Abstract
This phase I/II trial aims to evaluate the efficacy of concurrent atezolizumab with radiation therapy and temozolomide (TMZ) followed by adjuvant atezolizumab and TMZ in newly diagnosed glioblastoma (GBM) patients and to identify pre-treatment correlates with outcome (N = 60). Trial number: NCT03174197. The primary outcome was overall survival (OS) whereas secondary outcomes were retrospective global–omics analyses to identify pre-treatment immune and genetic tumor features that correlated with survival. Concurrent use of atezolizumab with radiation and TMZ demonstrated OS in line with published trials for newly diagnosed GBM. Tumor genomic (WES and/or targeted NGS panel), transcriptomic (RNAseq) and tissue microenvironment imaging, as well as fecal metagenomic sequencing were conducted. Gene set enrichment analysis of tumors identified multiple immune-based transcriptomic programs to distinguish patients with longer versus shorter survival (p ≤ 0.01). GBM immune enrichment was highly associated with the pre-treatment tumor mesenchymal subtype and patient gastrointestinal bacterial taxa profile.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is the most common primary malignant central nervous system tumor in adults and invariably carries a poor prognosis with a disease trajectory often marred by substantial neurologic morbidity and progressive disability. Despite optimized multimodality treatment that typically includes surgery, radiation, and alkylating chemotherapy, clinical trials including contemporary trials have reported a median overall survival (mOS) of only 14 to 16 months1,2,3,4. Recurrence is inevitable and at time of recurrence, mOS is on the order of 6 to 9 months5,6,7. Recent efforts have been focused on advancing immunotherapy and specifically investigating the role, if any, of checkpoint blockade to improve the outcomes for this devastating disease.
The majority of GBM patients (61%) have tumors with at least 1% or more Programmed death ligand 1(PD-L1)-positive cells, and 38% have at least 5% or greater PD-L1 expression. PD-L1 is commonly expressed on the GBM-infiltrating T cells of which there is a paucity in tumor tissue8. Expression of both PD-L1 and PD-1 are negative prognosticators for GBM outcome8. Unfortunately to date, PD-1 blockade has demonstrated limited efficacy in GBM patients9,10 with the exception of responses seen in patients with mismatch repair deficiencies11,12,13. The lack of efficacy has been ascribed to multiple factors including tumor heterogeneity14, systemic and local immunosuppression15,16 and the abundance of tumor-associated macrophages (TAMs) which are locally immunosuppressive and abundantly present in the GBM tumor microenvironment17,18. A limitation of prior CPI studies in GBM has been the lack of immune-predictive biomarkers and comprehensive genomic characterization of tumor samples. The role of checkpoint blockade as an efficacious therapy in GBM has been overall limited, but optimism was recently renewed following work performed by Cloughesy et al. which demonstrated that recurrent GBM patients who received neoadjuvant PD-1 monoclonal antibody blockade with pembrolizumab with continued adjuvant pembrolizumab following planned surgical resection had improved OS and progression free survival (PFS) in contrast to patients who did not receive neoadjuvant pembrolizumab19.
The benefit of the neoadjuvant PD-1 blockade is believed to be primarily driven by the re-activation of exhausted T cells and the possible modulation of the tumor microenvironment to influence non-T cell populations20,21. Single-cell RNA and TCR sequencing analysis suggests that neoadjuvant PD-1 blockade may expand the cytotoxic CD8 T cell population in the peripheral blood that traffic to the tumor which implies systemic T cell activation22. However, TCR analysis has revealed that the cytotoxic effector T cells transition into progenitor exhausted T cells that have lost their ability to exert effector function22.
Recent studies have highlighted the diverse immune profiles observed among GBM tumors23,24. Standard of care radiation and chemotherapy has been shown to increase antigen presentation and promote a pro-inflammatory tumor microenvironment25. The strategy of this clinical trial is to leverage these features of GBM by combining atezolizumab (PD-L1 inhibitor) with standard of care radiation therapy and temozolomide followed by continuation of atezolizumab with adjuvant temozolomide in newly diagnosed GBM patients to improve clinical outcomes in a safe and effective manner. To elucidate potential mechanisms and biomarkers associated with treatment benefit, we will perform whole exome sequencing (WES) with somatic mutation and somatic copy number alteration (SCNA) analysis, whole transcriptome sequencing, and multiplex immunofluorescence imaging on pretreatment tissue. Metagenomic sequencing of fecal samples will also be performed on a subset of patients.
Checkpoint inhibitor therapy has limited efficacy in the outcomes of patients with GBM underscoring the need to better understand factors that may contribute to resistance and treatment benefit. In this single arm prospective clinical trial, concurrent atezolizumab with radiation and temozolomide is safe with modest efficacy in patients with newly diagnosed GBM. Patients with GBM harboring an EGFR mutation is associated with a relatively worse mOS following treatment compared to patients with tumors enriched with a PTEN mutation. GBM immune enrichment is highly associated with the pre-treatment tumor mesenchymal subtype and fecal microbiome analysis identifies distinct bacteria differentially enriched by OS at the single taxa level and warrants further investigation.
Results
This study tested the safety and efficacy of atezolizumab administered in combination with radiation and TMZ (concurrent stage) followed by atezolizumab administered in combination with TMZ (adjuvant stage) in newly diagnosed GBM patients based on the WHO 2016 classification criteria26, unselected for MGMT status (Fig. 1A). In addition to scheduled MRI brain imaging, tumor and microbiome correlative molecular analyses were performed. A total of 78 GBM patients were screened and consented between August 2017 and July 2019. Eighteen patients were eliminated during screening, and 60 patients were treated (Fig. 1B). Baseline patient characteristics are shown in (Table 1) for age, sex, KPS, isocitrate dehydrogenase (IDH) mutation status, O6-methylguanine DNA methyltransferase (MGMT) methylation status. 28 of the 60 patients were on dexamethasone at time of trial registration (0–8 milligrams daily). Trial registration required that a baseline MRI be obtained no more than 14 days prior to study enrollment on a stable dose of steroids no greater than 4 mg a day of dexamethasone for at least 5 days prior to treatment start.
A Schematic of key clinical trial and correlative features. Created in BioRender. Woodman, S. (2024) https://BioRender.com/o72r244. B Cohort consenting and screening relevant to final correlative analysis.
Safety
Atezolizumab in combination with radiation and temozolomide was generally well tolerated. The administration of atezolizumab in our newly diagnosed GBM patients receiving standard of care (SOC) was not associated with any new, unreported toxicities. 56.7.4% (34/60) of patients experienced grade >3 possibly, or probably, or definitely related to treatment adverse event. The most common treatment related toxicities were lymphopenia (38.3%), thrombocytopenia (8.3%), alanine aminotransferase (ALT) increased (8.3%), fatigue, constipation and cerebral edema (5%) (Supplemental Table 1). One patient was removed during phase 1 due to a DLT, grade 3 hepatitis and grade 3 pneumonitis probably due to atezolizumab. Two patients were removed during phase II due to grade 4 hepatitis probably related to atezolizumab. The trial was not stopped early during the safety run in phase and continued to phase II and completed the full accrual of 60 patients.
