Abstract
The metabolic, immune, and endocrine systems show profound seasonal changes in animals, including humans. In addition, morbidity from cardiovascular and psychiatric diseases is more severe and mortality rate is higher in winter. However, their molecular mechanisms remain unknown. Here we report the seasonal transcriptome of 80 tissues collected over 1 year from male and female rhesus macaques kept in a semi-natural outdoor environment. We find seasonal changes in plasma metabolites and hormones. Transcriptome analysis identifies sex differences in seasonally oscillating genes (SOGs) in all tissues studied, and we generate the web database ‘Non-Human Primate Seasonal Transcriptome Atlas (NHPSTA).’ Transcriptional regulatory network analysis, siRNA knockdown, and mutant mouse analyses reveal regulation of SOGs by GA-binding protein (GABP). We also demonstrate seasonal oscillations in the expression of disease risk factor genes and drug interacting genes. NHPSTA provides a molecular resource for seasonally regulated physiology and targets for therapeutic interventions for seasonally regulated diseases.
Similar content being viewed by others
Introduction
The rhythms of life on Earth are shaped by celestial motion. For instance, to cope with seasonal changes in the environment, animals show profound alterations in various aspects of physiology and behavior, such as reproduction, molting, hibernation, and migration1. Seasonal differences in physiological processes are also evident in humans, including those related to hormone secretion, metabolism, sleep, immune function, and reproduction2,3,4,5,6,7,8. Furthermore, in humans there is seasonal variation in morbidity due to cardiovascular and cerebrovascular diseases, influenza, pneumonia, autoimmune diseases, depression, bipolar disorder, and schizophrenia, with most of these conditions becoming more severe and exhibiting higher mortality rates in winter5,6,7. Therefore, it is of critical importance to gain an in-depth understanding of the mechanisms that drive these seasonally regulated aspects of physiology and disease susceptibility. Although these seasonal rhythms are thought to arise from coordinated gene expression profiles in multiple tissues, the underlying molecular basis remains unknown.
Due to ethical constraints and limited access to disease-free human tissues, non-human primates (NHPs), which share similar genetic and physiological features with humans, are an ideal model. For instance, the rhesus macaque (Macaca mulatta) is phylogenetically close to humans and is one of the most widely studied NHPs. In addition, rhesus macaques exhibit robust seasonal changes in physiology and behavior, including seasonal reproduction, molting, and winter depression-like behavior3,9,10,11, making them an excellent model for understanding the molecular mechanisms of seasonally regulated physiology and diseases.
In the present study, we analyze seasonal transcriptome profiles of 924 samples of 80 neural and peripheral tissues collected every 2 months over 1 year from adult male and female rhesus macaques kept in a semi-natural outdoor environment. We establish the web database ‘Non-Human Primate Seasonal Transcriptome Atlas (NHPSTA)’ and demonstrate sex differences in seasonally oscillating genes (SOGs). Functional analyses uncover regulation of SOGs by GA-binding protein (GABP) and seasonal oscillations in the expression of disease risk factor genes and drug efficacy including enhanced alcohol metabolism in winter. NHPSTA provides a molecular basis for seasonally regulated physiology and diseases.
Results
Seasonal changes in hormone secretion
Macaques were kept in an open enclosure at the Center for the Evolutionary Origins of Human Behavior (EHUB), Kyoto University (Supplementary Fig. 1a, b). Toward a comprehensive understanding of the molecular mechanisms underlying seasonal physiological changes, samples of 80 tissues, specifically from 30 brain regions and 50 peripheral tissues, were collected from male and female adult macaques (ranging from 4 to 11 year-old) every 2 months from October 2019 to August 2020 at around 14:00 (Japan Standard Time) (Supplementary Fig. 1c, d). Macaques showed robust seasonal changes in reproductive phenotype. During macaque breeding season (i.e., December and February), males developed testes while females showed increased reddening of the anogenital skin (Fig. 1a). Metabolome analysis of plasma samples using capillary electrophoresis time-of-flight mass spectrometry (CE-MS) detected 222 metabolites (Supplementary Data 1) and revealed seasonal rhythmicity in 20 water-soluble metabolites, including glutamate (Glu), glutamine (Gln), cystine, and kynurenine (Fig. 1b). Interestingly, these neurotransmitters and metabolites have been proposed as biomarkers of inflammation and depression12,13,14.
a Seasonal changes in testicular length in males and red coloration score for hindquarter skin in females. Blue and orange backgrounds represent winter and summer, respectively. b Heatmap showing seasonal changes in plasma metabolites measured by CE-MS. M: male, F: female. c Seasonal secretion profiles of plasma testosterone and estradiol. d Principal component analysis of transcriptomes of 924 samples across 80 neural and peripheral tissues. Colors indicate different tissue types. e Seasonal expression profiles of genes involved in the hypothalamus-pituitary-gonadal axis as determined by RNA-seq analysis. RS: the coefficient of determination.
Consistent with previous reports15,16, levels of testosterone in males and estradiol in females were high during breeding season (Fig. 1c). However, the increase of testosterone in males was earlier (i.e., October) than that of estradiol in females (i.e., December). This result was consistent with a previous study that reported an earlier reproductive onset in males than females17. There was also a slight increase in the levels of testosterone in males and estradiol in females in summer (Fig. 1c). Of note, a recent human study reported bimodal secretion profiles of testosterone in males and estradiol in females, peaking in both winter and summer8.
We next extracted total RNA from 924 samples across 80 tissues, and conducted RNA-sequencing (RNA-seq) analysis based on Lasy-Seq protocol (average sequencing depth: 2,390,621). To address the issue of insufficient annotation, we merged three annotation files (Ensembl: Macaca_mulatta.Mmul_10.192.gtf; NCBI: GCF_003339765.1_Mmul_10_genomic.gtf; GSE128537_assembly.rheMac10.gtf)18, and assembled a total of 54,213 genes, including protein-coding genes, miRNA, lncRNA, misc_RNA, snRNA, snoRNA, rRNA, Y RNA, pseudogenes, and processed pseudogenes (Supplementary Data 2). Principal component analysis based on transcriptomes clearly separated neural and peripheral tissues (Fig. 1d). In addition, subclusters were formed depending on the functional classifications among peripheral tissues.
Vertebrate reproduction is governed by the hypothalamus-pituitary-gonadal (HPG) axis. RNA-seq analysis has elucidated the seasonal expression profiles of genes involved in the HPG axis. In male macaques in this study, there was a peak from August to October in each of the following genes: gonadotropin-releasing hormone (GNRH) in the hypothalamus; β-subunits of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) (e.g., LHB, FSHB) in the pituitary gland; and rate-limiting enzymes involved in testosterone production (steroidogenic acute regulatory protein [STAR] and hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 [HSD3B2]) in the testis (Fig. 1e). The seasonal expression profiles of these genes were less clear in females, probably due to the presence of estrous cycles. In contrast to males, in females LHB was high from December to April, followed by a delayed increase in the rate-limiting enzyme genes for estradiol synthesis (STAR and aromatase [cytochrome P450 family member 19 subfamily A member 1: CYP19A1]). Time lags in each process and sex differences in the seasonal expression profiles were also observed in other axes such as the hypothalamus-pituitary-thyroid axis for metabolism and thermoregulation (Supplementary Fig. 2a), the hypothalamus-pituitary-liver axis for growth (Supplementary Fig. 2b), and the hypothalamus-pituitary-adrenal axis for stress response (Supplementary Fig. 2c). Delays in the process of hypothalamus-pituitary-peripheral gland axes have previously been thought to occur on the order of minutes to hours8. However, we found much longer time lags (i.e., several months) in multiple axes at the transcriptional level. When we compared the seasonal expression profiles of sex steroid-regulated genes, males showed an earlier phase than females consistent with the differences in testosterone and estradiol secretion profiles (Supplementary Fig. 3).
Identification of seasonally oscillating genes (SOGs)
We next identified the SOGs in each tissue. In order to reduce false positives caused by the small sample size, strict parameters were applied at multiple steps during the analysis. First, fragments per million (FPM) values were log-transformed by log2 (FPM + 0.5) and genes with an average value > 2.5 across six sampling time points were designated as expressed genes. A total of 4,501-13,544 genes in males and 4,418-12,954 genes in females were detected depending on the tissue (Fig. 2a). A comparison of expressed genes between males and females showed a significant number of overlapping genes (82.25% - 96.48%: average 93.14% in male; 82.76% - 96.38%: average 92.20% in female) (Supplementary Fig. 4). To investigate the seasonal expression of individual genes, trends were extracted by fitting a cosine curve using the nonlinear least squares (nls) function in R, and the coefficient of determination (R-squared: RS) was used to evaluate the goodness of fit. SOGs were extracted if RS > 0.7 and the amplitude of seasonal oscillations was larger than one (specifically, 2-fold greater). This analysis identified 273−2,344 SOGs in males (Supplementary Data 3) and 342-1,943 SOGs in females (Supplementary Data 4), depending on the tissue (Fig. 2b). The expression profiles of all 54,213 genes in 80 tissues can be accessed and explored in the Non-Human Primate Seasonal Transcriptome Atlas Database (NHPSTA) (https://rhythm.itbm.nagoya-u.ac.jp/NHPSTA/).
