Abstract
Nuclear RNA decay has emerged as a mechanism for post-transcriptional gene regulation in cultured cells. However, whether this process occurs in animals and holds biological relevance remains largely unexplored. Here, we demonstrate that MTR4, the central cofactor of the nuclear RNA exosome, is essential for embryogenesis and spermatogenesis. Embryonic development of Mtr4 knockout mice arrests at 6.5 day. Germ cell-specific knockout of Mtr4 results in male infertility with a specific and severe defect in meiotic initiation. During the pre-meiotic stage, MTR4/exosome represses meiotic genes, which are typically shorter in size and possess fewer introns, through RNA degradation. Concurrently, it ensures the expression of mitotic genes generally exhibiting the opposite features. Consistent with these regulation rules, mature replication-dependent histone mRNAs and polyadenylated retrotransposon RNAs were identified as MTR4/exosome targets in germ cells. In addition, MTR4 regulates alternative splicing of many meiotic genes. Together, our work underscores the importance of nuclear RNA degradation in regulating germline transcriptome, ensuring the appropriate gene expression program for the transition from mitosis to meiosis during spermatogenesis.
Similar content being viewed by others
Introduction
RNA degradation is a vital mechanism for ensuring the precision and accuracy of gene expression. For a considerable period, nuclear RNA decay pathway was primarily perceived as a mechanism for disposing of unwanted and abnormally transcribed or processed RNAs1,2,3,4,5,6. However, recent discoveries have unveiled its capacity to degrade a significant population of normally transcribed and processed mRNAs3,7,8. This revelation has positioned nuclear RNA decay as an important layer in the regulation of gene expression. Nevertheless, most investigations into nuclear RNA degradation have been conducted in cultured cell lines. To date, it remains largely unclear whether the nuclear degradation of normal RNAs actually occurs in animals and, if so, how it influences embryo and animal development.
The RNA exosome complex (exosome) represents the primary RNA degradation machinery in the cell, operating in both the nucleus and the cytoplasm9,10,11,12,13. To achieve its full activity, the exosome requires the binding of multiple cofactors. Among these cofactors, MTR4, a DEIH RNA helicase encoded by Mtr4/Mtrex, is indispensable for various aspects of nuclear exosome functions1,4,5,14,15,16,17,18. MTR4, on its own, functions to prepare RNA substrates for exosome processing while also competing with RNA export factors to determine substrate specificity8,19,20,21,22. Moreover, MTR4 interacts with distinct proteins to form different complexes that recruit the exosome to various types of RNA substrates. In its association with RBM7 and ZCCHC8, MTR4 targets promoter upstream transcripts (PROMPTs) and enhancer RNAs (eRNAs) for degradation1,3,5. Conversely, it interacts with ZFC3H1 to facilitate exosomal degradation of processed transcripts, such as pre-mature terminated RNAs (ptRNAs)2,3. Despite its critical molecular functions, the biological function of MTR4 in animals remains to be investigated.
In this study, we generated Mtr4 knockout mice and found that their embryo development ceased at a very early stage. Male germ cell-specific Mtr4 knockout mice displayed male infertility and severe defects in meiotic initiation. Transcriptome and MTR4-binding profile analyses in pre-meiotic germ cells revealed its complicated roles in regulating RNA degradation and alternative splicing, shaping the transcriptome for the mitosis-to-meiosis transition. Meanwhile, MTR4/exosome functions to remove polyadenylated retrotransposon RNAs that possess gene repression effects and mutagenic threats.
Results
Mtr4 knockout leads to embryonic lethality in mice
To investigate the role of MTR4 at the animal level, we generated Mtr4 knockout (KO)-first, conditional-ready mice23,24. The Mtr4 KO-first embryonic stem cells have a large DNA construct inserted into intron 5, leading to inhibited expression of Mtr4 exons 6 to 27 (Fig. 1A). Consequently, only a small N-terminal region (1-170 aa) of the MTR4 protein, which lacks most functional domains, could be produced (Fig. 1B).
A Strategy for constructing the Mtr4 KO-first, conditional-ready allele of Mtr4tm1a(KOMP)Wtsi mice. En2 SA, mouse En2 splicing acceptor; IRES, internal ribozyme entry site; lacZ, lair conditioner Z; pA, poly A site; hBactP, human beta actin promoter; neo, promoter-driven neomycin resistant gene; FRT, flippase recognition target; loxP, locus of crossover in P1. B Schematic diagram for MTR4 full length (FL) and mutant proteins. C Bar plot showing the genotype ratio of Mtr4-cKO pups 3 weeks after birth from heterozygous parents. D The morphology of E6.0 and E6.5 Mtr4+/+ and Mtr4-/- mouse embryos. Scale bars, 200 μm. Source data are provided as a Source Data file.
Mtr4+/- mice appeared viable and fertile, displaying a normal appearance (Supplementary Fig. 1A). Intercrossing Mtr4+/- mice resulted in the expected Mendelian ratio of Mtr4+/+ and Mtr4+/- pups, but never Mtr4-/- pups (Fig. 1C), demonstrating that loss of Mtr4 results in embryonic lethality. Further analysis revealed that Mtr4-/- embryos with a normal appearance could only be found in utero until embryonic day 6.0 (E6.0) (Fig. 1D, Supplementary Fig. 1B). At E6.5 or later time points, only abnormal embryos or empty/debris-filled conceptuses were observed (Fig. 1D). These data demonstrate that MTR4 is essential for early murine embryogenesis.
Germ cell-specific deletion of Mtr4 results in male infertility
We then proceeded to investigate the biological functions of MTR4 in mouse tissue development. RT-PCR and western blot data demonstrated that Mtr4 mRNA and protein were highly expressed in testis compared to other tissues (Fig. 2A, B). Consistent with these data, publicly available human tissue data (www.proteinatlas.org) also demonstrate the enrichment of the human MTR4 mRNA in the testis (Supplementary Fig. 1C). We thus turned our attention to exploring the role of MTR4 in spermatogenesis and male fertility.
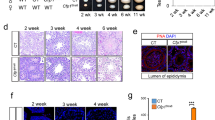
A RT-PCR to examine Mtr4 mRNA levels in different tissues from 8-week wild-type mice. 18S rRNA was used as a loading control. B Western blotting to examine MTR4 protein levels in different tissues from 8-week wild-type mice. GAPDH was used as a loading control. C Schematic diagram of STRA8 expression during mouse spermatogonia differentiation. D Schematic diagram for constructing Mtr4 conditional knockout allele. E Western blotting to examine MTR4 expression in testes in 8-week Cntl and Mtr4-cKO mice. Tubulin was used as a loading control. *: nonspecific band. F Summary of the fertility test of Cntl and Mtr4-cKO mice. G The morphology of 8-week Cntl and Mtr4-cKO mice testes. The bar plot shows the testicular/body weight ratio. Cntl, n = 4; Mtr4-cKO, n = 3. P-value is based on the two-sided unpaired student’s t test. Error bars, mean ± SEM, ***P = 3.82e-07. H H&E staining of the sections from 8-week Cntl and Mtr4-cKO testes (left) and epididymis (right). Cntl, n = 4; Mtr4-cKO, n = 3 biological repeats. Scale bars, 50 μm. I Immunofluorescence staining of DDX4 in 8-week Cntl and Mtr4-cKO testis sections. Cntl, n = 4; Mtr4-cKO, n = 3 biological repeats. Scale bar, 20 μm. Source data are provided as a Source Data file.
To examine the impact of MTR4 on spermatogenesis, we took advantage of Stra8-GFPCre mice, which induced recombination starting from type A1 spermatogonia (Fig. 2C). We generated Mtr4F/△;Stra8-GFPCre mice in the C57BL/6J background (Fig. 2D, Supplementary Fig. 1D, E), allowing for a conditional knockout (cKO) of Mtr4 specifically in differentiated spermatogonia. Western blot and RT-qPCR confirmed efficient Mtr4 knockout in the testes (Fig. 2E, Supplementary Fig. 1F). Hereafter, we referred to these mice as “Mtr4-cKO” mice.
Despite Mtr4-cKO mice maintaining a normal body size (Supplementary Fig. 1G), males exhibited complete infertility (Fig. 2F). The testes of adult Mtr4-cKO mice were significantly smaller and lighter than those of their littermate controls (Cntls) (Fig. 2G). In line with the infertility phenotype, hematoxylin and eosin (H&E) staining revealed an absence of sperm in the epididymis of Mtr4-cKO mice (Fig. 2H). Strikingly, the seminiferous tubules of Mtr4-cKO mice appeared narrow and almost devoid of germ cells, with only a single outer layer of germ cells near the basal lamina and a number of somatic Sertoli cells (Fig. 2H). DDX4 (also known as VASA) is expressed in various types of germ cells25 (Fig. 3B). Immunostaining using an anti-DDX4 antibody demonstrated a striking reduction in the number of DDX4+ cells in the Mtr4-cKO testes (Fig. 2I). Together, these data underscore the critical role of MTR4 in spermatogenesis and male fertility.
A H&E staining of the sections from Cntl and Mtr4-cKO testes at P8, P9, P10 and P12. Red arrows indicate the representative spermatocytes. P8, n = 2; P9, P10, P11, n = 3 biological repeats. Scale bars, 20 μm. B Schematics diagram of the expression of DDX4, PLZF, STRA8, c-KIT, γH2A.X, and SYCP3 during the early stage of mouse spermatogenesis process. Undiff spg, undifferentiated spermatogonia; A1, type A1 spermatogonia; A2-A4, type A2, type A3 and type A4 spermatogonia, respectively; In-B, intermediate and type B spermatogonia, respectively; pL, preleptotene spermatocyte; L/Z/P/D, leptotene, zygotene, pachytene and diplotene spermatocyte, respectively; MI-MII, meiosis 1 and meiosis 2 spermatocyte metaphase, respectively. C Immunofluorescence staining of STRA8 in P8 Cntl and Mtr4-cKO testis sections. Scale bar, 20 μm. The bar plot shows the quantification of STRA8+ cells per seminiferous tubule. Cntl, n = 47; Mtr4-cKO, n = 48. P-value is based on the two-sided unpaired student’s t test. Error bars, mean ± SEM, n.s. P = 0.5743. D Immunofluorescence staining of DDX4 in P8 Cntl and Mtr4-cKO testis sections. Scale bar, 50 μm. The bar plot shows the population of DDX4+ cells in each seminiferous tubule. Cntl, n = 43; Mtr4-cKO, n = 39. P-value is based on the two-sided unpaired student’s t test. Error bars, mean ± SEM, n.s. P = 0.2921. Source data are provided as a Source Data file.
