Abstract
Disruption of bile acid (BA) metabolism causes various liver diseases including hepatocellular carcinoma (HCC). However, the underlying molecular mechanism remains elusive. Here, we report that BA metabolism is directly controlled by a repressor function of YAP, which induces cholestasis by altering BA levels and composition via inhibiting the transcription activity of Fxr, a key physiological BA sensor. Elevated BA levels further activate hepatic YAP, resulting in a feedforward cycle leading to HCC. Mechanistically, Teads are found to bind Fxr in a DNA-binding-independent manner and recruit YAP to epigenetically suppress Fxr. Promoting BA excretion, or alleviating YAP repressor function by pharmacologically activating Fxr and inhibiting HDAC1, or overexpressing an Fxr target gene Bsep to promote BA exportation, alleviate cholestasis and HCC caused by YAP activation. Our results identify YAP’s transcriptional repressor role in BA metabolism as a key driver of HCC and suggest its potential as a therapeutic target.
Similar content being viewed by others
Introduction
An essential metabolic function of the liver is to produce bile acids (BAs), which emerge as hormone-like signaling molecules that govern a myriad of metabolic processes1, extending beyond their conventional roles as digestive detergents. Abnormal BA synthesis, transport, and metabolism cause chronic liver diseases (CLD), such as metabolic dysfunction-associated steatotic liver disease (MASLD), primary biliary cholangitis (PBC), and progressive familial intrahepatic cholestasis (PFIC)2,3,4,5. BAs are toxic when present in high concentrations; and induce liver injury and inflammation, leading to a poorer prognosis for advanced liver diseases, including hepatobiliary malignancy5,6. Cholestatic jaundice manifests in as many as 40% of patients diagnosed with hepatocellular carcinoma (HCC)7, underscoring the intimate link between disrupted BA metabolism and liver tumorigenesis. Hence, investigating the complex regulation of BA metabolism is crucial for acquiring novel perspectives on the mechanisms underlying liver tumorigenesis and shedding new light on liver cancer treatment.
BA production and export are tightly regulated by negative feedback mechanisms1,8. In the hepatocytes, the BA-sensing nuclear receptor, Farnesoid X receptor (Fxr), is the master transcriptional regulator controlling BA synthesis and excretion. BAs inhibit their synthesis by binding to Fxr, which suppresses the expression of Cyp7a19,10, the first and rate-limiting enzyme in the classic BA synthesis pathway, via activating the expression of Shp, a direct transcription target of Fxr9,10. Fxr activation also promotes BA export via another Fxr target gene, Bsep11. Therefore, as the central regulator of BA homeostasis, Fxr regulation by BAs critically controls BA production and excretion to avoid liver injury and inflammation induced by BA accumulation in cholestasis. In mice, Fxr deficiency leads to elevated serum BA levels, liver injury, and subsequent HCC12,13,14,15,16, but the mechanism underlying Fxr regulation in the liver remains elusive.
The transcriptional coactivators YAP and TAZ mediate transcriptional regulation of the conserved Hippo signaling pathway, in which a kinase cascade of Mst1/2 and Lats1/2 inhibits YAP and TAZ by phosphorylation-dependent cytoplasmic sequestration and degradation17. In hepatocytes, loss of Hippo kinases Mst1/2 induces HCC due to YAP and TAZ activation18. In mouse hepatocytes, YAP activation is sufficient to cause HCC, while YAP/TAZ loss abolishes HCC caused by Mst1/2 deficiency18. Emerging evidence suggests that elevated BA levels, while causing liver injury, also promote liver tumorigenesis. However, since YAP activation mediates many oncogenic pathways in the liver and induces fibrosis and inflammation as occurred in cholestasis18,19,20,21,22, it remains unclear whether and how YAP activation causes HCC by inducing cholestasis.
Here we identify a repressor function of YAP in regulating BA metabolism and export. Activated YAP induce severe cholestasis and fibrosis, preceding tumor formation, by inhibiting Fxr. YAP is recruited by TEAD transcription factors, which interact with Fxr in a DNA-binding-independent manner, to replace the transcription co-activator acetyltransferase, P300 with the histone deacetylase, HDAC1. Additionally, activating Fxr weaken the YAP-TEAD canonical transcriptional activators’ function, suggesting that Fxr plays an important role in the dynamic regulation of YAP transcriptional activation as well. Pharmacological activation of Fxr, HDAC1 inhibition, or Bsep overexpression, all reduce YAP-induced HCC in mice. Our results show that YAP activation induces a repressive epigenetic reprogramming of Fxr and suggest activating Fxr or its targets in BA metabolism as therapeutic targets of YAP-induced HCC.
Results
YAP activation in hepatocytes induces cholestasis
In hepatocytes, YAP activation causes hepatomegaly and HCC20,23,24,25. Interestingly, in the TetO-YAP mice23, 2 weeks after AAV-TBG-Cre injection and doxycycline (Dox) water feeding, we found induced hepatocyte expression of YAPS127A, a constitutively active form of YAP, induced severe cholestasis, characterized by enlarged gallbladder, jaundice, a gradual and drastic increase in serum BA concentration, apart from previously reported liver enlargement (Fig. 1a–e, Supplementary Fig. 1a). The cholestatic phenotype was accompanied by severe liver injury, indicated by elevated serum ALT activities, increased cell death, disrupted bile ducts, liver fibrosis, and inflammation (Fig. 1f–i, Supplementary Fig. 1b–d). These defects are also found in patients with cholestatic liver disease26,27,28. Interestingly, three days after YAP activation, the serum BA levels were already increased, preceding the increase in liver-to-body-weight ratio and the liver damage (Fig. 1d–f, h, Supplementary Fig. 1c). These results show that YAP-induced BA dysregulation is an early event before hepatomegaly.
