Abstract
Protective immunity, essential for brain maintenance and repair, may be compromised in Alzheimer’s disease (AD). Here, using high-dimensional single-cell mass cytometry, we find a unique immunometabolic signature in circulating CD4+ T cells preceding symptom onset in individuals with familial AD, featured by the elevation of CD38 expression. Using female 5xFAD mice, a mouse model of AD, we show that treatment with an antibody directed to CD38 leads to restored metabolic fitness, improved cognitive performance, and attenuated local neuroinflammation. Comprehensive profiling across distinct immunological niches in 5xFAD mice, reveals a high level of disease-associated CD4+ T cells that produce IL-17A in the dural meninges, previously linked to cognitive decline. Targeting CD38 leads to abrogation of meningeal TH17 immunity and cortical IL-1β, breaking the negative feedback loop between these two compartments. Taken together, the present findings suggest CD38 as an immunometabolic checkpoint that could be adopted as a pre-symptomatic biomarker for early diagnosis of AD, and might also be therapeutically targeted alone or in combination with other immunotherapies for disease modification.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder, and the most common cause of dementia in the elderly. Pathogenic genetic variants in one of three genes encoding for amyloid precursor protein (APP), presenilin-1 or presenilin-2 lead to a form of the disease known as early-onset familial AD, which accounts for 5–10% of AD cases1. Despite recent studies proposing amyloid-beta (Aβ)2, tau3,4, or neurofilament light chain (NfL)5 as markers of pathological changes in the brain, current approaches are not well suited for clinical screening during the pre-symptomatic stage.
Healthy life-long brain plasticity depends on the integrity and function of adaptive immunity6. During neurodegeneration, changes in the T cell immune landscape have been reported7,8,9,10, reinforcing the notion that the cellular state and effector function of T cells may reflect the neuropathological changes occurring in the brain, which could precede symptom manifestation11,12. Specialized niches within the brain’s borders, such as the meninges, host T cells that are needed for brain immunosurveillance13,14 and for remotely shaping behavior via cytokine-mediated neuromodulation15. Interleukin-17 (IL-17) is among the cytokines to which several reports have attributed a detrimental role in neurodegeneration10,16,17,18. Other findings suggested TH17 cell-intrinsic metabolic pathways19,20,21, such as those involving nicotinamide adenine dinucleotide (NAD+) and its related consuming-ectoenzyme CD38, as selective therapeutic targets for T cell metabolic rewiring22. While the elevation of CD38 in the AD brain was previously reported in the context of border-associated macrophages23, no reports described CD38 expression in connection with systemic immune dysfunction in peripheral blood mononuclear cells (PBMCs) in AD individuals.
Here, we report the elevation of metabolically impaired CD38-expressing CD4+ T cells in familial AD individuals, prior to symptom onset. Targeting CD38 restored T cell glucose metabolism and mitochondrial mass in vitro, improving metabolic fitness and cognitive performance in vivo in the 5xFAD mouse model of AD via dampening IL-1β-dependent TH17 pathogenic immunity. Our findings identified CD38 as an immunometabolic checkpoint and potential biomarker that could be therapeutically targeted in AD.
Results
Individuals carrying an APP pathogenic genetic variant harbor a metabolically dysfunctional subset of CD38-expressing CD4+ T cells prior to symptom onset
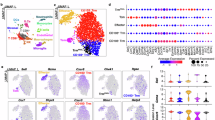
In the present study, we focused on individuals of a newly described highly consanguineous family, belonging to a minority ethnic group in northern Israel. These individuals carry a novel APP duplication rearrangement that leads to early-onset familial AD24 (Fig. 1a; Supplementary Fig. 1a; exact statistical values for all experiments are provided in Supplementary Data 1), involving pre-symptomatic (n = 16) and symptomatic individuals (n = 4), as evidenced by NfL assessment in plasma (Fig. 1b), compared to non-carrier healthy subjects from the same family (n = 17). Considering the small number of symptomatic patients, we focused on addressing if and how dysfunction of adaptive immunity occurs in pre-symptomatic family members (hereafter called APP carriers), prior to clinical manifestation of the disease. In order to determine a disease-associated signature, we performed immune profiling of PBMCs from healthy individuals and pre-symptomatic APP carriers, using high-dimensional single-cell mass cytometry by time-of-flight (CyTOF)25 with a standardized experimental workflow for exploration and discovery (Fig. 1a; Supplementary Fig. 1b). While no major differences were observed in the overall immune profile (Fig. 1c), we found an increase in the frequency of naive (TNaive) CD4+ and CD8+ T cells, accompanied by a decrease in both central (TCM) and effector memory (TEM) cells in pre-symptomatic APP carriers, as compared to their age- and ethnically-matched healthy counterparts (Fig. 1d, e). In silico analysis uncovered an expansion of cells presenting a TH2 profile (CXCR5− CXCR3− CCR6−), along with a decrease in activated CD4+ T cells in the APP carriers (Fig. 1f, g). No significant changes were detected in TH1, but a reduction in TH17 cells (Supplementary Fig. 1c, d). Moreover, at this pre-symptomatic stage, APP carriers presented an elevation in the frequency of circulating CD38-expressing CD4+ and CD8+ T cells (Fig. 1h, i), as well as an increase in CD38 median intensity (MdFI; Fig. 1j, k). In addition, APP carriers showed a significant elevation in CD38 cell-to-cell expression variation26 level, as assessed by the median absolute deviation (MAD; Supplementary Fig. 1e, f) and standard deviation (SD; Supplementary Fig. 1g, h) as variance metrics. Random Forest analysis successfully differentiated pre-symptomatic APP carriers from healthy individuals based on CD38-associated features, determined by the mean area under the receiver operating characteristic curves (ROC; Fig. 1l, m), suggesting CD38 as a potential blood-based biomarker in pre-symptomatic AD. A separate principal component analysis testing for sex-driven variability among the study participants revealed significant differences between female and male subjects within the healthy but not in the APP group (Supplementary Fig. 1i).
a Schematic representation showing study design. b Plasma neurofilament light chain (NfL) was determined in healthy subjects (n = 16), pre-symptomatic APP carriers (n = 15), and symptomatic AD individuals (n = 4). c PBMCs from healthy individuals (n = 17) and pre-symptomatic APP carriers (APP, n = 16) were analyzed by CyTOF. Frequency (%) of immune cells within circulating CD45+ cells. NK natural killer, NK T natural killer T, cDCs conventional dendritic cells, pDCs plasmacytoid dendritic cells. d, e T cells were subdivided into naive (TNaive), central memory (TCM), effector memory (TEM), and terminally differentiated effector memory (TEMRA) cells (Healthy, n = 17; APP, n = 16). Frequency (%) of the subsets within CD4+ and CD8+ T cells. Heatmaps represent the normalized median expression of differentiation markers. f Frequency (%) of circulating TH2 cells within CD4+ TH cells (Healthy, n = 17; APP, n = 16). g Frequency (%) of HLA-DR+ cells within CD4+ T cells (Healthy, n = 17; APP, n = 16). h, i Representative contour plots showing the frequency (%) of CD38hi cells within CD4+ and CD8+ T cells (Healthy, n = 17; APP, n = 16). j, k Median intensity (MdFI) of CD38 within CD4+ and CD8+ T cells (Healthy, n = 17; APP, n = 16). Results are pooled from three independent experiments. l Mean area under the receiver operating characteristic (ROC) curves illustrating the diagnostic performance of CD38-associated features following a Random Forest analysis. Inset presents the values for the three-fold cross-validation iterations (Healthy, n = 17; APP, n = 16). m Normalized variable importance (%) of the most discriminating features for diagnosis. P values are based on two-tailed Mann–Whitney U tests (b, g), multiple Mann–Whitney U tests with Holm-Šídák correction (c–e), and two-tailed unpaired t tests (f, h–k). All data show the mean ± SEM. *P < 0.05, **P < 0.01. SD standard deviation, MAD median absolute deviation. Exact P values are indicated in Supplementary Data 1. Source data are provided as a Source Data file.
We hypothesized that the observed immune alterations might be reflected in blood-circulating factors. Therefore, we conducted metabolic profiling of plasma (Fig. 2a). We found distinctive changes in the plasma metabolome of APP carriers relative to healthy individuals, characterized by a reduction in the NAD+ precursor trigonelline27 and allose, a stereoisomer of glucose (Fig. 2b; Supplementary Fig. 2a, b). Targeted metabolomics of those candidates involved in NAD+ metabolism, further confirmed the decrease in trigonelline, and an elevation in adenosine, with no changes in nicotinamide level (Fig. 2c–f). Expression of CD38 has been associated with metabolic dysfunction, particularly with respect to glucose homeostasis28,29. Therefore, we evaluated the metabolic and functional state of resting and in vitro activated T cells, by employing a modified multiplexed immunometabolic profiling pipeline, benchmarked against extracellular flux analysis30. Targeting key NAD+-consuming enzymes, glycolytic proteins, and components of the tricarboxylic acid cycle/electron transport chain (TCA/ETC), as well as markers of mitochondrial dynamics and signaling/transcription molecules/factors, revealed a metabolic dysfunction in CD4+, but not CD8+ T cells of APP carriers, as compared to healthy individuals (Fig. 2g–l; Fig. 2g, h and Supplementary Fig. 2c, d show PCA plots based on the indicated immune and metabolic markers, but not cell lineage identification markers). Breaking down the analysis to infer metabolically diverging subsets proved that our immunometabolic profiling strategy segregated naive, central memory, effector memory, terminally differentiated effector memory (TEMRA), and CD38-expressing T cells (Supplementary Fig. 2c, d). Although these different subsets might have individually shown small effects, they collectively did present a large one when APP carriers were compared to healthy subjects (Fig. 2g, j). Glucose uptake, through the GLUT family of transporters, serves as a key checkpoint in T cells by affecting T cell fate and orchestrating specialized adaptive immune responses21,31,32,33. Flow cytometry analysis of surface GLUT1 expression dynamics, following activation of isolated T cells, confirmed that CD4+, unlike CD8+ T cells, displayed a differential response when comparing APP carriers to healthy individuals (Fig. 2i, l). These results further validated a distinct cell-intrinsic glucose metabolic program for CD4+ T cells in pre-symptomatic AD individuals. When focusing on CD38-driven immunometabolic changes within APP carriers, CD38-expressing T cells exhibited increased CD39 and KLRG1 expression (Supplementary Fig. 2e, f) and altered glucose uptake of the fluorescent glucose analog 2-NBDG, relative to CD38− cells (Supplementary Fig. 2g, h). Follow-up analysis to rule out GLUT1-mediated uptake using the glucose analog 6-NBDG34, revealed a GLUT1-dependent deregulation of glucose metabolism in CD38-expressing CD4+, but not CD8+ APP T cells (Supplementary Fig. 2g, h). Further assessment of mitochondrial membrane potential employing the selective fluorescent probe MitoTracker Red CMXRos, showed depolarized mitochondria in resting CD38-expressing as compared to CD38− T cells (Supplementary Fig. 2i, j). Testing for responsiveness following in vitro activation of T cells from APP carriers, revealed mitochondrial dysfunction only in CD38-expressing CD4+, but not CD8+ T cells (Supplementary Fig. 2i, j). Taken together, we identified CD38 as a potential immunometabolic checkpoint in CD4+ T cells from pre-symptomatic APP carriers that could be targeted in AD.
