Abstract
The rapid accumulation of biomedical cohort data presents opportunities to explore disease mechanisms, risk factors, and prognostic markers. However, current research often has a narrow focus, limiting the exploration of risk factors and inter-disease correlations. Additionally, fragmented processes and time constraints can hinder comprehensive analysis of the disease landscape. Our work addresses these challenges by integrating multimodal data from the UK Biobank, including basic, lifestyle, measurement, environment, genetic, and imaging data. We propose UKB-MDRMF, a comprehensive framework for predicting and assessing health risks across 1560 diseases. Unlike single disease models, UKB-MDRMF incorporates multimorbidity mechanisms, resulting in superior predictive accuracy, with all disease types showing improved performance in risk assessment. By jointly predicting and assessing multiple diseases, UKB-MDRMF uncovers shared and distinctive connections among risk factors and diseases, offering a broader perspective on health and multimorbidity mechanisms.
Similar content being viewed by others
Introduction
The consistently increased accumulation of biomedical cohort data1,2 offers significant opportunities for robust and comprehensive modeling in clinical diagnosis and disease analysis3,4,5,6,7. However, many studies remain focused on predicting or assessing risks for specific diseases or broad disease categories3, which limits the exploration of risk factors and inter-disease correlations4. Additionally, the exploration process is often fragmented, lacking a standardized framework for integrating multiple tasks, leading to low data utilization and resource wastage8. Furthermore, time and cost constraints for data preprocessing and model implementation pose barriers to healthcare professionals9.
The health status of individuals is influenced by various factors such as demographics, lifestyle, environment, and genetics10. Leveraging this rich information can lead to more accurate predictions and deeper analyses11. While numerous methods have been developed to predict and assess single-type diseases using multimodal data3, the coverage of diseases remains insufficient. There has been a lack of research integrating multidimensional health information with various disease types in large-scale biomedical datasets. Some approaches model a large number of diseases simultaneously12, but the multitude of disease risk factors and complex connections between diseases pose significant challenges, making it difficult to comprehensively explore these relationships. Existing methods for constructing health information models may lack thorough consideration from data preprocessing through analysis and modeling stages, and they often lack further analysis and application extensions13.
In this work, we integrated rich multimodal data from the UK Biobank data1, encompassing basic information, lifestyle, measurements, environmental factors, genetics, and imaging data. We introduced UKB-MDRMF, a Multi-Disease Risk and Multimorbidity Framework designed for individual multi-disease prediction and health risk assessment across 1560 diseases. UKB-MDRMF is a comprehensive biomedical prediction and risk assessment model applied at a large scale. This comprehensive model outperforms single-category disease risk assessments and effectively uncovers potential connections among multiple risk factors and diseases, providing a broader perspective on health risk factors and multimorbidity mechanisms. Additionally, we streamlined the entire process from data input to model construction, enhancing the flexibility and convenience of using the UKB-MDRMF framework across various downstream tasks. We also developed an interactive platform (https://luminite.shinyapps.io/ukb-mdrmf/) to showcase detailed results of UKB-MDRMF, allowing exploration of disease predictions, variable importance, and comorbidities for specific risk factors and disease categories.
Results
Pipeline for the framework
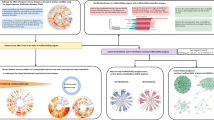
For the UK Biobank data, we developed a comprehensive standardized process to construct the UKB-MDRMF framework for the joint prediction and risk assessment of multiple diseases. The entire workflow encompasses data preprocessing, model construction, and applications, as shown in Fig. 1.
This pipeline utilizes input data from the diverse UK Biobank data, including six categories: basic, lifestyle, measurement, environment, genetic, and imaging data. Following field selection, data cleaning, and missing data preprocessing, predictors are generated. Response variables are derived from inpatient, self-reported, and primary care data, initially standardized to ICD-10 codes before conversion to Phecodes. After the temporal alignment of independent and dependent variables, the data is used to construct the UKB-MDRMF framework, encompassing disease prediction and risk assessment models. These models facilitate diverse applications, including establishing baseline conditions for multiple diseases, analyzing significant risk factors, exploring multimorbidity, and assessing survival risks. Icons are provided by Icons8 (https://icons8.com).
In the data processing phase, we integrated data from six categories: basic (e.g., demographics), lifestyle (e.g., activity, diet), measurement (e.g., biochemistry, blood tests), environment (e.g., social and natural environmental factors), genetic (e.g., genetic principal components and polygenic risk scores), and imaging (e.g., heart MRI, brain MRI). This integration forms the initial input for the workflow. We manually integrated and screened variables based on their meanings in each data type, referencing the collection of phenotypic information from genome-wide association studies (GWAS)14. The selected variables were categorized into three hierarchical levels (essential, detailed, and minor) based on the specific information they provide (see Supplementary Note 2.2, Supplementary Tables 1–3, and Supplementary Fig. 3 for more details). Next, we conducted data cleaning and imputation, integrating information from three sources (inpatient, self-report, and primary care data) into response variables. The data was randomly divided into training, validation, and test sets in an 8:1:1 ratio. To ensure consistency and independence among these sets, we partitioned the data randomly before data preprocessing. During preprocessing, the treatment of the training, validation, and test sets was entirely independent, eliminating the risk of data leakage between datasets. Following time alignment, we developed a modeling strategy for individual joint disease prediction and risk assessment that accounts for the complex interplay between different disease predictors and risk factors. The UKB-MDRMF framework serves as a basis for comprehensive analyses and investigating relationships between diverse health indicators and disease outcomes in the UK Biobank data. It can be used for establishing baseline predictions for specific diseases, exploring health risk factors, and conducting multimorbidity analyses. For detailed information about each step in the pipeline, refer to Section “Methods”.
UKB-MDRMF offers an efficient and flexible solution for leveraging large-scale biomedical data for analysis and modeling. It surpasses single-disease models by jointly predicting and assessing thousands of diseases (Phecodes), capturing potential multimorbidity mechanisms among numerous diseases and resulting in superior predictive performance. Additionally, we have developed an interactive platform for visualization and exploration, as detailed in Supplementary Note 1 and Supplementary Fig. 1.
Data preprocessing
To construct the multi-disease risk framework, we separately built and processed predictors and response variables. The first step in processing predictors was field screening. Including all variables in the model introduces noise and affects accuracy. Therefore, from 7228 phenotypes collected in GWAS, we subjectively selected 542 phenotypes based on their relevance and importance14, grouping them into six categories. The UK Biobank data, a large-scale biomedical data, includes diverse data types with unique encoding methods. We employed customized data cleaning and organization strategies to accommodate these diverse formats. Different encoding techniques were applied to enhance data quality and optimize model performance: continuous and integer variables were standardized or binned, and categorical variables were converted into binary or ordered categorical variables. As a result, we obtained 1106 input variables, with several phenotypes generating multiple variables during preprocessing.
