Abstract
Hepatic stellate cells (HSCs) are activated with chronic liver injury and transdifferentiate into myofibroblasts, which produce excessive extracellular matrices that form the fibrotic scar. While the progression of fibrosis is understood to be the cause of end-stage liver disease, there are no approved therapies directed at interfering with the activity of HSC myofibroblasts. Here, we perform a high-throughput small interfering RNA (siRNA) screen in primary human HSC myofibroblasts to identify gene products necessary for the fibrotic phenotype of HSCs. We find that depletion of ABHD17B promotes the inactivation of HSCs, characterized by reduced COL1A1 and ACTA2 expression and accumulation of lipid droplets. Mice deficient in Abhd17b are also protected from fibrosis in the setting of in vivo liver injury. While ABHD17B is a depalmitoylase, our data suggest that ABHD17B promotes fibrosis through pathways independent of depalmitoylation that include interaction with MYO1B to modulate gene expression and HSC migration. Together, our results provide an analysis of the phenotypic consequences for siRNAs targeting RNAs from >9500 genes in primary human HSCs and identify ABHD17B as a potential therapeutic target to inhibit liver fibrosis.
Similar content being viewed by others
Introduction
End-stage liver disease results from progressive fibrosis in the setting of chronic liver injury. While reversal of the underlying cause of injury can reduce the severity of fibrosis1, there are limited treatments for many sources of chronic injury, including non-alcoholic fatty liver disease/metabolic dysfunction-associated steatotic liver disease (NAFLD/MASLD)2. In other diseases of fibrotic injury, such as primary sclerosing cholangitis (PSC), there are no effective treatments beyond liver transplantation3.
HSC myofibroblasts are the primary cell type responsible for liver fibrosis4,5,6. In a healthy liver, HSCs are in a quiescent state and store vitamin A in lipid droplets4. With chronic injury, HSCs are activated and transdifferentiate into HSC myofibroblasts, characterized by the loss of lipid droplets and production of extracellular matrix (ECM) proteins that form the fibrotic scar7,8.
Resolution of liver fibrosis has been observed when the source of liver injury is removed9. In this setting, the HSC population is reduced through apoptosis, while 40–50% of HSC myofibroblasts revert to an inactive state, characterized by decreased collagen expression10,11. These results suggest that HSC myofibroblasts can be directed to an inactive phenotype with the correct signals.
Here, we performed an siRNA screen of the human druggable genome and >2000 long noncoding (lnc) RNAs to identify RNAs that could be targeted to promote HSC inactivation. We find that depletion of ABHD17B (FAM108B1) in primary human HSCs leads to a reduction in type I collagen expression and promotes HSC inactivation. These functions are independent of the depalmitoylase activity of ABHD17B and involve interaction with MYO1B. Furthermore, mice deficient in Abhd17b are protected from the development of liver fibrosis.
Results
Development of a high-throughput siRNA screen in primary human HSC myofibroblasts
We first optimized siRNA transfection conditions for human HSC myofibroblasts in a 384-well format (Supplementary Fig. 1a, b) by adapting approaches previously established for small molecule screens12,13. Next, we evaluated negative and positive controls. We transfected HSCs with non-targeting control siRNAs (NTC si) 1, 2, 3, 4, and 5 and performed Bodipy and Hoechst staining to evaluate the effect of the siRNAs on lipid accumulation and cell number. Compared to NTC si1 and si2, HSCs transfected with NTC si3, si4, and si5 demonstrated lower percentages of Bodipy positive cells with similar or reduced effect on cell numbers (Supplementary Fig. 1c, d). We compared NTC si3-si5 siRNAs with pooled siRNAs targeting GAPDH (GAPDH siP). Transfection with GAPDH siP did not cause a significant change in lipid accumulation (Supplementary Fig. 1e) and did not reduce cell numbers to the degree observed with NTC siRNAs (Supplementary Fig. 1f). Based on these results, we selected GAPDH siP as our negative control.
We then evaluated potential positive controls for induction of lipid droplets and transfection efficiency. siRNAs targeting ACTA2 and ASAH112 did not significantly increase the percentage of Bodipy-positive cells (Supplementary Fig. 1c). Depletion of PLK1 via siRNAs is established to cause death in multiple cell lines14, but despite ~70% depletion, reduction in HSC number was mild (~30–40% decrease) (Supplementary Fig. 1g, h). These results showed that depletion of ACTA2 or ASAH1 did not affect lipid accumulation, and depletion of PLK1 did not affect cell number sufficiently for these siRNAs to be used as controls.
To identify additional siRNA controls, we screened siRNA SMARTpools defined as hits in at least two different siRNA screens at the Institute of Chemistry and Cell Biology-Longwood (ICCB-L) (Supplementary Fig. 2a). We calculated the Z’ factor for each plate based on the percent positive cells of nortriptyline12 and GAPDH SMARTpool control wells and confirmed that the Z’ was acceptable (0.4-0.5). We identified potential positive hits, including AURKC, AKAP11, and TLR6 siRNAs (Supplementary Fig. 2a). We tested these siRNAs, and siRNAs targeting NFkB1, NFkB2, NR1H2, and NR1H3, which are annotated target genes shared by positive hits from our small molecule screen13, for potential positive controls. Unfortunately, none of these siRNAs showed a sufficiently robust phenotype to serve as a positive control for Bodipy staining (Supplementary Fig. 2b–j). Depletion of UBB dramatically reduced cell number (Supplementary Figs. 2a and 3a, b). We tested the Z’ of UBB siP versus GAPDH siP and nortriptyline versus GAPDH siP and confirmed that the Z’ values (0.25 and 0.60) were acceptable (Supplementary Fig. 3c, d). UBB pooled siRNA was selected as a control for transfection efficiency and nortriptyline12,13 as a positive control for Bodipy staining.
Primary siRNA screen identifies pooled siRNAs that inactivate HSC myofibroblasts
We screened the Dharmacon SMARTpool siRNA library targeting a total of 7641 mRNAs from the human druggable genome as well as siRNAs targeting 2237 lncRNAs in a one target/well format. Each SMARTpool was composed equally of four duplexes targeting the same RNA. The screen was conducted in technical triplicate (three distinct wells for each siRNA pool), with GAPDH SMARTpool siRNA as the negative control, UBB SMARTpool siRNA as the control for transfection efficiency, and 15 μM nortriptyline as the control for lipid accumulation and Bodipy staining (Fig. 1a, b). As indicated by the high toxicity of UBB siRNAs and high score of nortriptyline, the transfection was efficient and the staining worked as expected (Fig. 1b). The screening results for all siRNAs and controls are provided in Supplementary Data 1.
a Overview of the siRNA screen. b Results of the primary siRNA screen. Each dot represents the integrated score and toxicity value of three replicates of an experimental or control well. Data from all wells are shown. c The score and toxicity values for all experimental wells from the primary screen. d Summary of the validation screen results. e Summary of the siRNA knockdown efficiency in the cherrypick screen. f Grouping the final 71 hits by priority. g Expression of the 11 priority 1 genes in HSCs from the livers of CCl4-treated and control mice based on single-cell RNA sequencing results16. Circle size represents the fraction of cells expressing a gene, and color indicates mean expression level. Source data are provided as Supplementary Data 1–6 and a Source Data file.
We adapted our previous analysis method to calculate the score and toxicity for each siRNA pool13, based on the average percent Bodipy-positive cells for each siRNA pool compared to the baseline of the plate, the correlation among the three replicates, and total cell numbers. There was a small fraction of outliers, possibly due to inefficient depletion for a replicate or artifacts acquired during image acquisition. We developed a computational algorithm to correct outliers (Methods), which allowed us to retain variability while reducing the extreme variance that an outlying replicate could generate.
We identified 229 unique gene products as primary hits. This corresponded to 231 positive experimental wells because two wells contained the same siRNA pool, and one gene product was targeted by two different siRNAs. We selected primary hits based on the following criteria: 1. the score was significantly increased (FDR < 0.05); 2. toxicity was lower than the threshold determined by the nortriptyline wells; and 3a. transcripts per million (TPM) > 1 for mRNAs or TPM > 0.5 for lncRNAs in at least one of 18 HSC RNA sequencing samples12,15 or 3b. no sequencing data were available in these analyzed datasets (Fig. 1c). The screening results and expression analysis for all selected genes are provided in Supplementary Data 2.
Secondary screen with individual siRNAs
We next examined the phenotypic consequences of target depletion with individual duplexes in a one duplex/well, four wells/target format. This deconvolution secondary screen consisted of 920 experimental duplexes (targeting transcripts from 229 genes) tested in technical triplicate, relying on the same assay, controls, and analysis strategy as the primary screen (Supplementary Data 3). A total of 71 transcripts/gene products were validated as defined by at least two duplexes increasing Bodipy staining with an FDR < 0.05 and toxicity lower than the threshold determined by the UBB depletion (Fig. 1d and Supplementary Data 4).
Screening for hits that regulate ACTA2 and COL1A1
We next sought to quantify reduction in ACTA2 and COL1A1 as markers of HSC inactivation for the 71 genes identified in the deconvolution screen. For each condition we also quantified depletion of the mRNA targeted by each siRNA. To minimize technical variations, we measured ACTA2/COL1A1/target mRNA level in the same reaction as the housekeeping control PSMB213 by performing multiplexed qRT-PCR with probes labeled with different dyes. In addition to the two duplexes with the highest scores in the deconvolution screen, we included three additional siRNA duplexes from the Ambion siRNA library for 70 genes (one gene was not in the Ambion library). 352 siRNA duplexes were tested by qRT-PCR in quadruplicate.