Among the 80 tissues examined, the pancreas (PAN) had the lowest number of expressed genes and low number of SOGs in both males and females. Muscle tissues such as the abdominal muscle (MUA), gastrocnemius muscle (MUG), and heart (HEA) also had low numbers of expressed genes and SOGs (Fig. 2a, b). The low numbers of expressed genes in these tissues are consistent with baboon RNA-seq results19. In contrast, there were high numbers of SOGs in the stomach (STM) in both sexes, as well as in reproductive organs, specifically the prostate gland (PRO) in males and the uterus (UTE) in females (Fig. 2b). Only a weak correlation was observed between the number of expressed genes and the number of SOGs (correlation coefficient, male r = 0.25; female: r = 0.323).
The total numbers and peak phases of the SOGs in each tissue differed between males and females (Fig. 2b, c and Supplementary Fig. 5). Most tissue types had bimodal or unimodal peak phases in spring and/or autumn, and there were sex differences in the distribution of the peak phase (Supplementary Fig. 5). Furthermore, a comparison of male and female SOGs showed a limited number of overlapping SOGs [2.62 % (esophagus: OES) - 24.8 % (skin on the back: SKNB) (average: 9.53%) in males and 3.53% (arcuate nucleus: ARC) - 31.3% (stomach: STM) (average: 9.02%) in females depending on tissue] (Fig. 3). These results suggested the existence of a clear sex difference in seasonal physiological changes in various tissues.
Molecular bases of physiological changes
To identify seasonally regulated physiological functions that are conserved between sexes, we performed functional enrichment analysis of common overlapping SOGs between sexes using Metascape20. The top five enriched pathways and genes involved in representative pathways in different six tissues are highlighted in Fig. 4a, b. In brown adipose tissue (BAT), seasonal oscillation was observed in NDUFS2, IDH1, and COX1, and the pathways related to ‘response to oxidative stress’ and ‘generation of precursor metabolites and energy’ were enriched (Fig. 4a, b). In many mammalian species, including humans, hair follicles cycling is coordinated with yearly seasons21. Seasonal rhythms were observed in FOXN1, COL1A1, and KRT5 in the skin on the back (SKNB), highlighting the enrichment of ‘epidermis development’ (Fig. 4a, b). Seasonal changes in immune function have been reported5,6. We found seasonal changes in the expression of LILRB4, IL18, and IGF1 in the mesenteric lymph node (MEL), and LILRB4, CXCL10, and CCR2 in the spleen (SPL) (Fig. 4b). Accordingly, there was enrichment of ‘myeloid cell differentiation’ and ‘regulation of activated T cell proliferation’ in the MEL, and of ‘T cell cytokine production’ in the SPL (Fig. 4a). Susceptibility to winter depression is high in females, and patients are known to show augmented appetite and carbohydrate craving during winter22. Although macaques were fed the same diet throughout the year, genes involved in ‘carbohydrate metabolic process’ such as GLO1, LDHB, and UGDH were enriched and peaked during winter and spring in the duodenum (DUO) in females (Fig. 4a, b). Seasonal rhythms are driven by the endogenous circannual clock, which is entrained by seasonal changes in day length and temperature3,10,23. Persistence of the annual androgen rhythm has been reported in male rhesus macaques kept under a constant photoperiod, temperature, and food supply24. The pars tuberalis (PT) of the pituitary gland is the key regulator of seasonal rhythms25,26,27 and has been proposed to play the primary role in circannual rhythm28,29. Functional enrichment analysis of the SOGs in the PT revealed overrepresentation of genes involved in tissue remodeling, such as NOG, CCN3, and CAV1 (Fig. 4b). It has been proposed that seasonal clocks may arise from the generation of tissue remodeling30. Notably, recent discoveries of circannual genes underlying circannual rhythms of gonadal development in medaka fish also support the involvement of tissue remodeling in the regulation of the circannual clock31.
a GO enrichment analysis of common SOGs in both sexes. The top five enriched pathways in the following six tissues are shown: brown adipose tissue (BAT), skin on the back (SKNB), mesenteric lymph node (MEL), spleen (SPL), duodenum (DUO), and pars tuberalis of the pituitary gland (PT). b Heatmap showing the expression profiles of SOGs. Genes involved in representative pathways are highlighted.
Regulation of SOGs by GA-binding protein (GABP)
We next examined which genes were most widely detected as SOGs in multiple tissues in males and/or females (Fig. 5a). Among total 19,003 SOGs across 80 tissues, no gene was detected as an SOG in all 80 tissues (Fig. 5a). When we focused on the top 30 most common SOGs, we found enrichment of the genes related to the ‘aerobic electron transport chain,’ such as NDUFA1, SDHD, LDHB, COX5B, and COQ9, as well as ribosomal protein genes such as RPS27L and RPL35 (Fig. 5a, b and Supplementary Data 6).
a Distribution of SOGs ranked according to the number of tissues in which they show seasonal oscillation in male, female and both sexes. b GO enrichment analysis for the top 30 most common SOGs (one-sided Fisher’s exact test). c Distribution of potential transcription factors regulating SOGs ranked according to the number of tissues identified by transcriptional regulatory network analysis. d Effects of siRNA knockdown of GABPA and/or GABPB1 on GABPA and GABPB1 expression in macaque fibroblasts. (one-way ANOVA, Dunnett’s test, mean ± SEM, n = 4). e Effects of siRNA knockdown of GABPA and GABPB1 on the most common SOGs in macaque fibroblasts examined by qPCR (top) and RNA-seq (bottom) analyses. Numbers within the graph indicate the P value (unpaired two-tailed t-test, mean ± SEM, n = 4). f Comparison of 2,058 DEGs between GABPA and GABPB1 siRNA-treated and negative control siRNA-treated macaque fibroblasts (Benjamini-Hochberg correction adjusted P value (Padj) < 0.05 and fold change > 1.5). g Comparison of all 19,003 SOGs and 2058 GABP-regulated genes. h GO analysis of 1450 overlapping genes between SOGs and GABP-regulated genes (one-sided Fisher’s exact test).
To further understand the molecular mechanisms regulating these seasonal rhythms, we performed transcriptional regulatory network analysis using Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST version 2) and the ENCODE Transcription Factor Targets Dataset in males and/or females in the web-based portal Metascape20 (Fig. 5c and Supplementary Fig. 6). This analysis identified the E26 transformation specific (ETS)-family transcription factor, GABP and its interacting transcription factors and co-activators (e.g., SP1, YY1, E2F1, and CREB)32,33 as top hits (Fig. 5c and Supplementary Fig. 6, and Supplementary Data 7). GABP is a heterotetramer composed of two distinct proteins, GABPα and GABPβ. GABPα includes an ETS DNA binding domain, while GABPβ1 and GABPβ2 contain the transcriptional activation domain. Since the expression level of GABPB2 is approximately 2 log units lower than that of GABPB1 in various macaque tissues, we focused on GABPB1 in this study. To test whether GABP regulates the expression of SOGs, we investigated the effects of small interfering RNA (siRNA) knockdown of GABPA and GABPB1 using macaque fibroblasts. We first used quantitative real-time PCR (qPCR) to evaluate the effects of GABPA and GABPB1 knockdown on their expression. GABP regulates its own expression34, and we confirmed the effects of knockdown of each gene on its own expression as well as its autoregulatory effects (i.e., up-regulation of its partner) (Fig. 5d), demonstrating the validity of siRNA knockdown. We also used qPCR to demonstrate that GABP controlled the most common SOGs, such as NDUFA1, RPL35, SDHD1, and LDHB (Fig. 5e). To further explore SOGs targeted by GABP, we compared the results of RNA-seq analysis between macaque fibroblasts transfected with siRNA for both GABPA and GABPB1 and negative control siRNA. Although statistically significant knockdown of GABPA and GABPB1 was observed by its siRNA, the knockdown efficiencies for GABPA and GABPB1 were 50% and 69%, respectively (Fig. 5d). Therefore, we used an adjusted P value (Padj) threshold <0.05 and fold change > 1.5 to detect differentially expressed genes (DEGs) (Fig. 5f). This analysis identified 2,058 GABP-regulated genes (1,067 up-regulated and 991 down-regulated genes in GABP knockdown cells) (Fig. 5f and Supplementary Data 8). As fibroblasts populate all tissues35, we examined the overlap between GABP-regulated genes and all 19,003 SOGs. This analysis revealed that 70.4 % of the GABP-regulated genes in fibroblasts (1,450 genes) were SOGs in at least one tissue (Fig. 5g). Gene ontology (GO) analysis of these 1,450 overlapping genes highlighted pathways involved in cell cycles (Fig. 5h). Again, this is consistent with the idea that seasonal adaptive physiology involves changes in body composition, organ size, and tissue remodeling8,30,36.