MTR4 mutant spermatogenesis defects emerge at P9
To characterize the defects in Mtr4-cKO testes, we conducted H&E staining on testis sections from Cntl and Mtr4-cKO mice at P7, P14 and P21. We did not observe any discernible difference in the testicular tubules between the Cntl and Mtr4-cKO mice at P7 (Supplementary Fig. 1H), although Stra8 starts to express at P326,27. A remarkable reduction in cell counts was observed in the testes of P14 and P21 Mtr4-cKO mice (Supplementary Fig. 1H).
To pinpoint when the defects in spermatogenesis initially emerged in Mtr4 mutant mice, we examined additional time points between P7 and P14, specifically at P8, P9, P10, and P12. No apparent defect was observed in Mtr4-cKO mice at P8 (Fig. 3A). In Cntl testes, dark H&E-stained spermatocytes began to appear in the cavities of seminiferous tubules at P9 and accumulated further at P10 and P12 (Fig. 3A). In stark contrast, very few such dark-stained cells were visible in the testes of Mtr4-cKO mice from P9 through P12 (Fig. 3A). Given that meiotic initiation typically begins around P9, these data imply that Mtr4 mutant germ cells may exhibit defects either prior to or during the meiosis initiation stage.
MTR4 is required for meiotic initiation
Prior to meiosis initiation, undifferentiated spermatogonia proliferate and differentiate, followed by several rounds of mitosis (Fig. 3B). To further elucidate the role of MTR4 in spermatogenesis, we conducted immunostaining using germ cell development markers to dissect defects in Mtr4-cKO mice. In accordance with the fact that Stra8 expression initiates from type A1 spermatogonia28, there was no change in the number of undifferentiated spermatogonia stained with the PLZF antibody in Mtr4-cKO versus Cntl mice (Fig. 3B, Supplementary Fig. 2A).
STRA8 is abundantly expressed in both A1 spermatogonia and preleptotene (pL) spermatocytes28 (Fig. 3B). To examine whether MTR4 plays a role in spermatogonial differentiation, we conducted STAR8 immunostaining at a stage prior to the emergence of pL spermatocytes (P7 and P8). No difference in the number of STRA8+ cells between the Cntl and Mtr4 mutant testes was observed at this stage (Fig. 3C, Supplementary Fig. 2B), demonstrating that spermatogonial differentiation proceeded normally in Mtr4-cKO mice. Further, the population of DDX4+ cells remained unaffected in Mtr4-cKO mice at P8 (Fig. 3D), indicating that the mitotic progression of mutant germ cells was not apparently impaired.
In Mtr4 mutant testes, germ cell counts were reduced at P9, coinciding with the emergency of pL spermatocytes during regular spermatogenesis (Fig. 3A). To distinguish pL and A1 type germ cells that both abundantly express STRA8 (Fig. 3B), we performed double staining for STRA8 and γH2A.X, the latter of which is a marker for double-strand DNA breaks induced as part of the meiotic recombination process29 (Fig. 3B). In line with the undisturbed spermatogonial differentiation, STRA8+γH2A.X- (A1) spermatogonia were normally detected in Mtr4-cKO testes at P9 (Fig. 4A, Supplementary Fig. 2C). In contrast, although STRA8+γH2A.X+ cells were abundantly detected in the cavities of about 11% of seminiferous tubules in the Cntl testes, they were rarely observed in the mutant (Fig. 4A, Supplementary Fig. 2C). This indicates that meiotic initiation was severely impaired in the absence of MTR4.
A Co-staining of STRA8 and γH2A.X in P9 Cntl and Mtr4-cKO testis sections. Scale bar, 50 μm. The bar plots show the ratio of the STRA8+γH2A.X- (Cntl, n = 9; Mtr4-cKO, n = 8, left) and STRA8+γH2A.X+ (Cntl, n = 8; Mtr4-cKO, n = 8, right) tubules per section. P-values are based on the two-sided unpaired student’s t test. Error bars, mean ± SEM, n.s. P = 0.0586, ***P = 4.68e-07. B Chromosome spreads of 3-week Cntl spermatocytes and Mtr4-cKO spermatocyte-like cells were co-stained of SYCP3, γH2A.X and DAPI. Scale bars, 10 μm. The bar plot shows the quantification of leptotene/leptotene-like (Cntl, n = 35, 49; Mtr4-cKO, n = 10, 7), zygotene/ zygotene-like (Cntl, n = 82, 93; Mtr4-cKO, n = 1, 2), pachytene (Cntl, n = 117, 101; Mtr4-cKO, n = 0, 0) and diplotene (Cntl, n = 53, 56; Mtr4-cKO, n = 0, 0) spermatocytes. Error bars, mean ± SEM. Source data are provided as a Source Data file.
In agreement with impaired meiotic initiation, cells positively stained with SYCP3, a component of the synaptonemal complex formed particularly during meiosis30 (Fig. 3B), were plentiful in Cntl testes but hardly detected in Mtr4-cKO (Supplementary Fig. 2D). Terminal dUTP nick-end labeling (TUNEL) revealed that more germ cells were positively stained in Mtr4 mutant testes (Supplementary Fig. 2E), suggesting that mutant germ cells that failed to undergo meiotic initiation might be eliminated through the apoptotic pathway. Collectively, these data demonstrate that MTR4 plays an essential role in meiotic initiation during spermatogenesis.
The small population of pL/pL-like Mtr4 mutant spermatocytes cannot complete meiosis
In Mtr4-cKO mutant mice, we noticed that although most spermatogonia were unable to initiate meiosis, a small population exhibited a staining pattern characteristic of pL cells (Supplementary Fig. 2C). We thus examined whether these mutant germ cells could complete meiosis by tracking the progression of meiosis based on the staining patterns of SYCP3 and γH2A.X in spermatocyte nuclear spreads. In Cntl mice, as expected, meiosis progressed normally through leptotene, zygotene, pachytene, and diplotene stages (Fig. 4B). In contrast, in Mtr4-cKO mice, while a few leptotene- and zygotene-like spermatocytes could be observed, no pachytene or diplotene spermatocyte was detected (Fig. 4B). Therefore, although a few MTR4-deficient spermatogonia could progress into spermatocytes, they were unable to accomplish meiosis.
Evidence that MTR4 is involved in nuclear exosomal RNA degradation in mice
To understand how MTR4 functions in spermatogenesis, we examined whether it is involved in nuclear exosomal degradation in mice. Consistent with a previous study31, MTR4 predominantly localized in the nuclei of germ cells (Supplementary Fig. 3A), consistent with its location in HeLa cells8,10. Furthermore, immunoprecipitation (IP) with an anti-MTR4 antibody in RNase-treated mouse testis homogenate revealed its association with exosome components, such as EXOSC10 and EXOSC5, as well as nuclear exosome co-factor ZFC3H1 and nuclear cap-binding protein CBP802,8,32 (Supplementary Fig. 3B).
Moreover, consistent with findings in cultured cells7,21,33, we observed polyA RNA accumulation in the nuclei of Mtr4 mutant germ cells compared to Cntls using fluorescence in situ hybridization (FISH) with an oligo(dT) probe (Supplementary Fig. 3C). Notably, this accumulation was already evident at P8 in mutant testes (Fig. 5A), even though no apparent spermatogenesis defect was observed at this stage (Fig. 3A). These results collectively indicate that, similar to its roles in cultured cells, MTR4 is involved in nuclear exosomal RNA degradation in mice.
A FISH analysis of polyA RNA distribution using oligo(dT) probes in P8 Cntl and Mtr4-cKO testis sections. DAPI was stained to mark the nuclei. Cntl, n = 2; Mtr4-cKO, n = 2 biological repeats. Scale bars, 10 μm. B A diagram for the cell sorting and polyA RNA-seq experimental procedure. C Volcano plots showing the expression changes of all genes (18,168 genes analyzed, left) and protein-coding genes (14,558 genes analyzed, right) in Mtr4-cKO germ cells compared to the corresponding Cntls. D Gene Ontology (GO) term analysis of all differentially expressed (DE) genes. P-values are based on hypergeometric test corrected for multiple hypothesis tests using the Benjamini-Hochberg method. E Screenshots of RNA-seq signals of representative upregulated meiotic gene (Spo11, upper) and downregulated mitotic gene (Vcp, lower). Spo11: FC = 2.598, FDR = 8.68e-4; Vcp: FC = 0.435, FDR = 1.29e-4. F Schematics diagram of mitotic and meiotic stages of the mouse spermatogenesis process. G Heatmap showing the expression of DE genes in our RNA-seq dataset (left) and at individual spermatogenic stages from sc-RNA seq dataset (right). A1, type A1 spermatogonia; In, intermediate spermatogonia; BS, S phase type B spermatogonia; BG2M, G2/M phase type B spermatogonia; G1, G1 phase preleptotene spermatocyte; epL, early S phase preleptotene spermatocyte; mpL, middle S phase preleptotene spermatocyte; lpL, late S phase preleptotene spermatocyte. H MTR4/exosome represses meiotic gene expression and maintains mitotic gene expression directly or indirectly. Source data are provided as a Source Data file.