Large biomedical data often contain substantial amounts of missing information, and the UK Biobank is no exception. Following data cleaning, we examined the data collection procedures and systematically analyzed missing patterns. The UK Biobank provides details on data collection, including the participation subgroup for each variable and the time and location of data collection for each participant. By considering this detailed information, we understood the mechanisms behind missingness and handled missing values appropriately. We categorized missing variables into different types and filled in missing information using strategies such as deletion of low-quality individuals, imputation with special values, incorporation of missingness indicators, and imputation with models. High missing rates for many variables were due to the systematic design of data collection procedures. By leveraging the knowledge of the data collection process, we achieved accurate and interpretable imputation results, facilitating the UKB-MDRMF construction.
For response variables, we used 1560 Phecodes to systematically summarize diseases15. The corresponding data originated from four sources: hospital inpatient, death register, self-report, and primary care data. These data were encoded separately and standardized as ICD-10 codes. After integration, the standardized codes were mapped to Phecodes using Phecode Map 1.2 with ICD-10 Codes15 to serve as the final response variables.
Since our goal is disease prediction rather than imputation, it is crucial to ensure that responses occur after predictors. Therefore, after separately preprocessing the two major data parts, predictors and response variables, we performed time alignment before inputting them into the model, enhancing the study’s significance.
Model construction
To develop the UKB-MDRMF framework, we focused on two primary aspects: individual disease prediction and risk assessment. Here, the information from different disease types was not used as input variables in the model.
In the disease prediction task, we evaluated various approaches, including Logistic Regression16, Random Forest17, XGBoost18, LightGBM19, FCNN20, and specialized models such as POPDx12 and CATI21. A comprehensive comparative analysis was conducted to assess their performance across different disease predictions, using the AUC (area under the receiver operating characteristic curve) as the evaluation metric. The comparison between the training and testing sets is shown in Supplementary Fig. 15.
We employed a stepwise approach for predictor types, starting with basic information variables and gradually incorporating different feature types, ultimately including genetic variables. Figure 2a illustrates the performance of different disease prediction models across various data categories. The FCNN model performed best, achieving an overall median AUC exceeding 0.7 after the addition of measurement data. Traditional non-parametric machine learning methods such as Random Forest, XGBoost, and LightGBM exhibited moderate performance in multi-task learning, while methods like POPDx and CATI, which incorporate multi-disease semantic information, did not significantly improve predictive accuracy. Moreover, with more feature information for prediction, the predictive accuracy of all models consistently improved. Figure 2b shows the performance of disease prediction models considering varying numbers of positive individuals in the training set. For rare diseases (with positive individuals ranging from 0 to 200, where the occurrence rate is typically defined as 1 in 2000)22,23, we used 200 cases as the threshold, given the training set of 400,000 data points. CATI and FCNN achieved higher AUCs for rare diseases, while for common diseases, CATI, FCNN, and Logistic Regression all performed well. Figure 2c compares the performance of two different modeling strategies: individual disease modeling versus joint modeling for all diseases, where the latter directly outputs predictions for all Phecodes. For disease prediction tasks, the AUC in joint prediction across all disease categories is comparable to single-disease prediction. Focusing on the predictive accuracy of various types of diseases reveals that oral and intestinal diseases have relatively lower AUC values. In contrast, diseases related to the reproductive system demonstrate notably high predictive accuracy, with the AUC exceeding 0.95 for pregnancy-related diseases and 0.8 for genital diseases.
Model performance in disease prediction (a–c) and risk assessment (d–f) on the test set. a Performance of disease prediction models across different data categories. The prediction process initiates with basic information and gradually integrates additional categories. Seven machine learning and deep learning methods are compared. b The box plot illustrates model performance on the testing set with different numbers of positive patients in the training set (horizontal axis). c Disease prediction performance of the FCNN model using six data categories. Individual FCNNs were trained for each disease type and compared with FCNNs trained collectively for all Phecodes. The numerical values above each box plot represent the p values from two-sided Wilcoxon tests in each disease type, and no multiple comparison correction was applied. d Performance of risk assessment models (survival models). Testing set C-index comparisons across four models are used to assess risk assessment model performance, considering various input data categories. e Model performance on the testing set under different numbers of positive patients in the training set (horizontal axis). f Risk assessment performance of the DeepSurv model across 21 disease types. Similar to (c), the numerical values above each box plot represent the p values from two-sided Wilcoxon tests in each disease type, and no multiple comparison correction was applied. Box plots depict the median (central line), interquartile range (box), and whiskers extending to the minimum and maximum values, excluding outliers—defined as points beyond 1.5× the interquartile range from the first and third quartiles.
Similar to disease prediction, the risk assessment process followed a stepwise addition strategy for each category of predictors. We used traditional Cox proportional hazards (CoxPH) models24, DeepSurv25, and advanced models such as POPDxSurv12 and CATISurv21. Performance was evaluated using the C-index26. As shown in Fig. 2d, among the four survival models, DeepSurv outperformed others, followed by CATISurv, with their median C-index around or above 0.7. In comparison, POPDxSurv showed relatively poor performance, requiring all six data categories as input to achieve better predictive results. Figure 2e presents the predictive capabilities of different models for both rare and common diseases in risk assessment. Accuracy for the CoxPH model improved with more positive cases, and deep learning methods consistently outperformed across both rare and common diseases, especially DeepSurv. Similar to Fig. 2c, f shows the DeepSurv model’s risk assessment performance across 21 disease types in a survival analysis setting. In this scenario, joint predictions consistently yield higher C-index values for all disease types, significantly outperforming single-disease models at a significance level of 0.05. Consistent with the results from disease prediction models, the highest-performing disease types include pregnancy-related conditions, genital disorders, mental disorders, and lung diseases. For additional comparisons of disease prediction and risk assessment between single-disease models and joint disease predictions for high-prevalence diseases, refer to Supplementary Note 4.3.3 and Supplementary Figs. 13, 14.
For disease prediction and risk assessment tasks, we further investigate the incremental improvement in model performance for different types of diseases by gradually adding various data categories (basic information, lifestyle, measurement, environment, genetic, and imaging data). Figure 3 shows that for almost all disease types, both disease prediction and survival models improved with more data categories. After adding each data category, we calculated the p-value for the difference in the distribution of test set prediction metrics before and after adding this category. This allowed us to determine the significance of the improvement in prediction metrics brought by adding different categories of data, as shown in Supplementary Tables 6 and 7. Significant improvements were achieved after adding lifestyle, measurement, and imaging variables. For the disease prediction task, lifestyle and imaging variables contributed to significant performance improvements in 90.5% and 100% of disease types, with an average AUC increase of 2.3% and 2.4%, respectively. In contrast, environmental and genetic variables yielded fewer improvements. A significant improvement was achieved by adding lifestyle variables for digestive and nervous diseases, whereas for reproductive diseases, highly accurate predictions were obtained using only basic information.
The accuracy of disease prediction and survival modeling for each disease type gradually adding data categories. Med. AUC represents the median AUC of the best-performing disease prediction model, FCNN, for each disease type, using only basic information for prediction. Med. C-Index represents the median C-Index of the best-performing survival model, DeepSurv, for each disease type, using only basic information for survival modeling. All points in the plot represent the median values of the corresponding metrics, with the ends of the lines indicating the 25th and 75th percentiles of disease performance. The number of valid diseases in each category is recorded in parentheses following the disease type name. Additionally, models using image data show slight differences in the number of valid diseases due to variations in truncation times.