Tissue remodeling in mice occurs with changes in photoperiod and temperature
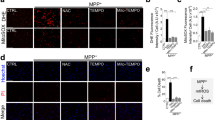
Since functional analyses in primates are costly and time-consuming, cross-species analyses are widely used in primate research19,37. A number of studies demonstrated that mice can model molecular mechanisms of seasonal physiology and behaviors26,27,38,39. Therefore, we next focused on mice for the functional analysis. In mammals, pineal melatonin is believed to play a crucial role in the control of many seasonal functions and behaviors40. As most commonly used strains of laboratory mice genetically lack melatonin41, they are generally considered to be non-seasonal animals. In our previous study, however, we showed that melatonin-proficient CBA/N mice could transform photoperiodic information at the hypothalamic-pituitary level26,39. Therefore, we examined the effects of 1-month exposure to winter-like short days and cool conditions (SC: 8 h light 16 h dark, 10 °C) or summer-like long days and warm conditions (LW: 16 h light 8 h dark, 30 °C) on organ weights in melatonin-proficient CBA/N and melatonin-deficient C57BL/6 N strains. Strikingly, we found robust changes in organ weights in the HEA, PAN, and kidney (KID), with higher weights in the SC condition in both CBA/N and C57BL/6 N mice (Fig. 6a). These results suggest that there could be seasonal changes in organ weight in mice, and melatonin is not required for this regulation. We then examined the effects of SC and LW conditions on tissue remodeling in C57BL/6 N mice using immunohistochemistry for Ki67, a marker of cell division. Preliminary analysis of mice exposed to SC or LW conditions for 2 weeks or 1 month revealed few Ki67-immunopositive cells. Therefore, we used mice exposed to SC or LW conditions for 1 week. The number of Ki67-positive cells was higher in the HEA and PAN under SC than LW conditions (Fig. 6b, c). Although the number of Ki67-positive cells was not different in the KID between the two conditions, the glomerular size was significantly larger under SC than LW conditions (Fig. 6b, c). When we performed hematoxylin and eosin (H&E) staining of the HEA, the cardiomyocyte diameter was larger under SC than LW conditions (Fig. 6b, d). Immunohistochemistry for β-catenin, which localizes to the acinar cell membrane, showed that acinar cell size was larger under SC than LW conditions (Fig. 6b, e). These results demonstrate that tissue remodeling occurs between the SC and LW conditions in mice. Given the significant differences in food and water intake between the SC and LW conditions (Fig. 6f), these results likely reflect the metabolic differences between summer- and winter-mimicking environments. Although the photoperiod and temperature changes between SC and LW conditions are components of seasonal changes, further work is required to confirm whether these changes occur seasonally under natural environmental conditions.
a Effects of 1-month exposure to short-day and cool (SC) or long-day and warm (LW) conditions on organ weight in CBA/N and C57BL/6 N mice. Numbers within the graph indicate the P value (unpaired two-tailed t-test, mean + SEM, n = 8). b (top) Effects of 1-week exposure to SC or LW conditions on Ki67-positive cell numbers in the heart (HEA), pancreas (PAN), and kidney (KID) of C57BL/N mice. Numbers within the graph indicate the P value (unpaired two-tailed t-test, mean + SEM, n = 4). (bottom) Effects of 1-week exposure to SC or LW conditions on cardiomyocyte diameter, pancreatic acinar cell size, and glomerular size. (t-test, mean + SEM, n = 4). c Representative images of Ki67 immunohistochemistry in the heart (HEA), pancreas (PAN), and kidney (KID) under SC and LW conditions. The number of Ki67 immuno-positive cells (red arrows) was higher under SC than LW conditions in the HEA and PAN, while glomerular size was larger under SC than LW conditions in the KID. d Representative images for the H&E staining of cardiomyocytes under SC and LW conditions. e Representative images of the immunohistochemistry of β-catenin, which localizes to the acinar cell membrane. Note that the nuclear density (i.e., number of nuclei in the field) is greater under LW than SC conditions, indicating a larger cytoplasmic area under SC conditions. L: Islet of Langerhans; G: Glomerulus. Scale bars: 50 μm. f Daily food and water intake in male C57BL/6 N mice kept under SC and LW conditions. Numbers within the graph indicate the P value (unpaired two-tailed t-test, mean + SEM, n = 6).
In vivo role of GABP in tissue remodeling
To test the in vivo role of GABP in photoperiod- and temperature-induced tissue remodeling, we aimed to generate GABP mutant mice. A loss-of-function mutation of Gabpa results in preimplantation lethality42. Gabpb1 gene gives rise to two alternatively spliced isoforms, Gabpβ1L and Gabpβ1S. These two isoforms differ in C-terminal lengths and Gabpβ1L has a longer C-terminal tail which is encoded entirely by exon 9. Deletion of exons 2 to 6 of Gabpb1, which contain the N-terminal ankyrin repeat domain that mediates interactions with Gabpα, results in early embryonic lethality43. However, mice with targeted deletion of exon 9 of Gabpb1, which contains the leucine zipper domain that contributes to the assembly of the Gabpα2β2 tetramer, were reported to be viable43. Therefore, we generated Gabpb1 exon 9 deletion mutants using the CRISPR/Cas9 genome editing technique (Fig. 7a). We co-injected crRNAs, tracrRNA and Cas9 protein into 223 fertilized eggs obtained from C57BL/6 N mice and then transferred injected embryos into the oviducts of pseudopregnant ICR recipients. We succeeded in obtaining 16 G0 generation offspring. Consistent with the previous study43, the G0 mosaic individuals had a weak hind limb phenotype (Supplementary Movie 1). In contrast, our Gabpb1 exon 9 deletion homozygous mutants failed to survive after weaning. In the previous study, exon 9 was replaced by a neomycin cassette43. Since retention of selectable marker cassettes in target loci is known to cause unexpected phenotypes due to disruption of expression of neighboring genes44, the mild phenotype reported in the previous study could have been due to be the result of neomycin cassette retention, however we cannot completely rule out possible CRISPR/Cas9 off-targets in our study. Since our homozygous Gabpb1 exon 9 deletion mutants could not survive after weaning, we analyzed the phenotypes of heterozygous mutant (+/Mut) and wild-type (+/+) littermates. When male wild-type and heterozygous mutant mice were exposed to SC or LW conditions for 1 week, Gabpb1 expression tended to be higher under SC than LW conditions in wild-type mice (Fig. 7b), whereas it was lower in heterozygous mutant mice compared to wild-type mice (Fig. 7b). The magnitude of the organ weight changes between SC and LW conditions (i.e., P values and fold changes) tended to be smaller in heterozygous mutant mice compared to wild-type littermates (Fig. 7b). Furthermore, differences in the numbers of Ki67-positive cells in the HEA and PAN were smaller in heterozygous mice than in wild-type mice (Fig. 7b, c, f). In addition, the magnitudes of changes in cardiomyocyte diameter, acinar cell size, and glomerular size were smaller in heterozygous mutant mice than the wild-type mice (Fig. 7c-f). These results suggest the involvement of Gabpb1 in regulating photoperiod- and temperature-induced changes in tissue remodeling.
a CRISPR/Cas9 targeting of the mouse Gabpb1 exon 9. Schematic representation of the mouse Gabpb1 gene illustrates crRNAs (blue characters) and the protospacer adjacent motif (PAM) sequences (red characters) tailored to exon 9. Sequencing of the deletion band shows ligation of the Cas9 target site. PCR across the genomic deletion region shows the smaller PCR product in mutants. Western blotting of Gabpβ1 with an antibody recognizing both long and short forms shows absence of long form of Gabpβ1 in the mutant homozygous (Mut/Mut) mice. β-actin was detected as loading control. b Effects of 1-week exposure to SC or LW conditions on Gabpb1 expression and organ weight in Gabpb1 exon 9 deletion mutant heterozygous mice and their wild-type littermates (two-way ANOVA, Šídák’s multiple comparisons test, mean + SEM, +/+: n = 3; +/Mut: n = 4). Numbers within the graph indicate the P value. c Immunohistochemistry of Ki67 in the heart (HEA), pancreas (PAN), and kidney (KID). In WT mice, the number of Ki67 immuno-positive cells (red arrows) was higher under SC than LW conditions in the HEA and PAN, while glomerular size was larger under SC than LW conditions in the KID. These differences were smaller in Gabpb1 heterozygous mutant mice. L: Islet of Langerhans; G: Glomerulus. d H&E staining of cardiomyocytes. Changes in cardiomyocyte diameter between SC and LW conditions were more obvious for WT than Gabpb1 heterozygous mice. e Immunohistochemistry of β-catenin, which localizes to the acinar cell membrane. Changes in the size of pancreatic acinar cells between SC and LW conditions were more obvious in WT than Gabpb1 heterozygous mice. Scale bars: 50 μm. f Effects of 1-week exposure to SC or LW conditions on Ki67-positive cell numbers, cardiomyocyte diameter, pancreatic acinar cell size, and glomerular size in Gabpb1 exon 9 deletion mutant heterozygous mice and their wild-type littermates (two-way ANOVA, Šídák’s multiple comparisons test, mean + SEM, +/+: n = 3; +/Mut: n = 4). Numbers within the graph indicate the P value.