Mtr4 mutant pre-meiotic spermatogonia demonstrate a transcriptome with mixed mitotic/meiotic features
To identify the causes of impaired meiotic initiation in Mtr4-cKO mice, we isolated c-KIT+ male germ cells (differentiated spermatogonia) from Cntl and Mtr4 mutant testes at P834, and carried out whole-cell polyA RNA-seq analysis to specifically detect changes in processed RNAs (Figs. 3B, 5B). We performed three biological replicates, which showed strong correlations for both Cntl and Mtr4-cKO samples (Supplementary Fig. 4A). RNA-seq signals showed that Mtr4 was nearly completely knocked out in Mtr4-cKO germ cells (Fig. 2D, Supplementary Fig. 4B). As expected, PROMPTs and ptRNAs, known nuclear exosome substrates in cultured cells2,5, were globally accumulated in the mutant germ cells (Supplementary Fig. 4C), further confirming the role of MTR4 in nuclear exosomal RNA degradation in mice.
We identified a total of 1145 upregulated genes and 2805 downregulated genes (1.5-fold change, FDR (Wald test, Benjamini-Hochberg adjusted) <0.05) (Fig. 5C). The majority of the upregulated genes appeared to be noncoding (784 in 1145), suggesting that MTR4 primarily targets noncoding RNAs (ncRNAs) for degradation in pre-meiotic cells (Fig. 5C). In contrast, over 95% of the downregulated genes are protein-coding (Fig. 5C). Notably, a higher population of upregulated genes showed more than a 2-fold change compared to the downregulated genes upon Mtr4 knockout (Supplementary Fig. 4D). Consistent with the meiosis initiation defects observed in the Mtr4-cKO germ cells, gene ontology (GO) analysis revealed that these differentially expressed genes (DEGs) were enriched in Biological Process (BP) pathways related to mitotic and meiotic regulation (Fig. 5D). For instance, Spo11, a gene essential for meiosis, was upregulated35; while Vcp, critical for mitosis, was downregulated in Mtr4-cKO germ cells36 (Fig. 5E).
To gain a comprehensive understanding of the impact of MTR4 deletion on the gene expression program of pre-meiotic spermatogonia, we analyzed the expression profiles of DEGs in Mtr4 mutant spermatogonia throughout regular spermatogenesis. For this, we analyzed a previously published single-cell RNA sequencing (scRNA-seq) dataset that encompassed various types of synchronized spermatogenic cells37. Based on comprehensive analysis (see details in “Methods”), genes specifically expressed at mitotic stages (A1-BG2M) were considered as mitotic genes, while those specifically expressed at meiotic stages (G1-lpL) were classified as meiotic genes (Fig. 5F).
Remarkably, a distinct pattern emerged for DEGs in Mtr4-cKO germ cells: most upregulated genes (54.4%) were induced after meiosis initiation, while the majority of downregulated (73.6%) genes were repressed as spermatogenesis progressed to meiotic stages (Fig. 5G). Thus, a portion of the pre-meiotic transcriptome pre-maturely exhibited meiotic characteristics in response to MTR4 deletion. This indicates that MTR4 functions in promoting mitotic gene expression while repressing meiotic genes (Fig. 5H).
MTR4 differentially regulates genes with distinct length and intron number
To understand how MTR4 affects pre-meiotic germ cell transcriptome, we systematically identified gene features correlating with gene expression changes in Mtr4-cKO germ cells. The features we used included gene length, mature RNA length, intron number, and guanine and cytosine (GC) content of the mature RNAs. We found that gene length and intron number are important features correlating with gene expression in Mtr4-cKO germ cells (Supplementary Fig. 4E).
Shorter genes tend to be upregulated, while longer genes are more likely to be downregulated (Fig. 6A). Similarly, genes with fewer introns tend to be upregulated, while those with more introns are downregulated (Fig. 6B). When genes were grouped by length, the impact of intron number on RNA level changes was noticeable across all categories (Supplementary Fig. 4F). Conversely, when genes were grouped by intron number, gene length was found to influence RNA expression only in gene groups with fewer introns (<9) (Supplementary Fig. 4G). The impacts of gene length and intron number became even more evident when separating genes into upregulated, unchanged and downregulated groups in Mtr4-cKO germ cells (Fig. 6C, D). Upregulated genes were generally shorter and possessed fewer introns compared to unchanged ones, while downregulated genes were longer and contained more introns (Fig. 6C, D). Thus, in pre-meiotic germ cells, MTR4 appears to repress shorter genes with fewer introns and ensure the expression of longer genes with more introns.
A Violin plot showing the expression changes of genes of varying length in Mtr4-cKO germ cells compared to the corresponding Cntls. n = 3625; 3625; 3625; 3625; 3624 from left to right. P-values are based on the two-sided unpaired Wilcoxon test. Medians are shown in the plot. B Violin plot showing the expression changes of genes with varying intron numbers in Mtr4-cKO germ cells compared to the corresponding Cntls. n = 2436; 4620; 3308; 3992; 3768 from left to right. P-values are based on the two-sided unpaired Wilcoxon test. Medians are shown in the plot. C Violin plot showing length of genes exhibiting upregulated, no-changed and downregulated expression. Up, n = 1145; n/c, n = 14,175; down, n = 2805. P-values are based on the two-sided unpaired Wilcoxon test. n: the number of differential expression genes. D Violin plot showing the intron number of genes exhibiting upregulated, no-changed and downregulated expression. Up, n = 651; n/c, n = 12,332; down, n = 2705. P-values are based on the two-sided unpaired Wilcoxon test. n: the number of differential expression genes. E Violin plot showing the length of upregulated meiotic genes and downregulated mitotic genes in Mtr4-cKO germ cells. Upregulated meiotic gene, n = 211; downregulated mitotic genes, n = 1389. P-value is based on the two-sided unpaired Wilcoxon test. F Violin plot showing intron number of upregulated meiotic genes and downregulated mitotic genes in Mtr4-cKO germ cells. Upregulated meiotic gene, n = 164; downregulated mitotic genes, n = 1,83. P-value is based on the two-sided unpaired Wilcoxon test. G Violin plot showing the length of specifically expressed genes at individual spermatogenic stages in sc-RNA seq dataset. BG2M, n = 1509; G1, n = 60. P-value is based on the two-sided unpaired Wilcoxon test. H Violin plot showing the intron number of specifically expressed genes at individual spermatogenic stages in sc-RNA seq dataset. BG2M, n = 1509; G1, n = 60. P-value is based on the two-sided unpaired Wilcoxon test. I Cumulative distribution of gene length of specifically expressed genes at BG2M and G1 stage. BG2M, n = 1509; G1, n = 60. P-value is based on the two-sided unpaired Wilcoxon test. J Cumulative distribution of intron number of specifically expressed genes at BG2M and G1 stage. BG2M, n = 1509; G1, n = 60. P-value is based on the two-sided unpaired Wilcoxon test. K Violin plots showing the expression level of Mtr4 (left) and Dis3 (right) at individual spermatogenic stages from sc-RNA seq dataset. BG2M, n = 58; G1, n = 204. P-values are based on the two-sided unpaired Wilcoxon test. L The expression of Mtr4/exosome reduces and the meiotic genes with shorter length and fewer introns activate during mitotic-meiotic transition. Data in (A–H, K) are presented as box and whisker plots. The central lines denote median value, while the bounds of box mark the 25th to 75th percentiles and whiskers refer to minimum and maxima values. Source data are provided as a Source Data file.
Mitotic and meiotic genes harbor distinct features
Given the roles of MTR4 in the differential regulation of mitotic and meiotic genes (Fig. 5H), we compared the length and intron number of upregulated meiotic and downregulated mitotic genes in Mtr4 mutant mice. Indeed, upregulated meiotic genes were shorter and had fewer introns than downregulated mitotic genes (Fig. 6E, F).
To further explore whether mitotic and meiotic genes generally possess distinct features, we analyzed the length and intron number of genes specifically expressed at each mitotic (A1, In, BS, and BG2M) and meiotic (G1, epL, mpL, and IpL) stage of regular spermatogenesis37. Intriguingly, we observed a transition at the boundary between mitosis and meiosis (BG2M-to-G1) (Fig. 6G, H). Germ cells exhibited a significant shift from predominantly expressing longer genes with more introns (BG2M in mitosis) to expressing shorter genes with fewer introns (G1 in meiosis) (Fig. 6I, J). Consistent with this shift, we also observed decreased expression levels of Mtr4 and exosome components during the transition from mitosis (A1-BG2M) to meiosis (G1-lpL) (Fig. 6K, Supplementary Fig. 5A–J). In contrast, RNA export factors did not display such downregulation (Supplementary Fig. 5K, L). These findings support the view that in pre-meiotic germ cells, MTR4/exosome represses meiotic genes, which are typically shorter and with fewer introns, while ensuring the expression of mitotic genes that are longer and contain more introns. As germ cells transition from mitosis to meiosis, the reduced expression of Mtr4 and exosome components might lead to increased expression of meiotic genes, facilitating meiosis initiation (Fig. 6L).
MTR4 binds to various types of RNA substrates
To gain further insights into how MTR4 differentially regulates mitotic and meiotic genes with different features, we aimed to profile MTR4 binding and examine the impact of gene length and intron number. Due to the insufficient yield of c-KIT+ spermatogonia at P8 for regular CLIP-seq, we utilized LACE-seq (Linear Amplification of cDNA Ends and sequencing), a technique developed to identify the binding profiles of RNA-binding proteins at single-cell level38,39 (Fig. 7A).