Baseline model for multi-disease prediction and risk assessment
UKB-MDRMF provided a baseline for multi-disease prediction and risk assessment. We conducted comparative analyses between our UKB-MDRMF and several existing methods, which were fine-tuned for specific medical applications via transfer learning. The results are summarized in Table 1. UKB-MDRMF, leveraging diverse data types, demonstrated superior predictive performance for specific diseases compared to other methods. For disease prediction, we employed the FCNN model, while the DeepSurv model was used for risk assessment. The comparative results for other methods were based on the best-performing strategies reported in their respective studies. Additional comparisons are provided in Supplementary Note 4.3.5 and Supplementary Table 8. Since some studies use ICD codes while others use Phecodes, the correspondence can be found in Supplementary Data 4.
Analysis of important risk factors
Our UKB-MDRMF utilized interpretable tools, such as SHAP (SHapley Additive exPlanations)27, to analyze risk factors and evaluate the impact of features on various diseases.
Figure 4 displays SHAP-based results. Figure 4a shows the normalized proportion of the top 30 significant risk variables across six categories for 21 diseases. Basic information was most predictive for pregnancy, vessel, and genital diseases. Lifestyle variables were significant for pregnancy and cortex neural diseases, while measurement variables impacted genital diseases. Environmental variables affected upper respiratory diseases, image variables influenced heart, blood, and urinary diseases, and genetic variables had a relatively uniform impact across several diseases, such as heart and eye diseases. Figure 4b–d highlight specific variable importance across different data categories. Figure 4b reveals that “Bipolar and major depression status” and “Age at recruitment” were consistently top factors. “Carotid ultrasound”, “Length of longest manic/irritable episode”, and “Body fat percentage” were also significant. Figure 4c, d illustrates the overall positive and negative effects of certain variables on disease outcomes. The absence of “Bipolar or Depression” reduced the incidence of all diseases. Conversely, increases in “Waist circumference”, “Body mass index”, “Cholesterol”, “Seated height”, and “Microalbumin in urine” promoted the occurrence of various diseases. Figure 4d indicated that a higher “Age at recruitment” significantly increased the risk of pregnancy-related diseases. To highlight the similarities and differences in importance between disease prediction and risk assessment models, we compared the top ten variables of the best-performing FCNN and DeepSurv models in Fig. 4e. Similarities were observed in the importance indicators from prediction and survival models, such as imaging-related, depression-related, fat-related, and age-related variables. Further details on the importance of risk assessment survival models are available in Supplementary Fig. 16.
a Normalized proportion of the top 30 significant risk factors among six categories of independent variables for each of the 21 disease types. b Frequencies for the top 5, top 10, and top 20 important variables in each of the 21 types of diseases. c Distribution of variable importance for all Phecodes, with colors ranging from blue (negative effect) to red (positive effect). d Average importance values for each variable category and disease type, with blue indicating negative effects and red indicating positive effects. e Comparison of risk factor importance for disease prediction (left, from FCNN) and risk assessment (right, from DeepSurv). Diseases were aggregated into nine major types derived from the 21 disease types. Thicker lines indicate greater importance. Icons provided by Icons8 (https://icons8.com).
Multimorbidity and trend analysis of disease risks
By exploring the capabilities of UKB-MDRMF, we harnessed the advantages of multi-disease joint prediction and assessment to analyze disease multimorbidity and trends in disease risk. Using a data-driven approach, we identified the best-performing neural network models. We extracted the weight matrices connecting the penultimate layer to the output layer, generating a corresponding vector for each output disease Phecode. The distance between vectors delineated the correlation between different diseases, effectively capturing multimorbidity patterns and revealing how certain diseases may co-occur or influence each other within the context of the model. Figure 5 illustrates the multimorbidity mechanisms and age-related risk trends across multiple diseases. In Fig. 5a, the projection of final layer features obtained by training with different network structures (FCNN and DeepSurv) characterized multimorbidity patterns of various diseases. Strong clustering effects in both sub-figures indicated consistency, notable in the six clusters highlighted in Fig. 5a. Figure 5b corresponds to the clustering within the two red circles in Fig. 5a. For projections across all categories, refer to Supplementary Fig. 17.
a Two-dimensional projection of the disease prediction model’s multimorbidity mechanisms using t-SNE, where each point represents a predicted Phecode, and each color represents a major disease type. Closer points indicate similar disease patterns. The six circles in the figure delineate specific multimorbidity patterns. b Multimorbidity patterns of selected clusters' internal Phecodes. The size of each data point indicates the number of affected individuals, with larger points representing higher frequencies of occurrence. The thickness of the lines represents the frequency of comorbidity, with thicker lines indicating higher frequencies. c Risk profiles of nine major disease types estimated by the DeepSurv model across different age groups. The size of the circles indicates the cumulative number of affected individuals, with larger circles representing higher numbers. The shading represents the magnitude of risk, with darker shades indicating higher risk levels. Icons provided by Icons8 (https://icons8.com).
The network on the left was predominantly composed of purple-blue dots representing genital diseases, along with bladder neck obstruction from the urinary category and testicular-related diseases from the endocrine category in Fig. 5b. The network on the right displayed the inherent associative disease mechanisms of mental illnesses. The thicker line connecting anxiety disorder with mood disorders, major depressive disorder, and bipolar indicated a higher frequency of their co-occurrence in practice. Figure 5c demonstrated the hazard function extraction for DeepSurv, revealing the incidence of different diseases across various ages. With increasing age, digestive and circulatory diseases darkened significantly, suggesting a faster increase in risk, while the risk of reproductive diseases remained almost unchanged with age. For detailed information on the clusters highlighted within the remaining four black circles in Fig. 5a, refer to Supplementary Fig. 18. In addition, the multimorbidity network was constructed using embeddings from neural network models (FCNN for disease prediction and DeepSurv for risk assessment) to identify clusters of diseases with strong comorbid relationships. Six primary clusters spanning various disease categories were identified, revealing shared mechanisms and comorbidity patterns. For detailed methodology and visualizations, refer to the Supplementary Figs. 10 and 11.
Multi-center validation using all of us data
The All of Us cohort provided an additional validation dataset to assess model performance across a broader, more diverse population28. Due to differences in variable definitions between the datasets, we retrained the FCNN and DeepSurv models from the UKB-MDRMF using the All of Us data. The UK Biobank field selections are detailed in Supplementary Data 1, while the All of Us data dictionary is provided in Supplementary Data 3. Our results indicated that, while introducing new variable categories enhanced prediction accuracy and survival metrics, similar to findings with the UK Biobank, the improvements were less marked. This was primarily because the basic information already achieved high predictive performance, as detailed in Supplementary Note 4.3.2 and Supplementary Fig. 12. Furthermore, joint modeling of all phecodes within the All of Us data yielded high prediction AUCs and survival C-Index values, with diseases of the reproductive system consistently showing the highest predictive accuracy. This validation demonstrates the robustness of UKB-MDRMF and its potential applicability to varied datasets.
Discussion
UKB-MDRMF represents a significant advancement in the field by offering a comprehensive solution for multi-disease prediction, risk assessment, and diverse applications. It not only achieves higher predictive accuracy but also effectively uncovers potential connections among multiple risk factors and diseases, as well as between diseases themselves. Its streamlined and user-friendly nature makes it accessible for various research and clinical settings. The versatility of UKB-MDRMF is a key strength, allowing for insightful results at every stage of the process through a combination of statistical analyses and deep learning modeling.
