Abstract
Immunotherapy (IO) is an effective treatment for various cancers; however, the benefits are modest for small cell lung cancer (SCLC). The poor response of SCLC to anti-PD-1/PD-L1 IO is due in part to the lack of cytotoxic T cells because of limited chemokine expression from SCLC tumors. Immunogenic radiosensitizers that enhance chemokine expression may be a promising strategy forward. Here, we show that the PARP inhibitors (PARPi), including olaparib, talazoparib and veliparib, in combination with radiotherapy (RT) enhance the immune activation and anti-tumor efficacy in SCLC cell lines, patient-derived xenograft (PDX) and syngeneic mouse models. The effect is further enhanced by continued delivery of adjuvant PARPi. The combination treatment (PARPi with RT) activates the cGAS-STING pathway and increases the mRNA levels of the T cell chemo-attractants CCL5 and CXCL10. In addition to upregulation of transcription, the combination treatment increases chemokine CXCL10 protein levels via stabilization of CXCL10 mRNA in an EIF4E2-dependent manner. The incorporation of anti-PD-L1 IO into the PARPi with RT combination therapy further improves the anti-tumor efficacy by increasing T cell infiltration and function. This study thus provides a proof of principle for the combination of PARP inhibitors, RT and anti-PD-L1 IO as a treatment strategy for SCLC.
Similar content being viewed by others
Introduction
Small cell lung cancer (SCLC) is a highly aggressive lung cancer subtype with poor prognosis1. Despite the use of chemotherapy and selective use of radiotherapy (RT) as standard treatments, the five-year survival rate for patients with extensive-stage SCLC (ES-SCLC) is less than 5%. Immunotherapy (IO) is effective against various cancers, including melanoma, non-small cell lung cancer, MSI-high colorectal cancer, head and neck cancer, and among other malignancies2,3,4,5,6. Most IO therapeutics block checkpoint proteins or their partners to prevent inhibitory signals for T cells, allowing T cells to access and perform their cytotoxic anti-tumor function7. In 2018, for patients with ES-SCLC, a landmark clinical trial of atezolizumab (anti-PD-L1) in combination with chemotherapy demonstrated the first improvement in overall survival (OS) in a phase III study of systemic agents in over 30 years8. However, the OS benefit was relatively small, with a median OS of 12.3 months with atezolizumab compared to 10.3 months with chemotherapy alone. Additionally, the median progression-free survival (PFS) gain was less than one month. Similar results were also observed in another trial of durvalumab (anti-PD-L1) in combination with platinum-etoposide, in which the median OS was 2 months longer than the platinum-etoposide group9. These results suggest that while the combination of IO with chemotherapy may be beneficial for some patients with SCLC, the overall survival benefit is small, highlighting the need for further strategies to decrease IO resistance and improve IO efficacy for patients with ES-SCLC.
Multiple factors contribute to immunotherapy resistance and one of the most important factors is the lack of T cell infiltration10,11. Chemokines, such as CCL5 and CXCL10, are critical for T cell infiltration12. DNA damaging agents, including RT, trigger chemokine transcription by the formation of micronuclei due to the mis-segregation of DNA during cell cycle progression followed by translocation of cGAS into these micronuclei13. This activates the cGAS-STING pathway in RT-treated cancer cells to induce chemokine transcription13,14. In addition to transcriptional control via the cGAS-STING pathway, chemokines are subjected to regulation by mRNA translation and degradation mechanisms, which are also central and interrelated steps for gene expression15,16. However, the mechanistic details of the degradation of relevant chemokine mRNA after DNA damage are poorly understood.
With respect to mRNA degradation, the mRNA 3′ untranslated region (UTR) contains several sequence elements, such as the adenylate-uridylate-rich element (AU-rich element), which can be targeted by microRNAs (miRNA) or AU-rich element binding proteins (AUBP)17,18,19. The binding of miRNAs or AUBPs to the AU-rich element (ARE) can lead to the degradation of the mRNA or the repression of translation, thus reducing the amount of chemokine protein produced. Previous studies have shown that EIF4E2, a translation initiation factor, plays an important role in mRNA translation suppression. EIF4E2 (4EHP), a homologous member of EIF4E, has been reported to destabilize target mRNA or participate in translation repression with AU-rich elements by interacting with tristetraprolin (TTP) or induce translation silence by competing with EIF4E and recruiting miRNA through the CCR4-NOT complex20,21,22,23. However, it is unknown whether EIF4E2 regulates cGAS-STING pathway-induced chemokines on the mRNA degradational level.
Here, we demonstrate that PARP inhibition induces radiosensitization increased CXCL10 mRNA level by activating the cGAS-STING pathway. We conduct unbiased whole transcriptome analysis of SCLC after treatment with the combination of RT and olaparib and identify the downregulation of EIF4E2 leading to increased protein levels of the T-cell chemoattractant CXCL10. The upregulated chemokine protein levels promote cytotoxic CD8+ T cell infiltration. Finally, the incorporation of anti-PD-L1 IO with PARPi (olaparib or talazopairb) and RT improves anti-tumor efficacy by increasing T cell infiltration. These findings reveal a mechanism of treatment-induced chemokine regulation where the combination of PARP inhibition and RT could improve tumor control and enhance IO response against SCLC.
Results
Adjuvant maintenance of PARP inhibitor olaparib enhances radiosensitization of SCLC
Previous work with PDX models demonstrated that PARPi is a radiosensitizer for SCLC where PARPi was delivered concurrent with and adjuvant to RT24. Clinically, most radiosensitizers are given concurrently with long-course RT (5 to 6 weeks) without a substantial adjuvant phase25,26,27. However, given that short-course RT (2 weeks) for consolidative thoracic RT for ES-SCLC patients is more common, we sought to examine whether RT concurrent with and followed by adjuvant maintenance PARPi is more effective than concurrent PARPi only28.
We initially characterized the radiosensitization of olaparib by short-term viability assays in the SBC5 human cell line (DMF37 = 1.25 with 100 nM) and the KP1 murine cell line (DMF37 = 1.34 with 10 nM; Supplementary Fig. 1a) that was confirmed by clonogenic survival assays (SBC5 DMF37 = 1.69 with 1 μM, KP1 DMF37 = 1.46 with 0.1 μM; Supplementary Fig. 1b). In contrast, in non-cancer lung fibroblast cell lines LF1 and Wi-38 PARPi radiosensitization was not observed suggesting this effect is cancer specific (Supplementary Fig. 2a,b).
To determine whether maintenance olaparib increases radiosensitization, we designed a 5-arm study (Fig. 1a). Cells were pulsed (PUL) for 3 h with 1 µM of olaparib that was withdrawn 1 h after RT was delivered. Cells in the combination (COM) arm were also exposed to 1 μM olaparib in a similar manner but olaparib was not withdrawn after RT but rather maintained (Fig. 1a). DMF37 of COM was 1.91 while the DMF37 of PUL was 1.49 (Fig. 1b) and demonstrated maintenance olaparib in COM enhanced PARPi radiosensitization.
a Schematic of experimental design with olaparib and RT treatments in vitro and in vivo (Created in BioRender. Ran, X. (2025) https://BioRender.com/e93s595). b Survival fraction of SBC5 treated with pulse olaparib (PUL) or continuous olaparib (COM) with RT. c The tumor growth curve of SBC5 xenograft treated with VEH/OLA (n = 6 mice), RT/PUL/COM (n = 7 mice). d Upper: Quantification of olive tail movement by alkaline comet assay in treated SBC5 cells (n = 3 biological replicates) for 3 and 48 h. Lower: Representative images of DNA damage detected by comet assay in SBC5 cells upon treatment for 48 h. e Upper: The quantification of γH2AX immunostaining in each group for 3 and 48 h (n = 3 biological replicates). Lower: Representative dsDNA damage detected by γH2AX nuclear foci immunostaining in SBC5 upon treatment for 48 h. f Upper: Quantification of the frequency of cGAS+ micronuclei per cell (n = 3 biological replicates). Lower: Representative cGAS+ micronuclei staining in SBC5 upon treatment for 2 days. g Upper: Quantification of the relative intensity of p-STING normalized by β-Actin. Lower: western blot of p-STING, STING, and cGAS in SBC5 and SW1271 upon treatment for 3 days from one of two independent experiments. h Heatmap of CCL5 and CXCL10 mRNA in SBC5(Y), H146(A), 82(N), H526(P), H1048(P) and KP1(A) cells after treatment for 2 days and in SBC5 xenograft and SCRX-Lu149(A) PDX for 15 days. i, j Upper: Western blot against cGAS or STING in EGFP KO control (Ctrl) or cGAS KO or STING KO cells for SBC5 and H1048. Lower: CCL5 or CXCL10 mRNA in Ctrl or cGAS/STING KO cells upon treatment for 48 h (n = 3 biological replicates). For (c), the data are presented as mean ± SD. For (d) and (e), mean counts of olive movement or γH2AX nuclear foci per cell are presented as a line. For (b), (d)–(f), (i), (j), the data are presented as mean ± SD of three replicates for each treatment. All comparisons were calculated with one-way or two-way ANOVA. (p: ****<0.0001 < *** < 0.001 < ** < 0.01 < * < 0.05). All exact p values are listed in the source data. Source data are provided as a Source Data file.