a Schematic diagram of the experimental procedure. YAP activation in hepatocytes is induced by AAV-TBG-Cre followed by doxycycline (Dox) water feeding. b Representative images of livers from the indicated mice. c Serum from the indicated mice. d Liver/body-weight ratios, ***P = 0.0001, ****P < 0.0001. e Serum BA concentration, *P = 0.049, **P = 0.0046, ****P < 0.0001. f Serum ALT, ***P = 0.0009, ****P < 0.0001. g Representative images of liver sections stained by H&E. h Representative images of TUNEL staining of liver samples. i Representative immunofluorescent images of liver samples. j, k qRT-PCR analysis of indicated genes in liver tissue. ****P < 0.0001, *P = 0.013 (j, Cyp7a1); **P = 0.003, *P = 0.011, ****P < 0.0001 (j, Cyp8b1); ****P < 0.0001 (j, Cyp7b1); **P = 0.006 (j, Cyp27a1); ****P < 0.0001 (k, Shp); ****P < 0.0001 (k, Bsep). l Western blotting analysis of liver tissue. Scale bars: 1 cm (b), 100 μm (g–i). n = 5 biological replicates in each group (d–f), n = 4 biological replicates in each group (j, k). Data are presented as Mean ± SD, P-values were determined by 1-way ANOVA with Sidak’s test (d–f, j, k). Similar results were obtained from three independent experiments (g–i, l). Source data are provided as a Source Data file. Image in (a) was created in BioRender (Jin, Y. (2025) https://BioRender.com/h79u623).
We found altered expression of genes encoding key enzymes involved in BA synthesis, especially Cyp7a1 and Cyp8b1, which were first upregulated, and then downregulated by prolonged YAP activation. Cyp27a1 or Cyp7b1 expression in the alternative BA pathway was also downregulated by prolonged YAP activation (Fig. 1j). The seeming paradox was due to hepatocyte dedifferentiation and cell fate transition to bile duct epithelial cell (BEC)-like cells induced by prolonged YAP activation20,23. We showed previously that Sox9 is required for YAP-induced hepatocyte-to-BEC-like cell transition20. We deleted Sox9 in the hepatocytes by generating the TetO-YAP; Sox9Δ mice. Indeed, after two weeks of Dox feeding, the liver in the TetO-YAP; Sox9Δ mice exhibited more severe cholestasis compared to that in the TetO-YAP mice, characterized by a drastically enlarged gallbladder and increased serum total BA levels (Supplementary Fig. 1e, f). Although Cyp7b1 was downregulated, the expression of Cyp7a1, Cyp8b1, and Cyp27a1 was preserved in the TetO-YAP; Sox9Δ liver (Supplementary Fig. 1g). In addition, while tumor cells in the TetO-YAP liver were Hnf4a- Sox9+, they maintained hepatocyte phenotypes, which were Hnf4a+ with normal Cyp7a1 expression in the TetO-YAP; Sox9Δ liver (Supplementary Fig. 1h). As Sox9 only has a limited role in YAP-induced hepatocyte dedifferentiation20, the expression of Cyp7a1, Cyp8b1, Cyp7b1 and Cyp27a1 was not further upregulated in the TetO-YAP; Sox9Δ liver because the hepatocytes were still dedifferentiated to a certain extent.
Since Fxr is a key nuclear receptor controlling BA homeostasis, we examined the expression of Fxr and its transcriptional targets, Shp and Bsep. While Fxr expression was unchanged, Fxr transcriptional activity rapidly declined three days after Dox feeding, as Shp and Bsep expression was downregulated (Fig. 1k). Consistently, Shp protein expression was drastically reduced, while Cyp7a1 expression was increased by YAPS127A expression 3 days after Dox feeding (Fig. 1l). In the TetO-YAP; Sox9Δ liver, Shp and Bsep exhibited further reduction compared to the TetO-YAP liver, consistent with our previous finding that Sox9 inhibits YAP activity (Supplementary Fig. 1g, i)20. These results indicate that YAP activation in hepatocytes led to BA accumulation due to both BA overproduction and impaired exportation. Therefore, the reduction of Fxr transcriptional activity is an early event in YAP-induced cholestasis, which precedes tumor formation.
YAP activation in hepatocytes alters BA production and composition
We then tested whether YAP directly regulates BA metabolism by regulating the BA sensor Fxr in hepatocytes. BA quantity and composition were determined by mass spectrometry. 2 weeks after YAP activation in the TetO-YAP mice, the total BA levels in both the liver and serum were significantly increased and BA composition was dramatically altered compared to controls (Fig. 1a, Fig. 2a, b). Importantly, similar to observations in the Fxr−/− mice16, the dominant BAs were TβMCA and TCA in the YAP-activated liver (Fig. 2b, c, Supplementary Fig. 2a, b). In addition, the mice with YAP activation resulted in a drastic increase of conjugated, hydrophilic TβMCA, in both their liver and serum (Fig. 2b, c). The accumulation of primary BAs in the liver indicates dysregulation of both BA synthesis and export, consistent with Fxr inhibition by YAP activation in hepatocytes. In the AAV-TBG- YAPS127A-injected mice (YAP* mice; Fig. 2d), the total BA levels were also increased, and BA composition was similarly altered in the liver and serum, compared to the Fxr−/− and TetO-YAP mice (Fig. 2e–g, Supplementary Fig. 2c, d). These results confirm that YAP activation inhibited Fxr activation. Notably, serum BAs, which are primarily reabsorbed from the ileum in the WT mice, were similar in composition to the liver BAs when YAP was activated (Fig. 2b, f), with TβMCA and TCA as the two major BAs (Fig. 2b, c, f, g, Fig. S2a–d). Conversion of the serum BA profile to a liver one (Fig. 2b, f) indicates severe BA leakage from the hepatocytes to the bloodstream, which was likely caused by liver injury, as indicated by elevated ALT levels and TUNEL signals in the YAP-activated liver (Fig. 1f, h).
