Abstract
Leafy (Lfy1) is a classical dominant mutant showing more leaf number above primary ear and later flowering time in maize, but the causal gene together with its underlying genetic mechanism are unknown. Here, we report the cloning of Lfy1 mutant, and find that a retrotransposon insertion leads to leafy phenotype by elevating expression of its neighboring gene ZmOM66. ZmOM66 encodes an AAA+ ATPase that locate in mitochondria and interacts with itself. ZmOM66 overexpression affects the starch degradation, as well as contents of glucose, pyruvic acid, trehalose-6-phosphate, and TCA cycle related amino acids, and influences expression patterns of circadian clock genes. Moreover, expressions of floral related genes, including photoperiod regulated gene ZmPHYB1, integrator genes ZCN7, ZNC8 and ZCN12, and floral meristem identity genes ZMM4, ZMM15, and MASD67, are also significantly decreased by ZmOM66 overexpression. These results deepen our understanding of the regulatory mechanism of floral transition and leaf number in plant.
Similar content being viewed by others
Introduction
Floral transition, the switch from vegetative to reproductive growth, is a critical stage in the life history of flowering plants, which determines the flowering time and leaf number1. In agronomy, timing of floral transition strongly affects biomass and yield production, and plant fitness2. In the past decades, more than 300 genes have been identified to regulate floral transition and leaf number in Arabidopsis, Oryza Sativa, and Triticum aestivum3. This regulation occurs through a complex network of genetic pathways, such as photoperiod, circadian clock, vernalization, hormones, sugar, aging, and autonomous pathways in leaves or shoot apical meristems (SAM)2. Then, these pathways converge to the integrator genes, such as SUPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and FLOWERING LOCUS T (FT). Finally, the integrators activate the downstream floral meristem identity genes, such as APETALA1 (AP1) and FRUITFULL (FUL), to complete floral transition1,2,3.
In maize, the floral transition is characterized by the cessation of leaf formation and the elongation of SAM to create tassel primordium4, whose genetic dissection started from identifications of three classical late-flowering mutants, indeterminate1(id1), delayed flowering1(dlf1) and Leafy (Lfy1)4. ID1 encodes a zinc-finger transcription factor that functions in the autonomous pathway by regulating sugar synthesis and transport5,6,7, and DLF1 is a bZIP transcription factor homologous to the Arabidopsis floral integrator FLOWERING LOCUS D (FD)8. Besides ID1 and DLF1, more than twenty genes have been identified to regulate floral transition in maize by quantitative trait locus (QTL) cloning and comparative genomics analysis, including the pathway-related regulatory gene ZmCCT9, ZmCCT10, ZmCOL3, ZmPHYB, ZmNF-YC2, ZmSPL13, ZmSPL29 and ZmGI9,10,11,12,13,14,15, integrator gene ZCN8, ZCN7, ZCN12 and ZmMADS116,17,18,19, and floral meristem identity gene ZMM4, ZMM15 and ZmMADS6719,20. However, Lfy1 has not yet been cloned.
Lfy1 is a spontaneous mutant obtained from a single cross hybrid in 1983, with the main feature of a greater leaf number above the primary ear (LNPE, 9–15 leaves) than common maize (4–7 LNPE)4,21,22. It is noteworthy that this Lfy1 was not named following the Arabidopsis LEAFY (LFY) gene, whose homologs in maize were Zea FLO/LFY 1(zlf1) and zlf223. These extra leaves expand the total leaf area of Lfy1, which increases light interception and photosynthetic capacity and subsequently contributes to crop productivity24,25. Moreover, Lfy1 hybrids tend to accumulate more digestible carbohydrates in the canopy than common maize26,27,28. Due to these desirable features, Lfy1 has been rapidly applied to produce silage maize hybrids in America, Canada, Hungary, Nigeria, and other countries since the 1980s29,30.
Due to its complex genetic mechanism with phenotype varied depending on genetic background4, the cloning of Lfy1 gene is challenging. The earliest report suggested that Lfy1 is a single gene-controlled dominant mutant22. Later, it was found that segregating populations from Lfy1 crosses exhibited a normal distribution of LNPE31,32,33. Together with the fact that using populations crossed from non-Lfy1 lines failed to identify any flowering-related QTL in this Lfy1 locus34,35, it indicated that Lfy1 might provide a distinctive allele for controlling floral transition and leaf number. In our previous work, we generated three F2 populations and found that the leafy phenotype was controlled by a single gene only in Lfy1×B73 population. Using this population, we narrowed the Lfy1 locus to a 55-kb interval according to the B73 reference genome32.
In this study, we clone the causal gene for the Lfy1 mutant and find that a Copia long terminal repeat (LTR) retrotransposon insertion elevates expression of its neighboring gene ZmOM66 to cause the leafy phenotype. ZmOM66 encodes a member of the AAA+ ATPase family protein, which localizes in mitochondria. ZmOM66 overexpression affects starch degradation, as well as several metabolites involved in carbon/nitrogen metabolism, which may subsequently regulate floral transition and determine leaf number. These findings not only provide a gene for floral transition and leaf number determination but also extend the molecular mechanism of nutrient-involved plant growth and development regulation.
Results
Cloning of the Lfy1 mutant
We previously fine-mapped the Lfy1 locus to a 55-kb interval on chromosome 3, using a population crossed from CO412, an inbred harboring the Lfy1 allele (Fig. 1a) and B73 line32. This region contained two genes, Zm00001d044411 and Zm00001d044412 (Fig. 1b). Sequencing, gene expression, and protein accumulation analyses excluded both genes as the causative gene for Lfy1 mutation (Supplementary Figs. 1 and 2). To test the possibility of an additional sequence responding to the leafy phenotype, we constructed a bacterial artificial chromosome (BAC) library and screened out three clones for sequencing, which successfully assembled the whole-genome sequence for Lfy1 locus (Supplementary Fig. 3). Compared with the 55-kb sequence in B73 genome, this sequence was 172-kb (Fig. 1b). We used Softberry to predict coding genes, but failed to find any other genes except Zm00001d044411 and Zm00001d044412 within this sequence (Supplementary Data 1).
a Pictures of CO412 (Lfy1 mutant) and Z58 plants. The arrows indicate the ears, Bar = 15 cm; b Sequence comparisons of the Lfy1 locus among B73, Z58, and Lfy1 mutant. The blue and red boxes indicate coding genes and the Copia LTR-RT, respectively. Gray lines indicate the same sequences between different genotypes; c Expression level of genes within 100 kb up- and down streams of the LTR-RT. The expression data were from the transcriptome analysis of NILB-Lfy1 and NILB73 three times independently. The error bars indicate mean ± SD. A two-tailed Student’s t-test was used to determine P-values between genotypes. ND, not detected (Fragments Per Kilobase Million (FPKM) < 0.5). Source data are provided as a Source Data file.
According to a PCR-based sequence comparison (Supplementary Figs. 2c and 4a), we speculated that the Z58 line might harbor a sequence more similar to Lfy1 mutant than B73 within the Lfy1 locus. Thus, we constructed a Fosmid library for the Z58 line (Supplementary Table 1) and screened out 5 clones for sequencing within the Lfy1 region, which successfully assembled a sequence of ~70 kb (Fig. 1b and Supplementary Fig. 4a, b). Sequence alignment revealed an extra sequence of 12,806-bp in the Lfy1 mutant compared with the Z58 line. Meanwhile, the remaining sequence was 99.9% identical to the right part of the 172-kb Lfy1 sequence (Fig. 1b).
According to the RepeatMasker website, this 12,806-bp sequence is a Copia LTR retrotransposon (Copia LTR-RT) with two short target site duplications (TSDs, CGTCG). And its 5’ and 3’ LTR sequences were each 1626 bp long, which surrounded a middle sequence of 9549 bp for encoding five transposition-related proteins (Supplementary Fig. 5 and Supplementary Table 2). In addition, the 5’ and 3’ LTR sequences were completely identical, indicating that this LTR insertion occurred in a recent year, which was consistent with the fact that Lfy1 mutant was first reported in 198321. Together, these results suggested that this LTR-RT insertion might relate to leafy phenotype.
