiently differentiate into adipocytes. Recognizing that MiT/TFE transcription factors might also regulate adipocyte differentiation, we used a doxycycline-inducible short hairpin (sh)-RNA system to induce gene knockdown after differentiation thereby minimizing confounding developmental effects (Fig. S4). We tested inducible knockdown of Pnlpa2 (ATGL), Lipa, and MiT/TFE transcription factors in parallel. Of the MiT/TFE transcription factors, we examined Tfeb, Mitf, and Tfe3, all of which were dynamically regulated at the mRNA and/or protein level in adipocytes from fasted mice or adipose tissue respectively, but not Tfec as it was not consistently expressed in 3T3L1 cells. Knockdown of Lipa and Tfeb demonstrated the most reproducible effect on NEFA release with exposure to nutrient-restricted conditions (Fig. S4). Only with the targeting of Tfeb was statistical significance reached in the glycerol assay, although trends were similar for the other MiT/TFE factors and Lipa (Fig. S4). Targeting of Tfeb also resulted in the strongest negative regulation of Lipa as assessed by qPCR (Fig. S4). At the protein level, targeting of Tfeb and Mitf had the strongest attenuation of LAL levels as measured by immunoblot (Fig. S4). As with pharmacological or genetic targeting of ATGL in vivo (Fig. S2), knockdown of Pnlpa2 in 3T3L1-derived cells did not modify release of lipolytic products under nutrient-restricted conditions. These data demonstrate that genetic targeting of Lipa or the MiT/TFE transcription factors in 3T3L1 derived adipocytes under conditions of nutrient restriction inhibits lipolysis.
Primary adipocyte progenitor cells are proliferative in ex vivo culture and are also amenable to viral transgenesis25,26. Therefore, we used a similar approach to achieve inducible knock down of lysosomal genes after adipogenic differentiation (Fig. S4). Targeting of Tfeb again had the most consistent inhibitory effect on NEFA release with nutrient restriction, although there was a directionally consistent trend for each of the MiT/TFE factors (Fig. S4). Assaying of the glycerol surge revealed consistent, though not statistically significant, trends in alignment with the NEFA result (Fig. S4). Targeting of both Tfeb and Tfe3 attenuated the fasting surge in Lipa expression by qPCR (Fig. S4). Collectively, our data in primary AP-derived adipocytes demonstrated overlapping functionality of multiple MiT/TFE factors in the regulation of Lipa expression and lipolysis with nutrient restriction, although Tfeb targeting resulted in the strongest and most consistent effect.
The profound inhibitory effect of targeting MiT/TFE factors with nutrient restriction in adipocyte culture provided an orthogonal strategy to target lysosomes in vivo, experiments of particular importance given that in vitro nutrient restriction does not replicate the integrated physiology of fasting. Amongst the MiT/TFE factors, we prioritized TFEB-targeting based on a consistent and potent effect on fasting-induced lipolysis with in vitro loss of function (Fig. S4), its dynamic regulation with fasting in adipose tissue by immunoblot (Fig. 1b), and precedent for TFEB regulation of lipid catabolism in other tissues27. We used the previously published Tfeb-floxed mouse and crossed it with the Adipoq-CreER mouse to facilitate adipocyte-specific loss of function in a temporally-controlled fashion (Fig. 3a)28. Adipoq+/+;Tfebf/f mice or floxed littermate controls were administered a standard tamoxifen protocol by I.P. injection, followed by sequential challenge with isoproterenol and a 24-hour fast, separated by a one-week recovery period. TFEB loss of function attenuated the NEFA and glycerol surge in serum after 24 h of fasting; however, isoproterenol-stimulated lipolysis was not attenuated and if anything, slightly augmented (Fig. 3b, c). Consistent with disruption of a TFEB-LIPA regulatory axis, we observed attenuation of both factors at mRNA level in isolated adipocytes (Fig. 3d) and at the protein level in adipose tissue (Fig. 3e). Residual TFEB in these samples may be due to signal from non-adipocyte stromal-vascular cells. There was no corresponding reduction in ATGL signal as might be expected if TFEB regulated both Lipa and Pnpla2, and at the mRNA level there was evidence of augmented canonical pathway gene expression in adipocytes, including Pnpla2 and Mggl, suggesting a possible compensatory effect.
a Schematic of tamoxifen-inducible adipocyte-specific Tfeb loss of function (KO) relative to controls (FLOX), using mice that were 8 weeks old at study start. Paired blood draws were used to assess the lipolytic response to adrenergic stimulus (isoproterenol) and fasting. For panels d and f–j, data show mean, S.D.M. b. Change in plasma non-esterified fatty acids (NEFA) with isoproterenol (ISO) or fast: FLOX n = 9 (6 male, 3 female); KO n = 13 (7 male, 6 female). Two-way ANOVA to assess genotype effect. c Change in plasma glycerol with isoproterenol (ISO) or fast. FLOX n = 9 (6 male, 3 female); KO n = 13 (7 male, 6 female). ****p < 0.0001, two-way ANOVA to assess genotype effect. d qPCR analyses of isolated adipocytes in control mice (FLOX) or adipocyte specific Tfeb loss of function (KO) (n = 9 FLOX; n = 13 KO). Two-way ANOVA to assess genotype effect. e Immunoblots of gonadal adipose tissues (gWAT) for LAL, TFEB, and ATGL collected at the 24-hour fasting timepoint. Note: tubulin control was run on a different gel. f qPCR analyses of glucose metabolism genes in isolated adipocytes from control mice (FLOX) or adipocyte specific Tfeb loss of function (KO) mice (n = 9 FLOX; n = 13 KO). Two-way ANOVA to assess genotype effect, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. g Terminal plasma glucose in control mice (FLOX) or adipocyte specific Tfeb loss of function (KO). FLOX n = 9 (6 male, 3 female); KO n = 13 (7 male, 6 female). Significance assessed by two-tailed t-test. h Terminal plasma ketones in control mice (FLOX) or adipocyte specific Tfeb loss of function (KO). FLOX n = 9 (6 male, 3 female); KO n = 13 (7 male, 6 female). Significance assessed by two-tailed t-test. i Change in body weight. FLOX n = 9 (6 male, 3 female); KO n = 13 (7 male, 6 female). Significance assessed by two-tailed t-test. j Terminal plasma fibroblast growth factor 21 (FGF21) levels measured by ELISA: FLOX n = 9 (6 male, 3 female). Significance assessed by two-tailed t-test.
Diverse metabolic processes are often inter-connected, particularly in the context of fasting where there is an orchestrated transition from glucose to lipid metabolism, providing rationale to also examine glucose metabolism and related pathways. We first moved beyond our focus on lipolysis and lipid metabolism genes (Fig. 3d) and examined a panel of glucose metabolic genes in adipocytes from the TFEB loss of function model (Fig. 3f). At the transcript level, we observed differential expression of several genes including Srebp2, Foxo1, Chrebp, and genes encoding glucose transporters, most evident in inguinal adipocytes. If TFEB was a direct transcriptional driver of these gene programs in adipocytes, one would expect their downregulation with TFEB loss of function. However, many of the differentially regulated genes were higher relative to control, which could be consistent with compensatory responses. Indeed, if the glucose metabolism genes were persistently active due to a delayed transition to lipid metabolism, we reasoned that there would be physiological evidence of persistent reliance on glucose metabolism at the systemic level. Indeed, adipocyte-specific TFEB loss of function resulted in lower systemic glucose levels with fasting (Fig. 3g). Since flux of adipocyte-derived fatty acids is a critical source of substrate for ketogenesis, we measured ketones, finding reduced levels in both males and females (Fig. 3h). TFEB loss of function also resulted in attenuated body weight loss (Fig. 3I) and reduced plasma FGF21 in male mice (Fig. 3J), though this protein effect was not reflected by qPCR analyses of the liver (Fig. S4). By contrast, we did observe a reduction in liver expression of Pparg and Hmgsc1, both of which are involved in lipid metabolism (Fig. S4). Despite dynamic changes in liver transcripts in response to adipocyte-specific Tfeb targeting, we did not detect compensatory changes in expression of Tfeb or lipolytic genes in the local stromal-vascular fraction of adipose tissue (Fig. S4). Therefore, through in vitro and in vivo genetic targeting of MiT/TFE transcription factors, we provide orthogonal data implicating lysosomes in the lipolytic mobilization of adipocyte lipid stores and in the systemic transition from glucose to lipid metabolism with fasting. As with targeting of any transcription factor, there are likely additional genes beyond those involved in lysosomal biogenesis and function that are either directly part of the TFEB-dependent transcriptional program or indirectly modulated. While we did not find evidence for concomitant ATGL loss of function with targeting of Tfeb as might be expected if TFEB was directly regulating Pnpla2, this nonetheless provided rationale for additional experiments focused directly on Lipa.