Efficacy
Concurrent use of atezolizumab with radiation and TMZ was tolerable and demonstrated efficacy in line with published trials for newly diagnosed GBM. As of the cutoff date of 11 May 2023, the Kaplan-Meier estimated median follow-up time was 51.7 months. Of the 60 patients, 56 with IDH1 wild-type and 4 with IDH1 mutant tumors, reached an mOS of 18.0 months (95% CI [14.2, 25.0]). Among the IDH1 wild-type group, 50 out of 56 patients had died, resulting in an mOS of 16.1 (95% CI [13.9, 24.6]) months. MOS for patients with MGMT methylated tumors was 25.4 (95% CI [10.9–40.9]) compared to 14.6 (95% CI [13.6–22.9]) in patients with MGMT unmethylated tumors. (Supplemental Fig. 1) Out of the 60 patients treated 6 (10%) had a complete response (CR) and 8 (13.33%) had a partial response (PR) with a duration of response of 13.91 months (95%CI [6.67–32.92])
Somatic mutation and copy number profiles were consistent with known aberrations in GBM
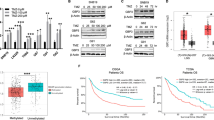
Somatic mutation testing was performed on pre-treatment IDH1 wildtype samples (n = 43 samples). The most frequent genes harboring somatic mutations were PTEN (53%, n = 20), TP53 (32%, n = 12), and EGFR (21%, n = 8), consistent with frequencies observed in prior independent GBM cohorts27,28 (Fig. 2A). The SCNA profile was also akin to prior reports, showing broad gains across chromosome 7, and narrower gains in 12q14.1 and 14q11.2, as well as, losses across chromosome 10 and narrower losses in 9p21.3/22.2 and 17p11.2 (Supplemental Fig. 2A)
A Oncoplot of recurrent somatic mutations. B Unsupervised clustering of tumors based on global gene mutation profiling. C Kaplan-Meier curve of OS for patients with EGFR vs. PTEN mutant GBM tumors. The Cox proportional hazards model-derived hazard ratio and p-value from the Wald test are displayed. Source data are provided as a Source Data file. D Unsupervised clustering of GBM tumors based on transcriptome analysis. E Kaplan-Meier curve of OS for patients with cluster 1 vs. cluster 2 GBM tumors. The Cox proportional hazards model-derived hazard ratio and p-value from the Wald test are displayed.
Unsupervised cluster-based analysis identifies key molecular features associated with anti-PD-L1-treatment patient overall survival
To better understand the relationship between molecular features in GBM tumors and clinical outcome, unsupervised hierarchical clustering was performed on pre-treatment GBM tumor samples using global gene mutation profiles from WES (n = 29). Samples separated into two major clusters: all cluster A tumors harbored mutations in PTEN, whilst no tumors in cluster 2 possessed PTEN mutations. Rather, cluster B was highly enriched in EGFR mutant tumors (Fig. 2B). A tumor harboring both a PTEN and an EGFR mutation was assigned to cluster A, indicating similarity to PTEN/non-EGFR mutant tumors compared to those with an EGFR/non-PTEN mutation status. Patients with cluster A tumors (i.e., PTEN mutant samples) showed a statistically significantly better mOS from the time of treatment initiation compared to patients with cluster B tumors (p = 0.013, HR = 0.325 (95% CI [0.128, 0.823]) (Fig. 2C). Notably, among the four patients with available PTEN/EGFR mutation status and still living at the most recent data cut-off date, three had tumors with a PTEN mutation, and none harbored an EGFR mutation.
We next assessed the unsupervised clustering of the transcriptome (Fig. 2D). Among the most highly differentially expressed genes in cluster 1, were C1QA, C1QB, FCER1G and Collagens 1A1, 1A2 and 3A1 (Supplemental Fig. 2B). Patients with cluster 1 tumors trended toward achieving a better mOS (p = 0.076, HR = 1.826 (95% CI [0.931, 3.583])) than those in cluster 2 (Fig. 2E).
An unsupervised clustering-based analysis, like that deployed for global gene mutation- and transcriptome data was performed using global SCNAs. Differential copy number alteration groups failed to demonstrate an OS difference (Supplemental Fig. 2C). For each of the molecular features analyzed (viz., gene mutation, SCNA and transcription profile), the patients’ age, sex, dexamethasone treatment and tumor MGMT methylation status failed to account for the clustering differences observed (p > 0.05).
Gene set enrichment analysis of tumors from patients with higher vs. lower mOS
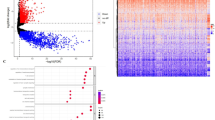
To determine pre-treatment gene expression programs associated with differential OS following treatment initiation, pretreatment GBM samples were divided into low and high OS groups (mOS = 16.1 months). Gene Set Enrichment Analysis (GSEA) performed on the entire set of transcribed genes in each tumor demonstrated a marked enrichment in immune-related genesets associated with a longer OS, notably expression programs associated with interferon gamma and inflammatory response, as well as antigen presentation were highly represented (p = 0.0015 - 0.038, FDR-corrected, Fig. 3A). A detailed analysis of specific immune marker transcription profiles showed elevated levels of CD14, CD86, CCR2, CCR5, HLA-A and HAVCR2 (aka TIM3) in the tumors of patients who achieved longer OS (Fig. 3B). The cytolytic score, key immunological features known to be associated with immune checkpoint inhibitor clinical benefit, were also elevated in GBM tumors of patients who achieved a higher mOS (Fig. 3C)29.
A Patients were separated into long vs. short OS groups based on a median threshold (red=deceased, green=alive). Patient age, sex, and tumor MGMT methylation status are as noted. Transcriptomic gene set enrichment was performed, the heatmap represents the relative expression levels of genes for each patient’s GBM tumor sample. Sankey plot connects specific genes with corresponding enriched Hallmark genesets. B Representative differentially expressed genes between GBM tumors from patients with short (n = 17) vs. long (n = 22) OS. The “box” shows the interquartile range (IQR) between the 25th and 75th percentiles, the “center line” marks the median, the “whiskers” extend to 1.5 times the IQR, and points beyond are outliers, with “minima” and “maxima” as the absolute lowest and highest values. C Differential cytolytic score between GBM tumors from patients with short (n = 17) vs. long (n = 22) OS. Source data are provided as a Source Data file.
