Abstract
Promoting endogenous cardiomyocyte proliferation is a promising strategy for cardiac repair. Identifying key factors that regulate cardiomyocyte proliferation can advance the development of novel therapies for heart regeneration. Here, we identify Foxk1 and Foxk2 as key regulators of cardiomyocyte proliferation, whose expression declines during postnatal heart development. Cardiomyocyte-specific knockout of Foxk1 or Foxk2 impairs neonatal heart regeneration after myocardial infarction (MI) injury. AAV9-mediated Foxk1 or Foxk2 overexpression extends the postnatal cardiomyocyte proliferative window and enhances cardiac repair in adult mice after MI. Mechanistically, Foxk1 and Foxk2 drive cardiomyocyte cell cycle progression by directly activating CCNB1 and CDK1 expression, forming the CCNB1/CDK1 complex that facilitates G2/M transition. Moreover, Foxk1 and Foxk2 promote cardiomyocyte proliferation by upregulating HIF1α expression, which enhances glycolysis and the pentose phosphate pathway (PPP), which further favors cardiomyocyte proliferation. These findings establish Foxk1 and Foxk2 as promising therapeutic targets for cardiac injury.
Similar content being viewed by others
Introduction
Ischemic heart disease, a leading cause of global mortality, imposes a significant burden on individuals and healthcare systems1. It is often driven by coronary artery occlusion, which typically progresses to myocardial infarction (MI), leading to myocardial ischemia and subsequent rapid, extensive cardiomyocyte death2. Furthermore, the lost cardiomyocytes cannot be replenished due to the limited proliferative capacity of adult cardiomyocytes3,4. As a result, the infarcted cardiac tissue was replaced by fibrotic scar tissue, which compromises the contractility of the remaining myocardium and leads to ventricular remodeling and, ultimately, heart failure5. In contrast, neonatal mammals are capable of fully restoring cardiac function in response to injury by facilitating endogenous cardiomyocyte proliferation and promoting heart regeneration through specific signaling pathways6,7,8,9. Studies on neonatal mouse heart regeneration have led to the identification of several key signaling pathways and targets involved in this process. These include cell cycle regulators10,11, epigenetic modifications12,13,14, transcriptional networks15,16,17, extracellular matrix18,19, angiogenic or lymphoangiocrine factors20,21,22, and immune cells and their derived cytokines23,24,25,26,27 among others. Identifying factors that promote cardiomyocytes to re-enter the cell cycle is therefore crucial for preventing cardiac remodeling following MI.
Forkhead box (Fox) proteins are a superfamily of evolutionarily conserved transcription factors characterized by their “forkhead” or “winged-helix” DNA binding domain (DBD)28. Among them, the Foxk family consists of two closely related members, Foxk1 and Foxk2, which share a similar structure and are ubiquitously expressed in various tissues and organs29. Foxk1 and Foxk2 have been implicated in regulating cell cycle progression by suppressing cell cycle inhibitors or promoting the expression of positive cell cycle regulators30,31. Specifically, Foxk1 has been shown to inhibit the expression of the cell cycle inhibitor P21, thereby enhancing the proliferation of myogenic progenitor cells and contributing to skeletal muscle regeneration32,33. In parallel, Foxk2 has been identified as a key regulator of cardiac development, where it activates cardiac lineage-specific genes and controls heart formation during human embryonic stem cell differentiation34. Furthermore, Foxk1 and Foxk2, in response to cellular stresses, promote aerobic glycolysis by upregulating genes involved in glycolytic enzyme expression and metabolic homeostasis, underscoring their pivotal role in maintaining cellular energy balance and facilitating adaptive responses to metabolic demands35,36. Given their essential functions in regulating the cell cycle and promoting glycolysis, both critical for cardiomyocyte proliferation, we hypothesize that Foxk1 and Foxk2 also play a key role in cardiomyocyte proliferation and heart regeneration.
In this study, we demonstrated that Foxk1 and Foxk2 are potent inducers of cardiomyocyte proliferation and heart regeneration. Cardiomyocyte-specific knockout of Foxk1 or Foxk2 impaired neonatal heart regeneration following MI. Upregulation of Foxk1 and Foxk2 extended the postnatal cardiomyocyte proliferative window and promoted heart regeneration in adult hearts after MI. Foxk1 and Foxk2 promoted cardiomyocyte proliferation via dual effects that enhance cell cycle re-entry and induce metabolic reprogramming. These findings highlight Foxk1 and Foxk2 as promising therapeutic targets for regenerative treatments in patients with ischemic heart diseases.
Results
Foxk1 and Foxk2 control cardiomyocyte proliferation
We analyzed the expression patterns of Foxk1 and Foxk2 across various developmental stages of mouse cardiomyocytes, including embryonic day 14.5 (E14.5), postnatal day 1 (P1), P4, P7, P14, and P56, using our previously obtained RNA-seq data37. The results showed that the expression levels of Foxk1 and Foxk2 were gradually decreased during heart development (Supplementary Fig. 1A). Western blot and qRT-PCR analysis also showed that the FOXK1 and FOXK2 expression in cardiomyocytes decreased with postnatal heart development (Fig. 1A and Supplementary Fig. 1B).
A Western blot and quantitative analysis of FOXK1 and FOXK2 expression in wild-type mouse cardiomyocytes at different ages (n = 3 per group). B, C Representative images and quantitative analysis of co-immunostaining of α-actinin (red) and pH3 (green) (B), as well as Ki67 (green) (C), indicated by yellow arrows. in primary cardiomyocytes transfected with siRNA to target Foxk1 (siFoxk1), Foxk2 (siFoxk2), or a scramble negative control (siNC) (n = 4 per group). D–I Representative images and quantitative analysis of co-immunostaining of α-actinin (red) and pH3 (green) (D, G), Ki67 (green) (E, H), and Aurora B (green) (F, I) in primary cardiomyocytes overexpressed Foxk1 or Foxk2 with adenovirus (n = 4 per group). Data are presented as mean ± SEM; p values determined by one-way ANOVA with Bonferroni multiple comparisons test (A–C and G–I). Source data are provided as a Source Data file.
To uncover whether Foxk1 and Foxk2 are involved in cardiomyocyte proliferation, we utilized siRNA to individually knock down these two genes in primary neonatal mouse cardiomyocytes (Supplementary Fig. 1C, E). Immunostaining of Ki67 and phosphorylated histone H3 Ser10 (pH3) were used to detect cardiomyocyte cell cycle entry and karyokinesis, respectively. Results showed that cardiomyocyte proliferation was significantly inhibited after 48 h of treatment with Foxk1 or Foxk2 siRNA (Fig. 1B, C). Conversely, overexpression of Foxk1 or Foxk2 using adenoviruses (Supplementary Fig. 1D, F) significantly increased the number of Ki67 and pH3 positive cardiomyocytes, demonstrating their role in promoting cardiomyocyte proliferation (Fig. 1D, E, G, H). Additionally, Aurora B immunostaining further confirmed the pro-proliferative role of Foxk1 and Foxk2 in cardiomyocytes by detecting cytokinesis (Fig. 1F, I). We also explored the synergistic effects of co-overexpressing Foxk1 and Foxk2, finding that Foxk1 and Foxk2 did not show an additive impact on enhancing cardiomyocyte proliferation (Supplementary Fig. 1G, H). This suggests that Foxk1 and Foxk2 regulate cardiomyocyte proliferation through a conserved mechanism.
Foxk1 and Foxk2 prolong the cardiomyocyte proliferative time window
To investigate whether Foxk1 and Foxk2 can sustain cardiomyocyte proliferation in vivo, we individually overexpressed these two genes in P1 mice with adeno-associated virus serotype 9 (AAV9) via subcutaneous injection of AAV9-Foxk1 and AAV9-Foxk2, in which target gene expression was driven by the cardiomyocyte-specific cTNT promoter (Fig. 2A). Western blot analysis revealed that FOXK1 and FOXK2 protein levels were significantly higher at P4, P7, and P14 in hearts infected with AAV9-Foxk1 and AAV9-Foxk2 compared to those infected with AAV9-GFP (Fig. 2B and Supplementary Fig. 2A, B). The cardiac morphology and the heart weight/body weight ratio showed no significant changes in the Foxk1 and Foxk2 overexpressing groups compared to the AAV9-GFP group (Fig. 2C and Supplementary Fig. 2C–E). Echocardiography also revealed no alterations in cardiac function and ventricular wall thickness (Supplementary Fig. 2F).