Temporal transition from ATGL-dependent to LAL-dependent adipocyte lipolysis with fasting
Our data implicating lysosomal lipolysis in fasting was collected using a 24-hour fasting model, which represents a prolonged fast in mice. Three questions are raised by the collective data implicating a lysosomal program in fasting adipocyte lipolysis: (1) is the lysosomal program operative at stages of the adaptive fasting response earlier than the 24-hour timepoint, (2) given prior evidence of a role for ATGL in fasting lipolysis, is an ATGL loss of function effect detectable at earlier stages in adaptive fasting, and (3) would genetic targeting of the specific lysosomal lipase (LAL) phenocopy adipocyte-specific targeting of TFEB, the putative master regulatory of lysosomal function? To investigate these questions, we first performed immuno-blots in adipose tissues collected from fasting mice, but at earlier timepoints (Fig. S5). In perigonadal and inguinal AT, the ATGL signal was progressively attenuated between 4 and 12 h of fasting, with detection of LAL increasing between 8 and 12 h of fasting. No clear pattern of fasting dependency was observed in brown adipose tissue (Fig. S5). This could indicate differential regulation of lysosomal programs with fasting in BAT reflecting the distinct physiological roles of BAT and WAT. Next, we repeated our pharmacologic inhibition protocol, administering vehicle, atglistatin, or lalistat2 to male mice prior to and during fasting, but focused instead on measurement of blood NEFA and glycerol at earlier timepoints (Fig. S5). We found attenuation of lipolytic products in blood with ATGL inhibition at baseline and after 4 h of fasting, an effect that was attenuated by 12 h of fasting. By contrast, we observed inhibition of lipolytic metrics at the 12-hour timepoint with pharmacologic targeting of LIPA. We further explored the LIPA role, finding lalistat2 dependent attenuation of lipolytic products in both control and adipocyte-specific ATGL knockout mice, suggesting a role for LIPA independent of ATGL. In this experiment, we also noted a trend toward attenuation of lipolytic metrics with ATGL loss of function after 8 h of fasting, an effect that was lost by the 24-hour timepoint.
We next used these time course data to inform design of an adipocyte-targeted genetic experiment, comparing ATLG and LIPA targeting. We generated two new mouse models by crossing the inducible adipocyte driver (Adipoq-CreER) with floxed Pnpla2 (ATGL KO) mice and with floxed Lipa (LAL KO) mice. We used tamoxifen to induce recombination prior to a sequential protocol of isoproterenol, one-week recovery, and then fasting (Fig. 4a). Because tamoxifen itself has been shown to have context-dependent metabolic effects29, we examined the effect of tamoxifen on lipolytic metrics, finding that fasting lipolysis was preserved after the 5-day tamoxifen protocol (Fig. S6). To further control for potential confounding effects of tamoxifen, both control and experimental mice used in inducible loss of function experiments were administered an identical tamoxifen protocol. Based on our time course data (Fig. S5), we selected an additional 8-hour intermediate timepoint after fasting onset for blood collection. Like prior genetic and pharmacological targeting of Pnpla2/ATGL, we observed marked attenuation of isoproterenol-stimulated lipolysis (Fig. 4b). With fasting, there was a detectable inhibitory effect at the 8-hour timepoint, which was lost by 24 h (Fig. 4b). By contrast, targeting of Lipa did not attenuate isoproterenol-mediated lipolysis and in the case of glycerol, there was a small but significant increase in the circulating surge with Lipa targeting (Fig. 4c). At the 8-hour fasting time-point, the NEFA surge was reduced with Lipa loss of function and by 24 h of fasting, significant reductions in both the glycerol and NEFA surge were evident. These results suggest that ATGL is functional during an acute adrenergic stimulus and in the early phase of fasting, but that there is a transition within a few hours of fasting in the mouse where a LAL-dependent mechanism becomes dominant.
a ATGL loss of function (KO) by targeting Pnpla2 (b, d, e, h,i, l,m, p); LAL loss of function (KO) by targeting Lipa (c, f,g, j,k, n,o, q); each shown relative to controls (FLOX). For panels b–o, data shown as mean, S.D.M. b Isoproterenol (ISO)-stimulated non-esterified fatty acids (NEFA) (left) and glycerol (right). FLOX n = 12 (8 male, 4 female); KO n = 15 (8 male, 7 female). For b–g: two-way ANOVA, genotype effect. c ISO-stimulated NEFA (left) and glycerol (right). FLOX n = 19 (9 male, 10 female); KO n = 18 (10 male, 8 female). d Body weight change with fasting. FLOX n = 12 (8 male, 4 female); KO n = 13 (8 male, 5 female). e Plasma glucose change with fasting. FLOX n = 12 (8 male, 4 female); KO n = 13 (8 male, 5 female). f Body weight change with fasting. FLOX n = 19 (9 male, 10 female); KO n = 18 (10 male, 8 female). g Plasma glucose change with fasting. FLOX n = 19 (9 male, 10 female); KO n = 18 (10 male, 8 female). h Terminal plasma ketones. FLOX n = 12 (8 male, 4 female); KO n = 13 (8 male, 5 female). For h–k, significance assessed by two-tailed t-test. i Terminal liver triglycerides. FLOX n = 12 (8 male, 4 female); KO n = 13 (8 male, 5 female). j Terminal plasma ketones. FLOX n = 19 (9 male, 10 female); KO n = 18 (10 male, 8 female). k Terminal liver triglycerides. FLOX n = 19 (9 male, 10 female); KO n = 18 (10 male, 8 female). l qPCR of inguinal (iWAT) adipocytes from ATGL KO (n = 12) or control (n = 13). For l–o Two-way ANOVA: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. m qPCR of gonadal (gWAT) adipocytes: ATGL KO (n = 12) or FLOX (n = 13). n qPCR of inguinal adipocytes: LIPA KO (n = 12) or FLOX (n = 12). o qPCR of gonadal adipocytes: LIPA KO (n = 12) or FLOX (n = 12). p Immunoblot of inguinal and gonadal adipose tissues from control or adipocyte-specific ATGL loss of function (KO) mice; 24-hour fasting timepoint. q Immunoblot of inguinal and gonadal adipose tissues from control or LIPA loss of function (KO) mice; 24-hour fasting timepoint. Note for p,q: tubulin controls run on different gels.
We next examined additional surrogate variables of relevance to the adaptive fasting response, focusing on the terminal 24 h fasting timepoint when we collected a full complement of tissue samples. With inducible adipocyte-targeted ATGL loss of function, we did not observe modulation of fasting weight loss or glucose reduction (Fig. 4d, e); whereas targeting of LAL attenuated loss of body weight and accentuated the glucose reduction (Fig. 4f, g). A similar divergence was observed for ketone levels in blood and liver triglycerides. With ATGL targeting, levels were either no different or increased relative to control mice at 24 h (Fig. 4h, i); whereas with Lipa knockout, ketone production and liver triglycerides were reduced consistent with attenuated flux of lipid to the liver (Fig. 4j, k).