Zm00001d044416 overexpression phenocopies the Lfy1 mutant
Previous studies showed that when a transposon is inserted in an intergenic region, it influences the mutant phenotype mainly by influencing expression of its neighboring genes36. Our previous study developed two NILs (NILB-Lfy1 and NILB73) by backcrossing CO412 into B7337. Both NILs showed similar vegetative meristems at V3 (3 expanded leaves, vegetable stage) and V5 stage (floral transition stage)37. But at the V6 stage, NILB73 completed the floral transition with a feature of elongated SAM, while NILB-Lfy1 still stayed at the vegetative stage37. Using transcriptome data of shoot tips (less than 1 cm length, containing SAM, leaf primordium and a small part of immature leaves) at V5 stage34, we investigated the expression of genes located within 100 kb up- and downstream of this LTR-RT (Fig. 1c). Since Lfy1 was a dominant mutant, we focused on upregulated genes in NILB-Lfy1 compared with NILB73, and found two candidate genes Zm00001d044411 and Zm00001d044416 (Fig. 1c and Supplementary Table 3). Zm00001d044416 directly neighbored to this LTR-RT and located at 26 kb downstream, while Zm00001d044411 located at 38 kb upstream (Fig. 1b). Both genes showed under-detectable expressions in NILB73 (Fragments Per Kilobase Million (FPKM) < 1), and increased their expressions to low levels in NILB-Lfy1, with FPKM of 2.86 for Zm00001d044416 and 1.26 for Zm00001d044411 (Fig. 1b, c).
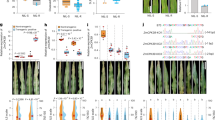
To determine whether Zm00001d044411 or Zm00001d044416 was the causative gene for Lfy1 mutant, we generated transgenic overexpression (OE) lines, and obtained 7 heterozygous OE lines for Zm00001d044416, with expressions increased by 189 to 1732 times compared to wild type (WT) (Fig. 2a, b and Supplementary Fig. 6). Phenotype observation showed delayed flowering time for all 7 transformants. In detail, OE1, with the highest Zm00001d044416 expression, had the severest phenotype that failed to start flower transition and remained in vegetative growth until it turned to senescence and died (Fig. 2c, d). OE2, with moderate Zm00001d044416 expression, had eight LNPE, but produced aberrant inflorescences with vegetative characteristics (Fig. 2e, f and Supplementary Fig. 7a, b). After crossing OE2 pollens to transgenic background Zong31, we obtained one transgenic positive OE2-F1 plant that show a similar phenotype to OE1 plant with failure of starting flower transition (Supplementary Fig. 7c). OE3 had a little lower Zm00001d044416 expression than OE2, and could produce tassel and pollen, but failed to produce ear (Fig. 2g). We crossed OE3 pollens to Zong31, and harvested one transgenic positive F1 plant that contained 11 LNPE, and could produce tassel and pollen, but produced abnormal ear with vegetative characteristic (Fig. 2h and Supplementary Fig. 8). We continued to cross OE3-F1 pollens to Zong31 to produce BC1F1 plants. Within this BC1F1 population, ~5% transgenic plants produced pollens, and showed a flowering time delayed by 15 to 21 days compared to their none-transgenic sisters (Fig. 2i). We also obtained three independent transformants for Zm00001d044411 gene, but they all showed similar LNPE to the non-transformants (Supplementary Fig. 9). These results concluded that Zm00001d044416 overexpression could phenocopy the Lfy1 mutant, indicating it the functional gene for Lfy1 mutant.
a The vectors used for Zm00001d044416 transformation; b Zm00001d044416 expression levels in overexpression (OE) line OE1, OE2, and OE3 with three times independently; c Picture for the transgenic background Zong31 (as wild type, WT); d Picture for Zm00001d044416 OE1-T0 plant; e, f Picture for Zm00001d044416 OE2-T0 plant (e) and its leaf number above primary ear (LNPE, f); g Picture for Zm00001d044416 OE3-T0 plant; h LNPE of OE3 × Zong31 F1 plant, NT, non-transgenic siblings; i Days to anthesis of OE3 × Zong31 transgenic (OE) and non-transgenic (NT) BC1F1 plants. OE-F, the fertile OE plants; OE-NF, the non-fertile OE plants. Bar (in c to g) = 15 cm. The error bars indicate mean ± SD. A two-tailed Student’s t-test was used to determine P-values between the OE line (5 plants) and WT (8 plants). Source data are provided as a Source Data file.
We then obtained an ethyl methanesulfonate (EMS) mutant from a mutant library38 for Zm00001d044416. This mutant had a nucleotide substitution (G to A) at 1056 bp position relating to the start codon ATG, which resulted in a premature termination (Supplementary Fig. 10a,b). Compared with WT plants, this mutation did not show any alteration in either flowering time or LNPE (Supplementary Fig. 10c, d). Moreover, we generated two knockout lines for Zm00001d044416 using the CRISPR-Cas9 method, with one deleting 20 bp from position 187 bp to 206 bp, and the other having 1 bp insertion at 198 bp position (Supplementary Fig. 11a, c). Phenotype observation also failed to find any changes in flowering time and LNPE by gene knockout (Supplementary Fig. 11d, e). These results suggested that the loss function of Zm00001d044416 did not affect floral transition.
The prolonged-expression period of Zm00001d044416 related to the leafy phenotype
To further explore the mutation reason for the leafy phenotype, we developed two NILs (NILZ-Lfy1 and NILZ58) by backcrossing the Lfy1 locus from CO412 into Z58. Both NILs showed similar Zm00001d044416 expression levels in all tissues, except in shoot tip (Fig. 3a). In shoot tip of NILZ58, Zm00001d044416 expression gradually increased from V1 to V3 stage, reached a peak at V3 stage, then decreased dramatically at V4 stage when the floral transition started. After that, it kept very low level at V4 to V5 (under floral transition) and V6 to V7 (after floral transition) stages (Fig. 3b). In NILZ-Lfy1, Zm00001d044416 expression also increased from V1 to V3 stage, but maintained at high levels from V3 to V7 stage (Fig. 3b). Moreover, Zm00001d044416 expression level was higher in NILZ-Lfy1 than in NILZ58 during all investigated stages (Fig. 3b).
a, b Expression levels of Zm00001d044416 in different tissues (a), and in shoot tips during plant growth from V1 (one expanded leaf) to V7 (b) in NILZ-Lfy1 and NILZ58. Tassel collected at tassel emergence stage (VT); unpollinated ear and husk collected at silking stage (R1); DAP12, 12 days after pollination; G5, 5 days after germination. Each experiment was repeated three times independently. The error bars indicate mean ± SD, and a two-tailed Student’s t-test was used to determine P-values between the two NILs; c, d In situ hybridization of Zm00001d044416 in shoot tips of NILZ58 (c) and NILZ-Lfy1 (d) at V4 stages. SAM, shoot apical meristem; the red arrows indicate primary vascular bundles. Bar = 200 μm. Source data are provided as a Source Data file.
To explore tissue-specific expression of Zm00001d044416, we conducted in situ hybridizations using shoot tips of NILZ58 and NILZ-Lfy1 at V4 stage (Fig. 3c). The results showed that Zm00001d044416 was majorly expressed in primary vascular bundles, SAM and leaf primordium in both NILs (Fig. 3c), indicating that expression position of this gene was similar between two NILs. However, Zm00001d044416 expression level seemed higher in NILZ-Lfy1 than in NILZ58, as indicated by the deeper staining. Together, these results indicated that Zm00001d044416 expression in the shoot tip was elevated and prolonged during floral transition in NILZ-Lfy1, which might respond to a leafy phenotype.