A Experimental design: neonatal mice were administered AAV9-GFP, AAV9-Foxk1, or AAV9-Foxk2 on postnatal day 1 (P1) to overexpress Foxk1 or Foxk2 specifically in cardiomyocytes. Heart tissue was subsequently collected at P14. B Western blot analysis of FOXK1 and FOXK2 expression in P14 mouse cardiomyocytes (n = 3 mice per group). C H&E-stained heart sections (n = 5 mice per group). D, E WGA staining of heart tissue sections and quantitative analysis of the cross-sectional area of cardiomyocytes (CM size) (n = 3 per group). F Analysis of total cardiomyocyte counts per heart across different experimental groups (n = 6 per group). G Representative images of cardiomyocytes isolated from the hearts of mice treated with AAV9-Foxk1 or AAV9-Foxk2 and quantification of nucleation at P14 (n = 3 per group). H–J Representative images of co-immunostaining and quantitative analysis for α-actinin (red) and Aurora B (green, indicated by yellow arrows) (H), pH3 (green) (I), and Ki67 (green) (J) in heart tissue sections (n = 4 per group). Data are presented as mean ± SEM. p values were determined by one-way ANOVA with Bonferroni multiple comparisons test (E, F, and H–J) and two-way ANOVA with Bonferroni multiple comparisons test (G). Source data are provided as a Source Data file.
Furthermore, wheat germ agglutinin (WGA) staining showed that overexpression of Foxk1 and Foxk2 reduced the cardiomyocyte cross-sectional area (CSA) at P14 (Fig. 2D, E). Despite the heart weight remaining unchanged, this reduction in CSA suggests an increase in the number of cardiomyocytes. Quantification of cardiomyocytes at P14 following collagenase digestion showed Foxk1 and Foxk2 overexpression increased the number of cardiomyocytes (Fig. 2F). There was also a significant increase in the fraction of mononucleated cardiomyocytes and a decrease in the fraction of multinucleated cardiomyocytes, as indicated by α-actinin and DAPI staining of isolated cardiomyocytes in Foxk1 and Foxk2 overexpressing hearts (Fig. 2G). An increase in the number of mononuclear cardiomyocytes indicates enhanced proliferative capacity in cardiomyocytes. Immunostaining showed that Foxk1 and Foxk2 overexpression increased Aurora B (Fig. 2H), Ki67 (Fig. 2I), and pH3 positive (Fig. 2J) cardiomyocytes at P14. Taken together, these findings demonstrate that the overexpression of Foxk1 and Foxk2 delays cardiomyocyte cell cycle arrest, thereby extending the postnatal window of cardiomyocyte proliferation.
Foxk1 and Foxk2 are required for neonatal heart regeneration
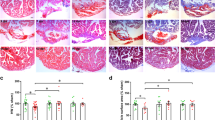
To explore the role of Foxk1 or Foxk2 in heart regeneration, we constructed cardiomyocyte-specific Foxk1 or Foxk2 knockout (cKO) mice by crossing Myh6-MerCreMer mice with Foxk1fl/fl or Foxk2fl/fl mice, respectively (Supplementary Fig. 3A, B). FOXK1 and FOXK2 expression was markedly reduced after tamoxifen administration for three consecutive days after birth (Fig. 3A–C). Neonatal Day-1 mice underwent MI injury, and the hearts were harvested 28 days post-MI (dpi). Masson staining revealed remarkably larger infarct scar areas in Foxk1 cKO and Foxk2 cKO mice compared to their littermates (Fig. 3D, E). WGA staining revealed that Foxk1 and Foxk2 deficient cardiomyocytes were larger in size than those of their littermates (Fig. 3F, G). Immunostaining showed that the numbers of pH3+ and Aurora B+ cardiomyocytes were reduced in the hearts of Foxk1 and Foxk2 cKO mice at 5 dpi compared with those of their littermates (Fig. 3H–K and Supplementary Fig. 3C–F). Moreover, we evaluated the protective effects of Foxk1 and Foxk2 by TUNEL assays. The result revealed that the loss of Foxk1 and Foxk2 did not affect cardiomyocyte apoptosis under ischemic conditions (Supplementary Fig. 3G, H).
A Experimental design: Tamoxifen was injected on postnatal day 0 (P0), P1, and P2 to induce Foxk1 or Foxk2 deletion in cardiomyocytes. Myocardial infarction (MI) was induced on P1. Heart tissues were collected at 5- and 28-days post-infarction (dpi), and echocardiography was performed at 28 dpi. B, C Western blot analysis of FOXK1 (B) and FOXK2 (C) expression in the hearts of Foxk1 cKO or Foxk2 cKO mice after tamoxifen injection (n = 3 mice per group). D, E Representative images of Masson staining and quantitative analysis of fibrotic scar size in Foxk1 cKO (D), Foxk2 cKO (E) and the littermate hearts at 28 dpi (n = 8 in Foxk1fl/fl group, n = 10 in Foxk1 cKO group. n = 11 in Foxk2fl/fl group, n = 7 in Foxk2 cKO group). F, G WGA staining and quantitative analysis of cardiomyocyte size in Foxk1 cKO (F), Foxk2 cKO (G), and littermate hearts at 28 dpi (n = 6 per group). H–K Representative images and quantitative analysis of pH3+ and Aurora B+ cardiomyocytes (green, indicated by yellow arrows) in Foxk1 cKO (H, J), Foxk2 cKO (I, K), and littermate hearts at 5 dpi (n = 3 per group) from the border zone. L, M Representative echocardiography images and quantification of ejection fraction and fractional shortening in Foxk1 cKO (L), Foxk2 cKO (M) and littermate hearts at 28 dpi (n = 9 in Foxk1fl/fl group, n = 11 in Foxk1 cKO group. n = 9 in Foxk2fl/fl group, n = 7 in Foxk2 cKO group). Data are presented as mean ± SEM. p values were determined by two-tailed unpaired t-tests (D–G, L, and M) and two-way ANOVA with Bonferroni multiple comparisons test (H–K). Source data are provided as a Source Data file.
Echocardiography showed that the ejection fraction (EF) and fractional shortening (FS) were significantly reduced in Foxk1 cKO and Foxk2 cKO mice at 28 dpi (Fig. 3L, M), which indicated that Foxk1 or Foxk2 specific deletion in cardiomyocytes was harmful to the recovery of heart function in neonatal mice after cardiac injury. Overall, these data suggested that Foxk1 and Foxk2 are required for neonatal heart regeneration and endogenous cardiomyocyte proliferation after MI injury.
Foxk1 and Foxk2 improve cardiac regeneration and repair in adult mice
We next evaluated the therapeutic potential of Foxk1 and Foxk2 for cardiac injury. Adult male mice (8–10 week-old) were induced MI and immediately administered AAV9-Foxk1 or AAV9-Foxk2 via tail vein injection (Fig. 4A). Western blot analysis showed the protein levels of FOXK1 and FOXK2 were significantly increased in the heart tissues below ligation suture at 14 dpi (Fig. 4B). Overexpression of Foxk1 or Foxk2 slightly improved survival rates after myocardial infarction in mice, but the difference was not statistically significant (Supplementary Fig. 4A). To test the functionality of Foxk1 and Foxk2 on cardiac function recovery on adult mice, serial echocardiographic assessments were performed over the following eight weeks. The results showed that, compared with mice treated with AAV9-GFP, administration of AAV9-Foxk1 and AAV9-Foxk2 led to a gradual and significant improvement in cardiac function, with higher left ventricular EF and FS (Fig. 4D, E), improved left ventricular dilation (Supplementary Fig. 4B–E), and inhibited ventricular wall thinning (Supplementary Fig. 4F, G). We collected hearts at 56 dpi and found that overexpressing Foxk1 and Foxk2 reduced both the heart weight and heart weight/body weight ratio (Supplementary Fig. 4H, I), without a decrease in total body weight (Supplementary Fig. 4J). In hearts treated with AAV9-Foxk1 and AAV9-Foxk2, WGA staining revealed decreased cardiomyocyte size at 56 dpi (Fig. 4F) and Masson staining showed a decreased fibrotic scar size (Fig. 4G), demonstrating the pro-regenerative effects of Foxk1 and Foxk2 against MI.
A Experimental design: AAV9-GFP, AAV9-Foxk1, or AAV9-Foxk2 was injected into adult mice via the tail vein after MI. Echocardiography was performed 1 day after MI (dpi), 7 dpi, 14 dpi, 28 dpi, 42 dpi, and 56 dpi. Heart tissue was collected at 14 dpi and 56 dpi. B Western blot analysis of FOXK1 and FOXK2 levels in mouse hearts infected with AAV9-GFP, AAV9-Foxk1, or AAV9-Foxk2 at 14 dpi (n = 3 per group). C–E Echocardiography analysis of heart function in mice administered AAV9-GFP, AAV9-Foxk1, or AAV9-Foxk2 at the indicated time points and quantification of EF and FS (n = 5 in sham group and n = 8 in MI group). F WGA staining of Foxk1- or Foxk2-overexpressing hearts at 56 dpi (n = 4 in sham group, n = 6 in AAV9-GFP group, n = 4 in AAV9-Foxk1 group, n = 5 in AAV9-Foxk2 group). G Masson staining and quantification of fibrotic scars at 56 dpi after LAD ligation and AAV9-GFP, AAV9-Foxk1 or AAV9-Foxk2 treatment (n = 4 in sham group, n = 8 in MI group). Data are presented as mean ± SEM. p values were determined by two-way ANOVA with Bonferroni multiple comparisons test (D, E) and one-way ANOVA with Bonferroni multiple comparisons test (F, G). Source data are provided as a Source Data file.