a Total BA concentration in the liver and serum of the TetO-YAP mice as measured by mass spectrometry, *P = 0.038, ****P < 0.0001. b Pie charts show percent composition of BA species in the liver and serum of the TetO-YAP mice. The color legend is located on the right side. c TCA, TβMCA concentration in the liver and serum of the TetO-YAP mice, *P = 0.041, **P = 0.0048, **P = 0.0067, ****P < 0.0001 in sequence. d Schematic diagram of the experimental procedure. e Total BA concentration in the liver and serum of the AAV-TBG-YAP * injected mice and paired control mice, as measured by mass spectrometry, ***P = 0.0005, **P = 0.002. f Pie charts showing the percent composition of BA species in the liver and serum of the AAV-TBG-YAPS127A injected mice and paired control mice. The color legend is the same as in (b). g TCA, TβMCA concentration in the liver and serum of the AAV-TBG-YAPS127A injected mice and paired control mice, **P = 0.0068, **P = 0.0032, *P = 0.026, *P = 0.014 in sequence. h Western blotting analysis of indicated genes in the mouse mature hepatocyte organoids (MHOs). i, j qRT-PCR analysis of indicated genes in the mouse MHOs. ****P < 0.0001 (i); **P = 0.0038, *P = 0.05 (j). k Western blotting analysis of indicated genes in the mouse MHOs. l, m qRT-PCR analysis of indicated genes in the mouse MHOs, ****P < 0.0001 (l); **P = 0.0045 (Shp, l); **P = 0.003, *P = 0.045 (Bsep, l); ****P < 0.0001, ***P = 0.0003 (Cyp7a1, m); ***P = 0.0006, **P = 0.0042, *P = 0.022 (Cyp8b1, m). data are presented as Mean ± SD, 1-way ANOVA with Sidak’s test. n = 3 biological replicates in Dox- group, n = 4 biological replicates in Dox+ 2weeks group (a–c), n = 3 biological replicates in each group (e, g), n = 3 independent experiments (i, j, l, m). Data are presented as Mean ± SD, p values were determined by two-sided unpaired t-test (a, c, e, g, i, j). Similar results were obtained from three independent experiments (h, k). Source data are provided as a Source Data file. Image in (d) was created in BioRender (Jin, Y. (2025) https://BioRender.com/h79u623).
In both TetO-YAP mice and YAP* mice, the levels of many BA species were increased, which would damage hepatocytes (Fig. 2c, g, Supplementary Fig. 2a–d). Previous studies showed that BAs can activate YAP15,29. To test how serum BAs altered by Yap activation might regulate BA metabolism, we utilized a mature hepatocyte organoid (MHO), which preserves the physiological regulation of BA metabolism30. Mouse MHOs treated with serum from the YAP* mice showed reduced Yap phosphorylation, Cyp7a1 expression, but activated Shp expression, compared to those treated with the wild-type serum (Fig. 2h, i), indicating that serum from YAP-activated mice activated YAP and Fxr. Indeed, expression of the YAP and Fxr target genes was upregulated, while expression of Cyp7a1 and Cyp8b1 was consistently albeit mildly downregulated (Fig. 2i, j). Of note, in serum from YAP-activated mice, TCA and TβMCA were the most abundant BAs (Fig. 2b, f). Interestingly, TCA not only activates FXR but also activates YAP through S1PR2 activation29. Conversely, TβMCA functions as an endogenous FXR antagonist31. To test the solo and collective function of these two BAs in regulating Yap and Fxr, mouse MHOs were treated with TCA and TβMCA, at a concentration similar to that in the serum (Fig. 2c, g). Consistent with previous findings15,29,31, while TCA activated Fxr and Yap, TβMCA inhibited Fxr and YAP activities (Fig. 2l, m), and consistently, Cyp7a1 and Cyp8b1 expression was inhibited by TCA but upregulated by TβMCA (Fig. 2m). Intriguingly, combined TβMCA and TCA treatment activated YAP similarly to TCA treatment alone, but TCA’s activity in activating Fxr was effectively antagonized by TβMCA (Fig. 2k–m). In addition, although the Fxr agonist GW406432 strongly induced Shp expression, it did not activate YAP (Supplementary Fig. 2e). These data indicate that different species of BAs differentially regulate Fxr, as well as YAP. Taken together, these data show that YAP activation in the liver inhibited Fxr, which in turn increased BA concentrations and altered BA composition. Altered BAs may further change Fxr and YAP activities.