a Schematic representation showing study design. b Volcano plot of significantly enriched metabolites (color coded; P < 0.05, two-tailed unpaired t test) is shown following metabolic profiling of plasma from healthy individuals (n = 16) and APP carriers (APP, n = 15). c–e Trigonelline (Healthy, n = 16; APP, n = 12), adenosine (Healthy, n = 15; APP, n = 16), and nicotinamide (Healthy, n = 18; APP, n = 16) were determined by targeted metabolomics of plasma. f NAD+ metabolism representation. g, h, j, k PBMCs from healthy individuals (n = 9) and APP carriers (n = 8) were analyzed by CyTOF. Isolated T cells from healthy individuals (n = 6) and APP carriers (n = 6) were evaluated by flow cytometry. g, j Heatmaps representing the normalized median expression of all assessed immunometabolic markers within circulating CD4+ and CD8+ T cells, as assessed by CyTOF (Healthy, n = 9; APP, n = 8). The mean within total, naive (TNaive), central memory (TCM), effector memory (TEM), terminally differentiated effector memory (TEMRA), and CD38-expressing T cells is shown. Principal component analyses (PCA) are presented for total CD4+ and CD8+ T cells; Padj. values show PERMANOVA with Benjamini-Hochberg correction. i, l Median fluorescence intensity (MdFI) of GLUT1s within CD4+ and CD8+ T cells, following activation, as determined by flow cytometry (Healthy, n = 6; APP, n = 6). Insets present the area under the curve (AUC). Results are pooled from two independent experiments. P values are based on two-tailed unpaired t tests (c, i, l), two-tailed Mann–Whitney U tests (d, e), and multiple Mann–Whitney U tests with no correction for multiple comparisons (g, j). All data show the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Exact P values are indicated in Supplementary Data 1. Source data are provided as a Source Data file. NAD+ nicotinamide adenine dinucleotide, TCA tricarboxylic acid cycle, ETC electron transport chain, TRL trigonelline, NMN nicotinamide mononucleotide, NAM nicotinamide, ADPR adenosine diphosphate ribose, AMP adenosine mononucleotide, ADO adenosine, GLUT1s surface GLUT1.
Targeting CD38 improves metabolic fitness and memory in 5xFAD mice via modulation of pathogenic TH17-mediated immunity
To address the potential mechanistic relationship between CD38 and altered T cell immunometabolic functions, we used the 5xFAD transgenic mouse model of AD. When compared to age-matched wild-type (WT), 5xFAD mice presented a modest increase in the frequency of CD38-expressing CD4+ T cells prior to symptom onset (Fig. 3a–d), with further elevation in CD38-associated features (including frequency, MdFI, MAD and SD) along chronological aging, as assessed by flow cytometry (Supplementary Fig. 3a–d). Symptomatic aged 5xFAD mice presented an expansion in both circulating CD4+ and CD8+ TNaive cells, and a contraction of the TEM pool, relative to WT mice (Supplementary Fig. 3e). Assessing glucose homeostasis, employing the oral glucose tolerance test (oGTT), showed an impaired glucose metabolism in WT relative to symptomatic aged 5xFAD mice (Supplementary Fig. 3f, g). To compare cellular versus whole-organismal metabolism, we tested glucose uptake and mitochondrial function, using 2-NBDG glucose uptake and MitoTracker Green multiplexed with MitoTracker Red CMXRos, as surrogate analyses for T cell metabolic fitness across conditions (Fig. 3e). Targeting CD38 in vitro, while differentially affecting the distinct T cell subsets, reversed the uptake of 2-NBDG and rescued mitochondrial mass in circulating AD T cells (Fig. 3f–h; Supplementary Fig. 4a–c). These results prompted us to test the therapeutic potential of targeting CD38 in 5xFAD mice.
a–d Peripheral blood from 2 to 6-month-old (young) and 8 to 12-month-old (aged) wild-type (WT) and 5xFAD mice was analyzed by flow cytometry. a, b Frequency (%) of CD38+ cells within CD4+ and CD8+ T cells from young WT (n = 8) and 5xFAD mice (n = 15). c, d Frequency (%) of CD38+ cells within CD4+ and CD8+ T cells from aged WT (n = 11) and 5xFAD mice (n = 13). e Schematic representation depicting in vitro assessment of glucose uptake, mitochondrial mass and membrane potential (∆ψm). f Glucose uptake was assessed in circulating T cells from 10 to 12-month-old WT (n = 5) and 5xFAD mice (n = 9), using 2-NBDG. Median fluorescence intensity (MdFI) of 2-NBDG within CD4+ and CD8+ T cells. Results are pooled from two independent experiments. g Mitochondrial (Mt) mass was assessed in peripheral T cells from 10 to 12-month-old WT (n = 5) and 5xFAD mice (n = 7), using MitoTracker Green. MdFI of MitoTracker Green within CD4+ and CD8+ T cells. h Mitochondrial membrane potential was assessed in circulating T cells from 10 to 12-month-old WT (n = 5) and 5xFAD mice (n = 7), using MitoTracker Red CMXRos. MdFI of MitoTracker Red CMXRos within CD4+ and CD8+ T cells. P values are based on two-tailed Mann–Whitney U tests (a, b), two-tailed unpaired t tests (c, d), and two-way RM ANOVA with Tukey’s multiple comparisons tests (f–h). All data show the mean ± SEM. *P < 0.05, ****P < 0.0001. Exact P values are indicated in Supplementary Data 1. Source data are provided as a Source Data file.
Mice were placed in metabolic cages for assessment of physical and metabolic parameters, including food intake, respiratory exchange ratio, energy expenditure, and locomotion activity, for 3 consecutive weeks during anti-CD38 antibody (αCD38) treatment (Fig. 4a; Supplementary Fig. 5a–d). A novel tensor component analysis method (TCAM)35, allowing to longitudinally represent the 3-week study as a single compressed dot per animal, revealed that targeting CD38 improved both physical and metabolic features in 5xFAD mice (Fig. 4b), restoring their metabolic fitness 2 weeks following initiation of αCD38 administration (Supplementary Fig. 5e–g), without modifying the body mass composition (Supplementary Fig. 5h, i). This analysis, however, revealed more lean mass in 5xFAD as compared to high fat-containing WT mice (Supplementary Fig. 5h, i).
a IgG or anti-CD38 administration regime. b Tensor component analysis (TCA) based on metabolic and physical parameters (5xFAD - IgG, n = 7; 5xFAD - αCD38, n = 8). TCA distance, computed after TCAM analysis, relative to wild-type (WT) - Vehicle mice (n = 4). Bounds of the box extend from the 25th to 75th percentiles, with an internal line plotted at the median, and whiskers defined by the minimum and maximum values. c Novel object recognition (NOR) cognitive performance of 5xFAD - IgG (n = 16) and 5xFAD - αCD38-treated mice (n = 17), 1 month after initiation of the treatment. WT - Vehicle mice (n = 7) are shown as technical controls. Results are pooled from four independent experiments. d Glucose level in hippocampi of 10 to 12-month-old 5xFAD - IgG (n = 15) and 5xFAD - αCD38-treated mice (n = 12). Results are pooled from five independent experiments. e IL-1β quantification in cortices of 10 to 12-month-old 5xFAD - IgG (n = 15) and 5xFAD - αCD38-treated mice (n = 13), across four independent experiments (average of mice/group/experiment is plotted) as assessed by ELISA. WT - Vehicle mice (n = 16) were used as technical controls. P values are based on one-way ANOVA with Dunnett’s multiple comparisons tests (b), one-way ANOVA with Bonferroni’s multiple comparisons tests (c), two-tailed Mann–Whitney U tests (d), and one-way RM ANOVA with Bonferroni’s multiple comparisons test (e). All data show the mean ± SEM. *P < 0.05, **P < 0.01. Exact P values are indicated in Supplementary Data 1. Source data are provided as a Source Data file.
To determine whether the observed physical and metabolic improvement might lead to attenuation in behavioral deficits, we next assessed recognition memory using the novel object recognition (NOR) test, and anxiety behavior in an open field. Working memory assessment of αCD38-treated 5xFAD mice showed an improvement in their recognition memory, 1 month after initiation of the treatment (Fig. 4c), with no effect on locomotor ability and anxiety-related parameters, such as the distance traveled, and the time spent in the center of the arena, respectively (Supplementary Fig. 5j). We further tested whether the attenuated cognitive impairment could be linked to amelioration of AD hallmarks, including amyloidosis36,37, altered glucose metabolism38,39, and IL-1β as a proxy of neuroinflammation40,41,42. We observed no effect on both Aβ plaques and soluble Aβ42 (Supplementary Fig. 6a, b), but a reduction in glucose level in the hippocampus of αCD38- as compared to IgG-treated 5xFAD mice (Fig. 4d). Bulk RNA-sequencing analysis of the hippocampi revealed an enrichment of genes involved in neuronal synaptic plasticity (homer1)43, autophagy (mTOR)44 and learning and memory (igf2r)45 in αCD38- relative to IgG-injected mice (Supplementary Fig. 6c). Evaluating the effect of αCD38 treatment on brain IL-1β, revealed a reduction at a protein level when analyzing the cortices of these same mice, a phenomenon that was also observed when each independent experiment was separately analyzed (Fig. 4e; Supplementary Fig. 6d). These results suggest leveraging IL-1β level as a potential readout for the efficacy of αCD38 immunotherapy in AD.
Recent studies identified adaptive immune cells within the brain’s borders13,14, including the choroid plexus and meninges, which remotely affect brain function46,47,48,49. Following a comprehensive cellular and cytokine profiling by flow cytometry across distinct immunological niches, we identified CD4+ T cells residing in the dural meninges as a source of disease-associated IL-17A in 5xFAD mice, which was downregulated by targeting CD38, with no effect on IFN-γ- or IL-4-producing T cells (Fig. 5a; Supplementary Fig. 7a–d). Since the cerebrospinal fluid (CSF), which comprises the brain secretome50, effluxes the dura51, we conducted a proteomic profiling of the CSF. We found that two key metabolic mediators, acetyl-CoA acetyltransferase 2 (Acat2) and ATP-citrate lyase (Acly), previously shown to control TH17 cell effector function through glycolytic-epigenetic reprogramming21, were downregulated in αCD38- compared to IgG-treated 5xFAD mice, 1 month after initiation of the treatment (Fig. 5b). Also the TAM receptor Mertk52 and the damage-associated molecular pattern Hmgb153, were downregulated in αCD38- relative to IgG-injected 5xFAD mice (Fig. 5b), likely reflecting an attenuation in neuroinflammation.
a Dural meninges, deep cervical lymph nodes (dcLNs), spleen, and peripheral blood from 10 to 12-month-old wild-type (WT) - Vehicle (n = 8), 5xFAD - IgG (n = 15) and 5xFAD - αCD38-treated mice (n = 15) were analyzed by flow cytometry, 1 month after initiation of the treatment. Results are pooled from three independent experiments. b Cerebrospinal fluid (CSF) from 10 to 12-month-old 5xFAD - IgG (n = 8) and 5xFAD - αCD38-treated mice (n = 7) were analyzed by proteomic profiling. Volcano plot of significantly enriched proteins (color coded; P < 0.05, two-tailed unpaired t test). Results are pooled from two independent experiments. c Schematic representation showing in vitro differentiation into non-pathogenic (np, - IL-1β) or pathogenic (p, + IL-1β) TH17 cells. d Representative density plots, showing the frequency (%) of IL-17A-producing cells within npTH17 (n = 8) and pTH17 cells (n = 8). e Frequency (%) of CD38-expressing cells within npTH17 (n = 8) and pTH17 cells (n = 8). Results are pooled from two independent experiments. f Pearson correlation analysis (r, two-tailed P value) between the frequency of IL-17A+ and CD38+ cells in npTH17 (n = 8) and pTH17 cells (n = 8), showing the 95% confidence bands of the best-fit line. g mRNA expression of il1β and il17ra in the cortices of 10 to 12-month-old 5xFAD - IgG (n = 8) and 5xFAD - αCD38-treated mice (n = 7) were determined by quantitative real-time PCR. Results are pooled from two independent experiments. P values are based on two-way RM ANOVA with Dunnett’s multiple comparisons test (a), two-tailed unpaired t tests (d, e), and multiple unpaired t tests with Holm-Šídák correction (g). All data show the mean ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001. Exact P values are indicated in Supplementary Data 1. Source data are provided as a Source Data file. Acat2 Acetyl-CoA acetyltransferase 2, Acly ATP-citrate lyase, Hmgb1 high-mobility group box 1, DAMP damage-associated molecular pattern.