Zm00001d044416 encodes an AAA+ ATPase that interacts with itself in mitochondria
Zm00001d044416 encoded a protein of 464 amino acids (aa), which contained a typical N-terminal domain of the AAA-type ATPase from 36 to 129 aa, and an AAA+ ATPase domain from 251 to 391 aa (Fig. 4a). Phylogenetic analysis showed that the closest homolog of Zm00001d044416 in Arabidopsis was AtOM6639 (Fig. 4b and Supplementary Fig. 12a). Thus, we named Zm00001d044416 as ZmOM66.
a The ZmOM66 protein contains an N-terminal domain and an AAA+ ATPase domain; b Phylogenetic relationships of ZmOM66 and its homologs in nematode (C. elegan), human (H. sapens), yeast (S. cerevisiae), Arabidopsis (At), rice (Os), and maize (Zm). ZmOM66 homologs were aligned using bootstrap consensus in MEGA 7.0 software. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Scale bar = average number of amino acid substitutions per site; c–e Subcellular localization of the ZmOM66 protein in tobacco leaf epidermal cells (c), maize leaf protoplast (d) and Arabidopsis root hairs (e); f Yeast two-hybrid assay of the interaction between two ZmOM66 proteins. The interactions between pGADT7-T and pGBKT7-53 and between pGADT7-T (AD) and pGBKT7-Lam (BD) were used as positive and negative controls, respectively. (g, h) Luciferase complementation imaging (LCI) (g) and bimolecular fluorescence complementation (BiFC) assay (h) of two ZmOM66 proteins in tobacco leaves. The fluorescence signals represent the interaction activities. Bar = 20 μm. In c–e and h, each experiment was repeated three times independently.
AAA+ ATPases had many members, and exhibit multiple subcellular localizations with diverse biological functions40,41,42. As AtOM66 protein is located in mitochondria39, we guessed ZmOM66 might have a same location. To solidify this speculation, we fused the open reading frame (ORF) of ZmOM66 to the yellow fluorescent protein (YFP) gene and transferred into tobacco leaves and maize protoplasts. Both revealed that ZmOM66 protein co-localized with a previously reported mitochondria-located protein DEK3643 (Fig. 4c, d). Moreover, the ZmOM66-YFP fusion vector was transferred into Arabidopsis, which showed YFP signal co-localizing with the red fluorescence of Mitotracker at mitochondria, confirming the mitochondria localization of this protein (Fig. 4e).
AAA+ ATPase family proteins often form homo-oligomers44. We cloned ZmOM66 ORF into both pGADT7 and pGBKT7 vectors to perform a yeast two-hybrid (Y2H) assay, which revealed that ZmOM66 protein interacted with itself (Fig. 4f). Bimolecular fluorescence complementation (BiFC) and luciferase complementation imaging (LCI) experiments were carried out in N. benthamiana to confirm the self-interaction of ZmOM66 and further showed that this interaction occurred in mitochondria (Fig. 4g, h). In addition, the interaction between ZmOM66 and Zm00001d039183, a close paralog of ZmOM66 in maize with high expression in shoot tip (Supplementary Figs. 12b and 13), was detected in mitochondria by BiFC and LCI experiments (Supplementary Fig. 14).
Contents of carbon and nitrogen metabolic components were abnormal in ZmOM66 overexpression line
To investigate how ZmOM66 triggered changes in flowering time and LNPE in maize, we conducted transcriptome analysis using shoot tips, newly developed immature leaves, and mature leaves (the fifth leaf) from V5-stage OE3-BC1F1 plants, which were harvested at zeitgeber time 0 (ZT0, 0 h after the start of a day, same to the end of a night, Fig. 5a). Compared with WT (transgenic negative OE3-BC1F1), 911 up- and 91 down-regulated genes (differentially expressed genes, DEGs) were identified from shoot tips of ZmOM66 OE lines (Fig. 5b and Supplementary Data 2). Similarly, 2005 up- and 276 down-, and 1363 up- and 621 down-regulated DEGs were identified from immature leaves and mature leaves, respectively (Fig. 5c, d and Supplementary Data 2).
a The sketch map of tissues used in transcriptome analysis; b–d Number of differentially expressed genes (DEGs) in shoot tips (b), immature leaves (c), and mature leaves (d) of ZmOM66 OE line compared with wild type (WT); (e-g) Major GO terms of the functionally annotated DEGs in shoot tips (e), immature leaves (f), and mature leaves (g); h Starch contents in WT and ZmOM66 OE line evaluated at the end of a day (ED, ZT16) and at the end of a night (EN, ZT0). ZT indicates the zeitgeber time, and the start of a day is set as ZT0; i Pathway for carbon and nitrogen metabolism; j Metabolites showing significant content changes in ZmOM66 OE line compared to WT at ZT0; k Contents of amino acid relating to TCA cycle in WT and ZmOM66 OE lines at ZT0. In h, j, and k, each experiment was repeated three times independently. The error bars indicate mean ± SD. A two-tailed Student’s t-test was used to determine P-values between OE and WT. Source data are provided as a Source Data file.
GO analysis revealed that the shoot tip DEGs were majorly enriched in metabolic activity-related GO terms, including response to carbohydrates, response to nitrogen compound, and respiratory burst (Fig. 5e). Both immature leaf and mature leaf DEGs showed similar enrichment to the shoot tip DEGs in these metabolic activity-related GO terms (Fig. 5f, g). In addition, immature leaf DEGs also enriched in carbohydrate binding and hexose transmembrane transporter activities (Fig. 5f), and mature leaf DEGs in carbohydrate biosynthetic process, response to high light intensity, chloroplast envelope, and cell growth (Fig. 5g). These results indicated that ZmOM66 overexpression majorly affected carbohydrate and nitrogen metabolic activity.
In mature leaves, starch commands a central role in carbon budget by acting as both a source to release carbon reserves for growth and development and a sink for either starch translocation or starch storage45,46. Starch was synthesized during the daytime, and degraded into disaccharide or monosaccharides for transporting during the night45,46. To explore how ZmOM66 regulates carbohydrate and nitrogen metabolism, we firstly evaluated starch content in mature leaves at ZT0 and ZT16 (the end of a day). The results showed that starch content significantly decreased in ZmOM66 OE lines compared to WT at both timepoints (Fig. 5h). However, the degree of content declining in ZT0 (63%) was greater than that in ZT16 (49%, Fig. 5h), suggesting that starch degradation might be more affected by ZmOM66 overexpression (Fig. 5h). To confirm this, we measured three photosynthesis indicators, namely photosynthetic efficiency of photosystem II (Phi2), photosynthetic efficiency of non-photochemical quenching (PhiNPQ), and photosynthetic efficiency of non-regulatory energy dissipation (PhiNO), and found similar levels for each indicator between ZmOM66 OE lines and WT at ZT8 (middle of a day) (Supplementary Fig. 15), indicating that starch synthesis is not visibly affected by ZmOM66 overexpression. These results suggested that ZmOM66 overexpression majorly influenced starch consumption.
To further investigate starch consumption efficiency, we performed metabolic profiling analyses in mature leaves using GC-TOF-MS and LC-ESI-MS/MS at ZT0, in which starch was degraded for a whole night, but not started to synthesize. The results showed that the content of glucose, a key starch consumption metabolite, significantly decreased in ZmOM66 OE lines compared to WT (Fig. 5i, j). Moreover, the downstream metabolites, such as pyruvic acid and α-ketoglutarate in the tricarboxylic acid (TCA) cycle47 also decreased (Fig. 5i, j). Trehalose-6-phosphate (T6P) plays a crucial role involving in sugar pathway during floral transition, and its content negatively correlated with the rate of starch consumption48,49. Metabolic profiling also found significant decrease in T6P content by ZmOM66 overexpression (Fig. 5i, j). Together, these results indicated that ZmOM66 overexpression accelerated starch consumption by decreasing starch content and carbohydrates in mature leaves.
Accelerated starch degradation always induces carbon starvation, which accelerates amino acid degradation to provide alternative carbon skeletons entering the TCA cycle50,51 (Fig. 5i). Thus, we evaluated the content of TCA cycle-related amino acids, and found significant decreases of leucine, serine, tyrosine, and citrulline, in ZmOM66 OE compared with WT (Fig. 5k), further reflecting that carbon/nitrogen metabolism changed due to ZmOM66 overexpression.