Changes in disease-related genes
To understand the molecular basis of seasonally regulated diseases, we used the human gene-disease association data source called DisGeNET database in the Metascape20 to search for disease risk factor genes in SOGs common to male and female macaques. We found that SOGs are closely related to several diseases (Fig. 8a), and identified their risk factor genes (Fig. 8b) in the lung (LUN), aorta (AOR), bronchus (BRO), and colon (COL). For example, seasonal rhythms in the expression of TLR3 and CCR2, and enrichment in ‘pneumonia and influenza’ were observed in the LUN (Fig. 8a, b). In the AOR, CD36 and CXCL16 showed seasonal transcription rhythms and ‘vascular inflammation’ and ‘acute coronary syndrome’ were enriched, while in the BRO, CD40 and C3 expression showed seasonal oscillation and ‘sinusitis’ was enriched (Fig. 8a, b). Furthermore, in the COL, ATF4 and IRF8 showed seasonal rhythms and ‘bacterial infection’ was enriched (Fig. 8a, b).
a DisGeNET enrichment analysis results for SOGs in the lung (LUN), aorta (AOR), bronchus (BRO), and colon (COL). b Heatmaps showing the expression profiles of SOGs. Genes involved in representative diseases are highlighted. c Seasonal expression profiles of ACE2 and TMPRSS2 in the BRO in males and females. d Heatmaps showing the expression profiles of SOGs. Risk factors for psychiatric diseases are highlighted.
The emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and its associated symptoms, named coronavirus disease 2019 (COVID-19), has caused a devastating pandemic worldwide. During the epidemic phase, the timing and magnitude of waves of SARS-CoV-2 infection are influenced by a variety of factors, such as vaccination, imposition and cessation of non-pharmaceutical interventions (e.g., social lockdown, travel restrictions, use of masks), and virus evolution leading to increased transmissibility. However, during the endemic period, and in the absence of new interventions, SARS-CoV-2 infection is expected to follow seasonal patterns similar to other seasonal coronaviruses such as OC43, NL63, and 229E45,46. Current evidence suggests that the sexual dimorphism in COVID-19, with higher severity and mortality in males than in females47. SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) and the serine protease TMPRSS2 to enter cells. Notably, dynamic seasonal fluctuations in TMPRSS2 were observed in the BRO in both sexes of macaques in this study, with the rhythmicity being more pronounced in males than in females (Fig. 8c). Clinical studies show more severe COVID-19 outcomes in patients with diabetes, obesity, cardiovascular disease and hypertension, and these comorbidities have been suggested to be associated with the expression of ACE2 and TMPRSS2 in peripheral tissues48. Indeed, we found expression of these receptors in various peripheral tissues, with particularly high expression in the digestive and respiratory systems (Supplementary Fig. 7).
Psychiatric diseases such as schizophrenia, depression, and bipolar disorder are also seasonally regulated3,6,7. We found seasonal oscillations in the expression profiles of genes associated with schizophrenia in multiple brain regions (Fig. 8d). For example, LRRTM1 and DLG2 showed seasonal oscillations in the medial prefrontal cortex, while PPP1R1B exhibited seasonal rhythms in the amygdala central nucleus and ventral tegmental area.
Drug efficacy could relate to seasonal changes
Accumulating evidence has demonstrated the time-of-day dependent efficacy of various drugs, and chronotherapy aims to optimize medical treatments by taking into account circadian rhythms49. However, the season-dependent efficacy of drugs remains unknown. To identify drugs whose efficacy is likely to be affected by the seasons, we compared all 19,003 SOGs in this study against the Drug Gene Interaction database (DGIdb v5.0)50. Among the top 50 drugs ranked by the number of targeted SOGs, ~75% were antineoplastics according to the USP Medicare Model Guidelines (Fig. 9a, b, Supplementary Fig. 8a, and Supplementary Data 9). As we demonstrated seasonally regulated tissue remodeling (Figs. 6 and 7), the enrichment of antineoplastics appears reasonable. In addition, steroid hormones showed seasonal variation (Fig. 1 and Supplementary Fig. 2), so identification of the hormonal agents dexamethasone and progesterone seems plausible (Fig. 9a, b). Cardiovascular disease, metabolism, immune function, and psychiatric disorders are seasonally regulated. It is noteworthy that the cardiovascular agent atorvastatin, the blood glucose regulator metformin hydrochloride, the immunological agents sirolimus and cyclosporine, and the antipsychotics haloperidol and olanzapine were detected in the top 50 drugs (Fig. 9a, b). It is also interesting to note that analgesics such as celecoxib, aspirin, and alcohol were found in the top 50 drugs. Among the top 50 drugs, we examined the effects of LW or SC conditions on the effects of EtOH administered by oral gavage in C57BL/6 N mice. Spontaneous locomotor activity, as measured by total distance traveled in the open field test, was significantly reduced at 1 h after EtOH administration under both SC and LW conditions compared with water-treated control mice. SC mice recovered at 3 h after administration, while LW mice continued to show reduced activity (Fig. 9c). To verify seasonal changes in alcohol efficacy, we also examined the effects of EtOH on motor coordination using the accelerating rotarod. As expected, EtOH-treated mice had worse rotarod performance than control mice at 4 h post-treatment under both SC and LW conditions. While the rotarod endurance time improved in SC mice at 5 h after dosing, at this time point it was still significantly shorter in LW mice compared to control mice (Fig. 9d). Plasma EtOH measurements further confirmed the reduced alcohol metabolism under the LW condition compared to the SC condition at 3 and 5 h post-treatment (Fig. 9e). These results demonstrate changes in alcohol efficacy can occur with changes in season-related variables. It is interesting to note that the monthly number of alcohol-related hospital admissions peaks in summer51.
a Distribution of drugs ranked according to the number of targeted SOGs. All 19,003 SOGs were compared against the Drug Gene Interaction database. b Therapeutic categories of the top 50 drugs ranked by the number of targeted SOGs. c Effects of alcohol on spontaneous locomotor activity of mice kept under SC or LW conditions. Graphs depict the distance traveled in the open-field test at 1 and 3 h after EtOH administration. Numbers within the graph indicate the P value (two-way ANOVA, Šídák’s multiple comparisons test, mean + SEM, LW water treated group: n = 7, other groups: n = 8). d Effects of EtOH on the rotarod performance of mice kept under SC or LW conditions. Graphs depict the latency to fall from the rotarod as determined across five trials at 4 and 5 h after the administration. Numbers within the graph indicate the P value (two-way ANOVA, Šídák’s multiple comparisons test, mean + SEM, LW water treated group: n = 7, other groups: n = 8). e Effects of SC and LW conditions on EtOH metabolism. Numbers within the graph indicate the P value (two-way ANOVA, Šídák’s multiple comparisons test, mean + SEM, n = 8). f Distribution of SOGs ranked according to the number of targeting drugs.
We also listed the SOGs ranked by the number of targeting drugs (Fig. 9f, Supplementary Fig. 8b, and Supplementary Data 10). Among the top 25 genes, we found receptors for seasonally oscillating testosterone and estrogen, AR and ESR1, respectively. Among the top 10 genes, NFE2L2, VDR, ALDH1A1, CYP2D6, and CYP2C19 are of particular interest. Our previous study showed that the NRF2 antioxidant pathway, encoded by the NFE2L2 gene, was involved in the winter depression-like behavior of medaka fish52. VDR encodes the receptor for vitamin D, and the relationship between vitamin D and seasonality has been discussed for a long time53. ALDH1A1 metabolizes acetaldehyde to acetate and is associated with alcohol-related phenotypes such as alcohol consumption levels, alcohol dependence risk, and blood alcohol metabolism54. This fits well with the results shown in Fig. 9c–e. CYP2D6 and CYP2C19 are known to be involved in drug metabolism. In particular, CYP2D6 metabolizes as many as 25% of common drugs, suggesting that the efficacy of these drugs could be change according to season.