In the search for a mechanism accounting for αCD38 modulation of TH17 immunity, we differentiated TH17 cells in vitro to assess their immune profile and effector function (Fig. 5c). We observed that recombinant IL-1β induced both an increase in the frequency of IL-17A-producing and CD38-expressing cells in polarized pathogenic (p) relative to non-pathogenic (np) TH17 cells (Fig. 5d, e), revealing a positive correlation between IL-17A and CD38 (Fig. 5f). Since IL-17 receptor A (IL-17RA) has been shown to mediate communication from the meninges to the cortex47, we next examined whether the beneficial effect of peripherally targeting CD38 could be mediated in part through modulation of cortical IL-17RA and IL-1β signaling, which we observed to drive TH17 pathogenicity. Real-time quantitative PCR (RT-qPCR) analyses showed a selective decrease of cortical il1β in αCD38- as compared to IgG-treated 5xFAD mice (Fig. 5g), which correlated with il17ra downregulation, with no effect on other tested cytokines (Supplementary Fig. 7e, f). To determine whether target engagement following αCD38 immunotherapy could result in cytotoxicity, we assessed cell viability by flow cytometry, observing no differences across conditions neither in vivo nor in vitro (Supplementary Fig. 7g, h). Further, by using a different antibody clone for flow cytometry, we ruled out the possibility that reduced CD38 expression in pTH17 cells could be attributed to an artifact due to prior treatment with αCD38 antibody (Supplementary Fig. 7i). While in the present study we did not include a group of untreated 5xFAD mice, our previous experience revealed no differences between untreated and IgG-injected mice, neither in the 5xFAD54 nor DM-hTAU40 mouse models. Altogether, targeting CD38 resulted in an improvement of metabolic fitness and cognitive performance potentially via abrogation of IL-1β-dependent TH17 pathogenic function in AD.
TH17 cells are most abundant at mucosal barriers, where exposure to environmental factors shape the microbiome55,56. To further support the negative impact of meningeal TH17 immunity on cognitive performance in AD, we assessed the level of TH17 cells in intestinal-associated tissues of specific-pathogen-free (SPF) relative to germ-free (GF) 5xFAD mice, previously reported to significantly outperform conventional 5xFAD mice in cognitive tests57. Peyer’s patches (Pps), but not mesenteric lymph nodes (mLNs) of 5xFAD presented a higher frequency of TH17 cells relative to WT mice, while Pps of GF 5xFAD mice showed a negligible level (Supplementary Fig. 8a). Meningeal immunity has been proposed to be evolutionary shaped to mirror that of the gut58,59. Comparing CD38+ TH subsets, identified by their canonical master regulators, showed reduced frequency of FOXP3 regulatory (Tregs)- but an increase of RORγt (TH17)-expressing T cells in dural meninges and Pps of 5xFAD mice relative to WT mice, further revealing a positive correlation between these distant, yet similar immunological niches (Supplementary Fig. 8b–d). These results strengthen the proposed pathogenic effect of TH17 cells on cognition during AD, suggesting that their accumulation is possibly dependent on commensal-derived signals.
Discussion
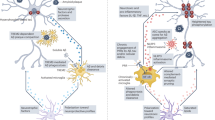
In the present study, we identified a metabolically dysfunctional CD38-expressing CD4+ T cell subset, emerging prior to symptom onset in individuals with dominantly inherited AD (Fig. 6a). Targeting CD38 in a mouse model of AD, resulted in an improvement of physical and metabolic parameters, together with an attenuation of memory deficits (Fig. 6b), likely due to reduction of local brain inflammation associated with modulation of meningeal pTH17 immunity (Fig. 6c).
a Identification of dysfunctional CD38-expressing CD4+ T cells before disease onset in individuals with familial Alzheimer’s disease (AD). b αCD38 immunotherapy in 5xFAD mice leads to attenuated neuroinflammation, and improved metabolic fitness and cognition. c Identification of TH17 cells accumulating in Peyer’s patches and dural meninges of 5xFAD mice, suggests that gut-educated CD4+ T cells may be a source of meningeal IL-17A during AD. Targeting CD38 abrogates disease-associated TH17 cell immunity, potentially by breaking meningeal to cortical communication via modulation of IL-17A/IL-1β crosstalk.
Previous reports have significantly contributed to the understanding of the NAD+-CD38 axis in aging28,60,61 and neurodegeneration62,63,64. However, much of this work was focused on peripheral macrophages60,61. Here, we revealed the emergence of a subset of CD38hi T cells in pre-symptomatic familial AD individuals, suggesting that disease-associated factors might contribute to the elevation of CD38 prior to cognitive decline. In line with a previous report26, we provide further evidence that CD38 cell-to-cell expression variation level in T cells, which directly reflects their cellular state and is seemingly conserved across mice and humans, could be potentially used as a biomarker in pre-symptomatic AD and other familial neurodegenerative conditions. However, it would be interesting to assess CD38 expression in T cells from symptomatic familial AD individuals, and whether CD38 could also serve as an early diagnostic marker in sporadic AD, which accounts for 90–95% of AD cases1. Even though evidence indicates that biological sex could be considered as a key variable in disease heterogeneity65, we did not detect sex-based differences within the APP group, possibly due to the limited number of study participants. We also showed that circulating T cells in AD are skewed towards a TH2 profile and that HLA-DR, an activation marker, is significantly reduced in CD4+ T cells. The fact that we observed a decrease in TH17 cells, might imply an accumulation of these cells in the human diseased brain10. Age-dependent thymic involution is manifested by a T cell naive/memory imbalance, featuring a reduction in naive cells with clonal expansion of memory T cells66. APP carriers presented an accumulation of naive T cells at the expense of the contraction of highly differentiated memory cells, accompanied by a remodeling of the plasma metabolome that favored a decrease in the NAD+ precursor trigonelline27, shown to restore memory function in 5xFAD mice67, along with an increase in adenosine, reported to mediate non-canonical CD38-dependent immunosuppression68. It is possible that changes in trigonelline and adenosine, but not nicotinamide level, were detected due to their higher bioavailability and stability in plasma. Whether this systemic milieu could result in a metabolic deviation of the peripheral immune system is not clear. Glucose transporter GLUT1 deficiency in the endothelium was reported to worsen AD pathology in a mouse model38. Accordingly, here we demonstrated a selective metabolic rewiring affecting GLUT1-dependent glucose metabolism and driving mitochondrial dysfunction in CD4+ T cells in pre-symptomatic AD individuals. It remains to be elucidated whether T cells from APP carriers might be directly affected by the potential overexpression of this pathogenic genetic variant. Together with previous studies11,40,54,69, these findings support the contention of systemic low-grade chronic inflammation as a potential risk factor in AD, which could be a reflection of the ongoing brain pathology. Similarly to APP carriers, 5xFAD mice presented a T cell naive/memory imbalance. Though modest, circulating CD4+ T cells featured an elevation in CD38 that preceded symptom onset, which increased with aging, in line with previous reports28,60,61. Notably, 5xFAD mice exhibited reduced body weight, with higher lean mass relative to age-matched WT mice. It is therefore possible that this phenotype, previously associated with a robust hyperactivity70, could account for the enhanced peripheral glucose tolerance.
Here, we found that targeting CD38 in 5xFAD mice contributed to restoration of the metabolic fitness that is lost in AD, and protection against cognitive decline. Mechanistically, targeting CD38 improved T cell glucose metabolism and mitochondrial mass in vitro, attenuated hippocampal glucose hypometabolism, as well as cortical IL-1β-driven inflammation in vivo, an AD hallmark associated with disease progression71,72, linking metabolism and immunity73. Glucose metabolism has been shown to modulate the effector function of TH17 cells by controlling a metabolic-transcriptional network, with Acly serving as a key rate-limiting mediator generating acetyl-CoA and dictating TH17 cell pathogenicity21, while restraining the development of FOXP3+ Tregs74. Accordingly, our results showed that CSF-derived Acly and Acat2, an enzyme downstream of Acly, are both downregulated following targeting CD38 in 5xFAD mice. Several independent reports have demonstrated a fundamental role for IL-17 beyond typical host defense at barrier sites17,47,49,75. The fact that of all tested tissues, we identified TH17 cells accumulating solely in Pps and dural meninges of 5xFAD mice, suggests that gut-educated CD4+ T cells may be a source of disease-associated IL-17A during AD, supporting a gut-brain axis in which TH17 cells, induced in intestine-associated tissues, could drive neuroinflammation76 and promote worsening of cognition77. In fact, GF 5xFAD mice, which are devoid of TH17 cells, were reported to show a superior cognitive performance relative to SPF 5xFAD mice57. In line with our results, additional evidence suggested a gut-brain axis in which an increase of TH17 cells, along with a decrease of Tregs, can drive neurodegeneration in 3xTg AD mice78. Since TH17 pathogenicity is limited by FOXP3+ Tregs, disruption of a TH17/Tregs cell balance contributes to the development of immune-mediated diseases79. By comparing CD38-expressing TH subsets in dural meninges and Pps, we also showed that loss of controlling Tregs might account for the expansion of TH17 cells, providing further evidence that meningeal immunity resembles that of the gut58,59. Distinct pathogenic regulators have recently emerged for selective targeting pTH17, while sparing protective npTH17 cells20,80,81,82,83,84,85,86,87. In this study, we found that in vitro exposure to IL-1β was sufficient to induce pTH17 cells, driving an increase in CD38-expressing cells, whereas in vivo targeting CD38 consistently decreased cortical IL-1β in the AD brain, suggesting that αCD38 treatment could lead to IL-1β-mediated modulation of TH17 cell pathogenicity. Therefore, even though a homeostatic axis whereby meningeal IL-17A release could signal in the cortex through Il17ra was previously proposed47, it is tempting to speculate that in AD, IL-1β derived from the brain parenchyma, could also signal back to the meninges, creating a feedback loop that sustains meningeal pTH17 cells.
Similar to cancer, in which CD38 has been proposed as an amenable target, both alone22 and in combination with PD-L1 blockade88, our findings support the contention that immune mechanisms might go awry in otherwise seemingly unrelated chronic diseases. Specifically, we provide evidence that CD38 could be considered as an immunometabolic checkpoint, joining the therapeutic promise of other immune checkpoints previously proposed to modify AD54 and other forms of dementia40. Though some of the effects observed following the treatment were modest, we believe further optimization of the therapeutic strategy, could yield a more robust beneficial effect. Designing novel therapeutic antibodies to target CD38 in combination with other immune checkpoints in T cells, is likely to increase functional selectivity and reduce non-specific side effects, ultimately enhancing the desired biological effect89. However, since CD38 could be expressed by other immune and non-immune cells60,61, our results suggest that the efficacy of targeting CD38 might not solely rely on CD38 expression level in T cells, opening up additional therapeutic opportunities.