ZmOM66 overexpression changed the expression of floral transition genes
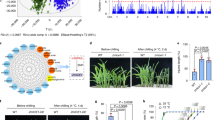
Starch degradation is often associated with circadian clock gene expressions51. Particularly, GIGANTEA (GI) and PSEUDO RESPONSE REGULATORs (PRRs) act as major mediators between the circadian clock and the master regulator of flowering time control1,2,3 (Fig. 6a). To test whether the prolonged flowering time and the elevated LNPE in Lfy1 mutant were associated with the abnormal expression of circadian clock genes, we conducted qRT-PCR to determine ZmGI2, ZmPRR5, and ZmPRR7 expressions in mature leaves in response to the circadian rhythm (Fig. 6a). The results revealed an expression peak of ZmGI2 at ZT8 in WT (OE3-BC2F1 negative plants), while this peak moved to ZT12 in ZmOM66 OE line (OE3-BC2F1 positive plants). In addition, the expression peak level of ZmGI2 in ZmOM66 OE line was higher than that in WT (Fig. 6b). ZmPRR7 expression was higher in ZmOM66 OE line than in WT during the daytime, especially at ZT8 and ZT16 (Fig. 6c). In contrast, ZmPRR5 showed an obvious expression peak in WT at the end of night, while only a weak peak in OE line (Fig. 6d). These results suggested that ZmOM66 overexpression disturbed expressions of circadian clock genes, by influencing ZmGI2 and ZmPRR7 expressions during the daytime, and ZmPRR5 expression during the night.
a The circadian clock genes, downstream integrator genes, and floral meristem identity genes in maize floral transition. Compared to WT, genes with altered expression in ZmOM66 OE lines are shown in red; b–d Expression of circadian clock gene ZmGI2 (b), ZmPRR7 (c), and ZmPRR5 (d) in response to rhythm change in leaves of ZmOM66 OE lines and WT. ZT indicates the zeitgeber time and the start of a day is set as ZT0; e, f Expression of previously reported floral transition genes10,18,19,20 in maize leaves (e) and shoot tip (f) of WT and ZmOM66 OE lines at ZT0. The error bars indicate mean ± SD. A two-tailed Student’s t-test was used to determine P-values between OE and WT. ND, not detected. In b to f, each experiment was repeated three times independently. Source data are provided as a Source Data file.
Besides GIs and PRRs, ZCN8 is a homolog of the Arabidopsis FT gene (Fig. 6a). Its expression significantly decreased in the ZmOM66 OE line compared to WT at ZT0 (Fig. 6e). Significantly decreased expressions were also detected for other previously reported maize floral transition genes10,18,19,20, including photoperiod regulated gene ZmPHYB110, integrator genes ZCN7 and ZCN1218, and floral meristem identity genes ZMM4, ZMM15, and MASD6719,20 (Fig. 6e, f). These results indicated that expression levels of the key floral transition genes significantly decreased under ZmOM66 overexpression.
Utilization of the Lfy1 allele in the breeding of high-yield maize
We crossed Lfy1 into B73, X178, and Z58 to determine whether it could be utilized in breeding maize with high biomass and grain yield. Lfy1-containing NILs in B73 and X178 backgrounds showed severely delayed anthesis time, while their silking times were less affected, which resulted in failure of self-fertilization37. However, NILs in the Z58 background had a moderately delayed anthesis time (7–14 days), and most plants could be self-pollinated to produce inbred seed (Supplementary Fig. 16).
As Z58 is a female parent of the widely used hybrid ZD958 in China32,33, we used NILZ58 to substitute Z58 for crossing with the maternal inbred Chang7-2 to produce a modified ZD958 hybrid (ZD958Lfy1, Fig. 7a, b). ZD958Lfy1 had three LNPE more than ZD958 (Fig. 7c) and showed silking and anthesis times four and five days later, respectively, when compared to ZD958 (Fig. 7d). With a prolonged vegetative growth period, the ear length and ear weight per plant of ZD958Lfy1 were 19.3% and 21.5% higher than those of ZD958, respectively (Fig. 7e, f). In addition, the plant height and ear height in ZD958Lfy1 were also increased by 14% (Fig. 7g).
a, b Ears of ZD958 (Z58 × Chang7-2; a) and ZD958Lfy1 (Lfy1 introgressed Z58 × Chang7-2; b); c–g Leaf number above primary ear (LNPE) and total leaf number (LN) (c), days to anthesis (DTA) and days to silk (DTS) (d), ear length (e), ear weight (f), plant height (PH) and ear height (EH) (g) in ZD958 and ZD958Lfy1. After pollen shedding, 25 plants were randomly selected for evaluating LN, LNPE, PH, and EH. More than 180 plants were selected for evaluating DTA and DTS. After harvest, 30 ears were randomly selected for measuring EL and EW. The error bars indicate mean ± SD. A two-tailed Student’s t-test was used to determine P-values between genotypes. Source data are provided as a Source Data file.
Discussion
Floral transition is a central phase in the life of all flowering plants1,2,3. Although regulatory pathways for floral transition had been well studied in the model plant Arabidopsis, temperate maize has developed some special regulatory pathways during its domestication from the tropical grass teosinte for adaptation to the high-latitude regions52,53. Thus, as a day-neutral plant, temperate maize can occur flowering irrespective of day-length, while flowering time in Arabidopsis mainly affected by photoperiod4. Therefore, cloning genes from temperate maize might help to reveal novel pathways for plant floral transition. Lfy1 is a classical late-flowering mutant in maize, which has been reported for more than forty years21. However, due to its complex genetic mechanism with phenotype varied depending on genetic background, the causal gene for Lfy1 has not been uncovered. In this study, we cloned this mutant and concluded that ZmOM66 responded for the leafy phenotype, basing on experiments of fine-mapping (Fig. 1), sequence and expression analyses (Figs. 1b and 3a, c), and transgenic verification (Fig. 2).
It is noteworthy that although Lfy1 mutant was classified as a late-flowering mutant, it also had a typical characteristic of more LNPE4,21. Concerning the mechanism of later flowering and more LNPE simultaneously occurred in Lfy1 mutant, we speculated that ZmOM66 might affect leaf initiation firstly, and then resulted in late-flowering time. This speculation could be supported by the evidence that different expressions of ZmOM66 expression in Lfy1 mutant and normal plant happened before and under floral transition stages. For detail, in normal plants, ZmOM66 expression gradually increased during leaf primordium development at V1 to V3 stages, while decreasing dramatically at V4 stage when floral transition started (Fig. 3b). In addition, no obvious flowering phenotype could be observed in ZmOM66 mutant and knockout plants (Supplementary Figs. 10 and 11), indicating that ZmOM66 is not necessary for floral transition, but high expression might have contributions. Thus, the early expression (before floral transition) of ZmOM66 in normal plants indicated that ZmOM66 function is likely to promote leaf primordium development, but not to floral organ development. And during floral transition, its expression was quickly suppressed, which is likely to stop leaf primordium, because in plants the SAM start to create tassel primordium for initiating flowering only after it ceases leaf formation4. In contrast, in Lfy1 mutant, ZmOM66 expression did not decrease at V4 stage after it increased from V1 to V3 (Fig. 3b), which might cause failure of stopping leaf initiation, subsequently resulted in extra leaves above ear. Then, more leaf initiations consequently postpone flowering time, because the rate of each leaf initiation is generally constant in plant4,54. Together, these results suggested that ZmOM66 expression should be precisely suppressed before floral transition for stopping leaf initiation to trigger floral transition, and in Lfy1 mutant, its prolonged high expression failed to stop leaf initiation.
Fine-mapping results revealed that part of ZmOM66 gene, including 431-bp coding sequence (from 1 to 431 bp in its first exon) and its upstream sequence, located in the candidate interval of the Lfy1 locus (Fig. 1b). PCR-based genome sequencing revealed that the 431 bp coding sequence and 9216 bp upstream sequence were identical between Lfy1 mutant and Z58, indicating that the changed expression of ZmOM66 in Lfy1 mutant might not due to its own sequence (Supplementary Fig. 4). Previous studies have shown that transposons play crucial roles for domestication of floral transition in maize11,53,55. For example, a MITE element in 70 kb upstream of ZmRap2.755, a Harbinger-like transposable element in 57 kb upstream of ZmCCT911, and L1-type retrotransposon and an LTR/Gypsy type retrotransposon in 7 and 31 kb upstream of ZmELF3.153, all participated flowering time regulation by affecting their neighboring gene expressions11,53,55. In this study, we also found an LTR-RT insertion in 26 kb upstream of ZmOM66, which is the main difference that we could found between Lfy1 and Z58, suggesting a similar phenomenon of transposon affecting flowering-related gene expression to previous works11,53,55 (Fig. 1b, c).
