Abstract
Treatment decisions for an incidental renal mass are mostly made with pathologic uncertainty. Improving the diagnosis of benign renal masses and distinguishing aggressive cancers from indolent ones is key to better treatment selection. We analyze 13261 pre-operative computed computed tomography (CT) volumes of 4557 patients. Two multi-phase convolutional neural networks are developed to predict the malignancy and aggressiveness of renal masses. The first diagnostic model designed to predict the malignancy of renal masses achieves area under the curve (AUC) of 0.871 in the prospective test set. This model surpasses the average performance of seven seasoned radiologists. The second diagnostic model differentiating aggressive from indolent tumors has AUC of 0.783 in the prospective test set. Both models outperform corresponding radiomics models and the nephrometry score nomogram. Here we show that the deep learning models can non-invasively predict the likelihood of malignant and aggressive pathology of a renal mass based on preoperative multi-phase CT images.
Similar content being viewed by others
Introduction
Widespread utilization of cross-sectional imaging has led to increased detection of asymptomatic incidental renal lesions1,2,3, accompanied by increasing surgeries and ablations performed for suspicious malignant renal masses4. However, over the same period, kidney cancer-specific mortality has not substantially decreased, suggesting possible overtreatment of benign renal masses and limited benefit from instant extirpative surgeries5,6. Around 20% of resected renal masses are reported to be benign7. Therefore, more accurate identification of benign lesions could potentially avoid unnecessary surgeries and associated risk and morbidity7. Nowadays, with evolving management strategies such as active surveillance and ablation, knowing whether a renal mass is benign or malignant is insufficient; knowledge of its indolent or aggressive histology is also important8,9. Even many malignant renal masses follow an indolent course10,11,12. The decision whether to offer active surveillance, ablation or surgery for a certain patient depends on an individualized balance between cancer-specific mortality risk and comorbidity-related mortality risk13.
Computed tomography (CT)14,15,16 and magnetic resonance imaging (MRI)17,18 are commonly used for the detection of renal masses, but they have limited accuracy in estimating the probability of malignancy and degree of aggressiveness. Percutaneous biopsy is another way to obtain histology information before treatment. However, percutaneous biopsy is found non-diagnostic in 20% of cases and differs from the final pathologic diagnosis of resected tissues in another 10% of cases19. It also demonstrates relatively low accuracy in predicting tumor nucleus grade, necrosis, and sarcomatoid differentiation probably due to high tumor heterogeneity in renal cell carcinoma (RCC)20. In addition, the invasive procedure puts patients at risk of complications such as hemorrhage, infection, and implantation metastasis21.
Under most circumstances, the decision between active surveillance, percutaneous ablation, and surgical resection is made without a reliable pathologic diagnosis22. Thus, there is an urgent need to improve the non-invasive diagnosis of benign renal masses and differentiate aggressive tumors (prompting treatment) from indolent tumors (allowing ablation or deferred treatment)23. Artificial Intelligence (AI)-driven quantitative analysis of preoperative medical images can play a critical role. Radiomics-based machine learning models have been used to evaluate renal masses with promising results24,25,26,27. However, hand-crafted radiomics features have intrinsic limitations, such as lacking contextual information and being sensitive to variations in tumor delineation. In contrast, deep learning models learn hierarchical features from original images in an end-to-end manner. There are some recent deep learning studies aimed at characterizing renal masses, but few have a large sample size or a comprehensive analysis including both histology subtype and ISUP grade28,29,30,31.
In this study, we develop deep learning models using preoperative CT scans to discriminate benign versus malignant renal masses, as well as indolent versus aggressive tumors. Previous diagnostic models solely predicted high-grade pathology of clear cell renal cell carcinoma (ccRCC), while our deep learning model incorporated most renal mass pathologic subtypes and comprehensively assessed tumor aggressiveness by integrating histologic subtype, tumor grade, and adverse pathologic features. We explore further differences in patient survival between AI-predicted indolent and aggressive tumors, which were less explored in previous studies.
Results
Flowchart of the recruitment and clinicopathologic characteristics of each cohort
Flowchart of patient recruitment was demonstrated in Fig. 1A. A total of 4557 patients undergoing nephrectomy with preoperative enhanced CT scans were identified. There were 2400 patients in the training set, 598 patients in the internal test set, 561 patients in the external test set, 610 patients in the prospective test set, and 388 patients in the TCIA test set. Detailed clinicopathologic information for each cohort were listed in Table 1. In the training set, benign and indolent renal masses, malignant and indolent tumors, and malignant aggressive tumors represent 16.8, 61.5, and 21% of cases, respectively. In the internal test set, they each represent 15.7, 62, and 21.4% of cases. In the external test set, 15.9% of patients had benign and indolent renal masses, 61.1% patients had malignant and indolent tumors, 19.8% patients had malignant and aggressive tumors. In the prospective test set, malignant and indolent tumors held 13.8%, malignant and aggressive tumors held 63.9%, and malignant and aggressive tumors held 21%. In the TCIA test set, the three subtypes held 3.6, 51, and 45.4%, respectively (Table 1). Percentages of each specific histologic classification across different datasets were illustrated in Fig. 1B.
Diagnostic accuracy of the deep learning model to differentiate benign from malignant renal masses
The segmentation network achieved satisfactory performance in delineating kidney tumor (Fig. 2B, E) with a dice similarity coefficient score (DICE) of 0.852 (Table S2). Based on the cropped images, a multi-phase convolutional neural network was established to predict the malignancy of renal mass. Representative CT images of a benign renal mass (metanephric adenoma) and a malignant renal mass (clear cell renal cell carcinoma) were listed in Fig. 2A, D, respectively. Class activation maps (CAM) of benign and malignant masses are presented in Fig. 2C, F, visualizing the import regions in the image for the neural network to predict the malignancy32.
A Representative CT images of a benign renal mass. B Automatic segmentation of the renal mass and kidney. C Class activation maps (CAMs) of the benign and malignant prediction for the benign renal mass, where red color represents a region more significant to the designated classification. D Representative CT images of a malignant renal mass. E Automatic segmentation of the renal mass and kidney. F CAMs of the benign and malignant prediction for the malignant renal mass. G The receiver operating characteristic (ROC) curves of the deep learning-based model, the radiomics model, and the nephrometry score nomogram on the internal test set. H The ROC curves of the deep learning-based model, the radiomics model, and the nephrometry score nomogram on the external test set. I The ROC curves of the deep learning-based model, the radiomics model, and the nephrometry score nomogram on prospective test set. J The ROC curves of the deep learning-based model and nephrometry score nomogram on TCIA test set. K The ROC curves of the deep learning-based model and performance of seven radiologists on prospective validation set. L The diagnostic accuracy of seven radiologists with (n = 7) or without (n = 7) the assistant of the deep learning-based model. M Clustered observer similarity matrix of the deep learning-based model and seven radiologists. N The diagnostic efficiency of seven radiologists with (n = 7) or without (n = 7) the assistant of the deep learning-based model. In Fig. 2B, E yellow areas represent renal lesions identified by the nnU-Net model, while blue areas represent the kidney tissue also identified by the nnU-Net model. In Fig. 2C, F, the color intensity represents the level of importance for the deep learning model in making its decision. Red (designated as point 1) indicates the most important regions, green (designated as point 0.5) signifies regions of lesser importance, and blue (designated as point 0) denotes regions that are not important for the model’s decision. Source data for ROC curves are provided as a Source Data file.