Highly immune infiltrated GBM tumors are associated with specific immune cell types, the mesenchymal GBM subtype, and fecal microbiome profiles
To characterize the immune enrichment observed in tumors, an immune metric, the ESTIMATE Immune Score (EIS) was derived for each GBM sample using the ESTIMATE algorithm (See Materials and Methods)30. Samples were median-divided into low versus high EIS and Kaplan-Meier analysis performed. Notably, patients with GBM tumors having a low EIS showed a 14.5 months mOS, while those with a high EIS displayed a 24.8 months mOS (p = 0.02, HR = 0.45 (95% CI [0.23–0.90]), Fig. 4A). In addition, 21% of the high EIS group was still alive at 48.7 months. A similar analysis of IDH1 wildtype GBM tumors (n = 143) from the Cancer Genome Atlas31 in which patients had not received immunotherapy, failed to show an association of EIS with OS (low EIS mOS = 13.6 months versus high EIS 11.7 months mOS, p = 0.77, HR = 1.06 (95% CI [0.73–1.53]) Fig. 4A). To further assess if the observed correspondence between baseline tumor EIS and OS in our immunotherapy-based study was distinct, we analyzed the baseline EIS relative to OS in another independent set of GBM tumors (n = 83) receiving standard of care therapy32. As with TCGA, there was no statistically significant difference between low versus high EIS with OS in this cohort (low EIS mOS = 19.2 months versus high EIS 24.6 months mOS, p = 0.25, HR = 0.77 (95% CI [0.48–1.21]), Fig. 4A).
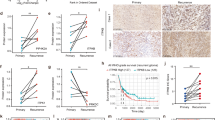
A KM analysis of patients with low vs. high Estimate Immune Score (EIS) tumors from three cohorts: 1) current study cohort, anti-PD-L1 plus standard of care 2) TCGA, standard-of-care only 3) GLASS, standard-of-care only. The Cox proportional hazards model-derived hazard ratio and p-value from the Wald test are displayed. Source data are provided as a Source Data file. B Patient tumor samples were grouped into low or high EIS based on median threshold score. The transcriptome of each sample was deconvoluted into specific inferred cell states, and the relative contribution of each cell state compared between low (n = 20) and high (n = 19) EIS groups. Right annotation indicates the -log10 p-value (unadjusted) from a two-sided Wilcoxon test. Source data are provided as a Source Data file. C Box plot representations of key differential inferred immune cell states present in low vs. high EIS GBM tumors. The “box” shows the interquartile range (IQR) between the 25th and 75th percentiles, the “center line” marks the median, the “whiskers” extend to 1.5 times the IQR, and points beyond are outliers, with “minima” and “maxima” as the absolute lowest and highest values. Source data are provided as a Source Data file. D Kaplan-Meier curve of OS for patients with low vs high PD-L1 expression faction in CD3CD8+ cells. The Cox proportional hazards model-derived hazard ratio and p-value from the Wald test are displayed. E GBM Mesenchymal Subtype is associated with high EIS and higher mOS compared to the proneural or classical subtypes. Source data are provided as a Source Data file.
To delineate the immune cell types that underlie the immune enrichment in tumors associated with improved survival outcome in anti-PD-L1 treated patients, the Xcell immune deconvolution algorithm was employed (See Materials and Methods)33. The majority of cell types in high EIS GBM tumors harbored myeloid lineage markers: activated dendritic cells, monocytes, macrophages, with notably a more statistically significant M1 macrophage profile. In addition, a lymphoid lineage cell type identified to be associated with high EIS GBM tumors was CD4+ effector memory T cells (Fig. 4B, C). Patients whose tumors had a higher percentage of cytotoxic (CD3 + CD8 + ) T cells that expressed PD-L1 achieved a longer mOS p = 0.036030, HR = 0.439 (0.205–0.940)) (Fig. 4D), not observed in PD-L1 expressing GFAP + , CD68 + or CD3 + CD8- cells.
A recent study by Lee et al.22, using scRNAseq to interrogate immune cell subsets in GBM identified 11 subclusters of myeloid cells. We leveraged these subcluster transcription profiles to analyze our cohort samples. Notably, the dendritic cell “DC” subcluster profile demonstrated a statistically significant mOS difference (Supplemental Fig. 3A), consistent with the immune deconvolution we performed (Fig. 4B) identifying activated myeloid dendritic cells to make up a statistically significant fraction of more highly immune infiltrated tumors, which corresponded with a better OS. Further subset analysis by Lee et al.22, identified a CCR7 + LAMP3+ “activated and/or migratory” DC subpopulation to be upregulated following neoadjuvant anti-PD-L1. We identified these two markers to be higher in tumors associated with increased OS in our cohort (Supplemental Fig. 3B). These data suggest that the dendritic cell profile of primary GBM pretreatment tumors associated with an improved clinical outcome may be better primed for effective anti-PD-L1 treatment.
We also leveraged the 11 scRNAseq derived subclusters composing the lymphoid cell compartment of GBM to assess pretreatment tumors in our cohort22. Among these lymphoid subclusters, only L4 and L5 were associated with better OS in our cohort (Supplemental Fig. 3C). A granular analysis of the genes within the L4 set revealed the expression of lymphocyte markers and antigen presenting genes (e.g., CD3E, CD4, HLA-A, PSMB8) to be more elevated in our cohort of GBM tumors associated with a higher OS, while the expression of multiple inhibitory genes (e.g., FOXP3, CD27, CD28, TNFRSF18, ICOS, CTLA4, TIGIT) showed no statistically significant difference. A more detailed examination of the genes within the L5 set revealed the expression of cytolytic enzyme genes, consistent with the cytolytic score (geometric mean of GZMA and PRF1 gene expression) (Fig. 3C), to be statistically higher in the tumors of patients in our study with greater OS. The CD4 compartment markers22 have multiple genes associated with the immune inhibitory system (e.g., CTLA4, PDCD1, LAG3, ICOS, TNFRSF9 (aka, 4-1BB), HAVCR2 (aka, TIM-3)). Of these, TNFRSF9 and HAVCR2 were more elevated in the tumors of patients who demonstrated a longer OS, whilst no statistically significant expression difference was observed in any of the other immune checkpoint genes (Supplemental Fig. 3D).
GBM tumors harboring wild-type IDH1 have been shown by transcriptional profiling to separate into pro-neural, classical and mesenchymal subtypes31. Pre-treatment GBM tumors in our cohort likewise parsed into these three subtypes (28.2%, 41.0%, 30.7%, respectively). Consistent with prior studies31, we observed that mesenchymal subtype GBMs were markedly enriched in immune cell content, with 11 of 12 (91.7%) of mesenchymal tumors having a high EIS, as compared to 27.2% of proneural and 31.3% of classical subtypes, respectively (Fig. 4D). Notably, anti-PD-L1 treated patients with mesenchymal subtype GBM tumors achieved a greater mOS compared to the proneural and classical subtypes (26.5 months vs. 15.5 and 15.6 months., respectively).