Although the alteration of gene expressions had been widely reported by transposon insertion, the alteration mechanism is complex and mostly under-uncovered36. To reveal the mechanism of elevated expression of ZmOM66 by the LTR-RT insertion, we analyzed the structure of this LTR-transposon and found that the 5’ and 3’ LTR sequences were completely identical, indicating that the insertion time occurred in a recent year, which was consistent with the fact that Lfy1 mutant was found in 198321. Then, we cloned this LTR-RT into a PGL3 vector to drive a luciferase reporter gene expression in maize leaf protoplasts, and found that it could increase luciferase activity by 3 times (Supplementary Fig. 17). This finding of enhancer function was similar to the insertions of a LTR retrotransposon in upstream of MdMYB1 in apple and a Hopscotch transposable element in upstream of TB1 in maize56,57. However, this 3 times enhancer function could not fully explain the 15 times increased expression of ZmOM66 in Lfy1 mutant compared with normal plants. This indicated some other mechanism also involving in the alteration of ZmOM66 expression. To investigate other mechanism, we conducted a whole-genome bisulfite sequencing (WGBS-seq) using the shoot tips of NILZ-Lfy1 and NILZ58 at V5 stage, since transposon insertion tends to affect methylation levels in its surrounding regions36. It found that the CG methylation level within 24 kb upstream region, promoter and genebody of ZmOM66 were all higher in NILZ-Lfy1 than in NILZ58, supporting changes of methylation level by this LTR insertion (Supplementary Fig. 18). Although the classical theories inferred that promoter DNA methylation was associated with transcriptional repression, growing evidence had suggested promoter methylation also contributed to high transcriptional activity, especially in rapidly differentiated tissues58,59. Therefore, this LTR-RT insertion may result in abnormal methylation and expression for the surrounding genes in shoot tips, which act as a classically rapid-differentiated tissue in maize. This speculation could be supported by the evidence that ZmOM66 only upregulated in shoot tips (Fig. 3a). Together, these results supposed that the LTR-RT insertion may cause a variety of changes, e.g. methylation level and enhancer effect, for an additive effect to elevate ZmOM66 expression.
The AAA+ ATPase contains many members in plants with diverse cellular functions, such as male meiosis, immune responses, pollen tube development and so on39,60,61,62. However, the role of AAA+ ATPase in regulating flowering time has been rarely reported, with exceptions of AtFtsH4, AaFRAT1, and AaFRAT1’ direct orthologue AtFRAT163,64. Loss of function of the Arabidopsis AtFtsH4 postpones flowering time mainly because of its floral organ defect63, and lesions in Arabis alpina AaFRAT1 gene correlated with earlier flowering performance, especially under the perpetual flowering 1 (pep1) mutant background64. AtFRAT1 acted as a weaker repressor to flowering in Arabidopsis under long day conditions61. In this study, we found an AAA+ ATPase gene, ZmOM66 contributing to Lfy1 mutation and floral transition in maize. Although both AaFRAT1 and ZmOM66 regulate flowering time, their proteins subcellular localization was different, with AaFRAT1 locating in interphase between endoplasmic reticulum and peroxisomes64, and ZmOM66 in mitochondria, indicating some differently functional mechanism between both proteins. On the other hand, sequence alignment revealed an Arabidopsis AtOM66 that showed a closer phylogenetic relation to ZmOM66 (Supplementary Fig. 12). Whereas, AtOM66 did not change flowering time by both reduced expression and overexpression39. Zm00001d039183 was a close phylogenetic paralog of ZmOM66 in maize (Supplementary Fig. 12). This gene also showed no influence on flowering time either by overexpression or knockout (Supplementary Fig. 19). These results indicated that ZmOM66 might have a distinct functional mechanism from its paralogs and orthologs in regulating floral transition. This speculation could be further supported by indirect evidence that all five ZmOM66 paralogs in maize showed no expression changes in ZmOM66 EMS mutant and knockout lines (Supplementary Fig. 20), indicating that they might not be functional redundant paralogs to ZmOM66, as functional redundant paralogs normally change their expressions for functional compensation for the disturbed function from others, through dosage balance mechanisms65.
To explore the mechanism of ZmOM66 regulating floral transition, we conducted transcriptome and metabolism analysis, and found that contents of carbon and nitrogen metabolic components, together with carbon and nitrogen metabolic activity-related genes, were changed under ZmOM66 overexpression (Fig. 5). Together with decreased levels of starch content (Fig. 5h), it could suggest that ZmOM66 may link metabolic changes and flowering regulation in maize. Another classical late-flowering mutant id1 in maize, had also been confirmed to link metabolic changes and flowering time regulation7. But unlike id1 mutant, in which sucrose and other soluble sugars significantly increased7, ZmOM66 overexpression decreased starch and carbohydrate contents (Fig. 5h, j), even they had similar variation trend in starch/sucrose ratio in the end of night (Supplementary Fig. 21). This indicated some different mechanism between ID1 and ZmOM66 for linking metabolic changes and flowering time. On the other hand, circadian clock genes were changed in id1, Lfy1 mutant and ZmOM66 OE lines37,66, indicating that carbohydrate content together with related circadian clock changes, are crucial for the endogenous floral inductive pathway in maize.
ZmOM66 protein is located in mitochondria. However, there is little consideration of how mitochondrial-localized proteins influence flowering time, making it challenging to build a direct bridge among ZmOM66, carbon and nitrogen metabolism, circadian clock and flowering time67. Previous work showed that ZmOM66 homolog gene AtOM66 was related to mitochondrial stress, and the starch content significantly decreased in AtOM66 overexpressed lines, even it has no late-flowering phenotype39. Thus, we evaluated expressions of three ALTERNATIVE OXIDASE (AOX) genes, and two NAD(P)H dehydrogenase genes, whose orthologous were thought to be markers for mitochondrial stress response in Arabidopsis67,68. We found increased expressions for all 5 genes under ZmOM66 overexpression (Supplementary Fig. 22a). In addition, β-amylase (BAM) gene has been reported to involved in stress response and starch degradation at night69. Five BAM genes showed raised expression levels in sink organs, including shoot tips and immature leaves in ZmOM66 OE lines (Supplementary Fig. 22b). Together with the fact that ZmOM66 mainly expressed in vascular bundle of shoot tips, we speculated that ZmOM66 overexpression may trigger the mitochondrial stress response in sink organs, and then transmit these signals to the source organ, i.e., mature leaf, to change its metabolic process, subsequently altered leaf initiation and flowering time (Supplementary Fig. 23).
In summary, we cloned the classical mutant of Lfy1, and revealed that the Lfy1 mutation was caused by abnormally elevated and prolonged expressions of ZmOM66 gene. ZmOM66 located in mitochondria, whose overexpression affected starch degradation, as well as contents of glucose and TCA cycle amino acids, and influenced expression patterns of circadian clock genes, which further resulted in increased leaf number and postponed flowering time. These results deepen our understanding of regulating flowering time and leaf primordium development in plants.
Methods
Plant material
CO412 is a popular inbred line generated in Canada with a pedigree of (391134 × CO255) CO255 × (A619L × A632)37. This inbred contained a dominant Lfy1 allele that contributed to more LNPE (10–12) and delayed pollen shedding time. Two BC6F2 NIL lines were developed from backcrosses between the Lfy1-containing line CO412 (donor parent) and either the B73 or Z58 inbred line (recurrent parent). In each cross-generation, plants with more LNPE ( > 7 in the Z58 background and >8 in the B73 background) were selected for genotyping using polymorphic markers in the Lfy1 candidate region32. Plants containing the Lfy1 locus were selected for the next crosses. After six backcrosses, heterozygous individuals were self-pollinated to generate the homozygotic NILZ58 and NILB73, and their introgressed lines NILZ-Lfy1 and NILB-Lfy1, respectively. All plant materials were grown in the field in Beijing in summer and in Sanya of Hainan province in winter in China. Arabidopsis Col-0 and Nicotiana benthamiana were grown in a chamber at 22 °C, with a 16 h/8 h light/dark photoperiod.