The data preprocessing phase in UKB-MDRMF was not merely a preparatory step but provided valuable insights for clinical research29. We categorized the input data into six categories and conducted modeling and analysis step by step, starting from easy to difficult. This approach facilitates the application of UKB-MDRMF to datasets from different centers, regardless of the amount of collected information. We also incorporated polygenic risk scores (PRS) into the genetic categories, which are crucial for disease prediction and further enhance the model’s performance30.
By delving into the data missingness mechanisms, we uncovered valuable information that influences the robustness of subsequent analyses. In large cohort studies such as the UK Biobank, various missing patterns are common31. We conducted a thorough investigation of the complex data missing patterns for each variable type. Various reasons contributed to missing data, and we did not blindly remove or impute data with high missing rates32. Instead, we considered the data collection process, enabling a comprehensive understanding of the reasons for missing data. This strategy not only enhances the model’s predictive performance but also mitigates potential biases in the data, making it more practically meaningful.
The advantage of our UKB-MDRMF lies in its simultaneous consideration of individual disease prediction and risk assessment. This approach outperforms methodologies that focus solely on one aspect, capturing the intricate interplay between diseases and providing a more comprehensive view of an individual’s health landscape. The flexibility embedded in our framework allows for the incorporation of diverse model types, making it adaptable to the distinct characteristics of various large-scale biomedical datasets. The provision of a swift baseline, compared across multiple methods, underscores the efficiency and competitiveness of our approach, making it an invaluable resource for researchers in diverse medical domains. Additionally, the adaptability of UKB-MDRMF enables the incorporation of disease-specific diagnostic models tailored to different medical conditions. This versatility empowers clinicians to efficiently diagnose and manage a wide spectrum of diseases, ultimately improving patient care and treatment outcomes33. Furthermore, our UKB-MDRMF facilitates the assessment of multiple risk assessment models, allowing for a rigorous evaluation of their performance. A comparison of multiple methods helped users identify the most appropriate model and modeling approach for their specific medical background. The results of method comparisons also indicated the advantages of deep learning methods, particularly in modeling large-scale biomedical data, due to their ability to capture latent information from heterogeneous and noisy datasets34. Interestingly, for the prediction of rare diseases, embedding matrix-based methods incorporating textual prior information12 did not improve model performance in our approach, contrary to some existing methods. This observation may be attributed to the substantial volume of data in the UK Biobank, allowing deep learning models to achieve superior performance without the need for additional textual information.
Given the complexity of predictors and diseases, UKB-MDRMF excels in identifying correlations and pinpointing key risk factors that significantly influence specific diseases. This not only aligned with the complexity of real-world clinical research but also had the potential to unveil insights into disease etiology and progression. The applications of UKB-MDRMF allowed for risk factor assessment and analysis for different diseases, such as the discovery of significant variables. We offered a straightforward means of invoking interpretable methods to elucidate variable importance27. This interpretability is essential for clinicians and researchers seeking insights into the factors35. Our results highlighted the significance of imaging information and the variable importance variations across different diseases.
Age, mental health, and obesity-related variables have emerged as key contributors to disease risk across multiple categories, underscoring their significant roles in disease progression. As depicted in Fig. 4, conditions such as bipolar disorder and depression are associated with increased susceptibility to various diseases. This heightened risk likely stems from disruptions in the neuroendocrine system, including imbalances in neurotransmitters like serotonin, dopamine, and norepinephrine, and immune dysfunction, which compromise immune responses and elevate disease vulnerability36,37. Similarly, aging is linked to accelerated declines in metabolism, immune regulation, and cellular repair, increasing susceptibility to chronic diseases38. Obesity-related variables, including body mass index (BMI), waist circumference, and body fat percentage, contribute to disease risk by promoting insulin resistance, chronic inflammation, and dyslipidemia, which are pivotal in the development of metabolic disorders, cardiovascular disease, and liver conditions39,40. Furthermore, lymphocyte percentage, a marker of immune function, is indicative of immune dysregulation and chronic inflammation, processes closely linked to autoimmune diseases, infections, and cancer progression41. Microalbumin in urine serves as an early indicator of kidney damage, signaling glomerular dysfunction and systemic vascular injury, especially in individuals with diabetes or hypertension42. Collectively, these findings highlight the critical contributions of mental health, aging, obesity, immune function, and early biomarkers in understanding disease risk and enhancing prediction accuracy.
The joint modeling of multiple diseases enhanced our method’s ability to characterize multimorbidity patterns, providing a data-driven exploration of how diseases co-occur and revealing the underlying mechanisms of mutual connections and influences among diseases within large-scale biomedical datasets. This understanding is crucial, offering patients and healthcare providers insights into potential complications that may arise in tandem with a primary disease diagnosis. Our modeling approach revealed similarities between different diseases and uncovered associations between coexisting diseases that might be challenging to identify through individual studies. Unlike multimorbidity analyses based on clinical experience or ICD-10 codes6, the multimorbidity captured by UKB-MDRMF depicted potential comorbidity situations within large-scale biomedical data, determined by similar risk factors. Figure 5 highlights several instances of diseases co-occurring outside their traditional categories, based on the hierarchical classification of disease coding. Additionally, by considering the community clustering effects among diseases, we identify several of the largest clusters (Supplementary Figs. 10 and 11), which align with findings in existing literature5,6. Our approach uncovers unconventional connections, such as linking urinary-related diseases at the intersection of cardiovascular and kidney systems (e.g., Hypertensive chronic kidney disease) and blood disorders (e.g., Anemia of chronic disease), consistent with real-world clinical outcomes43 and holding significant clinical implications. Furthermore, the age-dependent multi-disease risk analysis added another layer of sophistication to our framework. It enabled the categorization and targeted management of different disease patterns across various age groups, facilitating early interventions and treatments where they are most effective. This capability holds the promise of not only improving patient outcomes but also reducing the overall burden on healthcare systems.
The validation of the UKB-MDRMF framework using the All of Us data underscores its adaptability and potential for analyzing multimorbidity across diverse populations. After adjusting the variables to align with the All of Us data, we found that the results and interpretability of the UKB-MDRMF were effectively transferable. Notably, the inclusion of new variable types consistently improved prediction accuracy. Furthermore, the demonstration of results comparable to those observed with the UK Biobank data highlights the model’s capability to generalize key disease mechanisms across different populations. This robust performance confirms the framework’s utility in a broad array of clinical settings and underscores its potential for widespread application in multimorbidity analysis.
UKB-MDRMF represents a significant step toward more accurate and comprehensive disease prediction and risk assessment, but several limitations remain. Our framework is primarily based on UK Biobank data, and while validation using the All of Us dataset has demonstrated the effectiveness of the modeling process, population-specific biases may still affect generalizability, necessitating further validation across more diverse, multi-center datasets. Additionally, by excluding pre-baseline diseases, the model may overlook important correlations between past and future health conditions. While our joint learning approach captures disease interdependencies, it does not explicitly model competing risks, which could improve the precision of survival modeling. Future work should focus on enhancing generalization, integrating real-time data, providing a more detailed characterization of disease interactions, and fostering multi-center collaborations to maximize the clinical utility and impact of UKB-MDRMF in real-world healthcare settings.