Given recent evidence supporting the impact of the gastrointestinal microbiome on tumor immunology and clinical outcomes of immune checkpoint inhibitor therapy, we leveraged a set of fecal samples collected from a subset of trial patients. The resulting microbiome profiles derived from metagenomic shotgun sequencing processed with MetaPhLAn 3.034 were evaluated mainly based on survival metrics. Pre-treatment samples from patients with longer overall survival showed similar alpha diversity levels, a measurement of within sample diversity, as assayed by number of observed species and derived Inverse Simpson scores (Fig. 5A, B)
A The alpha-diversity of each patient’s pre-treatment fecal microbiome was determined and compared between patients with low (n = 9) versus high (n = 13) OS. The “box” shows the interquartile range (IQR) between the 25th and 75th percentiles, the “center line” marks the median, the “whiskers” extend to 1.5 times the IQR, and points beyond are outliers, with “minima” and “maxima” as the absolute lowest and highest values. Source data are provided as a Source Data file. B Delineation of the type and relative abundance of each species identified in each fecal microbiome between patients with low versus high OS. Source data are provided as a Source Data file. C Principal coordinate analysis of fecal microbiome beta-diversity in low versus high OS patients. Source data are provided as a Source Data file. D Prevalence of fecal microbiome bacterial species in patients with low vs. high OS. Source data are provided as a Source Data file.
We evaluated beta-diversity, a measurement of diversity across samples, by principal coordinate analysis to compare the structure of the fecal microbiomes in low versus high OS. Although not reaching statistical significance, pre-treatment samples from patients with longer overall survival clustered together and apart from those who experienced shorter survival (Fig. 5C). Due to the observed separation of centroids between short and long survivors, we aimed to determine if there were compositional differences that distinguished both groups. After running a differential abundance analysis using ANCOM-BC35,36, we observed bacteria mainly of the Bacillota phylum, also known as Firmicutes, enriched in long OS patients. (Fig. 5D).
The same analyses of alpha and beta diversity were performed comparing patients with low versus high EIS tumors. Notably, patients with a higher EIS had a higher observed alpha diversity (Supplemental Fig. 4A, B). As with OS, principal coordinate analysis to compare the structure of the fecal microbiomes in patients low versus high EIS GBM was not statistically significant, but a differential abundance analysis also showed Bacteroides thetaiotaomicron (or B. theta) to be enriched in patients with low EIS tumors (Supplemental Fig. 4C, D).
Immune infiltration and EGFR mutation are independent predictors of OS in anti-PD-L1 treated GBM
Many prognostic and predictive markers have been reported for GBM (viz, MGMT methylation, gross total resection (GTR) vs. subtotal resection (STR), age, sex and performance status)37,38,39,40. Univariate KM analysis in this study showed high EIS and EGFR mutation status to each be statistically significantly associated with better and worse OS, respectively. We thus performed a multivariate analysis to assess the relationship between OS and each known prognostic/predictive feature. Although, the HRs associated with methylated MGMT (0.55), male (1.41) and STR (3.76) did not reach statistical significance, they each trended in a manner consistent with prior reports. Patients with high EIS GBM showed a reduced hazard ratio (HR = 0.23, 95% CI [0.07–0.78], p = 0.019), while those with EGFR mutated tumors showed an elevated hazard ratio (HR = 6.27, 95% CI [1.20–32.62], p = 0.029) (Fig. 6A), making these two features the most statistically significant relative to anti-PD-L1 mediated OS in this cohort. Figure 6B shows a summary graphic of this study in which we identified newly diagnosed GBM patients who achieved a longer OS after anti-PD-L1 treatment to have transcription-based markers for activated myeloid dendritic, M1 macrophage, and effector CD4+ memory cells, as well as, a higher cytolytic score and CD8A/B levels, consistent with greater protein marker profiles for cytotoxic T cells. Among these IDH wild-type tumors, global gene mutation profiling separated tumors into two distinct groups, one group harboring PTEN mutations and achieving a longer OS compared to a group possessing EGFR mutations.
A Multivariate hazard ratio analysis was performed using univariate features (high transcriptome-based Immune Score (n = 19) and EGFR mutation status (n = 4)) identified in this study, as well as, known GBM prognostic and predictive variables (MGMT methylation (n = 9); KPS - Kanofsky Performance Score (n = 31); Age (n = 31); Male (n = 21); STR - Subtotal Resection (n = 6)) (n = 31). Line plots are presented as HR + /- 95% CI. P-values are provided by Wald test. B Graphical image of study results showing the cancer cell intrinsic (differential gene expression of markers associated with activated dendritic cells, M1 macrophages, helper T cells, cytolytic markers and PTEN/EGFR mutation status) and gut microbiome features associated with longer mOS following anti-PD-L1 treatment. Created in BioRender. Woodman, S. (2024) https://BioRender.com/b92g788.
Discussion
In this study the concurrent use of atezolizumab (anti-PD-L1) with radiation and TMZ was tolerable and demonstrated efficacy in line with published trials for newly diagnosed GBM. We leveraged genomic, transcriptomic and multiplex immunofluorescent approaches on tumors, as well as, metagenomic analysis on stool samples to identify correlates of differential clinical outcome to SOC plus anti-PD-L1 therapy. Our GBM cohort’s global pre-treatment molecular profiles were consistent with previously reported cohorts. However, with deeper interrogation, using both unsupervised and supervised analyses, we observed specific molecular features that distinguished patients who achieved a longer OS with anti-PD-L1 treatment.
Multiple studies indicate that GBM tumors with a mesenchymal subtype tend to display more aggressive behavior and greater resistance to standard-of-care approaches31,41. Consistent with prior studies, we observed that mesenchymal subtype tumors showed comparatively elevated markers of immune infiltration. Our study further demonstrated that patients whose tumors displayed a greater immune infiltrate achieved a longer OS with anti-PD-L1 treatment. Importantly, the evaluation of tumors in two distinct and independent GBM cohorts from The Cancer Genome Atlas (TCGA) and Glioma Longitudinal AnalySiS (GLASS) Consortium (Fig. 4A) who underwent a chemoradiation regimen, but did not receive anti-PD-L1 treatment, failed to show a statistically significant relationship between high EIS and OS. Thus, tumor immune infiltration alone appears insufficient to mediate a longer survival outcome, but the presence of an immune infiltrate may mediate the effects of anti-PD-L1 treatment resulting in the higher OS observed in a subgroup of our cohort. This potential signal of activity warrants further investigation in a future trial. Our study supports the hypothesis that the mesenchymal subtype of GBM may also particularly benefit from anti-PD-L1 approaches given its inherent immune status.