We sought to confirm the effect of maintenance olaparib in vivo, where NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were engrafted with SCRX-Lu149 chemonaive (CN) and SCRX-Lu149 chemoresistant (CR) PDX tumors and SBC5 cells and treated with olaparib as depicted in Fig. 1a. The median time to volumetric endpoint of COM treatment was 85 days for SBC5, 51.5 days for SCRX-Lu149 CN and 53 days for SCRX-Lu149 CR compared with PUL treatment of 43 days for SBC5, 32 days for SCRX-Lu149 CN, and 39 days for SCRX-Lu149 CR (log-rank p = 0.0002 for SBC5; 0.0012 for SCRX-Lu149 CN and 0.0009 for SCRX-Lu149 CR; Fig. 1c and Supplementary Fig. 1c,d).
Next, we sought to determine whether the radiosensitizing effect was a result of increased DNA damage. Here, we evaluated the DNA damage detected in PUL versus COM treatment at 2 time-points, 3 h or 48 h in SCLC cell lines. First, we evaluated the total DNA damage by an alkaline comet assay to assess single-stranded and double-stranded DNA breaks (DSB). Olive tail moment of COM (mean 7.2) was significantly (p < 0.0001) greater than PUL (mean 2.8) at the 48-h timepoint (Fig. 1d), which demonstrated that olaparib maintenance prevented DNA repair and also continued to generate DNA damage. Orthogonally, we assessed DSBs by visualizing sub-nuclear foci of γH2AX in SBC5 cells. Similar to comet assay, maintenance of olaparib following concurrent PARPi and RT (COM) demonstrated greater DSBs at 48 h as compared with PUL or RT alone (Fig. 1e). While in the non-tumor lung fibroblast cells, including LF1 and Wi-38, γH2AX were similar between RT alone and COM treatment, suggesting DNA damage from PARPi radiosensitization is cancer specific (Supplementary Fig. 2c).
PARPi with RT synergistically promotes cGAS-STING-dependent chemokine transcription
DNA damage induces the formation of micronuclei and triggers the translocation of cGAS into micronuclei13,14, to produce 2′3′-cGAMP (cyclic GMP-AMP) resulting in the phosphorylation of STING to stimulate downstream chemokine transcription, including CCL5 and CXCL1029,30. As we observed COM treatment (concurrent and maintenance olaparib combined with RT) induced greater DNA damage than PUL (pulsed concurrent olaparib combined with RT) treatment (Fig. 1d,e), we sought to define the immunogenic properties of the COM therapeutic strategy.
To determine COM effects on the cGAS-STING pathway, we evaluated the frequency of micronuclei formation, tracked cGAS translocation and detected the phosphorylation of STING across four conditions: (1) vehicle [VEH] control, (2) RT alone, (3) olaparib [OLA] alone, (4) COM treatment. OLA combined with RT significantly increased the fraction of micronucleated SCLC SBC5 cells (0.24 with COM, 0.14 RT, 0.06 OLA, 0.05 VEH, RT vs COM p = 0.00083, one-way ANOVA; Supplementary Fig. 2d, left) that was not apparent in fibroblast non-tumor cell lines, LF1 and Wi-38 (RT vs COM p = NS; Supplementary Fig. 2d, middle and right). We observed COM increased the frequency of cGAS translocation into these micronuclei (0.39 with COM, 0.25 RT, 0.12 OLA, 0.016 VEH; RT vs COM p = 0.00083, one-way ANOVA; Fig. 1f), and resulted in robust STING phosphorylation as compared to OLA or RT monotherapy in SCLC cell lines SBC5 and SW1271 (Fig. 1g). In addition, we observed that the other PARP inhibitors veliparib (VELI) and talazoparib (TALA) combined with RT also increased STING phosphorylation as compared to VELI, TALA or RT monotherapy in SW1271 (Supplementary Fig. 3). We then evaluated the downstream products of the cGAS-STING pathway, specifically mRNA levels of chemokines CCL5 and CXCL10 that mediate the immune response by recruiting leukocytes, including T cells31,32, across cell-lines representing 4 SCLC molecular subtypes33. SBC5 (Y), H82 (N), H526 (P) and H146 (A) cells were treated with VEH, OLA, RT, or COM treatment for 48 h and CCL5 and/or CXCL10 mRNA were quantified by qPCR (Fig. 1h). Compared to VEH control, we observed that the COM treatment induced higher mRNA levels of CCL5 (12.5-fold change) or CXCL10 (4- to 10-fold change) mRNA, than RT or OLA monotherapy across the 4 subtypes of SCLC cell-lines (Fig. 1h). Similarly, we also evaluated the potential effects on immune response by VELI or TALA in SCLC cell lines. Compared to VEH control, we observed that the COM treatment induced higher mRNA levels of CCL5 (4.5 to 5.5-fold change) or CXCL10 (5-fold change) by VELI and higher levels of CCL5 (6.5 to 8.5-fold change) or CXCL10 (13-fold change) by TALA, than RT or PARPi monotherapy across SCLC cell lines (Fig. 1h). We also observed in SBC5 cells, TALA (20 nM) induced higher CXCL10 mRNA levels than OLA (1 μM) or VELI (1.6 μM), indicating TALA to be more potent than OLA or VELI. We confirmed that COM (olaparib+RT) increased CCL5 and CXCL10 mRNA levels in vivo. SBC5 xenografts and SCRX-Lu149 CN PDXs were sacrificed with tumors collected at day 15 where higher levels of CCL5 (13.9-fold increase for SBC5 xenograft and 13.7-fold for SCRX-Lu149 CN PDX) and CXCL10 (11.3-fold for xenograft SBC5 and 15.3-fold for SCRX-Lu149 CN PDX) mRNAs were observed in COM as compared OLA or RT monotherapy (Fig. 1h).
To determine whether the enhanced chemokine transcription from OLA, RT or COM treatment was directly mediated by the cGAS-STING pathway, we knocked-out (KO) cGAS or STING by CRISPR-Cas9 in SBC5 and H1048 SCLC cell lines with 2 different sgRNAs per gene target. After confirming KO by western blot, we quantified changes in CCL5 or CXCL10 mRNA levels (Fig. 1i,j). We observed that cGAS-KO or STING-KO reversed and decreased CCL5 and CXCL10 mRNA levels following OLA, RT or COM treatment as compared to cGAS-wildtype or STING-wildtype sgRNA control (Ctrl) against EGFP. Taken together, we concluded the enhancement of chemokine levels by COM is cGAS-STING dependent.
The combination of PARP inhibition and radiotherapy downregulates the translational repressor EIF4E2
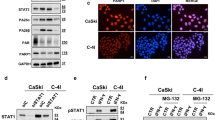
After observing that PARPi combined with RT increased chemokine levels synergistically, we investigated other biological pathways that could enhance immune activation in response to COM treatment. To achieve this, we treated cells with VEH, RT, OLA, or COM, then extracted RNA after 72 h for RNA-seq. Our analysis showed distinct gene expression patterns among these four conditions, with the most significant changes observed in the COM treatment, as illustrated in a heatmap (Fig. 2a). Gene set enrichment analysis (GSEA) with the REACTOME database identified significant pathways that are altered, including apoptosis and cell cycle, which are consistent with previous reports34. Additionally, we identified pathways related to the process of mRNA translation and degradation that were modulated in the COM treatment, but not in the OLA vs. VEH or RT vs. VEH comparisons (Fig. 2b). Among the downregulated genes, we found that members of the EIF4 family of translational suppressors, including EIF4E2, were significantly decreased (Fig. 2c). EIF4E2 (also named 4EHP) is a translational repressor that also participates in suppression of innate immune response, such as type I interferons35,36, and tristetraprolin-target mRNAs, including CXCL1021. To investigate the downregulation of EIF4E2 induced by COM, we confirmed a decrease in EIF4E2 protein levels in 4 SCLC subtypes by western blot. In H1048(P), H446(N), SBC5(Y), and KP1(A) cells, EIF4E2 protein levels were the lowest in the COM-treated cells, at 0.16, 0.11, 0.48, and 0.14 compared to VEH, respectively, after 3–5 days of treatment (Fig. 2d). Furthermore, we validated the downregulation of EIF4E2 in vivo using the genetically engineered mouse model KP1 cell line engrafted into B6129F mice. Western blot analysis showed that EIF4E2 protein levels were significantly diminished in KP1 tumors following COM treatment (0.33 compared to VEH control, n = 5, p = 0.0052; Fig. 2e).
a Heatmap showing hierarchical clustering of significantly changed genes in SBC5 cells treated with vehicle (VEH), olaparib (OLA), RT, or combination of olaparib and RT (COM) for 3 days. b Top 10 pathways significantly modulated by the COM. The p value was calculated by GESA with two-tailed permutation testing. c Fold change of EIF4 family mRNA level treated with OLA, RT, or COM treatment as normalized by the expression to VEH. d Upper: Quantification of the relative intensity of EIF4E2 protein level normalized by the intensity of β-Actin. Lower: Western blot analysis of EIF4E2 expression in H1048 (P), H446 (N), SBC5 (Y) and KP1 (A) cells treated with vehicle, OLA, RT or COM treatment from one of two independent experiments. e Left: Schematic of KP1 allografts experimental design (n = 5 mice for each treatment, Created in BioRender. Ran, X. (2025) https://BioRender.com/e93s595). Middle: Western blot against EIF4E2 and Actin. Right: Quantification of the relative intensity of EIF4E2 protein level normalized by the intensity of β-Actin (VEH vs COM p = 0.0052, OLA vs COM p = 0.001, RT vs COM p = 0.044). Mean counts are presented as a line and the comparison was calculated with one-way ANOVA. (p: 0.001 < ** < 0.01 < * < 0.05). All exact p values are listed in the source data. Source data are provided as a Source Data file.