We analyzed mRNA levels for ACTA2 and COL1A1 using two methods: a linear regression method and the standard ΔΔCt method with PSMB2 as the endogenous control (Supplementary Data 5, Methods). COL1A1 reduction did not always correlate with ACTA2 reduction. Approximately 66% of the siRNA duplexes showed a knockdown efficiency >50%, and ~69% of the siRNA duplexes reduced the expression of the target transcript by more than 40%. The knockdown efficiency for 11% of siRNA duplexes could not be determined due to low expression of the target transcript or low-quality melting curves (Fig. 1e). We prioritized candidates based on the following criteria: priority 1: at least two siRNAs reduced COL1A1 by at least 30% (based on the fold change calculated by either method) with at least 40% depletion of the target gene; priority 2: at least three siRNAs reduced ACTA2 by at least 60% (fold change calculated by either method) with at least 40% depletion of the target gene and candidate was not included in priority 1; priority 3: two siRNAs reduced ACTA2 by at least 60% (fold change calculated by either method) with at least 40% depletion of the target gene and candidate was not included in priority 1 or 2. We identified eleven genes as priority 1, four as priority 2, and eight as priority 3 (Fig. 1f and Table 1).
Of note, 37 lncRNAs were among the 229 hits in the primary screen, and three lncRNAs (AK2/NR_037592, C1orf140/NR_024236, and CLTB/NR_045724) were among the 71 hits in the deconvolution screen. Depletion of these lncRNA genes had some effect on ACTA2 but did not reduce COL1A1 mRNA levels, and no lncRNA genes were ranked as priority 1–3 candidates.
ABHD17B depletion increases lipid accumulation and induces HSC inactivation
We further analyzed the expression of the priority 1 genes in fibrotic livers based on single-cell RNA sequencing (scRNA-seq) data from control and CCl4-treated mice16. Only Abhd17b, Tgfbr2, and Psmc4 were expressed in more than 10% of HSCs and demonstrated an average increase in expression of greater than 1.5-fold in HSCs with CCl4 treatment (Fig. 1g and Supplementary Data 6). Interfering with Tgfbr2 activity or blocking TGF-β signaling in HSCs reduces liver fibrosis in vivo17,18, supporting inclusion of TGFBR2 in the final group of genes. We also found that depletion of PSMC4, a subunit of the 26S proteasome19, reduced ACTA2 and COL1A1 expression in primary human HSCs from a second donor (Supplementary Fig. 4), further confirming results of the screen.
ABHD17B encodes an enzyme with palmitoyl-hydrolase activity, and members of the ABHD17 family regulate palmitoylation of proteins including N-Ras20. The function of ABHD17B in HSCs is unknown, but in models of chronic injury, expression is induced to a higher degree in HSCs than hepatocytes or any other nonparenchymal cell type with the exception of lymphocytes (Supplementary Fig. 5). We confirmed the effect of ABHD17B depletion on lipid accumulation using primary HSCs from multiple donors. Transfection with two different ABHD17B siRNA duplexes significantly increased the accumulation of lipid droplets in cells from three donors. HSCs from the remaining donor (2) showed a significant increase in lipid accumulation with one ABHD17B siRNA (Fig. 2a). These results confirm that depletion of ABHD17B is sufficient to increase lipid accumulation, which suggests an inactive HSC phenotype.
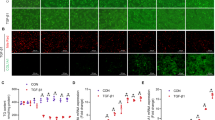
a Fraction of Bodipy positive (top) and total cells (bottom) after two days in the indicated conditions. DMSO and nortriptyline (Nor) are negative and positive controls for Bodipy, respectively. siRNAs targeting UBB are a control for transfection efficiency, and siRNAs targeting GAPDH are a negative control for Bodipy. Error bars represent mean ± SEM (n = 10 biological replicates). One-way ANOVA test. b mRNA expression quantified by qRT-PCR two days after HSCs were transfected with the indicated siRNAs. Data represent three independent experiments. Error bars represent mean ± SEM (n = 3 biological replicates). One-way ANOVA test. c ABHD17B expression quantified by qRT-PCR three days after HSC transfection. Data represent two independent experiments from two donors (5 and 6), which are further analyzed in (d–g). Each dot represents the mean of biological replicates for pooled non-targeing control (NTC siP; n = 4 biological replicates), single non-targeting control (NTC si5; n = 4), and other indicated groups (n = 2). ABHD17B siP are compared to NTC siP; ABHD17B si1 and ABHD17B si4 are compared to NTC si5. HSCs from donor 5 (d) and donor 6 (e) were transfected with the indicated siRNAs prior to serum starvation and stimulation with scar-in-a-jar conditions for 72 h. α−smooth muscle actin (αSMA, red) and collagen (orange) were visualized by immunofluorescence in the same field of view. Nuclei were stained with Hoechst (blue). Representative images from two independent experiments are shown. White bars indicate 50 µm. Quantification of αSMA fibers (f) and collagen area (g) from scar-in-a-jar assay. Data represent two independent experiments for each of the two donors (5 and 6). Each dot represents averaged result from n = 4 biological replicates. Data from donors 5 and 6 are combined for four independent experiments. Error bars represent mean ± SEM. One-way ANOVA test. h Members of the ABHD17 family were depleted using pooled siRNAs. COL1A1 levels were quantified by qRT-PCR three days after transfection. Error bars represent mean ± SEM (n = 8 biological replicates for control groups of donors 1 and 4; n = 7 for the control group of donor 3; n = 4 for all other groups). One-way ANOVA test. Source data are provided as a Source Data file.
Consistent with screening results in HSCs (Fig. 1), depletion of ABHD17B reduced ACTA2, COL1A1, and COL3A1 levels in HSCs from a second donor (Fig. 2b). Next, we tested if depletion of ABHD17B affects the formation of α-SMA fibers as a marker of HSC activation and collagen deposition in the ECM as an indication of fibrotic activity. Both the siRNA pool (siP) and two different siRNA duplexes targeting ABHD17B significantly reduced ABHD17B mRNA in HSCs from two different donors (Fig. 2c) without affecting cell number (Supplementary Fig. 6a), resulting in HSC inactivation as evident from reduced staining of α-SMA (encoded by ACTA2) and collagen type I (Fig. 2d–g and Supplementary Fig. 6b, c). siRNAs targeting TGFBR1 and ACTA2 were included as positive controls. The observed effect of ABHD17B depletion on HSC phenotype suggests that ABHD17B promotes maintenance of myofibroblast features, and depletion leads to inactivation.
ABHD17B promotes collagen expression in lung fibroblasts
Similar to liver fibrosis, idiopathic pulmonary fibrosis (IPF) is characterized by excessive ECM synthesized by activated fibroblasts and has limited treatment options21. We asked if ABHD17B also plays a role in promoting the fibrotic activity of primary human lung fibroblasts. We found that depletion of ABHD17B significantly reduced the expression of COL1A1 and ACTA2 (Supplementary Fig. 7), indicating that ABHD17B also plays a role in regulating the fibrotic activity of lung fibroblasts.
ABHD17B and ABHD17C promote collagen expression in human HSCs
ABHD17B belongs to the ABHD17 family, which includes ABHD17A and ABHD17C. All three family members are expressed in primary human HSCs, as indicated by RNA sequencing data12,13,15. Abhd17a and Abhd17b are also induced in mouse HSCs with fibrotic injury in vivo (Supplementary Fig. 8a)16. These data prompted us to ask whether other ABHD17 family members also affect collagen expression in human HSCs. We found that depletion of ABHD17B and ABHD17C each reduced COL1A1 expression in the three primary HSC donor lines tested, while depletion of ABHD17A was associated with modest reduction of COL1A1 in only one line (Fig. 2h). In addition, depletion of ABHD17B did not affect ABHD17A or ABHD17C expression except for a small decrease in ABHD17A observed in one line. (Supplementary Fig. 8b–d). These results suggest that the phenotypes observed with depletion of ABHD17B are not affected by changes in expression of other ABHD17 family members.
Analysis of ABHD17B structure
ABHD17 family proteins were identified as depalmitoylases in a screen for serine hydrolases that increase the turnover of palmitate20. ABHD17A has the strongest depalmitoylase activity, which was shown to be mitigated with a Ser to Ala mutation20. We performed structural analysis of ABHD17B with molecular dynamics (MD) simulations following de novo folding with AlphaFold22 or I-Tasser23 (Supplementary Fig. 9a). The backbone Cα atoms stabilize with lower root mean square deviations (RMSD) for the structure generated with AlphaFold, therefore we used this structure for further analysis to define the active site of ABHD17B. Structural analysis of ABHD17B identified Ser170 on a loop between an alpha helix and beta sheet proximal to Asp235. His264 is located on a second loop between a different alpha helix and beta sheet, capable of forming a catalytic triad representative of alpha beta hydrolases (Supplementary Fig. 9b, c)24. These findings suggested that Ser170 has a high likelihood of being the active residue for depalmitoylation and that mutation of this residue would disrupt depalmitoylase activity. We performed a serine activity based probe (SABP) assay to quantify the activity of wild-type (WT) ABHD17B and ABHD17B containing a Ser to Ala mutation at residue 170 (S170A) and found that the mutation resulted in reduced hydrolase activity (Fig. 3a). These findings indicate that S170 is required for serine hydrolase/depalmitoylase activity in ABHD17B.
a HEK-293 cells expressing wild-type (WT) ABHD17B-FLAG and ABHD17B-S170A-FLAG were lysed and treated with a desthiobiotin-labeled serine activity based probe (SABP). Serine activity was quantified by streptavidin (SA) HRP. Total ABHD17B protein was quantified by anti-FLAG antibody. Top: representative images. Bottom: each band was quantified three times using Fiji. Error bars represent mean ± SEM. Results are representative of two independent experiments. b COL1A1 and ABHD17B were quantified by qRT-PCR in HSCs transduced with lentivirus expressing GFP, ABHD17B-WT, or ABHD17B-S170A. Expression was normalized to PSMB2. Error bars represent mean ± SEM (n = 8 biological replicates). 2-tailed unpaired t-test between WT and S170A. Results are representative of two independent experiments. c HSCs were treated with nortriptyline (black) or palmostatin B (red) at indicated concentrations for 48 h. Cells were fixed and stained with Bodipy and Hoechst. Each dot represents the averaged value of ten biological replicates. Error bars represent mean ± SD (n = 10 biological replicates). d, e HEK-293 cells expressing wild-type ABHD17B-FLAG were treated with increasing concentrations of ABD957. Serine activity was measured following similar procedures as in (a). Band intensities were quantified three times using Fiji and normalized to FLAG signal (e). Error bars represent mean ± SEM. Results are representative of two independent experiments. f, g HSCs from donor 1 (f) and donor 3 (g) were treated with ABD957 for 48 h before Bodipy and Hoechst staining. Nanchangmycin (NCMC) served as a positive control13. Error bars represent mean ± SEM. f: n = 10 biological replicates for DMSO, n = 5 for the other groups; g: n = 3 for DMSO, n = 4 for NCMC, and n = 5 for the other groups. One-way ANOVA test. h, i HSCs from donor 1 were treated with ABD957 or NCMC for 48 h. ACTA2 (h) and COL1A1 (i) levels were quantified. Error bars represent mean ± SEM (n = 8 biological replicates for DMSO, n = 4 for the other groups). One-way ANOVA test. Source data are provided as a Source Data file.