With constitutive targeting of ATGL in adipocytes (Fig. 2j, k), we observed an increase in TFEB and LIPA protein levels in adipose tissue. In the current inducible ATGL loss of function experiment, the augmentation of ketone production (male and female, Fig. 4h) and liver triglycerides (female, Fig. 4l) also suggested the possibility of a similar compensatory lysosomal response. Therefore, we again examined the expression of canonical and lysosomal lipolysis genes by qPCR in isolated adipocytes and at the protein level in adipose tissue by immuno-blot, first finding evidence for successful targeting of the two genes (Fig. 4j–q). In addition, Lipa/LAL levels were augmented in adipose samples collected from mice with inducible ATGL loss of function, particularly in the perigonadal depot. Conversely, Lipa/LAL loss of function resulted in augmentation of ATGL in adipose tissues, although this effect was not seen at the mRNA level in isolated adipocytes. We also considered whether there might be lipases in addition to Lipa/LAL induced in the absence of ATGL. Indeed, in adipocytes from the ATGL loss of function model, we detected increased transcriptional activity of several genes linked to lipid digestion (Fig. S7). Several were also dynamically regulated in adipocytes from wild-type mice subjected to fasting and refeeding, although unlike Lipa, these genes appeared to be primarily induced after refeeding (Fig. S7). These collective data reinforce the role of canonical ATGL-dependent lipolysis in adipocytes with acute adrenergic stimulus and early fasting, during which catecholamines are known to exhibit a modest surge in circulation30,31,32,33. However, as fasting progresses beyond a few hours in mice, there is a transition with increasing dependency on the lysosomal lipase. None of the genetic interventions appeared to completely neutralize the fasting surge in lipolytic products. This could be consistent with lipolytic activity by non-adipocytes or reflect the action of lipases other than ATGL and LAL in adipocytes.
Lysosome-dependent lipolysis is operative in human adipose tissue with fasting
Our a priori hypothesis of fasting-induced lysosomal lipolysis in adipose tissue emerged from a longitudinal transcriptomic analysis of human adipose tissue during an inpatient 10-day fast where we highlighted transcript level data for lysosomal genes, including LIPA and LAMP118. The dynamic fasting changes in MiT/TFE transcription factors in murine adipose tissue and adipocytes provided rationale to re-examine the human RNA-seq data-set for corollary changes in human MiT/TFE factors. Two of the four factors demonstrated dynamic and significant modulation with fasting: MITF and TFEC (Fig. 5a). Moreover, the dynamic change in MITF positively predicted change in LIPA, with a non-significant, but directionally consistent trend for a TFEC-LIPA correlation (Fig. 5b, c). These data suggest a possible role for MiT/TFE factors in the human fasting response in adipose tissue.
a MiT/TFE factor fold change relative to day 0 from human transcriptomics analyses of subcutaneous adipose tissue during inpatient 10 day fast (n = 7). Differential expression using a linear model the likelihood ratio test against a reduced model that did not include the time factor (∼1) and with Benjamin-Hochberg multiple-test correction. b Two-sided Spearman correlation between TFEC fold change and LIPA fold change data-set in A (n = 7). c Two-sided Spearman correlation between MITF fold change and LIPA fold change data-set in A (n = 7). d Doxycycline-inducible targeting of lipolysis genes in adipocytes derived from human SGBS preadipocytes and subjected to nutrient restriction relative to scramble controls (shSCR). The dark gray bars with associated dashed lines denote non-nutrient restricted controls. One-way ANOVA/Dunnett’s. Each dot indicates the mean of technical replicates from n = 3 independent biological replicate experiments, and expressed as mean, S.D.M. NEFA=non-esterified fatty acids. e LIPA assessed by qPCR for knockdown experiments in F. Significance assessed with Friedman’s/Dunnett’s tests. Each dot indicates the mean of technical replicates from n = 3 independent biological replicate experiments, and expressed as mean, S.D.M. f Human adipose tissue explant culture and lipolytic response to nutrient restriction. Left=glycerol; right=NEFA. Pharmacologic inhibitors were used to assess lysosomal-dependent lipolysis (Bafilomycin, Lalistat2). Each dot indicates the mean of technical replicates from n = 4 independent biological replicate experiments, and expressed as mean, S.D.M. One-way ANOVA/Dunnett’s (glycerol) and Friedman’s/Dunnett’s tests (NEFA). Con=control. g Human adipose tissue explant culture and lipolytic response to in vitro adrenergic stimulus (catecholamines): left=glycerol; right=NEFA. Pharmacologic inhibitors were used to assess lysosomal-dependent lipolysis (Bafilomycin, Lalistat2). Each dot indicates the mean of technical replicates from n = 4 independent biological replicate experiments, and expressed as mean, S.D.M. Friedman’s/Dunnett’s tests. Con=control.
We next used human SGBS cells, which are amenable to viral transgenesis, as an in vitro source of human adipocytes34,35. This line was derived from the adipose tissue of an infant with Simpson-Golabi-Behmel Syndrome and there is precedent for its use in the modeling of human adipocyte biology36. We followed a similar protocol as used in murine 3T3L1 and AP cells, introducing doxycycline-inducible, shRNA constructs via lentivirus to proliferating SGBS cells prior to adipogenic differentiation. Targeting of LIPA, but not PNLPA2 (ATGL), attenuated NEFA release with nutrient restriction, as observed in murine cells, whereas targeting of the MiT/TFE factors had variable inhibitory effects on lipolytic metrics (Fig. 5d). Unlike in murine cells, TFEC was expressed, and its targeting resulted in attenuation of glycerol release. Targeting of TFEC also attenuated LIPA expression, even though the merger of biological replicates revealed a statistical trend after correction for multiple hypothesis testing (Fig. 5e; p = 0.14).
We also examined lysosomal lipolysis in human adipose tissue explant culture, analogous to our murine adipose tissue explant experiments (Figs. 5f, g, S8). We used subcutaneous adipose samples obtained from patients undergoing surgical abdominoplasty. For each donor, we performed three technical replicates for each culture condition, and tested the capacity of pharmacological inhibitors to attenuate NEFA and glycerol release with nutrient restriction. Fig. 5f, g show mean normalized responses for each of the participants and Fig. S8 shows individual participant level data. Both lysosomal inhibitors attenuated the release of lipolytic products with nutrient restriction. These data in human cells and human adipose tissue suggest that lysosomal lipolysis involving an MiT/TFE-LIPA axis may also be operative in humans, although the strength of this conclusion must be tempered by the limitations inherent to the in vitro model system where restriction of insulin and nutrients in the culture media does not recapitulate the complex physiology of organismal fasting. Moreover, while the core lysosomal mechanism may be conserved, there was also mouse-human divergence in functionality of the MiT/TFE factors, with evidence of more important roles for TFEC and MITF in human cells and tissues.
Network medicine identifies a core human fasting network associated with aging genes and diseases of aging
Decades of work in model organisms demonstrate that various forms of dietary restriction extend lifespan and suggests that fasting physiology may be of broader relevance to human health3. We next sought to further contextualize the relevance of the adaptive fasting response in adipose tissue. We hypothesized that dynamic molecular changes in adipose tissue would interact with pathways of relevance to aging but that this relationship is likely complex and, thus, reductionist analyses may not capture key insights. Therefore, we leveraged a network medicine approach to test for potential connections between fasting, aging pathways, and pathways involved in diseases of aging.