EIF4E2 represses CXCL10 mRNA stability to decrease CXCL10 protein level and tumor T cell infiltration
After demonstrating the downregulation of EIF4E2 by COM treatment, we measured EIF4E2 and CXCL10 protein levels in SBC5 VEH or COM treated cells from 3 h to 6 days. We observed an inverse correlation (Pearson’s correlation coefficient R = −0.9304, R2 = 0.8656, p = 0.0008, Cohen’s d = 0.8835) between EIF4E2 and CXCL10 protein levels with a decrease of EIF4E2 and an increase of CXCL10 in SBC5 COM treated cells over time (EIF4E2: 0.852 on day 3, 0.717 on day 4, 0.724 on day 5 and 0.544 on day 6, normalized to 0 h; CXCL10: 10.8-fold increase on day 3,44.8-fold on day 4, 48.2-fold on day 5 and 68.1-fold on day 6, normalized to 0 h; Fig. 3a).
a Left: EIF4E2 and CXCL10 protein level in SBC5 with VEH or COM treatment at indicated time points from one of two independent experiments. Right: Quantification of EIF4E2 and CXCL10 protein level normalized by Actin on different days relative to the protein level at 0 h. b Relative CXCL10 mRNA level in EGFP KO or EIF4E2 KO for HEK293 and SBC5 cells (n = 4 biological replicates). c CXCL10 mRNA stability in EGFP KO control (Ctrl) or EIF4E2 KO cells for HEK293 treated with Dactinomycin (DACT) for 0, 2, 4 h (n = 3 biological replicates). d Western blot and quantification of CXCL10 protein levels in Ctrl or EIF4E2 KO for HEK293 or SBC5 cell-lines from one of two independent experiments. e Relative CXCL10 mRNA level in empty vector (Empty) or EIF4E2 overexpression for HEK293 and SBC5 cells (n = 3 biological replicates). f Left: overexpression of FLAG-EIF4E2 in HEK293 EIF4E2 KO cells. Right: CXCL10 mRNA level in HEK293 EIF4E2 KO cells expressing empty vector or FLAG-EIF4E2 after 4 h’ DACT treatment (n = 3 biological replicates). g Western blot and quantification of CXCL10 protein levels in Empty or V5-EIF4E2 overexpression for HEK293 or SBC5 cell-lines from one of two independent experiments. h Left: Schematic of SBC5-Empty or V5-EIF4E2 overexpression tumor engraftment in PBMC humanized NSG mice (Created in BioRender. Ran, X. (2025) https://BioRender.com/e93s595). Middle: Representative flow cytometry data for total T cells. Right: Cumulative data for total T cell (CD3+CD45+) in tumors on day 35 (n = 12). For (a), data are presented as individual dot plot and the correlation was analyzed by two-tailed Pearson correlation test. For (b), (c), (e) and (f), data are presented as mean ± SD and statistically analyzed by two-tailed, unpaired t test. For (h), data are presented as dot plot and statistically analyzed by the two-tailed, unpaired t test. (p: ****<0.0001 < *** < 0.001 < ** < 0.01 < * < 0.05). All exact p values are listed in the source data. Source data are provided as a Source Data file.
Next, we sought to define the mechanism of EIF4E2 in downregulating CXCL10 protein levels and subsequent immune response. Previous studies have reported that EIF4E2 interacts with GIGYF2 and TTP to form a complex that degrades mRNA20,21,37,38. Therefore, we hypothesized that COM-induced downregulation of EIF4E2 stabilizes CXCL10 mRNA, leading to increased CXCL10 protein. To test this hypothesis, we measured CXCL10 mRNA and protein levels with EIF4E2 KO and wildtype Ctrl (EGFP sgRNA control) in HEK293 and SBC5 cell lines. We observed a significant increase in CXCL10 mRNA levels in EIF4E2 KO cells (>1.3-fold to >1.4-fold increase, respectively, each p < 0.001 by two-tailed unpaired t test; Fig. 3b) compared to wildtype Ctrl cells.
To investigate whether the EIF4E2 mediates an increase in CXCL10 mRNA through mRNA stabilization, we conducted a mRNA decay assay with CXCL10 in HEK293 cells. To prevent the production of new mRNA transcripts, we added the transcriptional inhibitor Dactinomycin (DACT), allowing us to examine mRNA degradation. In Ctrl (EGFP sgRNA control) cells, CXCL10 mRNA decayed to 0.40 at 4 h in the presence of actinomycin compared to baseline. In contrast, CXCL10 mRNA degradation was considerably slower in EIF4E2 KO cells, reaching only 0.70 at 4 h, indicating that EIF4E2 mediates rapid CXCL10 mRNA degradation (Fig. 3c). In addition, we confirmed that CXCL10 protein levels in HEK293 and SBC5 cells and EIF4E2 KO cells exhibited a 1.3-fold and 1.4-fold increase in CXCL10 protein levels, respectively, as compared to EIF4E2 wildtype Ctrl cells (Fig. 3d).
To further validate our hypothesis, we overexpressed EIF4E2 in HEK293 and SBC5 cells and measured CXCL10 mRNA level, mRNA stability and protein level, respectively. We found EIF4E2 overexpression led to decreased CXCL10 mRNA levels (decrease to 75% and 57% of Empty Ctrl) both in HEK293 and SBC5 cells, respectively (Fig. 3e). Additionally, we conducted a mRNA decay assay, in which we overexpressed EIF4E2 in HEK293 WT or EIF4E2 KO cells and examined the CXCL10 mRNA stability. We observed in the HEK293 WT cells, the presence of exogenous EIF4E2 led to a slight decrease in CXCL10 mRNA stability (decay to 0.6 in empty vector vs 0.5 in EIF4E2 overexpression, Supplementary Fig. 4a). While in EIF4E2 KO cells, the rescue with EIF4E2 overexpression resulted in a robust decrease of CXCL10 mRNA level (47% in EIF4E2 overexpression vs empty vector, Supplementary Fig. 4b) and greatly decreased mRNA stability significantly (decay to 0.86 in empty vector vs 0.44 in EIF4E2 overexpression, p < 0.05; Fig. 3f). Then we confirmed the CXCL10 protein level in HEK293 and SBC5 cells and EIF4E2 overexpression cells exhibited lower CXCL10 protein levels as compared to wildtype Empty Ctrl cells (decreased to 25% and 35%, respectively; Fig. 3g). Altogether these data provide evidence that EIF4E2 destabilizes CXCL10 mRNA to decrease CXCL10 mRNA and protein levels.
Previous studies have demonstrated that CXCL10 is a key chemokine for tumor T cell infiltration12. To demonstrate whether EIF4E2 regulation of CXCL10 affects T cell tumor infiltration, we engrafted PBMC humanized NSG mice with SBC5 cells expressing empty vector or EIF4E2 and monitored tumor T cell infiltration and tumor growth. SBC5 empty vector controls and EIF4E2 were implanted on opposite flanks of the same mouse to control for potential heterogeneity in T cell clones for each individual mouse. We found in vivo EIF4E2 overexpression led to decreased tumor T cell (CD3+CD45+) infiltration (11.37% in SBC5-Empty vs 5.30% in SBC5-EIF4E2, n = 12, p = 0.0087, two-tailed paired t test, Fig. 3h), demonstrating overexpression of EIF4E2 inhibits tumor T cell infiltration. In addition, EIF4E2 overexpression did not impact cell proliferation in vitro but significantly increased tumor growth in humanized NSG mice (Supplementary Fig. 5a,b). All these data indicated that EIF4E2 decreases tumor T cell infiltration and increases tumor growth in humanized mice. This suggests an excess of EIF4E2 may have pro-tumor effects in vivo in a humanized model system via interactions with the tumor-immune microenvironment. Overall, this supports the translational clinical benefit of EIF4E2 downregulation that PARPi and RT can improve anti-tumor effects partly through suppressing levels of EIF4E2, in addition to the intrinsic cytotoxic effects of PARPi and RT.