Next we asked how the S170A mutation affected gene expression in HSCs. Ectopic expression of ABHD17B-WT was associated with an increase in COL1A1 expression, and this induction was blunted with expression of comparable levels of ABHD17B-S170A (Fig. 3b). This trend was also observed for expression of the fibrotic markers TGFBR1 and TIMP1, while a significant change in ACTA2 expression was not observed (Supplementary Fig. 10a). These results suggest that S170 is required for ABHD17B to fully promote fibrotic gene expression, but there may also be mechanisms that function independent of depalmitoylation.
Depalmitoylase activity of ABHD17B is not required to promote HSC activation
To explore the role of depalmitoylation further, we treated HSCs with two depalmitoylase inhibitors. We treated HSCs with increasing concentrations of the inhibitor Palmostatin B (PalmB), which inhibits ~80% of ABHD17B depalmitoylase activity at 10 µM20. A modest increase in lipid droplet formation was observed at the highest concentration (100 µM), while there was no difference at lower concentrations (Fig. 3c). We next evaluated the effect of the depalmitoylase inhibitor ABD957, which selectively inhibits the activities of ABHD17A, ABHD17B, ABHD17C, CES2 and ABHD625. We found that the inhibitor also reduced the activity of ABHD17B by ~80% at 10 µM (Fig. 3d, e); however, even at concentrations up to 30 µM, ABD957 had no effect on lipid accumulation (Fig. 3f, g) and did not reduce ACTA2 or COL1A1 expression (Fig. 3h, i). Similar results were observed when treating HSCs overexpressing ABHD17B with ABD957 (Supplementary Fig. 10b). These results show that neither global inhibition of depalmitoylase activity by PalmB nor more specific inhibition of the depalmitoylase activity of the ABHD17 family proteins by ABD957 affects the fibrotic activity of HSCs.
Analysis of global gene expression and interacting partners
We next performed RNA-seq and differential expression analysis to define genes and pathways affected by depletion of ABHD17B that may not involve depalmitoylation. ABHD17B was depleted with two different siRNA duplexes, and each was compared to the non-targeting control (Supplementary Data 7). Results from both siRNAs were combined (Methods), and depletion of ABHD17B was associated with a decrease in expression of genes involving the cytoskeleton, adhesion, contractility, ECM, and spindle formation (Fig. 4a and Supplementary Fig. 11). Heatmaps are shown for genes associated with these Gene Ontology (GO) categories to highlight mRNAs most affected by depletion of ABHD17B (Fig. 4b and Supplementary Fig. 12). These results suggest that ABHD17B promotes expression of genes regulating ECM production along with contractility and adhesion to drive fibrosis.
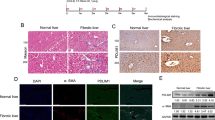
a RNA-seq and differential expression analysis were performed using nontargeting control (NTC) si5, and two siRNAs targeting ABHD17B (Supplementary Data 7). Dot plot displays the Gene Ontology (GO) terms most enriched in the genes repressed with depletion of ABHD17B. The color of each dot represents the adjusted p-value (hypergeometric test with Benjamini-Hochberg multiple testing correction), and the size of the dot represents gene count. b Heatmaps show expression of the repressed gene set for indicated GO categories for HSCs transfected with NTC (C), siRNA4 targeting ABHD17B (s4) and siRNA1 targeting ABHD17B (s1). Expression is centered and scaled by row (gene). c Proteins that interact with ABHD17B were identified by precipitation of ABHD17B-FLAG and GFP-FLAG followed by mass spectrometry (MS) (Supplementary Data 8). Enrichment of GO categories (Molecular Functions) for the 15 proteins showing the strongest interaction with ABHD17B (String-db.org) is shown. False discovery rate (FDR) was calculated by Benjamini-Hochberg procedure (String-db.org). d The interactions between the 15 proteins showing the strongest interaction with ABHD17B are displayed in addition to ABHD17B. Dark red lines indicate experimentally-determined interactions (String-db.org). e HSCs were transfected with lentivirus to express GFP-FLAG or ABHD17B-FLAG for 48 h before anti-FLAG precipitation followed by probing for MYO1B (top) and FLAG expression (bottom). Two precipitations were performed for each condition (#1 and #2) and compared to total lysates (L). Data are representative of two independent experiments. f Relative mRNA expression quantified by qRT-PCR in primary human HSCs (Donor 4) treated witih non-targeting siRNAs (NTC si5) and pooled siRNAs targeting MYO1B. Error bars represent mean ± SEM (n = 8 biological replicates). 2-tailed unpaired t-test. Data are representative of three independent experiments. g Wound healing assay was performed in HSCs (donor 4) transfected with siRNAs targeting MYO1B (green) and ABHD17B (blue). Normalized wound width was calculated at the indicated time points from three individual scratches. Error bars represent mean ± SEM (n = 3 biological replicates). One-way ANOVA test. Data are representative of two independent experiments. Source data are provided as Supplementary Data and a Source Data file.
We then evaluated the protein partners of ABHD17B. We ectopically expressed ABHD17B-FLAG and control GFP-FLAG in HSCs before performing precipitation and mass spectrometry (MS). Proteins showing the strongest enrichment with ABHD17B precipitation (Supplementary Data 8, bold) are associated with pathways (Fig. 4c) similar to those affected by depletion of ABHD17B (Fig. 4a). We next evaluated interactions among the 15 proteins showing the strongest interaction with ABHD17B (Fig. 4d). One large cluster was identified in which proteins are linked by experimental data supporting their interaction (red lines), while a second cluster contains the serine/threonine kinases STK38 and STK38L. ABHD17B is also included in this analysis, but there was no previous evidence of interaction. Together, RNA-seq and MS data indicate that ABHD17B promotes expression of gene products involved in ECM production, contractility, and adhesion, and ABHD17B also directly interacts with proteins in these pathways. ABHD17 proteins localize to early endosome and the plasma membrane20,26 in a pattern similar to that observed for MYO1B27. Given this co-localization, we focused on the interaction predicted between ABHD17B and MYO1B. HSCs were transduced with lentivirus expressing ABHD17B-FLAG or GFP-FLAG followed by anti-FLAG immunoprecipitation and probing for MYO1B, which demonstrated enrichment of MYO1B protein with precipitation of ABHD17B (Fig. 4e). We also investigated whether the S170A mutation affects binding of ABHD17B to MYO1B. In these co-immunoprecipitation experiments in HSCs with ectopic expression of ABHD17B-WT and ABHD17B-S170A, the S170A mutation did not mitigate interaction with MYO1B (Supplementary Fig. 13a), indicating that the S170 residue is not essential for the interaction between ABHD17B and MYO1B.
We then depleted MYO1B in human HSCs to determine if the effect was similar to that observed with depletion of ABHD17B. Reduction in MYO1B was associated with attenuated expression of COL1A1 (Fig. 4f and Supplementary Fig. 13b), and depletion of either ABHD17B or MYO1B resulted in reduced migration in a wound healing assay (Fig. 4g and Supplementary Fig. 13c). Together, these results show that ABHD17B and MYO1B interact, and each promotes COL1A1 expression and HSC migration, suggesting that they may work together to regulate these processes. We then asked how depletion of MYO1B affects the phenotype of ABHD17B over-expression (Supplementary Fig. 13d). We again found that depletion of MYO1B was associated with reduced expression of COL1A1, but COL1A1 was not significantly reduced in HSCs depleted of MYO1B when ABHD17B was overexpressed, suggesting that while MYO1B may act as a downstream effector of ABHD17B, the activity of ABHD17B is not mediated only through interaction with MYO1B.