Discussion
The molecular mechanisms underlying seasonal reproduction have been studied extensively in the past few decades, providing insight into conservation and divergence among taxa. For example, key regulators of seasonal reproduction, such as the springtime hormone TSH and thyroid hormone deiodinases, are evolutionarily conserved among various vertebrate species1,25,26,27,55,56. In contrast, diversity is observed in light input pathways. In non-mammalian vertebrates, light detected by deep brain photoreceptors regulates TSH and deiodinases, while light detected by the eyes regulates nocturnal melatonin secretion, and hence TSH and deiodinases in mammals. However, studies of seasonal reproduction have typically focused on a limited number of tissues, such as the hypothalamus, pituitary gland, and gonads1,25,26,27,55. To understand dynamic seasonal changes in various physiological processes, comprehensive analyses across various tissue types are essential. Our NHPSTA of 80 neural and peripheral macaque tissues identified SOGs in all tissues examined (Fig. 2). Analysis of common SOGs in each tissue successfully provided insights into the molecular basis of seasonally regulated physiology, including metabolism in the brown adipose tissue, epidermis development in the skin, immune function in lymph node tissue and spleen, and carbohydrate metabolism in the duodenum. Interestingly, a limited number of SOGs (average: ~9%) were common between males and females (Fig. 3). Because expressed genes in the two sexes showed significant overlap (average: ~92%) (Supplementary Fig. 4), we inferred that sex differences were specific to SOGs. However, it is noteworthy that we used the cosinor method to identify SOGs in the present study to minimize false positives. If we could detect SOGs with irregular curve shapes by improving the resolution of the sampling time points, the number of common SOGs between males and females might be higher. Nevertheless, differences in SOGs may contribute to sex differences in various seasonal phenotypes, such as animal reproduction and susceptibility to winter depression7,17.
To cope with seasonal environmental changes, various species, from unicellular organisms to vertebrates, have evolved an endogenous circannual clock of approximately 1 year. The circannual clock regulates the timing of various physiological processes, such as reproduction, molting, hibernation, and migration to improve fitness. The evolutionary conservation of a circannual rhythm supports its functional significance23. Although several hypotheses have been proposed regarding the mechanisms underlying circannual rhythms, the molecular basis remains unclear. Almost every tissue harbors an ~24 hr circadian clock, and core circadian clock genes exhibit clear daily oscillations in a wide range of tissues19. Therefore, highly conserved SOGs expressed in various tissues are of interest. The most frequently detected SOGs in multiple tissues were genes related to the aerobic electron transport chain (Fig. 5). This appears to reflect the dynamic seasonal changes in metabolism in various tissues. This was consistent with the results of recent analysis of seasonal genes across human tissues produced by the GTEx consortium, revealing functional enrichment for the aerobic electron transport chain57.
Our transcriptional regulatory network analysis and siRNA knockdown in macaque fibroblasts suggested the involvement of the ETS-family transcription factor GABP in the regulation of SOGs (Fig. 5). GO analysis of GABP-targeted SOGs highlighted the pathways involved in tissue remodeling. Interestingly, exposure to summer- and winter-mimicking conditions resulted in profound changes in organ weights and tissue remodeling in both melatonin-proficient CBA mice and melatonin-deficient C57BL mice (Fig. 6). Although melatonin plays a critical role in seasonal reproduction in mammals40, our results suggest that melatonin is not essential for seasonal tissue remodeling and that the commonly used C57BL strain could be a useful model for studying seasonal tissue remodeling. Indeed, heterozygous Gabpb1 mutant mice had attenuated responses to photoperiod and temperature changes compared with those of wild-type littermates with a C57BL genetic background (Fig. 7). These results suggest a molecular mechanism for whole-body seasonal adaptation. Since our recent monthly transcriptome analysis of medaka also suggested the involvement of tissue remodeling, dynamic seasonal tissue remodeling appears to be the key mechanism underlying seasonal rhythms31. Of note, seasonal gland mass changes have also been reported in humans8. GABP is intimately involved in critical cellular functions, including protein synthesis, development, and cellular differentiation, and is evolutionarily conserved from nematodes to humans32,33. Therefore, its functional significance in seasonal adaptation in other organisms is of interest.
In humans, there is substantial evidence for seasonal variation in disease risk with evolutionarily conserved mechanisms6. However, the molecular mechanisms underlying this seasonal variation remain unclear. Our analysis revealed seasonal oscillations in the expression of risk factor genes for several diseases (Fig. 8). Detailed analyses of seasonally regulated risk factor genes and the pharmacological manipulation of SOGs are promising for the development of circannual clock-based therapeutics.
We also observed marked sex differences in SOGs and in the distributions of their peak phases (Figs. 2–4). These observations may help to clarify the underlying mechanisms of sex differences in the susceptibility to various diseases. In addition, we demonstrated seasonal changes in alcohol efficacy, and identified drugs and target genes that are likely to be affected by the seasons. Therefore, tailoring treatments according to the time of year may maximize the efficacy of precision medicine and thus improve patient outcomes.
In conclusion, our study identified the molecular basis of whole-body seasonal adaptation, and NHPSTA provides a platform to discover potential biomarkers and targets for the development of therapeutic interventions and chronotherapies for seasonally regulated diseases.
Methods
Ethics
All experiments were carried out in accordance with the ARRIVE guidelines, the guidelines for Care and Use of Non-Human Primates (version 3, 2010) published by Kyoto University, and the guidelines of Nagoya University and RIKEN. All experiments were approved by the Animal Care and Use Committee of Kyoto University, the Animal Experimental Committee of Nagoya University, and the Institutional Animal Care and Use Committee of the RIKEN Kobe branch.
Animals
Twenty six adult rhesus macaques (Chinese origin) were kept in an open enclosure (524 m2) with environmental enrichment, including various feeders, climbing structures, and swings with natural light and temperature at the Center for the Evolutionary Origins of Human Behavior, Kyoto University, Inuyama, Japan (Supplementary Fig. 1). Monkeys were provided with two meals per day at the estimated ad libitum levels, and water was always available ad libitum. Keepers and veterinarians routinely monitor the health of rhesus macaques. The macaques did not have apparent injuries or clinical symptoms for several years prior to or on the day of euthanasia. In addition, we did not find any apparent organ abnormalities during sample collection. Nine week-old male CBA/N mice (n = 16) and C57BL/6 N mice (n = 52) were purchased from Japan SLC Inc. (Hamamatsu, Japan) and CLEA Japan Inc. (Tokyo, Japan), respectively. After 1 week of habituation, mice were housed under summer-mimicking long-day and warm conditions (LW: 16 h light 8 h dark, 30 °C) or winter-mimicking short-day and cool conditions (SC: 8 h light 16 h dark, 10 °C) using an animal housing system (LP-30CCFL-8CTAR, NK system; Nippon Medical & Chemical Instruments, Osaka, Japan) for 1 month for organ weight analysis and 1 week for histological analysis. Food (Labo MR Stock, Nosan Corp.) and water were provided to mice ad libitum.
Sample collection
All monkeys were moved from an open enclosure to individual cages 1-2 days prior to euthanasia. The individual cages were located in a room with a window, and the room light was turned on and off at the sunrise and sunset, respectively. The monkeys were fed 100 g pellets (AS, Oriental Yeast Co., Ltd., Tokyo, Japan) twice a day at approximately 11:00 and 15:00 prior to the day of euthanasia. Water was available ad libitum. The monkeys were immobilized with 10 mg/kg body weight (BW) ketamine hydrochloride (intramuscularly), and anaesthetized with 80 mg/kg BW pentobarbital (intravenously). Blood was collected from the HEA for hormone and metabolite measurements with 0.13% EDTA. Tissues were collected after perfusion with ice-cold Ames’ medium (Sigma, St. Louis, MO). Each brain region was punched out (2 mm diameter) from 2 mm brain slices generated using a monkey brain matrix (MBM-2500C, ASI Instruments, Warren, MI)58. Mice were euthanized by isoflurane anaesthesia (Wako 099-06571) followed by cervical dislocation at middle of the day under both SC and LW conditions. Tissues samples were collected for qPCR and histological analyses.
Hormone measurement
Plasma was obtained by centrifugation (1300 g, 24 °C, 10 min) and immediately frozen in liquid nitrogen. Measurement of the levels of plasma testosterone, E2, IGF-I, free T4, and cortisol levels was conducted by SRL Inc. (Tokyo, Japan) by electro chemiluminescence immunoassay (ECLIA) (Roche Diagnostics, Basel, Switzerland). The lower limit of detection for E2 was 5.0 pg/mL, and samples under the limit are labeled as ‘not detected (ND)’.