A A diagram for the cell sorting and LACE-seq experimental approach. B Violin plot showing the normalized MTR4 binding counts on RNAs of varying length expressed in Cntl germ cells. n = 2130; 2130; 2130; 2130; 2130 from left to right. P-values are based on the two-sided unpaired Wilcoxon test. Medians are shown in the plot. C Violin plot showing the normalized MTR4 binding counts on RNAs with varying intron numbers expressed in Cntl germ cells. n = 3422; 1161; 1944; 2031; 2022 from left to right. P-values are based on the two-sided unpaired Wilcoxon test. Medians are shown in the plot. D Metaprofile showing the MTR4 signals on mRNAs exhibiting upregulated, no-changed, and downregulated expression based on merged LACE-seq data. Up, n = 340; n/c, n = 11,115; down, n = 2661. E Same as (D), except that MTR4 signals on ptRNAs are shown. Up, n = 419; n/c, n = 396. F Same as (D), except that MTR4 signals on RDH and RIH mRNAs are shown. RDH, n = 65; RIH, n = 14. G Heatmap showing the expression of RDH and RIH genes in Cntl and Mtr4-cKO germ cells. H Screenshots of RNA-seq signals of representative upregulated RDH (left) and no-changed RIH (right) genes in Cntl and Mtr4-cKO germ cells. I Same as (D), except that MTR4 signals on histone mRNAs are shown. Up, n = 51; n/c, n = 18. J Bar plot showing the population of RDH and RIH genes exhibiting upregulated, no-changed, and downregulated expression in hMTR4 KD HeLa cells relative to the corresponding Cntls based on rRNA depleted RNA-seq. RDH, n = 61; RIH, n = 10. Data in (B, C) are presented as box and whisker plots. The central lines denote median value, while the bounds of box mark the 25th to 75th percentiles and whiskers refer to minimum and maxima values. Source data are provided as a Source Data file.
We conducted three replicates of MTR4 LACE-seq, which demonstrated a high level of reproducibility (Supplementary Fig. 6A). Based on LACE-seq data, we identified several GC-rich motifs near MTR4-binding sites (Supplementary Fig. 6B). MTR4 binding generally decreased as gene length and intron number increased (Fig. 7B, C), indicating a preference for shorter RNAs with fewer introns. This aligns with the preferential accumulation of short and intron-few RNAs in Mtr4 mutant germ cells (Fig. 6A, B). Genes with the longest lengths (>59.7 kb) or the highest number of introns (>10) also showed abundant MTR4 binding, likely due to the accumulated non-specific binding along the transcripts (Fig. 7B, C). When genes were grouped by length, the effect of intron number on MTR4 binding was specifically evident in shorter and few-intron genes (gene length <39 kb, intron number <5) (Supplementary Fig. 6C). Consistently, when grouped by intron numbers, gene length had a significant effect on MTR4 binding only in genes with shorter length and fewer introns (gene length <40 kb, intron number <3) (Supplementary Fig. 6D). These results suggest that intron number has a more significant impact on MTR4 binding than gene length.
We next analyzed MTR4 distribution patterns on mRNAs differentially regulated by MTR4. As expected, MTR4 showed a greater enrichment on upregulated mRNAs compared to downregulated and unchanged ones, with stronger enrichment at the middle and the 3’ regions compared to the 5’ region (Fig. 7D, Supplementary Fig. 6E). Interestingly, for ptRNAs, MTR4 consistently bound to the 5’ region, regardless of whether they were regulated by MTR4. However, we observed a specific enrichment at the 3’ region of upregulated ptRNAs in response to Mtr4 KO (Fig. 7E, Supplementary Fig. 6F), suggesting that this differential MTR4 binding may influence ptRNA degradation.
We also examined MTR4 binding on PROMPTs. Metagene profiles revealed that MTR4 was enriched at the 5’ of upregulated PROMPTs versus unchanged ones (Supplementary Fig. 6G). This 5’ enrichment could partly be attributed to the variable lengths of PROMPTs3,40. Supporting this possibility, the metaprofiles of RNA-seq reads from Mtr4 mutant also displayed a 5’ preference (Supplementary Fig. 6H). These findings collectively demonstrate that MTR4 binds to distinct regions of different degradation substrates in pre-meiotic germ cells.
A conserved role of MTR4 in the degradation of mature replication-dependent histone (RDH) mRNAs in the nucleus
RDH genes are a group of short, intronless genes whose mRNAs end with a stem-loop sequence at the 3’ end, instead of a polyA tail41,42. Given that these features are aligned with typical MTR4/exosome targets, we examined MTR4 binding on these RNAs.
MTR4 predominantly bound to the middle and the 3’ region of RDH mRNAs (Fig. 7F). In contrast, its binding to replication-independent histone (RIH) mRNAs, which contain multiple introns and are polyadenylated, was significantly less profound (Fig. 7F). This suggests that MTR4/exosome might play a role in degrading RDH mRNAs in the nucleus. Indeed, we found that RDH mRNAs were greatly accumulated upon Mtr4 depletion, while RIH mRNA levels did not change substantially (Fig. 7G, H).
Interestingly, for RDH genes, RNA signals upstream of the canonical cleavage site (CCS), but not downstream, were significantly elevated in Mtr4-cKO germ cells (Supplementary Fig. 7A), suggesting that the properly processed forms undergo nuclear exosomal degradation. To validate whether properly cleavage RDH mRNAs were polyadenylated and degraded by the exosome, we specifically analyzed RNA-seq reads with at least two extra consecutive As compared to the reference genome (extraAA reads, see details in “Methods”). The IGV snapshots revealed that these reads were preferentially mapped to or near the canonical 3’ cleavage site (Supplementary Fig. 7B). Moreover, MTR4 showed a preference for binding to the middle and 3’ region of upregulated histone mRNAs, compared to unchanged ones (Fig. 7I). Although the mechanism for polyadenylation of these mRNAs remains unclear, these data suggest that a subset of properly processed histone mRNAs are polyadenylated and degraded by the nuclear exosome.
To further validate the nuclear degradation of mature histone mRNAs and assess whether this mechanism is conserved, we revisited our previous RNA-seq data from rRNA-depleted RNAs in nuclear fractions of Cntl and hMTR4 knockdown HeLa cells8. Once again, we observed that most RDH, but not RIH, mRNAs were substantially upregulated in hMTR4 KD cells compared to Cntls (Fig. 7J, Supplementary Fig. 7C). Thus, MTR4 plays a conserved role in the degradation of mature RDH mRNAs in the nucleus.
Gene upregulation in Mtr4 mutant is mainly due to inhibited RNA degradation
Our LACE-seq data showed that 65% of upregulated mRNAs in Mtr4-cKO germ cells displayed apparent MTR4 binding (Fig. 8A), indicating they are bona fide nuclear exosome degradation targets. However, how do the remaining 35% of upregulated mRNAs without MTR4 binding accumulate in these cells? PROMPTs have been reported to promote the transcription of downstream genes by facilitating chromatin accessibility43,44. In line with this view, protein-coding genes with higher PROMPT accumulation in Mtr4-cKO germ cells showed moderately more pronounced increases in expression compared to those with lower PROMPT accumulation (Fig. 8B). One-seventh of upregulated mRNAs without apparent MTR4 binding exhibited upregulated PROMPTs (Fig. 8C, D). The upregulation of these mRNAs may result from the accumulation of corresponding PROMPTs. However, it is also possible that these mRNAs and their PROMPTs are upregulated due to increased transcription from shared promoters. Thus, gene upregulation in Mtr4 mutant pre-meiotic germ cells is primarily attributed to inhibited RNA degradation, with additional contributions potentially arising from other mechanisms, including the accumulation of PROMPTs.
A Pie plot showing the population of upregulated mRNAs with MTR4 binding. n = 361. B Cumulative distribution of the expression changes of mRNAs in Mtr4-cKO germ cells compared to the corresponding Cntls. mRNAs are categorized into those with highly-upregulated, mildly-upregulated and no-changed PROMPT. Highly-up, n = 1367; mildly-up, n = 1631; n/c, n = 613. P-values are based on the one-sided Kolmogorov-Smirnov test. C Pie plot showing the population of upregulated mRNAs without MTR4 binding possessing upregulated PROMPT. D Screenshots of RNA-seq signals of upregulated Zfp397 with upregulated PROMPT. E Pie plot showing the distribution of MTR4 binding reads mapped to the indicated RNA classes. Categories are mutually exclusive. n = 1,631,508. F Pie plot showing the distribution of MTR4 binding reads mapped to the indicated repeat elements (RE). Categories are mutually exclusive. G Bar plot showing the distribution of reads mapped to the indicated RNA classes in Cntl and Mtr4-cKO RNA-seq data. Categories are mutually exclusive. Cntl, n = 94,489,710; Mtr4-cKO, n = 96,925,892. H Volcano plots showing the expression changes of SINEs (38,475 genes analyzed, left), LINEs (4890 genes analyzed, middle) and LTRs (6144 genes analyzed, right) in Mtr4-cKO germ cells compared to the corresponding Cntls. I Metaprofile showing MTR4 signals on SINEs exhibiting upregulated and no-changed or downregulated expression based on merged LACE-seq data. Up, n = 606; n/c or down, n = 4231. J Cumulative distribution of the expression changes of mRNAs in Mtr4-cKO germ cells compared to the corresponding Cntls. mRNAs are categorized into those with highly-upregulated, mildly-upregulated and no-changed B2 SINE. Highly-up, n = 361; mildly-up, n = 527; n/c, n = 1098. P-values are based on the one-sided Kolmogorov-Smirnov test. K Pie plot showing the population of downregulated mRNAs possessing upregulated B2 SINE.
Polyadenylated retrotransposon RNAs are accumulated in Mtr4 mutant germ cells
Eukaryotic genome contains plentiful retrotransposons, whose transposition requires the production and reverse transcription of retrotransposon RNAs with a polyA tail45,46,47. In germ cells, to avoid spontaneous mutation, it is particularly important to repress these retrotransposons. The mouse genome harbors three major types of retrotransposons, including long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and long terminal repeats (LTRs)47,48.
We noticed that 52.4% of MTR4 binding reads were mapped to repeat elements (RE) (Fig. 8E), and among these, more than half were mapped to SINEs, LINEs, and LTRs (Fig. 8F), suggesting these retrotransposon RNAs might be removed by the nuclear exosome in male germ cells. In agreement with this possibility, similar to that of PROMPTs, RNA-seq read population of RE increased in Mtr4-cKO germ cells in comparison to Cntl cells (Fig. 8G).