EIF4E2 destabilizes CXCL10 mRNA via the AU-rich element of the CXCL10 mRNA 3′UTR region
To interrogate the mechanism responsible for CXCL10 mRNA degradation by EIF4E2, we directly investigated whether the CXCL10 3′UTR region harboring a adenylate-uridylate(AU)-rich region is the mechanistic target for EIF4E2. We transfected a dual-luciferase reporter assay where a Renilla luciferase (Rluc) construct was fused to the CXCL10 3′ UTR into Ctrl (EGFP sgRNA control) or EIF4E2 KO HEK293 cells. The Rluc-CXCL10-3′UTR reporter expression was increased in EIF4E2-KO by 30% relative to Ctrl (p = 0.0002, two-way Anova; Fig. 4aright). With quantitative qPCR, we also found that Rluc mRNA with CXCL10 3′UTR region was more stable in EIF4E2 KO cells (50% increase relative to Ctrl, p < 0.0001, Fig. 4amiddle). In an orthogonal approach, SBC5 SCLC cells were transduced with GFP-CXCL10 3′UTR lentiviral particle and flow-sorted for monoclonal expansion. After confirming stable and consistent GFP levels, the SBC5-GFP-CXCL10-3′UTR monoclonal cell-line was then used to generate LacZ-KO control (Ctrl) and EIF4E2 KO cell lines (Fig. 4bleft). To quantify the stability of the CXCL10-3′UTR construct with and without EIF4E2, GFP signal was measured in these EIF4E2-KO and Ctrl cell lines by flow cytometry. We observed that GFP in EIF4E2-KO cells was increased ~1.3-fold (Fig. 4bright). Altogether, these data from the dual-luciferase (Fig. 4a) and GFP (Fig. 4b) reporter assays demonstrated that EIF4E2 suppresses an early step of CXCL10 expression through targeting the transcribed CXCL10 mRNA for degradation by its 3′UTR region.
a Top: Schematic of the psicheck2-Rluc-CXCL10 3′UTR reporter. Left: Western blot against EIF4E2 in Ctrl or EIF4E2 KO HEK293 cells. Quantification of relative Renilla luciferase mRNA level (middle) and protein level (right) in EGFP KO control (Ctrl) or EIF4E2 KO HEK293 cells transfected with psicheck2 or psicheck2-CXCL10 3′UTR plasmids (n = 3 biological replicates for mRNA detection and n = 4 biological replicates for protein detection). b Top: Schematic of the GFP-CXCL10 3′UTR reporter. Left: Western blot against EIF4E2 in LacZ KO control (Ctrl) or EIF4E2 KO for SBC5. Right: Quantification of GFP intensity in Ctrl or EIF4E2 KO for SBC5 expressing GFP-CXCL10 3′UTR (n = 4 biological replicates). c Top: Schematic of the psicheck2-Rluc-CXCL10 3′UTR WT and 3′UTR △ARE reporter. Bottom: Quantification of relative Renilla luciferase mRNA stability in Ctrl or EIF4E2 KO HEK293 cells transfected with psicheck2-Rluc-CXCL10 3′UTR WT or △ARE reporter plasmid for 24 h and then treated with Dactinomycin (DACT) for 4 h (n = 3 biological replicates). d Top: Schematic of the CXCL10-ORF + 3′UTR WT or 3′UTR △ARE plasmid and the mRNA produced (Created in BioRender. Ran, X. (2025) https://BioRender.com/v38l510). (Left) Relative level of CXCL10 genomic DNA (n = 4 biological replicates), (Middle) CXCL10 mRNA stability (n = 3 biological replicates), and (Right) western blot and quantification of CXCL10 protein levels (from one of two independent experiments) in SBC5 expressing CXCL10-ORF + 3′UTR WT or 3′UTR △ARE e Left: Schematic of SBC5-3′UTR WT or SBC5-3′UTR △ARE tumor engraftment in PBMC humanized NSG mice (Created in BioRender. Ran, X. (2025) https://BioRender.com/e93s595). Middle: Representative flow cytometry data for total T cells. Right: Cumulative data for total T cell (CD3+CD45+) in tumors (n = 6) on day 35. For (a–d), data are presented as mean ± SD. For (a) data are analyzed by two-way ANOVA. For (b–d), data are analyzed by two-tailed, unpaired t test with an exception for (e) by two-tailed, paired t test. (p: **** < 0.0001 < *** < 0.001 < ** < 0.01 < * < 0.05). All exact p values are listed in the source data. Source data are provided as a Source Data file.
Upon demonstrating CXCL10-3′UTR is important for EIF4E2 induced CXCL10 mRNA destabilization, we further investigated whether the AU-rich element (ARE) is essential for EIF4E2-induced mRNA destabilization. We transfected Rluc-CXCL10-3′UTR WT or △ARE reporter plasmids into HEK293 Ctrl or EIF4E2 KO cells and measured Rluc mRNA stability by an mRNA decay assay. We found in the Ctrl cells, ARE deletion significantly increased Rluc mRNA stability (decay to 0.6 in 3′UTR WT vs 0.85 in 3′UTR △ARE, p < 0.01; Fig. 4c). In contrast, in the EIF4E2 KO cells, ARE deletion did not significantly increase Rluc mRNA stability (decay to 0.87 in 3′UTR WT vs 0.8 in 3′UTR △ARE, ns; Fig. 4c). In an orthogonal approach, we generated SBC5 cells, which express CXCL10-ORF + 3′UTR WT or △ARE mRNA under the control of a CMV promotor. We found that both cell lines contained similar CXCL10 genomic DNA levels (Fig. 4dleft) but 3′ UTR △ARE increased CXCL10 mRNA stability (decay to 0.64 in 3′UTR WT vs 0.78 in 3′UTR △ARE, p < 0.01; Fig. 4dmiddle) and protein level (1.3-fold increase; Fig. 4dright). In summary, these data demonstrated that the ARE region of the CXCL10-3′UTR is mechanistically essential for EIF4E2-induced CXCL10 mRNA destabilization.
To investigate whether ARE deletion regulates T cell tumor infiltration and tumor growth, we engrafted PBMC humanized NSG mice with SBC5 expressing CXCL10-ORF + 3′UTR WT or 3′UTR △ARE on either side of the same mouse and measured tumor T cell infiltration and tumor growth. We found SBC5 3′UTR △ARE cells exhibited an increase of T cell tumor infiltration in comparison to SBC5 3′UTR WT cells (16% in 3′UTR WT vs 35% in 3′UTR △ARE), indicating that loss of ARE increases the stability of CXCL10 mRNA and protein levels to enhance tumor T cell infiltration (Fig. 4e). In addition, 3′UTR △ARE did not exhibit any effects on cancer cell proliferation in vitro or in vivo (Supplementary Fig. 6a,b). Altogether, these data lead us to conclude that the CXCL10-3′UTR-ARE region is the mechanistic target of EIF4E2 that regulates CXCL10 mRNA levels and subsequent tumor T-cell infiltration mediated by CXCL10.
Olaparib combined with RT increases T-cell tumor infiltration
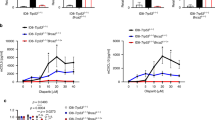
After establishing COM treatment increased the CXCL10 mRNA levels via cGAS-STING (Fig. 1) and induced loss of EIF4E2 prevented CXCL10 mRNA degradation leading to greater CXCL10 protein levels (Figs. 2–4), we sought to investigate the implications in vivo (schematic shown in Fig. 5a). We interrogated COM treatment on T cell infiltration with an immune-component syngeneic murine model of SCLC. We observed that COM treatment directly enhanced antitumor efficacy in immune-competent B6129F mice bearing KP1 tumors. Compared with RT alone, COM treatment showed tumor growth inhibition on day 12 (68%, p < 0.001, COM vs VEH; 67%, p < 0.001, COM vs OLA; 39%, p < 0.01, COM vs RT) demonstrating PARPi radiosensitization in this syngeneic KP1 murine model (Fig. 5b).
a Schematic of experimental design with KP1 allografts (Created in BioRender. Ran, X. (2025) https://BioRender.com/e93s595). b Tumor growth curve and Kaplan Meier analysis of KP1 allograft (n = 6 mice for each treatment) treated with olaparib (OLA), RT, or combination (COM) therapy. OLA or vehicle (VEH) was delivered by oral gavage 5 days per week and 8 Gy of RT was delivered on day 2. c Left: Representative of IHC staining for total T cells (CD3+) in KP1 tumors upon treatment (n = 3 mice for each treatment, scale bar 250 μm). Right: The quantification of CD3+ cells in KP1 tumors. d Schematic of gating strategy for immune profiling. e Left: Representative flow cytometry data for total T cells. Right: Cumulative data for total T cell (CD3+CD45+, n = 6). f–j Cumulative data for CD4+T cell (CD3+CD45+CD4+, n = 6), CD8+ T cell (CD3+CD45+CD8+, n = 6), memory CD8+ T cell (CD3+CD45+CD8+CD44highCD62Llow, n = 6), exhausted T cell (CD3+CD45+TIM-3+PD-1+, n = 5) and exhausted CD8+ T cell (CD3+CD45+CD8+TIM-3+PD-1+, n = 5). k Cumulative data for PD-L1+ cells from KP1 tumors (n = 5). For (b), tumor growth curve data are presented as mean ± SD and statistically analyzed by two-way ANOVA. Kaplan-Meier data are analyzed by log-rank (Mantel-Cox) test. For (c), data are presented as floating bars (min to max) with line at median and statistically analyzed with one-way ANOVA. For (e–k), data are presented as a mean of replicates and statistically analyzed with one-way ANOVA. (p: ****<0.0001 < *** < 0.001 < ** < 0.01 < * < 0.05). All exact p values are listed in the source data. Source data are provided as a Source Data file.
To determine COM treatment effects on T cell infiltration, tumors were harvested and disassociated on day 12 following treatment for IHC staining and flow cytometry (flow gating strategy shown in Fig. 5d and representative plots shown in Fig. 5eleft). By IHC staining, we observed a significant increase of CD3+ T cell infiltration in KP1 tumors by COM treatment, in comparison to RT monotherapy (RT 0.39% vs COM 5.5%, p < 0.05; Fig. 5c). With immune profiling we found that COM treatment had the greatest significant increase in tumor infiltrated CD3+CD45+ T cells (3.98% of total live cells, Fig. 5eright), whereas minimal differences of tumor infiltrated T cells were observed between VEH control (0.59%), OLA monotherapy (1.30%) and RT monotherapy (0.47%). Similarly, when examining the T cell populations for CD4+ and CD8+, COM induced a significant decrease in CD4+ T cell (35.8%) and increase of CD8+ T cell (45.8%) infiltration (Fig. 5f,g, respectively) with modest differences between VEH control (57.8% CD4+, 19.1% CD8+), OLA monotherapy (56.2% CD4+, 17.5% CD8+), RT monotherapy (64.0% CD4+, 16.9% CD8+). We further distinguished memory T cells (CD62L-low, CD44-high) in these CD8+ T cells and observed the greatest proportion of memory CD8+ cells (% of memory CD8+ T cells in total live cells) with COM treatment (1.92%) compared to VEH, OLA, RT conditions (0.04–0.28%; Fig. 5h). While COM treatment recruited T cells into tumor, we observed that most exhibited exhaustion cell surface markers (TIM-3+, PD-1+). The greatest exhausted T cell population (Fig. 5i) was observed in the COM group (62.66% of total T cells) as compared to RT (29.2%), OLA (44.4%) or VEH (36.6%) comprised primarily of exhausted CD8+ T cells (Fig. 5j) most evident in COM (89.9%) when compared to RT (53.3%), OLA (69.4%), and VEH (48.6%).