Our UKB-MDRMF stands as a powerful and versatile tool poised to make substantial contributions to the healthcare system. With its comprehensive approach to multi-disease prediction and risk assessment, UKB-MDRMF offers insights that can inform more effective healthcare strategies and interventions. Its ability to uncover connections among risk factors and diseases provides a deeper understanding of health dynamics, paving the way for more targeted and personalized approaches to patient care.
Methods
Data description
This study was conducted using the UK Biobank Resource under Application Number 98327, with ethical approval from the North West Multi-Center Research Ethics Committee as a Research Tissue Bank. The UK Biobank study protocol is available online (https://www.ukbiobank.ac.uk/), and all participants provided written informed consent for data provision and linkage. UK Biobank is a large-scale biomedical resource comprising 502,467 individuals aged 37 to 72, recruited between 2006 and 2010 in the United Kingdom. The dataset includes demographic, biological, lifestyle, environmental, measurement, mental health, and imaging data. To assess the generalizability of our model beyond UK Biobank, we performed external validation using the All of Us dataset28, a diverse U.S. cohort designed to include underrepresented populations in biomedical research. The dataset integrates electronic medical records and participant surveys, with all participants providing informed consent. Ethical oversight is provided by the All of Us Institutional Review Board. This dataset was used for model validation, leveraging its broad demographic and clinical diversity to evaluate performance across populations.
Overview of UKB-MDRMF
UKB-MDRMF developed using the UK Biobank data, aims to forecast and assess disease risk. The data undergoes rigorous preprocessing to ensure data quality, predictive accuracy, and generalizability. The prediction pipeline of the UKB-MDRMF framework consists of two main components: data preprocessing and data modeling, as shown in Fig. 1.
To build a robust prediction model, we first constructed and preprocessed independent and dependent variables, denoted as X and Y, respectively. For X, we carefully selected fields, cleaned and transformed the data, and handled missing values. For Y, we used Phecodes as labels extracted and transformed from in-hospital, self-reported, and primary care data. Time alignment was performed to synchronize data points for practical value and significance in constructing the prediction model.
After data preprocessing, we employed advanced machine learning models, including deep learning techniques, to aid in auxiliary disease prediction and future disease risk prediction. These models were trained on the preprocessed data to evaluate an individual’s risk of developing a disorder. The output included predictions such as individual disease prediction, classification of disease types, or comprehensive predictions covering all Phecodes. The detailed process is outlined as follows.
Data preprocessing
Data filtering
Data screening is divided into two parts: GWAS data and non-GWAS data. For GWAS data, we utilized a total of 7228 phenotypes14. First, to prevent information leakage of disease information (Y) and ensure the robustness of the UKB-MDRMF framework, we excluded all health-related outcomes for creating the predictor variables (X). This includes disease diagnoses, medication records, prescription data, surgery details, and hospital registration summaries. Second, online follow-up data from around 2016 were omitted due to the need for temporal alignment. After initial screening, 1011 phenotypes remained. In the subsequent filtering step, composite data (such as the Albumin/Globulin ratio), ECG (with low participation), and family history (strongly related to disease but not relevant) were removed. Other extraneous factors (mostly early life factors such as birth month, birthplace, and some test results) were also excluded, leaving 542 phenotypes mapped to 365 data fields. Additionally, there are 12 meaningful measurement fields for carotid artery ultrasound measurements. Among them, the Index of Multiple Deprivation (England, Wales, Scotland) (Data-Fields 26410, 26426, 26427) represents the same data for different regions. Therefore, the data from these three regions were combined into a single Index of Multiple Deprivation.
For the non-GWAS part, specifically including MRI and ultrasound data, we mainly focused on the imaging data of Heart MRI and Brain MRI for some participants in the UK Biobank, which are structured data analyzed beforehand. After retaining meaningful measurement data, there are 27 for Heart MRI and 884 for Brain MRI. Some fields required related data fields for imputation, such as the number of live births (Data-Field 2734) and whether the participant ever had a stillbirth, spontaneous miscarriage, or termination (Data-Field 2774). These fields were not initially included in the GWAS data but were introduced later. For further details on genetic and imaging data processing, refer to Supplementary Notes 2.5, 2.6 and Supplementary Fig. 5.
Finally, based on their specific contents, the data were categorized into six groups: basic characteristics (height, weight, waist-to-hip ratio, age, gender, ethnicity, occupation, education, appearance, etc.), measures (blood, urine, biochemical indicators, auditory and visual assessments, bodily pain perception, physical measurements, etc.), lifestyle (smoking and drinking habits, mental health, exercise, electronic device usage, sleep patterns, diet, early life factors, etc.), natural and social environment (working environment, living environment, early life environment, air/noise pollution, poverty index, etc.), imaging (MRI and ultrasound data), and genetics (genetic principal components and PRS information). The comprehensive screening procedure is shown in Fig. 6. We also empirically assigned a priority level to each of these data fields according to their importance to our downstream tasks. Specifically, the selected variables were prioritized as essential information, detailed information, and minor information, respectively. A brief summary of selected data fields is presented in Supplementary Fig. 2. For more details on the specific selection of variables, refer to Supplementary Data 1.
Based on 7228 phenotypes from the GWAS, we selected 542 phenotypes after excluding treatment diagnoses and some that are either irrelevant or difficult to measure. These 542 phenotypes are then grouped into six categories based on their nature. Additionally, principal component analysis (PCA) is performed on the structural data of Heart MRI, Brain MRI, and Ultrasound retaining the top 11, top 89, and top 5 principal components for each, respectively. Icons provided by Icons8 (https://icons8.com).
Data cleaning
Although the UKB raw data is structured, several preprocessing steps are still necessary. We adopted a targeted approach to clean and preprocess variables based on their specific characteristics, as illustrated in Fig. 7.
For continuous and integer variables, we first apply special encoding techniques and then determine whether to handle them as continuous or discrete based on whether they have more than 20% identical values. For categorical variables, unordered encodings are transformed into binary variables. Ordered encodings undergo special encoding techniques before discrete handling. Icons provided by Icons8 (https://icons8.com).
For continuous and integer variables, if the data field was encoded in a non-quantitative manner, we created binary indicator variables for each special code and then reassigned them as “NA” or their corresponding quantity. For instance, for Data-Coding 100290, indicator variables were created for codes − 1 and − 3, denoting “Do not know” and “Prefer not to answer”, respectively, and assigned as “NA”. Code − 10, signifying “Less than a year”, was reassigned to 0.5. Subsequently, we checked whether more than 20% of the values in the data were identical. If so, we performed equal frequency binning (n = 3) and represented the variable with its “level” as an ordered categorical variable. Otherwise, we standardized the variable by subtracting the mean and dividing it by the standard deviation.