Both LINE and SINE RNAs are known to be polyadenylated47,48,49. Although LTRs are not known to be strongly polyadenylated, they were quite abundantly detected in our polyA RNA-seq (Supplementary Fig. 8A). All three kinds of retrotransposon RNAs were globally accumulated in Mtr4-cKO germ cells compared to Cntl cells (Fig. 8H, Supplementary Fig. 8B–D). Among these RNAs, the population of SINEs detected by polyA RNA-seq was elevated in Mtr4-cKO germ cells in comparation to Cntl cells (Supplementary Fig. 8A), indicating that they were most upregulated among retrotransposon RNAs in response to Mtr4 deletion.
Many retrotransposons are located in the middle of protein-coding genes50,51,52,53. To confirm that retrotransposon RNAs were indeed expressed and polyadenylated, we selectively analyzed those retrotransposon regions with 5-fold more enrichment of RNA-seq signals than flanking regions, with RNA-seq reads containing at least two additional non-templated adenines compared to the reference genome (extraAA reads, see details in “Methods”). This strict selection yielded fewer retrotransposon RNAs, but their upregulation in Mtr4-cKO germ cells remained significant (Supplementary Fig. 8E). Thus, the nuclear exosome functions to prevent the accumulation of polyadenylated retrotransposon RNAs that have potential mutagenic threat45,54.
We also examined MTR4 binding on retrotransposon RNAs. MTR4 was modestly enriched on upregulated SINEs compared to downregulated or unchanged ones (Fig. 8I, Supplementary Fig. 8F). Same as mRNAs and ptRNAs, MTR4 binding was more enriched at the 3’ region of upregulated SINEs as compared to the downregulated or unchanged ones (Fig. 8I, Supplementary Fig. 8F). We could not detect apparent enrichment of MTR4 on upregulated LINEs or LTRs, compared to the corresponding downregulated or unchanged ones for unclear reason (Supplementary Fig. 8G, H). Thus, these data indicate that at least polyadenylated SINEs are degradation substrates of the nuclear exosome.
Accumulated B2 SINEs may contribute to the repression of a subset of genes in Mtr4 mutant spermatogonia
The widespread downregulation of protein-coding genes in Mtr4-cKO germ cells is unexpected (Fig. 5C), as the major role of MTR4 is to degrade nuclear RNAs. How could MTR4 deletion cause reduced gene expression? Mouse B2 SINEs and human Alu elements have been reported to repress transcription55,56. We speculated that in MTR4-deficient germ cells, accumulated B2 SINE RNAs might repress the transcription of the corresponding host protein-coding genes in cis (Supplementary Fig. 8I). To examine this, we separated intragenic B2 SINEs into highly upregulated, mildly upregulated and unchanged groups, and analyzed the expression changes of the corresponding host genes in Mtr4 KO germ cells. As shown in Fig. 8J, although there was no difference in expression changes between host genes with unchanged and mildly upregulated B2 SINEs, a slightly more pronounced reduction in expression was observed in host genes with highly upregulated B2 SINEs, compared to those with mildly upregulated ones. This suggests a potential role of B2 SINEs in inhibiting gene expression in cis. However, this putative in cis role cannot explain the global downregulation of protein-coding genes, as only 9% of downregulated genes showed accumulated intragenic B2 SINEs in Mtr4 KO germ cells (Fig. 8K). B2 SINEs was reported to inhibit transcription through direct binding to RNAP II55,56. It is possible that the global repression of protein-coding genes might be mainly due to the in trans effect of accumulated B2 SINEs or other unclear mechanisms. Notably, recent studies also found that deletion of core exosome component in mouse embryonic stem cells leads to global transcription repression57,58.
Mtr4 KO leads to alternative splicing changes to mitotic and meiotic genes
When analyzing MTR4 binding distribution on protein-coding genes, we found that a significant population was located on introns (Fig. 9A), suggesting a role of MTR4 in pre-mRNA splicing. To examine this, we analyzed alternative splicing (AS) events in Mtr4-cKO samples compared to Cntls. Interestingly, we identified a total of 2021 AS events that showed significant changes in response to Mtr4 KO (Fig. 9B). The majority of AS events were exon skipping, with no significant bias in terms of exon exclusion or inclusion (Fig. 9B). Approximately 85% of the AS genes exhibited MTR4 binding within introns (Supplementary Fig. 8J). Notably, several splicing factors, including Sf3a1, Sf1, Rbm22, Snrnp2559, are downregulated upon MTR4 depletion (Supplementary Data 3). Thus, it is possible that MTR4 may regulate AS through both direct and indirect mechanisms.
A Pie plot showing the distribution of MTR4 binding reads mapped to the indicated regions of protein-coding genes. Categories are mutually exclusive. n = 1,081,322. B Bar plot showing the number of alternative splicing events in Mtr4-cKO germ cells compared to the corresponding Cntls. SE, skipped exons; RI, retained introns; MXE, mutually exclusive exons; A5SS, alternative 5’SS; A3SS, alternative 3’SS. C GO term analysis of the genes with significant SE events. P-values are based on hypergeometric test corrected for multiple hypothesis tests using the Benjamini-Hochberg method. D Sashimi plot of RNA-seq signals of altered SE event of Sycp1. E Sashimi plot of RNA-seq signals of altered SE event of Chfr. F RT-PCR analysis of altered SE events shown in (D, E). n = 3 biological repeats. G A model summarizing the role of MTR4 in shaping pre-meiotic transcriptome, ensuring proper meiosis initiation of germ cells and maintaining male fertility. Source data are provided as a Source Data file.
Significantly, GO term analysis of SE genes apparently enriched pathways related to meiotic regulation (Fig. 9C). This suggests that in pre-meiotic cells, MTR4 regulates meiotic gene expression through modulating alternative splicing as well. To validate splicing changes in Mtr4-cKO germ cells, we isolated c-KIT+ cells from Cntl and mutant testes at P8 and carried out RT-PCRs for two selective genes, Sycp1 and Chfr, with functions in meiosis and/or mitosis regulation60,61. Consistent with RNA-seq data, Sycp1 showed increased inclusion of exon 6 (Fig. 9D, F), whereas Chfr displayed increased exclusion of exon 9 in Mtr4-cKO germ cells (Fig. 9E, F). Notably, exon 6 of Sycp1 encodes a region (amino acids 95-149) critical for head domain dimerization of SYCP1, which is essential for stabilizing the synaptonemal complex structure60,62. On the other hand, exon 9 skipping (155 bp) in Chfr causes frameshift, disrupting Chfr expression and its role in preventing inappropriate entry into mitosis or meiosis metaphase63. Taken together, our study indicates that MTR4 safeguards meiosis initiation through complicated regulation of RNA metabolism (Fig. 9G, Supplementary Fig. 9).
Discussion
Nuclear RNA degradation is increasingly considered to be an important post-transcriptional regulatory mechanism. However, its biological relevance, especially in animals, remains largely unclear. In this work, we found that nuclear RNA degradation is essential for both embryogenesis and spermatogenesis. With a focus on spermatogenesis, we provide compelling evidence that nuclear RNA degradation is particularly critical for the mitosis-to-meiosis transition through regulating RNA abundance and processing of meiotic and mitotic genes.
We demonstrated that specific KO of Mtr4 in male germ cells results in complete male fertility and severe meiosis initiation defect. Our work aligns with a recent study that reported a significant decrease in male fertility in mice with a whole-body KO of Zcchc8, another nuclear exosome co-factor64. However, it is worth noting that Zcchc8 KO mice exhibited a reduced birth rate and demonstrated significant developmental abnormalities. Thus, it remains plausible that the observed reduction in male fertility was a secondary consequence of these broader developmental defects. We provided compelling and direct evidence that MTR4/exosome-mediated nuclear RNA degradation plays a crucial role in shaping the transcriptome for initiating meiosis during spermatogenesis. MTR4/exosome preferentially degrades RNAs produced from short genes with fewer introns, while maintaining the expression of long genes with a higher number of introns. In line with this regulation rule and the essential role of MTR4 in meiosis initiation, a distinct shift is observed as cells transition from spermatogonia to spermatocytes: gene expression shifts from longer genes with more introns to shorter genes with fewer introns. Further, during this transition, the expression levels of MTR4/exosome are generally reduced. Thus, nuclear RNA degradation appears to function in shaping the transcriptome by differentially regulating the expression of meiotic and mitotic genes. Our data also suggest that the initiation of meiosis is a continuous process that necessitates tight regulation of nuclear RNA degradation. Many meiotic genes are prematurely induced during the mitotic phase of differentiated spermatogonia, but their expression levels need to be precisely regulated through nuclear exosomal degradation. In addition to changes in mRNA expression, we observed that ncRNAs were primarily upregulated upon MTR4 deletion. Increasing evidence suggests the involvement of ncRNAs in the regulation of spermatogenesis65,66. Thus, the dysregulation of ncRNAs may contribute to meiotic initiation defects in Mtr4 mutant mice.
Apart from previously known nuclear exosome substrates, we have identified polyadenylated retrotransposon RNAs as targets of the nuclear exosome in germ cells. A recent study demonstrated that in mouse stem cells, NEXT/exosome selectively represses the expression of non-polyadenylated, short retrotransposon RNAs67. Given that retrotransposon RNAs require a polyA tail for transposition45,54, the accumulation of polyadenylated retrotransposon RNAs detected here in Mtr4 deleted germ cells implies that nuclear RNA degradation could be vital for safeguarding the integrity of the germline genome by preventing spontaneous mutations. Additionally, we found that MTR4/exosome plays a role in removing mature histone mRNAs. Remarkably, this function appears to be conserved between mice and humans, and it is observed in both germ cells and cultured somatic cells8,10. Previously, NEXT was suggested to be involved in the removal of unsuccessfully processed histone mRNAs68. Different from that, our data here in mouse germ cells and in HeLa cells together imply that a significant population of properly processed histone mRNAs are poly- or oligo-adenylated and removed in the nucleus. While the precise functional significance of this role of MTR4/exosome in somatic cells remains uncertain, in the context of germ cells, it may contribute to the initiation of meiosis.