As T cell exhaustion could be mediated by the PD-1/PD-L1 axis39, we sought to determine whether COM treatment associated with exhausted T cell population was due to upregulated PD-L1. Through in vitro experiments, we quantified COM treatment effect on CD274 (PD-L1) mRNA levels by qPCR and PDL1 cell surface protein levels by flow cytometry. We found PD-L1 gene expression was most prominently upregulated in COM treatment (Supplementary Fig. 7a). Consistent with this, we found a higher proportion of cells with detectable PD-L1 protein following COM treatment (14.0% SBC5; 23.6% KP1) compared to RT (8.9% SBC5; 12.9% KP1), OLA (7.3% SBC5; 11.6% KP1), or VEH control (5.5% SBC5; 4.5% KP1; Supplementary Fig. 7b). Following these in vitro findings, we sought to confirm COM treatment affecting PD-L1 upregulation in vivo. We found COM significantly increased PD-L1 protein levels (25.8%) compared to RT (1.6%), OLA (8.4%), or VEH control (6.0%; Fig. 5k). To investigate the mechanism of PD-L1 upregulation, we stimulated SBC5 cells with a cGAS activation product, cGAMP and measured the PD-L1 mRNA level. We found cGAS-STING activation upregulates PD-L1 mRNA level (Supplementary Fig. 7c). On the other hand, we also detected PD-L1 mRNA and protein level in SBC5 Ctrl or EIF4E2 KO cells and we found EIF4E2 KO downregulates PD-L1 mRNA and protein level but not affect PD-L1 mRNA stability (Supplementary Fig. 7d,e).
Taken together, COM treatment effectively increased tumor infiltration of CD8+ T cells, however, most exhibited an exhaustion phenotype that was concomitant with increased PD-L1 levels. Therefore, we sought to investigate if the incorporation of anti-PDL1 immunotherapy would enhance COM-induced immunogenic effects to improve antitumor IO response against SCLC.
Anti-PD-L1 immunotherapy with PARP inhibitor and RT improves anti-tumor efficacy
To investigate COM treatment (OLA + RT) with anti-PD-L1, we treated B6129F mice bearing KP1 tumors with olaparib (50 mg/kg, 5 of 7 days per week), 8 Gy RT (day 2), and anti-mouse PD-L1 antibody (10 mg/kg, twice weekly; Fig. 6a). Anti-PD-L1 therapy with COM treatment compared to COM alone led to significant tumor growth inhibition at day 12, 15 and 19 (60%, p < 0.0001 on day 12, 64%, p < 0.0001, on day 15 and 63%, p < 0.0001, on day 19, respectively; Fig. 6bleft) and freedom from 1000 mm3 volumetric endpoint (median time to progression 17 vs. 24 days; p < 0.01; Fig. 6bright).
a Schematic of experimental design with KP1 allografts to olaparib, RT, and anti-PD-L1 antibody (ab) combination treatment (Created in BioRender. Ran, X. (2025) https://BioRender.com/e93s595). b Tumor growth curve and Kaplan Meier plot of KP1 allografts treated with VEH+IgG (n = 6), VEH+anti-PD-L1 ab (n = 6), COM (OLA + RT)+IgG (n = 6) and COM (OLA + RT)+anti-PD-L1 ab (n = 8). COM is olaparib 50 mg/kg day 1-5 per week plus RT 8 Gy, day 2. IgG or anti-PD-L1 ab 10 mg/kg was delivered by IP injection twice a week. c–e The cumulative data for immune profiling of KP1 tumor (n = 6) treated with VEH+IgG, VEH+a-PD-L1 ab, COM (OLA + RT)+IgG or COM (OLA + RT)+a-PD-L1 ab, including total T cell (CD3+CD45+), CD4+T cell (CD3+CD45+CD4+), and CD8+ T cell (CD3+CD45+CD8+). f Schematic of experimental design with KP1 allografts to TALA, RT, TALA + RT, and TALA + RT+anti-PD-L1 ab combination treatment (Created in BioRender. Ran, X. (2025) https://BioRender.com/e93s595). g Tumor growth curve and Kaplan Meier plot of KP1 allografts treated with TALA (n = 6), RT (n = 6), TALA + RT (n = 6) and TALA + RT+anti-PD-L1 ab (n = 6). TALA 0.2 mg/kg was delivered day 1–5 per week plus RT 8 Gy, day 2. Anti-PD-L1 ab 10 mg/kg was delivered by IP injection twice a week. h–j The cumulative data for immune profiling of KP1 tumor (n = 5) treated with TALA, RT, TALA + RT or TALA + RT+a-PD-L1 ab, including total T cell (CD3+CD45+), CD4+T cell (CD3+CD45+CD4+), and CD8+ T cell (CD3+CD45+CD8+). For (b) and (g), tumor growth curve data are as mean ± SD and statistically analyzed by two-way ANOVA. Kaplan-Meier data are analyzed by log-rank (Mantel-Cox) test. For (c–e) and (h–j), data are presented as mean of n = 6 tumors and statistically analyzed with one-way ANOVA. (ns, no significance; p: ****<0.0001 < *** < 0.001 < ** < 0.01 < * < 0.05). All exact p values are listed in the source data. Source data are provided as a Source Data file.
The flow analysis conducted on the tumor microenvironment after treatment with COM+anti-PD-L1 antibody showed a significant decrease in exhausted total T cells (39.8% in COM+IgG vs 21.1% in COM+anti-PD-L1 ab) and exhausted CD8+ T cells. Anti-PD-L1 with COM treatment reversed by ~2-fold the exhausted total T cell and CD8+ T cell population (21.1% for exhausted total T cell and 37.8% for exhausted CD8+ T cells in COM+anti-PD-L1 ab treatment) that COM alone (39.8% for exhausted total T cell and 68.5% for exhausted CD8+ T cells in COM+IgG treatment) induced, indicating that the anti-PD-L1 antibody prevented T cell exhaustion (Supplementary Fig. 8a,b). Increased total T cells infiltrated the tumor in the COM+anti-PD-L1 antibody treatment group as compared to the COM+IgG treatment group (Fig. 6c). There was no significant difference in proportions of CD4+T cell and CD8+ T cells in the COM+anti-PD-L1 antibody treatment group in comparison to the COM+IgG treatment group (Fig. 6d, e).
In addition to olaparib, we also investigated the anti-tumor efficacy of combination of talazoparib (TALA) + RT+anti-PD-L1 antibody. We treated B6129F mice bearing KP1 tumors with TALA (0.2 mg/kg, 5 of 7 days per week), 8 Gy RT (day 2), and anti-mouse PD-L1 antibody (10 mg/kg, twice weekly; Fig. 6f). Anti-PD-L1 therapy with COM treatment compared to COM alone led to significant tumor growth inhibition at day 8, 10, 12, 14 and 16 (67.3%, p < 0.01 on day 12; 72.4%, p < 0.0001 on day 14; 71.6%, p < 0.0001 on day 16; Fig. 6gleft) and freedom from 1000 mm3 volumetric endpoint (median time to progression 21 vs. 31 days; p = 0.0013,**; Fig. 6gright).
The flow analysis conducted on the tumor microenvironment after treatment with TALA + RT+anti-PD-L1 antibody showed a significant increase of total T cell tumor infiltration in the TALA + RT+anti-PD-L1 antibody treatment group as compared to the TALA + RT treatment group (Fig. 6h). There was a significant decrease in the proportion of CD4+T cell and an increase in the proportion of CD8+ T cells in the TALA + RT+anti-PD-L1 antibody treatment group in comparison to the TALA + RT treatment group (Fig. 6i, j).
These findings provide evidence that the addition of anti-PD-L1 antibody to COM treatment with various PARP inhibitors enhanced T cell infiltration, ultimately leading to improved immune-mediated anti-tumor efficacy.
Discussion
In this study, we demonstrated the greatly enhanced radiosensitizing and immunogenic effect with continuous PARPi and RT. We found the combination treatment (PARPi and RT) increased DNA damage, activated the cGAS-STING pathway, and upregulated CCL5 and CXCL10 mRNA levels across all SCLC molecular subtypes. By whole transcriptome sequencing, we identified the translational repressor EIF4E2 as one of the top-ranking genes that significantly decreased following combination treatment. Mechanistically, the downregulation of EIF4E2 after combination treatment increased the T cell attractant chemokine CXCL10 protein levels via stabilization of CXCL10 mRNA that is dependent on the AU-rich element in the 3′UTR region of CXCL10 mRNA. In vivo immune profiling of syngeneic SCLC mouse models revealed the combination treatment significantly increased CD8+ T cell infiltration, created an inflamed tumor microenvironment, and augmented the anti-tumor efficacy of anti-PD-L1 immunotherapy (Fig. 7).