BAC and Fosmid library construction and sequencing
The DNA of CO412 was extracted from leaves, digested by EcoR I and Hind III, and used for BAC library construction. Three markers, locating within the Lfy1 locus with one targeting to each of Zm00001d044410, Zm00001d044412, and Zm00001d044416 genes, were used to screen clones that contained the Lfy1 region. DNA from positive clones was extracted using an Omega BAC/PAC DNA Maxi Kit (Biolink Biotechnology, Beijing, China). After digestion by EcoR I, Hind III, and Pst I, DNA fragments were separated on an agarose gel using the fingerprint method. According to fragment distributions between different clones, the whole candidate region of Lfy1 in CO412 was sequenced and assembled.
Z58 DNA was extracted from its leaves, broken into 38–48 kb DNA fragments, and then inserted into a Fosmid vector to construct a Fosmid library by Takara Bio Inc. (Takara, Dalian, China). This library contained ~100,000 clones.
Sequencing of BAC and Fosmid clones was conducted by the Beijing Genomics Institute (BGI; Beijing, China) using an Illumina HiSeq 2500 System. The coding genes in the Lfy1 confidential region were predicted by Softberry (http://www.softberry.com/), and the insertion was predicted on the Repeatmasker website (https://www.repeatmasker.org).
Maize transformation and EMS mutation identification
For overexpression, the ORF of ZmOM66 was inserted into the binary vector pCambia3301 driven by the CaMV 35S promoter and then transformed into immature embryos of maize inbred line Zong31 according to the Agrobacterium-mediated transformation method using Agrobacterium strain LBA4404. The positive lines were determined by PCR analysis with primers listed in Supplementary Data 3. As harvesting shoot tip is lethal to plant, we used leaves for expression analysis of the target gene in OE lines. The OE2, OE3, and OE3-F1 pollens were crossed to transgenic background Zong31 to generate OE2-T1, OE3-F1, and OE3-BC1F1 for analyzing their LNPE and anthesis data, respectively.
For the knockout line, gRNA was designed using CRISPR-P (http://crispr.hzau.edu.cn/CRISPR2/), introduced into the CRISPR/Cas9 binary vector VK005-02, and then transformed into immature embryos of maize inbred line CAL with the same method for generating OE lines. An EMS mutant was obtained from an EMS mutant library38. The gene editing patterns and EMS mutant site were determined using PCR amplification and sequencing with primers listed in Supplementary Data 3.
The EMS mutation was crossed into B73 (the mutant background) to generate F2 population. Each F2 individual was sequenced to distinguish its genotype, and the homozygous mutant (AA genotype) and homozygous WT (GG genotype) were used for phenotyping. For knockout lines, the leaf number above ear and days to anthesis were compared to their non-transformed individuals using T1 plants which separated from the same T0 heterozygous plants. A two-tailed Student’s t-test was used to determine the significant differences between WT and mutants/ knockout lines.
RT-PCR analysis and in situ hybridization of ZmOM66 in NILZ-Lfy1 and NILZ58
The tassel at emergence stage, unpollinated ear and husk at R1 stage, root and leaf at V2 stage, seed at 12 days after pollination (DAP12), coleoptile at 5 days after germination were used to compared ZmOM66 expression level in different tissues. The shoot tips from V1 to V7 stages were harvested for analyzing ZmOM66 expression during floral transition. For detecting circadian clock gene expressions, newly developed mature leaves were collected from ZT0 to ZT24 with an interval of 4 h at the V5 stage. All samples from five plants were pooled for each biological replication, and three replications were sampled for RNA extraction (Tiangen, Beijing, China) and Real-Time PCR analysis with primers listed in Supplementary Data 3. The quantification method (2-ΔCt) was used to calculate gene expression, and the ACTIN gene was used as an internal control.
The probe sequences for in situ hybridization were listed in Supplementary Data 3. The shoot tips at V4 stage were immediately placed in the in situ hybridization fixative (Wuhan Servicebio Technology Co., Ltd), and was pumped and stored at 4 °C. The tissue was dehydrated by increasing alcohol solutions (30%, 50%, 75%, 85%, 90%, 95% and 100%), and embedded in wax. Then, the tissue was sliced by 6 μm thick, and the slices were added in pre-hybridization solution and incubated at 65 °C for 25 min. After discarding the pre-hybridization solution, slices were added to the hybridization solution with probes, and hybridized overnight at 40 °C. In the next day, the hybridization solution was removed, and the slices were washed by 2×SSC, 1×SSC, and 0.5×SSC in turn. Adding the corresponding branch probe and corresponding signal probe, slices were washed by SSC buffer in turn. After blocking, the slices were added anti-DIG-AP and incubated at 40 °C for 50 min. After washing with PBS and adding BCIP/NBT solution, signals were observed in microscope.
Subcellular localization
The ORFs of ZmOM66 and dek36 were amplified from B73, which were further cloned into pEarleyGate101 and pCAMBIA1300 to construct ZmOM66-YFP and DEK36-mCherry fusion vectors, respectively. Both vectors were transformed into Agrobacterium strain GV3101 and co-infiltrated into tobacco leaves. After 48 h of incubation, fluorescence signals were observed using a laser confocal microscope LSM 980 (Zeiss, Jena, Germany). ZmOM66-YFP fusion vector was transformed into maize leaf protoplasts by polyethylene glycol-mediated DNA transfection70. Fluorescence signals were observed using the LSM 980.
Agrobacterium strain GV3101 with the ZmOM66-YFP fusion vector was transformed into Arabidopsis Col-0 using the flower dip method. The root hairs of transgenic plants were used for YFP observation before and after staining with MitoTracker orange (Invitrogen, Carlsbad, CA, USA). Fluorescence signals were observed under the LSM 980.
Yeast two-hybrid, BiFC and LCI assays
For Y2H assay, ZmOM66 ORF was cloned into pGBKT7 and pGADT7 vectors, and Zm00001d039183 ORF into pGBKT7. The vector pairs were co-transformed into yeast strain Y2HGold using a Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA, USA). The yeast cells were grown on a base medium lacking Trp and Leu (SD/-Trp-Leu) and then transformed to a medium lacking Trp, Leu, Ade, and His (SD/-Trp-Leu-Ade-His). Protein interactions were observed after 72 h incubation at 30 °C.
For BiFC assay, ZmOM66 ORF was cloned into pEarleygate201 (with nYFP) and pEarleygate202 (with cYFP), and Zm00001d039183 ORF into pEarleygate202. Agrobacterium strain GV3101 containing infused nYFP and cYFP vectors was co-infiltrated into tobacco leaves. After 48 h incubation, fluorescence signals were observed using the LSM980.
For LCI assay, ZmOM66 ORF was cloned into pCAMBIA1300-nLUC and pCAMBIA1300-cLUC, and Zm00001d039183 ORF into pCAMBIA1300-cLUC. Agrobacterium strain GV3101 containing nLUC and cLUC was co-infiltrated into tobacco leaves. After 48 h incubation, 100 mM luciferin was spread on the leaf surface, and firefly luciferase activity was detected using a NightShade LB985 In Vivo Plant Imaging System (Berthold Technologies, Bad Wildbad, Germany). The primers for vector construction are shown in Supplementary Data 3.
Transcriptome sequencing
At V5 stage, the shoot tips, newly developed immature leaves, and mature leaves were harvested at ZT0. Samples from five plants were pooled for each biological replication, and three replications were sampled for RNA extraction (Tiangen, Beijing, China). RNA sequencing was conducted using DNBSEQ-T7 (Annoroad Gene Technology, Beijing, China).
All sequenced reads from each sample were aligned to the B73 reference genome using HISAT2. The raw read number was used to calculate the FPKM for each gene. DEGs were identified using the DESeq2 R package with a |log2fold change | ≥ 1 and a False Discovery Rate (FDR)-adjusted P-value < 0.05. The agriGO online website (http://systemsbiology.cau.edu.cn/agriGOv2/) was used to perform gene ontology analysis under a threshold of FDR < 0.05.