We focused on our previously published adipose tissue fasting human transcriptome, consisting of 5077 differentially expressed transcripts with fasting18; however, we applied a higher level of statistical stringency yielding a subset of 946 transcripts after Bonferroni correction. This gene set was mapped to the consolidated human protein-protein interactome (PPI), an in silico network based on experimentally validated interactions37. Of 946 genes, 732 (77%) were present in the human interactome. Unmapped transcripts were either non-coding or coded for proteins for which a PPI has not been demonstrated. We first tested whether the fasting transcriptome forms a discrete module (i.e., subnetwork) within the larger human interactome. The 732 mapped proteins exhibited dense interconnectivity (419 nodes and 596 PPIs) that could not be explained by random chance (p = 0.0012) and therefore is consistent with a discrete subnetwork or functional module (Figs. 6a, S9).
a Differentially expressed (DE) transcripts from a longitudinal human fasting study were mapped to the consolidated human interactome, which includes >230,000 physical protein-protein interactions (PPIs). DE fasting transcripts formed a statistically distinct subnetwork (module) in the interactome. b Interconnectivity of the fasting genes and aging genes from the Aging Atlas that mapped to the interactome. c Betweenness centrality (BC), a measure of importance in information transfer across a network based on shortest paths was used to identify candidate regulators of the interaction between the fasting and aging phenotypes. This unbiased in silico analysis identified MITF as a critical node. d Network proximity between the fasting module and aging disease modules or cardiometabolic disease modules. The node size is proportional to the number genes/proteins in the module. The width of the edge is proportional to proximity significance.
We next determined if the fasting module included genes that are relevant to endophenotypes associated with aging pathobiology. Indeed, we observed significant overlap between the fasting module and genes related to inflammation (p = 2.54e-10), reactive oxygen species (p = 7.05e-4), and genomic instability (p = 0.002) (Table S1). We used aging genes from the Aging Atlas as a second resource to validate the relevance of fasting module to aging38. Of 394 aging-related genes, 387 (98%) mapped to the interactome, which significantly overlapped with the 732 fasting genes that mapped to the human interactome (p = 5.574e-5). More importantly, however, we found a high degree of interconnectivity between the aging genes and the fasting genes in the human PPI network (Fig. 6b, p = 1.0e-16).
We used betweenness centrality (BC) to rank candidate mediators of the interaction between fasting and aging phenotypes in silico, an approach previously demonstrated to discover functionally important regulators39. Numerous known factors of importance ranked highly based on BC scoring, including the cell cycle/senescence regulator CDK1, oncogenic proteins RB1 and RET, and the insulin/insulin like growth factor (IGF)1 effector IRS1 (Table S2). Importantly, however, this analysis also ranked MITF highly, a MiT/TFE family member that regulated fasting lipolysis in human adipocytes (Fig. 6c).
Having established a new fasting module in the human interactome inclusive of differentially expressed transcripts and functional PPI of relevance to aging pathobiology, we next tested for proximity between the fasting module and aging-related disease modules in the human interactome. We considered disease modules for cardiometabolic diseases, for which adipose tissue and/or lipid metabolism are known to play an important role. We also included an additional swath of diseases of aging. We found proximity in the interactome between the fasting module and cardiometabolic disease modules, including type 2 diabetes (1.31, p = 0.01), non-alcoholic steatohepatitis (1.26, p = 3.68e-07), coronary artery disease (1.33, p = 1.23e-07), and lipodystrophy (1.21, p = 1.25e-09) (Table S3). However, additional aging diseases demonstrated a similarly high degree of network proximity to the fasting module, including Alzheimer’s disease (1.32, p = 1.56e-08), breast (p = 2.43e-12) and endometrial (p = 2.49e-12) neoplasms, and progeria (p = 3.87e-5) (Table S4). Lastly, we considered whether the targets of drugs that have been considered as potential anti-aging therapeutics demonstrated network proximity to the fasting network module (Table S5). The targets of dasatinib and quercetin, which have been advanced as senolytic agents and the mTor inhibitor rapamycin demonstrated network proximity to the fasting module, whereas metformin did not (p = 0.32). These collective network medicine analyses demonstrate that fasting drives a discrete functional program in human adipose tissue that is highly interconnected with aging biology, diseases of aging, and candidate anti-aging drugs and that the MiT/TFE factor, MITF, is a lead candidate mediator of the fasting-aging axis. This orthogonal and unbiased discovery underscores the potential relevance of the MiT/TFE family of lysosomal regulators and provides rationale for future study of MiT/TFE-mediated fasting responses in aging disease models. Moreover, the identification of potential functional links between fasting networks in adipose tissue and diseases ranging from Alzheimer’s disease to cancer, suggests that the relevance of this work may extend beyond cardiometabolic diseases already known to be tightly linked to energy balance and adipose tissue biology.
Discussion
The canonical view of lipolysis in adipocytes invokes adipocyte lipases including the rate-limiting action of ATGL5,6,11,14,40. In this study, orthogonal evidence points to an alternative lysosomal mechanism of fasting-induced lipolysis: (1) lysosomal genes and proteins were dynamically regulated in the adipose tissue and adipocytes of fasting mice and humans; (2) targeting of lysosomes with two different pharmacological inhibitors, though not specific for adipocytes, attenuated fasting lipolysis in mice; (3) genetic targeting of MiT/TFE transcription factors that regulate lysosomal biogenesis and function reduced expression of the lysosomal lipase (LAL) and neutralized fasting lipolysis metrics in cultured mouse and human adipocytes under nutrient restricted conditions and in fasting mice; (4) in a head-to-head comparison of inducible, adipocyte-targeted loss of function of the canonical ATGL enzyme versus the lysosomal lipase, LAL, we observed a transition from ATGL to LAL dependency as fasting progressed beyond a few hours in mice. As such, we propose that an alternative lysosomal pathway is a critical mechanism of adipocyte lipolysis with fasting.
One question raised by our study is how can a new lipolysis pathway in adipocytes be reconciled with prior studies of the canonical lipolysis pathway? First, foundational work characterizing ATGL-dependent lipolysis was largely conducted with adrenergic stimuli as proxy for physiological stressors6,11,41,42. Similarly, pharmacological evidence against a role for lysosomes in adipocyte lipolysis also relied on the adrenergic stimulus model43. However, the increase in catecholamines with fasting is modest (less than two-fold)30,31,32,33 and therefore the quantitative role of adrenergic signaling to fasting lipolysis may have been over-estimated. Second, most studies of ATGL functionality have utilized constitutive loss of function approaches, which risks confounding by basal differences in adipose tissue and/or whole-body metabolic function, including differences in basal lipolysis, early fasting lipolysis (4–5 h)6,19,44,45, and fed-state metabolic cage measurements14. In studies that examined fasting, single timepoint comparisons between wild-type and knockout mice has been the norm11,46,47. Without repeated within-animal measurements, it can be difficult to distinguish the true fasting delta from basal differences. Third, prior studies are not uniform in demonstrating the supremacy of ATGL-dependent lipolysis. ATGL transgenic gain of function did not augment fasting lipolysis48. Conversely, with ATGL loss of function driven by Fabp4-Cre where repeated measures were employed, the NEFA surge between early (5 h) and late (48 h) was similar in both wild-type mice and ATGL KO (both approximately 0.25 mM delta)19. A small study of human patients with genetic ATGL loss of function suggests attenuation of lipolysis with adrenergic stimulus, but no effect with fasting49. Therefore, we propose that literature predating our study already suggested the possibility of contributions from non-canonical lipases, as shown here with Lipa/LAL, and perhaps extending to additional lipases.
A striking theme emerged across experimental models of the temporal relationship between the canonical and the alternative lysosomal lipolytic pathways. Our study reaffirmed the critical role of ATGL in the rapid lipolytic response to an adrenergic stimulus and in the early stage of fasting where modest increases in circulating catecholamines are evident6,14,19,41. By contrast, the adaptive fasting response proceeds through several stages, including depletion of glycogen stores, a surge in gluconeogenesis to support glucose metabolism, and finally lipolytic release of adipose lipid stores to provide substrate for ketogenesis by the liver or for direct oxidation by tissues with the requisite machinery1,50. It is this final adipocyte-dependent stage that marks the transition to lipid metabolism where sustained adipocyte lipolysis is critical, and this final transition is also crucial for the long-term survival of humans during fasting. Even though there is temporal overlap in the adaptive stages of fasting51, the time-scale of the transition to lipid metabolism is considerably longer than what is required of an adrenergic response. Therefore, we speculate that these two lipolytic systems evolved as complementary systems with the canonical ATGL mechanism tuned to respond rapidly to hormonal stimuli and the lysosomal system as a backup system to address energetic stress when fasting is prolonged.