The classification performance is summarized in Fig. 2 and detailed in Table S4. The AUCs for differentiating malignant from benign renal masses were 0.898, 0.853, 0.871, and 0.881 in the internal test set (Fig. 2G), external test set (Fig. 2H), prospective test set (Fig. 2I), and TCIA test set (Fig. 2J), respectively. Alexander Kutikov et al. constructed two nomograms for predicting malignant and high-grade pathology using R.E.N.A.L. nephrometry scores with 525 renal masses33. Besides, in previous studies many radiomics models were proposed for differentiating malignant from benign renal lesions26,34. In this study, we also developed two radiomics models as benchmarks for the two deep learning models respectively. Our deep learning-based diagnostic model outperformed the nephrometry score nomogram and the radiomics model for predicting malignancy across three test sets (Fig. 2G–I) encompassing all three CT phases.
Seven radiologists with over 5 years of experience reading abdominal CT imaging from tertiary academic medical centers, given clinical information of each patient and blind to histopathologic data, evaluated CT images of the renal masses in the prospective test set. The deep learning-based model showed better performance than six of the seven expert radiologists (Fig. 2K). Specifically, when the model matched the average sensitivity of these radiologists, its specificity surpassed the average specificity of them. Conversely, when it matched their average specificity, its sensitivity outperformed the average sensitivity of them (Fig. 2K). We then compared the sensitivities of the AI system and the reader performance at specificities fixed to match reader performance, and found that the deep learning model significantly outperformed four radiologists (Table S5). When the sensitivities were matched, the deep learning model demonstrated higher specificity than six radiologists, but none of them reached a statistical significance (Table S5). We further asked seven radiologists to revisit and re-evaluate CT scans from our prospective validation cohorts with a minimum washout period of two months between assessments. This time, however, we provided the radiologists with the predictive results from our deep learning model, emphasizing that it had outperformed the average diagnostic capabilities of seven radiologists. With the additional insights offered by the deep learning model, we observed a significant improvement in the diagnostic accuracy of the radiologists (Fig. 2L). Analysis of diagnostic correlation revealed that seven radiologists exhibited stronger diagnostic similarity among themselves compared to the deep learning model (Fig. 2M), and this may partly explain why the diagnostic accuracy of radiologists improved with the assistance of the deep learning model. We also measured the time taken by the radiologists to evaluate CT scans from the 101st to the 200th case and compared it to their initial evaluation time, noticing no significant differences in time spent on CT scan evaluation (Fig. 2N).
The diagnostic model also showed robust performance in subgroup analyses of solid tumors (Fig. S3A–D), cystic tumors (Fig. S3E–H), and small renal masses (SRMs) (Fig. S3I–L), and outperformed both the radiomics model and the nephrometry score nomogram. In addition, we have developed additional diagnostic models that can handle various combinations of CT phases. We have showcased the AUCs of our two deep learning models, evaluating their performance with different CT phase combinations in the internal test set: N (non-contrast), A (arterial phase), V (venous phase), as well as N + A, N + V, and A + V (Table S6).
Diagnostic accuracy of the deep learning model to differentiate aggressive from indolent renal tumors
Besides malignancy, aggressiveness is of growing importance in the treatment of renal tumors. Thus, we sought to develop a second diagnostic model to differentiate between indolent (benign and indolent, malignant and indolent) and aggressive (malignant and aggressive) renal tumors. Classification of indolent malignant tumors and aggressive malignant tumors were mainly based on 2022 WHO classifications and previous reports. To confirm the reasonability of the classification and find out whether the proposed aggressiveness could translate into actual survival differences, we analyzed the survival differences between indolent and aggressive malignant tumors in the Zhongshan cohort and Zhejiang cohort. Detailed information on the two cohorts were listed in Table S7. As expected, indolent tumors were associated with significantly longer survival compared with aggressive tumors in both the Zhongshan cohort (p < 0.001 for DSS, RFS, and OS) and Zhejiang cohort (p < 0.001 for DSS, RFS, and OS) (Fig. S4).
Sharing the same network architecture, we developed another multi-phase convolutional neural network to predict the aggressiveness of renal tumors. The classification performance is summarized in Fig. 3 and detailed in Table S8. Representative CT images of an indolent renal mass (mucinous tubular and spindle cell RCC) and an aggressive renal mass (renal cell carcinoma NOS) are demonstrated in Fig. 3A, D, respectively. Class activation maps of indolent and aggressive tumors are presented in Fig. 3C, F, visualizing the import regions in the image for predicting the invasiveness.
A Representative CT images of an indolent renal mass. B Automatic segmentation of the renal mass and kidney. C CAMs of the indolent and aggressive prediction for the indolent renal mass, where red color represents a region more significant to the designated classification. D Representative CT images of an aggressive renal mass. E Automatic segmentation of the renal mass and kidney. F CAMs of the indolent and aggressive prediction for the aggressive renal mass. G The receiver operating characteristic (ROC) curves of the deep learning-based model, the radiomics model, and nephrometry score nomogram on the internal test set. H The ROC curves of the deep learning-based model, the radiomics model, and nephrometry score nomogram on the external test set. I The ROC curves of the deep learning-based model, the radiomics model, and nephrometry score nomogram on the prospective test set. J The ROC curves of the deep learning-based model and nephrometry score nomogram on TCIA test set. In Fig. 2B, E yellow areas represent renal lesions identified by the nnU-Net model, while blue areas represent the kidney tissue also identified by the nnU-Net model. In Fig. 2C, F, the color intensity represents the level of importance for the deep learning model in making its decision. Red (designated as point 1) indicates the most important regions, green (designated as point 0.5) signifies regions of lesser importance, and blue (designated as point 0) denotes regions that are not important for the model’s decision. Source data for ROC curves are provided as a Source Data file.
In addition to the nephrometry score nomogram, we also developed a second standardized radiomics model as another benchmark model. The AUCs for predicting aggressive pathology are 0.792 in the internal test set (Fig. 3G), 0.763 in the external test set (Fig. 3H), 0.783 in the prospective test set (Fig. 3I), and 0.755 in the TCIA test set (Fig. 3J), higher than the radiomics model and the nephrometry score nomogram to predict high-grade pathology in all test cohorts. In subgroup analyses we found that the deep learning model achieved robust performance in solid tumors, cystic tumors, and SRMs. It also outperformed the radiomics model and the nephrometry score nomogram in all subgroup analyses (Fig. S5). Interestingly, AUCs of the radiomics model and the nephrometry score for SRMs dropped significantly compared with all cases or other subgroups (Fig. S5). Classification performance across different CT phases were listed in Table S6.
Associations between radiological aggressiveness and survival outcomes
We have validated the diagnostic model discriminating aggressive tumors from indolent tumors across different cohorts, proving the robustness and accuracy of the diagnostic models. Another important aspect of aggressiveness evaluation is still survival outcomes. We, therefore, compared the oncologic outcomes between patients with tumors predicted to be indolent by the AI-based diagnostic model (AI-predicted indolent) and patients with tumors predicted to be aggressive (AI-aggressive). We found profound survival differences between AI-predicted indolent tumors and AI-predicted aggressive tumors in both internal test sets (DSS, p < 0.001, HR = 5.48; RFS, p < 0.001, HR = 4.11; OS, p = 0.005, HR = 2.66) and external test (DSS, p < 0.001, HR = 20.61; RFS, p < 0.001, HR = 9.69; OS, p < 0.001, HR = 13.19) in terms of DSS, RFS, and OS (Fig. 4A–F). A similar trend was observed in the training set as well (Fig. S6). Baseline characteristics of patients with malignant renal tumors in training, internal test, and external test sets for survival analyses were listed in Table S7. The 5-year survival rates were 97.9% (DSS), 94.5% (RFS), and 96.8% (OS) for AI-predicted indolent tumors; 86% (DSS), 77.2% (RFS), and 85.1% (OS) for AI-predicted aggressive tumors in the internal test set. In the external test set, the 5-year survival rates were 99.6% (DSS), 96.8(RFS), and 98.9 (OS) for AI-predicted indolent tumors; 79.6% (DSS), 74.6% (RFS), and 79% (OS) for AI-predicted aggressive tumors.