For single-response and multiple-response categorical variables, if the coding lacked a natural order, we created a binary indicator variable for each code (including “NA”). If there was a natural order, we followed the same procedure as for continuous and integer variables, creating binary indicator variables for any special codes and reassigning the variable as an ordered categorical variable. For instance, for Data-Coding 100327, we created indicator variables for codes −1 and −3 and assigned them as “NA” until the next subsection to perform missing imputation. Additionally, the meanings of 7 ("No friends/family outside household”) and 6 ("Never or almost never”) were nearly identical, so we reassigned 7 as 6 to reduce coding redundancy.
Process of phecodes
The health-related records of UKB participants come from multiple sources, including hospital inpatient data, mortality records, coded primary care data, and self-reported health conditions. These sources provide a comprehensive view of participants’ health status, but the heterogeneity of diagnostic codes across these sources poses a challenge for analysis. Phecodes offer a recognized approach to grouping ICD codes to capture clinically meaningful concepts for research15. To address this, we transformed inpatient, death register, primary care, and self-report data into 1560 Phecodes, creating a unified set of codes to represent Y, as shown in Fig. 8. First, we used coding mapping tables to transform self-report and primary care data into ICD-10 codes. We then integrated multi-source ICD-10 diagnosis codes for each subject and recorded the corresponding disease occurrence time for each ICD-10 code for time alignment. Finally, the ICD-10 codes were transformed into one-hot encoded Phecodes to ensure consistency and clinical relevance44. For additional details on Phecode processing, refer to Supplementary Note 2.4 and Supplementary Table 4.
Data originates from three sources: hospital inpatient, self-report, and primary care data. They were separately encoded and standardized as ICD-10 codes. After integration, the standardized codes were mapped to Phecodes to serve as the final response variables. Icons provided by Icons8 (https://icons8.com).
Missing data processing
The UK Biobank data includes numerous variables, many with missing values. We analyzed and handled the missingness in various ways, detailed in Supplementary Note 4.1. Here, we outline our procedure for dealing with missing data.
We first removed 3792 participants whose data was collected in the pilot assessment center due to high missing rates and low data quality. After examining the data collection procedure, we assumed missing completely at random (MCAR) or missing at random (MAR) mechanisms for our selected variables, making multiple imputations viable. It is important to note that there is no empirical way to distinguish MCAR from MAR. Our assumption is based on data collection knowledge. As shown in Supplementary Note 4.1, high missing rates are mainly due to related data fields, collection time, and assessment centers.
Moreover, we carefully identified and handled missingness related to other variables during data collection. This process is summarized in Supplementary Fig. 4. Specifically, some missingness was associated with other variables. We categorized such patterns into three types. Pattern-I involves instances where missing data can be precisely imputed using related variables, informed by data collection procedures. For example, participants were exempt from the query regarding “weekly usage of red wine” (Data-Field 1568) due to negligible or no alcohol consumption, as determined by Data-Field 1558. Such missingness was directly imputed based on other variables. Pattern-II missingness occurs when data is absent because the variable is not applicable to certain subgroups. For instance, sex-specific factors relevant only to one gender would be missing for the other. To address this in our analysis, we implement two distinct strategies: For categorical variables affected by Pattern-II missingness, we introduce a new category labeled ‘inapplicable’. This modification helps maintain the integrity of the dataset without distorting the analysis due to missing values. For continuous variables, we opt to impute missing values using the mean of the observed data. This approach aligns with the popular missing indicator method45, facilitating consistent statistical treatment across different types of data. Unlike the conventional missing indicator method, where missing indicators are added separately in the prediction model, we integrate these indicators directly within our existing variable set to avoid redundancy. For example, the sex variable itself serves as the missing indicator for sex-specific variables. This integrated approach streamlines the model by reducing unnecessary complexity and enhancing the efficiency of our analyses. Pattern-III encompasses missingness associated with other variables but does not meet the criteria of the previous types. For this type, we constructed binary indicator variables for the remaining missing values to account for potentially informative missingness. We provide a detailed classification of missing patterns in Supplementary Table 5 and Supplementary Data 2.
Finally, we used MissForest, implemented in the R package MissRanger, to impute the remaining missing values. MissForest46 is a robust imputation method widely adopted in various scientific fields32. MRI data were collected for a subgroup of about 50,000 participants. We treated this subgroup as our population when incorporating MRI data, directly imputing the imaging data using MissForest. For additional results on missing data analysis, refer to Supplementary Note 4.1 and Supplementary Figs. 6–9.
Model construction
Model construction process
Our model construction focused on two primary objectives: disease prediction and risk assessment. For disease prediction, when not considering the timing of covariates and response variables, the problem becomes a straightforward prediction task. In this case, the model for X and Y disregards time, making it useful for disease diagnosis. For risk assessment, we considered each variable’s recording time and built a survival analysis model. Each patient’s age served as the timescale.
As shown in Fig. 9, the modeling process begins by aligning the input variables based on their types and recording times. These variables are grouped into six categories: basic information, lifestyle, measurement, natural and social environment, genetics, and imaging, and are used as covariates in the model. For disease prediction, we employed machine learning algorithms such as logistic regression, boosting, and deep learning to predict over 1560 Phecodes as a multi-label classification problem. In contrast, for risk assessment, we incorporated each variable’s recording time and built a survival analysis model, using each patient’s age as the timescale. The UKB data used in our study has a follow-up period extending until September 2023, with events occurring at the end of this period treated as right-censored to ensure that individuals still at risk by that time remain appropriately accounted for. Additionally, we considered the impact of deaths occurring post-enrollment in survival analysis. If a participant died before developing a specific disease of interest, their time-to-event data for that disease was censored at the time of death. This adjustment ensures that mortality effects are properly integrated into the survival models.
First, a time alignment process is applied to ensure that disease occurrences post-date the baseline data. The red-shaded section at the bottom illustrates the alignment process for different individuals, where the red dashed line represents the enrollment time. Features such as basic, lifestyle, and other characteristics are collected at the time of enrollment. Phecodes recorded before enrollment are marked in gray and treated as missing values during model training, ensuring they are not used for training purposes. After integrating Phecode data from multiple sources, only the earliest occurrence of the same Phecode post-enrollment is retained. Next, various multi-disease prediction and risk assessment models are applied for comprehensive evaluation. These models are trained separately using distinct loss functions. Finally, model interpretability analysis is performed, incorporating associations between different diseases and risk factors for integrated analysis. Variable importance results are derived from all model weights, while multimorbidity relationships are inferred from the embeddings in the penultimate network layer. Icons provided by Icons8 (https://icons8.com).
In the final modeling framework, we adopted neural network-based approaches that demonstrated the best performance for both disease prediction (FCNN) and risk assessment (DeepSurv), which were subsequently applied to various downstream analyses. Ensuring strict temporal alignment of input variables was a critical aspect of model construction. Specifically, covariates (X) must occur before the diagnosis of the target disease (Phecode, Y) to maintain causality. As depicted in Fig. 9, disease occurrences before the enrollment time are marked in gray and excluded from training. Additionally, if a disease appears multiple times in the records, only the earliest diagnosis post-enrollment is retained for time alignment. This setup ensures that the framework allows forward-looking disease risk assessment rather than merely identifying associations between features and diseases retrospectively.
Following this, multi-disease prediction and risk assessment models are trained separately using distinct loss functions. During network training, the backpropagation algorithm optimizes model weights, enabling further downstream analyses such as variable importance evaluation and multimorbidity analysis. Variable importance is derived from SHAP-based weight analysis27, while multimorbidity relationships are inferred from the embeddings in the penultimate network layer. Further details can be found in Supplementary Note 4.3.1.