The complementarity of the canonical ATGL-dependent and lysosomal lipolysis systems may extend beyond differences in the temporal phases of maximal functionality during fasting. First, while we did not detect evidence of directionally concordant loss of one pathway when targeting the other, we did observe evidence of compensatory pathway upregulation with disruption of the complementary system. This was particularly evident with ATGL loss of function where MiT/TFE factors and Lipa/LAL were augmented and may in part explain the lack of a consistent phenotype when targeting ATGL with fasting in this study, particularly when prolonged. It is possible that this compensatory response is variable and context dependent, in which case it could explain variability in observed phenotypes with ATGL loss of function and prolonged fasting19,47. Another potential way these two systems could interact is by the lysosomal digestion of the products of ATGL activity, a scenario in which LAL would in essence replace the activity of canonical downstream lipases such as HSL and MGGL. The analyses contained in this study cannot exclude such low-level direct cooperativity between ATGL and LAL; however, if LAL function was downstream from and entirely dependent on ATGL, then one would not expect the temporally distinct phenotypes that we observed, specifically a potent effect of LIPA loss of function at 24 h, a fasting timepoint when ATGL targeting had limited effect. It will be important in the future to understand any direct mechanism(s) by which these two systems communicate.
We also discovered a consistent signal of MiT/TFE regulatory involvement in lipolysis across in vitro and in vivo models, providing orthogonal evidence for lysosomal programs in fasting lipolysis. We selected TFEB for further in vivo study because it was the most potent and consistent regulator of lipolysis with nutrient restriction in murine adipocytes, in vitro, finding that inducible adipocyte-specific targeting of TFEB was sufficient to attenuate the systemic NEFA surge with fasting. The involvement of TFEB is itself not surprising, as it is the most established lysosomal regulator with precedent for its role in lipolysis in other tissues such as the liver27,52. However, independent targeting of more than one MiT/TFE factor was sufficient to attenuate the lipolytic surge with nutrient restriction, suggesting cooperativity. Indeed, the MiT/TFE family members regulate gene expression as homo- or hetero-dimers, resulting in regulatory mechanisms, including examples of redundancy and cooperativity that vary in a cell-type-specific manner53,54. Our data add to this complexity in suggesting murine-human divergence in the roles played by specific MiT/TFE factors such as TFEC, which was lowly expressed in murine cells but was a potent regulator of LIPA expression and lipolysis in human SGBS-derived adipocytes. The variable contributions of the MiT/TFE factors across experimental models provide rationale in the future to dissect the mechanisms of regulatory cooperativity and contextual specificity.
Our study suggests that lysosome-dependent lipolysis in adipocytes plays a role in the adaptive starvation response, even though we did not subject mice to a formal survival study. Beyond survival during starvation, human genetics studies have established that loss of function of lipolytic genes, including canonical lipases and LIPA, drive systemic metabolic derangements mediated in part by pathological responses in tissues most sensitive to lipotoxicity, such as the liver and striated muscle12,13,55. Even in adipose tissue which is defined by its capacity to absorb massive lipid fluxes, loss of function of lipolytic genes leads to adipose tissue dysfunction, including lipoatrophy and adipose tissue inflammation14,15. GWAS also point to LIPA as risk locus for coronary artery disease, suggesting that more subtle regulatory disruptions of LIPA and lysosomal lipolytic function may also be important to metabolic homeostasis56,57,58.
In summary, we propose a model of lipolysis in adipocytes with fasting whereby transcriptionally regulated lysosomal lipolysis is critical to the adaptive fasting response by complementing the acute activity of the canonical system that is activated by adrenergic signaling and rapid signal transduction. Furthermore, we provide evidence for human conservation of this core lysosomal mechanism of lipolysis in adipocytes, and its dependency on MiT/TFE factors, even though there may be interspecies divergence in the specific family member(s) most operative and definitive establishment of causality in humans is challenging because ex vivo nutrient restriction of human cells and adipose tissue explants cannot recapitulate the integrated physiology of organismal fasting. The life-extending effects of dietary restriction in model organisms suggest that feeding-fasting transitions might be of broader importance to the biology of human aging, a concept reinforced by our unbiased network medicine approach linking the molecular response to fasting in adipose tissue to diseases of aging, including a diversity of diseases beyond cardiometabolic diseases that are already known to be directly modulated by energy balance and adipose tissue function. Future mechanistic dissection of this fasting-aging axis may reveal opportunities for the development of therapies targeting the pathobiology of aging and diseases of aging.
Methods
Regulatory approval
Human explant cultures utilized discarded and deidentified subcutaneous adipose tissue obtained from standard clinical care panniculectomy procedures and therefore was deemed IRB exempt by the University of Pittsburgh Institutional Review Board. The human fasting study was previously published and approved by the Mass General Brigham (formerly Partners) HealthCare Institutional Review Board59. The fasting protocol complied with the guidelines of the Health Insurance Portability and Accountability Act, and written informed consent was obtained from all participants. Animal experiments were approved by and in compliance with the University of Pittsburgh Institutional Animal Care and Use Committee.
Murine studies
Mice were maintained under a 12-hour dark/light cycle at 22 °C ± 2 °C receiving food and water ad libitum unless specified. For some experiments, male mice were used for initial testing, however, the a priori goal of the genetic experiments was to use approximately equal numbers of male and female mice. For fasting experiments, fasting was initiated in the morning and continued for up to 24 h. Wild-type C57Bl/6 mice were purchased from Jackson Laboratory and Charles River and either directly used or bred in our colony. Adiponectin-Cre, Adiponectin-CreER, and Pnlpa2-floxed mice were purchased from Jackson Laboratories. Tfeb-floxed mice were obtained from Professor A. Ballabio, Telethon Institute of Genetic Medicine, Naples, Italy28.
The Lipa conditional knockout allele (Lipa-flox) was created by inserting LoxP sites in the introns flanking exon 4 using CRISPR/Cas9 gene-editing-assisted system (Fig. S8)60. The Cre-mediated recombination of the LoxP sites causes the deletion of exon 4. The splicing of exon 3 with exon 5 is predicted to result in a frameshift and a premature stop codon that would induce nonsense-mediated mRNA decay61. SpyCas9 target sites in the region surrounding the exon 4 of the Lipa gene were identified using the CRISPOR website http://crispor.tefor.net/62. Two target sequences “Lipa-416rev” (5’-CTAAACAGCCAACTGTTAGG-3’, chr19:34,511,084-34,511,103 of the mm10 assembly) and Lipa 752forw” (5’-CTGTATCGTTCGCATCTTGA-3’, chr19:34,510,771-34,510,790) were selected based on their location, their limited number of potential off-targets and various favorable efficiency and specificity scores. Cas9 single guide RNAs were produced as described60. LoxP sites, with an adjacent EcoRI diagnostic site, were positioned so that they would disrupt the SpyCas9 target sequences. For the donor DNA template, an Alt-R™ HDR Donor Blocks “Lipa-flox Alt-R HDR” was used, this is a modified double-strand DNA custom synthesized by Integrated DNA Technologies, Inc.