Starch content quantification and photosynthesis measurements
At V5 stage, newly developed mature leaves were collected for starch analysis at ZT0 and ZT16, and for photosynthesis analysis at ZT8. Starch was determined using a Total Starch Assay Kit (Megazyme, Wicklow, Ireland), and Phi2, PhiNPQ, and PhiNO were measured using Photosynq MultispeQ (Photosynq, Michigan, USA). All measurements were conducted with three replicates, and three plants were harvested for each replicate. A two-tailed Student’s t-test was used to determine the significant differences between genotypes.
Metabolite profiling
At the V5 stage, newly developed mature leaves were collected at ZT0 for metabolite analysis. Twenty milligrams of powder were diluted into a 500 μL solution of methanol: isopropanol: water (3:3:2 V/V/V), vortexed for 3 min, and ultrasound for 30 min. The sample was mixed with 100 μL of 15 mg/mL methoxyamine hydrochloride in pyridine. The mixture was incubated at 37 °C for 2 h. Then, 100 μL N,O-Bis (trimethylsilyl) trifluoroacetamide was added to the mixture, kept at 37 °C for 30 min after vortex mixing, and then used for GC-MS analysis in an Agilent 7890B gas chromatograph coupled to a 7000D mass spectrometer with a DB-5MS column (J & W Scientific, Folsom, CA, USA). Significant differences in sugar content between ZmOM66 OE and WT were determined using a two-tailed Student’s t-test analysis.
For energy metabolite analysis, 0.05 g powder mentioned above was mixed with 500 µL 70% methanol (precooled at −20 °C). After vertexing for 5 min at 587 × g and being kept on ice for 5 min, the mixture was centrifuged at 13,523 × g for 5 min at 4 °C. Then, 400 μL supernatant was transferred into a new centrifuge tube and kept at −20 °C for 30 min. The supernatant was centrifuged again at 21,130 × g for 20 min at 4 °C. Then, 200 μL supernatant was collected for LC-MS analysis using the QTRAP 6500 LC/MS/MS System (SCIEX, Framingham, MA, USA). Significant differences in metabolites between ZmOM66 OE lines and WT were determined by Variable importance in Projection (VIP) ≧ 1.
Phenotypic analysis of ZD958 and ZD958Lfy1
The ZD958 and ZD958Lfy1 were sown in Zhuozhou, Hebei Province, China. Each hybrid was over-seeded in an isolated plot and manually thinned at the V5 stage to ensure an expected plant density. The row space was 60 cm, and plant distance within row was 25 cm. After pollen shedding, 25 plants for each trail were randomly selected for evaluating leaf numbers, LNPE, plant height, and ear height. More than 180 plants were selected for evaluating days to silking and days to anthesis. After harvest, 30 ears in the middle of each trial were randomly selected for measuring grain weight and ear length. A two-tailed Student’s t-test was used to determine the significant differences between genotypes.
Protoplast transient assays
The Copia LTR-RT sequence was amplified from the Lfy1 mutant and inserted into the pGL3 vector in front of the cauliflower mosaic virus (CaMV) 35S minimal promoter to drive firefly luciferase gene expressions. Transient expression assays were performed in maize protoplasts by polyethylene glycol-mediated DNA transfection70, and fluorescence signals were measured using the Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA). The empty pGL3 vectors were used as controls, and six biological replicates were assayed for each vector.
Whole-genome bisulfite sequencing (WGBS-seq)
Shoot tips were harvested from plants at the V5 stage for genome DNA extraction using the cetyl-trimethyl-ammonium bromide method. Samples from five plants were pooled for each biological replication, and two replications were sampled for WGBS-seq using Illumina Novaseq6000 (Annoroad Gene Technology, Beijing, China). The adapter sequences and low-quality reads were trimmed with Trimmomatic. The clean data was aligned to the reference genome by Bismark with default options. The uniquely mapped reads were retained for further analysis. After removing the cloned reads generated by PCR amplification, only the C sites covered with at least 4 reads were considered to estimate the methylation level. The CG, CHG, and CHH methylation levels of the targeted region were estimated as the weighted methylation71. The differential methylation analysis was performed with Metilene72.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-seq and WGBS-seq datasets are available from the China National Center for Bioinformation with BioProject accession of PRJCA022542. The genomic DNA sequences of ZmOM66 gene and the Copia LTR retrotransposon in the candidate region in Lfy1 mutant are accessible under NCBI GenBank accession PV178999 and PV179000. Source data are provided in this paper.
References
Freytes, S., Canelo, M. & Cerdán, P. Regulation of flowering time: when and where? Curr. Opin. Plant. Biol. 63, 102049 (2021).
Bluemel, M., Dally, N. & Jung, C. Flowering time regulation in crops-what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32, 121–129 (2015).
Bouché, F., Lobet, G., Tocquin, P. & Périlleux, C. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic. Acids. Res. 44, D1167–D1171 (2016).
Colasanti, J., & Muszynski, M. in Handbook of Maize: its Biology. 41–55 (Springer, 2009).
Colasanti, J., Yuan, Z. & Sundaresan, V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93, 593–603 (1998).
Kozaki, A., Hake, S. & Colasanti, J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic. Acids. Res. 32, 1710–1720 (2004).
Coneva, V., Guevara, D., Rothstein, S. J. & Colasanti, J. Transcript and metabolite signature of maize source leaves suggests a link between transitory starch to sucrose balance and the autonomous floral transition. J. Exp. Bot. 63, 5079–5092 (2012).
Muszynski, M. G. et al. delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142, 1523–1536 (2006).
Hung, H. et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 109, E1913–E1921 (2012).
Dong, Z. et al. A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS ONE 7, e43450 (2012).
Huang, C. et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 115, E334–E341 (2018).
Jin, M. et al. ZmCOL3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J. Integr. Plant Biol. 60, 465–480 (2018).
Stephenson, E. et al. Over-expression of the photoperiod response regulator ZmCCT10 modifies plant architecture, flowering time and inflorescence morphology in maize. PLoS ONE 14, e0203728 (2019).
Su, H. et al. Identification of ZmNF-YC2 and its regulatory network for maize flowering time. J. Exp. Bot. 72, 7792–7807 (2021).
Yang, J. et al. ZmSPL13 and ZmSPL29 act together to promote vegetative and reproductive transition in maize. New Phytol. 239, 1505–1520 (2023).
Danilevskaya, O. N., Meng, X., Hou, Z., Ananiev, E. V. & Simmons, C. R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146, 250–264 (2008a).
Alter, P. et al. Flowering time-regulated genes in maize include the transcription factor ZmMADS1. Plant Physiol. 172, 389–404 (2016).
Castelletti, S. et al. Maize adaptation across temperate climates was obtained via expression of two florigen genes. PLoS Genet. 16, e1008882 (2020).
Sun, H. et al. dlf1 promotes floral transition by directly activating ZmMADS4 and ZmMADS67 in the maize shoot apex. New Phytol. 228, 1386–1400 (2020).
Danilevskaya, O. N. et al. Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol. 147, 2054–2069 (2008b).
Shaver, D. Genetics and breeding of maize with extra leaves above the ear. in Proc. 38th Annual Corn and Sorghum Industry Research Conference 161–180 (Chicago, IL: American Seed Trade Association,1983).
Muirhead, R. C., & Shaver, D. L. Genetic factor capable of altering leaf number and distribution in maize. US Patent US4513532A (1985).
Bomblies, K. et al. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130, 2385–2395 (2003).
Subedi, K. D. & Ma, B. L. Ear position, leaf area, and contribution of individual leaves to grain yield in conventional and leafy maize hybrids. Crop Sci. 45, 2246–2257 (2005).
Zsubori, Z. T., Hegyi, I. P. Z. & Marton, C. L. Genotype and year effects on morphological and agronomical traits of silage maize (Zea mays L.) hybrids. Acta. Antiq. Hung. 58, 81–89 (2010).
Andrews, C. J., Dwyer, L. M., Stewart, D. W., Dugas, J. A. & Bonn, P. Distribution of carbohydrate during grainfill in leafy and normal maize hybrids. Can. J. Plant Sci. 80, 87–95 (2000).