In comparison to RT treatment, PARPi olaparib combined with RT leads to increased DNA damage and subsequent chemokine CXCL10 mRNA increase via cGAS-STING pathway. The combination treatment triggers EIF4E2 downregulation to increase CXCL10 protein levels via stabilizing CXCL10 mRNA. The upregulated CXCL10 level recruits more T cells to the tumor sites and reshapes the tumor microenvironment (Created in BioRender. Ran, X. (2025) https://BioRender.com/k72k169).
Previous studies have shown that DNA targeting therapeutics, including RT or PARPi, induce DNA damage and activate chemokine mRNA expression via the cGAS-STING pathway in various cancers, including breast cancer40,41, melanoma14, ovarian cancer42, NSCLC43 and SCLC44,45,46,47. However, the understanding of whether and how DNA targeting therapeutics affect chemokine mRNA stability is limited. To our knowledge, our study demonstrated for the first time a mechanism for how DNA targeting therapeutics can enhance the immune response by regulating chemokine mRNA stability. Beyond cGAS-STING mediated chemokine transcriptional activation, we identified a mechanism of chemokine regulation where a combination PARPi and RT significantly downregulated EIF4E2 to stabilize CXCL10 mRNA and increase CXCL10 protein levels to enhance T cell tumor infiltration. Furthermore, we determined the AU-rich element in 3′UTR region of CXCL10 is the specific regulatory region for the EIF4E2 mechanism of action in mRNA stability. These findings demonstrate that mRNA stability regulation can play an important role in immune activation induced by DNA-directed therapeutics.
There are some limitations in our study. Although we demonstrated the downregulation of EIF4E2 across four subtypes of SCLC in vitro and GEMM model KP1 (A subtype) in vivo, whether EIF4E2 is downregulated in vivo has not been directly examined for the other subtypes (N, P, Y, inflamed subtypes)33,48. Additionally, while we were able to leverage an allograft syngeneic model for our in vivo studies, we were not able to examine spontaneous primary syngeneic models. Nonetheless, we presented complementary in vivo and in vitro data to support our interpretation that combination PARPi and RT treatment downregulated EIF4E2 to further enhance CXCL10 chemokine protein levels.
In addition to cGAS-STING mediated chemokine transcription activation, our results demonstrated that PARPi and RT combination treatment decreased EIF4E2 to increase T cell attractant chemokine levels and enhanced immunotherapy efficacy in preclinical models of SCLC. Beyond PARPi and RT as a combinatorial strategy24,49,50, EIF4E2 itself is a potential therapeutic target to enhance immunotherapy in SCLC and warrants further investigation. Future directions could be directed toward deciphering the upstream regulation of EIF4E2 in the context of DNA-directed therapeutics and its potential as an immunogenic enhancer. In conclusion, this study identified the combination of PARPi and RT in SCLC as radiosensitizing and immunogenic through a regulatory mechanism of chemokine mRNA stability with therapeutic implications.
Methods
Cell lines and reagents
Human SCLC cell lines were purchased from the American Type Culture Collection (ATCC; H446, H82, H526, H1048, SW1271). SBC5 cell line was gifted from Dr. Kazuhiro Yasufuku lab (University Health Network). SBC5, H446, H82, and H526 cell lines were maintained with RPMI-1640 medium supplemented with 10% FBS. H1048 cells were cultured with HITES medium supplemented with 5% FBS. SW1271 cells were maintained with Leibovitz’s L-15 medium supplemented with 10% FBS. Murine SCLC p53−/− Rb1−/−p130−/− (RPP) cell line KP1 was gifted from Dr. Julien Sage and cultured with RPMI1640 medium supplemented with 10% BGS51. (The source of cells is listed in the Supplementary Table 1)
Olaparib, veliparib and talazoparib were diluted in DMSO and kept at −20 °C. For in vivo dosing, the vehicle was 10% DMSO in PBS with 10% 2-hydroxy-propylb-cyclodextrin, and olaparib or talazoparib were diluted in PBS with 10% 2-hydroxy-propylb-cyclodextrin. Anti-murine PD-L1 antibody (Clone80, mIgG 1 D265A, AstraZeneca) and IgG (Bio X Cell) were diluted in PBS52.
Short-term cell viability assay
Cell lines were seeded into 96-well plates 24 h prior to irradiation and treated with olaparib or DMSO 2 h prior to irradiation. Cells were exposed to the indicated dose of radiation (Gamma ray) with irradiator Gammacell 40. After 7 days, cell proliferation was assessed with Alamar Blue or Cell Titer-Glo in a microplate reader (Synergy neo2 multi-mode reader, Biotek).
Clonogenic assay
Cell lines were seeded into six-well plates 24 h prior to irradiation and treated with olaparib or DMSO 2 h prior to irradiation. Cells were exposed to the indicated dose of radiation (Gamma ray) with irradiator Gammacell 40. After 14–21 days, colonies of at least 50 cells were fixed, stained with crystal violet, and counted.
Immunofluorescence
Cells were seeded into 6-well plates 24 h prior to irradiation and treated with olaparib 2 h prior to irradiation. At selected timepoints after irradiation, cells were washed with PBS twice and fixed with 4% paraformaldehyde for 10 min. After the fixation, cells were permeabilized with 0.5% Triton-100 in PBS for 2 min and blocked with 5% BSA for 30 min. Following that, cells were blotted with primary antibody (anti-γH2AX, Cell signaling technology 9718S; anti-cGAS, Cell signaling technology 15102S) for 1 h and anti-rabbit secondary antibody conjugated with Fluorescence Alex 488 for 30 min. Nuclear DNA was stained with DAPI for 10 min. Images were captured by Leica SP8 confocal microscope and the number of γH2AX or cGAS+ micronuclei and primary nuclei were quantified with ImageJ. (The source of antibodies is listed in Supplementary Table 4).
Generation of CRISPR knockout cell lines
The design of sgRNA targeting cGAS, STING, or EIF4E2 was performed using public software CRISPICK or CRISPOR (primers are listed in Supplementary Table 2). Guide pairs were synthesized, annealed, and inserted into Crispr V2 plasmid backbone and then validated by sequencing53,54. The HEK293T cells were transfected with constructed sgRNA plasmids and packaging plasmids and the supernatant containing lentiviral particles was collected after 48 h. SBC5, H1048, and HEK293 cells were transduced with the lentiviral particles and underwent puromycin, or G418 selection.
Plasmid construction
pFLAG-EIF4E2 and pV5-EIF4E2 were generated by Dr Nahum Sonenberg lab22. pCXCL10-ORF + 3′UTR WT and pCXCL10-ORF + 3′UTR △ARE fragments were amplified from plasmids and inserted into pLenti-CMV/TO Egfp puro plasmid backbone between Sal I and BamH I digestion sites55.
Western blot
Cell lysates were separated on 10–15% SDS–polyacrylamide gels with Laemmli buffer and Tricine buffer system. The proteins were then transferred onto the nitrocellulose membrane or PVDF membrane at 250 mA for 2 h or 100 mA for 8–12 h at 4 °C. The membrane was incubated with blocking buffer (5% BSA or milk in TBST) for 1 h at room temperature followed by incubation with respective antibodies (antibodies are listed in Supplementary Table 4) at 4 °C overnight. After washing with TBST three times, the membrane was incubated with IRDye goat anti-rabbit 800CW and goat anti-mouse 680RD secondary antibodies (LI-COR Biosciences) or HRP-conjugated anti-rabbit or mouse antibody. Imaging was performed by Odyssey Clx infrared imaging system (LI-COR) or Chemidoc system (BIO-RAD) or iBright FL1500 and bands were quantified and normalized using ImageJ.
RNA isolation and reverse transcription and real-time quantitative PCR (RT-qPCR)
The total RNA was isolated using the High Pure RNA Isolation Kit (Roche Life Science, cat#11828665001) according to the manufacturer’s instructions. RNA concentration was measured by NanoDrop 2000 UV–Vis spectrophotometer (Thermo Scientific).
Reverse transcription reactions were carried out using SuperScript III First-Strand Synthesis SuperMix (Invitrogen, cat# 18080-400) according to the manufacturer’s instructions.
The obtained cDNA was diluted with H2O and used for qPCR. Gene-specific primers were used for PCR amplification and detection (primers listed in Supplementary Table 2). The relative gene expression was normalized by the level of human GAPDH or murine Gapdh and calculated with RQ = 2−△△Ct.
Total RNA sequencing
Total RNA-seq library was prepared from 1 μg RNA with Illumina TruSeq stranded total RNA library kit (catalog 20020596) according to manufacturer’s protocol. Library quality and yield were determined by Agilent TapeStation and Qubit fluorometer, respectively. The library was sequenced on Illumina Hiseq X-10 as 2 × 150 bp pair-end reads for around 100 million reads per sample.