Genetically engineered mice were generated by the Mouse Embryo Services Core (University of Pittsburgh, Department of Immunology). Briefly, fertilized embryos (C57BL/6 J, The Jackson Laboratory) produced by natural mating, were microinjected, in the pronuclei with a mixture of 0.67 µM EnGen Cas9 protein (New England Biolabs, Cat. No. M0646T), the two Cas9 guides RNA: Lipa-416rev and Lipa-752forw (21.23 ng/µl each) and the dsDNA donor template “Lipa-flox Alt-R HDR” (2.5 ng/µl). The injected zygotes were cultured overnight, the next day the embryos that developed to the 2-cell stage were transferred to the oviducts of pseudopregnant CD1 female surrogates.
Potential founders were genotyped by PCR of each of the target regions (Fig. S8). The PCR products were then digested with EcoRI to identify fragments potentially carrying LoxP sites. To confirm that LoxP sites were in on the same allele PCR (with Lipa-F2 x Lipa-R2) was performed and Sanger sequencing of TOPO-cloned PCR product confirmed the correct sequence of the conditional allele. Furthermore, since the sequences of the primers Lipa-F2 and Lipa-R2 are beyond the donor template, this also validates the proper integration in the endogenous Lipa locus.
Pharmacological inhibitors of ATGL or lysosomes were administered by I.P. injection, twice daily in the following doses: Atglistatin (Sigma; 60 μM/kg); Bafilomycin (Sigma; 1 mg/kg); Lalistat2 (Sigma; 1 mg/kg). Isoproterenol was administered after a 4-hour fast as a one-time dose by I.P. injection to stimulate lipolysis (Sigma; 10 mg/kg).
Adipocytes were isolated from murine adipose tissues with a modified version of the classic Rodbell method24 as previously described63,64. Adipose tissues were minced and digested in an enzyme cocktail of 1 mg/mL collagenase type D (Roche) and 2 mg/mL Dispase 2 (Invitrogen) in PBS. The suspension was incubated in a shaking water bath (37 °C, 225 rpm, 40 min). The digest was filtered (300 μm nylon mesh, Spectrum Labs) and centrifuged at 400 g for 10 min. The supranatants containing floating adipocytes were collected.
Human samples
Transcriptomics data obtained from longitudinal sampling of periumbilical human adipose tissue during an inpatient 10 day fast59 was previously published18. Of note, individual transcript data as shown in Fig. 5 was not previously presented in figure form. Human explant cultures utilized discarded subcutaneous adipose tissue obtained from abdominoplasty surgeries as part of standard clinical care and was deemed IRB exempt. As such, only limited demographic data shown in Fig. S8 was linked to each sample. Of note, the clinical care of these patients includes a standard overnight fast before surgery and therefore the samples were collected from fasting individuals.
In vitro lipolysis
Concentrations of pharmacological inhibitors were used following published protocols at 10 μm (Atglistatin) and 50 nM (Bafilomycin and Lalistat2). Mice used as a source of APs were 4–6 weeks old. Inguinal adipose tissue depots were excised, minced, and digested in an enzyme cocktail, consisting of 1 mg/mL collagenase D (Roche) and 2 mg/mL Dispase II (Invitrogen), as described previously and similar to the Rodbell method24,25. The stromal vascular fraction was subjected to negative selection by column-based magnetic-assisted cell sorting (MACS). Monoclonal anti-CD31 microbeads (Miltenyi Biotec) were utilized to negatively select endothelial cells. A biotin-conjugated monoclonal anti-lineage cocktail, followed by anti-biotin MicroBeads (Miltenyi Biotec), was used to negatively select hematopoietic cells. The cell isolate was cultured in DMEM-F/12 GlutaMAX medium (Gibco) with 10% Premium FBS (Corning) and Pen/Strep. Medium was changed for the first time 2 days after plating. APs were cultured to confluence, then exposed to dexamethasone (1 μM), insulin (10 μg/ml), and isobutylmethylxanthine (0.5 mM) in standard culture medium (DMEM-F/12 GlutaMAX, 10% FBS). 72 h after induction, cells were switched to a maintenance medium, consisting of a standard culture medium supplemented with insulin (10 μg/ul). The maintenance medium was changed every other day.
3T3-L1 preadipocytes (Zen-Bio, #SP-L1-F) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 100 units/ml of Penicillin-Streptomycin (DMEM) containing 10% fetal bovine serum (FBS, Gibco). Differentiation of 3T3-L1 preadipocytes into adipocytes was achieved by supplementing the media with dexamethasone (1 μM), insulin (10 μg/ml), and isobutylmethylxanthine (0.5 mM) a cocktail of 10 μg/ml insulin (Sigma-Aldrich), 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich), and 1 μM dexamethasone (Sigma-Aldrich) for 3 days; after which cells were maintained in DMEM containing 10% FBS and 10 μg/ ml insulin for 6 additional days. The culture medium was changed every other day.
SGBS human preadipocytes were cultured in DMEM/F12 supplemented with 10% FBS and differentiated based on previously published protocols for 21 days36. Differentiation was initiated for four days with media containing 33 µM biotin, 17 µM pantothenic acid, 100U/ml penicillin/streptomycin, 2 µM rosiglitazone, 10 µg/ml human apo-transferrin, 20 nM human insulin, 25 nM dexamethasone, 500 µM 3-isobutyl-1-methylxantine, 100 nM cortisol, 200 pM triiodothyronine. Thereafter, the media was supplemented with the same additives except for rosiglitazone, 3-isobutyl-1-methylxantine and dexamethasone. The differentiation medium was replaced every other day.
Human or murine adipose tissue (200 mg/well) was minced to pieces of approximately 1 mm in diameter and cultured in DMEM (mouse) or DMEM/F12 (human) supplemented with 10% FBS. Adrenergic stimulation of cultured adipocytes and adipose tissue explants was achieved with addition of catecholamines (25 μm/ml) to the media. For in vitro cultures, nutrient restriction was achieved by lowering the glucose concentration of the media to 200 μg/ml or 1 mM, lowering the FBS concentration to 0.1%, removing glutamate and sodium pyruvate, and in the case of adipocyte cultures, removing insulin.
Plasmids
For loss-of-function assays, validated shRNA (Sigma) or scramble sequences were subcloned into a lentiviral vector (tet-pLKO-puro, addgene-21915). Knockdown efficiency was confirmed by qPCR in 3T3-L1 cells. shRNA TRC Numbers were mouse: shPnpla2-TRCN0000249777, shLipa-TRCN0000076829, shTfeb-TRCN0000076829, shTfe3-TRCN0000084670, shMitf-TRCN0000305603. Human: shPNPLA2- TRCN0000222744, shLIPA- TRCN0000350924, shMITF- TRCN0000019120, shTFEC- TRCN0000016098, shTFE3- TRCN0000232151, shTFEB- TRCN0000013110.
Biochemical assays
Plasma non-esterified free fatty acids (NEFA) were measured using an in vitro enzymatic colorimetric assay (Wako Chemicals). Glycerol, glucose, and MTT assays (Sigma) were measured with enzymatic colorimetric assays according to the manufacturer’s protocols.
Protein assays
Tissues were homogenized in RIPA buffer (ThermoFisher Scientific) with Protease Inhibitor Cocktail (100X) (ThermoFisher Scientific) to extract total protein. Protein was quantified and electrophoresed in equal quantities (45 ng per well) on 10% SDS poly acrylamide gels and transferred to Immun-Blot PVDF Membranes (Bio-rad). Membranes were blocked in 5% nonfat dry milk (Bio-rad) prepared in 0.1% tween-20 for 1 h followed by primary antibody (1:1000, unless otherwise stated) incubation at 40C overnight. Membranes were washed three times with 0.1% tween-20 in tris base saline and incubated with secondary antibody Horseradish peroxidase–conjugated IgG anti-mouse (Invitrogen), anti-rabbit (Cell Signaling Technology) for 1 h. After incubation, membranes were washed three times with 0.1% tween-20 in tris base saline and developed with SuperSignal West Pico plus Chemiluminescent Substrate (ThermoFisher Scientific). Blots were imaged with Chemdoc (Biorad). Antibody performance was validated by comparing the resultant band placement to those reported by the manufacturer and found either on their website or in the accompanying package insert literature. FGF21 in plasma was measured with a mouse FGF-21 ELISA (Abcam) according to the manufacturer’s protocol. Lysosomes were isolated from adipocytes (Abcam) and perilipin concentrations were measured by ELISA (LS Bio).