A Kaplan–Meier analysis of disease-specific survival (DSS) in the internal test set. B Kaplan–Meier analysis of recurrence-free survival (RFS) in the internal test set. C Kaplan–Meier analysis of overall survival (OS) in the internal test set. D Kaplan–Meier analysis of DSS in the external test set. E Kaplan–Meier analysis of RFS in the external test set. F Kaplan–Meier analysis of OS in the external test set. G Forest plot of AI-aggressiveness score cox regression analyses performed in different subgroups in the external test set. The centers for the error bars represent the hazard ratios. H C-indexes of AI-aggressiveness score, radiomics model, the nephrometry nomogram, TNM stage, and ISUP grade in predicting DSS, RFS, and OS in the internal and external test set. The error bands in the survival curves in Fig. 4A–F represent the 95% confidential intervals. P values were two-sided and not adjusted for multiple comparisons. Source data for survival analyses are provided as a Source Data file.
In multivariate analyses after adjustment for TNM stage and ISUP grade, AI-aggressiveness subtypes (AI-predicted aggressive versus AI-predicted indolent) remained an independent adverse risk factor for patient survival in DSS (p = 0.008, HR = 5.80), RFS (p = 0.003, HR = 3.78), and OS (p = 0.007, HR = 4.31) analyses in external test set (Table S9). Interestingly, we found that AI-aggressiveness score as a continuous variable ranging from 0 to 10 was also an adverse independent risk factor for DSS (p = 0.002, HR = 2.433), RFS (p < 0.001, HR = 2.928), and OS (p = 0.002, HR = 2.231) in external test set (Table S9), which revealed a potential link between the likelihood of a renal mass being aggressive and the risk of relapse of the renal tumor. If a renal tumor is more likely to be an aggressive tumor predicted by the AI-based model, the tumor is more likely to metastasize in the end. AI-aggressiveness score functions more than predicting the aggressiveness of a renal tumor. In tumors of same aggressiveness subtype, indolent or aggressive, AI-aggressiveness score is still an adverse risk factor for DSS (indolent, p = 0.001, HR = 5.239; aggressive, p < 0.001, HR = 2.255), RFS (indolent, p < 0.001, HR = 3.802; aggressive, p < 0.001, HR = 2.214), and OS (indolent, p < 0.001, HR = 3.806; aggressive, p < 0.001, HR = 1.982) (Fig. 4G). Forest plots showed that the AI-aggressiveness score was significantly associated with dismal survival across different subgroups, including TNM stages, ISUP grades, and histologic subtypes (Fig. 4G). All these results indicated that the AI-aggressiveness score was significantly associated with patient survival and had great potential to be a reliable prognostic biomarker. We compared the prognostic value between the AI-aggressiveness score, the radiomics model, the nephrometry score nomogram, the TNM stage, and ISUP grades and surprisingly found that the AI-aggressiveness score outperformed all the other risk factors in both internal and external test set in terms of DSS, RFS, and OS (Fig. 4H).
We conducted further analyses to evaluate the model’s performances specifically in subsets of high clinical significance, namely T1a and T1 solid tumors. Our findings indicate that even within this subset, where imaging overlap between lesions is most pronounced, the diagnostic models exhibit robust performance in the two primary diagnostic tasks (Table S10). Additionally, we stratified the model’s performance based on tumor size and stage (T1aN0M0, T1bN0M0, T2-T4N0M0&T1-T4N1M0), as outlined in Table S11. These analyses not only reinforce our initial findings but also demonstrate that, beyond considering tumor size, our models possess strong predictive capabilities, making the analyses less biased. The spatial resolution of CT scans was provided in Table S12.
Distinct genetic and immune landscapes of aggressive and indolent renal tumors
Afterward we analyzed the genetic, transcriptomic, and immune landscapes in aggressive versus indolent tumors to explore the underlying biological mechanisms of the dismal survival in aggressive tumors. Between indolent versus aggressive tumors, we found no difference in TMB. Among the top 28 mutant genes, AHNAK2 mutation increased in aggressive tumors with statistical significance (p = 0.029) (Fig. 5A). On the transcriptomic level, we observed a heavily infiltrated but immunosuppressive tumor microenvironment (Fig. 5, Fig. S7, and Table S13), which is further illustrated in supplementary materials. To confirm the differences in immune infiltrations, we performed immunohistochemistry and toluidine blue staining on tissue microarrays of 229 patients. There were more CD8 + T cells (p = 0.051) and Tregs (p = 0.041) in aggressive tumors, more mast cells (p = 0.019) in indolent tumors. Detailed analyses could be found in the supplementary materials.
A Waterfall plot demonstrating clinical information and genetic mutations in indolent and aggressive renal tumors. B Gene ontology analyses of aggressive versus indolent tumors. C Heatmap displaying expressions of immune-related gene clusters, angiogenesis gene clusters, and prognostic gene clusters in indolent and aggressive tumors. D Raincloud plot of immune score in indolent (n = 93) and aggressive tumors (n = 127). E Raincloud plot of immune suppression score in indolent (n = 93) and aggressive tumors (n = 127). F Immune cell fractions in indolent (n = 93) and aggressive tumors (n = 127). G Violin plot of immune cell infiltrations in indolent and aggressive tumors (B cells, p = 0.702; CD8 + T cells, p = 0.006; CD4 + T cells, p = 0.200; Tregs, p = 0.028; NK cells, p = 0.221; Monocytes, p = 0.304; Macrophages, p = 0.127; Dendritic cells, p = 0.731; Mast cells, p < 0.001; Eosinophils, p = 0.129; Neutrophils, p = 0.328). H Representative images of FOXP3 staining and raincloud plot of Treg densities in indolent (n = 171) and aggressive tumors (n = 51). I Representative images of CD8 staining and raincloud plot of CD8 + T cell densities in indolent (n = 176) and aggressive tumors (n = 52). J Representative images of toluidine blue staining and raincloud plot of Mast cell densities in indolent (n = 175) and aggressive tumors (n = 52). The components of the box plots in the raincloud plots comprise the minimum and maximum non-outlier values in the dataset, the median (center), the first quartile (Q1, 25th percentile), and third quartile (Q3, 75th percentile) defining the box bounds, and whiskers extending to 1.5 times the interquartile range (IQR) beyond Q1 and Q3, with outliers plotted separately. The error bars in the violin plot represented the upper and lower quartiles. P values were two-sided and not adjusted for multiple comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001.
Discussion
Over the past decades, increased use of medical imaging has led to higher detection of renal masses, along with diagnosis at smaller tumor size35,36. The decision to offer surgical resection is often made without pathologic diagnosis, which is within standard treatment37. However, this can sometimes lead to overtreatment of benign renal lesions7. Percutaneous biopsy yields relatively high accuracy in determining histologic subtype, but it’s invasive and time-consuming8, with risks of complications such as significant pain, biopsy tract seeding, perinephric hematoma, etc. The non-diagnostic rate can be as high as 22.6%38. Meanwhile, accurate diagnosis of fat-poor angiomyolipoma, oncocytoma, and complex renal cysts using CT or MRI by radiologists is challenging23. There exists a gap between the clinical need for a simple yet accurate characterization of renal masses and current diagnostic tools.