Functional recovery of AAV9-Foxk1 and AAV9-Foxk2 treated groups suggested increased cardiomyocyte proliferation. Further, we determined the cycling cardiomyocytes in different groups at 14dpi with immunostaining of pH3 and Ki67. Results showed that both pH3 and Ki67-positive cardiomyocytes dramatically increased in AAV9-Foxk1 and AAV9-Foxk2 treated hearts (Fig. 5A, B). We also found that AAV9-Foxk1 and AAV9-Foxk2 treatment increased Aurora B positive and the fraction of mononucleated and diploid cardiomyocytes (Fig. 5C, D), which indicated that Foxk1 and Foxk2 drive adult cardiomyocyte proliferation. In addition, we evaluated fibroblast activation and inflammatory response with immunostaining. We observed that the overexpression of Foxk1 or Foxk2 inhibited the activation of fibroblasts (Supplementary Fig. 5A) but without affecting macrophages or reparative macrophages infiltration (Supplementary Fig. 5B, C), suggesting that the enhanced cardiomyocyte proliferative capacity after Foxk1 and Foxk2 overexpression may contribute to reduced myocardial fibrosis. The evidence thus far suggested that Foxk1 and Foxk2 effectively enhance cardiac recovery post-MI by promoting cardiomyocyte proliferation and myocardial regeneration in adult mice.
A–C, Representative images of co-immunostaining and quantitative analysis for α-actinin (red) and pH3 (green) (A), Ki67 (green) (B), and Aurora B (green, indicated by yellow arrows) (C) positive cardiomyocytes after Foxk1 or Foxk2 overexpression at 14 dpi (n = 3 per group). D Representative images and quantification of cardiomyocyte nucleation and ploidy of isolated cardiomyocytes after Foxk1 or Foxk2 overexpression at 56 dpi (n = 4 per group). Data are presented as mean ± SEM. p values determined by two-way ANOVA with Bonferroni multiple comparisons test (A, B, and D (nucleation state)) and one-way ANOVA with Bonferroni multiple comparisons test (C, D (ploidy)), ns: not significant. Source data are provided as a Source Data file.
Foxk1 and Foxk2 induce cardiomyocyte proliferation by activating CCNB1/CDK1 complex
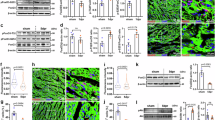
To unveil the mechanisms by which Foxk1 and Foxk2 induce cardiomyocyte proliferation, we performed Cleavage Under Targets and Tagmentation (CUT&Tag) and RNA-seq on primary neonatal cardiomyocytes treated with Ad-GFP, Ad-Foxk1 or Ad-Foxk2 for 48 h. CUT&Tag results showed that a total of 7310 genomic FOXK1 binding sites and 9170 genomic FOXK2 binding sites were identified. Among these sites, 15% of FOXK1 binding sites were localized to promoter regions (Supplementary Fig. 6A), while 56.74% of FOXK2 binding sites were localized to promoter regions (Supplementary Fig. 6B). Gene Ontology (GO) analysis of the binding genes in CUT&Tag showed significant enrichment in positive regulation of the cell cycle (Fig. 6A, B), suggesting that Foxk1 and Foxk2 may directly promote cardiomyocyte proliferation by promoting cell cycle-related gene expression.
A, B GO analysis of direct target genes from CUT&Tag-seq of Foxk1- or Foxk2-overexpressing cardiomyocytes. C Sankey diagram showing the direct target genes of FOXK1 from an overlay of CUT&Tag and RNA-seq profiles. D FOXK1, and FOXK2 CUT&Tag signals (normalized reads) in the Ccnb1 and Cdk1 locus, respectively. Filled boxes indicate peaks. E, F ChIP-PCR demonstrating the binding of FOXK1 to the Ccnb1 promoter region (E) and FOXK2 to the Cdk1 promoter region (F) (n = 3 per group). G, H qRT-PCR analysis showing the expression levels of Ccnb1 and Cdk1 in cardiomyocytes following overexpression of Foxk1 or Foxk2 (n = 4 per group). I, J Ccnb1, and Cdk1 transcript levels after knockdown of Foxk1 or Foxk2 in cardiomyocytes (n = 3 per group). K, L Western blot analysis of CCNB1 and CDK1 in cardiomyocytes following overexpression (K) and knockdown (L) of Foxk1 or Foxk2 (n = 3 per group). M, N Flow cytometry analysis of cell cycle distribution in Foxk1- and Foxk2-overexpressing cardiomyocytes (n = 4 per group). O, P Representative images and quantitative analysis of co-immunostaining of α-actinin (red) and pH3 (green, indicated by yellow arrows) in cardiomyocytes overexpressing Foxk1 with Ccnb1 knockdown (O), and in cardiomyocytes overexpressing Foxk2 with Cdk1 knockdown (P) (n = 3 per group). Data are presented as mean ± SEM. p values were determined by a two-sided Hypergeometric test (A, B), two-tailed unpaired t-test (E–J), two-way ANOVA with Bonferroni multiple comparisons test (N) and one-way ANOVA with Bonferroni multiple comparisons test (O, P). Source data are provided as a Source Data file.
Integrating CUT&Tag and RNA-seq data revealed that FOXK1 and FOXK2 directly targeted 1011 and 514 genes, respectively. Combining GO functional annotations, we identified Ccnb1 as a downstream target of FOXK1 within GO terms related to cell cycle regulation (Fig. 6C, D). Using the same analytical approach, we determined that Cdk1 was a downstream target of FOXK2 (Fig. 6D, Supplementary Fig. 6C). ChIP-qPCR confirmed that FOXK1 and FOXK2 could bind to the promoter regions of Ccnb1 and Cdk1, respectively (Fig. 6E, F). Adenovirus-mediated overexpression of Foxk1 in primary cardiomyocytes increased CCNB1 expression (Fig. 6G, K), while Foxk1 knockdown decreased it (Fig. 6I, L). Correspondingly, Foxk2 exhibited a similar regulatory pattern on CDK1 expression (Fig. 6H, J). CCNB1 binds to CDK1 to form a complex that phosphorylates target substrates, inducing the cell cycle G2-M transition and promoting cell cycle progression38. Immunoblot analysis indicates that overexpression of both Foxk1 and Foxk2 leads to an increase of global phosphorylation of CDK substrates, as detected by antibodies that recognize phosphorylated serine residues within the CDK substrate consensus motif, (K/H)pSP (Supplementary Fig. 6E). Cell cycle analysis by flow cytometry following propidium iodide (PI) staining revealed that overexpression of Foxk1 and Foxk2 increased the percentage of cardiomyocytes in S and G2/M phases (Fig. 6M, N). Additionally, we found that Foxk1 and Foxk2 did not alter CCNE1 expression but significantly increased CCNE2 expression and suppressed the expression of P27 and P21 (Supplementary Fig. 6F, G), which indicates that Foxk1 and Foxk2 can not only promote cardiomyocyte division, but also enable them to overcome quiescence and enter the cell cycle31,32.
To investigate the role of the CCNB1/CDK1 complex in mediating the proliferative effects of Foxk1 and Foxk2, we knocked down these genes in cardiomyocytes overexpressing Foxk1 or Foxk2. We found that knocking down Ccnb1 significantly reduced the proliferative effects of Foxk1, while knocking down Cdk1 blocked the effects of Foxk2 (Fig. 6O, P and Supplementary Fig. 6D). Furthermore, we found that CCNB1 and CDK1 exhibited cross-regulation in the context of Foxk1- and Foxk2-mediated cardiomyocyte proliferation. Specifically, knocking down Cdk1 in cardiomyocytes overexpressing Foxk1, as well as knocking down Ccnb1 in cardiomyocytes overexpressing Foxk2, similarly reduced their pro-proliferative effects (Supplementary Fig. 6H, I). These results indicate that Foxk1 and Foxk2 can not only enhance the proliferative capacity of cardiomyocytes by promoting the expression of the CCNB1/CDK1 complex but also enable cardiomyocytes to overcome their quiescent state.
Foxk1 and Foxk2 induce cardiomyocyte metabolic reprogramming
Foxk1 and Foxk2 play an essential role in promoting aerobic glycolysis35. Reactome enrichment analysis of our RNA-seq data also showed that overexpression of Foxk1 and Foxk2 led to downregulation of genes associated with the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) (Supplementary Fig. 7A, B). To investigate their effects on cardiomyocyte metabolism, we overexpressed Foxk1 and Foxk2 individually using adenovirus and performed Seahorse analysis. Extracellular acidification rate (ECAR) and mitochondrial oxygen consumption rate (OCR) were measured to assess glycolytic and OXPHOS activity, respectively. The results showed that overexpression of Foxk1 or Foxk2 significantly enhanced cardiomyocyte glycolysis while suppressed OXPHOS (Fig. 7A–D). Conversely, siRNA-mediated knockdown of Foxk1 or Foxk2 resulted in increased OXPHOS and reduced glycolysis (Supplementary Fig. 7C–F). These findings suggest that Foxk1 and Foxk2 promote a metabolic shift toward glycolysis in cardiomyocytes.