In this study, we identified a metabolically dysfunctional subset of disease-associated CD38-expressing CD4+ T cells before symptom onset in AD which, when targeted in 5xFAD mice, resulted in an improvement of metabolic fitness and cognition. Even though it is possible that αCD38 might directly affect the brain, targeting CD38 abrogated disease-associated TH17 cell immunity, potentially by breaking meningeal to cortical communication via modulation of IL-17A/IL-1β crosstalk (Fig. 6). Taken together, these findings suggest an immunometabolic checkpoint that could be adopted as a pre-symptomatic biomarker and may also be therapeutically targeted alone or in combination with other immunotherapies for disease modification in AD.
Methods
Human observational study
The human observational study was approved by the Galilee Medical Center Review Board (IRB approval No. 2019-11735) and the Weizmann Institutional Review Board (IRB approval No. 1595-1). Written informed consent was obtained from all individuals. Participants were not compensated for their participation in the study.
Non-fasting fresh whole human peripheral blood samples, from a cohort of individuals belonging to a seven-generation pedigree including females and males aged 29–68 years, were collected in the morning using EDTA-coated collection tubes (BD). While members of this family carry an autosomal dominant APP duplication rearrangement that leads to early-onset familial AD, including pre-symptomatic (do not fulfill yet the diagnostic criteria) and symptomatic (with clinically diagnosed AD) patients, subjects from the same family in which the genetic testing did not show the APP duplication were considered as non-carrier healthy controls. Cognitive impairment was previously assessed by the Mini-Mental State Examination (MMSE) and the Montreal Cognitive Assessment (MoCA)24.
Blood samples were centrifuged for 10 min at 2000 × g, 4 °C. Plasma supernatant was collected, divided into aliquots, and frozen at −80 °C until further NfL determination, metabolic profiling, or targeted metabolomics. Isolation of PBMCs was performed with Ficoll-Paque® density gradient (GE Healthcare). Following the isolation, cells were resuspended in Cryostor® cell cryopreservation media (Merck), aliquoted into cryovials, and placed in a Mr. Frosty freezing container (Nalgene). Subsequently, PBMCs were transferred to a liquid nitrogen tank and stored in the vapor phase until further ex vivo reactivation and processing for CyTOF and flow cytometry.
NfL determination
NfL was measured with Simoa® Human Neurology 4-Plex B assay (N4PB) on a Quanterix® SIMOA HD-1 platform, according to manufacturer’s instructions.
NfL was determined in plasma from healthy subjects (females, n = 7; males, n = 9; average age, 47.38 years), pre-symptomatic APP carriers (females, n = 10; males, n = 5; average age, 34.53 years), and symptomatic AD individuals (females, n = 3; males, n = 1; average age, 54.75 years).
Metabolic profiling
Extraction and analysis of polar metabolites were performed as previously described90,91, with some modifications: 100 μL of plasma were mixed with 1 mL of a pre-cooled (-20 °C) homogenous methanol (MetOH):methyl-tert-butyl-ether (MTBE) 1:3 (v/v) mixture. The tubes were vortexed and then sonicated for 30 min in an ice-cold sonication bath (briefly vortexed every 10 min). Then, UPLC-grade water DDW:MetOH (3:1, v/v) solution (0.5 mL) containing internal standards: 13C- and 15N-labeled amino acids standard mix (Sigma-Aldrich) was added to the tubes, followed by centrifugation. The upper organic phase was transferred into 2 mL Eppendorf tube. The polar phase was reextracted as described above, with 0.5 mL of MTBE. Both organic phases were combined and dried in a speedvac and then stored at −80 °C. The lower polar phase used for polar metabolite analysis was lyophilized, dissolved in 150 μL of 1:1 DDW:MetOH (v/v), centrifuged at 20,000 × g for 5 min, transferred to a new tube, and centrifuged again. For analysis, 80 μL were transferred into HPLC vials.
Metabolic profiling of the polar phase was done as previously described91, with minor modifications. Briefly, analysis was performed using Acquity I class UPLC System combined with mass spectrometer Q Exactive Plus Orbitrap™ (Thermo Fisher Scientific) operated in a negative ionization mode. The LC separation was done using the SeQuant Zic-pHilic (150 × 2.1 mm) with the SeQuant guard column (20 × 2.1 mm, Merck). The composition of mobile phase B was acetonitrile, and the composition of mobile phase A was acetonitrile and 20 mM ammonium carbonate with 0.1% ammonium hydroxide in DDW:acetonitrile (80:20, v/v), respectively. The flow rate was kept at 200 μL/min, and the gradient was as follows: 0-2 min 75% of B, 14 min 25% of B, 18 min 25% of B, 19 min 75% of B, for 4 min.
Data processing was done using TraceFinder (Thermo Fisher Scientific). The compounds were identified by accurate mass, retention time, isotope pattern, and fragments and verified using an in-house generated mass spectra library. Peaks were quantified by calculating the area under the curve (AUC) and then normalizing the AUC values by internal standards and original sample volume.
Metabolic profiling was performed in plasma from healthy participants (females, n = 7; males, n = 9; average age, 47.69 years), and pre-symptomatic APP carriers (females, n = 10; males, n = 5; average age, 34.53 years).
Targeted metabolomics
The LC-MS/MS instrument consisted of Acquity I-class UPLC system (Waters) and Xevo TQ-S triple quadrupole mass spectrometer (Waters) was used for the analysis. MassLynx and TargetLynx software (Waters) were applied for the acquisition and analysis of data. Chromatographic separation was done on a 100 × 2.1 mm i.d. 1.7 μm UPLC Atlantis Premier BEH C18 AX column (Waters) with 0.2% formic acid as mobile phase A and 0.2% formic acid in acetonitrile as B at a flow rate of 0.3 mL/min and column temperature 25 °C. A gradient was as follows: 0.5 min the column was hold at 4% B, then linear increase from 1 to 15% B in 1 min, then to 40% B in 3 min, and to 100% B in 0.5 min, and hold at 100% B for 1 min. Then back to 1% B during 0.5 min, and equilibration for 1 min. Samples kept at 8 °C were automatically injected in a volume of 1 μL. The mass spectrometer equipped with an electrospray ion source and operated in negative ion mode was used, with 0.10 mL/min of argon as a collision gas flow. The capillary voltage was set to 1.85 kV, cone voltage 30 V, source offset 12 V, source temperature 120 °C, desolvation temperature 500 °C, desolvation gas flow 600 l/h, cone gas flow 150 l/h.
Targeted metabolomics was performed in plasma from healthy individuals (females, n = 9; males, n = 9; average age, 44.72 years), and pre-symptomatic APP carriers (females, n = 11; males, n = 5; average age, 34.56 years).
Mass cytometry
Heavy-metal conjugation of antibodies
Antibodies were conjugated to heavy-metal ions with commercially available Maxpar® (Fluidigm) or MIBItag (Ionpath) reagents according to manufacturer’s instructions, and as previously described30 (see Supplementary Data 2). In short, antibodies were reduced with 4 mM TCEP (Thermo Fisher Scientific) for 30 min at 37 °C and washed two times. For conjugations using Maxpar® reagents, metal chelation was performed by adding metal solutions (50 mM) to chelating polymers and incubating for 40 min at 37 °C in a water bath. Metal-loaded polymers were washed twice using a 3 kDa microfilter (Millipore) by centrifuging for 25 and 30 min at 12,000 × g, room temperature (RT). For conjugations using MIBItag reagents, pre-loaded polymers were obtained, and no loading reactions were needed.
For both approaches, antibody buffer exchange was performed by washing purified antibodies through a 50 kDa microfilter (Millipore) and centrifuging for 10 min at 12,000 × g, RT. Partially reduced antibodies and metal-loaded polymers were incubated together for 90 min at 37 °C. Conjugated antibodies were washed three times for 10 min at 12,000 × g, RT. The concentration of the conjugated antibody was then determined by NanoDrop (Thermo Fisher Scientific) measurement, after which the antibody was diluted with stabilization buffer (Candor® Bioscience) to a final concentration of 0.5 mg/mL. The conjugated antibody was collected following a brief centrifugation (2 min, 1000 × g, RT) into an inverted column in a fresh tube and stored at 4 °C.
Barcoding and staining with heavy-metal-conjugated antibodies
Most antibodies were obtained pre-conjugated to heavy-metal isotopes from Fluidigm (see Supplementary Data 2). This is case for the Maxpar® Human Immune Monitoring (HIM) panel kit, which has been designed and optimized for deep immune profiling of human PBMCs, through comprehensive identification and characterization of key immune cell populations, including all major T cell subsets (CD4+ and CD8+ naive, central memory, effector, and effector memory), T helper (TH) cell subsets (TH1, TH2, and TH17), CD4+ regulatory T cells, natural killer cells, natural killer T cells, γδ T cells, B cell subsets (naive and memory), plasma cells, dendritic cells (conventional and plasmacytoid), monocytes, and neutrophils. The kit was used according to manufacturer’s instructions. In short, cells were labeled with Cell-ID™ cisplatin viability stain (Fluidigm), before incubating all individual samples with Human TruStain FcX™ (BioLegend, Cat. # 422302, 1:10) for 10 min at RT. Cell surface antibody master mix was added, and samples were incubated for 30 min at RT. To eliminate technical variability during acquisition, individual samples within one experiment were fixed with Maxpar® Fix I Buffer for 10 min at RT, permeabilized with Maxpar® Barcode Perm Buffer, and barcoded with the Cell-ID™ palladium (Pd) barcoding kit (Fluidigm) for 45 min at RT. Samples were then combined and washed once with Maxpar® Barcode Perm Buffer.
For the Immunometabolic panel, targets were allocated to specific heavy-metal isotopes following the sensitivity of the mass cytometer (e.g., placing lower abundance targets on higher sensitivity channels) and to avoid potential spillover, as previously outlined92. Briefly, all individual samples were incubated with Human TruStain FcX™ (BioLegend, Cat. # 422302, 1:10) for 10 min at RT. To eliminate technical variability during staining or acquisition, individual samples within one experiment were barcoded with a unique combination of cadmium (Cd)-tagged CD45 antibodies93 (Fluidigm) for 30 min at RT and combined into a composite sample. Cell surface antibody master mix was added to the composite sample. After incubation for 30 min at RT, cells were washed once prior to being labeled with Cell-ID™ cisplatin viability stain. To enable intracellular staining, cells were fixed with Pierce™ 1.6% formaldehyde (Thermo Fisher Scientific) in Maxpar® PBS for 10 min at RT and washed twice with Maxpar® Cell Staining Buffer. Cells were then permeabilized with ice-cold MeOH for an additional 10 min on ice and washed again two times with Maxpar® Cell Staining Buffer to remove any residual MeOH. Intracellular antibody master mix was added to the composite sample and incubated for 60 min at RT. Cells were washed once with Maxpar® Cell Staining Buffer.
After washing, composite samples were incubated overnight in Pierce™ 4% formaldehyde in Maxpar® PBS at 4 °C. The following day and prior to acquisition, samples were incubated with Cell-ID™ Intercalator-Ir for 20 min, washed once with Maxpar® Cell Staining Buffer, once with Maxpar® PBS, and once with Maxpar® mass cytometry grade water. The composite sample was then resuspended in Maxpar® mass cytometry grade water supplemented with 1:10 EQ™ Four Element Calibration Beads (Fluidigm), filtered through a 40 μm pore size cell strainer (Corning) and acquired on a Helios™ CyTOF® system (Fluidigm).