Our study primarily focuses on two primary tasks. The first primary task centers on developing a deep learning model capable of differentiating between malignant and benign renal masses. This diagnostic model has consistently demonstrated high accuracy across multiple cohorts. The R.E.N.A.L. nephrometry score-based nomogram33 have been used to evaluate the risks of a renal mass being malignant with an AUC of 0.76. Some radiomics models have been proposed as well. The largest CT-based radiomics model was developed by Yap et al. from CT images of 735 patients26 and achieved an AUC of 0.75. Nassiri N et al. proposed a radiomic-based machine learning algorithm using CT images from 684 patients and achieved an AUC of 0.8434. This nephreometry score nomogram, along with other radiomics models were all validated in small internal cohorts without sufficient external testing, while our models have been validated externally and prospectively across diverse cohorts with large sample size26,29,39. To validate its accuracy, we first compared our model against a standardized radiomics model and the nephrometry score nomogram, observing that our deep learning model outperformed the others. Secondly, through subgroup analyses, we observed robust performance in SRM subgroups, solid renal mass subgroups, and cystic renal mass subgroups. Furthermore, our model outperformed the average diagnostic capabilities of seven experienced radiologists from tertiary medical centers. We also conducted an exploration of how our deep learning model for differentiating between benign and malignant renal masses could be integrated into clinical workflows and potentially impact current practices. With the assistance of the deep learning model, the diagnostic accuracy of radiologists significantly improved. We observed that seven radiologists displayed a higher degree of diagnostic consistency amongst themselves when compared to the deep learning model. This suggests that the deep learning model and the radiologists may be approaching the classification of renal masses from different yet complementary perspectives. This divergence in diagnostic approach could explain why, when radiologists were informed of the model’s results, their diagnostic accuracy increased significantly. There were instances where the deep learning model provided accurate diagnoses while the radiologists erred, and vice versa, further highlighting the complementary nature of their diagnostic capabilities.
In the modern era, with growing evidence suggesting a non-inferior survival outcome for active surveillance and image-guide ablation compared with instant curative nephrectomy, there has been an increased adoption of ablation and active surveillance in the treatment of renal cell carcinoma9,10. Predicting aggressive versus indolent tumor histology could better assess oncologic risk relative to competing risks, which is important for precision medicine. For example, decision-making can be difficult in some elderly or co-morbid patients. Our findings may guide such patients with AI-predicted aggressive tumors towards prompt surgeries while patients with AI-predicted indolent tumors could consider active surveillance or ablation. Besides, nephron-sparing surgeries are recommended as the standard of care for localized renal tumors less than 7 cm when technically feasible, while the risk of complications and recurrences increases with anatomical complexity40. Preoperative aggressiveness estimation for challenging tumors may support surgical decisions, favoring radical nephrectomy for aggressive tumors. However, no radiology-related studies have comprehensively evaluated tumor aggressiveness taking stage, grade, histologic subtype, necrosis, and sarcomatoid differentiation into account. In previous studies aiming to predict the aggressiveness of renal tumors, researchers only focused on predicting high versus low ISUP or Fuhrman grade in ccRCC patients based on CT or MRI images, achieving AUCs ranging from 0.62 to 0.9141. However, many lacked external validation or involved small test cohorts, which raises concerns about model generalizability and bias. Small training sets in earlier studies also increased the risk of model overfitting. In a study with one of the largest cohorts, Demirjian et al. developed a CT-based radiomics signature to discriminate low-grade from high-grade ccRCC in 587 patients. The signature achieved an AUC of 0.7042.
The second primary task of our study involves developing a deep-learning model to distinguish between indolent and aggressive renal masses. In our study, we enrolled patients from six Chinese medical centers and five public imaging datasets. Our model was tested in an independent external cohort and a prospective multi-center cohort, further proving its generalization ability. Another important point is that our analysis includes most histologic subtypes, which reflects the real clinical scenario than studies focused on ccRCC subtype only43,44,45,46. Different histologic subtypes of renal lesions have varied prognosis. For example, collecting duct RCC and renal cell carcinoma, NOS are aggressive renal cell carcinoma with poor prognosis, while subtypes such as chromophobe RCC, clear cell papillary renal cell tumors and mucinous tubular and spindle cell RCC follow an indolent course with good prognosis47,48,49. In this study we predict the overall aggressiveness of renal tumors using a deep learning model based on radiological imaging, where the labels incorporate extensive pathological information, including tumor nuclear grade and histologic subtype, to determine the aggressiveness of renal masses. Other aggressive histology, such as sarcomatoid differentiation and coagulative necrosis are included in radiological aggressiveness evaluation50,51. The R.E.N.A.L. nephrometry score-based nomogram33 have been used to evaluate the risks of a renal mass being high-grade with AUCs of 0.76. Our second deep-learning diagnostic model has exhibited strong discriminatory capabilities and has outperformed a standardized radiomics model and the nephrometry score nomogram, maintaining robust performance in renal mass subgroups.
We further focused on subgroups of utmost clinical significance, specifically solid T1a and T1 renal masses. Our deep learning model demonstrated resilient performance across most subset cohorts. Upon analysis of the external test set, it was observed that our diagnostic model, designed to distinguish between indolent and aggressive tumors, achieved an AUC of 0.619. In the case of T1a tumors, aggressiveness was predominantly determined by the ISUP grade. However, it’s important to note that there exists a notable disparity in grading among pathologists from different hospitals. Even with slide reviews by pathologists, their grading is still significantly influenced by the initial grading outcomes. Larger lesions, particularly those categorized as >T2, typically exhibit more pronounced histological markers of aggressiveness. Notably, our diagnostic models maintained impressive performances across various size categories, highlighting that, apart from tumor size, our models possess substantial predictive abilities.
Notably, existing studies have not investigated the prognosis of radiographically aggressive vs. indolent renal tumors. It remains unknown whether tumors predicted as high grade by previous radiographic models actually have inferior outcomes43,44,45,46. We analyze the oncologic outcomes of tumors predicted as aggressive or indolent based on radiographic features. In our study, AI-predicted aggressive tumors demonstrated significantly worse outcomes compared to AI-predicted indolent tumors, confirming the higher malignancy of the aggressive subgroup. Interestingly, the AI-aggressiveness score also showed a higher C-index in predicting survival than the radiomics model and nephrometry score nomogram, further confirming its accuracy in identifying aggressive renal tumors from a distinct perspective of survival.
We also analyzed differences in biological behavior between aggressive and indolent tumors, which demonstrated distinct genetic and immune landscapes. Aggressive tumors were associated with more frequent SETD2 and AHNAK2 mutations. As the main methyltransferase for H3K36me3, SETD2 plays important roles in active transcription, alternative splicing, transcriptional repression, and DNA damage repair52. SETD2 mutations are a key molecular feature in renal cell carcinoma and are associated with recurrence and poor prognosis53. AHNAK2 was reported to be a novel prognostic marker and oncogenic protein by mediating hypoxia pathway-driven epithelial-mesenchymal transition and stem cell properties in renal cell carcinoma54. Further transcriptomic analyses revealed that markers, pathways, and immune cells referring to immune suppression significantly elevated in aggressive renal tumors. We found there were significantly higher infiltrations of CD8 + T cells and Tregs in aggressive tumors. In contrast to the majority of tumors, high densities of CD8 + T cells were associated with poor prognosis in RCCs. Tregs play a crucial role in tumor immune evasion. Tregs suppress antitumor T cell responses through multiple mechanisms, such as secreting inhibitory cytokines like TGFβ and IL-10, expressing the immune-inhibitory receptor such as CTLA4, etc55. A graphic abstract (Fig. S8) provides a visual representation of our study’s framework.