A, B Seahorse analysis detailing maximal respiration and spare respiratory capacity in cardiomyocytes overexpressing Foxk1 (A, n = 5) or Foxk2 (B, n = 6). Oxygen consumption rate (OCR) responses to mitochondrial stressors—oligomycin (Oligo), fluoro-carbonyl cyanide phenylhydrazone (FCCP), and a combination of antimycin A (Ant) and rotenone (Rot)—are depicted. C, D Glycolytic capacity and reserve, as measured by extracellular acidification rate (ECAR) during Seahorse analysis. This analysis assessed the response of Foxk1- or Foxk2-overexpressing cardiomyocytes to glucose, Oligo, and 2-deoxy-D-glucose (2-DG) to evaluate changes in cellular energy metabolism (n = 5 per group). E, F Quantitative results of U-[13C]-glucose-labeled glycolytic metabolites, pentose phosphate pathway metabolites, and TCA cycle metabolites in adult hearts after the treatment of AAV9-GFP, AAV9-Foxk1 and AAV9-Foxk2 at day 14 (n = 3 per group). G Representative images and quantitative results of pH3 positive cardiomyocytes in the indicated group (n = 4 per group). H Representative images and quantitative results of Ki67 positive cardiomyocytes in the indicated group (n = 4 per group). Data are presented as mean ± SEM. p values were determined by two-tailed unpaired t-test (A–D) and one-way ANOVA with Bonferroni multiple comparisons test (F–H). Source data are provided as a Source Data file.
To further explore the role of Foxk1 and Foxk2 on cardiomyocyte metabolism in vivo, we conducted glucose flux analysis using 13C-labeled glucose in the hearts of 8-week-old adult mice treated with AAV9-Foxk1 or AAV9-Foxk2 (Fig. 7E). This analysis revealed significant increases in glycolytic and pentose phosphate pathway (PPP) metabolites, accompanied by a decreasing trend in tricarboxylic acid (TCA) cycle intermediates (Fig. 7F). The metabolic reprogramming observed suggests increased glucose utilization through glycolysis and PPP. This reprogramming provides essential energy and biosynthetic precursors for cardiomyocyte proliferation.
Stimulation of glycolysis promotes cardiomyocyte proliferation by providing both energy and key biosynthetic intermediates necessary for cell cycle progression and growth39,40,41. To explore the functional consequences of enhanced glycolysis, we inhibited glycolytic flux in cardiomyocytes overexpressing Foxk1 and Foxk2 using 2-Deoxy-d-glucose (2-DG). Immunostaining showed significant decreases in cardiomyocyte proliferation, as marked by pH3 (Fig. 7G) and Ki67 (Fig. 7H). These results underscore the role of Foxk1 and Foxk2 in driving cardiomyocyte proliferation through glycolytic enhancement. Collectively, Foxk1 and Foxk2 act as metabolic regulators that reprogram cardiomyocyte metabolism by enhancing glycolysis and PPP, thereby creating a metabolic environment conducive to proliferation.
Foxk1 and Foxk2 induce cardiomyocyte glycolysis via HIF1α
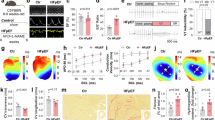
To elucidate the molecular mechanisms through which Foxk1 and Foxk2 regulate cardiomyocyte metabolism, we analyzed the aforementioned CUT&Tag-seq data and found that both Foxk1 and Foxk2 showed strong enrichment in the promoter region of Hif1α (Fig. 8A), a key regulator of cell metabolic reprogramming that enhances glycolysis42. ChIP-qPCR confirmed the findings from the CUT&Tag-seq analysis (Fig. 8B). Western blotting and qRT-PCR showed that both Foxk1 and Foxk2 overexpression enhanced HIF1α expression, while the opposite tendency was observed in Foxk1 or Foxk2 knockdown cardiomyocytes (Fig. 8C, D).
A FOXK1 and FOXK2 CUT&Tag signal (normalized reads) in the Hif1α locus. Filled boxes indicate peaks. B ChIP-qPCR illustrating the specific binding of FOXK1 and FOXK2 to the Hif1α promoter region (n = 3 per group). C qRT-PCR analysis of the mRNA levels of Hif1α in neonatal cardiomyocytes from Foxk1- or Foxk2-overexpressing and Foxk1- or Foxk2-knockdown cardiomyocytes (n = 4 per group). D Western blot analysis of HIF1α in cardiomyocytes following overexpression and knockdown Foxk1 or Foxk2 (n = 3 per group). E–H Seahorse analysis of glycolytic capacity (E, F) and maximal respiration (G, H) of cardiomyocytes with the indicated treatment (n = 5 per group). I, Quantitative analysis of the percentage of Ki67+ cardiomyocytes in the indicated groups (n = 4 for Ad-Ctrl+siHif1α group and n = 3 for other groups). J Immunostaining and quantitative analysis of pH3+ cardiomyocytes (green, yellow arrows) in the indicated groups (n = 3 per group). Data are presented as mean ± SEM. p values were determined by two-tailed unpaired t-test (B) and one-way ANOVA with Bonferroni multiple comparisons test (C, E–J). Source data are provided as a Source Data file.
To investigate whether HIF1α mediates Foxk1 and Foxk2-induced cardiomyocyte metabolic reprogramming, we knocked down Hif1α with siRNA in Foxk1 or Foxk2 overexpressing primary neonatal cardiomyocytes. Seahorse analysis revealed that both Foxk1 and Foxk2 induced glycolysis enhancement and mitochondrial OXPHOS suppression was suspended when Hif1α was knocked down (Fig. 8E–H). Immunofluorescence staining showed that Foxk1 or Foxk2-induced cardiomyocyte proliferation was suppressed by HIF1α inhibition (Fig. 8I, J). Taken together, our results demonstrate that HIF1α serves as the downstream regulator of Foxk1 and Foxk2 to promote glycolytic activity and proliferation. In addition to directly targeting CCNB1 and CDK1, metabolic reprogramming by HIF1α is also a key mechanism by which Foxk1 and Foxk2 regulate cardiomyocyte proliferation (Fig. 9).
Foxk1 or Foxk2 promote cardiomyocyte proliferation and heart regeneration through dual mechanisms: they stimulate cell cycle progression and induce metabolic reprogramming. As transcription factors, FOXK1 and FOXK2 bind to the promoter regions of Ccnb1 and Cdk1, key cell cycle regulators, upregulating their expression. CCNB1 and CDK1 form a complex that promotes the G2/M phase transition, crucial for cardiomyocyte proliferation. Additionally, both FOXK1 and FOXK2 interact with the promoter of Hif1α, a master regulator of the hypoxic stress response, and promote its expression, thus inducing glycolysis and metabolic reprogramming in cardiomyocytes, which create a conducive environment for cell proliferation. Gene therapy using AAV9-mediated overexpression of Foxk1 or Foxk2 can improve cardiac regeneration and repair in adult mice, highlighting their potential as promising therapeutic targets for interventions in ischemic heart disease. Created in BioRender. Cai, D. (2025) https://BioRender.com/m31y568.
Discussion
Stimulating cardiomyocyte proliferation represents a promising strategy for heart regeneration following cardiac injury18,19,43,44,45,46. In this study, we identify the transcription factors Foxk1 and Foxk2 as critical regulators of cardiomyocyte proliferation and cardiac repair. We observed that the expression of Foxk1 and Foxk2 declines during postnatal heart development, paralleling the loss of proliferative capacity in cardiomyocytes. Cardiomyocyte-specific knockout of either Foxk1 or Foxk2 in neonatal mice impairs heart regeneration after MI injury. Conversely, AAV9-mediated overexpression of Foxk1 and Foxk2 extends the postnatal proliferative window of cardiomyocytes and enhances heart regeneration following MI in adult mice. These findings position Foxk1 or Foxk2 as potential therapeutic targets for promoting cardiac repair following injury. Mechanistically, we show that Foxk1 and Foxk2 promote cardiomyocyte proliferation by activating the CCNB1/CDK1 complex and enhancing glycolysis through upregulating HIF1α transcription in cardiomyocytes.