Loss of Abhd17b protects against the development of liver fibrosis in vivo
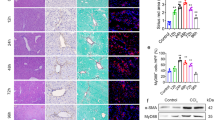
Abhd17b+/− mice were intercrossed, and Abhd17b+/+, Abhd17b+/−, and Abhd17b−/− offspring were generated. Abhd17b−/− mice bred normally, and liver histology appeared no different from that of Abhd17b+/+ mice (Fig. 5a). Mice were treated with carbon tetrachloride (CCl4) to induce liver fibrosis, and Abhd17b−/− mice showed reduced fibrosis as measured by hydroxyproline (Fig. 5b) and collagen proportionate area (Fig. 5c, d). Abhd17b−/− mice also showed reduced expression of Col1a1, Acta2, Tgfb1, Timp1, and Il1b (Fig. 5e–i). Reduction in αSMA (encoded by Acta2) and Pdgfrb was also observed in Abhd17b−/− mice along with a decrease in ductular reaction, as measured by Ck19 immunohistochemistry (IHC) (Fig. 5j). Abhd17b−/− mice also showed mild weight gain over 4 weeks of treatment, while Abhd17b+/+ mice did not gain weight (Supplementary Fig. 14). These results suggest that Abhd17b is necessary for a robust fibrotic response in the setting of chronic liver injury.
a Representative Sirius red staining results of livers from Abhd17b+/+ and Abhd17b−/− mice (n = 3 mice) receiving control olive oil via oral gavage 3 times a week for 4 weeks. Scale bar indicates 50 μm. b Hydroxyproline analysis was performed using mouse liver samples. Error bars represent mean ± SEM (n = 3 mice for olive oil groups, n = 14, 13 mice for CCl4 treatment); 2-tailed unpaired t-test. c Collagen proportionate area (CPA) was calculated based on Sirius red staining. Error bars represent mean ± SEM (n = 3 mice for olive oil groups, n = 14, 12 mice for CCl4 treatment); 2-tailed unpaired t-test. d Representative Sirius red staining results of livers from Abhd17b+/+ (n = 14 mice) and Abhd17b−/− (n = 13 mice) mice recieving CCl4 via oral gavage 3 times a week for 4 weeks. Scale bar indicates 50 μm. e–i Relative mRNA expresion was analyzed for the indicated gene products by qRT-PCR from liver samples. Error bars represent mean ± SEM (n = 3 mice for olive oil groups, n = 14, 13 mice for CCl4 treatment); 2-tailed unpaired t-test. j Representative IHC staining for Pdgfrb, Acta2 and Ck19 in livers from Abhd17b+/+ (n = 2 median mice based on hydroxyproline level) and Abhd17b−/− (n = 2 median mice based on hydroxyproline level) mice recieving either control olive oil or CCl4 via oral gavage 3 times a week for 4 weeks. Scale bar indicates 50 μm. Red arrows indicate examples of Ck19 positive cells. Source data are provided as a Source Data file.
Discussion
Extent of fibrosis remains the strongest predictor of mortality in the setting of chronic liver disease28. While treating the underlying causes of liver injury has been successful in halting progression and even promoting regression of fibrosis9,29, effective treatments are not available for many sources of disease2,3,30. HSC activation and transdifferentiation into HSC myofibroblasts as a result of chronic injury is the primary process leading to accumulation of collagen and other components of the ECM that form the fibrotic scar4,5,6. Here, we performed an siRNA screen to identify gene products necessary to maintain the fibrotic phenotype of HSC myofibroblasts, with the goal of defining new therapeutic targets to inhibit fibrosis. The screen identified ABHD17B, a member of the ABHD17 family of proteins, which have depalmitoylase activity20,25. ABHD17 proteins accelerate palmitate turnover on PSD95 and N-Ras20, and ABD957, an inhibitor of ABHD17 depalmitoylase activity, impairs N-Ras depalmitoylation in human acute myeloid leukemia cells25. ABHD17B was also reported to attenuate TGF-β-induced palmitoylation of hexokinase 1 (HK1) in LX-2 cells, leading to reduced HK1 secretion31. From these previous studies, it was not clear if and how ABHD17B is involved in HSC activation and liver fibrosis, prompting further investigation.
Through structural and mutation analysis we found that Ser 170 is the active residue for protease/depalmitoylase activity of ABHD17B. While we did not find depalmitoylase activity to be required for its profibrotic function, palmitoylation could still play a role in ABHD17B function, as its N-terminal region is itself palmitoylated to mediate membrane interactions20,26 and could affect binding to other proteins at membrane surfaces, including MYO1B27.
RNA-seq and MS studies were performed to better understand how ABHD17B may function independent of depalmitoylase activity. Depletion of ABHD17B affects expression of genes that control ECM production, adhesion, contractility, and the cytoskeleton, while MS studies revealed that ABHD17B associates with proteins involved in cell adhesion/cadherin binding and microfilament motor activity linked to actin in a network that includes MYO1B and MYO1C (Fig. 4d). We found that MYO1B interacts with ABHD17B by co-IP and regulates expression of COL1A1 and HSC migration (Fig. 4f, g). Myo1c-deficient mice are also protected from liver fibrosis32, suggesting that controlling the activity of type I myosins is one mechanism by which ABHD17B may regulate the activity of HSCs. We also observed that ectopic expression of ABHD17B at least partially rescues COL1A1 expression following depletion of MYO1B, suggesting that other partners, such as MYO1C could be involved in this pathway.
Additional interacting partners identified by MS could also contribute to similar pathways. CAVIN1/PTRF is not part of any cluster, but in addition to a role in transcription terminiation33, it is also part of the caveolin complex34, which regulates actin cytoskeleton and adhesion in HSCs35. Multiple nuclear functions are attributed to HNRPNK36,37, but it also associates with calponin38, a protein that binds actin in a complex regulating contractility39. STK38 and STK38L (NDR1/2) are serine-threonine kinases that form a separate cluster. These proteins are related to LATS1/2 in the Hippo signaling pathway40, and STK38 regulates MHY6 and sarcomere assembly in cardiomyocytes41, suggesting that it might modulate other non-muscle myosins in fibroblasts. The significance of interaction with mitochondrial proteins (ATP5F1A) is unclear, but it is linked in the protein network to MHY9 (Fig. 4d).
Environmental stiffness regulates the actin cytoskeleton and collagen expression in fibroblasts42. The interacting partners identified by MS, and confirmed with additional analysis, in the case of MYO1B, suggest ABHD17B may promote the fibrotic activity of HSCs by mediating the dynamic response to signals of environmental stiffness transmitted to the cystoskeleton through adhesion molecules and interpreted by type I myosin molecules. Type I myosins can sense tension on actin filaments43,44, and interactions with ABHD17B on membrane surfaces may modulate myosin-mediated endosomal transport45,46, including glucose transporters and TGF-β receptors that promote fibrosis17,47,48,49,50.
In summary, we performed an siRNA screen to identify genes that could be targeted to promote inactivation of HSCs, leading to the identification of ABHD17B. Depletion of ABHD17B in primary HSCs decreased collagen expression and modulated pathways involving the actin cytoskeleton, contractility, focal adhesion, and ECM. Furthermore, mice deficient in Abhd17b are protected from liver fibrosis. While we could not exclude the possible impact from Abhd17b depletion in other cell types from these in vivo experiments, the antifibrotic phenotype of Abhd17b-deficient mice should be mediated at least partially via HSC inactivation, as demonstrated by our in vitro experiments with primary HSCs. Together, these findings suggest that targeting ABHD17B-dependent pathways in HSCs may be a viable approach to reduce fibrosis. Current small molecules inhibiting the depalmitoylase activity of ABHD17B do not affect this process, and further studies will be required to develop approaches to inhibit the activity of ABHD17B.
Methods
Animals
All mouse experiments were approved by the IACUC of the Massachusetts General Hospital (2017000074) and were performed in compliance with all relevant ethical regulations. Abhd17b+/− (C57BL/6N-Abhd17bem1(IMPC)J/Mmucd) mice were purchased from the Mutant Mouse Resource and Research Center (MMRRC) and were maintained on C57BL/6 background. The mice were maintained under controlled housing conditions of 21.9 ± 0.8 °C, 45 ± 15% humidity, and 12 h light cycle 7 a.m.–7 p.m. Male mice received 40% carbon tetrachloride (CCl4) diluted in olive oil or olive oil control by oral gavage (100 μl total volume) 3 times a week for 4 weeks51. Abhd17b−/− mice and Abhd17b+/+ mice were generated from Abhd17b−/− and Abhd17b+/+ siblings produced from Abhd17b+/− parents. Studies were initiated on age matched Abhd17b−/− and Abhd17b+/+ mice at 8–9 weeks of age.
Cell culture and compound treatments
Primary human hepatic stellate cells (HSCs) were purchased from Lonza and Zen-Bio and cultured as described13. Donor information is provided below. There is limited availability of samples from individual donors. The cells used for the screening (Donors 1–4, Lonza) were from donated tissues of deceased donors with consent or legal authorization. The screen was conducted using primary HSCs at passage 8. Cells at passages 8–10 were used for the other experiments.
Donor | Vendor ID | Sex | Race |
|---|---|---|---|
1 | Lonza, HUCNP, ID: 4105 | M | Caucasian |
2 | Lonza, HUCNP, ID: 4270 | M | Caucasian |
3 | Lonza, HUCLS, ID:180761 | F | African American |
4 | Lonza, HUCLS, ID: 182821 | F | African American |
5 | Tebubio (source: Zen-Bio), HP-F-S, ID: HSC070218 | M | African American |
6 | Tebubio (source: Zen-Bio), HP-F-S, ID: HSC042617 | M | Caucasian |
Primary lung fibroblasts were collected from deidentified discarded excess tissue from a healthy donor (699) through the MGH Fibrosis Translational Research program with approval from the Massachusetts General Brigham Institutional Review Board. Cells were cultured as described previously52. HEK-293 cells were obtained from ATCC (CRL-1573) and cultured in DMEM supplemented with 10% fetal calf serum and 1% Penicillin/Streptomycin.
Palmostatin B was purchased from Calbiochem (Cat. # 178501). ABD957 was synthesized by chemists from Boehringer Ingelheim (purity: 93% as determined by HPLC-MS; MS (ESI+): m/z = 628.6 [M + H]+, 628.2 calculated). Nanchangmycin was purchased from Adooq (Cat. # A10621). Nortriptyline was purchased from Sigma (Cat. # N7261). HSCs were treated with compounds for 48 h at concentrations as indicated before fixed or lysed.