Metabolomics
Metabolomics analysis of plasma samples was conducted by Human Metabolome Technologies (Tsuruoka, Japan)59. Twelve plasma samples were mixed with methanol and extra-filtered (9100 g, 4 °C, 120 min). Capillary electrophoresis-mass spectrometry (CE-MS) was performed using an Agilent CE-MS system (Agilent Technologies, Santa Clara, CA) equipped with a time-of-flight mass spectrometer system. Identified metabolites were quantified by comparing their peak areas with those of authentic standards using MasterHands ver.2.17.1.11 software. The following criteria were applied to extract the seasonally oscillating plasma metabolites: i) Jonckheere–Terpstra–Kendall (JTK_CYCLE) algorithm, Padj <0.2 in all samples; ii) Welch’s t-test, P < 0.05 for February and April vs. August and October, April and June vs. October and December, and June and October vs. December and February.
RNA extraction and sequencing of macaque tissues
A total of 80 tissues, specifically from 30 brain regions and 50 peripheral tissues, were collected at around 14:00 (JST) from one male and one female every 2 months from October 2019 to August 2020. All samples were snap-frozen in liquid nitrogen and kept at -80 °C before RNA preparation. Total RNA was extracted using the RNeasy Plus Universal Mini Kit (Qiagen, Venlo, Netherland). RNA concentrations were measured with a Quant-iTTM RNA Assay kit (ThermoFisher Scientific, Waltham, MA) using a Qubit Fluorometer (Thermo Fisher Scientific), and integrity was assessed with an Agilent RNA 6000 Nano Kit on a Bioanalyzer (Agilent Technologies). Each 3’RNA-Seq library was prepared by the Lasy-Seq v1.1 protocol (https://sites.google.com/view/lasy-seq/). A maximum of 96 tagged libraries were mixed and sequenced with an Illumina HiSeqX Ten (Illumina, San Diego, CA) using paired-end sequencing (PE150). Mapping and quantification were performed by bowtie (v1.2.3) and RSEM (v1.3.3) using the Macaca mulatta genome (Genome assembly: Mmul_10) as a reference. To compensate for insufficient annotation, we merged three annotation files (Ensembl: Macaca_mulatta.Mmul_10.192.gtf; NCBI: GCF_003339765.1_Mmul_10_genomic.gtf; GSE128537_assembly.rheMac10.gtf), and assembled a total of 54,213 genes. Gene expression was presented as the fragments per million (FPM).
Identification of SOGs
Since our preliminary analysis revealed significant variations in the seasonal oscillation of transcriptomes between tissues and sexes, SOGs were separately extracted from each tissue and sex. To reduce false positives caused by small sample sizes, strict parameters were applied at multiple steps during the analysis, including filtering out the low expressed genes and SOG identifications. First, fragments per million (FPM) values were log-transformed by log2 (FPM + 0.5) and genes with an average value > 2.5 across six sampling time points were designated as expressed genes. Then, a cosine curve was fitted for each expressed gene with a period of 365 days using the nonlinear least squares (nls) function in R, and the coefficient of determination (R-squared, RS) was used to evaluate the goodness of fit. The amplitude (alpha) of the fitted curve was defined as the difference between the maximum and minimum of the seasonal oscillation and the day at the maximum of the fitted curve was defined as the phase (phi), which represents the expression peak of the gene. Finally, SOGs were extracted if RS > 0.7 and alpha > 1 representing a two-fold difference between the maximum and minimum expressions.
GO analysis
In order to investigate the pathways showing seasonal oscillations, the web-based portal Metascape 3.518 was used to perform GO analysis of the SOGs extracted from each tissue and sex. Gene symbols for the SOGs were specified in the program, and SOGs were analyzed as human genes using the Express Analysis pipeline with default parameters.
Cell culture and siRNA knockdown
Macaque fibroblast cells established from cultures of a tiny block of ear skin were maintained in high-glucose Dulbecco’s modified Eagle medium (DMEM) supplemented with 12% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (ThermoFisher Scientific) at 37 °C with 5 % CO260. Commercial siRNA targeting human GABPA and GABPB1, as well as negative control siRNA, were purchased from Qiagen (FlexiTube GeneSolution siRNA). A total of 40 pmoles siRNA composed of equal amounts of the four different siRNAs from the same FlexiTube GeneSolution product was used for the co-transfection experiments, and the final transfection mixture contained 40 pmoles for single-gene knockdown and 80 pmoles for knockdown of both GABPA and GABPB1. Cells were seeded in a six-well plate at a density of 1.2 × 105 cells/well the day before transfection. On the next day, each well was transfected with siRNA or negative control siRNA using the jetPRIME transfection reagent (Polyplus, Illkirch, France). Forty-eight hours after transfection, cells were harvested and total RNA was prepared using the RNeasy Lipid Tissue Mini Kit (Qiagen).
RNA-seq analysis of macaque fibroblasts
mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and was then fragmented, and the fragments were used to synthesize cDNA with random N6 primers. PE100 strand-specific sequencing libraries were constructed from the prepared cDNA and subsequently sequenced on the DNBseq platform with a yield of 50 – 60 million reads for each library (BGI, Shenzhen, China). Clean reads for the downstream analysis were obtained by excluding reads with adaptors or unknown bases [(N) > 0.1%] or low-quality reads that contained > 40% of the bases with a Phred score <20 as determined using the BGI internal program SOAPnuke. Then Bowtie261 was used to map the clean reads to our custom reference transcriptome that was previously prepared for the SOG analysis. Gene expression was quantified using the RSEM program62 and presented as the fragments per kilobase of transcript per million mapped reads (FPKM). Because knockdown efficiencies for GABPA and GABPB1 were relatively low, DEGs between control and double knockdown of GABPA/GABPB1 were detected using the DEseq2 program63 with an Padj <0.05 and fold change > 1.5. Overlapping genes between the SOGs and DEGs were identified using the intersect() function in R64 (https://www.R-project.org/) and subsequent GO annotations were performed using Metascape with the same parameters for the SOGs.
qPCR validation
RNA was reverse transcribed into cDNA using the ReverTra Ace qPCR RT Master Mix kit (Toyobo, Osaka, Japan). The effect of siRNA on gene expression was evaluated by qPCR in a 20 μL mixture containing SYBR™ Green PCR Master Mix (ThermoFisher Scientific) and 0.25 μM primers. The reaction was performed with a program consisting of an initial 2 min at 50 °C and 10 min at 95 °C, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 1 min on a QuantStudio 3 Real-Time PCR instrument (Applied Biosystems, Carlsbad, CA). We screened several housekeeping genes, including the housekeeping gene primer set (TaKaRa, Otsu, Japan), to find the most suitable internal control genes, and GAPDH and Rps18 were used as internal control genes for macaque fibroblasts and mouse tissues, respectively. Relative expression levels were calculated using the ΔΔCt method. All the qPCR primers used in this study are listed in Supplementary Data 11.
Histological analysis
Tissues were fixed with 4% paraformaldehyde phosphate (09154-85, Nacalai Tesque, Kyoto, Japan) for 24 h, then transferred to phosphate-buffered saline (PBS) and 70% ethanol, dehydrated and embedded in paraffin. Sections of 5 μm thickness were prepared for H&E, Ki67, and β-catenin staining. Tissue sections were deparaffinized and rehydrated with xylene and an ethanol series, and subjected to antigen retrieval by citrate buffer (ab93678, Abcam, Cambridge, UK) at 95 °C for 10 min. After blocking, the sections were incubated with anti-Ki67 at 1:200 dilution (ab16667, Abcam) and anti-β-catenin at 1:100 dilution (610154, BD Transduction Laboratories, Franklin Lakes, NJ) overnight at 4 °C. Signals were detected using DAB substrate provided in the Mouse and Rabbit Specific HRP/DAB IHC Detection Kit - Micro-polymer (ab236466, Abcam). Cell nuclei were additionally stained with Mayer’s hematoxylin (131-09665, Wako, Osaka, Japan), sequentially dehydrated, and mounted using VectaMount permanent mounting medium (H−5000, Vector Laboratories, Newark, CA). Glomerular size, cardiomyocyte diameter, and pancreatic acinar cell size were measured using image analysis software (BZ-H4M, BZ-H4C, and BZ-H4CM, Keyence, Osaka, Japan) by two researchers in a blinded manner.
Generation of Gabpb1 exon 9 deletion mutant mice
Targeted deletion of exon 9 of the Gabpb1 gene was performed in fertilized mouse eggs by microinjection using the CRISPR-Cas9 System. Ribonucleoprotein complex (crRNA:tracrRNA duplex 30 ng/μL, Cas9 protein 100 ng/μL, Alt-R CRISPR-Cas9 System [Integrated DNA Technologies: IDT, Coralville, IA]) was microinjected into the cytoplasm of C57BL/6NJcl (CLEA) fertilized eggs. After microinjection, embryos were transferred into the oviducts of 14- to 18-week-old pseudopregnant Slc:ICR female mice (Japan SLC) weighing 36-45 g. A combined anesthetic (0.3 mg/kg of medetomidine (Zenoaq, Fukushima, Japan), 4.0 mg/kg of midazolam (Astellas Pharma, Tokyo, Japan), and 5.0 mg/kg of butorphanol (Meiji Seika Pharma, Tokyo, Japan) was administered to recipient mice by intraperitoneal injection. After surgery, 0.3 mg/kg of atipamezole hydrochloride (Zenoaq) was administered by intramuscular injection65.