Foxk1 and Foxk2, two highly structured and function-conserved transcription factors, have been found to regulate a wide spectrum of biological processes, including cell cycle progression, and metabolic reprogramming35,47,48. Foxk1 has been shown to regulate skeletal muscle regeneration by regulating myogenic progenitor cell proliferation and differentiation33,49, and it can also promote cardiogenesis during heart development50. In this study, we report the novel function of Foxk1 and Foxk2 in promoting cardiomyocyte proliferation and heart regeneration. Employing CUT&Tag-Seq and RNA-seq, we demonstrate that Foxk1 and Foxk2 drive cardiomyocyte re-entry into the cell cycle by enhancing the expression of CCNB1 and CDK1, respectively. Forced expression of CCNB1 and CDK1 can induce cardiomyocyte division and heart regeneration10,51,52. Coincidentally, CCNB1 and CDK1, as the key regulatory molecules in the cell cycle, bind together to form the CCNB1/CDK1 complex, which promotes the G2/M phase transition38,53. On the other hand, cell cycle inhibitors such as P21 and P27 bind to the CCNB1/CDK1 complex and inhibit its activity54. Interestingly, we observed that both P21 and P27 are downregulated in cardiomyocytes overexpressing Foxk1 and Foxk2. These results indicate that Foxk1 and Foxk2 regulate cardiomyocyte cell cycle progression through a conserved and complementary mechanism. Specifically, our findings identify Foxk1 and Foxk2 as novel upstream regulators that activate the CCNB1/CDK1 complex, offering new insight into how cardiomyocytes maintain and exit the cell cycle.
In addition to identifying Foxk1 and Foxk2 as drivers of the cell cycle, we found that these transcription factors also induce cardiomyocyte metabolic reprogramming. Through the overexpression of Foxk1 and Foxk2 in cardiomyocytes, we observed a significant suppression of OXPHOS alongside a marked increase in glycolysis, as evidenced by Seahorse analysis. These results were further substantiated by the knockdown of Foxk1 or Foxk2, which resulted in a shift toward increased OXPHOS and decreased glycolytic activity. To investigate the metabolic effects of Foxk1 and Foxk2 overexpression in vivo, we performed glucose flux analysis with 13C-labeled glucose in the hearts of adult mice, revealing increased glycolytic intermediates and pentose phosphate pathway metabolites. These in vitro and in vivo data provide compelling evidence that Foxk1 and Foxk2 drive metabolic reprogramming in cardiomyocytes by enhancing glycolysis, consistent with recent reports that these transcription factors induce aerobic glycolysis by upregulating glycolytic enzymes35. Furthermore, we revealed that inhibition of glycolysis with 2-Deoxy-d-glucose (2-DG) in Foxk1- and Foxk2-overexpressing cardiomyocytes markedly reduced the enhanced proliferative capacity. Our study identifies Foxk1 and Foxk2 as key regulators of glycolytic reprogramming in cardiomyocytes, driving metabolic shifts that enhance cell proliferation. Metabolic state is closely related to cardiomyocyte proliferative capacity55. In a relatively hypoxic environment, fetal cardiomyocytes primarily depend on glycolysis as their main energy source56. Glycolytic flux not only generates ATP but also supports cardiomyocyte proliferation by providing glucose-derived metabolic intermediates for nucleotide, lipid, and protein biosynthesis57. Postnatally, as oxygen levels increase, cardiomyocyte metabolism gradually shifts from cytoplasmic glycolysis to oxygen-dependent mitochondrial OXPHOS, which ensures more efficient ATP production55. This increased glycolytic and proliferative capacity of cardiomyocytes may lead to an immature state in metabolism and structure that could impair the contractility of cardiomyocytes39,58,59. Therefore, when targeting Foxk1 and Foxk2 for cardiac regeneration therapies, it is crucial to carefully optimize the dosage and duration of treatment to mitigate potential adverse effects associated with excessive cardiomyocyte glycolysis.
HIF1α is an evolutionarily conserved transcription factor in response to hypoxic stress response. Under hypoxic conditions, the ubiquitination of HIF1α is disrupted, leading to its stabilization and allowing it to enter the nucleus to promote target gene transcription60,61. Deletion of Hif1a in cardiomyocytes causes the downregulation of genes related to glycolytic metabolism and cell cycle, which leads to the activation of the p53 pathway and excessive ROS generation62,63. Here, we discovered that Foxk1 and Foxk2 bind directly to the promoter region of Hif1α in cardiomyocytes. This interaction appears to be critical, as knocking down Hif1α significantly impairs the ability of Foxk1 and Foxk2 to promote glycolysis and proliferation in cardiomyocytes. These findings reveal a novel regulatory mechanism of HIF1α activity.
In summary, we reveal that Foxk1 and Foxk2 are critical transcription factors driving cardiomyocyte proliferation and heart regeneration. These factors exert dual pro-proliferative effects by promoting cell cycle progression via upregulation of the CCNB1/CDK1 complex and enhancing metabolic reprogramming through HIF1α activation. Gene therapy using AAV9-mediated overexpression of Foxk1 or Foxk2 is sufficient to improve heart functional recovery through cardiomyocyte renewal after cardiac injury in adult mice. These findings establish Foxk1 and Foxk2 as promising therapeutic targets for cardiac repair, offering new insights into the interplay between cell cycle control and metabolic regulation in heart regeneration.
Methods
Experimental animals
All experiments involving animals were conducted in accordance with the Guide for the Use and Care of Laboratory Animals. All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC), Fuwai Hospital, Chinese Academy of Medical Sciences. The investigators responsible for histological analyses were blinded to group information. Mice were randomly assigned to the experimental groups.
Mice
Neonatal 1-day-old mice of both sexes were used in the study. WT C57BL/6J mice were obtained from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Myh6-MerCreMer (JAX 005657) mice were acquired from the Jackson Laboratory (Bar Harbor, ME, USA). Foxk1fl/fl mice were constructed with the help of BIOCYTOGEN (Shanghai, China). Foxk2fl/fl mice were constructed with the help of Cyagen (Suzhou, China). Cardiomyocyte-restricted deletion of Foxk1 (Foxk1 cKO) or Foxk2 (Foxk2 cKO) was achieved by crossing Foxk1fl/fl or Foxk2fl/fl mice with Myh6-MerCreMer mice. The littermates were used as the negative controls. Tamoxifen (Sigma‒Aldrich, St Louis, USA) was administered by intraperitoneal injection at the indicated timepoints (at 0.1–0.15 mg/gram of mouse body weight) according to the protocol of the Jackson Laboratory. All the animals were kept at 23 °C and 50% humidity with a 12 h light/dark cycle. Mice had ad libitum access to food and water.
Neonatal and adult mouse MI
For the construction of the MI model, MI was induced in neonatal and adult mice by ligating the left anterior descending coronary artery as previously described in ref. 18,23. Neonatal mice (P1) were anesthetized by cooling on ice for 2 min 30 s, while adult mice were anesthetized by intraperitoneal injection of pentobarbital sodium (40 mg/kg; Solarbio, Beijing, China). After opening the chest in the fourth intercostal space, the LAD was ligated with a suture to induce left ventricle myocardial ischemia. Then, the incision was sutured with 6-0 nonabsorbable silk thread, and the mice were warmed until consciousness was restored. For the sham operation, we performed the same operation without suturing the LAD coronary artery. The video documentation of the P1 MI surgery procedures can be found in Supplementary Movie 1. Hearts were collected at different time points for immunochemical and histological analyses. The infarct size was quantified using a previously described method25.
Echocardiography
Left ventricle systolic function was evaluated by transthoracic echocardiography using a digital ultrasound system (Vevo 2100 Imaging System, VisualSonics, Toronto, Canada). Animals were anesthetized with 2.5% isoflurane, and the hair in the measurement area was removed. To measure EF and FS, short-axis images were obtained at the level of the papillary muscles in M-mode, and the left ventricular inner dimension (LVID) and left ventricular volume were determined in both diastole and systole.
Histology
The collected hearts were fixed in 4% paraformaldehyde at room temperature for 48 h, dehydrated in an ethanol and xylene series, and finally embedded in paraffin. For the MI model, hearts were sliced continuously from the ligation site to the apex of the heart, with an interval of 200 μm between each slice (5 μm thick), and four slices were collected for each heart. Hematoxylin and eosin (H&E) and Masson’s trichrome staining were performed using standard procedures. To determine scar size, we used the midline method as previously described in ref. 64. Briefly, the length of the midline that is affected by the infarct, which includes at least 50% of the whole thickness of the myocardial wall, is measured. Infarct size is calculated by dividing the sum of midline infarct lengths from all sections by the sum of midline circumferences from all sections.
Neonatal cardiomyocyte isolation and culture
Primary cardiomyocytes were isolated from the hearts of WT mice at P1 using a neonatal dissociation kit and isolation kit (Miltenyi Biotec, Teterow, Germany) according to the instructions. The cardiomyocytes were plated in DMEM supplemented with 10% FBS at 37 °C and 5% CO2 at a concentration of 5 × 105 cells/ml. For the virus infection experiments, cardiomyocytes were infected with 100 MOI of Ad:CMV:Foxk1 (Ad-Foxk1), Ad:CMV:Foxk2 (Ad-Foxk2), or Ad:CMV:Control (Ad-Ctrl) (Vigene Biosciences, Shangdong, China) in DMEM as previously described in ref. 19. The purity of neonatal cardiomyocytes was assessed using immunofluorescence staining analysis (Supplementary Fig. 8A). For siRNA experiments, cardiomyocytes were transfected with siRNA-Foxk1, siRNA-Foxk2, siRNA-Hif1α, siRNA-Ccnb1, siRNA-Cdk1 or negative control (NC) using Lipofectamine 3000 (Invitrogen, Waltham, USA) transfection reagent as previously described in ref. 27. After 48 h, cardiomyocytes were harvested for Western blotting, quantitative real-time polymerase chain reaction, Seahorse XF24 analysis, or proliferation assessment. The siRNA sequences used for targeting genes are listed in Supplementary Table 1 in the Supplementary information.