Dual-luciferase assay
Dual-luciferase reporters from the Sonenberg lab were generated from the psicheck v2 plasmid, in which a firefly luciferase (Fluc) gene is regulated by an HSV-TK promotor and a renillna luciferase (Rluc) gene is regulated by a T7 promotor. In the psicheck-CXCL10 3′UTR plasmid (Rluc-CXCL10 3′UTR), the CXCL10 3′UTR region was inserted between XhoI and NotI digestion sites following the Rluc gene. Rluc-CXCL10 3′UTR △ARE plasmid was generated based on the psicheck-CXCL10 3′UTR plasmid by deleting AU rich element (ATTTA). Cells were seeded in six-well plate and transfected with 1 μg of psicheck v2-empty plasmid or psicheck-CXCL10 3′UTR plasmid. After 24 h, cells were washed with PBS followed by the addition of 300 μL of lysis buffer to each well for 40 min. Rluc signal was detected with a microplate reader (Synergy neo2 multi-mode reader, Biotek) and normalized by Fluc. The expression level was calculated for the psicheck2-Rluc-CXCL10 3′UTR normalized by the psicheck2 reporter level for each condition.
While the total RNA was isolated from the similarly treated cells (cells in 10 cm dish and transfected with 5 μg plasmids) and Renilla luciferase mRNA level was normalized by the firefly luciferase (F) mRNA level. The relative Rluc mRNA level was calculated for the psicheck2-Rluc-CXCL10 3′UTR normalized by the psicheck2 reporter level for each condition.
Patient-derived and cell-line xenograft in vivo studies
Animal studies were conducted following the approved protocols by the Animal Care Committee at the Princess Margaret Cancer Centre (Toronto, ON, Canada). The animal holding room is specific-pathogen free and it is maintained at 21–22 °C with relative humidity between 40% and 60% and a 12–12 h light and dark photoperiod. The experimental and control mice were co-housed. SCLC PDX models (SCRX-Lu149 CN and SCRX-Lu149 CR) generously provided by Dr. Charles Rudin at Memorial Sloan Kettering Cancer Center (MSKCC; New York, NY, USA), were used56. Chemoresistant (CR) models were previously established in the Rudin laboratory, from a chemo-naïve (CN) SCRX-Lu149 PDX model as described, where SCRX-Lu149 CN PDXs were exposed to repeated cycles of chemotherapy to select PDX tumor cells56. SBC5 xenograft, SCRX-Lu149 CN and CR PDX models were engrafted as previously described using collagenase type IV. 1 million tumor cells were mixed with matrigel and engrafted into the right flank of 6–8 week old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; Jackson Laboratory; JAX005557) mice (male/female)57. Tumors were measured using digital caliper and tumor volumes were calculated using the (lw2)/2, where l is length and w is width of the tumor. Animal’s weight and tumor volumes were measured twice a week. After engraftment and tumor formation of about 100–150 mm3, mice were randomized into their respective treatment groups: vehicle (VEH), olaparib (OLA), radiation (RT), pulse olaparib and radiation (PUL), and combination of radiation and continuous olaparib (COM). Control mice were treated with the vehicle (10% 2-hydroxypropyl-β-cyclodextrin in PBS) for 90 days or until the tumor reached the volumetric end point (1000 mm3). Mice were humanely euthanized with CO2 when tumors reach the endpoint. Olaparib dissolved in the vehicle (50 mg/kg) was administered orally from Monday to Friday for 90 days or until the tumor reached the volumetric endpoint (1000 mm3). For RT, mice were anesthetized using 2-5% isoflurane and the tumor was irradiated once daily with 2 Gy per fraction of radiation using XRAD320 X-ray irradiation (Precision X-Ray). The mice were restrained in a custom-built lead shield and tumors were irradiated from day 2 to 5 after randomization. For PUL and COM arms, olaparib was administered 3 h prior to RT. Mice in PUL and COM groups were treated with olaparib for 5 days and received 8 Gy over 4 daily fractions, and the mice in the COM arm also received adjuvant olaparib for a total of 90 days of olaparib treatment or until volumetric endpoint (1000 mm3).
To generate PBMC humanized mice, 8–10-week-old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; Jackson Laboratory; JAX005557) mice were purchased from Jackson Laboratory and given an initial 2 Gy whole-body radiation dose (GC40). 3 × 106 density-gradient isolated PBMCs from a healthy donor were injected intravenously via tail vein 1 h after irradiation. On the second day, 2.5 × 106 SCLC cells were engrafted as previously described58. Tumors were measured twice a week with a digital caliper and tumor volumes were calculated using the (lw2)/2.
Syngeneic in vivo studies
The experiments were performed in female B6129F/Tac (Taconic Bioscience) mice that were 6–8 weeks old at time with allograft KP1 cells right flank implantation. Tumor volumes were calculated from digital caliper measurements using the formula volume = (lw2)/2. In most experiments, mice were randomized at a tumor volume of ~100–150 mm3, except for the experiment to measure immune profiling of COM+anti-PD-L1 antibody treatment, in which mice were randomized into treatment groups at a tumor volume of 200–250 mm3. Olaparib or talazoparib was administrated daily from Monday to Friday every week. The mice were anesthetized with isoflurane and tumors were irradiated in 8 Gy*1 fractions with an X-ray irradiator (XRAD 320) on day 2 after randomization. Anti-murine PD-L1 antibody (clone 80, D265A; AstraZeneca) was administrated twice a week with IP injection. Mouse weights and tumor volumes were measured twice weekly until mouse euthanasia at a tumor size of 1000 to 1500 mm3.
Immune profiling
For single-cell suspensions, tumors were excised and divided into 1 × 1 mm pieces. The tumor homogenate was incubated with a digestion medium at 37 °C for 30 min and mechanically passed through a strainer. The suspension was added into Ficoll and subjected to centrifuge at 930 × g for 15 min. The cell suspension in the layer of Ficoll was moved to a new tube and treated with red blood cell (RBC) lysis buffer for 10 min. The cells were washed with PBS and resuspended for further analysis.
For cell staining, 1 × 106 cells from single cell suspensions were blocked with BD anti-mouse Fc block (BD 553142) for 15 min on ice. After centrifuge, supernatant was removed and cells were stained with primary antibody conjugated with fluorescence as well as viability dye according to the standard protocol at 4 °C, avoiding light (All antibodies are listed in the Supplementary Table 3). Cells were washed with staining buffer twice and fixed with BD fixation buffer at 4 °C for 10 min without light. The fixation buffer was removed, and cells were resuspended in staining buffer for flow cytometry (LSRII and Symphony A3, BD) analysis with the resultant data analyzed by FlowJo v10. The gating strategy was shown in Supplementary Fig. 9.
Immunohistochemical (IHC) staining
Mouse tissues were fixed in formalin for 24 h and rehydrated in 70% ethanol for 48 h. Mouse tissues were embedded in paraffin and sectioned into 4 μm slices. Sections were deparaffinized and rehydrated in a series of decreasing ethanol solutions. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 15 min at room temperature. Antigen retrieval was completed using a Na Citrate (pH6) solution for CD3 slides. Nonspecific binding was reduced by a 10-min incubation with an animal-free blocker (Vector SP-5030). Sections were incubated for 45 min at room temperature with anti-CD3 antibody (Rabbit host; 1:400; Abcam; Cat No. ab5690). Sections were then incubated for 35 min at room temperature with HRP conjugated secondary antibody (Vector; Cat No. MP-7401). Slides were visualized using a 3,3′-diaminobenzidine solution (VECTSK4100) and counterstained with CAT hematoxylin (BioCare). Finally, slides were scanned at a 20X magnification using the Hamamatsu Photonics NanoZoomer 2.0-HT.
Digital imaging scan and IHC quantification
Scanned IHC images were quantified using the HALO software (Indica Labs). Individual cell data was reported for each slide with quantification presented as a percentage of CD3 positive over total number of cells detected (CD3+cells/Total cells).
Statistical analysis
DMF37 analysis
Dose modification factor 37 was modeled by the linear quadratic survival function (Prism). The dose of radiation for 37% of survival was calculated using the following equation: Y = Y0*exp(−1*(A*X + B*X^2)). Y is the percent of alive cells and X is the dose of radiation.
Tumor growth inhibition analysis
Tumor growth inhibition was calculated by comparing the average tumor volume (TV) of the treatment group with that of the control group at the timepoint.
Kaplan Meier analysis
The time to reach a volumetric endpoint of 1000 mm3 was defined as the event. Log-rank tests were used to evaluate differences between groups.
All the statistical analyses were done with two-tailed unpaired t test, one-way ANOVA, or two-way ANOVA analysis by Graphpad Prism 9.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-sequencing data generated in this study are available in NIH Gene Expression Omnibus under accession number GSE233820. Source data are provided in this paper.
References
Rudin, C. M., Brambilla, E., Faivre-Finn, C. & Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Prim. 7, 3 (2021).
Huang, A. C. & Zappasodi, R. A decade of checkpoint blockade immunotherapy in melanoma: understanding the molecular basis for immune sensitivity and resistance. Nat. Immunol. 23, 660–670 (2022).
Galvis, M. M. et al. Immunotherapy improves efficacy and safety of patients with HPV positive and negative head and neck cancer: a systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 150, 102966 (2020).
Xu, Y. et al. Programmed death-1/programmed death-ligand 1-axis blockade in recurrent or metastatic head and neck squamous cell carcinoma stratified by human papillomavirus status: a systematic review and meta-analysis. Front. Immunol. 12, 645170 (2021).
Gandhi, L. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018).
André, T. et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 383, 2207–2218 (2020).
Wei, S. C., Duffy, C. R. & Allison, J. P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018).
Horn, L. et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N. Engl. J. Med. 379, 2220–2229 (2018).
Paz-Ares, L. et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 394, 1929–1939 (2019).
Bai, R. et al. Mechanisms of cancer resistance to immunotherapy. Front. Oncol. 10, 1290 (2020).
Liu, Y.-T. & Sun, Z.-J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics 11, 5365–5386 (2021).