Distinct immune programs were observed to be associated with improved OS in a statistically significant manner. The tumors of patients who achieved a higher mOS showed enrichment for activated myeloid dendritic cell, M1 macrophage, and CD4 effector memory cell-states, indicative of an active immunogenic milieu. In addition, the cytotoxic T cell protein markers and molecular executors of immune-mediated cytotoxicity were notably elevated in the GBM tumors of patients with longer mOS, whereas tumors that showed no or low immune infiltration tended to be more enriched in neurodevelopmental markers. We leveraged recently published single cell gene expression profiles of immune cell types in GBM22,42 to better characterize the pretreatment transcriptomics of lymphoid and myeloid profiles in our cohort. Despite the lymphoid single cell profiles having multiple immune checkpoint markers, most genes did not display differential expression between survival groups. Differences were observed in lymphocyte markers and antigen presenting genes, indicating that the separation in survival outcomes within our cohort was being driven more by the presence of positive anti-tumor features rather than the inherent absence of immunosuppressive factors. Among pretreatment myeloid subsets, only the dendritic cell, specifically the CCR7 + DC profile, was associated with longer OS in our cohort, further supporting the prominence of an antigenic and immune responsive phenotype for improved clinical outcome. This more favorable overall lymphoid and myeloid immune milieu, in addition to a higher percentage of cytotoxic T cells expressing PD-L1, may account for the separation in mOS to be observed with the addition of single-agent anti-PD-L1.
Given recent data showing that specific gastrointestinal microbiome profiles affect tumor immune status and immune checkpoint inhibitor therapy, we analyzed samples collected from a subset of trial patients. The observation that pre-treatment samples from patients whose tumors demonstrated greater immune infiltration tended to have more diverse and enriched gastrointestinal microbiota, and that patients with these characteristics tended to have a longer OS, is consistent with a growing body of evidence that host, tumor and microbiome each contribute to the efficacy of immune checkpoint blockade therapy43,44.
Unsupervised global gene mutation tumor profiling separated patients into two subgroups with markedly different mOS. Further analysis showed all the PTEN mutant tumors to be in the subgroup with a better OS, whilst EGFR mutations were enriched in the other subgroup. These results are intriguing, as both PTEN and EGFR mutations have been proposed to have immune suppressive effects in a variety of cancers. Notably, the PTEN mutant tumor cohort also showed a statistically significantly better OS compared to the PTEN/EGFR wildtype group, suggesting that the difference in survival between the two subgroups may entail more than the relative effect of their respective PTEN or EGFR mutations, and each may serve as a marker of an overall cellular state with divergent immune responses. These findings provide a further impetus to investigate the functional relevance of PTEN and EGFR mutations on GBM tumor immunity and anti-PD-L1 efficacy.
Multiple factors have been shown to be associated with clinical outcome in the standard-of-care chemo-radiation setting (MGMT methylation, stereotactic intervention, high KPS, age). In this atezolizumab-treatment trial, MGMT methylation trended toward a greater OS, while less than a gross total resection and being biologically male trended to a lesser OS benefit, consistent with prior studies. Neither KPS nor age were statistically significant factors, likely given the entrance requirements of the study and relative cohort size. Notably, tumor immune infiltration and PTEN/EGFR mutation status were the only statistically significant, independent factors associated with OS in this study.
Our study shows that a substantial subset of GBM patients have tumors that may be more amenable to immune-based therapy and suggests potential future target opportunities. First, given that GBM tumors display a range of myeloid/macrophage milieus, deploying agents that shift profiles toward an even greater pro-inflammatory, anti-tumor phenotype can enhance the effects of checkpoint blockade therapy. Second, among the immune checkpoint molecules, TIM3 (aka, HAVC2) showed near ubiquitously elevated gene expression in more infiltrated tumors. TIM3 protein expression is usually an indicator of T cell dysfunction. However, pre-clinical studies indicate that combining TIM3 inhibition with PD-L1/PD1 axis inhibitors has a synergistic effect on inhibiting cancer cell growth and tumor antigen-specific T-cell response. Third, observations from the gut microbiome analysis are particularly intriguing in a CPI-treated GBM cohort. Correlation of the gut microbiome profile with OS, as well as the tumor immune infiltration is consistent with observations in other tumor types and warrants further investigation of the potential impact of the gut microbiome in GBM patients treated with IO and combination therapy. Finally, our study supports the view that the timing and duration of anti-PD-L1 treatment may be pivotal in the newly diagnosed setting, consistent with the efficacy observed when neoadjuvant anti-PD-L1/PD1 inhibition was employed in recurrent GBM disease19.
Methods
Study design and participants
This research complies with all relevant ethical regulations. The Institutional Review Board (IRB) at the University of Texas MD Anderson Cancer Center approved the study protocol. NCT03174197 was a phase I/II trial in patients with newly diagnosed GBM which was conducted at The University of Texas MD Anderson Cancer Center (MD Anderson). Patients were unselected for MGMT status. MGMT is a well-established clinical prognostic and predictive biomarker in glioblastoma in regards to response to alkylating therapy. During the Phase I safety run in component (N = 10), atezolizumab was administered intravenously every 2 weeks in combination with radiation and oral temozolomide (concurrent stage) followed by atezolizumab intravenously every 2 weeks administered in combination with oral temozolomide (adjuvant stage) days 1–5 out of a 28 day or 4 week cycle. With acceptable toxicity profile, the trial continued from the safety run in phase to Phase II, during which a larger number of patients (N = 50) were treated with atezolizumab in combination with radiation and temozolomide during the concurrent stage and atezolizumab in combination with temozolomide during the adjuvant stage. Patients had to be at least 18 years old, have histologically confirmed glioblastoma or gliosarcoma according to WHO 2016 criteria, and a Karnofsky Performance Scale (KPS) score ≥60. A baseline brain MRI obtained no more than 14 days prior to study enrollment on a stable or tapering dose of steroids no greater than 4 mg a day of dexamethasone for at least 5 days was required prior to treatment start. Approval for the study was granted through the IRB at MD Anderson, and all patients had provided written informed consent before study entry. Use of human material was approved. This trial was performed according to the Declaration of Helsinki’s principles.
Toxicity monitoring and futility monitoring of overall survival
Toxicity was monitored using Bayesian continuous monitoring45 where the toxicity evaluation endpoint was defined as treatment-related unmanageable toxicities, including grade 3 or 4 AEs that require termination of the treatment during the first 10 weeks of treatment. It is assumed that median time to DLT followed an Inverse Gamma distribution and the individual times to DLT followed an exponential distribution. Let mE represent the median in the newly accrued (experimental) patients and let mS represent the median to be used for comparison. For the comparison median we specify an Inverse Gamma prior with mean = 19.43 weeks and SD = 0.10 weeks – essentially a constant 30% 10-week toxicity rate. For the experimental median we specify an Inverse Gamma prior with mean of 19.43 weeks and SD = 10.0 weeks. Monitoring was continuous with the rule to stop if Pr(mE > mS | data) <0.05. Meanwhile, we applied the same method to monitor time to death continuously starting with the 10th patient assuming the median time to death follows an Inverse Gamma distribution and that the individual death times follow an exponential distribution. Let mE represent the median in the newly accrued (experimental) patients and let mS represent the historical median to be used for comparison. Based on historical data the median death time is about 15 months. For the historical median we specify an Inverse Gamma prior with alpha = 58.25 and beta = 858.75 (which has mean = 15.0 and SD = 2.0). For the experimental median we specify an Inverse Gamma prior with alpha = 4.25 and beta = 48.75 (which has a mean of 15.0 and SD = 10.0). The maximum sample size is 60 and the expected accrual rate is 2.5 per month. Monitoring will be continuous with the rule to stop if Pr(mE > mS + 6 | data) <0.05.