BA accumulation and Yap activation form a feed-forward loop in promoting liver tumorigenesis
The Fxr target-gene, Bsep (Abcb11), encodes the bile salt export pump (BSEP), which transports conjugated BAs across the hepatocyte canalicular membrane33. In humans, BSEP deficiency causes Progressive Familial Intrahepatic Cholestasis (PFIC), a major risk factor for childhood HCC4,34,35,36. Bsep-deficient mice develop hepatomegaly and intrahepatic cholestasis37, phenocopied by the YAP-activated liver. Since YAP activation suppressed Bsep expression, we asked whether restoring BA export by Bsep expression via AAV-TBG-Bsep could prevent YAP-induced liver tumor formation (Fig. 3a). Indeed, AAV-TBG-Bsep reduced YAP-induced serum BA levels and ALT activities, alleviated hepatomegaly and liver tumor formation (Fig. 3b–f, Supplementary Fig. 3a). YAP-induced liver injury, inflammation, and fibrosis were also reduced (Fig. 3g–i, Supplementary Fig. 3b). These results indicate that BA accumulation due to YAP-induced Fxr inhibition in hepatocytes played a major role in liver injury and tumor formation. To determine the contribution of increased serum BAs in causing liver injury and ultimately liver tumors, we reduced serum BAs by feeding the YAP* mice with the cholestyramine resin, which promotes BA excretion from the gut38,39 (Supplementary Fig. 3c). BA excretion by cholestyramine resin feeding indeed reduced serum BA levels, tumor sizes, and hepatic YAP activity, confirming that excessive serum BAs in the YAP* mice promoted tumor growth (Supplementary Fig. 3d–h). However, Bsep expression more drastically reduced the liver/body weight ratio and liver tumor formation (Supplementary Fig. 3i) compared to cholestyramine resin feeding, which only reduced the BA levels outside of hepatocytes. Taken together, these data indicate that efficient BA exportation from the liver is more critical for inhibiting YAP-induced liver tumor formation. Interestingly, YAP activation was observed in HCC that spontaneously formed in the Fxr−/− mice with cholestasis15,40. We further tested whether YAP activation is required for HCC induced by excessive BA in the Fxr−/− mice. We deleted Yap by injecting the AAV-TBG-Cre to the Fxr−/−; Yapfloxed mice (Fig. 3j) and found that Yap removal was sufficient to suppress HCC formation in the Fxr−/− mice, with reduced serum BA and alleviated liver damage (Fig. 3k–n). These data show that BA accumulation and YAP activation form a feed-forward loop in promoting HCC.
a Schematic diagram of the experimental procedure. b Representative images of liver of the indicated mice. c Liver/body weight ratios, ****P < 0.0001. d Serum BA concentration, ****P < 0.0001. e Serum ALT, ****P < 0.0001. f, h qRT-PCR analysis of indicated genes in the liver tissue, ****P < 0.0001 (f, h), **P = 0.0098 (Shp, f); **P = 0.0023 (Bsep, f); ***P = 0.0004, **P = 0.0026 (Ccl2, g); ***P = 0.0009, **P = 0.0048 (Tnf, g); ***P = 0.0004 (Acta2, h). i Representative immunofluorescent images of liver samples. j Schematic diagram of the experimental procedure. k Representative images of livers from the indicated mice. l Liver/body-weight ratios, ****P < 0.0001. m Serum BA concentration, ****P < 0.0001. n Serum ALT, ****P < 0.0001, **P = 0.003. Scale bars: 1 cm (b, k), 100 μm (i). n = 5 biological replicates in each group (c–e, l–n), n = 3 biological replicates in each group (f–h). Data are presented as Mean ± SD, P-values were determined by 1-way ANOVA with Sidak’s test (c–h, l–n). Similar results were obtained from three independent experiments (i). Source data are provided as a Source Data file. Image in (a, j) was created in BioRender (Jin, Y. (2025) https://BioRender.com/h79u623).
YAP activation inversely correlates with hepatic Fxr activity in both mice and humans
To further show that YAP activation inhibits Fxr transcriptional activity, YAP was removed from mouse hepatocytes by injecting an AAV-TBG-Cre into the YAPfl/fl mice (YAP∆Hep). Two weeks later, YAP transcriptional activity was reduced, while Fxr activity increased, as shown by Bsep and Shp expression (Fig. 4a, Supplementary Fig. 4a). We then determined whether YAP can regulate Fxr without the involvement of the gut-derived Fibroblast growth factor 15 (Fgf15)1. We cultured the MHOs with hepatocytes from the Fxr−/− mice (Fxr KO), or hepatocytes transduced by AAV-TBG-YapS127A (YAP*); or YAPfl/fl hepatocytes transduced by AAV-TBG-Cre (YAP KO). In the YAP* MHO, YAP activation phenocopied Fxr KO, in that it reduced Bsep and Shp expression, which was increased in the YAP KO MHO (Fig. 4b). In addition, when Fxr was activated by the agonist GW4064, or obeticholic acid (OCA)41, Bsep and Shp expression increased, which was blunted in the YAP* MHOs (Fig. 4c). Furthermore, BA production was suppressed by YAP activation, but increased by YAP KO (Fig. 4d). These results show that BA changes caused by Yap activation or loss can be recapitulated in vitro in MHOs, which have no connection to the gut where Fgf15 is produced. To further validate this conclusion, RNA-seq analyses of mouse MHOs were performed to test whether YAP inhibits expression of Fxr transcriptional target genes genome-wide. Gene-set enrichment analysis (GSEA) revealed that the Fxr target gene expression42,43 (Supplementary Data 1) was strongly enriched in control cells compared to YAP activation (Fig. 4e, Supplementary Fig. 4b), and in the YAP−/− cells, when compared with the control hepatocytes (Fig. 4f).