Gene expression
Total RNA was isolated from tissues or cells using Qiazol (Qiazen). cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies) from 500 ng of RNA. Real-time quantitative PCR was performed with Power SYBR Green PCR mixture (Applied Biosystems) on the QuantStudio™ 5 Real-Time PCR System (Applied Biosystems). Gene expression was normalized to GAPDH. The ΔΔCt method was used to calculate the fold change in transcript levels.
Immunofluorescence microscopy
Adipose tissue specimens for confocal imaging were analyzed with minor modifications to previously published methods25. Adipose tissues were labeled with anti-Lamp1, anti-Perilipin, and DAPI (Cell Signaling Technology). Wholemount adipose tissues were fixed and imaged using excitation from a 680- or 488-nm laser line of an argon laser at a 63×, 1.2-NA oil immersion objective on a laser scanning confocal microscope (SP8 LIGHTNING Leica).
Network medicine analyses
The consolidated human protein-protein interactome were assembled from different resources as described before which contains 16,470 proteins and 233,957 interactions37,65,66,67. We mapped differentially expressed transcripts from human fasting adipose tissue samples to the consolidated human interactome and constructed a network of 419 proteins, and -597 interactions. Genes associated with aging diseases, cardiometabolic diseases and inflammation were compiled from Phenopedia and DisGeNET68,69. Gene associated with endophenotypes except for inflammation were compiled from the Gene Ontology database (geneontology.org/). Drug targets were retrieved from DrugBank (go.drugbank.com). We used network proximity to quantify the closeness of the novel fasting module to endophenotype modules or disease-specific modules37,65,66. Network proximity is defined as the average minimum shortest path length in the interactome:
where ps is the minimum shortest path length from a fasting gene s to a disease gene d. The network proximity from drug targets to the fasting module was calculated in the same way. The significance of network proximity was evaluated by creating 1000 random modules of the same size and comparing the observed proximity value with the null model (random control) through fitting normal distributions.
Statistics and reproducibility
Experiments were repeated with a minimum of 3 biological replicates unless otherwise stated. Unless otherwise stated, the data contained in the graphs in this manuscript show either measurements from individual mice or from samples collected from individual mice (biological replicates) or from consecutive experiments/biological replicates, where each data point represents the normalized mean of the technical replicates for that experiment. Two-sided t-tests were used for comparison of two experimental groups and ANOVA (with correction for multiple hypotheses) when analyzing more than two groups. Two-way ANOVA was used for time-course data or when accounting for potential sex differences. Paired or repeated measures testing was done when comparing normalized biological replicate experiments. Corresponding non-parametric tests were used if the data was not normally distributed. Significance was assigned for p < 0.05. Unless otherwise indicated error bars indicate standard deviation of the mean (S.D.M.). For wild-type mouse experiments, mice were randomly assigned to each experimental group. For experiments using genetically engineered mice, the genotype determined assignment to the experimental group. No data points were excluded. For in vivo experiments where some analyses contain reduced n, this is either due to a technical problem (e.g. insufficient sample) or because a randomly selected subset of samples was analyzed (e.g. tissue immunoblots). For data points involving quantitation of a qualitative measure, specifically the interpretation of microscopy images, assessments were conducted by an observer blind to experimental group. Prism versions 9 and 10 (Graphpad) were used for statistical analyses. The networks (Fig. 6) were visualized using Cytoscape 3.7.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings from this study are available within the manuscript and its supplementary information. The raw sequencing files from the previously published adipose tissue transcriptomics study, which were used to generate a gene list for the network analyses, are protected and are not available due to data privacy laws. The gene list used as input for the network analyses are provided in the Source Data file. Deidentified and normalized count data is available without restrictions upon request to the corresponding author within 2 weeks of request (msteinhauser@pitt.edu). Source data are provided with this paper.
Code availability
Code used for network analyses are available at: https://github.com/bwh784/FastingModule/.
References
Cahill, G. F. Starvation in man. N. Engl. J. Med 282, 668–675 (1970).
Stewart, W. K. & Fleming, L. W. Features of a successful therapeutic fast of 382 days’ duration. Postgrad. Med J. 49, 203–209 (1973).
Longo, V. D. & Anderson, R. M. Nutrition, longevity and disease: From molecular mechanisms to interventions. Cell 185, 1455–1470 (2022).
Perry, R. J. et al. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell 160, 745–758 (2015).
Sakers, A., De Siqueira, M. K., Seale, P. & Villanueva, C. J. Adipose-tissue plasticity in health and disease. Cell 185, 419–446 (2022).
Zimmermann, R. et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386 (2004).
Vaughan, M., Berger, J. E. & Steinberg, D. Hormone-Sensitive Lipase And Monoglyceride Lipase Activities In Adipose Tissue. J. Biol. Chem. 239, 401–409 (1964).
Yang, A. & Mottillo, E. P. Adipocyte lipolysis: from molecular mechanisms of regulation to disease and therapeutics. Biochem J. 477, 985–1008 (2020).
Osuga, J. et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl Acad. Sci. USA 97, 787–792 (2000).
Taschler, U. et al. Monoglyceride lipase deficiency in mice impairs lipolysis and attenuates diet-induced insulin resistance. J. Biol. Chem. 286, 17467–17477 (2011).
Haemmerle, G. et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737 (2006).
Albert, J. S. et al. Null mutation in hormone-sensitive lipase gene and risk of type 2 diabetes. N. Engl. J. Med 370, 2307–2315 (2014).
Fischer, J. et al. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet 39, 28–30 (2007).
Schoiswohl, G. et al. Impact of reduced ATGL-mediated adipocyte lipolysis on obesity-associated insulin resistance and inflammation in male mice. Endocrinology 156, 3610–3624 (2015).
Cinti, S. et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res 46, 2347–2355 (2005).
Flaherty, S. E. et al. A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science 363, 989–993 (2019).
Sztalryd, C. & Kraemer, F. B. Regulation of hormone-sensitive lipase during fasting. Am. J. Physiol. 266, E179–E185 (1994).
Fazeli, P. K. et al. Prolonged fasting drives a program of metabolic inflammation in human adipose tissue. Mol. Metab. 42, 101082 (2020).
Wu, J. W. et al. Fasting energy homeostasis in mice with adipose deficiency of desnutrin/adipose triglyceride lipase. Endocrinology 153, 2198–2207 (2012).
Defour, M., Michielsen, C. C. J. R., O’Donovan, S. D., Afman, L. A. & Kersten, S. Transcriptomic signature of fasting in human adipose tissue. Physiol. Genomics 52, 451–467 (2020).
Lass, A. et al. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3, 309–319 (2006).
Kersten, S. The impact of fasting on adipose tissue metabolism. Biochim Biophys. Acta Mol. Cell Biol. Lipids 1868, 159262 (2023).
Nielsen, T. S. et al. Fasting, but not exercise, increases adipose triglyceride lipase (ATGL) protein and reduces G(0)/G(1) switch gene 2 (G0S2) protein and mRNA content in human adipose tissue. J. Clin. Endocrinol. Metab. 96, E1293–E1297 (2011).
Rodbell, M. Metabolism of Isolated Fat Cells. I. Effects of Hormones on Glucose Metabolism and Lipolysis. J. Biol. Chem. 239, 375–380 (1964).
Zhang, Y. et al. Targeting nuclear receptor NR4A1-dependent adipocyte progenitor quiescence promotes metabolic adaptation to obesity. J. Clin. Invest 128, 4898–4911 (2018).
Chu, A. Y. et al. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat. Genet 49, 125–130 (2017).