Western blot analysis
Western blot analysis was performed with specific antibodies as previously described in ref. 65. Left ventricular heart tissues and cardiomyocytes were harvested and lysed with RIPA lysis buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF) (Beyotime Institute of Biotechnology, Beijing, China). After homogenization and incubation at 4 °C for 40 min, all protein samples were mixed with SDS loading buffer and boiled for 10 min at 70 °C. Subsequently, 20 μg of total protein was loaded on a 10% sodium lauryl sulfate‒polyacrylamide gel for electrophoresis, and the separated proteins were transferred to a nitrocellulose membrane. After blocking, the membrane was incubated at 4 °C overnight with the following primary antibodies: anti-FOXK1 (1:1000, Abcam ab18196, UK), anti-FOXK2 (1:1000, Novus NBP1-87700, USA), anti-HIF1α (1:1000, Abcam ab1, UK), anti-CCNB1 (1:1000, Proteintech 55004-1-AP, USA), anti-CDK1 (1:1000, Proteintech 19532-1-AP, USA), anti-CCNE1 (1:1000, Proteintech 11554-1-AP, USA), anti-CCNE2 (1:1000, Proteintech 11935-1-AP, USA), anti-P27 (1:1000, Proteintech 25614-1-AP, USA), anti-P21 (1:1000, Proteintech 82669-2-RR, USA), and anti-Phospho-CDK Substrate Motif [(K/H)pSP] MultiMab (1:1000, Cell Signaling Technology 9477, USA). anti-Flag (1:1000, Sigma Aldrich F7425), and anti-β-tubulin (1:1000, Cell Signaling Technology 2146, USA). On the next day, the membrane was washed with Tris-buffered saline/0.1% Tween 20 (TBST) and incubated with the secondary antibody for 1 h at room temperature. The signals were detected with Pierce ECL Western Blot Substrate (Thermo Fisher Scientific, Waltham, USA).
qRT-PCR
To quantify gene expression, total RNA was extracted from cardiomyocytes using TRIzol reagent (Invitrogen, Waltham, USA), and then 1 μg of the total mRNA was reverse-transcribed into cDNA using PrimeScript RT Master Mix (Takara, RR036A, Japan). qRT-PCR was repeated three times using SYBR Green qPCR Master Mix (Applied Biosystems, Waltham, USA) and amplified on a Vii7 Real-Time PCR System (Applied Biosystems, Waltham, USA). The primer sequences used for targeting genes are listed in Supplementary Table 2 in the Supplementary information.
Metabolic assays
The OCR and ECAR were measured with a Seahorse Bioscience XF24 extracellular flux analyzer with Mito Stress and Glycolysis Stress kits (Agilent Technologies, California, USA), respectively, as previously described in refs. 35,36. A total of 70,000 cardiomyocytes were plated into a coated XF24 cell culture microplate with 100 μl of the standard medium, and then the cells were treated as described. Subsequently, metabolic analysis was performed according to the instructions, and the data were analyzed in seahorse wave software and normalized to 70,000 cells. Maximal Respiration was calculated as (Maximum rate measurement after FCCP injection) − (Non-Mitochondrial Respiration). Spare Respiratory Capacity was calculated as (Maximal Respiration) − (Basal Respiration). Glycolytic Capacity was calculated as (Maximum rate measurement after Oligomycin injection) − (Last rate measurement before Glucose injection). Glycolytic Reserve was calculated as (Glycolytic Capacity) − (Glycolysis).
Metabolic flux assays and targeted metabolic analysis
Two-month-old WT mice were injected with AAV9-GFP, AAV9-Foxk1, and AAV9-Foxk2 via the tail vein. After 14 days, the heart was quickly excised and intubated to the Langendorff perfusion apparatus maintained at 37 °C using a temperature-controlled bath. The isolated hearts were perfused retrogradely with Krebs–Henseleit (KH) buffer containing U-13C-glucose (8 mM) for 60 min. Subsequently, the heart was rapidly frozen and pulverized in liquid nitrogen. The heart samples were sonicated and extracted with an 80% methanol solution. The mixtures were centrifuged at 18,000×g for 15 min at 4 °C, then the supernatant was concentrated by centrifugation, and 100 μl of 80% methanol was added for LC-MS analysis. The metabolites were analyzed with an ultrahigh-pressure liquid chromatography-triple quadrupole mass spectrometer ACQUITY UPLC-Xevo TQ-S (Waters Corp., Milford, MA, USA). For data processing, the raw data files generated by UPLC-MS/MS were processed using MassLynx software (v 4.1, Waters Corp., Milford, MA, USA for peak extraction, integration, identification, and quantification of each metabolite. R language (v4.1.1) was used for subsequent statistical analysis.
Immunofluorescence staining
Isolated hearts were fixed in 4% paraformaldehyde and embedded in paraffin. Serial paraffin sections (5 μm) were prepared for immunostaining. After deparaffinization, the sections were subjected to antigen retrieval with Tris-EDTA buffer and then blocked with 5% donkey serum as previously described in ref. 27. The sections were incubated with the indicated primary antibodies overnight at 4 °C. After washing three times with PBS, the slides were incubated with appropriate fluorescently labeled secondary antibodies and shielded from light for 1 h at room temperature. Subsequently, the slides were washed three times in PBS and then counterstained with DAPI (Sigma‒Aldrich, St Louis, USA). The primary antibodies used were as follows: anti-sarcomeric alpha actinin (1:200, Abcam ab9465, UK), anti-phosphorylated Histone 3 (1:1000, Millipore 3377, USA), anti-Ki67 (1:200, Abcam ab16667, UK), anti-Aurora B (1:400, Sigma A5102, USA) and anti-WGA conjugated with Alexa Fluor 647 (20 mg/ml, Invitrogen W32466, USA). The secondary antibodies used were as follows: donkey anti-rabbit Alexa Fluor 488 (1:400, Invitrogen A-21206, USA), donkey anti-mouse Alexa Fluor 594 (1:500, Invitrogen A-21203, USA), and donkey anti-goat Alexa Fluor 647 (1:500, Invitrogen A21447, USA). Fluorescence was observed under a ZEISS LSM800 confocal laser scanning microscope (Carl Zeiss, Inc., Jena, Germany).
Cardiomyocyte isolation and quantification
Hearts were freshly collected and fixed in 4% PFA at room temperature for 2 h. The auricles were removed. Then, the hearts were cut into small pieces and incubated in the enzyme solution (collagenase II: 1.8 mg/ml; collagenase IV: 2.4 mg/ml, dissolved in PBS) at 37 °C on an end-over-end shaker. The supernatants were collected every 12 h, and the procedure was repeated until the tissue was completely digested. The total number of cardiomyocytes was counted with a hemocytometer. Isolated cardiomyocytes were co-stained with sarcomere actinin and DAPI (Sigma, D9542; 0.2 μg/ml), and their purity was assessed (Supplementary Fig. 8B). For nucleation counts, at least 300 cardiomyocytes per sample were quantified.
Wheat germ agglutinin staining and cardiomyocyte size quantification
Cardiomyocyte size was measured with wheat germ agglutinin (WGA) and DAPI staining as previously reported method44,66. The slides were boiled in EDTA buffer (1 mM EDTA, pH 8.0) for 2 min, followed by cooling down for 20 min under running water. The slides were permeabilized with 0.3% Triton X, and then washed with PBS three times. WGA working solution (10 μg/ml, conjugated to Alexa Fluor 647, Life Technologies) was incubated for 1 h away from light, and the slides were washed with PBS three times. The slides were mounted in an antifade mounting medium with DAPI. Subsequently, three independent samples were counted for each group, and images were captured at 400× magnification, encompassing fields of view from the left ventricle, the right ventricle, and the septum. For the myocardial infarction (MI) group, three fields of view from each side of the border zone, totaling six fields of view, were acquired for measurement. Cell boundaries were delineated through WGA staining. The ImageJ software was used to quantify the size of each cell. At least 500 cells per sample were quantified.