In addition to its direct involvement in removing mRNAs, MTR4/exosome also exerts regulatory control over the expression of protein-coding genes. It accomplishes this by rapidly eliminating unstable transcripts, such as PROMPTs, which promote gene expression by facilitating chromatin opening, and B2 SINE RNAs, which can repress gene expression, possibly both locally (in cis) and globally (in trans)43,44,55,56,69. Furthermore, MTR4 also plays a role in modulating pre-mRNA splicing of several meiotic genes. This multi-faceted regulation contributes to shaping of the gene expression program necessary for initiating meiosis.
Methods
Antibodies
Antibodies against DDX4/MVH (Abcam, ab13840), GAPDH (Proteintech, 60004-1-Ig), Tubulin (Proteintech, 66031-1-Ig), STRA8 (Abcam, ab49405), SYCP3 (Abcam, ab97672), SYCP3 (Abcam, ab15093), γH2A.X (Millipore, 05-636), SOX9 (Millipore, AB5535), CD117/c-KIT antibody (1:50, eBioscience, 11-1171-82) were purchased. The MTR4 antibodies were described previously8. The HRP conjugated secondary antibodies were purchased from Beyotime. The Alexa Fluor 488, Alexa Fluor 546 and Alexa Fluor 647 conjugated secondary antibodies were purchased from Invitrogen.
Generation of Mtr4 knockout (KO)-first, conditional-ready mice
We generated Mtr4+/- mice with Mtr4 knockout (KO)-first, conditional-ready embryo purchased from KOMP (#CSD79286). A cassette containing mouse En2 SA, lacZ, neo, FRT and loxP sites, was inserted in intron 5 and a loxP site was inserted in intron 6 of Mtr4 gene. The knockout allele (-) was initially an Mtr4 non-expressive form. Mtr4+/- mice were intercrossed to generate Mtr4-/- mice. Mtr4F/+ mice were produced by crossing Mtr4+/- with a globe FlpE transgenic mouse line. Mtr4+/Δ mice were generated by mating Mtr4F/F with Stra8-GFPCre knock-in mouse line. The FlpE and Stra8-GFPCre transgenic mouse lines were described previously37,70. Mtr4-koF and Mtr4-R PCR primers were used to detect the knockout allele (543 bp). Mtr4-F and Mtr4-R PCR primers were used to detect the wildtype allele (534 bp) and the floxed allele (667 bp). Primers of genotyping were listed in Supplementary Data 1. All animal experiments were performed according to the guidelines of the Animal Care and Use Committee at Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences. The experiments performed in this study were approved by the Ethics Committee of Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (2022-151).
RNA extraction and RT-qPCR
RNA was extracted from whole tissues or whole c-KIT+ cells using TRIzol reagent (Invitrogen, 15596018CN) following the manufacturer’s instructions. Genomic DNA was removed and cDNAs were synthesized from 1 μg of RNAs with random primer using the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme Biotech Co.,Ltd, #R212-01) according to the manufacturer’s protocol.
Quantitative PCR was carried out using SYBR qPCR SuperMix Plus (Novoprotein, E096-01B) according to the manufacturer’s instructions. Primer sequences were listed in Supplementary Data 1.
Western blotting
Protein samples were prepared using protein gel buffer (125 mmol/L Tris (pH 6.8), 4% SDS, 20% glycerol, 0.004% Bromophenol blue, 20 mM DTT) and lysis in 95 °C for 10 min. Protein samples were separated in a 10% SDS-PAGE gel and transferred onto PVDF membranes. After blocking with 5% non-fat milk for 1 h at room temperature, the membranes were incubated with diluted primary antibodies at 4 °C overnight. After washes with PBSTw (PBS with 0.1% Tween-20), the membranes were incubated with secondary antibodies at room temperature for 1 h. The signals were developed with Pierce ECL Substrate (Thermo Fisher Scientific, 34080). The uncropped and unprocessed scans of the western blotting are provided in the Source Data file or as a Supplementary Fig. in the Supplementary Information (Supplementary Figs. 10–12).
Histological and immunofluorescence analysis
For histological analysis, the testes were fixed overnight in Bouin’s buffer, embedded in paraffin, and sectioned. The sections were then deparaffinized, rehydrated in a graded ethanol series, and stained with hematoxylin and eosin (H&E) according to routine methods.
For immunofluorescence analysis, the testes were fixed with 4% paraformaldehyde (PFA), embedded, and sectioned. The sections were then subjected to boiling in 10 mM sodium citrate buffer (pH 6.0) for 18 min, brought to room temperature, and washed in PBST (PBS with 0.1% Triton X-100). Following this, the sections were incubated with blocking buffer (10% donkey serum (Jackson, 017-000-121) and 0.1% Triton X-100 in PBS) for 60 min at room temperature, and subsequently incubated with primary antibodies in the blocking buffer overnight at 4 °C. On the following day, the slides were washed three times for 10 min each in PBST, and then fluorescent dye-conjugated secondary antibodies were added at a 1:1000 dilution. After incubated for 1 h at room temperature, the sections were washed three times for 10 min each in PBST. Finally, DNA was counterstained with mounting medium containing DAPI (Abcam).
TUNEL assay was performed using the TUNEL BrightRed Apoptosis Detection Kit (Vazyme Biotech Co.,Ltd, #A113), according to the manufacturer’s instructions.
Fluorescence in situ hybridization
To stain polyA RNAs, the testes were fixed with 4% paraformaldehyde (PFA), embedded in O.C.T. Compound (Epredia, 6502) and sectioned. The sections were washed with PBS three times and permeabilized with PBST for 20 min, followed by washes with 2 × saline-sodium citrate buffer (SSC) twice and incubated at 37 °C with an Alexa 546-conjugated oligo dT (50 nt Ts) probe for 12–16 h. The sections were then washed with 2 × SSC twice, 0.5 × SSC once and 1 × PBS once, followed by DAPI staining.
Chromosome spreading and immunofluorescence
Chromosome spreading and immunofluorescence were performed as described71. The spreading nuclei were subjected to immunostaining with anti-SYCP3 and anti-γH2A.X antibodies, allowing for the classification of spermatocytes in spreading into preleptotene, leptotene, zygotene, and pachytene stages. Following washed with PBST, secondary antibodies were applied to the spreading nuclei and incubated for 1 h at room temperature. DNA was stained with DAPI for 10 min at room temperature.
Isolation of c-KIT+ spermatogonia
Single-cell suspensions derived from P8 mice testes were prepared according to the previously described method72 with minor modifications. Briefly, testicular tissue was digested with 120 U/ml of Collagenase Type I (Gibco) for 3 min at 37 °C. Following this, centrifugation facilitated the removal of interstitial cells. The residual tissue was subsequently resuspended in 0.25% trypsin/EDTA and agitated for 5 min at 37 °C. The digestion was halted by adding DMEM with 10% fetal bovine serum (FBS), followed by a 5-minute centrifugation. After discarding the supernatant, the resultant single-cell suspensions were collected in 200 μl of chilled DMEM. The suspensions were then incubated with anti-CD117/c-KIT antibody for 30 min at 4 °C. Flow cytometric analysis was executed employing the FACSAria Fusion (BD) system. Single-cell suspensions from P8 wildtype mouse testes without staining were use as negative control to define the gate, whose fluorescence intensity were about 1000. The cells whose intensity were over 1100 were collected as positive cells.
PolyA RNA-seq
The RNA-seq was performed based on SMART-seq2 with some modification. c-KIT+ cells (~8000 cells) from Cntl and Mtr4-cKO mice testes were sorted in DMEM, then centrifuged at 2000 g. The supernatant was removed as clean as possible. The whole-cell lysate were produced using lysis buffer from Single Cell Full Length mRNA-Amplification Kit (Vazyme Biotech Co.,Ltd, #N712) for mRNA amplification. The mRNAs were firstly reverse transcribed by Oligo(dT)VN Primer and then amplified for 11-12 cycles. cDNA libraries were then prepared for sequencing using the TruePrepTM DNA Library Prep Kit V2 (Vazyme Biotech Co.,Ltd, #TD503). The libraries were quantified by Qubit 2.0 and Agilent 2100, and sequenced in the Illumina Novaseq Pe150 Platform.
LACE‑seq
c-KIT+ cells (~8000 cells) from P8 Cntl mice testes were sorted in DMEM. LACE-seq was performed as described previously38,39. Briefly, the sorted cells were firstly irradiated twice with UV on ice at 400 mJ. Then RNA immunoprecipitation of the samples was performed using MTR4 antibody. The immunoprecipitated RNAs were then fragmented by MNase and dephosphorylated. Then a series of steps were performed to include, reverse transcription, first-strand cDNA capture by streptavidin beads, poly(A) tailing, pre-PCR, IVT, RNA purification, RT and PCR barcoding. The libraries were quantified by Qubit 2.0 and sequenced in the Illumina NextSeq 500 Platform.
Data analysis
RNA-seq analysis
All RNA-seq data that have passed the quality control using fastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) were used in the downstream analysis. The raw reads were filtered to remove the sequencing adaptors and low-quality bases using Cutadapt (v1.18) (https://cutadapt.readthedocs.io/en/stable/) with parameters: -a CTGTCTCTTATACACATCTCCGAGCCCACGAGAC -A CTGTCTCTTATACACATCTGACGCTGCCGACGA -m 20 –max-n 5 -q 30. Then, the clean reads were mapped using STAR (v2.7.10a)73 to the mouse genome (mm10, UCSC) with GENCODE gene annotation (vM24) with the default parameter. The SAMtools package (v1.9)74 was used to extract uniquely mapped reads from the BAM file. RNA-seq signal tracks were generated by using bam2wig.py provided in the RSeQC package (v3.0.0) (https://rseqc.sourceforge.net/), and visualized in UCSC genome browser with a custom track hub. Statistics of mapping reads were listed in Supplementary Data 2.