We employed state-of-the-art AI algorithms for the automatic segmentation of kidney structure and detection of renal masses, which is necessary for large-scale imaging studies. In traditional radiomics studies, tumor boundaries are first manually drawn for feature extraction by experienced radiologists which is time-consuming and less reproducible. In this study, we adapted the pre-trained nnU-net and fine-tuned it on our multi-center dataset, achieving satisfactory segmentation accuracy comparable to experienced radiologists. This makes the analysis pipeline more efficient and suitable for clinical application as radiologists only need to perform quality control and correct problematic results.
The deep learning models for renal mass classification utilized multi-phase CT images containing complementary information. To avoid the potential registration error from patient motion, a late fusion strategy was used where image features were extracted individually for each phase and then integrated through a cross-attention module. Moreover, in practical clinical scenarios, it’s not uncommon for some patients to have missing CT phases. To deal with the missing-phase problem, we trained single-phase and dual-phase candidates for easy adaption in real-world datasets. By adapting to situations where specific phases are absent, our models can enhance their clinical applicability and prove even more valuable in real-world settings. Compared to the pre-defined image features in radiomics studies, deep learning models have more expressive power and achieve better performance. The advantage is more profound in large-scale datasets like the multi-center cohort in our study. The CAM visualization also provides some interpretability of the deep learning models which are essential for reliable application in future clinical practice.
Several limitations remained. First of all, this is a retrospective study in nature, with inevitable selection bias. But we have validated our retrospective model in a multi-centered prospective cohort and multiple external test cohorts, which suggested a strong robustness and clinical applicability. Secondly, we only included patients who received nephrectomy, because in medical centers included in this study, needle biopsy was scarcely performed for resectable suspicious renal masses in actual clinical practice. Most doctors and patients opt for direct surgeries over biopsies. In most cases, needle biopsies are performed in unresectable or metastatic RCCs requiring further systematic treatment in these medical centers. This may bring bias but the proportion is extremely low compared to the large sample size, thus is unlikely to significantly affect the model performance. Another limitation of our study was that the training set included patients from Chinese hospitals, most of whom were Asian. Despite this, our model still achieved good performance in the TCIA cohort, where the majority of patients were white. Specifically, the model achieved an AUC of 0.726 for differentiating between indolent and aggressive renal tumors in White patients from the TCIA cohort. Additionally, our approach of using the maximal tumor slice to represent the lesion inevitably overlooks certain spatial heterogeneities within the tumor.
We have retrospectively developed and prospectively validated deep learning-based diagnostic models discriminating benign versus malignant renal masses and indolent lesions versus aggressive tumors in a multi-centered cohort. Our models demonstrated high AUCs in distinguishing benign from malignant, indolent from aggressive renal lesions across different cohorts. Patients with AI-predicted aggressive tumors have significantly worse RFS, DSS, and OS compared to patients with AI-predicted indolent tumors. The AI-aggressiveness score showed a higher C-index in predicting survival than the TNM stage and ISUP grade. Aggressive tumors are associated with higher mutations of SETD2 and AHNAK2, and demonstrate a heavily immune infiltrated but immunosuppressive TME with increased CD8 + T cell and Treg infiltrations.
Methods
Study participants
The study followed the Declaration of Helsinki and complies with all relevant ethical regulations. The study was undertaken with approvals from the ethics committee of Zhongshan Hospital, the First Hospital of Zhejiang Province, Quanzhou First Hospital, the First People’s Hospital of Lianyungang, Xiamen Branch of Zhongshan Hospital, and Zhangye People’s Hospital. Patients who received nephrectomy from January 2009 to June 2021 in Zhongshan Hospital, from February 2017 to June 2021 in Quanzhou First Hospital (Quanzhou Hospital), from October 2016 to June 2021 in the First People’s Hospital of Lianyungang (Lianyungang Hospital), from November 2017 to June 2021 in Xiamen Branch of Zhongshan Hospital (Xiamen Hospital) and from July 2013 to June 2021 in Zhangye People’s Hospital (Zhangye Hospital) with non-contrast, arterial and venous phase preoperative CT scans were screened for the training and internal test sets. Patients who underwent nephrectomy from January 2016 to June 2021 in the First Hospital of Zhejiang Province (Zhejiang Hospital) were screened for the external test set. Patients who received nephrectomy from October 2021 to June 2022 from Zhongshan Hospital, Zhejiang Hospital, Lianyungang Hospital, and Xiamen Hospital were prospectively collected for the prospective test set. Patients with either arterial or venous phase preoperative CT scans from The Cancer Imaging Archive (TCIA) were screened for the TCIA test set56. We aimed to evaluate the performance of deep learning models in a TCIA test set under real-world conditions where certain CT phases may be absent. Exclusion criteria included bilateral, multiple, or metastatic renal tumors; renal lesions proved to be lymphoma or metastasis from other sites; prior history of nephrectomy. We adhered to the APA Style Guide for Bias-Free Language, the JAMA Network Updated Guidance, and the NASEM guidelines in reporting race. The race information for the TCIA cohort was obtained directly from cBioPortal. For patients from other hospitals, race was self-reported. Sex and/or gender of participants was determined based on self-report. Given the lack of appreciable differences in key tumor characteristics, including tumor size, stage, grade, and outcome between males and females, we did not carry out sex or gender analyses. Informed consents for patients from retrospective cohorts were waived, and those for patients from the prospective test cohort were obtained. The informed consent for patients from retrospective cohorts was waived due to the fact that the study involved only the analysis of pre-existing CT images, posing no risk of harm to patients and not constituting an intervention. In the end, there were 4169 patients in the above six medical centers and 388 cases in the TCIA dataset included in our study. Clinical and radiological information of the TCIA test set were downloaded from the National Cancer Institute, the Cancer Imaging Program (CIP) (https://www.cancerimagingarchive.net). The TCIA cohort was composed of four independent public datasets, the Climb 4 Kidney Cancer—Kidney and Kidney Tumor Segmentation Challenge (C4KC-KiTS) cohort of the University of Minnesota Medical Center, the Cancer Genome Atlas (TCGA)—Kidney Clear Cell Carcinoma (KIRC) cohort, TCGA—Kidney Papillary Cell Carcinoma (KIRP) cohort, TCGA—Kidney Chromophobe (KICH) cohort and the Clinical Proteomic Tumor Analysis Consortium—Clear Cell Renal Cell Carcinoma (CPTAC-CCRCC) cohort. Patients with malignant renal masses who received curative nephrectomy from 2009 to 2019 in Zhongshan Hospital (Zhongshan cohort) or from 2012 to 2017 in Zhejiang Hospital (Zhejiang cohort) had follow up information. The last follow up time for the two cohorts was September, 2021. These two subsets of patients with follow up information were termed Zhongshan cohort and Zhejiang cohort. The tumor stage at the time of surgery was determined according to the 8th edition of the AJCC cancer staging system. R.E.N.A.L. scores were evaluated by seven experienced radiologists. To avoid potential confusion with data combinations and cohort information, we have provided a summary in Table S1, detailing the patient composition, types of data, and analyses conducted for each cohort.