Loss function for neural network-based methods
In the multi-disease prediction task, we construct a specific loss function for multiple outcomes (Phecodes). For the FCNN model, the loss function L(FCNN) is defined as follows:
where p is the total number of Phecodes (diseases), N is the total number of samples, yij indicates whether the i-th sample has the j-th disease (1 indicates occurrence after enrollment, 0 means no occurrence after enrollment), and \({\hat{g}}_{\theta }^{(j)}({x}_{i})\) represents the output of the neural network for the j-th disease prediction given the input features xi. For instances where the disease occurrence predates the covariate collection time, the corresponding terms in the loss function are set to “NA”, effectively excluding those cases from contributing to the model’s learning.
In the multi-task survival analysis, the goal is to model the time-to-event data for multiple diseases. We employ a survival function with the following form:
The expression describes the hazard function for individual i with disease j, where λ0j(t) is the baseline hazard function for the j-th disease, and \({f}_{\theta }^{(j)}({x}_{i})\) is the output risk score for disease j from the neural network. Rj(t) represents the set of individuals still at risk for disease j at time t, and eij is an indicator variable that equals 1 if the individual i develops disease j, and 0 otherwise. tij denotes the time of disease onset for individual i with disease j. The survival loss function L(DeepSurv) is then defined as
The function is a weighted negative log-likelihood loss, where diseases with a higher number of cases contribute more to the gradient updates during backpropagation, thus receiving larger weights in the model training.
Missing labels in multi-disease tasks
In our study, missing labels occur when an individual’s disease information (Phecode) is unavailable, often because the disease occurred before data collection or enrollment. During data preprocessing, these missing Phecodes are marked accordingly. To address missing labels during model training, we adapt our approach to exclude these entries from the loss function calculation. Specifically, for any Phecode labeled as missing for an individual, that term is omitted from the corresponding loss calculation. Consequently, these missing labels do not participate in the backpropagation process, thereby preventing any influence on the gradient updates. This ensures that only valid labels contribute to the model’s training, allowing the model to focus on learning from available and accurate disease information. By doing so, we maintain the integrity of the training process and avoid introducing bias from missing labels.
Multi-task DeepSurv risk calculation
The Multi-Task DeepSurv model is designed to handle multiple survival tasks simultaneously, with the primary objective being to estimate the hazard function for each disease. For individual i and disease j, the hazard function hij(t) is defined as:
which quantifies the instantaneous rate at which an event (disease) occurs for individual i at time t. The baseline hazard function for disease j, \({\hat{\lambda }}_{0j}(t)\), is estimated by:
where the numerator counts the number of individuals experiencing disease j within a small interval from t to t + δ, and the denominator aggregates the risk scores of all individuals at risk of disease j at time t. To evaluate the cumulative risk over time T, we calculate the cumulative hazard function for disease j as:
This integrates the baseline hazard over time to provide a comprehensive estimate of disease occurrence risk up to time T. Such an approach allows the model to dynamically capture risks across multiple time points and for different disease outcomes concurrently. This functionality makes the model especially suitable for long-term risk assessments in settings involving multiple diseases.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Individual-level UK Biobank data are available under restricted access due to participant confidentiality and data governance policies. Researchers can apply for access through the UK Biobank Access Management System (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). Access is granted to approved researchers affiliated with recognized institutions, following a formal application and approval process. The expected timeframe for access approval varies based on the review process and project requirements. Once granted, access remains available for the approved project duration as per UKB policies. The All of Us dataset used for validation is available on the All of Us Researcher Workbench (v7 release) under restricted access to protect participant privacy. Researchers can request access at https://www.researchallofus.org/. The All of Us Research Program follows a tiered access model, where individual-level data are available in the Registered Tier and Controlled Tier to approved researchers from eligible institutions. The access process involves registration, identity verification, and completion of research ethics training. The expected response time for access requests depends on the review process and approved access remains valid as long as compliance with All of Us data policies is maintained. Source data are provided with this paper.
Code availability
UKB-MDRMF is implemented in Python, and the code for our study is available on GitHub at https://github.com/kannyjyk/UKB-MDRMF and stored at https://doi.org/10.5281/zenodo.1503224847. The repository is organized into four parts: data preprocessing, missing data processing, model construction, and comparative analysis. Each part corresponds to a specific folder in the repository. This open-access resource promotes transparency, facilitates replication, and encourages further research in this domain.
Change history
28 January 2026
A Correction to this paper has been published: https://doi.org/10.1038/s41467-026-68888-1
References
Sudlow, C. et al. Uk biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Tan, K. H. X. et al. Cohort profile: the Singapore multi-ethnic cohort (mec) study. Int. J. Epidemiol. 47, 699–699j (2018).
Wan, E.Y.F. et al. Association of covid-19 with short-and long-term risk of cardiovascular disease and mortality: a prospective cohort in UK biobank. Cardiovasc. Res. 119, 1718–1727 (2023).
Cui, H. et al. scgpt: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024)
Beaney, T. et al. Identifying multi-resolution clusters of diseases in ten million patients with multimorbidity in primary care in England. Commun. Med. 4, 102 (2024).
Dong, G., Feng, J., Sun, F., Chen, J. & Zhao, X. M. A global overview of genetically interpretable multimorbidities among common diseases in the UK biobank. Genome Med. 13, 1–20 (2021).
Sriram, V. et al. Netmage: a human disease phenotype map generator for the network-based visualization of phenome-wide association study results. GigaScience 11, giac002 (2022).
Im, J., Lee, J., Lee, S. & Kwon, H. Y. Data pipeline for real-time energy consumption data management and prediction. Front. Big Data 7, 1308236 (2024).
Battenberg, K. et al. A flexible cross-platform single-cell data processing pipeline. Nat. Commun. 13, 6847 (2022).
Olsson, T., Barcellos, L. F. & Alfredsson, L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 13, 25–36 (2017).
You, J. et al. Plasma proteomic profiles predict individual future health risk. Nat. Commun. 14, 7817 (2023).
Yang, L., Wang, S. & Altman, R. B. Popdx: an automated framework for patient phenotyping across 392 246 individuals in the UK biobank study. J. Am. Med. Inf. Assoc. 30, 245–255 (2023).
Cataldo-Ramirez, C.C., Haddad, D., Amenta, N.& Weaver, T.D. Developing an automated skeletal phenotyping pipeline to leverage biobank-level medical imaging databases. Am. J. Biol. Anthropol. 181, 413–425 (2023)
rkwalters, D. Palmer. Nealelab/ukbb_ldsc: v2.0.0 (round 2 gwas update) https://doi.org/10.5281/zenodo.7186871 (2022).
Bastarache, L. Using phecodes for research with the electronic health record: from phewas to phers. Annu. Rev. Biomed. Data Sci. 4, 1–19 (2021).
Hosmer Jr, D.W., Lemeshow, S. & Sturdivant, R.X. Applied logistic regression, vol. 398 (John Wiley & Sons, 2013)
Breiman, L. Random forests. Mach. Learn 45, 5–32 (2001).