Gene expression was calculated by featureCounts (v1.6.0)75 with default parameters. Differentially expressed genes were called by the DESeq2 package (v1.20.0)76 with the following criteria: FDR (Wald test, Benjamini-Hochberg adjusted) < 0.05 and at least 1.5-fold change (Supplementary Data 3).
Functional enrichment analysis was performed on the list of differentially expressed genes by using the clusterProfiler package (v3.14.3)77 to determine if the genes are enriched for specific terms in Gene Ontology (GO) annotation of biological processes. The level of significance for the enrichment was calculated by a hypergeometric test for each term using all expressed genes as background. The p-values were corrected for multiple hypothesis tests using the Benjamini-Hochberg method to control the false discovery rate.
The RNA-seq data from Cntl and hMTR4 KD conditions in HeLa cell were described in GSE776418.
scRNA-seq analysis
We quantitated the expression of each stage in scRNA-seq as previously described37. In brief, Seurat was used to analyze the single-cell sequencing data78. The high-quality cells were retained by the default parameters, except for setting the “number of genes detected” threshold to over 2000. Subsequently, the read counts in single cells were normalized and quantified based on UMI using default parameters to obtain the relative expression levels of different genes in each cell. The final expression level of a gene at a specific stage was determined by averaging its abundance across all cells at the same stage.
The stage at which a gene was most abundantly expressed was then computed, followed by assigning each gene to its most abundantly expressed stage. To better designate each gene to a specific stage, we calculated the RNA abundance of each gene at each stage and evaluated its expression specificity according to the IGIA (Integrative Gene Isoform Assembler) score, with the cut-off set as 0.379. For genes with a cut-off value over 0.3, those mostly expressed at mitotic stages (A1-BG2M) were considered mitotic genes, and those mostly expressed at meiotic stages (G1-lpL) were classified as meiotic genes. For genes with a cut-off value below 0.3, if both the highest and second highest expression levels occurred in either the meiotic or mitotic stages, the gene was classified as meiotic or mitotic, respectively. Conversely, if these two stages corresponded to different phases, the gene was classified as non-stage-specific.
extraAA reads definition
To define these reads, we aligned all uniquely mapped RNA-seq reads to the genome and filtered those with imperfect alignments (mismatch > 0). Since our RNA-seq was non-strand-specific, we further selected these reads with two consecutive unmapped As (AA) or Ts (TT) at either end, designating them as extraAA reads.
PROMPT definition and quantification
We quantitated the PROMPTs as previously described with slight modifications40,80. We focused our analysis on protein-coding genes and therefore considered reads mapped to the −3 kb (kilobase) to 0 kb regions of 18,168 gene promoters from GENCODE (vM24) gene annotation by the BEDtools coverage (v2.28.0)81. Differentially expressed PROMPTs were called by the DESeq2 package (v1.20.0) with the following criteria: FDR (Wald test, Benjamini-Hochberg adjusted) < 0.05 and at least 1.5-fold change (Supplementary Data 3).
Differentially expressed PROMPTs used in cumulative distribution function plots were filtered based on the following criteria: P < 0.05 and at least 1.5-fold change.
ptRNA definition and quantification
To quantitate the ptRNAs, we defined the ptRNAs based on GENCODE annotated genes referring to previous studies with slight modifications2,82. In detail, we first filtered out genes that intersect with other genes and retained 33,742 GENCODE (vM24) genes as the candidate host genes of ptRNAs. Then, we defined the transcription end site of ptRNAs in the downstream region of the transcription start site (0–4 kb) for candidate host genes according to previous polyadenylation site (PAS) profiling data82. For a candidate gene with multiple PAS sites, we considered that multiple ptRNAs might be involved. In addition, to distinguish reads of ptRNAs from those of host genes, we selectively analyzed those possessing RNA-seq reads with at least two additional adenines compared to the reference genome (extraAA reads). Finally, we defined 975 potential ptRNAs, which were used in subsequent analysis. Differentially expressed ptRNAs were called by the DESeq2 package (v1.20.0) with the following criteria: FDR (Wald test, Benjamini-Hochberg adjusted) < 0.05 and at least 1.5-fold change (Supplementary Data 3).
Repeat elements detection and quantification
Annotation of retrotransposons (including divergence information for the SINE, LINE, LTR subfamilies) was downloaded from RepeatMasker (http://www.repeatmasker.org/). Reads were counted using the R package GenomicAlignments (mode = “Union”, inter.feature = FALSE) by R 3.5.1. For many reads may be mapped to diverse repeat elements locus on the genome, only uniquely mapped and the best-mapped ones were used to quantify expression. Differentially expressed retrotransposons were analyzed by the DESeq2 package (v1.20.0) with the following criteria: FDR (Wald test, Benjamini-Hochberg adjusted) < 0.05 and at least 1.5-fold change (Supplementary Data 3).
To confirm that retrotransposon RNAs are indeed expressed and polyadenylated, we selectively analyzed those displaying five-fold more enrichment of RNA-seq signal in the repeat region than flanking sequences and possessing RNA-seq reads with at least two additional unmapped adenines at the most 3’-end of RNA-seq reads compared to the reference genome (extraAA reads). Differentially expressed retrotransposons were analyzed by the DESeq2 package (v1.20.0) with the following criteria: FDR (Wald test, Benjamini-Hochberg adjusted) < 0.05 and at least 1.5-fold change (Supplementary Data 4).
Differentially expressed B2SINEs used in cumulative distribution function plots were filtered based on the following criteria: (1) possessing RNA-seq reads with extraAA reads; (2) P < 0.05 and at least 1.5-fold change.
LACE-seq analysis
All LACE-seq data that had passed the quality control using fastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) were used in the downstream analysis. The adapter sequences and poly(A) tails at the 3’ end of raw reads were removed using Cutadapt (v.1.15) with two parameters: -a ATCTCGTATGCCGTCTTCTGCTT -n 10 --minimum-length 18 -e 0.1., and -f fastq -a A -m 18 -n 2. Clean reads were first aligned to mouse pre-rRNA using Bowtie (v1.0.0), and the remaining unmapped reads were then aligned to the mouse reference genome (mm10). Before peak calling, the mapped reads were extended 30 nt to the 3’ end. Peaks were identified by Piranha (v1.2.1) with parameters: -s -b 20 -p 0.05. MTR4 binding genes were defined as those with three or more mapped LACE-seq reads. LACE-seq read counts and peaks are provided within the Source Data folder.
Metagene analysis
To determine the MTR4 binding profile on substrate RNAs more precisely, we conducted a metagene analysis of MTR4 binding sites, defined as the 3’ most nucleotide of MTR4 LACE-seq reads by DeepTools83. For visualization, biological replicates were pooled using the ‘merge’ command in SAMtools package (v1.9), and the signal intensity of MTR4 at each position of an mRNA was calculated by the computeMatrix function. To rule out the influence of outliers, any abnormal signal point that fell below the lower bound (1st quartile – 2×interquartile) or above the higher bound (3rd quartile + 2×interquartile) of the data set was replaced by the mean value of all points excluding the outliers. Aggregate metagene profiles were created with a bin size of 100 bp using the plotProfile functions, followed by visualization with the ggplot2 R package (v3.5.0, https://ggplot2.tidyverse.org/).
Alternative splicing analysis
The alternative splicing (AS) events between Cntl and Mtr4-cKO cells were identified using rMATS (v3.2.5)84. Five types of AS events were identified, including SE, MXE, RI, A3SS, and A5SS. The significantly differentially spliced events were filtered with FDR less than 0.05 (Supplementary Data 5). A ΔIncLevel cutoff of 0.2 was further used to get significant AS events for GO-term analysis. The level of significance for the differentially spliced events was calculated by Student’s t test for each term. The p-values were corrected for multiple hypothesis tests using the Benjamini-Hochberg method to control the false discovery rate.
Quantification and statistical analysis
Student’s t test was performed to compare the differences between Mtr4-cKO relative to Cntls. All results were presented as the mean ± SEM and P < 0.05 were considered significant.
Statistics and reproducibility
For all histology, immunofluorescence and FISH experiments, we performed at least two independent biological replicates. All uncropped and unprocessed scans of the histology, immunofluorescence, western blots and PCR gels were supplied in the Supplementary Information and Source Data files.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) with the accession code GSE253154 (RNA-seq) [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253154] and GSE253156 (LACE-seq) [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253156]. The previously published scRNA-seq [https://doi.org/10.1038/s41422-018-0074-y] and RNA-seq [https://doi.org/10.15252/embj.201696139] datasets used in this study were downloaded from accession code GSE107644 and GSE77641, respectively. The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files. The pipelines used in this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Andersen, P. R. et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 20, 1367–1376 (2013).
Ogami, K. et al. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 31, 1257–1271 (2017).
Meola, N. et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520–533 (2016).
LaCava, J. et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 (2005).
Lubas, M. et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637 (2011).
Fasken, M. B. et al. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J. Biol. Chem. 286, 37429–37445 (2011).
Fan, J. et al. mRNAs are sorted for export or degradation before passing through nuclear speckles. Nucleic Acids Res. 46, 8404–8416 (2018).
Fan, J. et al. Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 36, 2870–2886 (2017).
Januszyk, K. & Lima, C. D. The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 24, 132–140 (2014).
Schmid, M. & Jensen, T. H. The nuclear RNA exosome and its cofactors. Adv. Exp. Med. Biol. 1203, 113–132 (2019).
Schmid, M. & Jensen, T. H. Controlling nuclear RNA levels. Nat. Rev. Genet. 19, 518–529 (2018).
Garland, W. & Jensen, T. H. Nuclear sorting of RNA. Wiley Interdiscip. Rev. RNA 11, e1572 (2020).
Chlebowski, A., Lubas, M., Jensen, T. H. & Dziembowski, A. RNA decay machines: the exosome. Biochim. Biophys. Acta 1829, 552–560 (2013).