Costa, C., Dwyer, L. M., Stewart, D. W. & Smith, D. L. Nitrogen effects on grain yield and yield components of leafy and nonleafy maize genotypes. Crop Sci. 42, 1556–1563 (2002).
Subedi, K. D., Ma, B. L. & Smith, D. L. Response of a leafy and non-leafy maize hybrid to population densities and fertilizer nitrogen levels. Crop Sci. 46, 1860–1869 (2006).
Modarres, A. et al. Leafy reduced-stature maize for short-season environments: morphological aspects of inbred lines. Euphytica 96, 301–309 (1997).
Pinter, J. et al. Utilizing Leafy genes as resources in quality silage maize breeding. Maydica 56, 243–250 (2011).
Oishi, H., Cai H. W. & Saruwatari, Y. Effects of Lfy1 gene on some characteristics of an inbred line and hybrids. in Paper Presented at 51st Maize Genetics Conference Abstracts 190 (St. Charles, IL 2009).
Du, X. et al. Fine mapping of Leafy, a dominant mutant conferring extra leaves above the ear in maize. Euphytica 206, 49–56 (2015).
Cui, M. et al. Genetic mapping of the leaf number above the primary ear and its relationship with plant height and flowering time in Maize. Front. Plant Sci. 8, 1437 (2017).
Li, D. et al. The genetic architecture of leaf number and its genetic relationship to flowering time in maize. New Phytol. 210, 256–268 (2016).
Li, Z., Li, K., Yang, X., Hao, H. & Jing, H. Combined QTL mapping and association study reveals candidate genes for leaf number and flowering time in maize. Theor. Appl. Genet. 134, 3459–3472 (2021).
Wei, L. & Cao, X. The effect of transposable elements on phenotypic variation: insights from plants to humans. Sci. China Life Sci. 59, 24–37 (2016).
Du, X. et al. Transcriptome analysis of leafy near-isogenic lines provides molecular insights into floral transition in maize (Zea mays). Plant Breed. 139, 883–891 (2020).
Lu, X. et al. Gene-indexed mutations in maize. Mol. Plant 11, 496–504 (2018).
Zhang, B. et al. The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J. 80, 709–727 (2014).
Nagahama, M. et al. NVL2 is a nucleolar AAA-ATPase that interacts with ribosomal protein L5 through its nucleolar localization sequence. Mol. Biol. Cell. 15, 5712–5723 (2004).
Okreglak, V. & Walter, P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. USA 111, 8019–8024 (2014).
Zhang, G., Li, S., Cheng, K. W. & Chou, T. F. AAA ATPases as therapeutic targets: Structure, functions, and small-molecule inhibitors. Eur. J. Med. Chem. 219, 113446 (2021).
Wang, G. et al. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 214, 1563–1578 (2017).
Pan, Y., Zhan, J., Jiang, Y., Xia, D. & Scheuring, S. A concerted ATPase cycle of the protein transporter AAA-ATPase Bcs1. Nat Commun. 14, 6369 (2023).
Smith, A. M., Zeeman, S. C. & Smith, S. M. Starch degradation. Annu. Rev. Plant. Biol. 56, 73–98 (2005).
MacNeill, G. J. et al. Starch as a source, starch as a sink: the bifunctional role of starch in carbon allocation. J. Exp. Bot. 68, 4433–4453 (2017).
Buchanan, B. B., Gruissem, W., & Jones, R. L. Biochemistry and Molecular Biology of Plants. (John Wiley & sons, 2015).
Martins, M. C. M. et al. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol. 163, 1142–1163 (2013).
Wahl, V. et al. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339, 704–707 (2013).
Fernie, A. R., Carrari, F. & Sweetlove, L. J. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant. Biol. 7, 254–261 (2004).
Viana, A. J. et al. The sugar-responsive circadian clock regulator bZIP63 modulates plant growth. New Phytol. 231, 1875–1889 (2021).
Buckler, E. S. et al. The genetic architecture of maize flowering time. Science 325, 714–718 (2009).
Zhao, Y. et al. The evening complex promotes maize flowering and adaptation to temperate regions. Plant Cell 35, 369–389 (2023).
Colasanti, J. & Coneva, V. Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiol. 149, 56–62 (2009).
Salvi, S. et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104, 11376–11381 (2007).
Studer, A., Zhao, Q., Ross-Ibarra, J. & Doebley, J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 43, 1160–1163 (2011).
Zhang, L. et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat Commun. 10, 1494 (2019).
Smith, J., Sen, S., Weeks, R. J., Eccles, M. R. & Chatterjee, A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer 6, 392–406 (2020).
Xu, J. et al. Cellular Heterogeneity-Adjusted cLonal Methylation (CHALM) improves prediction of gene expression. Nat Commun. 12, 400 (2021).
Girard, C. et al. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11, e1005369 (2015).
Liu, Q. et al. NEEDLE1 encodes a mitochondria localized ATP-dependent metalloprotease required for thermotolerant maize growth. Proc Natl Acad Sci USA 116, 19736–19742 (2019).
Riglet, L. et al. KATANIN-dependent mechanical properties of the stigmatic cell wall mediate the pollen tube path in Arabidopsis. Elife 9, e57282 (2020).
Dolzblasz, A. et al. The mitochondrial protease AtFTSH4 safeguards Arabidopsis shoot apical meristem function. Sci Rep. 6, 28315 (2016).
Viñegra de la Torre, N. et al. FLOWERING REPRESSOR AAA+ ATPase 1 is a novel regulator of perennial flowering in Arabis alpina. New Phytol. 236, 729–744 (2022).
Iohannes, S. D. & Jackson, D. Tackling redundancy: genetic mechanisms underlying paralog compensation in plants. New Phytol. 240, 1381–1389 (2023).
Minow, M. A. A. et al. Distinct gene networks modulate floral induction of autonomous maize and photoperiod-dependent teosinte. J. Exp. Bot. 69, 2937–2952 (2018).
Welchen, E., Canal, M. V., Gras, D. E. & Gonzalez, D. H. Cross-talk between mitochondrial function, growth, and stress signalling pathways in plants. J. Exp. Bot. 72, 4102–4118 (2021).
Popov, V. N. et al. The uncoupling of respiration in plant mitochondria: keeping reactive oxygen and nitrogen species under control. J. Exp. Bot. 72, 793–807 (2021).
Nag, M., Lahiri, D., Garai, S., Mukherjee, D. & Ray, R. R. Regulation of β-amylase synthesis: a brief overview. Mol. Biol. Rep. 48, 6503–6511 (2021).
Lyznik, L. A. & Hodges, T. K. Polyethylene Glycol-mediated Dna Uptake Into Maize Protoplasts. 603–609 (Springer New York, 1994).
Schultz, M. D., Schmitz, R. J. & Ecker, J. R. Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28, 583–585 (2012).
Jühling, F. et al. Metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 26, 256–262 (2016).
Acknowledgements
We thank Dr. Lana M. Reid and Xiaoyang Zhu from Eastern Cereal and Oilseed Research Centre of Agriculture & Agri-Food Canada for providing CO412 inbred line; Dr. Yan Liu from CAAS for assisting with maize transformation; LetPub (www.letpub.com) for linguistic assistance and pre-submission expert review. This work was supported by the National Key Research and Development Program of China (2021YFD1200705), the National Natural Sciences Foundation of China (32201843 & 32372161), and the earmarked fund for China Agriculture Research System-Maize (CARS-02-13), the Chinese Universities Scientific Fund (2023TC188), and the Innovation Program of Chinese Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Contributions
W.G., D.X., G.R., and W.J. designed experiments. D.X. and X.Z. performed most of the experiments. D.X., G.R., and W.G. wrote the manuscript. L.J., G.X., Z.J., H.C., W.X., and A.J. analyzed the data. Chen Y., H.L., G.W., Cui Y., L.L., C.Y., L.Y., and F.J. interpreted the results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Joseph Colasanti and Jianbing Yan for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Du, X., Xu, Z., Lu, J. et al. A LTR retrotransposon insertion leads to leafy phenotype in maize by elevating ZmOM66 expression. Nat Commun 16, 3152 (2025). https://doi.org/10.1038/s41467-025-57811-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57811-9