The HIM panel kit was used for immune profiling of PBMCS from healthy individuals (females, n = 9; males, n = 8; average age, 46.18 years) and pre-symptomatic APP carriers (females, n = 11; males, n = 5; average age, 34.56 years). The Immunometabolic panel was used to evaluate the metabolic and functional state of PBMCS from healthy subjects (females, n = 6; males, n = 3; average age, 45.78 years) and pre-symptomatic APP carriers (females, n = 3; males, n = 5; average age, 34.88 years).
Data pre-processing
Raw CyTOF data were first bead normalized to remove acquisition sensitivity-related influences on marker expression using Fluidigm’s software. Next, normalized data were uploaded to the Cytobank (Beckman Coulter) analysis platform94 to identify single, live cells by excluding remaining beads (140Ce and 153Eu) and manually gating on DNA (193Ir) and viability (195Pt) channels before gating on CD45+ immune cells95. Barcoded cells were assigned back to their initial samples using their unique Pd (HIM panel) or Cd (Immunometabolic panel) barcode combination using Fluidigm’s software93 or an open-source single-cell debarcoder96, respectively. Data were subsequently imported into FlowJo software (BD) for downstream analysis (Supplementary Fig. 9). When necessary, data were further normalized using MetaboAnalyst 5.097,98, and scaled using GraphPad Prism (version 9.5.1, GraphPad Software).
Ex vivo reactivation of PBMCs
Short-term reactivation of cryopreserved PBMCs and subsequent CyTOF were performed as previously described99. Briefly, PBMCs were thawed at 37 °C in a water bath for 3 min. Cells were gently resuspended in tubes containing 10 mL of pre-warmed RPMI 1640 media (Gibco™) supplemented with 10% FBS (Biological Industries), L-glutamine (2 mM), penicillin (100 U/mL, Biological Industries), streptomycin (0.1 mg/mL, Biological Industries) and Benzonase® Nuclease (1:10,000, Millipore), and centrifuged for 7 min at 400 × g, RT. Washing was repeated once before cells were gently resuspended and rested overnight in 1 mL of pre-warmed RPMI 1640 media (Gibco™) supplemented with 10% FBS (Biological Industries), L-glutamine (2 mM), penicillin (100 U/mL, Biological Industries) and streptomycin (0.1 mg/mL, Biological Industries), in an incubator at 37 °C and 5% CO2.
For GLUT1s expression dynamics, isolated T cells from healthy (females, n = 4; males, n = 2; average age, 36 years) and pre-symptomatic APP (females, n = 4; males, n = 2; average age, 34.83 years) individuals, were analyzed by flow cytometry. Briefly, pan T cells were enriched following negative selection using magnetic beads (Miltenyi Biotec). Isolated CD4+ and CD8+ T cells were activated or not for up to 1, 3, or 6 days in a reverse time course with Dynabeads™ (1:1 cell-to-bead ratio, Gibco™), magnetic beads similar in size to antigen-presenting cells and coupled with anti-CD3 and anti-CD28 antibodies, so that their total time in culture would be identical and to ensure that all cells would finish their indicated activation period the same day. Samples were acquired in a CytoFLEX S cytometer (Beckman Coulter) and analyzed (Supplementary Fig. 10a–c) using FlowJo software (BD).
Mice
10 to 12-month-old (unless otherwise stated) heterozygous 5xFAD female mice on a mixed C57BL/6 and SJL genetic background (Tg6799, MMRRC_034840-JAX; The Jackson Laboratory), overexpressing familial AD mutant forms of human APP and PS1 transgenes under the transcriptional control of neural-specific elements of the mouse Thy1 promoter100, were used with a paternal transgenic inheritance101. Mice were bred and maintained at the Lockey preclinical research facility of the Weizmann Institute of Science. 8 to 9-month-old GF female mice were bred in the GF facility of the Weizmann Institute of Science, housed in gnotobiotic isolators (Park Bioservices), and routinely monitored for sterility. 2 to 3-month-old WT female mice were purchased from ENVIGO specifically for TH17 differentiation, and acclimated before initiation of the experiments.
In all experiments, age-matched mice were used (unless otherwise stated), WT littermates served as controls, and treatment was randomly assigned to control for cage effect. Experiments were blinded where observer bias could affect the results, including open field, novel object recognition tests, and immunohistochemistry. All mice were kept on a strict 24 h reverse light-dark cycle (with lights being turned on from 22:00 to 10:00), at an ambient temperature of 22 °C, 50% humidity and access to ad libitum food and water (unless otherwise stated). Genotyping was performed by PCR analysis of ear DNA, as previously described100. All experiments detailed here complied with the regulations formulated by the Institutional Animal Care and Use Committee (IACUC, No. 07290822-3 and No. 01800223-1) of the Weizmann Institute of Science.
Therapeutic antibody
For CD38 in vivo targeting, employing a modified administration regime22, CD38-specific antibody (anti-CD38, rat isotype IgG2a kappa; BioXCell, clone NIMR5) and isotype control immunoglobulin (rat IgG2a; BioXCell) were administered intraperitoneally (i.p.) three times a week (every second day) for 3 weeks, at a dose of 50 μg per mouse. For CD38 in vitro targeting, αCD38 and IgG2a (IgG) control antibodies were used at a dose of 10 μg/mL.
Mouse tissue collection and processing
Mice were deeply anesthetized with an i.p. injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). For CSF collection, the skin of the neck was shaved, and the mouse was placed prone to secure the head. The surgical site was swabbed with 70% ethanol, and a sagittal incision of the skin was made inferior to the occiput. Subcutaneous tissue and muscles were separated by blunt dissection with forceps, the dura mater was blotted dry with a sterile cotton swab, and CSF was collected using a sterile pulled capillary to penetrate the cisterna magna (through the dura mater, lateral to the arteria dorsalis spinalis) and immediately snap-frozen in liquid nitrogen and stored at −80 °C.
Blood was withdrawn by retro-orbital sinus puncture through the medial canthus of the eye using glass capillaries. Blood samples were collected into heparin-coated tubes and treated with eBioscience™ RBC Lysis Buffer for optimal removal of erythrocytes. Mice were then transcardially perfused with ice-cold PBS (Biological Industries) before spleens, deep cervical lymph nodes, Peyer’s patches, and mesenteric lymph nodes were excised and mashed with the plunger of a syringe against a 100 μm pore size cell strainer (Corning). Splenocytes were finally treated with eBioscience™ RBC Lysis Buffer for optimal removal of erythrocytes.
Following brain dissection, the skull cap was removed, and dural meninges collected and processed as previously described102. Briefly, meninges were incubated for 30 min at 37 °C in RPMI 1640 media (Gibco™) supplemented with 5% FBS (Biological Industries), collagenase IV (1.2 mg/mL, Sigma-Aldrich), DNAse (75 μg/mL, Roche), and HEPES (6 mM, Biological Industries). At the end of the incubation, the homogenate was filtered with a 70 μm pore size cell strainer (Corning) to obtain a single-cell suspension. Following dissection of the dura, brains were cut following a sagittal plane into two halves, and hemi-cortices and hippocampi were dissected, immediately snap-frozen in liquid nitrogen, and stored at −80 °C until further processing.
Oral glucose tolerance test
Mice were fasted for 6 h during the dark phase with free access to water, and tail blood glucose was measured for 0 time point for basal glucose level assessment (Bayer Contour® blood glucose meter and test strips). Subsequently, mice were gavaged with 10 μL/gr of a 20% glucose (Sigma-Aldrich) solution in PBS (Biological Industries) and blood glucose was measured at 15 min, 30 min, 45 min, 60 min, 90 min, and 120 min, following glucose oral feeding.
Body mass composition analysis
Mice were weighed right before inserted into the sampling compartment of a Bruker’s Minispec LF50 Lean/Fat Analyzer for the whole-body compositional analysis by nuclear magnetic resonance of fat tissue, lean tissue, and free fluid in a non-invasive, non-destructive, and stress-free manner, prior and following metabolic studies.
Metabolic phenotyping
Metabolic cages (PhenoMaster system, TSE-Systems) were used to continuously measure food intake, respiratory exchange ratio (RER), energy expenditure, and locomotion activity in singled-housed mice. The system consists of a combination of sensitive feeding sensors for automated measurements, gas calorimetry to calculate the energy expenditure of each mouse, and a photobeam-based activity monitoring system for detection and recording of ambulatory movements including rearing and climbing, in each cage. Mice were acclimated to metabolic cage accommodation with a 1-week daily training session of 10 h in identical cages prior to data acquisition, after which mice were returned to their original cages. All parameters were measured continuously and simultaneously for three weeks after initiation of the treatment, only briefly paused for a weekly calibration and retrieval of the generated data. Metabolic and physical parameters shown were measured during the dark phase.
Open field
Open field was performed as previously described103, 1 month after initiation of the treatment. In short, mice were allowed to freely explore a gray square box (50 × 50 × 50 cm) for 10 min. Total distance traveled by each mouse and time spent in the center of the arena were calculated using EthoVision XT 14 automated tracking system (Noldus).
NOR test
Novel object recognition test was performed during dark hours, in a dimly lighted room as previously described104, 1 month after initiation of the treatment. Briefly, mice were placed in a gray square box (50 × 50 × 50 cm) with visual cues on the walls. On the training day, mice were given four sessions of 6 min; during the first session, mice were allowed to explore the arena without objects (habituation), and in the following three trials, two objects of different color, shape, and texture were present in defined locations in the box (training, day 1). After 24 h, mice were returned to the arena, in which one of the objects was placed in a new location (testing day, day 2). Time spent exploring each object on each day was manually scored using EthoVision XT 14 automated tracking system (Noldus), and percentage of preference for the displaced object was calculated for each mouse, for each day, by dividing the time spent exploring the displaced object by the total exploration time of both objects, and multiplying the result by 100%, according to the formula: Percentage of preference = [(displaced object exploration time)/(displaced object exploration time + non-displaced object exploration time)] x 100%. The result of the calculation was ~50% on day 1 (training) and 60 - 90% on day 2 for mice with normal memory.
Immunohistochemistry
Immunohistochemistry was performed on paraffin-embedded mouse brain sections of 6 μm thickness. For Aβ staining, the M.O.M.® (Mouse On Mouse) Immunodetection kit (Vector labs) was used according to manufacturer’s instructions. The following primary antibodies were used: anti-Aβ (Covance, clone 6E10, Cat. # SIG-39320, 1:150), anti-Iba-1 (Abcam, polyclonal, Cat. # ab107159, 1:250) and anti-GFAP (Dako, polyclonal, Cat. # Z033429-2, 1:25). Secondary Alexa Fluor 488® (polyclonal, Cat. # 711-545-152, 1:150), Cy™3 (polyclonal, Cat. # 715-165-150, 1:150), and Cy™5 (polyclonal, Cat. # 705-175-147, 1:150)-conjugated antibodies (Jackson ImmunoResearch) were used. Slides were exposed to Hoechst nuclear staining (Invitrogen, 1:2000), before being sealed with Aqua-Poly/Mount (Polysciences) and glass covers.
Images were acquired using a PANNORAMIC™ Midi Slide Scanner (3DHISTECH), and quantitative analyses performed using ImageJ (NIH). For each animal, quantified values were averaged. Representative immunofluorescence images were equally optimized and processed for all experimental conditions displayed using Photoshop (Adobe).