a–c qPCR analysis of Bsep and Shp expression (a) in the indicated liver samples, ***P = 0.0002, ***P = 0.0003 in sequence; (b) in the indicated mouse MHOs, ****P < 0.0001; (c) in MHOs were treated with 5 µM Fxr agonist GW4064 or OCA for 48 h, ****P < 0.0001. d Total BA concentration in the medium of MHOs, ****P < 0.0001, ***P = 0.0002. e, f Gene-set enrichment analyses (GSEA) for Fxr target gene expression by comparing TetO-YAP MHOs with and without Dox treatment (e), and control with YAP knockout MHOs (f). g qPCR analysis for BSEP and SHP expression in the human MHOs. Cells were treated for 48 h, with 3 µM TDI-011536 (TDI) to activate YAP, 5 µM GW4064 to activate FXR, and 0.5 µM Verteporfin (VP) to inhibit YAP-TEAD binding, **P = 0.0016 (BSEP), ***P = 0.0003 (SHP), ****P < 0.0001. h Total BA concentration in the medium of human MHOs with the indicated treatment, ****P < 0.0001, **P = 0.0028, ***P = 0.0003. i The predictions for HCC samples with ‘liver tumor YAP signature’ (red and blue) and ‘Hepatic FXR signature’ (orange and green) in LIRI-JP cohort (n = 236). Heatmap shows the expression of listed genes, r values determined by Pearson correlation comparing YAP activity with gene expression. j Expression of selected genes in YAP ‘high’ or ‘low’ groups of HCC samples from the cohort, n = 31 in YAP ‘high’ group, n = 68 in YAP ‘low’ group, box extends from the 25th to 75th percentiles, line in the middle of the box is plotted at the median, whiskers down to the minimum and up to the maximum value, and p-values were determined by two-sided unpaired t-test with Welch’s correction, ***P = 0.0001, *P = 0.026, ****P < 0.0001. k, l GSEA for Fxr target gene expression by comparing YAPHigh HCC samples with all non-tumor samples (k), YAPlow HCC with all non-tumor samples in the cohort (l). n = 4 independent experiments (a), n = 3 independent experiments (b−d, g, h), n = 3 biological replicates (e, f). Data are presented as Mean ± SD, p-values were determined by two-sided unpaired t-test (a). Data are presented as Mean ± SD, p-values were determined by 1-way ANOVA with Sidak’s test (b–d, g, h). Source data are provided as a Source Data file.
In hepatocytes, loss of the Hippo kinase Mst1/2 causes BA metabolic abnormalities by altering Fgf15 signaling44. Since Mst1/2 loss causes liver tumors by activating YAP/TAZ19, Mst1/2 might regulate BA metabolism via YAP. To test this, YAP was dose-dependently removed in the Mst1/2 double knock out (DKO) hepatocytes by AAV-TBG-Cre injection into the Mst1/2fl/fl (Mst1/2 DKO), Mst1/2fl/fl; Yap fl/+ (Mst1/2 DKO, YAP het) and Mst1/2fl/fl; Yap fl/fl mice (Mst1/2/YAP triple knock-out (TKO) (Supplementary Fig. 4c). Consistent with previous reports using the Alb-Cre to delete Mst1/245,46,47, Mst1/2 DKO mice showed hepatomegaly, with boosted serum BA levels and ALT activities (Supplementary Fig. 4d–g). Notably, the Mst1/2 DKO; YAP het mice showed largely restored phenotypes while the Mst1/2/YAP TKO mice showed complete phenotypic rescue (Supplementary Fig. 4d–h). Importantly, expression of the Fxr target genes was reduced in the Mst1/2 DKO mice and restored to control levels in the TKO mice (Supplementary Fig. 4i). These results demonstrate that, independently of Mst1/2 regulation of Fgf15 signaling, YAP activation plays a pivotal role in BA metabolism.
We then tested whether YAP activation in human hepatocytes also inhibits FXR. Human MHOs (hMHOs) were cultured and treated with the Lats kinase inhibitor TDI-011536 (TDI) to activate YAP48, or conversely, the Verteporfin (VP) to inhibit YAP49, while GW4064 was used to activate FXR. Again, YAP activation drastically suppressed expression of the FXR target genes, BSEP and SHP, with or without GW4064, while disruption of the YAP-TEAD interaction by VP treatment partially restored BSEP and SHP expression (Fig. 4g, Supplementary Fig. 4k). Consistently, both YAP activation and FXR activation reduced BA excretion to the medium (Fig. 4h). To test whether YAP inhibition of FXR is also found in human HCC, human liver tumors were assayed for YAP activity by a previously established gene set that tracks the ‘liver tumor YAP signature.’ In the LIRI-JP cohort (which contains transcriptomic profiling of HCC and normal liver samples retrieved from the International Cancer Genome Consortium data portal), this YAP-signature gene set accurately predicted YAP activity50, and negatively correlated with FXR activity20 shown by expression of “hepatic FXR target genes” (Supplementary Data 1). In our analysis, 14.3% (n = 30) of human HCC had high YAP activity, whereas 32% (n = 67) had low YAP activity. Indeed, expression of classical YAP target genes, such as CTGF, CYR61, and AMOTL2, positively correlated with the ‘liver tumor YAP signature’ and classical FXR target genes, such as SHP (NR0B2), BSEP (ABCB11), GLYCTK positively correlated with expression of “hepatic FXR target genes” (Fig. 4i). Notably, the false discovery rate (FDR) ranks of the ‘liver tumor YAP signature’ and activation of ‘hepatic FXR target genes’ (which rank the transcriptional activity of YAP and FXR, respectively) inversely correlated in both paired tumor and non-tumor samples (Fig. 4i, Supplementary Fig. 4l). Consistent with this observation, expression of FXR target genes, such as SHP, BSEP and GLYCTK, was higher in YAP-low tumors, while FXR expression was unchanged (Fig. 4j). In addition, GSEA analyses with the ‘hepatic FXR target genes’ showed that the ‘hepatic FXR target genes’ were strongly enriched in the YAP-low group (Fig. 4k, l, Supplementary Fig. 4m, n). These data show YAP may also inhibit FXR activities in human HCC as we found in mice.