RNA-seq and data analysis
Total mRNA was extracted from cardiomyocytes after adenovirus infection using TRIzol reagent (Life Technologies) and incubated with DNase I for 30 min at 37 °C. RNA quantity and RNA integrity (RNA integrity number, RIN) were determined using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, USA), respectively. All samples were confirmed to be of acceptable quality as indicated by the A260/280 OD ratio (between 1.8 and 2.2) and RIN (≥7). Approximately 3 μg of the total RNA was subjected to poly(A) mRNA isolation using poly-T attached magnetic beads. Following purification, the mRNA was fragmented into small pieces, and the average insert size for the paired-end libraries was 250 bp. TruSeq library construction and sequencing were performed on the Illumina HiSeq X Ten sequencing platform (Illumina, San Diego, CA, USA) with the PE-150 module by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Raw reads generated for the samples have been submitted to the BioProject database of the National Center for Biotechnology Information (NCBI) with the accession number PRJNA886416. The clean reads were mapped to the mm10 mouse reference genome using the STAR software (Version: 2.5.1b) with the default parameters. The read counts were determined with featureCounts (version: 1.5.0), and differentially expressed genes (DEGs) between the Ad-Ctrl and Ad-Foxk1 or Ad-Foxk2 groups were identified using the R package DESeq2 with an adjusted p value threshold of ≤0.05.
CUT&Tag-Seq and data analysis
A CUT&Tag assay was performed according to the instructions of the commercial kit (Hyperactive® In-Situ ChIP Library Prep Kit for Illumina (pA-Tn5), TD902, Vazyme). Briefly, isolated cardiomyocytes were treated with adenovirus for 36 h and washed with ice-cold PBS twice. Then, 1 × 105 cells were washed with 1 ml of wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine (Sigma‒Aldrich), 1× protease inhibitor cocktail (5056489001, Sigma‒Aldrich)) and centrifuged at 300×g for 5 min at room temperature. The cell pellets were resuspended in 1 ml of wash buffer. Concanavalin A-coated magnetic beads (Bangs Laboratories) were washed twice with binding buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM MnCl2, 1 mM CaCl2). Next, 10 μl of activated beads were added, and the beads and cells were incubated at room temperature for 15 min. The bead-bound cells were resuspended in 50 μl of antibody buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.05% digitonin (Sigma‒Aldrich), 2 mM EDTA, 0.1% BSA, 1× protease inhibitor cocktail). Then, 1 μg of primary antibody (mouse monoclonal anti-Flag antibody (SAB4301135, Sigma‒Aldrich) or normal mouse IgG (sc-2025, Santa Cruz) was added, and the cells were incubated for 2 h at room temperature with slow rotation. The primary antibody was removed using a magnetic stand. Secondary antibody (1 μg) (rabbit anti-mouse IgG H&L (ab6709, Abcam) was diluted in 50 μl of Dig-wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.05% digitonin, 1× protease inhibitor cocktail), and the cells were incubated at room temperature for 1 h. The cells were washed three times with Dig-wash buffer to remove unbound antibodies. A hyperactive pA-Tn5 transposase adapter complex (TTE mix, 4 μM, Vazyme) was diluted 1:100 in 100 μl of Dig-300 buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine, 0.01% digitonin, 1× protease inhibitor cocktail). The cells were then resuspended in 300 μl of tagmentation buffer (10 mM MgCl2 in Dig-300 buffer) and incubated at 37 °C for 1 h. To terminate tagmentation, 10 μl of 0.5 M EDTA, 3 μl of 10% SDS, and 5 μl of 10 mg ml−1 Proteinase K were added to 300 μl of the sample, and the mixture was incubated at 50 °C for 1 h. DNA was purified using phenol‒chloroform-isoamyl alcohol extraction and ethanol precipitation as well as RNase A treatment.
For library amplification, 24 μl of DNA was mixed with 1 μl of TruePrep Amplify Enzyme (TAE, Vazyme), 10 μl of 5× TruePrep Amplify Enzyme buffer, 5 μl of ddH2O, and 5 μl of uniquely barcoded i5 and i7 primers from a TruePrep Index Kit V2 for Illumina (Vazyme). A total volume of 50 μl of the sample was placed in a thermocycler and run using the following program: 72 °C for 3 min; 98 °C for 30 s; 17 cycles of 98 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s; 72 °C for 5 min; and hold at 4 °C. To purify the PCR products, 1.2× volumes of VAHTS DNA Clean Beads (Vazyme) were added, and the mixtures were incubated at room temperature for 10 min. The libraries were washed twice with 80% ethanol and eluted in 22 μl of ddH2O. The libraries were sequenced on an Illumina NovaSeq platform, and 150-bp paired-end reads were generated.
Trimmed clean reads were aligned to the mouse reference genome(mml0) using Bowtie2 (version of 2.3.5.1) with the default parameters. The raw mapped reads were sorted using the Picard SortSAM function. MACS2 (version 2.2.6) was used to identify reads-enriched regions, and peaks with a false discovery rate lower than 5% were saved to detect chromosomal regions for further analysis. Visualization of peak distribution along genomic regions of genes of interest was performed with IGV, Genes with a peak within 3 kb of the transcription start site (TSS) were considered target genes, and an area of 6 kb surrounding each TSS was selected to obtain the CUT&Tag profiles of FOXK1/FOXK2 using bed tools. This area was also used to annotate the peaks with ChIPsseket 1.20.0.
Chromatin immunoprecipitation (ChIP)‒qPCR
ChIP assays were performed using a ChIP kit from Cell Signaling Technology according to the manufacturer’s instructions. Briefly, cardiomyocytes were isolated using the Neonatal Heart Dissociation Kit (Miltenyi Biotec) mentioned above. After transfection with Ad-Foxk1 or Ad-Foxk2 for 36 h, cardiomyocytes were then fixed with 37% formaldehyde for 10 min at room temperature and then incubated for 10 min in the presence of glycine. The cells were washed twice with ice-cold PBS and then resuspended in ChIP Sonication Cell Lysis Buffer supplemented with protease inhibitor cocktails after centrifugation. The cells were sonicated to obtain chromatin fragments of approximately 500–1500 bp. A total of 7–25 mg of sheared chromatin was used for each ChIP sample and incubated with 1–3 mg of appropriate antibodies overnight on an end-to-end rotator at 4 °C. After incubation, the immunoprecipitated DNA was quantified by RT-PCR on an ABI Prism 7500 detection system. The antibodies used in the ChIP assays were as follows: anti-Flag (Sigma‒Aldrich, F7425) and polyclonal IgG (Santa Cruz, sc-805). The primer sequences used for targeting genes are listed in Supplementary Table 3 in the Supplementary information.
Statistical analysis
All data are expressed as the mean ± standard error of the mean (SEM). A two-tailed unpaired Student’s t-test was used to assess statistical differences between the two groups. One-way or two-way ANOVA followed by Bonferroni’s multiple comparison test was performed for multiple groups comparison. The results with p values < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data presented in the study has been provided in the main text, supplementary materials, and source data files. The datasets generated for the transcriptome and CUT &Tag analysis are available through the National Center for Biotechnology Information (NCBI) with the accession number PRJNA886416. RNA-Seq data of the isolated cardiomyocytes at different ages was obtained from our published work37 (accession number PRJNA798841). Mass spectrometry data are available and have been uploaded to the MetaboLights repository with accession code MTBLS12191. Source data are provided in this paper.
References
Tsao, C. W. et al. Heart disease and stroke statistics-2023 update: a report from the American Heart Association. Circulation 147, e93–e621 (2023).
Prabhu, S. D. & Frangogiannis, N. G. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ. Res. 119, 91–112 (2016).
Bergmann, O. et al. Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 (2009).
Ali, S. R. et al. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc. Natl. Acad. Sci. USA 111, 8850–8855 (2014).
Frangogiannis, N. G. The inflammatory response in myocardial injury, repair, and remodelling. Nat. Rev. Cardiol. 11, 255–265 (2014).
Weinberger, M. & Riley, P. R. Animal models to study cardiac regeneration. Nat. Rev. Cardiol. 21, 89–105 (2024).
Haubner, B. J. et al. Functional recovery of a human neonatal heart after severe myocardial infarction. Circ. Res. 118, 216–221 (2016).
Zhu, W. et al. Regenerative potential of neonatal porcine hearts. Circulation 138, 2809–2816 (2018).
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011).
Mohamed, T. M. A. et al. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104–116.e112 (2018).
Chaudhry, H. W. et al. Cyclin A2 mediates cardiomyocyte mitosis in the postmitotic myocardium. J. Biol. Chem. 279, 35858–35866 (2004).
Trivedi, C. M., Lu, M. M., Wang, Q. & Epstein, J. A. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J. Biol. Chem. 283, 26484–26489 (2008).
Ai, S. et al. Divergent requirements for EZH1 in heart development versus regeneration. Circ. Res. 121, 106–112 (2017).
Yue, Z. et al. PDGFR-β signaling regulates cardiomyocyte proliferation and myocardial regeneration. Cell. Rep. 28, 966–978.e964 (2019).
Xin, M. et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 110, 13839–13844 (2013).
Aharonov, A. et al. ERBB2 drives YAP activation and EMT-like processes during cardiac regeneration. Nat. Cell. Biol. 22, 1346–1356 (2020).