Vanacova, S. & Stefl, R. The exosome and RNA quality control in the nucleus. EMBO Rep. 8, 651–657 (2007).
Wyers, F. et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737 (2005).
San Paolo, S. et al. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 5, e1000555 (2009).
Shcherbik, N., Wang, M., Lapik, Y. R., Srivastava, L. & Pestov, D. G. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 11, 106–111 (2010).
Hallais, M. et al. CBC-ARS2 stimulates 3’-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 20, 1358–1366 (2013).
Weick, E. M. et al. Helicase-Dependent RNA decay illuminated by a Cryo-EM structure of a human nuclear RNA exosome-MTR4 Complex. Cell 173, 1663–1677 (2018).
Giacometti, S. et al. Mutually exclusive CBC-containing complexes contribute to RNA fate. Cell Rep. 18, 2635–2650 (2017).
Silla, T., Karadoulama, E., Makosa, D., Lubas, M. & Jensen, T. H. The RNA exosome adaptor ZFC3H1 functionally competes with nuclear export activity to retain target transcripts. Cell Rep. 23, 2199–2210 (2018).
Schuller, J. M., Falk, S., Fromm, L., Hurt, E. & Conti, E. Structure of the nuclear exosome captured on a maturing preribosome. Science 360, 219–222 (2018).
Skarnes, W. C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011).
Lloyd, K. C. A knockout mouse resource for the biomedical research community. Ann. N. Y Acad. Sci. 1245, 24–26 (2011).
Xu, C., Cao, Y. & Bao, J. Building RNA-protein germ granules: insights from the multifaceted functions of DEAD-box helicase Vasa/Ddx4 in germline development. Cell Mol. Life Sci. 79, 4 (2021).
Zhou, Q. et al. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol. Reprod. 78, 537–545 (2008).
Griswold, M. D. Spermatogenesis: the commitment to meiosis. Physiol. Rev. 96, 1–17 (2016).
Zhou, Q. et al. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol. Reprod. 79, 35–42 (2008).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Page, S. L. & Hawley, R. S. The genetics and molecular biology of the synaptonemal complex. Annu Rev. Cell Dev. Biol. 20, 525–558 (2004).
Osman, B. A. et al. Localization of a novel RNA-binding protein, SKIV2L2, to the nucleus in the round spermatids of mice. J. Reprod. Dev. 57, 457–467 (2011).
Silla, T. et al. The human ZC3H3 and RBM26/27 proteins are critical for PAXT-mediated nuclear RNA decay. Nucleic Acids Res. 48, 2518–2530 (2020).
Fujiwara, N. et al. MPP6 stimulates both RRP6 and DIS3 to degrade a specified subset of MTR4-sensitive substrates in the human nucleus. Nucleic Acids Res. 50, 8779–8806 (2022).
Schrans-Stassen, B. H., van de Kant, H. J., de Rooij, D. G. & van Pelt, A. M. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 140, 5894–5900 (1999).
Johnson, D. et al. Concerted cutting by Spo11 illuminates meiotic DNA break mechanics. Nature 594, 572–576 (2021).
Zhu, K. et al. The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle. Cell Death Differ. 29, 2070–2088 (2022).
Chen, Y. et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 28, 879–896 (2018).
Su, R. et al. Global profiling of RNA-binding protein target sites by LACE-seq. Nat. Cell Biol. 23, 664–675 (2021).
Lei, W. L. et al. SRSF2 is required for mRNA splicing during spermatogenesis. BMC Biol. 21, 231 (2023).
Chen, Y. et al. Principles for RNA metabolism and alternative transcription initiation within closely spaced promoters. Nat. Genet. 48, 984–994 (2016).
Dominski, Z. & Marzluff, W. F. Formation of the 3’ end of histone mRNA: getting closer to the end. Gene 396, 373–390 (2007).
Marzluff, W. F., Wagner, E. J. & Duronio, R. J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9, 843–854 (2008).
Preker, P. et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008).
Liu, J. et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020).
Doucet, A. J., Wilusz, J. E., Miyoshi, T., Liu, Y. & Moran, J. V. A 3’ Poly(A) Tract Is Required for LINE-1 Retrotransposition. Mol. Cell 60, 728–741 (2015).
Dewannieux, M. & Heidmann, T. Role of poly(A) tail length in Alu retrotransposition. Genomics 86, 378–381 (2005).
Sultana, T., Zamborlini, A., Cristofari, G. & Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 18, 292–308 (2017).
Fueyo, R., Judd, J., Feschotte, C. & Wysocka, J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 23, 481–497 (2022).
Kramerov, D. A. & Vassetzky, N. S. SINEs. Wiley Interdiscip. Rev. RNA 2, 772–786 (2011).
Liu, N. et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553, 228–232 (2018).
Robbez-Masson, L. et al. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 28, 836–845 (2018).
Hancks, D. C. & Kazazian, H. H. Jr. Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9 (2016).
Zhang, Y., Romanish, M. T. & Mager, D. L. Distributions of transposable elements reveal hazardous zones in mammalian introns. PLoS Comput Biol. 7, e1002046 (2011).
Ustyantsev, I. G. et al. [Polyadenylation of Sine transcripts generated by RNA Polymerase III dramatically prolongs their lifetime in cells]. Mol. Biol. 54, 78–86 (2020).
Walters, R. D., Kugel, J. F. & Goodrich, J. A. InvAluable junk: the cellular impact and function of Alu and B2 RNAs. IUBMB Life 61, 831–837 (2009).
Allen, T. A., Von Kaenel, S., Goodrich, J. A. & Kugel, J. F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 11, 816–821 (2004).
Han, X. et al. Nuclear RNA homeostasis promotes systems-level coordination of cell fate and senescence. Cell Stem Cell 31, 694–716 (2024).
Torre, D. et al. Nuclear RNA catabolism controls endogenous retroviruses, gene expression asymmetry, and dedifferentiation. Mol. Cell 83, 4255–4271 (2023).
Wan, R., Bai, R., Zhan, X. & Shi, Y. How is precursor messenger RNA spliced by the spliceosome? Annu. Rev. Biochem. 89, 333–358 (2020).
Dunce, J. M. et al. Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly. Nat. Struct. Mol. Biol. 25, 557–569 (2018).
Lu, L. Y. & Yu, X. CHFR is important for the survival of male premeiotic germ cells. Cell Cycle 14, 3454–3460 (2015).
Crichton, J. H. et al. Structural maturation of SYCP1-mediated meiotic chromosome synapsis by SYCE3. Nat. Struct. Mol. Biol. 30, 188–199 (2023).
Scolnick, D. M. & Halazonetis, T. D. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature 406, 430–435 (2000).
Wu, Y. et al. Nuclear exosome targeting complex core factor Zcchc8 regulates the degradation of LINE1 RNA in early embryos and embryonic stem cells. Cell Rep. 29, 2461–2472 (2019).
Sahlu, B. W. et al. Long noncoding RNAs: new insights in modulating mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 11, 16 (2020).
Joshi, M. & Rajender, S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod. Biol. Endocrinol. 18, 103 (2020).
Garland, W. et al. Chromatin modifier HUSH co-operates with RNA decay factor NEXT to restrict transposable element expression. Mol. Cell 82, 1691–1707 (2022).
Lubas, M. et al. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 10, 178–192 (2015).
Ichiyanagi, T. et al. B2 SINE copies serve as a transposable boundary of DNA methylation and histone modifications in the mouse. Mol. Biol. Evol. 38, 2380–2395 (2021).
Lin, Z. et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 27, 1216–1230 (2017).
Scherthan, H. et al. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol. Biol. Cell 11, 4189–4203 (2000).
Gaysinskaya, V., Soh, I. Y., van der Heijden, G. W. & Bortvin, A. Optimized flow cytometry isolation of murine spermatocytes. Cytom. A 85, 556–565 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Wang, K. et al. Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nat. Commun. 10, 4714 (2019).
Ntini, E. et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 20, 923–928 (2013).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Li, W. et al. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 14, 6 (2016).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Shen, S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. USA 111, E5593–E5601 (2014).
Acknowledgements
We thank Dr. Mofang Liu for insightful discussions. We thank Dr. Naihe Jing and members of the Jing Lab, Dr. Guizhong Cui for embryo experimental support. We thank Dr. Hengyu Fan for sharing results prior to publication. We thank members of the Cheng lab for useful discussion. We thank the Genome Tagging Project (GTP) Center, Shanghai Institute of Biochemistry and Cell Biology, CAS for technical support. We thank the Histology, Flow Cytometry and Vivarium services at CEMCS, SIBCB. We thank the staff members of the Integrated Laser Microscopy System at the National Facility for Protein Science in Shanghai (NFPS), Shanghai Advanced Research Institute, Chinese Academy of Sciences, China for sample preparation, data collection and analysis. This work was supported by the National Key R&D Program of China (2022YFA1303300 [H.C.]), National Natural Science Foundation of China (31925008 [H.C.], 32230022 [H.C.], 32430023 [H.C.], 82341023 [Y.Z.], 32025008 [Y.X.], 32130064 [Y.X.]), the Strategic Priority Research Program of the Chinese Academy of Science (XDB0570100 [H.C.]), Fundamental Research Funds for the Central Universities (2042022dx0003 [Y.Z.]), Postdoctoral Fellowship Program of CPSF (GZC20233352 [Z.T.]).
Author information
Authors and Affiliations
Contributions
H.C. and M.T. conceived the project and designed the experiments. J.W. and L.Z. performed most experiments with help from Z.L. R.S. performed LACE-seq and provided technical assistance. Z.T. and Y.Z. performed most bioinformatics analyses with help from N.H. and Y.T. G.G., J.F., M.T., Y.X., and Y.Z. provided conceptual and experimental guidance. H.C., J.W. and L.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interest.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, L., Wang, J., Tang, Z. et al. The nuclear exosome co-factor MTR4 shapes the transcriptome for meiotic initiation. Nat Commun 16, 2605 (2025). https://doi.org/10.1038/s41467-025-57898-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57898-0