In the CRISPR/Cas9 system, four crRNA targeting sequences, specifically CACAGCAAGCCACTGTTACG, GTCTTGAACTGTACACAGTA, GAGACTGTTCATTAATTCCA, and CTCATAATTCAGACGAGAAC, were designed on both sides of the Gabpb1 exon 9 coding region using the CRISPR-Cas9 guide RNA design checker (IDT; https://sg.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE), a web tool for assessing on- and off-targeting potential.
Genotyping was performed by direct PCR. Briefly, an ear punch sample was lysed using 2 μL of 20 mg/mL Proteinase K in 125 μL of Tris-EDTA buffer at 55 °C overnight. The lysed samples were heat-inactivated at 95 °C for 5 min and used as a template for direct PCR using KOD FX Neo DNA polymerase (Toyobo, Osaka, Japan). A primer set, TGGTCTAAGAGACAACAACCACG and TCAGTATATAAAGAAGTACCGGTGTGC, was used to amplify the targeted region. The PCR products were supplemented with Loading Dye Brilliant Color (6×) (Nacalai Tesque, Kyoto, Japan) and analyzed by electrophoresis in 1.5% agarose gel (STAR Agarose; Rikaken, Tokyo, Japan). The DNA sequences of the PCR products were confirmed by direct sequencing. Briefly, the PCR product in a slice of agarose gel was extracted using the FastGene Gel/PCR Extraction Kit (Nippon Genetics, Tokyo, Japan). Sanger sequencing was performed by FASMAC Co., Ltd. (Atsugi, Japan) using the extracted PCR product with the corresponding forward or reverse primer.
For western blotting, frozen kidney collected from mouse pups was lysed with ice-cold radio-immunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors. Sonicated lysate was centrifuged (10,000 g, 4 °C, 20 min), and the supernatant was collected. Protein samples were boiled in 1 × sodium dodecyl sulfate (SDS) sample buffer for 5 min, separated by SDS-poly acrylamide gel electrophoresis (PAGE) on 15% gels and transferred to activated polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were blocked with 5% skim milk in Tris-buffered saline with Tween 20 (TBST) for 1 h at room temperature. GABPβ1 primary antibody (1:3000 dilution) (12597-1-AP; Proteintech, Chicago, IL)66, dissolved in 5% skim milk in TBST, was incubated 2 h at room temperature. Membranes were washed with TBST. Secondary rabbit antibody (1:5000 dilution) (NA934; Cytiva, Tokyo, Japan), dissolved in 5% skim milk in TBST, was incubated for 2 h at room temperature. Washed membranes with TBST was incubated with ECL prime (Cytiva) for 5 min at room temperature and imaged on a LuminoGraph II EM instrument (ATTO, Tokyo, Japan). β-actin primary antibody (1:1,000 dilution) (sc-47778; Santa Cruz Biotechnology, Dallas, TX) was used as an internal control after GABPβ1 signals were eliminated by an incubation in H2O2. The sizes of the protein were determined by Amersham ECL Rainbow Marker - Full range (Cytiva).
Mouse behavior analyses
Ethanol was used at a concentration of 4 g/kg BW based on a previous study67. Mice were kept under summer-mimicking LW conditions or winter-mimicking SC conditions for at least 1 month before the behavioral experiments. EtOH diluted in MilliQ water or MilliQ water alone were administered orally by gavage immediately after the light offset. Open field test: One and three h after a single oral gavage of EtOH, mice were placed in the center of the open field arena and behavior was recorded for 6 min using an over-head video camera under dim red light. Total distance traveled was calculated by Smart 3.0 software (Panlab Harvard Apparatus, Cornellà, Spain) to quantify spontaneous locomotor activity. Rotarod test: Effects of EtOH on motor coordination were analyzed by rotarod (Panlab Harvard Apparatus). All mice were habituated and trained to remain in the rotarod for 2 d prior to EtOH administration. The rotarod speed was set to increase gradually from 4 to 40 rpm over the course of 5 min, according to the previous study67. Four and five hours after a single oral gavage of EtOH, mice were submitted to testing session consist of five trials. We collected blood samples from the jugular vein 3 and 5 h after EtOH administration, and plasma EtOH levels were measured using the EnzyChrom Ethanol assay kit ECET-100 (BioAssay Systems, Hayward, CA).
Statistics
Statistical analyses were performed using GraphPad Prism10 (Graphpad Software Inc., San Diego, CA). Results are presented as the mean ± SEM. Data were analyzed using one-way or two-way ANOVA with Dunnett’s test or Šídák’s multiple comparisons tests where appropriate.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data sets generated in this study have been deposited in the NCBI’s Gene Expression Omnibus and are available through GEO series accession numbers GSE201986 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi] and GSE201987 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi]. To facilitate exploration of this resource, we have created NHPSTA web database (https://rhythm.itbm.nagoya-u.ac.jp/NHPSTA/). Source Data are provided with this paper. All codes used in the analysis are available at https://github.com/animalphysiology/ script_medaka2023.git. Source data are provided with this paper.
Code availability
This study did not use new codes.
References
Nakane, Y. & Yoshimura, T. Photoperiodic regulation of reproduction in vertebrates. Annu. Rev. Anim. Biosci. 7, 173–194 (2019).
Roenneberg, T. & Aschoff, J. Annual rhythm of human reproduction: I. Biology, sociology, or both? J. Biol. Rhythms 5, 195–216 (1990).
Wehr, T. A. Photoperiodism in humans and other primates: Evidence and implications. J. Biol. Rhythms 16, 348–364 (2001).
Foster, R. G. & Roenneberg, T. Human responses to the geophysical daily, annual and lunar cycles. Curr. Biol. 18, R784–R794 (2008).
Dopico, X. C. et al. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun. 6, 7000 (2015).
Stevenson, T. J. et al. Disrupted seasonal biology impacts health, food security and ecosystems. Proc. R. Soc. B 282, 20151453 (2015).
Wirz-Justice, A. Seasonality in affective disorders. Gen. Comp. Endocrinol. 258, 244–249 (2018).
Tendler, A. et al. Hormone seasonality in medical records suggests circannual endocrine circuits. Proc. Natl. Acad. Sci. USA 118, e2003926118 (2021).
Walker, M. L., Wilson, M. E. & Gordon, T. P. Endocrine control of the seasonal occurrence of ovulation in rhesus monkeys housed outdoors. Endocrinology 114, 1074–1081 (1984).
Chik, C. L. et al. Photoperiod-driven changes in reproductive function in male rhesus monkeys. J. Clin. Endocrinol. Metab. 74, 1068–1074 (1992).
Qin, D. et al. The first observation of seasonal affective disorder symptoms in Rhesus macaque. Behav. Brain Res. 292, 463–469 (2015).
Patel, R. S. et al. Novel biomarker of oxidative stress is associated with risk of death in patients with coronary artery disease. Circulation 133, 361–369 (2016).
Fujigaki, H., Yamamoto, Y. & Saito, K. L-Tryptophan-kynurenine pathway enzymes are therapeutic target for neuropsychiatric diseases: Focus on cell type differences. Neuropharmacology 112, 264–274 (2017).
Wang, L. et al. Disturbance of neurotransmitter metabolism in drug-naïve, first-episode major depressive disorder: a comparative study on adult and adolescent cohorts. Eur. Arch. Psychiatry Clin. Neurosci. 272, 1283–1296 (2022).
Robinson, J. A., Scheffler, G., Eisele, S. G. & Goy, R. W. Effects of age and season on sexual behavior and plasma testosterone and dihydrotestosterone concentrations of laboratory-housed male rhesus monkeys (Macaca mulatta). Biol Reprod 13, 203–210 (1975).
Wilson, M. E., Gordon, T. P., Blank, M. S. & Collins, D. C. Timing of sexual maturity in female rhesus monkeys (Macaca mulatta) housed outdoors. J. Reprod. Fertil. 70, 625–633 (1984).
Stephens, S. B. & Wallen, K. Environmental and social influences on neuroendocrine puberty and behavior in macaques and other nonhuman primates. Horm. Behav. 64, 226–239 (2013).
Yin, S. et al. Transcriptomic and open chromatin atlas of high-resolution anatomical regions in the rhesus macaque brain. Nat. Commun. 11, 474 (2020).
Mure, L. S. et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359, eaao0318 (2018).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Randall, V. A. & Ebling, F. J. Seasonal changes in human hair growth. Br. J. Dermatol. 124, 146–151 (1991).
Rosenthal, N. E., Genhart, M., Jacobsen, F. M., Skwerer, R. G. & Wehr, T. A. Disturbances of appetite and weight regulation in seasonal affective disorder. Ann. NY Acad. Sci. 499, 216–230 (1987).