Settembre, C. et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15, 647–658 (2013).
Settembre, C. et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012).
Ye, R. et al. Impact of tamoxifen on adipocyte lineage tracing: Inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Mol. Metab. 4, 771–778 (2015).
Chan, J. L., Mietus, J. E., Raciti, P. M., Goldberger, A. L. & Mantzoros, C. S. Short-term fasting-induced autonomic activation and changes in catecholamine levels are not mediated by changes in leptin levels in healthy humans. Clin. Endocrinol. (Oxf.) 66, 49–57 (2007).
Leiter, L. A., Grose, M., Yale, J. F. & Marliss, E. B. Catecholamine responses to hypocaloric diets and fasting in obese human subjects. Am. J. Physiol. 247, E190–E197 (1984).
Patel, J. N., Coppack, S. W., Goldstein, D. S., Miles, J. M. & Eisenhofer, G. Norepinephrine spillover from human adipose tissue before and after a 72-hour fast. J. Clin. Endocrinol. Metab. 87, 3373–3377 (2002).
Young, J. B., Rosa, R. M. & Landsberg, L. Dissociation of sympathetic nervous system and adrenal medullary responses. Am. J. Physiol. 247, E35–E40 (1984).
Wabitsch, M. et al. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int J. Obes. Relat. Metab. Disord. 25, 8–15 (2001).
Chen, Z. et al. Functional screening of candidate causal genes for insulin resistance in human preadipocytes and adipocytes. Circ. Res 126, 330–346 (2020).
Tews, D. et al. 20 Years with SGBS cells - a versatile in vitro model of human adipocyte biology. Int J. Obes. (Lond.) 46, 1939–1947 (2022).
Menche, J. et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 347, 1257601 (2015).
Consortium, A. A. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res 49, D825–D830 (2021).
Samokhin, A. O. et al. NEDD9 targets COL3A1 to promote endothelial fibrosis and pulmonary arterial hypertension. Sci. Transl. Med. 10 https://doi.org/10.1126/scitranslmed.aap7294 (2018).
Grabner, G. F., Xie, H., Schweiger, M. & Zechner, R. Lipolysis: cellular mechanisms for lipid mobilization from fat stores. Nat. Metab. 3, 1445–1465 (2021).
Ahmadian, M. et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 13, 739–748 (2011).
Mottillo, E. P. et al. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic β3-adrenergic receptor activation. J. Lipid Res 55, 2276–2286 (2014).
Soussi, H. et al. DAPK2 downregulation associates with attenuated adipocyte autophagic clearance in human obesity. Diabetes 64, 3452–3463 (2015).
Schreiber, R. et al. Hypophagia and metabolic adaptations in mice with defective ATGL-mediated lipolysis cause resistance to HFD-induced obesity. Proc. Natl Acad. Sci. USA 112, 13850–13855 (2015).
Huijsman, E. et al. Adipose triacylglycerol lipase deletion alters whole body energy metabolism and impairs exercise performance in mice. Am. J. Physiol. Endocrinol. Metab. 297, E505–E513 (2009).
Kienesberger, P. C. et al. Adipose triglyceride lipase deficiency causes tissue-specific changes in insulin signaling. J. Biol. Chem. 284, 30218–30229 (2009).
Fougerat, A. et al. ATGL-dependent white adipose tissue lipolysis controls hepatocyte PPARα activity. Cell Rep. 39, 110910 (2022).
Ahmadian, M. et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes 58, 855–866 (2009).
Natali, A. et al. Metabolic consequences of adipose triglyceride lipase deficiency in humans: an in vivo study in patients with neutral lipid storage disease with myopathy. J. Clin. Endocrinol. Metab. 98, E1540–E1548 (2013).
Steinhauser, M. L. et al. The circulating metabolome of human starvation. JCI Insight 3 https://doi.org/10.1172/jci.insight.121434 (2018).
Schupp, M. et al. Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues. BMC Genomics 14, 758 (2013).
Sardiello, M. et al. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009).
Hemesath, T. J. et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8, 2770–2780 (1994).
Fisher, D. E., Carr, C. S., Parent, L. A. & Sharp, P. A. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 5, 2342–2352 (1991).
Bernstein, D. L., Hülkova, H., Bialer, M. G. & Desnick, R. J. Cholesteryl ester storage disease: review of the findings in 135 reported patients with an underdiagnosed disease. J. Hepatol. 58, 1230–1243 (2013).
Wild, P. S. et al. A genome-wide association study identifies LIPA as a susceptibility gene for coronary artery disease. Circ. Cardiovasc Genet 4, 403–412 (2011).
Consortium, C. A. D. C. D. G. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet 43, 339–344 (2011).
Consortium, I. K. C. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet 7, e1002260 (2011).
Fazeli, P. K. et al. FGF21 and the late adaptive response to starvation in humans. J. Clin. Invest 125, 4601–4611 (2015).
Pelletier, S., Gingras, S. & Green, D. R. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity 42, 18–27 (2015).
Popp, M. W. & Maquat, L. E. Leveraging rules of nonsense-mediated mrna decay for genome engineering and personalized medicine. Cell 165, 1319–1322 (2016).
Concordet, J. P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46, W242–W245 (2018).
Monji, A. et al. A cycle of inflammatory adipocyte death and regeneration in murine adipose tissue. Diabetes https://doi.org/10.2337/db20-1306 (2022).
Kim, S. M. et al. Loss of white adipose hyperplastic potential is associated with enhanced susceptibility to insulin resistance. Cell Metab. 20, 1049–1058 (2014).
Samokhin, A. O. et al. NEDD9 targets. Sci. Transl. Med. 10 https://doi.org/10.1126/scitranslmed.aap7294 (2018).
Wang, R. S. & Loscalzo, J. Network module-based drug repositioning for pulmonary arterial hypertension. CPT Pharmacomet. Syst. Pharm. 10, 994–1005 (2021).
Wertheim, B. M. et al. Proline and glucose metabolic reprogramming supports vascular endothelial and medial biomass in pulmonary arterial hypertension. JCI Insight https://doi.org/10.1172/jci.insight.163932 (2023).
Yu, W., Clyne, M., Khoury, M. J. & Gwinn, M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics 26, 145–146 (2010).
Piñero, J. et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxf.) 2015, bav028 (2015).
Acknowledgements
We would like to thank Chunming Bi and Zhaohui Kou of the Mouse Embryo Services Core (University of Pittsburgh, Department of Immunology) for the microinjection of zygotes and production of Lipaf/f mouse. This work was supported by the NIH award KL2 TR002542 (PKF) and NIH award R01DK137913 (MLS).
Author information
Authors and Affiliations
Contributions
Conceptualization: G.V.N.K. and M.L.S. Methodology: G.V.N.K., R.S.W., M.W., S.G., A.E., B.A.M., P.K.F., and M.L.S. Formal analysis: G.V.N.K., R.S.W., and M.L.S. Investigation: G.V.N.K., A.X.S., N.L.D., T.A., D.S.S., N.K.D., P.K.F., and M.L.S. Resources: M.W., P.J.R., and A.E. Writing original draft: G.V.N.K. and M.L.S. Visualization: G.F.N.K. and M.L.S. Project administration: M.L.S. Funding acquisition: P.K.F. and M.L.S. All authors reviewed the manuscript and provided critical feedback.
Corresponding author
Ethics declarations
Competing interests
PKF is a consultant for Regeneron. She has also served on advisory boards Camurus, Crintetics, and Amryt/Chiesi and has research funding from Crinetics, Corcept, and Quest. These disclosures are not related to the content of the study. The other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kumar, G.N., Wang, RS., Sharma, A.X. et al. Non-canonical lysosomal lipolysis drives mobilization of adipose tissue energy stores with fasting. Nat Commun 16, 1330 (2025). https://doi.org/10.1038/s41467-025-56613-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56613-3