Histologic classification
Renal tumors were classified as benign indolent, malignant indolent, and malignant aggressive according to previous histologic research47,57 (Table 1). Benign and indolent renal masses included angiomyolipoma, complex renal cyst, oncocytoma, adult cystic nephroma, mixed epithelial and stromal tumor, metanephric adenoma, leiomyoma, haemangioma, schwannoma, juxtaglomerular cell tumor, inflammation, and hematoma. Papillary adenoma and renomedullary interstitial cell tumor were clinically silent and discovered as incidental findings, thus they were not included in our study. Malignant and indolent renal tumors included indolent ccRCC (ccRCC without invasion into major veins or perinephric tissues, grade 3–4 components, necrosis or sarcomatoid differentiation), indolent papillary RCC (papillary RCC without invasion into major veins or perinephric tissues, grade 3–4 components or sarcomatoid differentiation), chromophobe RCC, clear cell papillary renal cell tumors, multilocular cystic renal neoplasm of low malignant potential, succinate dehydrogenase-deficient RCC, mucinous tubular and spindle cell carcinoma, tubulocystic RCC, nephroblastoma, epithelioid angiomyolipoma, solitary fibrous tumor, other oncocytic tumors of the kidney and well-differentiated neuroendocrine tumor. Any indolent subtype with sarcomatoid differentiation were considered aggressive. Malignant and aggressive subtypes included aggressive ccRCC (non-indolent ccRCC), aggressive papillary RCC (non-indolent papillary RCC), TFE3-rearranged renal cell carcinoma, renal cell carcinoma NOS, collecting duct carcinoma, medullary carcinoma, any subtypes with sarcomatoid differentiation and sarcomas such as leiomyosarcoma, rhabdomyosarcoma, synovial sarcoma, Ewing sarcoma, etc. Tumor nucleus grades were recorded in clear cell renal cell carcinoma (ccRCC) and papillary renal cell carcinoma only. Three experienced pathologists re-valuated tumor subtypes according to the 2022 WHO classification for renal tumors and tumor nucleus grades according to the WHO/ISUP grading system for cases with available slides in the Zhongshan cohort and Zhejiang cohort. For few tumor samples graded according to Fuhrman grade before 2016 unavailable for re-evaluation, Fuhrman grades were matched to ISUP grades.
Automatic detection of renal masses
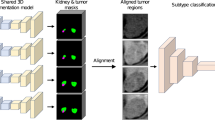
Firstly, nnU-Net was used to segment the kidney structure and renal masses on arterial phase CT images58. It is a convolutional neural network with an encoder-decoder structure that integrates multi-scale image features for medical image segmentation. The segmentation model takes 3D arterial phase images as the input and outputs the segmentation map of kidneys and tumors. The model was pre-trained on the official KiTS challenge dataset and fine-tuned on 100 cases from the training set of this study where ground-truth segmentations were provided by an experienced radiologist59. The detailed architecture of nnU-Net and benchmark of segmentation performance are provided in the Supplementary Material (Fig. S1 and Table S2). Next, the mass center of the detected tumor was calculated, and a volume of interest (VOI) with a fixed size of 14\(\times\)14\(\times\)16 cm was generated. To minimize potential errors in localization, a quality control step was implemented, wherein a radiologist reviewed and, if necessary, manually corrected the mass center. For the cases without arterial phase imaging in the TCIA dataset, the mass center was manually drawn at the axial image slice with maximum renal masses on either venous or non-contrast CT images. With the generated VOI, the multi-phase CT images were cropped and registered to the arterial phase image. Furthermore, the axial slices with maximum tumor were extracted from the registered multi-phase imaging volumes and resampled to a spatial resolution of 0.625\(\times\)0.625 mm.
Deep learning models for tumor diagnostics
We first investigated which image feature extraction backbone to employ. In our preliminary exploration of the malignancy classification task based on arterial phase CT, ResNet-18 performed the best, while increasing the complexity of network design did not lead to better classification results, as depicted in Table S3. Based on the pre-processed imaging slices, we developed a multi-phase convolutional neural network to integrate multi-phase CT images and predict tumor diagnostics. The ResNet-18 was used as the backbone network to extract image features for non-contrast, arterial, and venous phase images, respectively60. The weights of ResNet-18 were initialized using a pre-trained ImageNet model and fine-tuned on our curated dataset of CT images. This fine-tuning process allowed the model to adapt to domain-specific features present in medical images, enhancing its generalization capability for tumor diagnostics.
Then the individual features were fused to predict the final diagnostics. The detailed architecture of each network module and implementation details are provided in the Supplementary Material (Fig. S2). To deal with the missing phases in the TCIA dataset, in addition to the multi-phase model, we also built three models with single-phase input and three models with dual-phase input. The prediction of tumor malignancy and invasiveness were formulated as two binary classification tasks, and we trained two separate networks to predict corresponding probabilities.
Radiological interpretation
Seven experienced radiologists (Linpeng Yao, Xue Zhang, Risheng Huang, Xiaoxia Li, Jianjun Zhou, Feng Chen, and Chenchen Dai), each were fellowship trained in abdominal imaging with over 5 years of experience from prominent academic medical centers, evaluated the cases. Prior to the evaluation, a urological imaging expert (Professor Jianjun Zhou, who has reviewed over 5000 renal mass cases) conducted a training session for all observers. This training encompassed the explanation of common imaging characteristics through 50 representative cases from our internal test set. Additionally, standard scoring criteria from the literature, including the Ottawa Criteria61, and the 2019 version of the Bosniak classification62, were reviewed. However, the radiologists were not compelled to strictly adhere to these criteria, allowing for a more realistic clinical approach. The triple-phase CT images were then presented anonymously and in random order through our local picture archiving and communication system. The radiologists, blinded to any clinical or pathological information, were tasked with predicting whether the renal mass was malignant or benign. The radiologists were asked to provide binary predictions of malignancy (malignant or benign) for each case.
Computational analyses of genetic and immune landscapes
RNA-seq and mutational data of TCGA cohorts and CPTAC-CCRCC cohort were downloaded from the GDC Data Portal (https://portal.gdc.cancer.gov). We performed Gene Ontology (GO) analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses for upregulated genes in aggressive renal tumors in the TCIA cohort. Gene Set Enrichment Analysis was performed by applying the GSEA63 “weighted” enrichment statistics on a score for enrichment or depletion. For GSEA, we utilized pathways contained in Hallmark, Reactome, BioCarta, PID, GO, and KEGG databases. We calculated infiltrations of B cells, CD8 + T cells, CD4 + T cells (non-regulatory), Tregs, NK cells, monocytes, macrophages, dendritic cells, mast cells, eosinophils, and neutrophils in tumors from TCGA cohort CIBERSORT, a computational method for evaluating leukocyte representation in bulk tumors64. Infiltrations of major types of immune cells were also evaluated by single sample gene set enrichment analysis (ssGSEA)63,65. Stromal score, immune score, and estimate score were calculated with ESTIMATE for inferring stromal cell admixture, immune cell admixture, and tumor purity, respectively. The metagene value for immune suppression was measured by summarizing the gene signature as the mean expression of each gene in the category66.
Immunohistochemistry
For evaluation of infiltrations of Tregs, CD8 + T cells, and mast cells, we collected formalin-fixed, paraffin-embedded surgical specimens from patients who received nephrectomy from August 2020 to June 2021 with malignant tumors in Zhongshan cohort and constructed tissue microarrays for immunohistochemistry, and we term this cohort the IHC cohort. A total of 229 cases with obtainable specimens were collected, of which 177 were indolent and 52 were aggressive. Tregs were stained with FOXP3 antibody (ab22510, Abcam, diluted 1:100) and CD8 + T cells were stained with CD8 antibody (ab245118, Abcam, diluted 1:1000). We used toluidine blue stains to evaluate mast cell infiltrations. The densities of each immune cells were recorded as cells/mm2.