Gwinner, E. Circannual rhythms (Springer-Verlag, Berlin, 1986).
Michael, R. P. & Bonsall, R. W. A 3-year study of an annual rhythm in plasma androgen levels in male rhesus monkeys (Macaca mulatta) in a constant laboratory environment. J. Reprod. Fertil. 49, 129–131 (1977).
Nakao, N. et al. Thyrotrophin in the pars tuberalis triggers photoperiodic response. Nature 452, 317–322 (2008).
Ono, H. et al. Involvement of thyrotropin in photoperiodic signal transduction in mice. Proc. Natl. Acad. Sci. USA 105, 18238–18242 (2008).
Ikegami, K., Refetoff, S., Van Cauter, E. & Yoshimura, T. Interconnection between circadian clocks and thyroid function. Nat. Rev. Endocrinol. 15, 590–600 (2019).
Wood, S. & Loudon, A. The pars tuberalis: The site of the circannual clock in mammals? Gen. Comp. Endocrinol. 258, 222–235 (2018).
Lincoln, G. A. Brief history of circannual time. J. Neuroendocrinol. 3, e12694 (2019).
Hazlerigg, D. G. & Lincoln, G. A. Hypothesis: Cyclical histogenesis is the basis of circannual timing. J. Biol. Rhythms 26, 471–485 (2011).
Nakayama, T. et al. A transcriptional program underlying the circannual rhythms of gonadal development in medaka. Proc. Natl. Acad. Sci. USA 120, e2313514120 (2023).
Sharrocks, A. D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell. Biol. 2, 827–837 (2001).
Rosmarin, A. G., Resendes, K. K., Yang, Z., McMillan, J. N. & Fleming, S. L. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol. Dis. 32, 143–154 (2004).
Patton, J., Block, S., Coombs, C. & Martin, M. E. Identification of functional elements in the murine Gabpα/ATP synthase coupling factor 6 bi-directional promoter. Gene 369, 35–44 (2006).
Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature 593, 575–579 (2021).
Dufty, A. M. Jr, Clobert, J. & Møller, A. P. Hormones, developmental plasticity and adaptation. Trend. Ecol. Evol. 17, 190–196 (2002).
Enard, W. et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137, 961–971 (2009).
Masumoto, K. H. et al. Acute induction of Eya3 by late-night light stimulation triggers TSHβ expression in photoperiodism. Curr. Biol. 20, 2199–2296 (2010).
Ikegami, K. et al. Tissue-specific posttranslational modification allows functional targeting of thyrotropin. Cell Rep 9, 801–810 (2014).
Reiter, R. J. The pineal and its hormones in the control of reproduction in mammals. Endocr. Rev. 1, 109–131 (1980).
Ebihara, S., Marks, T., Hudson, D. J. & Menaker, M. Genetic control of melatonin synthesis in the pineal gland of the mouse. Science 231, 491–493 (1986).
Ristevski, S. et al. The ETS transcription factor GABPα is essential for early embryogenesis. Mol. Cell. Biol. 24, 5844–5849 (2004).
Xue, H. H. et al. Targeting the GA binding protein β1L isoform does not perturb lymphocyte development and function. Mol. Cell. Biol. 28, 4300–4309 (2008).
Pham, C. T., MacIvor, D. M., Hug, B. A., Heusel, J. W. & Ley, T. J. Long-range disruption of gene expression by a selectable marker cassette. Proc. Natl. Acad. Sci. USA 93, 13090–13095 (1996).
Antia, R. & Halloran, M. E. Transition to endemicity: Understanding COVID-19. Immunity 54, 2172–2176 (2021).
Cohen, L. E., Spiro, D. J. & Viboud, C. Projecting the SARS-CoV−2 transition from pandemicity to endemicity: Epidemiological and immunological considerations. PLoS Pathog 18, e1010591 (2022).
Bechmann, N. et al. Sexual dimorphism in COVID-19: potential clinical and public health implications. Lancet Diabetes Endocrinol 10, 221–230 (2022).
Lazartigues, E., Qadir, M. M. F. & Mauvais-Jarvis, F. Endocrine significance of SARS-CoV−2’s reliance on ACE2. Endocrinology 161, 1–7 (2020).
Bicker, J., Alves, G., Falcão, A. & Fortuna, A. Timing in drug absorption and disposition: The past, present, and future of chronopharmacokinetics. Br. J. Pharmacol. 177, 2215–2239 (2020).
Cannon, M. et al. DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res 52, D1227–D1235 (2024).
Poikolainen, K. Seasonality of alcohol-related hospital admissions has implications for prevention. Drug Alcohol Depend 10, 65–69 (1982).
Nakayama, T. et al. Seasonal changes in NRF2 antioxidant pathway regulates winter depression-like behavior. Proc. Natl. Acad. Sci. USA 117, 9594–9603 (2020).
Wirz-Justice, A., Skene, D. J. & Münch, M. The relevance of daylight for humans. Biochem. Pharmacol. 191, 114304 (2021).
Lind, P. A. et al. Association between in vivo alcohol metabolism and genetic variation in pathways that metabolize the carbon skeleton of ethanol and NADH reoxidation in the alcohol challenge twin study. Alcohol Clin. Exp. Res. 36, 2074–2085 (2012).
Yoshimura, T. et al. Light-induced hormone conversion of T4 to T3 regulates photoperiodic response of gonads in birds. Nature 426, 178–181 (2003).
Nakane, Y. et al. The saccus vasculosus of fish is a sensor of seasonal changes in day length. Nat. Commun. 4, 2108 (2013).
Wucher, V., Sodaei, R., Amador, R., Irimia, M. & Guigó, R. Day-night and seasonal variation of human gene expression across tissues. PLoS Biol 21, e3001986 (2023).
Paxinos, G., Huang, X. F., Petrides, M. & Toga, A. W. The rhesus monkey brain in stereotaxic coordinates, second ed. (Academic Press, 2008).
Sugiura, Y., Taguchi, R. & Setou, M. Visualization of spatiotemporal energy dynamics of hippocampal neurons by mass spectrometry during a kainate-induced seizure. PLoS One 6, e17952 (2011).
Hirai, H. et al. Cytogenetic differentiation of two sympatric tree shrew taxa found in the southern part of the Isthmus of Kra. Chrom. Res. 9, 313–327 (2002).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference 376 genome. BMC Bioinformat 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2021). URL https://www.R-project.org/.
Sumiyama, K., Kawakami, K. & Yagita, K. A simple and highly efficient transgenesis method in mice with the Tol2 transposon system and cytoplasmic microinjection. Genomics 95, 306–311 (2010).
Wu, H. et al. The Ets transcription factor GABP is a component of the hippo pathway essential for growth and antioxidant defense. Cell Rep 3, 1663–1677 (2013).
Krahe, T. E. et al. Energy drink enhances the behavioral effects of alcohol in adolescent mice. Neurosci Lett 651, 102–108 (2017).
Acknowledgements
We thank Naoyuki Yamamoto for helpful discussion. This work was supported by the JSPS KAKENHI Grant-in-Aid for Specially Promoted Research 26000013 (TYoshimura), Grant-in-Aid for Scientific Research (S) 19H05643 (TYoshimura) and 24H00058 (TYoshimura), Grant-in-Aid for Transformative Research Areas (A), Chronoproteinology 24H02303 (TYoshimura), Cooperative Research Program of Primate Research Institute, Kyoto University (2018-C−28, 2019-B-9, 2020-A−2, 2021-A-4) (TYoshimura). WPI-ITbM is supported by the World Premier International Research Center Initiative (WPI), MEXT, Japan.
Author information
Authors and Affiliations
Contributions
T.Yoshimura conceived the study. J.C., K.O., L.R., Y.N., Y.C., K.F., M.M., T.M.N., T.O., H.I. and T.Yoshimura designed the research. J.C., K.O., L.R., Y.N., T.Nakayama, Y.C., K.F., S.S., T.M.N., T.O., Y.K., M.Z., K.K., S.M., Y.J.G., K.S., H.I., and T.Yoshimura conducted the experiment. J.C., K.O., L.R., Y.C., K.F., T.Yamaguchi, D.X.S., A.J.N., and T.Yoshimura analyzed the data. T.Natsume, N.S.H., M.M., I.T., T.N.O., and Y.M. provided new materials. J.C., K.O., L.R., Y.C., K.F., Y.K., K.S., and T.Yoshimura wrote the manuscript. All authors discussed the results and commented on the manuscript. T.Yoshimura supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, J., Okimura, K., Ren, L. et al. Non-human primate seasonal transcriptome atlas reveals seasonal changes in physiology and diseases. Nat Commun 16, 3906 (2025). https://doi.org/10.1038/s41467-025-57994-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57994-1