YAP-Tead binds Fxr and inhibits its transcription activity
YAP regulates transcription mainly by binding to the DNA-binding Tead family of transcription factors51, and such YAP/TEAD-DNA interaction can also inhibit transcription52,53,54. Therefore, we hypothesized that YAP and/or Tead may interact with Fxr to inhibit its transcription activity. The published YAP and Tead4 Chromatin Immunoprecipitation Sequencing (ChIP-seq) data from the TetO-YAP mouse liver55 and Fxr ChIP-seq data from normal liver42 were retrieved and analyzed. Fxr and Tead4 both bind to DNA directly. Notably, in normal liver, only a small fraction of Tead4 and Fxr-binding peaks overlapped (Fig. 5a); however, three days after YAP activation, the number of overlapping Tead4 and Fxr binding peaks significantly increased (Fig. 5a). In addition, after YAP activation, YAP and Tead4 binding signals on the Fxr binding sites increased, suggesting that YAP activation might induce a YAP/Tead4/Fxr interaction (Fig. 5a, b). Fxr and YAP/Tead4 binding was also analyzed for the previously identified Shp enhancer56, Bsep promoter, and a putative Bsep enhancer, as well as transcription regulatory sequences of other Fxr target genes, such as Abcb4 and Slc51b (Fig. 5c, Supplementary Fig. 5a). In the WT liver, consistent with the genome-wide chromatin binding patterns of Fxr, YAP, and Tead4 (Fig. 5a, b), only Fxr, but not Tead4 or YAP, bound to these regions (Fig. 5c, Fig. S5a). Nevertheless, after YAP activation, both YAP and Tead4 gained access to them (Fig. 5c, Supplementary Fig. 5a). We then investigated whether YAP and Tead inhibit Fxr by competing with Fxr for chromatin binding using ChIP-PCR assays in mouse liver samples. After YAP activation, Tead4 binding increased on the Fxr binding regions, while Fxr binding to these regions was also increased (Fig. 5d). Therefore, it is unlikely that YAP-Tead4 competes with Fxr for DNA binding. ChIP-seq data analyses showed that the Tead4 DNA-binding element (TBE) was not required for Tead4/Fxr binding and, notably, the number and ratio of Tead4/Fxr co-binding peaks without TBE increased drastically in the YAP-activated liver (Fig. 5e). In addition, this type of peaks was also relatively abundant in human HepG2 cells (Supplementary Fig. 5b), suggesting Tead4/Fxr co-binding independent of TBE is common in both mouse and human cells. For example, the putative mouse Bsep enhancer contains a tandem, Fxr-binding motif (IR-1), without discernible TBE (Supplementary Fig. 5c). To test whether this putative enhancer regulates Bsep expression, this region was targeted in mouse MHOs by CRISPRi57, and Bsep expression was strongly inhibited, confirming the validity of the Bsep enhancer and the importance of Fxr in regulating Bsep expression (Fig. 5f). To investigate further how YAP and/or Tead regulate Fxr activity, a luciferase reporter was designed to track how the Bsep enhancer activity is regulated by FXR. In the Huh-7 cells, treatment with GW4064, strongly induced the Bsep reporter (3xIR1) activity (Fig. 5g), which was inhibited by activated YAP (YapS5A) and/or TEAD4 (Fig. 5g). Moreover, YAP5SA/S94A, which cannot interact with TEAD451, failed to suppress FXR transcriptional activity and abolished the additive suppressive effect with TEAD4 (Fig. 5g). These results indicate that TEAD4 critically inhibits FXR, and YAP inhibits FXR by binding to TEAD4. Moreover, while the full-length TEAD4 (FL-TEAD4) and the TEAD4 N-terminal domain (TEAD4 1-220; with the DNA-binding domain) inhibited FXR activity, the TEAD4 C-terminal domain (TEAD4 220-435) with the YAP-binding domain that acts as a dominant negative TEAD4 (dnTEAD4)58, further increased Fxr transcriptional activity, likely by reducing the YAP recruitment to FXR protein by endogenous TEADs (Fig. 5h).
a Venn diagrams showing the overlap of ChIP-seq peaks of Fxr and Tead4 in the indicated mouse liver. b Heatmaps summarizing the signal intensity of the Tead4 and Yap ChIP-seq on Fxr binding sites in the indicated mouse liver. c ChIP-seq signal tracks of the indicated gene locus in the indicated mouse liver. In the Bsep gene locus, the green-boxed region is the promoter, and the blue-boxed region is the putative enhancer. In the Shp gene locus, the green-boxed region is the enhancer. d ChIP-qPCR analysis of Fxr and Tead4 binding at the indicated gene locus in the freshly isolated hepatocytes, ****P < 0.0001. **P = 0.0057, ***P = 0.0006, **P = 0.0046 in sequence (Bsep promoter); **P = 0.0059, ***P = 0.0001 (Bsep enhancer); *P = 0.035, **P = 0.0024 (Shp enhancer). e Venn diagrams showing the total number of Tead4/Fxr co-binding of ChIP-seq peaks and the subset of peaks without TBE in mouse liver. f qRT-PCR analysis of Bsep expression in the mouse MHOs for enhancer validation using CRISPRi, data are presented as Mean ± SD, p-values were determined by two-sided unpaired t-test ***P = 0.0006. g–i Luciferase assays in the Huh-7 cells with the indicated reporters. All conditions used co-transfected with the Fxr expression plasmid, and GW4064 or VP treatment was 24 h, ****P < 0.0001. **P = 0.0015, ***P = 0.0009 (g); ***P = 0.0002 (h). n = 3 independent experiments (d, f, g–i). Data are presented as Mean ± SD, p-values were determined by 1-way ANOVA with Sidak’s test (d, g–i). Source data are provided as a Source Data file.