Imaging protocol- and imaging response-assessment criteria
MRI scans were acquired within 14 + /- 3 days of registration, 21-28 + /- 2 days following completion of the concurrent stage and then every 8 weeks or after every 2 cycles of adjuvant treatment of atezolizumab and temozolomide on a 1.5 or 3.0 Tesla MRI scanner using the standard protocol: axial T1-weighted sequence (T1WI) (repetition time [TR], 700 ms; echo time [TE], 12 ms; slice thickness, 5 mm; acquisition matrix 352 × 224); axial fluid attenuation inversion recovery (FLAIR) sequence (TR, 10000 ms; TE, 140 ms; slice thickness, 5 mm; acquisition matrix, 256 × 256); and axial post-contrast T1WI, acquired 5 min after the contrast injection (TR, 750 ms; TE, 13 ms; slice thickness, 5 mm; acquisition matrix, 384 × 256). The conventional T1WI and FLAIR sequences were used to assess response to therapy via the iRANO criteria46. Contrast-enhancing lesions with bidimensional measurements of >1 cm were considered as measurable (index) lesions, whereas smaller lesions and those with non-enhancing T2/FLAIR hyperintensity were considered to be non-measurable (non-index) lesions. Patients were categorized based on best response as having: 1) progressive disease; 2) stable disease; 3) partial response; or 4) complete response.
Correlative analyses
An abundance of data show isocitrate dehydrogenase (IDH) mutant GBM tumors to be molecularly and phenotypically distinct and associated with markedly better survival outcomes47. Thus, the correlative analyses focused on patients with IDH wildtype GBM consistent with the updated WHO 2021 criteria, and the IDH mutant tumors were not carried forward.
Sequencing and data possessing
BCL (raw output of Illumina HigSeq) files are processed using Illumina’s Consensus Assessment of Sequence and Variation (CASAVA) tool (http://support.illumina.com/sequencing/sequencing_software/casava.html) for demultiplexing/conversion to FASTQ format, which is the standard input for most aligners and downstream analytic tools. For whole exome sequencing, FASTQ files are aligned to the reference genome (human Hg19) using BWA48 with 3 mismatches with 2 in the first 40 seed regions for sequences less than 100 bp or using BWA mem with 31 bp seed length for sequences over 100 bp. The aligned BAM files are subjected to mark duplication, re-alignment, and re-calibration using Picard and GATK49 before any downstream analyses. For RNA sequencing, FASTQ files of RNA samples are processed using both STAR50 following the two-step alignment procedure and TopHat51 and Cufflinks52.
Somatic copy number analysis
Somatic copy number alterations were identified from WES data using ExomeLyzer53, followed by Circular Binary Segmentation54. Segmented copy number values were corrected by tumor purity using the In Silico Admixture Removal (ISAR) procedure55, where the tumor sample purity and ploidy estimates were calculated using sequenza algorithms56. Subsequently, statistically significant focal copy number alterations were identified from the ISAR-corrected segmented data using GISTIC 2.0.225. The following parameters were used: amplifications threshold: 0.3, deletions threshold: 0.3, focal length cutoff: 0.70, gene gistic: yes, confidence level: 0.99, q-value threshold: 0.25, join segment size: 4, remove X: no, cap val: 1.5, run broad analysis: yes, max sample segs: 2000, arm peel: yes, gene collapse method: extreme. Tumors were then clustered into a 2-cluster solution based on thresholded copy number at reoccurring alteration peaks from GISTIC output “all_lesions.conf_99.txt file”. Function GisticChromePlot implemented in P package “maftools” were used to plot gistic score in file “scores.gistic” along linearized chromosome.
Mutation analysis
Mutations are identified from two sources: 1) Samples collected for research purpose were subjected to whole exome sequencing. 2) Samples collected for clinical mutation identification were subject to sequencing by one of these four next generation sequencing (NGS)-based panels: “Solid Tumor Genomic Assay 2018” (coding sequence of 134 genes and copy number variations in 47 genes), “Solid Tumor Genomics Assay v1” (coding sequence of 128 genes and selected copy number variations (amplifications) in 49 genes), “50-Gene Somatic Mutation Analysis Panel Report “ (coding sequence of a total of 50 genes) and “FoundationOne”. The first three tests were developed and their performance characteristics determined by the Molecular Diagnostic Laboratory (MDL) at the M.D. Anderson Cancer Center.
For WES data, somatic mutations were called using MuTect57; Short insertions/deletions (indels) were called using Pindel (Ye et al. 2009). Following variant calling, we implemented a series of post-call quality filters. These filters were designed to retain somatic variants that met the following criteria: 1) Locate within the targeted sequencing region, 2) Pass mapping quality threshold of 25. 3) Exhibit a statistically significant different alternate/reference allele ratio (p ≤ 0.05) from that of the matched normal sample by proportion test. 4) Attained adequate coverage (≥10) in both the tumor and normal samples. 5) pass the tumor event log likelihood threshold of 6.3.
Public GBM datasets
The Cancer Genome Atlas (TCGA) GBM dataset: Normalized gene transcript abundances (data_mrna_seq_v2_rsem.txt), mutation calls (data_mutations.txt) and clinical data (data_clinical_sample.txt, data_clinical_patient.txt) of GBM samples contained within the TCGA cohort (Firehose Legacy) were accessed from cBioPortal (https://www.cbioportal.org). Transcription profiles of GBM samples (n = 144) that are IDH wildtype as determined by mutation calls and collected as primary tumor were used for downstream analysis.
The Glioma Longitudinal AnalySiS (GLASS) GBM dataset: Clinical data as well as normalized gene transcript abundance matrices of GLASS dataset were acquired from Synapse (https://www.synapse.org/glass)32. To ensure meaningful comparison, we limited our analysis to baseline GLASS GBM samples (n = 83) that are IDH wildtype, and had received full treatment, as indicated by a value of “1” in columns labeled “TMZ + RT, TMZ,” “treatment_tmz,” “treatment_concurrent_tmz” and “treatment_radiotherapy.”