Chen, T. & Guestrin, C. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, pp. 785–794 (2016).
Ke, G. et al. Lightgbm: a highly efficient gradient boosting decision tree. Adv. Neural Inf. Process Syst. 30, 3149–3157 (2017)
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Shen, Y. et al. Cati: a medical context-enhanced framework for diagnosis code assignment in the UK biobank study (2024).
Ferreira, C. R. The burden of rare diseases. Am. J. Med. Genet. Part A 179, 885–892 (2019).
Schieppati, A., Henter, J. I., Daina, E. & Aperia, A. Why rare diseases are an important medical and social issue. Lancet 371, 2039–2041 (2008).
Cox, D. R. Regression models and life-tables. J. R. Stat. Soc: Ser. B 34, 187–202 (1972).
Katzman, J. L. et al. Deepsurv: personalized treatment recommender system using a cox proportional hazards deep neural network. BMC Med. Res. Methodol. 18, 1–12 (2018).
Lawrence, I. & Lin, K. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45, 255–268 (1989)
Lundberg, S.M. & Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 30, 4768–4777 (2017)
Ramirez, A.H. et al. The all of us research program: data quality, utility, and diversity. Patterns 3, 100570 (2022)
Hou, J. et al. Generate analysis-ready data for real-world evidence: tutorial for harnessing electronic health records with advanced informatic technologies. J. Med. Internet Res. 25, e45662 (2023).
Lewis, C. M. & Vassos, E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 12, 1–11 (2020).
Xu, L. & Qiu, A. In: 2022 6th Annual International Conference on Data Science and Business Analytics (ICDSBA) (2022), pp. 72–82. https://doi.org/10.1109/ICDSBA57203.2022.00026 (2022).
Alaa, A. M., Bolton, T., Di Angelantonio, E., Rudd, J. H. & Van der Schaar, M. Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK biobank participants. PloS One 14, e0213653 (2019).
Strating, T., Hanjani, L.S., Tornvall, I., Hubbard, R., Scott, I.A. Navigating the machine learning pipeline: a scoping review of inpatient delirium prediction models. BMJ Health Care Inform. 30, e100767 (2023).
Miotto, R., Wang, F., Wang, S., Jiang, X. & Dudley, J. T. Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinformatics 19, 1236–1246 (2018).
Vellido, A. The importance of interpretability and visualization in machine learning for applications in medicine and health care. Neural Comput. Appl. 32, 18069–18083 (2020).
Matt, S. & Gaskill, P. Where is dopamine and how do immune cells see it?: dopamine-mediated immune cell function in health and disease. J. Neuroimmune Pharmacol. 15, 114–164 (2020).
Baldessarini, R. J., Vázquez, G. H. & Tondo, L. Bipolar depression: a major unsolved challenge. Int. J. Bipolar Disord. 8, 1 (2020).
Shen, X. et al. Nonlinear dynamics of multi-omics profiles during human aging. Nat. Aging 4, 1619–1634 (2024)
Hotamisligil, G. S. Inflammation and metabolic disorders. Nature 444, 860–867 (2006).
Després, J. P. & Lemieux, I. Abdominal obesity and metabolic syndrome. Nature 444, 881–887 (2006).
Furman, D. et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 25, 1822–1832 (2019).
Perkins, B. A., Ficociello, L. H., Roshan, B., Warram, J. H. & Krolewski, A. S. In patients with type 1 diabetes and new-onset microalbuminuria the development of advanced chronic kidney disease may not require progression to proteinuria. Kidney Int. 77, 57–64 (2010).
Jayamanna, U. & Jayaweera, J. S. Childhood anemia and risk for acute respiratory infection, gastroenteritis, and urinary tract infection: a systematic review. J. Pediatr. Infect. Dis. 18, 061–070 (2023).
Wu, P. et al. Mapping icd-10 and icd-10-cm codes to phecodes: workflow development and initial evaluation. JMIR Med. Inf. 7, e14325 (2019).
Allison, P.D. Missing data. In: The SAGE handbook of quantitative methods in psychology, pp. 72–89 (2009)
Stekhoven, D. J. & Bühlmann, P. Missforest—non-parametric missing value imputation for mixed-type data. Bioinformatics 28, 112–118 (2012).
Jiang, Y. et al. Ukb-mdrmf: a multi-disease risk and multimorbidity framework based on UK biobank data. BMJ Evid. Based. Med. 29, e112518 (2025).
Mamouei, M. et al. A comparative study of model-centric and data-centric approaches in the development of cardiovascular disease risk prediction models in the UK biobank. Eur. Heart J. Digit. Health 4, 337–346 (2023).
Blass, I. et al. Revisiting the risk factors for endometriosis: a machine learning approach. J. Pers. Med. 12, 1114 (2022).
Petrazzini, B. O. et al. Coronary risk estimation based on clinical data in electronic health records. J. Am. Coll. Cardiol. 79, 1155–1166 (2022).
Papadopoulou, A., Harding, D., Slabaugh, G., Marouli, E., Deloukas, P. Prediction of atrial fibrillation and stroke using machine learning models in UK biobank. medRxiv https://www.medrxiv.org/content/10.1101/2022.10.28.22281669v1 (2022).
Mars, N. et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med. 26, 549–557 (2020).
Markovitz, A. R. et al. Does pregnancy complication history improve cardiovascular disease risk prediction? findings from the hunt study in norway. Eur. Heart J. 40, 1113–1120 (2019).
Sun, L. et al. Polygenic risk scores in cardiovascular risk prediction: a cohort study and modelling analyses. PLoS Med. 18, e1003498 (2021).
Acknowledgements
We would like to acknowledge the UK Biobank for providing the dataset used in this study. This work was supported by the National Key R&D Program of China (No. 2022YFA1003803) and the National Natural Science Foundation of China (no. 12231017, 72171216, 71921001, and 71991474), all awarded to X.W. (Xueqin Wang).
Author information
Authors and Affiliations
Contributions
J.Y., X.W. (Xueqin Wang), and H.Z. proposed the concept and designed the methodology. J.Y., X.W. (Xueqin Wang), and H.Z. supervised the project. Y.J., B.Z., X.W. (Xiaopu Wang), B.T., H.P., and Z.L. performed the statistical analysis, visualized the results, and developed the software. Y.J., B.Z., X.W. (Xiaopu Wang), B.T., H.P. acquired and/or preprocessed the data. Y.J. drafted the initial manuscript. Y.J., X.W. (Xiaopu Wang), B.Z., B.T., H.P., Z.L., Y.S., Z.W., Z.J., J.W., J.Y., X.W. (Xueqin Wang), and H.Z. suggested revision ideas and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Vivek Sriram, Bhautesh Jani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, Y., Zhao, B., Wang, X. et al. UKB-MDRMF: a multi-disease risk and multimorbidity framework based on UK biobank data. Nat Commun 16, 3767 (2025). https://doi.org/10.1038/s41467-025-58724-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58724-3
This article is cited by
-
Genomic structural equation modeling decodes skeletal aging: novel loci discovery and multisystem genetic crosstalk
Journal of Translational Medicine (2025)
-
UKB-MDRMF: a multi-disease risk and multimorbidity framework based on UK biobank data
Nature Communications (2025)