Reverse transfection of siRNAs in the screen and follow-up experiments
For the screening, the transfection was performed in 384-well plates (Corning, 3764) in a high-throughput manner using Bravo Automated Pipettor (Agilent) and Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific). For each well in a 384-well plate, 1.25 µL siRNAs (in 1 µM stocks) were diluted in 8.5 µL Opti-MEM (to make the final concentration of 25 nM) in the wells, and then 10 µL diluted Dharmafect-1 (Horizon Discovery, T-2001-02, 9.95 µL Opti-MEM + 0.05 µL Dharmafect-1) was added. After a 40 min incubation, 750 HSCs in 30 µL antibiotic-free transfection media (DMEM + 16% FBS) were added to the siRNA and Dharmafect-1 mixture in each well. Cells were fixed and stained with Bodipy and Hoechst 72 h after transfection. Cells-to-CT 1-Step Taqman Kit was used to quantify mRNA expression. ACTA2, COL1A1 and target mRNA were each quantified in the same well with the endogenous control PSMB2 using QuantStudio 7 Flex (LifeTechnologies). The gene-specific TaqMan Real-time PCR Assays for qPCR analysis are provided in the qRT-PCR analysis section.
The siRNAs used in the screen were provided by the Institute of Chemistry and Cell Biology (ICCB)-Longwood screening facility, and the relevant information is provided in the Supplementary Data.
For the validation experiments, the transfection in HSCs was scaled up to 96-well plates (for staining) and 12-well plates (for qRT-PCR) proportionally based on surface area from the protocol described above. For transfection of siRNAs in primary lung fibroblasts, cells were reverse transfected using lipofectamine 2000 (Thermo Fisher Scientific, 11668019) according to manufacturer’s manual at 5000 cells/cm2 with 20 nM siRNA. Cells were lysed 72 h after transfection. The siRNAs used in the validation experiments were purchased from Horizon Discovery, and the relevant information is provided below.
Full name | Abbreviation | Cat. # |
|---|---|---|
siGENOME non-targeting control siRNA 1 | NTC si1 | D-001210–01 |
siGENOME non-targeting control siRNA 2 | NTC si2 | D-001210–02 |
siGENOME non-targeting control siRNA 3 | NTC si3 | D-001210–03 |
siGENOME non-targeting control siRNA 4 | NTC si4 | D-001210–04 |
siGENOME non-targeting control siRNA 5 | NTC si5 | D-001210–05 |
siGENOME human ACTA2 siRNA 1 | ACTA2 si1 | D-003450-01 |
siGENOME human ACTA2 siRNA 2 | ACTA2 si2 | D-003450-02 |
siGENOME human ASAH1 siRNA 1 | ASAH1 si1 | D-005228-01 |
siGENOME human ASAH1 siRNA 4 | ASAH1 si4 | D-005228-04 |
siGENOME human PSMC4 siRNA 1 | PSMC4 si1 | D-009261-17 |
siGENOME human PSMC4 siRNA 2 | PSMC4 si2 | D-009261-02 |
siGENOME human PLK1 SMARTpool | PLK1 siP | M-003290-01 |
siGENOME human KIF11 SMARTpool | KIF11 siP | M-003317-01 |
siGLO RISC-free siRNA | siGLO | D-001600-01 |
siGENOME human GAPDH SMARTpool | GAPDH siP | M-004253-02 |
siGENOME human NFkB1 SMARTpool | NFkB1 siP | M-003520-01 |
siGENOME human NFkB2 SMARTpool | NFkB2 siP | M-003918-02 |
siGENOME human NR1H2 SMARTpool | NR1H2 siP | M-003412-03 |
siGENOME human NR1H3 SMARTpool | NR1H3 siP | M-003413-01 |
siGENOME human UBB SMARTpool | UBB siP | M-013382-01 |
siGENOME human TLR6 SMARTpool | TLR6 siP | M-005156-01 |
siGENOME human AURKC SMARTpool | AURKC siP | M-019573-04 |
siGENOME human AKAP11 SMARTpool | AKAP11 siP | M-009277-01 |
siGENOME human ABHD17B siRNA 1 | ABHD17B si1 | D-005809-01 |
siGENOME human ABHD17B siRNA 4 | ABHD17B si4 | D-005809-04 |
siGENOME human ABHD17A SMARTpool | ABHD17A siP | M-005947-02 |
siGENOME human ABHD17C SMARTpool | ABHD17C siP | M-005929-02 |
siGENOME human MYO1B SMARTpool | MYO1B siP | M-023110-00 |
ON-TARGETplus human ABHD17B pool | ABHD17B siP | L-005809-01 |
ON-TARGETplus non-targeting control pool | NTC siP | D-001810-10 |
Lipid accumulation assay based on Hoechst and Bodipy staining
HSCs transfected with siRNAs or treated with compounds were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Cat. # 15710) for 15 min at room temperature (RT). Cells were washed with DPBS and stained with Bodipy 493/503 (0.25 μg/mL, Invitrogen, Cat. # D3922) and Hoechst (5 μg/mL, Invitrogen, Cat. # H1399) for 45 min at RT. Images were taken with the ImageXpress Micro Confocal (Molecular Devices) at the Institute of Chemistry and Cell Biology (ICCB)-Longwood screening facility and analyzed using the MetaXpress software. Cells with a cytoplasmic Bodipy staining intensity higher than the cutoff were defined as Bodipy-positive cells. Cells were counted and normalized to the total nuclei count in the same microscopic field to calculate the percentage of Bodipy positive cells. The cutoff was adjusted for each plate, so that there are about 10-20% Bodipy-positive cells in the DMSO condition and 80–90% Bodipy-positive cells in the nortriptyline-treated positive control wells. For the primary screen, the results were scanned for outliers, which were then corrected based on the method described in the following section. The score was then calculated based on the averaged percent positive cells for the siRNA compared to the baseline of the plate, as well as the correlation among three replicates. Toxicity was calculated based on total cell numbers.
Outlier correction in the primary and validation screens
To determine if the replicates of a specific library well contain outliers, the interquartile range (IQR) was calculated using all the replicates for each siRNA. The minimum IQR that was considered an outlier among all IQRs was used to set up the threshold to correct for outliers. In the primary screen, siRNAs that had an IQR larger than 11.3891 were corrected. To correct an outlier, Tukey’s method was applied, which uses a cleaning parameter to detect outliers. The cleaning parameter is 1.5 × 11.3891, following the same logic as the outlier detection when plotting a boxplot. If any value was larger than the median ± the cleaning parameter, that value was set to be exactly the median ± the cleaning parameter. This method allows us to keep some of the variability while reducing the extreme variance that an outlier can generate.
High-throughput evaluation of mRNA expression and knockdown efficiency
HSCs were transfected with siRNAs as described above. 72 h after transfection, cells were harvested using the Cells-to-CT 1-Step Taqman Kit (Invitrogen, Cat. # A25603) as previously described13 except that lysis was performed with 15 µL lysis buffer (plus DNase) for 5 min at RT, and then the reaction was stopped by adding 1.5 µL stop solution and incubating for 2 min at RT. To quantify the depletion of each target gene, customized 384-well plates arrayed with Taqman probes for each mRNA target gene were purchased from Thermo Fisher Scientific. Reverse transcription and amplification of cDNA was performed with TaqMan 1-Step qRT-PCR mix supplied with the kit. ACTA2, COL1A1 and target mRNA were each quantified in the same well with the endogenous control PSMB2. ACTA2, COL1A1, and target mRNA Taqman probes are FAM-labeled, while the PSMB2 probe is VIC-labeled and primer-limited. qPCR analysis was run on QuantStudio 7 Flex (LifeTechnologies) at ICCB-Longwood Screening Facility.
The mRNA expression results for ACTA2 (normalized to PSMB2) and COL1A1 (normalized to PSMB2) were analyzed using two methods: the linear regression strategy and the standard ΔΔCt method. For the linear regression strategy, the data was fit to a linear model: Ct_ACTA2 (or COL1A1) ~ Ct_PSMB2 + plate + siRNA. The estimate for each experimental siRNA was compared to that of the control siRNA to calculate the relative fold change. The knockdown efficiency for each target gene was calculated using the standard ΔΔCt method with PSMB2 as the endogenous control.
Analysis of mice single-cell RNA-sequencing data
To delineate mouse Abhd17b expression across various cell types, single-cell RNA-sequencing data from control and CCl4-treated mice were obtained from the NCBI GEO database (GSE171904 dataset)16. Cell Ranger (version 7.0.1; 10× Genomics) was used to process the raw sequencing data with default parameters, generating expression matrices using the cellranger count pipeline. These matrices from the two conditions were then aggregated with the cellranger aggr pipeline. The aggregated data was subsequently imported into R and analyzed with the Seurat package (version 5.0.0) to perform quality control and downstream analyses. Cell viability was ensured by retaining only cells with more than 200 expressed genes, and only genes present in at least three cells were included. Dot plots were generated by the “scanpy” Python package, and gene expression levels in dot plots represent mean Unique Molecular Identifiers (UMI) count across all cells in a group.