Maimela, N. R., Liu, S. & Zhang, Y. Fates of CD8+ T cells in tumor microenvironment. Comput. Struct. Biotechnol. J. 17, 1–13 (2019).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Harlin, H. et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 69, 3077–3085 (2009).
Ivanov, P. & Anderson, P. Post-transcriptional regulatory networks in immunity. Immunol. Rev. 253, 253–272 (2013).
Fabian, M. R., Sonenberg, N. & Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 (2010).
von Roretz, C., Di Marco, S., Mazroui, R. & Gallouzi, I.-E. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip. Rev. RNA 2, 336–347 (2011).
Otsuka, H., Fukao, A., Funakami, Y., Duncan, K. E. & Fujiwara, T. Emerging evidence of translational control by AU-rich element-binding proteins. Front. Genet. 10, 332 (2019).
Tao, X. & Gao, G. Tristetraprolin recruits eukaryotic initiation factor 4E2 to repress translation of AU-rich element-containing mRNAs. Mol. Cell Biol. 35, 3921–3932 (2015).
Fu, R., Olsen, M. T., Webb, K., Bennett, E. J. & Lykke-Andersen, J. Recruitment of the 4EHP-GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA 22, 373–382 (2016).
Chapat, C. et al. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl. Acad. Sci. USA 114, 5425–5430 (2017).
Peter, D. et al. GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression. Genes Dev. 31, 1147–1161 (2017).
Laird, J. H. et al. Talazoparib is a potent radiosensitizer in small cell lung cancer cell lines and xenografts. Clin. Cancer Res. 24, 5143–5152 (2018).
Morris, M. et al. Pelvic radiation with concurrent chemotherapy compared with pelvic and para-aortic radiation for high-risk cervical cancer. N. Engl. J. Med. 340, 1137–1143 (1999).
Curran, W. J. et al. Sequential vs. concurrent chemoradiation for stage III non-small cell lung cancer: randomized phase III trial RTOG 9410. J. Natl. Cancer Inst. 103, 1452–1460 (2011).
Adelstein, D. J. et al. An intergroup phase III comparison of standard radiation therapy and two schedules of concurrent chemoradiotherapy in patients with unresectable squamous cell head and neck cancer. J. Clin. Oncol. 21, 92–98 (2003).
Slotman, B. J. et al. Use of thoracic radiotherapy for extensive-stage small-cell lung cancer: a phase 3 randomised controlled trial. Lancet 385, 36–42 (2015).
Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019).
Decout, A., Katz, J. D., Venkatraman, S. & Ablasser, A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 21, 548–569 (2021).
Appay, V. & Rowland-Jones, S. L. RANTES: a versatile and controversial chemokine. Trends Immunol. 22, 83–87 (2001).
Dufour, J. H. et al. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 168, 3195–3204 (2002).
Rudin, C. M. et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer 19, 289–297 (2019).
Baskar, R., Dai, J., Wenlong, N., Yeo, R. & Yeoh, K.-W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 1, 24 (2014).
Ruscica, V. et al. Direct role for the Drosophila GIGYF protein in 4EHP-mediated mRNA repression. Nucleic Acids Res. 47, 7035–7048 (2019).
Zhang, X. et al. microRNA-induced translational control of antiviral immunity by the cap-binding protein 4EHP. Mol. Cell 81, 1187–1199.e5 (2021).
Tiedje, C. et al. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 44, 7418–7440 (2016).
Carreño, A. & Lykke-Andersen, J. The conserved CNOT1 interaction motif of tristetraprolin regulates ARE-mRNA decay independently of the p38 MAPK-MK2 kinase pathway. Mol. Cell Biol. 42, e0005522 (2022).
Han, Y., Liu, D. & Li, L. PD-1/PD-L1 pathway: current researches in cancer. Am. J. Cancer Res 10, 727–742 (2020).
Pantelidou, C. et al. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019).
Vanpouille-Box, C. et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 15618 (2017).
Shen, J. et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCaness. Cancer Res 79, 311–319 (2019).
Zhao, X. et al. Irradiation combined with PD-L1-/- and autophagy inhibition enhances the antitumor effect of lung cancer via cGAS-STING-mediated T cell activation. iScience 25, 104690 (2022).
Sen, T. et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 9, 646–661 (2019).
Zhang, N. et al. PARP inhibitor plus radiotherapy reshapes an inflamed tumor microenvironment that sensitizes small-cell lung cancer to the anti-PD-1 immunotherapy. Cancer Lett. 545, 215852 (2022).
Krebs, M. G. et al. Olaparib and durvalumab in patients with relapsed small cell lung cancer (MEDIOLA): an open-label, multicenter, phase 1/2, basket study. Lung Cancer 180, 107216 (2023).
Thomas, A. et al. Durvalumab in combination with olaparib in patients with relapsed SCLC: results from a phase II study. J. Thorac. Oncol. 14, 1447–1457 (2019).
Gay, C. M. et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 39, 346–360.e7 (2021).
Owonikoko, T. K. et al. Poly (ADP) ribose polymerase enzyme inhibitor, veliparib, potentiates chemotherapy and radiation in vitro and in vivo in small cell lung cancer. Cancer Med. 3, 1579–1594 (2014).
Willers, H., Krause, M., Faivre-Finn, C. & Chalmers, A. J. Targeting PARP for chemoradiosensitization: opportunities, challenges, and the road ahead. Int J. Radiat. Oncol. Biol. Phys. 112, 265–270 (2022).
Schaffer, B. E. et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 70, 3877–3883 (2010).
Schofield, D. J. et al. Activity of murine surrogate antibodies for durvalumab and tremelimumab lacking effector function and the ability to deplete regulatory T cells in mouse models of cancer. mAbs 13, 1857100 (2021).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Stringer, B. W. et al. A reference collection of patient-derived cell line and xenograft models of proneural, classical and mesenchymal glioblastoma. Sci. Rep. 9, 4902 (2019).
Campeau, E. et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529 (2009).
Gardner, E. E. et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell 31, 286–299 (2017).
Daniel, V. C. et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 69, 3364–3373 (2009).
Lin, S. et al. Establishment of peripheral blood mononuclear cell-derived humanized lung cancer mouse models for studying efficacy of PD-L1/PD-1 targeted immunotherapy. mAbs 10, 1301–1311 (2018).
Acknowledgements
This work was supported by the Conquer Cancer Foundation of American Society of Clinical Oncology (ASCO), the International Association for the Study of Lung Cancer (IASLC), and AstraZeneca. Research in the B.H. Lok Laboratory is supported by the Canada Foundation for Innovation, Canadian Institutes of Health Research (CIHR), National Institute of Health/National Cancer Institute (U01CA253383), Terry Fox Research Institute Program Project Grant (TFRI Project #1124), Clinical and Translational Science Center at Weill Cornell Medical Center, MSKCC (UL1TR00457). X.R. was supported by STARS 21 scholarship from 2020 to 2021. Research in the H.H.H. lab is supported by CIHR Operating grants (142246, 152863, 152864, and 159567 to H.H.H.), Terry Fox New Frontiers Program Project Grant (PPG19-1090 to H.H.H.). M.T. was supported by a CIHR Doctoral Award. Research in the N.S. lab is supported by a CIHR Foundation grant. We would like to thank Dr. Charles M. Rudin lab for gifting the SCRX-Lu149 chemonaive and chemoresistant PDX model. We are grateful to Dr. Julien Sage lab for providing the KP1 cell line. We would like to thank the Flow Cytometry Core Facility and Advance Optical Microscopy Facility (AOMF) department at UHN for assisting in immune profiling and immunofluorescence visualization work.
Author information
Authors and Affiliations
Contributions
X. Ran: conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing—original draft, writing—review and editing. B. Wu: investigation, validation, and data analysis V. Venkatasubramanian: investigation, validation, and data analysis. L.Song: validation, investigation and methodology in animal work. M.Teng: investigation, validation methodology and investigation in RNA-seq. W.B. Alawi: data analysis, visualization. X. Zhang and R.J. Ladak: investigation, methodology. V.Philip: methodology, writing—review and editing, supervision. N. Sonenberg and H He: resource, data curation, supervision, funding acquisition, methodology, writing—review and editing. B.H.Lok: conceptualization, resource, data curation, supervision, funding acquisition, validation, visualization, methodology, writing—original draft, project administration, writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
B.H.L. reports grants from Pfizer; and grants, personal fees, and non-financial support from AstraZeneca. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ran, X., Wu, B.X., Vidhyasagar, V. et al. PARP inhibitor radiosensitization enhances anti-PD-L1 immunotherapy through stabilizing chemokine mRNA in small cell lung cancer. Nat Commun 16, 2166 (2025). https://doi.org/10.1038/s41467-025-57257-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57257-z
This article is cited by
-
Zalcitabine induces ferroptosis in multiple myeloma through the TFAM–cGAS–STING–SLC7A11 axis
Journal of Translational Medicine (2026)
-
Targeting shared mechanisms of cisplatin resistance and metastasis in lung cancer for novel therapeutic strategies
Discover Oncology (2026)
-
Implications of DNA damage response and immunotherapy in tumor therapy
Cell Communication and Signaling (2025)
-
Immunotherapy for small cell lung cancer: current challenges and prospects
Experimental Hematology & Oncology (2025)
-
Overcoming resistance to anti-PD-L1 immunotherapy: mechanisms, combination strategies, and future directions
Molecular Cancer (2025)