Statistical analyses
For statistical analyses, SPSS Statistics 21.0 and R software 3.51 were used. We used Welch’s t-test for comparisons between continuous variables. Measurements were taken from distinct samples. For categorical variables, Pearson’s chi-square test or Fisher’s exact test were performed. The prediction performance of the deep learning models was evaluated using receiver operating characteristic curve (ROC) analysis and by estimating the area under the curve (AUC). In addition, quantitative classification metrics were calculated, including accuracy, sensitivity, and specificity. For each reader sensitivity and specificity values, the corresponding specificity and sensitivity of the deep learning model at that sensitivity or specificity settings are computed. A significant difference is determined by means of the McNemar test. The Kaplan–Meier analyses and univariate Cox regression analyses were applied for comparisons of patient survival. To further determine the contribution of AI-aggressiveness score compared to other risk factors as predictors of survival, they were all combined in a multivariate Cox proportional hazards model. Survival outcomes included disease-specific survival (DSS), recurrence-free survival (RFS), and overall survival (OS). Disease-specific survival was defined as the time of surgery to the time of death from renal tumors. Recurrence-free survival was calculated from the time of surgery to local recurrence, distant metastasis, or death from renal tumors. Overall survival represents time from surgery to death from all causes. For all survival analyses, we included patients with malignant tumors only. Values of p < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The imaging datasets utilized in this study, with the exception of those from the TCIA cohort, are currently restricted from public release due to data privacy laws and the policies of the respective institutional review boards. The TCIA cohort data can be downloaded from the Cancer Imaging Archive (https://www.cancerimagingarchive.net/collections/) and the GDC Data Portal (https://portal.gdc.cancer.gov/). If the interested researchers want to achieve the restricted imaging datasets for non-commercial use, they can request for the corresponding author S.W. (shuowang@fudan.edu.cn). Corresponding author will review their requests and ask for consent from each center, requestors will receive a response within 4 weeks. Source data are provided with this paper.
Code availability
A valid OSI-approved open-source license, the Apache License 2.0, is applied to our code. The code repository includes a clear LICENSE file specifying that the code is released under the Apache License 2.0. Codes are publicly available at (https://github.com/shuowang26/renal-mass-ai). Other information is available from the corresponding author (S.W.) upon request.
References
Sun, M. et al. Age-adjusted incidence, mortality, and survival rates of stage-specific renal cell carcinoma in North America: a trend analysis. Eur. Urol. 59, 135–141 (2011).
Znaor, A., Lortet-Tieulent, J., Laversanne, M., Jemal, A. & Bray, F. International variations and trends in renal cell carcinoma incidence and mortality. Eur. Urol. 67, 519–530 (2015).
Jayson, M. & Sanders, H. Increased incidence of serendipitously discovered renal cell carcinoma. Urology 51, 203–205 (1998).
Simone, G. et al. Trends in the use of partial nephrectomy for cT1 renal tumors: analysis of a 10-yr European multicenter dataset. Eur. J. Surg. Oncol. 42, 1729–1735 (2016).
Wong, M. C. S. et al. Incidence and mortality of kidney cancer: temporal patterns and global trends in 39 countries. Sci. Rep. 7, 15698 (2017).
Turner, R. M. 2nd, Morgan, T. M. & Jacobs, B. L. Epidemiology of the small renal mass and the treatment disconnect phenomenon. Urol. Clin. North Am. 44, 147–154 (2017).
Gill, I. S. et al. Comparison of 1,800 laparoscopic and open partial nephrectomies for single renal tumors. J. Urol. 178, 41–46 (2007).
Sanchez, A., Feldman, A. S. & Hakimi, A. A. Current management of small renal masses, including patient selection, renal tumor biopsy, active surveillance, and thermal ablation. J. Clin. Oncol. 36, 3591–3600 (2018).
Bhagavatula, S. K. et al. Percutaneous CT- and MRI-guided cryoablation of cT1 renal cell carcinoma: intermediate- to long-term outcomes in 307 patients. Radiology 296, 687–695 (2020).
Pierorazio, P. M. et al. Five-year analysis of a multi-institutional prospective clinical trial of delayed intervention and surveillance for small renal masses: the DISSRM registry. Eur. Urol. 68, 408–415 (2015).
Jewett, M. A. et al. Active surveillance of small renal masses: progression patterns of early stage kidney cancer. Eur. Urol. 60, 39–44 (2011).
Uzosike, A. C. et al. Growth kinetics of small renal masses on active surveillance: variability and results from the DISSRM registry. J. Urol. 199, 641–648 (2018).
Finelli, A. et al. Management of small renal masses: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 35, 668–680 (2017).
Young, J. R. et al. Clear cell renal cell carcinoma: discrimination from other renal cell carcinoma subtypes and oncocytoma at multiphasic multidetector CT. Radiology 267, 444–453 (2013).
Lee-Felker, S. A. et al. Qualitative and quantitative MDCT features for differentiating clear cell renal cell carcinoma from other solid renal cortical masses. Am. J. Roentgenol. 203, W516–W524 (2014).
Jinzaki, M. et al. Double-phase helical CT of small renal parenchymal neoplasms: correlation with pathologic findings and tumor angiogenesis. J. Comput. Assist. Tomogr. 24, 835–842 (2000).
Cornelis, F. et al. Combined late gadolinium-enhanced and double-echo chemical-shift MRI help to differentiate renal oncocytomas with high central T2 signal intensity from renal cell carcinomas. Am. J. Roentgenol. 200, 830–838 (2013).
Rosenkrantz, A. B. et al. MRI features of renal oncocytoma and chromophobe renal cell carcinoma. Am. J. Roentgenol. 195, W421–W427 (2010).
Silverman, S. G., Gan, Y. U., Mortele, K. J., Tuncali, K. & Cibas, E. S. Renal masses in the adult patient: the role of percutaneous biopsy. Radiology 240, 6–22 (2006).
Bjurlin, M. A. et al. Influence of renal biopsy results on the management of small kidney cancers in older patients: Results from a population-based cohort. Urol. Oncol. 35, 604 e601–604 e609 (2017).
Rasmussen, R. G. et al. Association of clear cell likelihood score on MRI and growth kinetics of small solid renal masses on active surveillance. Am. J. Roentgenol. 218, 101–110 (2022).
Leppert, J. T. et al. Utilization of renal mass biopsy in patients with renal cell carcinoma. Urology 83, 774–779 (2014).
Schieda, N. et al. Active surveillance of renal masses: the role of radiology. Radiology 302, 11–24 (2022).
Feng, Z. et al. Machine learning-based quantitative texture analysis of CT images of small renal masses: differentiation of angiomyolipoma without visible fat from renal cell carcinoma. Eur. Radiol. 28, 1625–1633 (2018).
Yan, L. et al. Angiomyolipoma with minimal fat: differentiation from clear cell renal cell carcinoma and papillary renal cell carcinoma by texture analysis on CT images. Acad. Radiol. 22, 1115–1121 (2015).
Yap, F. Y. et al. Shape and texture-based radiomics signature on CT effectively discriminates benign from malignant renal masses. Eur. Radiol. 31, 1011–1021 (2021).
Bektas, C. T. et al. Clear cell renal cell carcinoma: machine learning-based quantitative computed tomography texture analysis for prediction of Fuhrman nuclear grade. Eur. Radiol. 29, 1153–1163 (2019).
Xi, I. L. et al. Deep learning to distinguish benign from malignant renal lesions based on routine MR imaging. Clin. Cancer Res. 26, 1944–1952 (2020).
Xu, Q. et al. Differentiating benign from malignant renal tumors using T2- and diffusion-weighted images: a comparison of deep learning and radiomics models versus assessment from radiologists. J. Magn. Reson. Imaging 55, 1251–1259 (2021).
Zhao, Y. et al. Deep learning based on MRI for differentiation of low- and high-grade in low-stage renal cell carcinoma. J. Magn. Reson. Imaging 52, 1542–1549 (2020).
Lin, F. et al. A CT-based deep learning model for predicting the nuclear grade of clear cell renal cell carcinoma. Eur. J. Radiol. 129, 109079 (2020).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. In 16th IEEE International Conference on Computer Vision (ICCV) 618–626 (IEEE, 2017).