Differential gene expression and gene set enrichment
Gene expression levels were measured as log2-transformed transcripts per million (TPM) (log2(TPM + 1)). For identifying the differentially expressed genes (DEGs), we employed the “limma trend” mode available in the limma package58. This method uses a linear model to estimate a trend over all genes and adjusts the differential expression tests accordingly58. P-values are adjusted for multiple testing using the FDR method. Genes with an adjusted p-value below 0.05 were considered as DEGs.
Gene set Enrichment Analysis (GSEA) was conducted with fast gene set enrichment algorithm from R package “fgsea” (version 1.16.0)59,60 with default parameters. The gene set used for enrichment analysis is the “hallmark” gene set, which includes 50 gene sets that represent well-defined biological states or processes. Gene sets with an FDR corrected p-value below 0.05 were considered statistically significant.
ESTIMATE score, immune deconvolution and enrichement analysis of scrnaseq-derived immune profiles
We used the R package “immunedeconv” (version 2.1.0) and its “deconvolute_estimate” function to calculate ESTIMATE score61. The function integrates the ESTIMATE algorithm, which predicts tumor purity, and the presence of infiltrating stromal/immune cells in tumor tissues using gene expression data, through GSEA30.
Deconvolution of the immune microenvironment was conducted via function “deconvolute” in R package “immunedeconv”, with method parameter set as “xCell” and the rest as default. Wilcoxon test (two sided) was used to examine the difference in cell type abundance between groups.
Markers of various immune cell populations identified by Lee et al. were used as gene sets to calculate corresponding enrichment scores for each sample. Enrichment was performed using single-sample Gene Set Enrichment Analysis (ssGSEA) via “ssgsea” function from R package “corto” (version 1.2.4) with default parameters [PMID: 32232425].
GBM transcriptional subtype classification
Glioblastoma transcriptional subtyping was conducted on RNAseq TPM using the “ssgsea.GBM.classification” R package31 with glioblastoma subtype (Classic, Mesenchymal, and Proneural) signatures. This method quantified an enrichment score for each glioblastoma subtype in a sample using ssgsea algorithm, as well as a permuation-based P-value indicating the statistical significance. For assignment of a single subtype to each sample, the subtype with the lowest P-value would be assigned to each sample.
Multiplex immunofluorescence imaging
Tissue image immunoprofiling was conducted using the Opal chemistry and multispectral microscopy Vectra system (Perkin-Elmer) with 2 multiplex immunofluorescence (mIF) panels: (1) The first panel targeted CD3, CD8, CD68, PD-1, PD-L1, GFAP. (2) The second panel targeted CD3, CD8, Granzyme B, FOXP3, CD45RO, CD68, and GFAP. Sources, catalogue numbers and species of antibodies used are provided in Supplemental Table 2. Marker expression was then determined using image analysis software InForm™ 2.2.1. Mutually exclusive combinations of markers on a per cell basis were created from the InForm cell segmentation files. Of 60 patients enrolled, image immunoprofiling was performed successfully on pre-treatment tissue in 47 (IDH-wildtype) patients.
Metagenomic sequencing and microbiome profiling
MetaPhlAn version 3.0 (ref) classified taxonomy (CHOCOPhlan database version 20190134) data was imported into R environment (version 4.4.0) using Phyloseq R Package version 1.48.062 at species level. Alpha diversity was accessed using three metrics – Observed, Shannon, and Inverse Simpson. Beta diversity was quantified using Bray-Curtis distance and statistical significance was evaluated using Adonis2 test from Vegan package version 2.6-6.1 (https://github.com/vegandevs/vegan, https://vegandevs.github.io/vegan/) with 1000 permutations. Differentially abundant species was detected using ANCOMBC R package version 1.6.235,36.
Survival analysis
Survival was measured from the time of diagnosis commensurate with the initial study design, as displayed in Supplemental Fig. 1. To analyze the association between survival and molecular/correlative features, time from initial treatment was employed, allowing for the comparison with other studies that used this same initial metric. Kaplan-Meier curves were generated via ggsurvplot implemented in R package “survminer,” with statistical significance assessed using the log-rank test and hazard ratio estimated via Cox proportional hazards regression model implemented in R package “survival” (v 3.4.0). The Cox proportional-hazards model was also used for multivariate analysis to determine relative risk and independent statistical significance. Analyses were performed with R package “survivalAnalysis” (v 0.3.0).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The sequencing data, mutation calls and TPM matrix generated in this study and relevant non-identifiable clinical metadata have been deposited in the European Genome-Phenome Archive (EGA) under accession [EGAD50000001154]. https://ega-archive.org/studies/EGAS50000000784. These data are available upon request to the corresponding author for academic cancer research purposes in accordance with the conditions of consent agreed to by the source participants. Requests will be addressed within 8 weeks and, if approved, access will be made available for a oneyear period, renewable upon additional request. Processed data generated in this study are available via https://github.com/WoodmanLab/GBMnat that has been archived as https://doi.org/10.5281/zenodo.13931489 or provided in supplemental materials. Source data are provided with this paper.
Code availability
The custom code for genomic, transcriptomic, and multiplex immunofluorescence imaging analysis is available at https://github.com/WoodmanLab/GBMnat that has been archived as https://doi.org/10.5281/zenodo.13931489, and the code for microbiome analysis can also be found at https://github.com/mda-primetr/Weathers-GBM.IO-2024.
References
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Gilbert, M. et al. Dose-Dense Temozolomide for Newly Diagnosed Glioblastoma: A Randomized Phase III Clinical Trial. J. Clin. Oncol. 31, 4085–4091 (2013).
Weller, M. et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 18, 1373–1385 (2017).
Wen, P. Y. et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 22, 1073–1113 (2020).
Lamborn, K. R. et al. Progression-free survival: an important end point in evaluating therapy for recurrent high-grade gliomas. Neuro Oncol. 10, 162–170 (2008).
Wick, W. et al. Lomustine and Bevacizumab in Progressive Glioblastoma. N. Engl. J. Med. 377, 1954–1963 (2017).
Wu, W. et al. Joint NCCTG and NABTC prognostic factors analysis for high-grade recurrent glioma. Neuro Oncol. 12, 164–172 (2010).
Nduom, E. K. et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro. Oncol. (2015).
Omuro, A. et al. Nivolumab plus radiotherapy with or without temozolomide in newly diagnosed glioblastoma: Results from exploratory phase I cohorts of CheckMate 143. Neurooncol. Adv. 4, vdac025 (2022).
Lim, M. et al. Phase 3 Trial of Chemoradiotherapy With Temozolomide Plus Nivolumab or Placebo for Newly Diagnosed Glioblastoma With Methylated MGMT Promoter. Neuro Oncol. 24, 1935–1949 (2022).
Blumenthal, D. T. et al. Pembrolizumab: first experience with recurrent primary central nervous system (CNS) tumors. J. Neurooncol. 129, 453–460 (2016).