Magadum, A. et al. Live cell screening platform identifies PPARδ as a regulator of cardiomyocyte proliferation and cardiac repair. Cell. Res. 27, 1002–1019 (2017).
Bassat, E. et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547, 179–184 (2017).
Feng, J. et al. Versican promotes cardiomyocyte proliferation and cardiac repair. Circulation 149, 1004–1015 (2023).
Liu, X. et al. Lymphoangiocrine signals promote cardiac growth and repair. Nature 588, 705–711 (2020).
Das, S. et al. A unique collateral artery development program promotes neonatal heart regeneration. Cell 176, 1128–1142.e1118 (2019).
Wang, Y. et al. Mydgf promotes cardiomyocyte proliferation and neonatal heart regeneration. Theranostics 10, 9100–9112 (2020).
Li, J. et al. Regulatory T-cells regulate neonatal heart regeneration by potentiating cardiomyocyte proliferation in a paracrine manner. Theranostics 9, 4324–4341 (2019).
Li, Y. et al. gp130 controls cardiomyocyte proliferation and heart regeneration. Circulation 142, 967–982 (2020).
Han, C. et al. Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res. 25, 1137–1151 (2015).
Li, R., Xiang, C., Li, Y. & Nie, Y. Targeting immunoregulation for cardiac regeneration. J. Mol. Cell. Cardiol. 177, 1–8 (2023).
Chen, Z. et al. Triiodothyronine induces a proinflammatory monocyte/macrophage profile and impedes cardiac regeneration. J. Mol. Cell. Cardiol. 191, 7–11 (2024).
Golson, M. L. & Kaestner, K. H. Fox transcription factors: from development to disease. Development 143, 4558–4570 (2016).
Liu, Y. et al. FOXK transcription factors: regulation and critical role in cancer. Cancer Lett. 458, 1–12 (2019).
Li, L., Gong, M., Zhao, Y., Zhao, X. & Li, Q. FOXK1 facilitates cell proliferation through regulating the expression of p21, and promotes metastasis in ovarian cancer. Oncotarget 8, 70441–70451 (2017).
Yu, Y. et al. FOXK2 amplification promotes breast cancer development and chemoresistance. Cancer Lett. 597, 217074 (2024).
Hawke, T. J., Jiang, N. & Garry, D. J. Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J. Biol. Chem. 278, 4015–4020 (2003).
Garry, D. J. et al. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. USA 97, 5416–5421 (2000).
Ji, Z., Li, Y., Liu, S. X. & Sharrocks, A. D. The forkhead transcription factor FOXK2 premarks lineage-specific genes in human embryonic stem cells for activation during differentiation. Nucleic Acids Res. 49, 1345–1363 (2021).
Sukonina, V. et al. FOXK1 and FOXK2 regulate aerobic glycolysis. Nature 566, 279–283 (2019).
Sakaguchi, M. et al. FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism. Nat. Commun. 10, 1582 (2019).
Feng, J., Li, Y. & Nie, Y. Methods of mouse cardiomyocyte isolation from postnatal heart. J. Mol. Cell. Cardiol. 168, 35–43 (2022).
Huang, R. et al. BECN1 promotes radiation-induced G2/M arrest through regulation CDK1 activity: a potential role for autophagy in G2/M checkpoint. Cell. Death. Discov. 6, 70 (2020).
Fukuda, R. et al. Stimulation of glycolysis promotes cardiomyocyte proliferation after injury in adult zebrafish. Embo. Rep. 21, e49752 (2020).
Wang, X. et al. TLR3 mediates repair and regeneration of damaged neonatal heart through glycolysis dependent YAP1 regulated miR-152 expression. Cell. Death. Differ. 25, 966–982 (2018).
Honkoop, H. et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife 8, e50163 (2019).
Schönenberger, M. J. & Kovacs, W. J. Hypoxia signaling pathways: modulators of oxygen-related organelles. Front. Cell. Dev. Biol. 3, 42 (2015).
Ahmed, M. S. et al. Identification of FDA-approved drugs that induce heart regeneration in mammals. Nat. Cardiovasc. Res. 3, 372–388 (2024).
Nguyen, N. U. N. et al. A calcineurin-Hoxb13 axis regulates growth mode of mammalian cardiomyocytes. Nature 582, 271–276 (2020).
Zhou, W. et al. Pharmacologically inducing regenerative cardiac cells by small molecule drugs. elife 13, RP93405 (2024).
Li, J., Zhu, D., Hu, S. & Nie, Y. CRISPR-CasRx knock-in mice for RNA degradation. Sci. China Life. Sci. 65, 2248–2256 (2022).
Yu, M. et al. The function of FoxK transcription factors in diseases. Front. Physiol. 13, 928625 (2022).
Tang, M. et al. FOXK1 participates in DNA damage response by controlling 53BP1 function. Cell. Rep. 32, 108018 (2020).
Shi, X. et al. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J. Cell Sci. 125, 5329–5337 (2012).
Sierra-Pagan, J. E. et al. FOXK1 regulates Wnt signalling to promote cardiogenesis. Cardiovasc. Res. 119, 1728–1739 (2023).
Bicknell, K. A., Coxon, C. H. & Brooks, G. Forced expression of the cyclin B1-CDC2 complex induces proliferation in adult rat cardiomyocytes. Biochem. J. 382, 411–416 (2004).
Borden, A. et al. Transient Introduction of miR-294 in the heart promotes cardiomyocyte cell cycle reentry after injury. Circ. Res. 125, 14–25 (2019).
Jackman, M., Lindon, C., Nigg, E. A. & Pines, J. Active cyclin B1–Cdk1 first appears on centrosomes in prophase. Nat. Cell. Biol. 5, 143–148 (2003).
Besson, A., Dowdy, S. F. & Roberts, J. M. CDK inhibitors: cell cycle regulators and beyond. Dev. Cell 14, 159–169 (2008).
Puente, B. N. et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 157, 565–579 (2014).
Maroli, G. & Braun, T. The long and winding road of cardiomyocyte maturation. Cardiovasc. Res. 117, 712–726 (2021).
Zhu, J. & Thompson, C. B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 20, 436–450 (2019).
Karbassi, E. et al. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 17, 341–359 (2020).
Doenst, T., Nguyen, T. D. & Abel, E. D. Cardiac metabolism in heart failure: implications beyond ATP production. Circ. Res. 113, 709–724 (2013).
Jaakkola, P. et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Knutson, A. K., Williams, A. L., Boisvert, W. A. & Shohet, R. V. HIF in the heart: development, metabolism, ischemia, and atherosclerosis. J. Clin. Invest. 131, e137557 (2021).
Compernolle, V. et al. Cardia bifida, defective heart development and abnormal neural crest migration in embryos lacking hypoxia-inducible factor-1alpha. Cardiovasc. Res. 60, 569–579 (2003).
Guimarães-Camboa, N. et al. HIF1α represses cell stress pathways to allow proliferation of hypoxic fetal cardiomyocytes. Dev. Cell 33, 507–521 (2015).
Takagawa, J. et al. Myocardial infarct size measurement in the mouse chronic infarction model: comparison of area- and length-based approaches. J. Appl Physiol. 102, 2104–2111 (2007).
Cai, D. et al. Balasubramide derivative 3C attenuates atherosclerosis in apolipoprotein E-deficient mice: role of AMPK-STAT1-STING signaling pathway. Aging 13, 12160–12178 (2021).
Cardoso, A. C. et al. Mitochondrial substrate utilization regulates cardiomyocyte cell cycle progression. Nat. Metab. 2, 167–178 (2020).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) (82425006 to Y.N., 82300448 to L.B., and 82300325 to J.F.), the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CAMS-I2M, 2024-I2M-TS-003 to Y.N. and 2021-I2M-1-072 to Y.N.), the National Key Research and Development Project of China (2022YFA1104503 to Y.N.), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2024-RC320-04 to Y.N. and 2023-PT310-03 to Y.N.), the National High-Level Hospital Clinical Research Funding (2023-GSP-ZD-2-01 to Y.N.), the Guangdong Basic and Applied Basic Research Fund Shenzhen Joint Fund of China (2022A1515111197 to C.L.).
Author information
Authors and Affiliations
Contributions
D.C. and C.L. conducted the experiments, analyzed data, and drafted the manuscript. H.Li., C.W., Z.C., and J.Z. constructed the mouse model and performed the echocardiographic analysis. S.S. and Y.X performed the bioinformatic analysis. L.B., J.F., M.H., H.W. H. Lian., Z.Y., and Y.Z. provided important comments, analyzed the data, and edited the manuscript. Y.N. conceived the study, designed the experiments, and wrote the manuscript. All authors contributed to the discussion and critical review of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cai, D., Liu, C., Li, H. et al. Foxk1 and Foxk2 promote cardiomyocyte proliferation and heart regeneration. Nat Commun 16, 2877 (2025). https://doi.org/10.1038/s41467-025-57996-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57996-z