Soluble Aβ42 ELISA
Hippocampal samples were weighed and homogenized, accordingly, in Tris Buffered Saline EDTA (TBSE) solution (50 mM Tris, 150 mM NaCl, and 2 mM EDTA, pH 7.4) with the addition of 1% protease inhibitor cocktail (1 mL/100 mg tissue, Sigma-Aldrich) using a microtube homogenizer with plastic pestles. The homogenates were then centrifuged for 40 min at 350,000 × g in 500 μL polycarbonate centrifuge tubes (Beckman Coulter) at 4 °C in an Optima MAX-XP Ultracentrifuge with a TLA 120.1 rotor (Beckman Coulter). The supernatant (TBSE-soluble fraction) was collected, aliquoted, and stored at −80 °C. BCA assay (Pierce BCA Protein Assay Kit) was performed to determine the total protein amount for normalization. To quantify Aβ42 peptides, the human Aβ42 Ultrasensitive ELISA Kit (Invitrogen) was used according to manufacturer’s instructions. Data were acquired using a Spark microplate reader (Tecan).
IL-1β ELISA
Cortical samples were weighed and homogenized with metal beads in a volume of Cell Lysis Buffer 2 (R&D Systems) according to their weight, using Qiagen’s TissueLyser II. To quantify IL-1β, the mouse IL-1β Quantikine™ ELISA kit was used according to manufacturer’s instructions. Data were acquired using a Spark microplate reader (Tecan).
Glucose quantification
The supernatant (TBSE-soluble fraction) from the processed hippocampal and cortical samples was further tested for glucose level using a Roche Cobas 111 analyzer according to manufacturer’s instructions.
Proteomic profiling
Mouse CSF samples (±10 μL) were added to ± 190 μL of an 8 M urea buffer and loaded onto 3 kDa cut-off spin columns (Waters). Volume was reduced to 25 μL by centrifugation at 14,000 × g for 10 min. Filters were reversed and centrifuged to extract the proteins. Proteins were reduced with 5 mM dithiothreitol (Sigma-Aldrich) for 1 h at RT and alkylated with 10 mM iodoacetamide (Sigma-Aldrich) in the dark for an additional 45 min at RT. Samples were diluted to 2 M urea with 50 mM ammonium bicarbonate. Proteins were then subjected to digestion with 250 ng trypsin (Promega) overnight at 37 °C at a 50:1 protein:trypsin ratio, followed by a second trypsin digestion (250 ng) for 4 h. The digestions were stopped by addition of trifluroacetic acid (1% final concentration). Following digestion, peptides were desalted using Oasis HLB (μElution format, Waters). The samples were vacuum-dried and stored at −80 °C until further analysis.
ULC/MS grade solvents were used for all chromatographic steps. Dry digested samples were dissolved in 97:3% H2O/acetonitrile + 0.1% formic acid. Each sample was loaded using split-less nano-Ultra Performance Liquid Chromatography (10 kpsi nanoAcquity, Waters). The mobile phase was as follows: A) H2O + 0.1% formic acid and B) acetonitrile + 0.1% formic acid. Desalting of the samples was performed online using a reversed-phase Symmetry C18 trapping column (180 µm internal diameter, 20 mm length, 5 µm particle size, Waters). The peptides were then separated using a T3 HSS nano-column (75 µm internal diameter, 250 mm length, 1.8 µm particle size, Waters) at 0.35 µl/min. Peptides were eluted from the column into the mass spectrometer using the following gradient: 4% to 30% B in 105 min, 30% to 90% B in 10 min, maintained at 90% for 7 min, and then back to initial conditions.
The nanoUPLC was coupled online through a nanoESI emitter (10 μm tip, New Objective) to a quadrupole orbitrap mass spectrometer (Orbitrap Exploris 480, Thermo Fisher Scientific) using a FlexIon nanospray apparatus (Proxeon). Data was acquired in data-dependent acquisition (DDA) mode, using a 2-sec cycle time method. MS1 resolution was set to 120,000 (at 200 m/z), mass range of 380–1500 m/z, normalized AGC of 200% and maximum injection time was set to 50 msec. MS2 resolution was set to 15,000, quadrupole isolation 1.4 m/z, normalized AGC of 75%, dynamic exclusion of 40 sec, and maximum injection time mode set to “Auto”.
Raw data were processed with MaxQuant v2.0.1.0105. The data were searched with the Andromeda search engine against the mouse (Mus musculus) protein database as downloaded from Uniprot (www.uniprot.com) and appended with common lab protein contaminants. Enzyme specificity was set to trypsin, and up to two missed cleavages were allowed. Fixed modification was set to carbamidomethylation of cysteines, and variable modifications were set to oxidation of methionines, asparagine and glutamine deamidation, and protein N-terminal acetylation. Peptide precursor ions were searched with a maximum mass deviation of 4.5 ppm and fragment ions with a maximum mass deviation of 20 ppm. Peptide and protein identifications were filtered at an FDR of 1% using the decoy database strategy (MaxQuant’s “Revert” module). The minimal peptide length was 7 amino acids, and the minimum Andromeda score for modified peptides was 40. Peptide identifications were propagated across samples using the match-between-runs option checked. Searches were performed with the label-free quantification option selected. The quantitative comparisons were calculated using Perseus v1.6.0.7. Decoy hits were filtered out, and only proteins that were detected in at least 50% of the samples of at least one experimental group were kept.
Bulk RNA-sequencing analysis
Hippocampi were processed as previously described106. llumina output files were demultiplexed with bcl2fastq v.2.17. Resulting FASTQ files were analyzed using the zUMIs pipeline107. Reads were aligned with STAR (v.2.5.3a) to the GRCm38 genome (release 84; Ensembl), and exonic unique molecular identifier (UMI) counts were used for downstream analysis. Data was further normalized by dividing each sample by its sum of UMIs, resulting in normalized expression levels corresponding to the gene’s fraction in the sample.
RNA purification, cDNA synthesis, and quantitative real-time PCR analysis
Cortices were homogenized in Tri Reagent (Bio-Lab). RNA was purified using a standard chloroform extraction. Two micrograms of total RNA were used to generate cDNA (High-Capacity cDNA Reverse Transcription kit, Applied Biosystems). The expression of specific mRNAs was assayed by fluorescence-based quantitative real-time PCR (RT-qPCR) using a Fast-SYBR PCR Master Mix (Applied Biosystems). Quantification reactions were performed in triplicate for each sample using the ∆∆Ct method with hypoxanthine-guanine phosphoribosyltransferase (hprt) serving as the reference housekeeping gene. At the end of the assay, a melting curve was constructed to verify the specificity of the reaction. To determine the expression level of some cytokines, cDNA was pre-amplified for 14 PCR cycles, according to manufacturer’s instructions (PreAmp Master Mix Kit, Applied Biosystems), thereby increasing the sensitivity of the subsequent real-time PCR reaction. The TaqMan Assays-on-Demand probes Mm00446968_m1 (hprt1), Mm01178820_m1 (tgfb1), Mm00446190_m1 (il6), and Mm00518984_m1 (il23) were used. For other genes examined, the following primers were used: hprt forward 5’-TCAGTCAACGGGGGACATAAA-3’ and reverse 5’-GGGGCTGTACTGCTTAACCAG-3’; il17ra forward 5’-AGTGTTTCCTCTACCCAGCAC-3’ and reverse 5’-GAAAACCGCCACCGCTTAC-3’; il1b forward 5’-GCAACTGTTCCTGAACTCAACT-3’ and reverse 5’-ATCTTTTGGGGTCCGTCAACT-3’. RT-qPCR reactions were performed and analyzed using StepOne Real-Time PCR system and software (Applied Biosystems).
Flow cytometry and sorting
Flow cytometry staining was performed as follows. Briefly, isolated human T cells were stained with monoclonal antibodies to anti-CD45 (BioLegend, clone HI30, Cat. # 304023, 1:200), anti-CD11b (BioLegend, clone ICRF44, Cat. # 301328, 1:200), anti-CD19 (BioLegend, clone HIB19, Cat. # 302237, 1:200), anti-CD3 (BioLegend, clone UCHT1, Cat. # 300459, 1:200), anti-CD4 (BioLegend, clone OKT4, Cat. # 317416, 1:200), anti-CD8 (BioLegend, clone SK1, Cat. # 344743, 1:200), anti-GLUT1 (R&D, clone 202915, Cat. # FAB1418P; Cat. # FAB1418F, 1:200), anti-CD38 (BioLegend, clone HIT2, Cat. # 303525, 1:200), anti-CD45RA (BioLegend, clone HI100, Cat. # 304125, 1:200), or anti-CD45RO (BioLegend, clone UCHL1, Cat. # 304227, 1:200) in BD HorizonTM Brilliant Stain buffer for 30 min at RT, in the presence of Human TruStain FcX™ (BioLegend, Cat. # 422302, 1:10) to prevent unspecific binding. Cells were then washed and stained with LIVE/DEAD™ Fixable Aqua Dead Cell Stain (Invitrogen, 1:1000) for 30 min.
Murine cells were stained with monoclonal antibodies to anti-CD31 (BioLegend, clone 390, Cat. # 102409, 1:200), anti-CD45 (BioLegend, clone 30F11, Cat. # 103132, 1:200), anti-CD11b (BioLegend, clone M1/70, Cat. # 101224, 1:200), anti-B220 (BioLegend, clone RA3-6B2, Cat. # 103244; Cat. # 103241, 1:200), anti-TCRγδ (BioLegend, clone GL3, Cat. # 118107, 1:200), anti-TCRβ (BioLegend, clone H57-597, Cat. # 109220, 1:200), anti-CD4 (BioLegend, clone GK1.5, Cat. # 100410, 1:200), anti-CD8a (BioLegend, clone 53-6.7, Cat. # 100706; Cat. # 100744, 1:200), anti-CD25 (BioLegend, clone PC61, Cat. # 102006, 1:200), anti-CD44 (BioLegend, clone IM7, Cat. # 103026, 1:200), anti-CD62L (BioLegend, clone MEL-14 Cat. # 104418, 1:200), anti-PD-1 (BioLegend, clone RMP1-30, Cat. # 109110, 1:200), anti-CD38 (BioLegend, clone 90, Cat. # 102730, 1:200), anti-CD39 (BioLegend, clone Duha59, Cat. # 143804, 1:200), anti-CD73 (R&D Systems, clone 496406, Cat. # FAB4488A, 1:200), anti-CCR6 (BioLegend, clone 29-2L17, Cat. # 129814, 1:200), anti-CD49b (BioLegend, clone DX5, Cat. # 108905, 1:200), or anti-LAG-3 (BioLegend, clone C9B7W, Cat. # 125227, 1:200) in BD HorizonTM Brilliant Stain buffer for 30 min at RT, in the presence of anti-CD16/32 TruStain FcX™ (BioLegend, clone 93, Cat. # 101320, 1:25) to prevent unspecific binding. Cells were then washed and stained with LIVE/DEAD™ Fixable Aqua Dead Cell Stain (Invitrogen, 1:1000) for 30 min.
For intracellular cytokine staining, the murine cells were stimulated for 5 h at 37 °C with PMA (500 ng/mL) plus ionomycin (500 ng/mL), in the presence of brefeldin A and monensin. Cells were then fixed in formaldehyde 4% (when the intracellular staining was performed the following day) or BD Fixation/Permeabilization solution (when the intracellular staining was performed on the same day) for 20 min at 4 °C. When the staining workflow was resumed the following day, cells were permeabilized with BD Perm/Wash buffer for 15 min prior to being stained for 30 min at RT with anti-IL-17A (BioLegend, clone TC11-18H10.1, Cat. # 506904, 1:100), anti-IL-22 (eBioscience, clone IL22JOP, Cat. # 17-7222-82, 1:100), anti-IFN-γ (BioLegend, clone XMG1.2, Cat. # 505824, 1:75), anti-IL-4 (BioLegend, clone 11B11, Cat. # 504127, 1:100), or anti-IL-10 (BioLegend, clone JES5-16E3, Cat. # 505022, 1:50) in BD Perm/Wash buffer with 2% normal donkey serum (Jackson ImmunoResearch).