Johanns, T. M. et al. Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discov. 6, 1230–1236 (2016).
Bouffet, E. et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J. Clin. Oncol. 34, 2206–2211 (2016).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Razavi, S. M. et al. Immune Evasion Strategies of Glioblastoma. Front Surg. 3, 11 (2016).
Chongsathidkiet, P. et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat. Med. 24, 1459–1468 (2018).
Chen, Z. & Hambardzumyan, D. Immune Microenvironment in Glioblastoma Subtypes. Front Immunol. 9, 1004 (2018).
Zhao, J. et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 25, 462–469 (2019).
Cloughesy, T. F. et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med. 25, 477–486 (2019).
Strauss, L. et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci. Immunol. 5, eaay1863 (2020).
Rao, G. et al. Anti-PD-1 Induces M1 Polarization in the Glioma Microenvironment and Exerts Therapeutic Efficacy in the Absence of CD8 Cytotoxic T Cells. Clin. Cancer Res. 26, 4699–4712 (2020).
Lee, A. H. et al. Neoadjuvant PD-1 blockade induces T cell and cDC1 activation but fails to overcome the immunosuppressive tumor associated macrophages in recurrent glioblastoma. Nat. Commun. 12, 6938 (2021).
White, K. et al. Identification, validation and biological characterisation of novel glioblastoma tumour microenvironment subtypes: implications for precision immunotherapy. Ann. Oncol. 34, 300–314 (2023).
Masuda, T. et al. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 30, 1271–1281 (2020).
Rajani, K. R. et al. Harnessing Radiation Biology to Augment Immunotherapy for Glioblastoma. Front Oncol. 8, 656 (2018).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 (2016).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Zehir, A. et al. Erratum: Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 1004 (2017).
Rooney, M. S. et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Wang, Q. et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 32, 42–56.e6 (2017).
Varn, F. S. et al. Glioma progression is shaped by genetic evolution and microenvironment interactions. Cell 185, 2184–2199.e16 (2022).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017).
Beghini, F. et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10, e65088 (2021).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021).
Lin, H. & Peddada, S. D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514 (2020).
Katsigiannis, S. et al. MGMT-Positive vs MGMT-Negative Patients With Glioblastoma: Identification of Prognostic Factors and Resection Threshold. Neurosurgery 88, E323–E329 (2021).
Stark, A. M. et al. Glioblastoma: clinical characteristics, prognostic factors and survival in 492 patients. Clin. Neurol. Neurosurg. 114, 840–845 (2012).
Lamborn, K. R., Chang, S. M. & Prados, M. D. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro-oncol. 6, 227–235 (2004).
Colopi, A. et al. Impact of age and gender on glioblastoma onset, progression, and management. Mech. Ageing Dev. 211, 111801 (2023).
Kim, Y. et al. Perspective of mesenchymal transformation in glioblastoma. Acta Neuropathol. Commun. 9, 50 (2021).
Arrieta, V. A. et al. Immune checkpoint blockade in glioblastoma: from tumor heterogeneity to personalized treatment. J. Clin. Invest 133, e163447 (2023).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Park, E. M. et al. Targeting the gut and tumor microbiota in cancer. Nat. Med. 28, 690–703 (2022).
Thall, P. F., Wooten, L. H. & Tannir, N. M. Monitoring event times in early phase clinical trials: some practical issues. Clin. Trials 2, 467–478 (2005).
Okada, H. et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol. 16, e534–e542 (2015).
Miller, J. J. et al. Isocitrate dehydrogenase (IDH) mutant gliomas: A Society for Neuro-Oncology (SNO) consensus review on diagnosis, management, and future directions. Neuro Oncol. 25, 4–25 (2023).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Kim, D. & Salzberg, S. L. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 12, R72 (2011).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Zhang, J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346, 256–259 (2014).
Olshen, A. B. et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557–572 (2004).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Favero, F. et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 26, 64–70 (2015).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Sturm, G., Finotello, F. & List, M. Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol. Biol. 2120, 223–232 (2020).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013).
Acknowledgements
The study was supported by Genentech. The authors acknowledge all members of the Platform for Innovative Microbiome & Translational Research (PRIME-TR), the Moon Shots Program, Rare Tumor Initiative, and the support of the High Performance Computing for research facility at the University of Texas MD Anderson Cancer Center for providing computational resources that have contributed to the research results reported in this paper.
Author information
Authors and Affiliations
Contributions
Conception and design: SPW, SEW, JdG. Collection of data and patient recruitment: S.P.W., S.E.W., X.L., B.O., R.H., M.L., W.K.Y., M.P.P., C.K.M., J.dG. Data analysis and interpretation: S.P.W., X.L., H.L., H.Z., M.K., S.E.W., N.A., A.D., J.dG. Manuscript writing: S.P.W., X.L., S.E.W., J.dG., N.A., H.Z., A.D. Final review and approval of manuscript: S.P.W., X.L., H.Z., A.V.D., M.K., B.M., H.L., R.A.H., N.K.M., B.J.O., M.P.P., M.L., C.K.M., W.K.Y., L.M.S., D.M.M., I.W., E.R.P., S.H., A.F., J.A.W., K.S., W.C.D., N.J.A., S.E.W., J.dG. Accountable for all aspects of the work: S.P.W., X.L., H.Z., A.V.D., M.K., B.M., H.L., R.A.H., N.K.M., B.J.O., M.P.P., M.L., C.K.M., W.K.Y., L.M.S., D.M.M., I.W., E.R.P., S.H., A.F., J.A.W., K.S., W.C.D., N.J.A., S.E.W., JdG.
Corresponding authors
Ethics declarations
Competing interests
K.S. and W.C.D. are members of the M.D. Anderson Genentech Joint Steering Committee and provided input on the design of the translational research and review of data and their scientific interpretation. K.S. and W.C.D. participated in funding decisions for the translational research and manuscript review. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Weathers, SP., Li, X., Zhu, H. et al. Improved overall survival in an anti-PD-L1 treated cohort of newly diagnosed glioblastoma patients is associated with distinct immune, mutation, and gut microbiome features: a single arm prospective phase I/II trial. Nat Commun 16, 3950 (2025). https://doi.org/10.1038/s41467-025-56930-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56930-7
This article is cited by
-
A phase I-II study of niacin in patients with newly diagnosed glioblastoma: safety and interim phase II analysis
Journal of Neuro-Oncology (2026)
-
Immunotherapies for Central Nervous System Tumors and their Neurologic Complications
Current Neurology and Neuroscience Reports (2026)
-
From Classical to Emerging Biomarkers of Brain and Central Nervous System Tumors. An Evidence-Based Review with a Focus on Gliomas
Cellular and Molecular Neurobiology (2026)
-
Current strategies and novel immunotherapeutic approaches for overcoming immune resistance in glioblastoma
Discover Oncology (2025)