Analysis of α-SMA fibers and extracellular collagen deposition (scar-in-a-jar assay)
Primary human HSCs were seeded in 384-well plates (CellCarrier-384 Ultra plates, Cat. # 6057300) in stellate cell medium (ScienCell/Innoprot, Cat. # 5301-b) supplemented with 2% FCS (Thermo Fisher Scientific), 1% penicillin/streptomycin, and stellate cell growth supplement (ScienCell/Innoprot, Cat. # 5352), and transfected using JetPrime transfection reagent (Polyplus, Cat. # 101000027). The final siRNA concentration per well was 16 nM with 0.8 µl transfection reagent per well. Cells were serum starved the next day, followed by stimulation with TGF-β (10 ng/ml, R&D, Cat. # 240-B) in conditions of molecular crowding (37.5 mg/ml Ficoll 70 and 25 mg/ml Ficoll 400, GE Healthcare, Cat. # 17-0310-10 and # 17-0300-10) and vitamin C supplementation (0.2 mM; Sigma, Cat. # A8960) to promote collagen deposition. 72 h post TGF-β stimulation, cells were fixed in 80% Methanol for 30 min, washed in PBS, and permeabilized using 1% Triton X-100 in PBS. Cells were blocked in 3% BSA for 30 min at RT and incubated with anti-alpha-SMA antibody (1:1000; Sigma, Cat. # A2547) and anti-collagen type I antibody (1:1000; Sigma, Cat. # SAB4200678) in PBS at 37 °C for 1.5 h. Cells were washed with PBS and incubated with secondary antibodies for 30 min at 37 °C (1:1000 AF568 goat anti-mouse IgG1, Thermo Fisher Scientific, Cat. # A-21124; and 1:1000 AF647 goat anti-mouse IgG2a, Thermo Fisher Scientific, Cat. # A-21241). Nuclei were stained with Hoechst (1 µM, Molecular Probes, Cat. # H-3570), cells were stained with HCS CellMaskTM Green at 0.2 μg/ml (Invitrogen, Cat. # H32714). Nuclear, cytoplasm, αSMA and collagen-I images were acquired using the IN Cell Analyzer 2200. Images were transferred to Perkin Elmer’s Columbus Image Storage and Analysis system, and analyzed using a custom image analysis protocol with total cell number, number of α-SMA fibers/cell, and collagen area/total cell number being quantified, as described53.
Lentiviral transduction of HSCs
Lentivirus was produced in HEK-293 cells by transfecting plasmids expressing either EGFP_FLAG, ABHD17B-FLAG, or S170A-ABHD17B-FLAG (nucleotide sequences for ABHD17B and S170A are provided in Supplementary Fig. 15) along with pMD2.G (Addgene plasmid # 12259) and psPAX2 (Addgene plasmid # 12260) using X-tremeGENE 9 transfection reagent13. Lentivirus was administered with 10 μg/mL polybrene (Sigma-Aldrich, Cat. # TR-1003-G) for 24 h before the first media change. After 24 h, cells were selected with puromycin (1 µg/mL, Cat. # A1113803) for 4 days. RNA was harvested in Trizol, and cell lysates for Western blot were harvested in 1μg/ml Pierce IP Lysis buffer. For experiments involving siRNA followed by lentiviral transduction, siRNA transfections were performed as described. 24 h after transfection, media was changed and cells were transduced with lentivirus with 1 μg/mL polybrene (to reduce toxicity). Cells were not treated with puromycin and were harvested 48 h after lentiviral transduction.
Serine activity probe-based assays
Thermo Scientific™ ActivX™ Desthiobiotin-FP Serine Hydrolase Probe was purchased from Thermo Fisher (Cat. # 88317). Anti-FLAG® M2 Magnetic Beads (Cat. # M8823-1ML) were purchased from Millipore Sigma. Blots were developed with either anti-biotin, HRP-linked antibody (1:1000, Cell Signaling Technology, Cat. # 7075, Lot # 36), streptavidin (SA) HRP (1:1000, Cell Signaling Technology, Cat. # 3999) or anti-FLAG antibody (1:1000, Cell Signaling Technology, Cat. # 14793, Lot # 7). HEK-293 cells were transiently transfected using X-tremeGENE™ 9 DNA Transfection Reagent (Roche, Cat. # XTG9-R0) with 6 µL of transfection reagent to 2 µg plasmid for 48 h Cells were harvested in 300 µL Pierce™ IP Lysis Buffer (Thermo Scientific, Cat. # 87787) and frozen at −80 °C. Lysates were cleared at 10,000 × g to remove cell debris and placed in new tubes, and an equal amount of Pierce Lysis Buffer equilibrated Millipore Sigma Anti-FLAG® M-2 Magnetic Beads (Cat. # M8823) was added (~5 µL beads per reaction), allowing anti-FLAG protein to bind for 30 min on a rocker. Beads were then washed twice in 1 mL Pierce Lysis Buffer using a magnetic rack. Beads were resuspended in 50 µL of Pierce Lysis Buffer and labeled with addition of 1 µL of ActivX™ Desthiobiotin-FP stock (at 100 µM in DMSO) for 30 min on bead, rocking at RT with occasional vortexing. Supernatant was removed and proteins were eluted in 50 µL 3X FLAG protein (Sigma-Aldrich, Cat. # SAE0194) at 100 µg/mL for 30 min on a rocker. Supernatant was removed using magnetic rack and placed in new tubes along with NuPAGE™ Sample Reducing Agent (10X) (Invitrogen, Cat. # NP0009) and NuPAGE™ LDS Sample Buffer (4X) (Invitrogen, Cat. # NP 0007). Samples were then incubated at 95 °C for 5 min. Samples were loaded on Bolt™ 4–12%, Bis-Tris, 1.0 mm, Mini Protein Gels (Cat. # NW04120BOX) in NuPAGE™ MOPS SDS Running Buffer (20X) (Cat. # NP 0001002). SeeBlue™ Plus2 Pre-stained Protein Standard (Cat. # LC5925) was used as a molecular weight marker. After electrophoresis, material was transferred to iBlot™ 2 Transfer Stacks, nitrocellulose (Cat. # IB23001) using an iBlot™ 2 Gel Transfer Device (Cat. # IB21002S). Nitrocellulose membrane was blocked in 1% BSA (Thermo Scientific, Cat. # 37520) and probed using anti-biotin antibody or SA-HRP overnight at 4 °C. Blots were washed 3 times with Tris Buffered Saline Tween (TBST) buffer (Boston BioProducts, Cat. # IBB-181–6) and developed using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, Cat. # 34580). Blots were then stripped according to manufacturer’s specifications with Thermo Fisher Scientific Restore™ Western Blot Stripping Buffer (Cat. # 21059), washed in TBST 3 times, re-blocked in 1% BSA and re-probed with anti-FLAG antibody overnight at 4 °C. Nitrocellulose membrane was then washed 3 times in TBST and probed with goat-anti-Rabbit IgG (H + L) secondary antibody, HRP (1:200, Invitrogen, Cat. # 34260, Lot # XE 353750).
qRT-PCR analysis
RNA samples were extracted using TRIzol (Invitrogen, Cat. # 15596026). Using 1 μg total RNA as input, reverse transcription was performed with the iScript gDNA Clear cDNA Synthesis Kit (BIO-RAD, Cat. # 1725035) according to manufacturer’s instructions. TaqMan Universal PCR Master Mix (Applied Biosystems, Cat. # 4305719) and TaqMan Real-time PCR Assays (ThermoFisher Scientific) were used for the quantitative real-time PCR analysis of the cDNA samples. For each qRT-PCR experiment, each dot represents the mean value for a single sample analyzed in duplicate to quadruplicate and PSMB2 as the endogenous control13. The gene-specific TaqMan Real-time PCR Assays used in this study are listed below.
Assay ID | |
|---|---|
Human ACTA2 | Hs00426835_g1 |
Human COL1A1 | Hs00164004_m1 |
Human PSMB2 | Hs01002946_m1 |
Human ASAH1 | Hs00602774_m1 |
Human PLK1 | Hs00983227_m1 |
Human UBB | Hs00430290_m1 |
Human ABHD17B | Hs00925211_m1 |
Human GAPDH | Hs02786624_g1 |
Human COL3A1 | Hs00943809_m1 |
Human ABHD17A | Hs07290154_g1 |
Human ABHD17C | Hs01593305_m1 |
Human AURKC | Hs00152930_m1 |
Human AKAP11 | Hs01568654_m1 |
Human TLR6 | Hs04975840_m1 |
Human NFkB1 | Hs00765730_m1 |
Human NFkB2 | Hs01028890_g1 |
Human NR1H2 | Hs01027208_m1 |
Human NR1H3 | Hs00172885_m1 |
Human POLR2A | Hs00172187_m1 |
Human MYO1B | Hs00362654_m1 |
Human PSMC4 | Hs00197826_m1 |
Human TIMP1 | Hs00171588_m1 |
Human TGFBR1 | Hs00610320_m1 |
Mouse Gapdh | Mm99999915_g1 |
Mouse Col1a1 | Mm00801666_g1 |
Mouse Acta2 | Mm00725412_s1 |
Mouse Timp1 | Mm01341361_m1 |
Mouse Tgfb1 | Mm01178820_m1 |
Mouse Il1b | Mm00434228_m1 |
Mouse Abhd17b | Mm01197077_m1 |
The sequences of the primers used for analyzing the overexpression level of wild-type and mutant ABHD17B are 5′-ggactgaagatgaagtcattgacttttcacatgg-3′ (forward), and 5′-cttatcgtcgtcatccttgtaat cg-3′ (reverse), and the primers were used to recognize both ABHD17B-WT and ABHD17B-S170A transcripts.
For quantification of siRNA-mediated silencing in the scar-in-a-jar assay, cells were transfected and treated as described, lysed in RLT Plus buffer, and RNA was isolated with the RNeasy Plus Kit according to the manufacturer’s protocol (Qiagen, Cat. # 74134). To obtain sufficient material for qRT-PCR, lysates from 6 to 12 wells were pooled prior to RNA isolation. 150 ng RNA were reverse transcribed using the High-capacity cDNA Archive Kit (Applied Biosystems, Cat. # 4322169). qPCR assays were performed using TaqMan Universal PCR Master Mix (Applied Biosystems, Cat. # 4318157), probes listed in the table above, and the ViiA 7 Real-Time PCR System (Thermo Fisher Scientific).