For intracellular nuclear staining, the eBioscience Foxp3/Transcription Factor Staining Buffer set was used. Murine cells were simultaneously fixed and permeabilized in Foxp3 Fixation/Permeabilization working solution for 60 min at RT while protected from light. Cells were then incubated in Permeabilization buffer for 15 min prior to being stained for 30 min at RT with anti-RORγt (eBioscience, clone B2D, Cat. # 12-6981-82, 1:40), anti-FOXP3 (BioLegend, clone MF-14, Cat. # 126406, 1:100), anti-T-bet (eBioscience, clone eBio4B10 (4b10), Cat. # 50-5825-82, 1:20), or anti-GATA-3 (eBioscience, clone TWAJ, Cat. # 25-9966-42, 1:10) in Permeabilization buffer with 2% normal donkey serum. Samples were acquired in a CytoFLEX S cytometer (Beckman Coulter) and analyzed (Supplementary Fig. 10a–c) using FlowJo software (BD).
Glucose uptake
Glucose uptake was assessed using the fluorescent glucose analog 2-NBDG (Abcam) or 6-NBDG (Invitrogen). Cells were initially stained for T cell immune profiling. Following starvation for 15 min in glucose-free RPMI 1640 media (Gibco™) supplemented with 10% dialyzed FBS (Sigma-Aldrich), cells were incubated with 100 μM 2-NBDG or 6-NBDG for 30 min at 37 °C and washed. Samples were acquired in a CytoFLEX S cytometer (Beckman Coulter) and analyzed (Supplementary Fig. 10a–c) using FlowJo software (BD).
For human T cells, isolated CD4+ and CD8+ cells from pre-symptomatic APP carriers (females, n = 4; males, n = 3; average age, 33.14 years), were analyzed. For murine T cells, glucose uptake was assessed following 24 h incubation with either αCD38 or IgG control antibodies at a dose of 10 μg/mL.
Mitochondrial mass and membrane potential (∆ψm)
Mitochondrial mass and membrane potential were assessed using the fluorescent selective probes MitoTracker™ Green FM Dye (Invitrogen) and MitoTracker™ Red CMXRos Dye (Invitrogen), respectively. Cells were incubated in Maxpar® PBS with the reactive dye for 15–30 min at 37 °C, washed, and additionally stained for T cell immune profiling. Samples were acquired in a CytoFLEX S cytometer (Beckman Coulter) and analyzed (Supplementary Fig. 10a–c) using FlowJo software (BD).
For human T cells, isolated CD4+ and CD8+ cells from pre-symptomatic APP carriers (females, n = 4; males, n = 2; average age, 34.83 years), were analyzed. Mitochondrial membrane potential was assessed following 24 h activation with Dynabeads™ (1:1 cell-to-bead ratio, Gibco™). For murine T cells, mitochondrial mass and membrane potential were assessed following 24 h incubation with either αCD38 or IgG control antibodies at a dose of 10 μg/mL.
TH17 differentiation
Splenic naive CD4+ TCRβ+ CD25− CD44lo CD62L− cells (Supplementary Fig. 10d) from 2 to 3-month-old WT female mice were FACS-sorted with a SORP-FACSAria Cell Sorter (BD Biosciences), and 5 × 105 cells were activated with Dynabeads™ (1:1 cell-to-bead ratio, Gibco™). Naive CD4+ T cells were polarized into non-pathogenic (np) TH17 cells in the presence of recombinant human (rh) TGF-β1 (0.4 ng/mL), recombinant murine (rm) IL-6 (20 ng/mL), rhIL-23 (20 ng/mL), and antibodies to IFN-γ (polyclonal, 10 μg/mL) and IL-4 (polyclonal, 10 μg/mL). To differentiate naive CD4+ T cells into pathogenic (p) TH17 cells, rmIL-1β (50 ng/mL) was included in the cytokine cocktail. All cytokines and antibodies were purchased from PeproTech. Click’s media (Sigma-Aldrich) was supplemented with 10% FBS (Biological Industries), L-glutamine (2 mM), penicillin (100 U/mL, Biological Industries), streptomycin (0.1 mg/mL, Biological Industries) and β-mercaptoethanol (40 nM, Biological Industries). After 5 days of culture at 37 °C and 5% CO2, cells were harvested, stained, and acquired in a CytoFLEX S cytometer (Beckman Coulter). Samples were analyzed (Supplementary Fig. 10a–c) using FlowJo software (BD).
Statistical analysis
No statistical methods were used to predetermine sample size, which was chosen with adequate statistical power based on literature and past experience11,40,43,54,108. Exact sample sizes are indicated in the Figure legends. Statistical analyses were performed using GraphPad Prism (version 9.5.1, GraphPad Software), with statistical tests, comparisons, exact P values, and data exclusion criteria for all experiments provided in Supplementary Data 1. Experiments were repeated two to six times, unless otherwise stated. For pooled analysis of results from different independent experiments, all participants/mice from the same experimental group were pooled, and a new statistical comparison was made for the entire pooled experiment, as performed for the individual experiments. For comparisons between two groups, two-tailed unpaired and paired t tests or Mann–Whitney U tests were performed. One- or two-way ANOVA was used for comparisons between more than two groups with Holm-Šídák, Tukey, or Dunnett tests for correction of multiple comparisons. Non-parametric hypothesis testing methods were used for cases when the measured quantity could not be assumed to follow a normal distribution and failed to pass one or more normality tests. Multivariate hypothesis testing was done using permutational analysis of variance (PERMANOVA). Benjamini-Hochberg procedure was applied when the false discovery rate adjustment was applied. P values < 0.05 were considered significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The hippocampi bulk RNA-sequencing data generated in this study have been deposited to the NCBI database under accession code GSE290076. The CSF proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under accession code PXD060761. Source data are provided with this paper.
References
Bateman, R. J. et al. Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res Ther. 3, 1 (2011).
Nakamura, A. et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature 554, 249–254 (2018).
Mattsson-Carlgren, N. et al. Longitudinal plasma p-tau217 is increased in early stages of Alzheimer’s disease. Brain 143, 3234–3241 (2020).
Barthélemy, N. R. et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat. Med. 26, 398–407 (2020).
Preische, O. et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat. Med. 25, 277–283 (2019).
Ziv, Y. et al. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat. Neurosci. 9, 268–275 (2006).
Sulzer, D. et al. T cells from patients with Parkinson’s disease recognize α-synuclein peptides. Nature 546, 656–661 (2017).
Gate, D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020).
Campisi, L. et al. Clonally expanded CD8 T cells characterize amyotrophic lateral sclerosis-4. Nature 606, 945–952 (2022).
Gate, D. et al. CD4 + T cells contribute to neurodegeneration in Lewy body dementia. Science 374, 868–874 (2021).
Baruch, K. et al. Breaking immune tolerance by targeting Foxp3(+) regulatory T cells mitigates Alzheimer’s disease pathology. Nat. Commun. 6, 7967 (2015).
Lindestam Arlehamn, C. S. et al. Synuclein-specific T cell reactivity is associated with preclinical and early Parkinson’s disease. Nat. Commun. 11, 1875 (2020).
Castellani, G., Croese, T., Peralta Ramos, J. M. & Schwartz, M. Transforming the understanding of brain immunity. Science 380, eabo7649 (2023).
Rustenhoven, J. & Kipnis, J. Brain borders at the central stage of neuroimmunology. Nature 612, 417–429 (2022).
Salvador, A. F., de Lima, K. A. & Kipnis, J. Neuromodulation by the immune system: a focus on cytokines. Nat. Rev. Immunol. 21, 526–541 (2021).
Zenaro, E. et al. Neutrophils promote Alzheimer’s disease-like pathology and cognitive decline via LFA-1 integrin. Nat. Med. 21, 880–886 (2015).
Brigas, H. C. et al. IL-17 triggers the onset of cognitive and synaptic deficits in early stages of Alzheimer’s disease. Cell Rep. 36, 109574 (2021).
Rosenzweig, N. et al. Sex-dependent APOE4 neutrophil-microglia interactions drive cognitive impairment in Alzheimer’s disease. Nat. Med. 30, 2990–3003 (2024).
Karmaus, P. W. F. et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 565, 101–105 (2019).
Wu, L. et al. Niche-selective inhibition of pathogenic Th17 cells by targeting metabolic redundancy. Cell 182, 641–654.e20 (2020).
Hochrein, S. M. et al. The glucose transporter GLUT3 controls T helper 17 cell responses through glycolytic-epigenetic reprogramming. Cell Metab. 34, 516–532.e11 (2022).
Chatterjee, S. et al. CD38-NAD+axis regulates immunotherapeutic anti-tumor T cell response. Cell Metab. 27, 85–100.e8 (2018).
Mrdjen, D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395.e6 (2018).
Kalfon, L. et al. Familial early-onset Alzheimer’s caused by novel genetic variant and APP duplication: a cross-sectional study | Bentham Science. Current Alzheimer Research.
Hartmann, F. J. et al. Comprehensive immune monitoring of clinical trials to advance human immunotherapy. Cell Rep. 28, 819–831.e4 (2019).
Lu, Y. et al. Systematic analysis of cell-to-cell expression variation of T lymphocytes in a human cohort identifies aging and genetic associations. Immunity 45, 1162–1175 (2016).
Membrez, M. et al. Trigonelline is an NAD+ precursor that improves muscle function during ageing and is reduced in human sarcopenia. Nat. Metab. 6, 433–447 (2024).
Camacho-Pereira, J. et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 23, 1127–1139 (2016).
Tarragó, M. G. et al. A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD+ decline. Cell Metab. 27, 1081–1095.e10 (2018).
Hartmann, F. J. et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat. Biotechnol. 39, 186–197 (2021).
Macintyre, A. N. et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 20, 61–72 (2014).
Fu, H. et al. The glucose transporter 2 regulates CD8 + T cell function via environment sensing. Nat. Metab. 5, 1969–1985 (2023).
Qiu, J. et al. Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074.e5 (2019).
Barros, L. F. et al. Kinetic validation of 6-NBDG as a probe for the glucose transporter GLUT1 in astrocytes. J. Neurochem. 109, 94–100 (2009).
Mor, U. et al. Dimensionality reduction of longitudinal’omics data using modern tensor factorizations. PLoS Comput. Biol. 18, e1010212 (2022).
Wang, Y. et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015).
Griciuc, A. et al. TREM2 acts downstream of CD33 in modulating microglial pathology in alzheimer’s disease. Neuron 103, 820–835.e7 (2019).
Winkler, E. A. et al. GLUT1 reductions exacerbate Alzheimer’s disease vasculo-neuronal dysfunction and degeneration. Nat. Neurosci. 18, 521–530 (2015).
Clarke, J. R. et al. Alzheimer-associated Aβ oligomers impact the central nervous system to induce peripheral metabolic deregulation. EMBO Mol. Med. 7, 190–210 (2015).
Rosenzweig, N. et al. PD-1/PD-L1 checkpoint blockade harnesses monocyte-derived macrophages to combat cognitive impairment in a tauopathy mouse model. Nat. Commun. 10, 465 (2019).