In our studies, the FXR transcriptional activity was more suppressed by FL-TEAD4 than TEAD4 1-220, and treatment with VP abolished this difference (Fig. 5h). Again, these results support that TEAD recruited YAP to inhibit FXR. Since no discernable TBE can be detected in the Bsep enhancer region, the TEAD-FXR binding must be distinct from TEAD-DNA binding. This hypothesis was tested with a TEAD4 DNA-binding mutant (TEAD4S100A/Q103A) with significantly reduced DNA binding ability59. Here, while as expected, YAP-induced TBE luciferase activity was drastically reduced by the mut TEAD4 (8xGTII)60 (Supplementary Fig. 5d), the mut TEAD4 showed similar suppression of Fxr compared to that of WT TEAD4 (Fig. 5i), indicating that DNA binding is not required for TEAD to inhibit FXR transcriptional activity.
We next investigated whether TEAD4 inhibits FXR by forming a protein complex. Overexpressed Fxr and TEAD4 co-immunoprecipitated (co-IP) (Fig. 6a, Supplementary Fig. S6a), and the TEAD4 N-terminal domain (1-220), but not the C-terminal domain (220-435), bound Fxr (Fig. 6b). Furthermore, in mouse primary hepatocytes with YAP activation, both Tead and YAP co-IPed Fxr from the nuclear extracts, suggesting Yap and Tead form a complex with Fxr to suppress Fxr transcriptional activity in vivo (Fig. 6c, Supplementary Fig. 6b).
a, b Immunoprecipitation of indicated proteins in Huh-7 cells. c Immunoprecipitation of indicated proteins in the nuclear-enriched fraction of mouse hepatocytes. d Immunoprecipitation of HA-FXR in the nuclear-enriched fraction of Huh-7 cells, TDI-011536 treatment was 24 h. e Immunoprecipitation of HA-FXR in the nuclear-enriched fraction of Huh-7 cells. Similar results were obtained from three independent experiments (a–e). Source data are provided as a Source Data file.
YAP recruits transcriptional repressors to inhibit FXR
YAP-TEAD might inhibit FXR transcriptional activity by recruiting the Nucleosome Remodeling and Deacetylase (NuRD) complex52,53,54. Since Fxr promotes Shp expression by interacting with p300 acyltransferase61, YAP-TEAD may inhibit FXR by inducing a switch from a FXR-P300 transcriptional activator complex(es) to a FXR-TEAD-YAP-NuRD transcriptional inhibitor complex, which includes type I HDACs (HDAC1/2). We performed luciferase assays of the Bsep enhancer reporter in the Huh-7 cells, with a P300/CBP inhibitor, A48562 or a HDAC1/2 inhibitor, Entinostat63. P300 inhibition by A485 led to a dose-dependent decrease in FXR transcriptional activity (Supplementary Fig. 6c), while HDAC1 inhibition by Entinostat not only elevated the baseline level of GW4064-induced FXR transcriptional activity, but also partially abolished FXR inhibition by YAP-TEAD. These results suggest that HDAC1/2 mediates the YAP-TEAD inhibition of FXR transcriptional activity, which is promoted by P300/CBP.
To determine the mechanism through which YAP-TEAD disrupted the FXR/p300 interaction, the FXR complex in the nuclei-enriched fraction from Huh-7 cells was analyzed via co-IP. In a YAP-dose dependent manner, YAP activation increased FXR binding to the NuRD core components, including HDAC1, MTA1 and CHD4, at the expense of p300 (Fig. 6d). Conversely, disrupting the YAP/TEAD interaction with VP treatment or YAP5SA/S94A expression enhanced co-IP of FXR-p300 but reduced co-IP of FXR-YAP/HDAC1/MTA1/CHD4 (Fig. 6e, Supplementary Fig. 6d-e). These data indicate that TEAD recruits YAP to inhibit FXR transcriptional activity by recruiting the NuRD complex to replace P300.
YAP-induced hepatic tumor formation is inhibited by Fxr activation or by inhibition of YAP-mediated transcriptional repression
To test whether YAP-induced BA metabolic defects are indeed critical for HCC formation due to Fxr inhibition, YAP* mice were fed with GW4064 and/or Entinostat (Fig. 7a). GW4064 treatment reduced liver and serum BA levels, ALT activities, inflammation, and fibrosis, as well as YAP*-induced hepatomegaly and tumor formation (Fig. 7b–i, Supplementary Fig. 7a–c). YAP activity and the BA concentrations in the serum and liver were also reduced (Fig. 7e, Supplementary Fig. 7c–e). Entinostat treatment alone had a limited effect in reducing serum BA levels or in activating Fxr (Fig. 7e, f), but a combination of Entinostat and GW4064 further enhanced the effect of GW4064 in reducing liver abnormalities and preventing HCC formation induced by YAP activation (Fig. 7b–i, Supplementary Fig. 7a, b). Combined GW4064 and Entinostat treatment further reduced BA concentrations (Fig. 7e, Supplementary Fig 7c–e). To test further whether GW4064 and Entinostat act similarly in human hepatocytes, YAP-activated hMHOs were treated with GW4064 and/or Entinostat (Fig. 7j). Consistent with data in mice, Entinostat treatment alone had limited effect in the hMHOs in regulating FXR activity and BA export (Fig. 7k–o), whereas the combination of GW4064 with Entinostat enhanced GW4064-induced FXR activation, BA export, and reduction of YAP activity (Fig. 7k–o). Taken together, these data show that Fxr inhibition by the inhibitory role of activated YAP plays a major role in HCC formation. FXR activation and inhibition of HDAC1/2-mediated FXR suppression provide novel insights into suppressing YAP-induced liver tumorigenesis.