RNA sequencing
HSCs from donor 3 were transfected with NTC si5, ABHD17B si1, and ABHD17B si4 in triplicate. Cells were harvested in Trizol reagent, and RNA was purified through a Direct-zol RNA Miniprep Kit (Cat. #R2050). All samples had RIN scores greater than 9 (Agilent 4150 TapeStation System, G2992 AA) and underwent PolyA-selection and stranded library preparation prior to sequencing at 150 paired end reads (Genewiz from Azenta Life Sciences). RNA-seq counts were generated by bcbio-nextgen using salmon54. Counts were imported into R using tximport55 and DESeq256. Gene annotations for pseudo-alignment were obtained from Ensembl, version Homo_sapiens.GRCh38.98. AnnotationHub was used to obtain annotations for the Rscripts. Data manipulation and plots were done using Tidyverse57. Differential expression analysis was performed using DESeq256 with lfcShrinkage using [apeglm]58 with the design specified as design = ~treatment_short. P values were calculated using the Wald test and corrected using the Benjamini-Hochberg multiple testing procedure. Genes were defined as repressed by the following criteria: Adjusted p value < 0.05 for both si1 and si4 and (log2FC-siRNA1_vs_ctrl ≤ −0.585 AND log2FC-siRNA4_vs_ctrl ≤ 0) or (log2FC-siRNA1_vs_ctrl ≤ 0 AND log2FC-siRNA4_vs_ctrl ≤ −0.585). Heatmaps were generated by pheatmap59. For functional analysis, clusterProfiler60 with GOSemSim61,62 and DOSE63 were used.
Precipitation and mass spectrometry
Precipitation experiments involving ABHD17B-FLAG (wild-type) and GFP-FLAG were performed following transduction of HSCs with lentivirus. HSCs were selected with puromycin (1 μg/ml) for 4 days and grown to confluence before harvest in Pierce Lysis Buffer. Lysates were frozen at −80° C, thawed at 4 °C, and cleared by centrifugation at 4500 × g for 10 min at 4 °C. Lysates were incubated with anti-FLAG (M2) beads on a rocker for 30 min at 4 °C with vortexing at 10 min intervals. Beads were washed two times with 1 mL Pierce Lysis Buffer using a magnetic rack and eluted in 100 μl Pierce lysis buffer containing 3X FLAG® Peptide (Sigma, Cat. # F4799) at 20 μM. Eluted proteins were boiled at 95 °C, and gel electrophoresis was performed before probing for indicated proteins. For precipitation experiments involving ABHD17B-WT and ABHD17B-S170A, HSCs were transduced with lentivirus expressing ABHD17B-FLAG, ABHD17B-S170A-FLAG, and empty vector for 48 h (without puromycin) prior to harvest. Cells were lysed and analyzed as describe above. The antibodies used for western blot include anti-FLAG antibody (1:500–1:1000, Cell Signaling Technology, Cat. # 14793, Lot # 7), anti-MYO1B antibody (1:200; AbClonal, Cat. # A21113, Lot # 4100000017), anti-β actin antibody (1:10000, Santa Cruz Biotechnology, Cat. # sc-47778, Lot # J1916), and anti-GAPDH antibody (1:1000; Cell Signaling Technology, Cat. #8884, Lot # 3). For MS experiments, after electrophoresis, proteins were visualized with Coomassie Blue. Bands were excised above and below the location of GFP protein for MS analysis at the Taplin Mass Spectrometry following approaches previously described64. Data from two biological experiments, each in duplicate were analyzed. DESeq256 (with the design specified as design = ~group) was applied to spectral counts to obtain a list of proteins showing differential abundance between ABHD17B IP samples and control GFP IP samples. P values were calculated using the Wald test and corrected using the Benjamini-Hochberg multiple testing procedure.
Protein structure prediction, molecular dynamics based analysis of ABHD17B
Full-length human ABHD17B structures were obtained either from deposit of de novo Deepmind Alphafold Structure Database65 or prepared de novo through I-Tasser23. All models of full-length ABHD17B structure were prepared in Schrödinger Maestro and minimized using Schrodinger Protein Preparation Wizard using OPLS200566. Minimized structures were exported in .pdb format and prepared using solution builder from CHARMM-GUI67 with a salt concentration of 150 mM NaCl68. Molecular dynamics simulations were performed using NAMD 2.1269 with the CHARMM36m force field70 on full-length ABHD17B generated from either i-Tasser71 or AlphaFold222. Visualization and analyses were performed using VMD 1.9.372. The system was equilibrated for 10 ns restraining the Cα atoms of the protein (1.0 kcal/mol/A2) to allow for solvation. Root mean square deviations (RMSD) using backbone Cα atoms were calculated using VMD RMSD trajectory tool and exported and plotted using Graphpad Prism version 9. Three-dimensional representations were generated in Schrödinger66.
Generation of ABHD17B S170A mutant
DNA sequences that encode the wild-type amino acid sequence of human ABHD17B and encode a mutation of Ser 170 to Ala in ABHD17B were synthesized by GENScript. The cDNAs also contain additional identical nucleotide modifications that do not affect amino acid sequence (Supplementary Fig. 15).
Wound healing analysis
HSCs were transfected with nontargeting control siRNAs and those targeting ABHD17B or MYO1B as described above using confluent cells split 1:1. One day after transfection, a p-20 pipette tip was used to generate a wound field. Cells were monitored and the width of the wounds were visualized via microscopy at indicated times. Images were taken using EVOS FL microscope and exported into ImageJ to measure relative wound width.
Hepatic hydroxyproline assay
To quantify collagen level, mouse liver samples were isolated from the same region of the left liver lobe73. Isolated samples were homogenized and processed to evaluate hydroxyproline concentration using hydroxyproline assay kits (Sigma-Aldrich, Cat. # MAK008) following manufacturer’s instruction.
Collagen proportionate area (CPA) and immunohistochemistry (IHC)
Livers were fixed in 4% paraformaldehyde at 4 °C. After dehydration through graded ethanol and paraffin embedment, samples were sectioned at 5 μm. CPA was measured as described previously74. Pico-Sirius red staining was performed using the left liver lobe from formalin fixed paraffin embedded sections (iHisto). Whole sections were scanned and loaded into ImageJ to calculate the ratio of collagen positive area against the total parenchyma area and expressed as a percentage. One Abhd17b−/− sample was not included in the analysis due to very weak staining in multiple sections that appeared to be the result of an error with the paraffin embedding process. Immunohistochemistry (IHC) was conducted on paraffin sectioned samples following standard IHC procedure with antigen recovery. The following antibodies were used: PDGF Receptor β (28E1) antibody (1:100, Cell Signaling Technology, Cat. # 3169, Lot # 13), α-Smooth Muscle Actin (D4K9N) antibody (1:300, Cell Signaling Technology, Cat. # 19245, Lot # 6), and Cytokeratin 19 antibody [EP1580Y] (1:500, Abcam, Cat. # ab52625, Lot # 1011985-1).
Statistical analysis and figure preparation
Data involving comparisons of more than two groups were analyzed in GraphPad Prism version 9 using one-way ANOVA analysis and Dunnett’s multiple comparison test. Mouse experiments and qRT-PCR results comparing two groups of data were analyzed in Excel by unpaired two-tailed student’s t-test. For RNA sequencing and mass spectrometry data analysis, p values were calculated using the Wald test and corrected using the Benjamini-Hochberg multiple testing procedure. For the GO analysis of RNA sequencing results, a hypergeometric test with Benjamini-Hochberg multiple testing correction was applied. Enrichment of GO categories for the 15 proteins showing the strongest interaction with ABHD17B was analyzed on String-db.org, and false discovery rate (FDR) was calculated by Benjamini-Hochberg procedure. For main figures where applicable, the exact p-values are labeled in the figure. For Supplementary Figs. where applicable, ns indicates not significant, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, and **** indicates p < 0.0001. The n in figure legends indicates number of biological replicates. Error bars represent mean ± SEM or mean ± SD as indicated in the figure legends. Results are representative of 2–3 independent experiments as indicated in the figure legends. Dotmatics Vortex was used for visualizing screening results. Adobe Illustrator 2024 was used for preparing figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-seq data generated in this study have been deposited at Gene Expression Omnibus under accession number GSE240123. Human datasets GSE78853 and GSE68108 and mouse dataset GSE171904 were also used in this study. The screening results, RNA-seq data, mass spectrometry analysis data are provided in the Supplementary Data. The MS data have been deposited to the ProteomeXchange Consortium through the MassIVE partner respository with the identifier PXD060068. The qPCR data and other quantification results generated in this study are provided in the Source Data files. Source data are provided with this paper.
References
Poynard, T. et al. Impact of pegylated interferon alfa-2b and ribavirin on liver fibrosis in patients with chronic hepatitis C. Gastroenterology 122, 1303–1313 (2002).
Musso, G., Gambino, R., Cassader, M. & Pagano, G. A meta‐analysis of randomized trials for the treatment of nonalcoholic fatty liver disease. Hepatology 52, 79–104 (2010).
Karlsen, T. H., Folseraas, T., Thorburn, D. & Vesterhus, M. Primary sclerosing cholangitis – a comprehensive review. J. Hepatol. 67, 1298–1323 (2017).
Friedman, S. L., Roll, F. J., Boyles, J. & Bissell, D. M. Hepatic lipocytes: the principal collagen-producing cells of normal rat liver. Proc. Natl. Acad. Sci. USA 82, 8681–8685 (1985).
Maher, J. J. & McGuire, R. F. Extracellular matrix gene expression increases preferentially in rat lipocytes and sinusoidal endothelial cells during hepatic fibrosis in vivo. J. Clin. Investig. 86, 1641–1648 (1990).
Mederacke, I. et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 4, 1–11 (2013).
Friedman, S. L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 88, 125–172 (2008).
Bataller, R. & Brenner, D. A. Liver fibrosis. J. Clin. Investig. 115, 209–218 (2005).
Falize, L. et al. Reversibility of hepatic fibrosis in treated genetic hemochromatosis: a study of 36 cases. Hepatology 44, 472–477 (2006).
Kisseleva, T. et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc. Natl. Acad. Sci. USA 109, 9448–9453 (2012).
Troeger, J. S. et al. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology 143, 1073–1083.e22 (2012).
Chen, J. Y. et al. Tricyclic antidepressants promote ceramide accumulation to regulate collagen production in human hepatic stellate cells. Sci. Rep. 7, 44867–13 (2017).