Heneka, M. T. et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678 (2013).
Venegas, C. et al. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature 552, 355–361 (2017).
Dvir-Szternfeld, R. et al. Alzheimer’s disease modification mediated by bone marrow-derived macrophages via a TREM2-independent pathway in mouse model of amyloidosis. Nat. Aging 2, 60–73 (2022).
Ulland, T. K. et al. TREM2 maintains microglial metabolic fitness in alzheimer’s disease. Cell 170, 649–663.e13 (2017).
Pascual-Lucas, M. et al. Insulin-like growth factor 2 reverses memory and synaptic deficits in APP transgenic mice. EMBO Mol. Med. 6, 1246–1262 (2014).
Derecki, N. C. et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J. Exp. Med. 207, 1067–1080 (2010).
Alves de Lima, K. et al. Meningeal γδ T cells regulate anxiety-like behavior via IL-17a signaling in neurons. Nat. Immunol. 21, 1421–1429 (2020).
Filiano, A. J. et al. Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature 535, 425–429 (2016).
Ribeiro, M. et al. Meningeal γδ T cell-derived IL-17 controls synaptic plasticity and short-term memory. Sci. Immunol. 4, eaay5199 (2019).
Tüshaus, J. et al. An optimized quantitative proteomics method establishes the cell type-resolved mouse brain secretome. EMBO J. 39, e105693 (2020).
Rustenhoven, J. et al. Age-related alterations in meningeal immunity drive impaired CNS lymphatic drainage. J. Exp. Med. 220, e20221929 (2023).
Fourgeaud, L. et al. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016).
Gaikwad, S. et al. Tau oligomer induced HMGB1 release contributes to cellular senescence and neuropathology linked to Alzheimer’s disease and frontotemporal dementia. Cell Rep. 36, 109419 (2021).
Baruch, K. et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat. Med. 22, 135–137 (2016).
Ivanov, I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Atarashi, K. et al. ATP drives lamina propria T(H)17 cell differentiation. Nature 455, 808–812 (2008).
Mezö, C. et al. Different effects of constitutive and induced microbiota modulation on microglia in a mouse model of Alzheimer’s disease. Acta Neuropathol. Commun. 8, 119 (2020).
Fitzpatrick, Z. et al. Gut-educated IgA plasma cells defend the meningeal venous sinuses. Nature 587, 472–476 (2020).
Di Marco Barros, R., Fitzpatrick, Z. & Clatworthy, M. R. The gut-meningeal immune axis: priming brain defense against the most likely invaders. J. Exp. Med. 219, (2022).
Chini, C. C. S. et al. CD38 ecto-enzyme in immune cells is induced during aging and regulates NAD+ and NMN levels. Nat. Metab. 2, 1284–1304 (2020).
Covarrubias, A. J. et al. Senescent cells promote tissue NAD+ decline during ageing via the activation of CD38+ macrophages. Nat. Metab. 2, 1265–1283 (2020).
Blacher, E. et al. Alzheimer’s disease pathology is attenuated in a CD38-deficient mouse model. Ann. Neurol. 78, 88–103 (2015).
Hou, Y. et al. NAD+ supplementation normalizes key Alzheimer’s features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl Acad. Sci. USA 115, E1876–E1885 (2018).
Hou, Y. et al. NAD+ supplementation reduces neuroinflammation and cell senescence in a transgenic mouse model of Alzheimer’s disease via cGAS-STING. Proc Natl Acad Sci USA 118, e2011226118 (2021).
Ferretti, M. T. et al. Sex differences in Alzheimer disease - the gateway to precision medicine. Nat. Rev. Neurol. 14, 457–469 (2018).
Mittelbrunn, M. & Kroemer, G. Hallmarks of T cell aging. Nat. Immunol. 22, 687–698 (2021).
Farid, M. M., Yang, X., Kuboyama, T. & Tohda, C. Trigonelline recovers memory function in Alzheimer’s disease model mice: evidence of brain penetration and target molecule. Sci. Rep. 10, 16424 (2020).
Boison, D. & Yegutkin, G. G. Adenosine metabolism: emerging concepts for cancer therapy. Cancer Cell 36, 582–596 (2019).
Chen, X. et al. Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy. Nature 615, 668–677 (2023).
Oblak, A. L. et al. Comprehensive evaluation of the 5XFAD mouse model for preclinical testing applications: a MODEL-AD study. Front. Aging Neurosci. 13, 713726 (2021).
Jack, C. R. et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 12, 207–216 (2013).
Ou, Y.-N. et al. FDG-PET as an independent biomarker for Alzheimer’s biological diagnosis: a longitudinal study. Alzheimers Res. Ther. 11, 57 (2019).
Dror, E. et al. Postprandial macrophage-derived IL-1β stimulates insulin, and both synergistically promote glucose disposal and inflammation. Nat. Immunol. 18, 283–292 (2017).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Douglas, A., Stevens, B. & Lynch, L. Interleukin-17 as a key player in neuroimmunometabolism. Nat. Metab. 5, 1088–1100 (2023).
Schnell, A. et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell 184, 6281–6298.e23 (2021).
Faraco, G. et al. Dietary salt promotes neurovascular and cognitive dysfunction through a gut-initiated TH17 response. Nat. Neurosci. 21, 240–249 (2018).
Guo, X. et al. Chiral nanoparticle-remodeled gut microbiota alleviates neurodegeneration via the gut-brain axis. Nat. Aging https://doi.org/10.1038/s43587-023-00516-9 (2023).
Huber, S. et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3− and Foxp3+ regulatory CD4 + T cells in an interleukin-10-dependent manner. Immunity 34, 554–565 (2011).
Damasceno, L. E. A. et al. STING is an intrinsic checkpoint inhibitor that restrains the TH17 cell pathogenic program. Cell Rep. 39, 110838 (2022).
Wang, C. et al. CD5L/AIM regulates lipid biosynthesis and restrains th17 cell pathogenicity. Cell 163, 1413–1427 (2015).
Wagner, A. et al. Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell 184, 4168–4185.e21 (2021).
Gaublomme, J. T. et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163, 1400–1412 (2015).
Sie, C. et al. IL-24 intrinsically regulates Th17 cell pathogenicity in mice. J. Exp. Med. 219, e20212443 (2022).
Gerriets, V. A. et al. Metabolic programming and PDHK1 control CD4. + T cell Subsets Inflamm. J. Clin. Invest. 125, 194–207 (2015).
Gao, Y. et al. Transcription factor EGR2 controls homing and pathogenicity of TH17 cells in the central nervous system. Nat. Immunol. 24, 1331–1344 (2023).
Endo, Y. et al. 1-Oleoyl-lysophosphatidylethanolamine stimulates RORγt activity in TH17 cells. Sci. Immunol. 8, eadd4346 (2023).
Chen, L. et al. CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov. 8, 1156–1175 (2018).
Carter, P. J. & Rajpal, A. Designing antibodies as therapeutics. Cell 185, 2789–2805 (2022).
Malitsky, S. et al. Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol. N. Phytol. 210, 88–96 (2016).
Zheng, L. et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun. 6, 6001 (2015).
Takahashi, C. et al. Mass cytometry panel optimization through the designed distribution of signal interference. Cytom. A 91, 39–47 (2017).
Iyer, A., Hamers, A. A. J. & Pillai, A. B. Cytof® for the masses. Front. Immunol. 13, 815828 (2022).
Kotecha, N., Krutzik, P. O. & Irish, J. M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. Chapter 10, Unit10.17 (2010).
Bagwell, C. B. et al. Automated data cleanup for mass cytometry. Cytom. A 97, 184–198 (2020).
Zunder, E. R. et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333 (2015).
Xia, J. & Wishart, D. S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 6, 743–760 (2011).
Pang, Z. et al. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022).
Hartmann, F. J. et al. High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J. Exp. Med. 213, 2621–2633 (2016).
Oakley, H. et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Sasmita, A. O. et al. Parental origin of transgene modulates amyloid-β plaque burden in the 5xFAD mouse model of Alzheimer’s disease. Neuron https://doi.org/10.1016/j.neuron.2024.12.025 (2025).
Scheyltjens, I. et al. Single-cell RNA and protein profiling of immune cells from the mouse brain and its border tissues. Nat. Protoc. 17, 2354–2388 (2022).
Hsiao, E. Y., McBride, S. W., Chow, J., Mazmanian, S. K. & Patterson, P. H. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc. Natl Acad. Sci. USA 109, 12776–12781 (2012).
Oliveira, A. M. M., Hemstedt, T. J. & Bading, H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 15, 1111–1113 (2012).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Ben-Moshe, S. et al. The spatiotemporal program of zonal liver regeneration following acute injury. Cell Stem Cell 29, 973–989.e10 (2022).
Parekh, S., Ziegenhain, C., Vieth, B., Enard, W. & Hellmann, I. zUMIs - a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7, giy059 (2018).
Tsitsou-Kampeli, A. et al. Cholesterol 24-hydroxylase at the choroid plexus contributes to brain immune homeostasis. Cell Rep. Med. 4, 101278 (2023).
Acknowledgements
We thank the members of the Schwartz lab and Weizmann Institute of Science for insightful discussions, and apologize to colleagues whose work was not included due to space constraints. We especially thank R. Valdés-Mas for providing critical feedback on bioinformatic analyses, C. Raanan, I. Biton, O. Rosenberg, N. Stettner, T. Mehlman, and A. Brandis for technical support, and S. Schwarzbaum for proofreading the manuscript. J.M.P.R. and G.C. received a Senior Postdoctoral Fellowship from the Weizmann Institute of Science. M.A.A. received a postdoctoral fellowship from Fundación Ramón Areces. The work was supported by an Advanced European Research Council grant (N° 741744), the Israel Science Foundation (ISF, grant No. 991/16), the ISF-Legacy Heritage Biomedical Science Partnership (grant No. 135 4/15), the Thompson Foundation and the Adelis Foundation (given to M.S.).
Author information
Authors and Affiliations
Contributions
J.M.P.R., G.C., D.K., and T.C. equally contributed to the study. J.M.P.R. conceived the study, designed, performed, and interpreted the experiments, prepared the figures, and wrote the manuscript. G.C., D. K., and T. C. designed, performed, and interpreted the experiments and reviewed the manuscript. D. K. created the artwork. A.T.K. performed the open field and novel object recognition tests. C.B. performed the quantitative real-time PCR analyses and aided with in vivo experiments. M.A.A. helped with in vivo experiments and reviewed the manuscript. L.C. assisted with the soluble Aβ42 ELISA. S.P.C. performed the immunohistochemistry and assisted with in vivo experiments. T.M.S. aided with FACS sorting, mass, and flow cytometry experiments. Y.K. assisted with the metabolic cages’ experiments. A.S. performed the proteomic profiling. M.I. and S.M. performed the metabolomic profiling. S.O. oversaw mice breeding and care and coordinated key animal experimentation. S.F. and L.R. provided support with bioinformatic analyses. L.K., S.K., N.S., R.P., J.A.P., and T.C.F.Z. were involved in the collection of human samples and clinical characterization of the patients. R.F. performed NfL determination and provided insights on the project. M.S. conceived the study, interpreted the experiments, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.S. is co-founder and CSO of ImmunoBrain. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peralta Ramos, J.M., Castellani, G., Kviatcovsky, D. et al. Targeting CD38 immunometabolic checkpoint improves metabolic fitness and cognition in a mouse model of Alzheimer’s disease. Nat Commun 16, 3736 (2025). https://doi.org/10.1038/s41467-025-58494-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58494-y