Kutikov, A. et al. Anatomic features of enhancing renal masses predict malignant and high-grade pathology: a preoperative nomogram using the RENAL Nephrometry score. Eur. Urol. 60, 241–248 (2011).
Nassiri, N. et al. A radiomic-based machine learning algorithm to reliably differentiate benign renal masses from renal cell carcinoma. Eur. Urol. Focus 8, 988–994 (2022).
Hollingsworth, J. M., Miller, D. C., Daignault, S. & Hollenbeck, B. K. Rising incidence of small renal masses: a need to reassess treatment effect. J. Natl Cancer Inst. 98, 1331–1334 (2006).
Kane, C. J., Mallin, K., Ritchey, J., Cooperberg, M. R. & Carroll, P. R. Renal cell cancer stage migration: analysis of the National Cancer Data Base. Cancer 113, 78–83 (2008).
Campbell, S. C. et al. Guideline for management of the clinical T1 renal mass. J. Urol. 182, 1271–1279 (2009).
Pierorazio, P. M. et al. Management of renal masses and localized renal cancer: systematic review and meta-analysis. J. Urol. 196, 989–999 (2016).
Lee, H. S., Hong, H., Jung, D. C., Park, S. & Kim, J. Differentiation of fat-poor angiomyolipoma from clear cell renal cell carcinoma in contrast-enhanced MDCT images using quantitative feature classification. Med. Phys. 44, 3604–3614 (2017).
Simhan, J. et al. Objective measures of renal mass anatomic complexity predict rates of major complications following partial nephrectomy. Eur. Urol. 60, 724–730 (2011).
Kuusk, T., Neves, J. B., Tran, M. & Bex, A. Radiomics to better characterize small renal masses. World J. Urol. 39, 2861–2868 (2021).
Demirjian, N. L. et al. CT-based radiomics stratification of tumor grade and TNM stage of clear cell renal cell carcinoma. Eur. Radiol. 32, 2552–2563 (2022).
Yasar, S., Voyvoda, N., Voyvoda, B. & Ozer, T. Using texture analysis as a predictive factor of subtype, grade and stage of renal cell carcinoma. Abdom. Radiol. 45, 3821–3830 (2020).
Cornelis, F. et al. Multiparametric magnetic resonance imaging for the differentiation of low and high grade clear cell renal carcinoma. Eur. Radiol. 25, 24–31 (2015).
Yi, X. et al. Computed tomography radiomics for predicting pathological grade of renal cell carcinoma. Front. Oncol. 10, 570396 (2020).
Cui, E. et al. Predicting the ISUP grade of clear cell renal cell carcinoma with multiparametric MR and multiphase CT radiomics. Eur. Radiol. 30, 2912–2921 (2020).
Bhindi, B. et al. The probability of aggressive versus indolent histology based on renal tumor size: implications for surveillance and treatment. Eur. Urol. 74, 489–497 (2018).
Massari, F. et al. The tumor entity denominated “clear cell-papillary renal cell carcinoma” according to the WHO 2016 new classification, have the clinical characters of a renal cell adenoma as does harbor a benign outcome. Pathol. Oncol. Res. 24, 447–456 (2018).
Zhao, M., He, X. L. & Teng, X. D. Mucinous tubular and spindle cell renal cell carcinoma: a review of clinicopathologic aspects. Diagn. Pathol. 10, 168 (2015).
Zhang, B. Y. et al. A novel prognostic model for patients with sarcomatoid renal cell carcinoma. BJU Int. 115, 405–411 (2015).
Leibovich, B. C. et al. Predicting oncologic outcomes in renal cell carcinoma after surgery. Eur. Urol. 73, 772–780 (2018).
Wagner, E. J. & Carpenter, P. B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 (2012).
Hakimi, A. A. et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin. Cancer Res. 19, 3259–3267 (2013).
Wang, M. et al. AHNAK2 is a novel prognostic marker and oncogenic protein for clear cell renal cell carcinoma. Theranostics 7, 1100–1113 (2017).
Speiser, D. E., Ho, P. C. & Verdeil, G. Regulatory circuits of T cell function in cancer. Nat. Rev. Immunol. 16, 599–611 (2016).
Clark, K. et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013).
Jang, A. et al. Multiple growth periods predict unfavourable pathology in patients with small renal masses. BJU Int. 121, 732–736 (2018).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Heller, N. et al. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: results of the KiTS19 challenge. Med. Image Anal. 67, 101821 (2021).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (IEEE, 2016).
Al Nasibi, K. et al. Development of a multiparametric renal CT algorithm for diagnosis of clear cell renal cell carcinoma among small (</= 4 cm) solid renal masses. Am. J. Roentgenol. 219, 814–823 (2022).
Silverman, S. G. et al. Bosniak classification of cystic renal masses, version 2019: an update proposal and needs assessment. Radiology 292, 475–488 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Gentles, A. J. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015).
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013).
Kardos, J. et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight 1, e85902 (2016).
Acknowledgements
This study was funded by grants from National Natural Science Foundation of China [81902563 (Y.X.), 81974393 (J.G.), 82102967 (Y.Q.), 82202106 (C.D.), Outstanding Youth Scholars Foundation of Zhongshan Hospital [2021ZSYQ15 (Y.X.)], Science and Technology Guided Project of Fujian Province [2019D025 (J.Z.)], Scientific Research Cultivation and Medical Innovation Project of Fujian Province [2019CXB33 (J.Z.)], Hexi University President Fund Innovation Team Project [CXTD2022012 (J.Y.)], Funding Program for Young Research Projects in the Scientific and Technological Plan for High-quality Development of Health in Xiamen[2024GZL-QN027 (J.L.)], Natural Science Foundation of Fujian Province [2024J011442 (C.D.)], International Science and Technology Cooperation Program under the 2023 Shanghai Action Plan for Science [(23410710400) (S.W.)], and Shanghai Sailing Programs of Shanghai Municipal Science and Technology Committee [22YF1409300 (S.W.)]. All these study sponsors have no roles in the study design, in the collection, analysis and interpretation of data. We would like to thank Dr. Jianming Zeng (University of Macau) and all the members of his bioinformatics team for their generous help in bioinformatics analysis. We thank pathologists Qi Sun (Fudan University) and Haiyue Lin (the First People’s Hospital of Lianyungang) for re-evaluating pathologic slides. We thank radiologist Xiaoxia Li (Fudan University) for assessing tumor malignancy and R.E.N.A.L. score. The computations in this research were performed using the CFFF platform of Fudan University.
Author information
Authors and Affiliations
Contributions
Y.X., L.Y., J.L., J.Y., and Q.B. for acquisition, analysis and interpretation of data, statistical analysis, and drafting of the manuscript; Y.H., X.Z., R.H., R.W., Y.Q., L.L., and P.Z. for technical and material support, as well as statistical analyses; K.W., H.W., and S.W. for algorithm development and statistical analyses; J.Z., J.G., F.C., C.D., and S.W. for study design, interpretation of data, drafting of the manuscript, obtaining funding, and study supervision. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Issam El Naqa, Shangqi Gao and Sadhna Nandwana for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xiong, Y., Yao, L., Lin, J. et al. Artificial intelligence links CT images to pathologic features and survival outcomes of renal masses. Nat Commun 16, 1425 (2025). https://doi.org/10.1038/s41467-025-56784-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56784-z
This article is cited by
-
Fully automated 3D multi-modal deep learning model for preoperative T-stage prediction of colorectal cancer using 18F-FDG PET/CT
European Journal of Nuclear Medicine and Molecular Imaging (2025)