Li, W. et al. Nanchangmycin regulates FYN, PTK2, and MAPK1/3 to control the fibrotic activity of human hepatic stellate cells. Elife 11, e74513 (2022).
Johnston, S. M., Shamu, C. E. & Smith, J. A. Automation considerations for RNAi library formatting and high throughput transfection. eBook. eISBN 978-1-60805-940-9. Front. Rna. 1, 21–39 (2014).
Zhou, C. et al. Long noncoding RNAs expressed in human hepatic stellate cells form networks with extracellular matrix proteins. Genome Med. 8, 31 (2016).
Yang, W. et al. Single‐cell transcriptomic analysis reveals a hepatic stellate cell–activation roadmap and myofibroblast origin during liver fibrosis in mice. Hepatology 74, 2774–2790 (2021).
Qi, Z., Atsuchi, N., Ooshima, A., Takeshita, A. & Ueno, H. Blockade of type β transforming growth factor signaling prevents liver fibrosis and dysfunction in the rat. Proc. Natl. Acad. Sci. USA 96, 2345–2349 (1999).
Dooley, S. et al. Smad7 prevents activation of hepatic stellate cells and liver fibrosis in rats. Gastroenterology 125, 178–191 (2003).
Tanahashi, N. et al. Chromosomal localization and immunological analysis of a family of human 26s proteasomal atpases. Biochem. Biophys. Res. Commun. 243, 229–232 (1998).
Lin, D. T. S. & Conibear, E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife 4, e11306 (2015).
Wynn, T. A. Integrating mechanisms of pulmonary fibrosis. J. Exp. Med. 208, 1339–1350 (2011).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Rauwerdink, A. & Kazlauskas, R. J. How the same core catalytic machinery catalyzes 17 different reactions: the serine-histidine-aspartate catalytic triad of α/β-hydrolase fold enzymes. ACS Catal. 5, 6153–6176 (2015).
Remsberg, J. R. et al. ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth. Nat. Chem. Biol. 17, 856–864 (2021).
Martin, B. R. & Cravatt, B. F. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135–138 (2009).
Almeida, C. G. et al. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat. Cell Biol. 13, 779–789 (2011).
Hagström, H. et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J. Hepatol. 67, 1265–1273 (2017).
Bonis, P. A. L., Friedman, S. L. & Kaplan, M. M. Is liver fibrosis reversible? J. New Engl. J. Med. 344, 452–454 (2001).
Sanyal, A. J. et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N. Engl. J. Med. 362, 1675–1685 (2010).
Chen, Q. et al. HK1 from hepatic stellate cell–derived extracellular vesicles promotes progression of hepatocellular carcinoma. Nat. Metab. 4, 1306–1321 (2022).
Arif, E. et al. Targeting myosin 1c inhibits murine hepatic fibrogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 320, G1044–G1053 (2021).
Jansa, P., Mason, S. W., Hoffmann‐Rohrer, U. & Grummt, I. Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. Embo J. 17, 2855–2864 (1998).
Hill, M. M. et al. PTRF-cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 (2008).
Ilha, M. et al. Exogenous expression of caveolin‐1 is sufficient for hepatic stellate cell activation. J. Cell. Biochem. 120, 19031–19043 (2019).
Expert-Bezançon, A., Caer, J. P. L. & Marie, J. Heterogeneous nuclear ribonucleoprotein (hnRNP) K Is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken β-tropomyosin pre-mRNA. J. Biol. Chem. 277, 16614–16623 (2002).
Takimoto, M. et al. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem. 268, 18249–18258 (1993).
Laury‐Kleintop, L. D., Tresini, M. & Hammond, O. Compartmentalization of hnRNP‐K during cell cycle progression and its interaction with calponin in the cytoplasm. J. Cell. Biochem. 95, 1042–1056 (2005).
Reynaert, H. Hepatic stellate cells: role in microcirculation and pathophysiology of portal hypertension. Gut. 50, 571–581 (2002).
Zhang, L. et al. NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Curr. Biol. 25, 296–305 (2015).
Liu, J. et al. Stk38 modulates rbm24 protein stability to regulate sarcomere assembly in cardiomyocytes. Sci. Rep. 7, 44870 (2017).
Mochitate, K., Pawelek, P. & Grinnell, F. Stress relaxation of contracted collagen gels: Disruption of actin filament bundles, release of cell surface fibronectin, and down-regulation of DNA and protein synthesis. Exp. Cell Res. 193, 198–207 (1991).
Laakso, J. M., Lewis, J. H., Shuman, H. & Ostap, E. M. Myosin I can act as a molecular force sensor. Science 321, 133–136 (2008).
Greenberg, M. J., Lin, T., Goldman, Y. E., Shuman, H. & Ostap, E. M. Myosin IC generates power over a range of loads via a new tension-sensing mechanism. Proc. Natl. Acad. Sci. USA 109, E2433–E2440 (2012).
Raposo, G. et al. Association of myosin I alpha with endosomes and lysosomes in mammalian cells. Mol. Biol. Cell 10, 1477–1494 (1999).
Brandstaetter, H., Kendrick-Jones, J. & Buss, F. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J. Cell Sci. 125, 1991–2003 (2012).
Bose, A. et al. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature 420, 821–824 (2002).
Chandrashekaran, V. et al. Purinergic receptor X7 mediates leptin induced GLUT4 function in stellate cells in nonalcoholic steatohepatitis. Biochim. Biophys. Acta Mol. Basis Dis. 1862, 32–45 (2016).
Chung, C. et al. Pentachloropseudilin inhibits tansforming growth factor‐β (TGF‐β) activity by accelerating cell‐surface Type II TGF‐β receptor turnover in target cells. ChemBioChem. 19, 851–864 (2018).
Chung, C.-L., Tai, S.-B., Hu, T.-H., Chen, J.-J. & Chen, C.-L. Roles of myosin-mediated membrane trafficking in TGF-β signaling. Int. J. Mol. Sci. 20, 3913 (2019).
Alsamman, S. et al. Targeting acid ceramidase inhibits YAP/TAZ signaling to reduce fibrosis in mice. Sci. Transl. Med. 12, eaay8798 (2020).
Ramos, C. et al. Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am. J. Resp. Cell Mol. 24, 591–598 (2001).
Weigle, S., Martin, E., Voegtle, A., Wahl, B. & Schuler, M. Primary cell-based phenotypic assays to pharmacologically and genetically study fibrotic diseases in vitro. J. Biol. Methods 6, e115 (2019).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000research 4, 1521 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wickham, H. et al. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Kolde & Heatmaps., Raivo. 2015. pheatmap: P.
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 16, 284–287 (2012).
Yu, G. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol. Biol. 2117, 207–215 (2020).
Yu, G. et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978 (2010).
Yu, G., Wang, L.-G., Yan, G.-R. & He, Q.-Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609 (2015).
Chantranupong, L. et al. Rapid purification and metabolomic profiling of synaptic vesicles from mammalian brain. Elife 9, e59699 (2020).
Varadi, M. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2021).
Schrödinger Release 2024-2: Maestro, Schrödinger, LLC, New York, NY, 2024.
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM‐GUI: a web‐based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Sparks, R. P. et al. An allosteric binding site on sortilin regulates the trafficking of VLDL, PCSK9, and LDLR in hepatocytes. Biochemistry 59, 4321–4335 (2020).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinforma. 9, 40 (2008).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Ikenaga, N. et al. Selective targeting of lysyl oxidase-like 2 (LOXL2) suppresses hepatic fibrosis progression and accelerates its reversal. Gut. 66, 1697 (2017).
Sojoodi, M. et al. Peroxidasin deficiency re-programs macrophages toward pro-fibrolysis function and promotes collagen resolution in liver. Cell Mol. Gastroenterol. Hepatol. 13, 1483–1509 (2022).
Acknowledgements
This work was funded through a grant to A.C.M. from Boehringer Ingelheim Pharma GmbH Co KG. We thank the team from the ICCB-Longwood Screening Facility, and especially Richard Siu, for their assistance. We thank Nicola Zimmermann and Sabine Weigle at Boehringer Ingelheim for conducting in vitro analysis of α-SMA fibers and extracellular collagen deposition. We thank members of the Mullen lab, including Raza Ur Rahman and colleagues at Boehringer Ingelheim, including John Broadwater, Peter Seither, and Christofer Tautermann for helpful discussion, as well as Ross Tomaino and the Taplin Mass Spectrometry Core at Harvard Medical School.
Author information
Authors and Affiliations
Contributions
W.L. and A.C.M. conceived the study with C.B-K., J.F.R., J.F.D, and D.M.S. W.L., R.P.S., and C.S. performed the experiments with assistance from J.Y.C., J.S., S.P.M., M.S., Y.Y., N.A., B.J.T., A.W., B.D.M, and M.L.B., and D.W. supported processing of imaging data. Protein structural analysis was performed by R.P.S. with support from A.W. and G.A., and G.A. also oversaw synthesis and analysis of ABD957. All other computational analyses were performed by L.P., R.K., and V.B. under the direction of S.J.H.S., and Z.L. and P.Z. with support from C.Z. The manuscript was written by W.L., R.P.S., C.S., D.M.S., and A.C.M. with input from all other authors.
Corresponding author
Ethics declarations
Competing interests
A.C.M. has received research funding from GSK for unrelated projects. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, W., Sparks, R.P., Sun, C. et al. Screening the human druggable genome identifies ABHD17B as an anti-fibrotic target in hepatic stellate cells. Nat Commun 16, 2109 (2025). https://doi.org/10.1038/s41467-025-56900-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56900-z
This article is cited by
-
Palmitoylation: an emerging therapeutic target bridging physiology and disease
Cellular & Molecular Biology Letters (2025)
-
Ultrasound-activated dual-nanozyme platform for targeted ROS scavenging and NO delivery in liver fibrosis therapy
Advanced Composites and Hybrid Materials (2025)