a Schematic diagram of the experimental procedure. b Representative liver images in the indicated mice. c Liver/body weight ratios of the indicated mice, ****P < 0.0001, **P = 0.0023. d Serum ALT of the indicated mice, ****P < 0.0001, *P = 0.04. e Serum BA concentration of the indicated mice, ****P < 0.0001. *P = 0.026, *P = 0.047 in sequence. f–h qRT-PCR analysis of indicated genes in the liver tissue, ****P < 0.0001. **P = 0.017, **P = 0.023, ***P = 0.0003 in sequence (Ctgf, g); **P = 0.0036 (Cyr61, g); ***P = 0.0003, **P = 0.0017, *P = 0.049 (Col1a1, g); **P = 0.0089 (Acta2, g); ***P = 0.0003, *P = 0.041 (Ccl2, h); **P = 0.001, *P = 0.012 (Il1b, h); ***P = 0.0005 (Tnf, h). i Representative immunofluorescent images of the indicated liver samples. j Schematic diagram of the experimental procedure. k, l Intracellular (k), and medium (l) BA concentrations in hMHOs after treatment, ****P < 0.0001. ***P = 0.0002 (k); ***P = 0.0002, *P = 0.034, ***P = 0.0002 in sequence (l). m–o qRT-PCR analyses of hMHOs after treatment, ****P < 0.0001. **P = 0.0019, *P = 0.047 (CYP7A1, m); **P = 0.0087 (CYP8B1, m); **P = 0.0049 (CTGF, n); **P = 0.0013, **P = 0.0062 in sequence (CYR61, n); *P = 0.017 (SHP, o); *P = 0.0008 (BSEP, o). Scale bars: 1 cm (b), 100 μm (i). n = 5 biological replicates in each group (c–e), n = 3 biological replicates in each group (f–h), n = 3 independent experiments (k–o). Data are presented as Mean ± SD, p values were determined by 1-way ANOVA with Sidak’s test (c–h, k–o). Similar results were obtained from three independent experiments (i). Source data are provided as a Source Data file. Image in (a, j) was created in BioRender (Jin, Y. (2025) https://BioRender.com/h79u623).
FXR activation inhibits YAP
Reduction of TBE-dependent YAP transcriptional activator function by treatment with GW4064 and Entinostat raises the possibility that FXR activation may convert YAP from a transcriptional activator to a transcriptional repressor in the canonical YAP-TEAD complex that promotes cell proliferation and survival by binding TBE. To test this hypothesis, a TBE luciferase assay (8xGTII) monitored YAP-TEAD-dependent transcriptional activation, showing YAP-activated TEAD transcriptional activity was indeed inhibited by activated FXR (Fig. 8a). Since TEAD DNA-binding activity is not required for FXR transcriptional suppression (Fig. 5i), the DNA-binding-deficient TEAD4S100A/Q103A was tested for its ability to form a complex with HDAC1. Notably, compared with the TEAD4, the TEAD4S100A/Q103A enhanced YAP-TEAD4-HDAC1 complex formation (Fig. 8b). In addition, co-expression of TBE reduced YAP/TEAD4/HDAC1 binding to FXR (Fig. 8c), suggesting TBE also binds the YAP/TEAD4/HDAC1 complex and might compete with FXR binding sites. To investigate the relationship between the TBE-bound, YAP-TEAD-P300 activator and the YAP-TEAD-HDAC1 repressor, co-IP assays were performed in the Huh-7 cells to evaluate YAP transcriptional regulation by FXR. Consistent with our data showing FXR activation inhibits the TBE-dependent canonical transcriptional activation by YAP-TEAD (Fig. 8a), in the FXR complex, GW4064 drastically reduced FXR binding to YAP, pan-TEAD, and HDAC1, but increased binding to P300 (Fig. 8d). Nevertheless, in the YAP or TEAD TBE complex, GW4064 treatment reduced P300, TEAD, and YAP binding, suggesting FXR activation by GW4064 reduced the YAP-TEAD interaction as well as YAP-TEAD activation by P300 when bound to TBE (Fig. 8d). To further test whether reduced YAP-TEAD activation is caused by increased HDAC1 binding to the TBE, we performed ChIP-PCR for TEAD4 and HDAC1 binding to the 8X TBE site using the 8xGTII plasmid. While GW4064 did not change the YAP5SA-induced increase in TEAD4 binding to the 8X TBE site, we observed that YAP5SA expression resulted in a significant reduction in HDAC1 binding to the 8X TBE site, which was reversed by GW4064 treatment (Fig. 8e). This observation was confirmed in mouse liver samples (Fig. 8f, Supplementary Fig. 8a), revealing an important scenario in BA metabolic regulation—elevated BA activates both FXR and YAP, which play interactive and antagonistic roles in BA metabolism, liver injury and growth. While YAP activation inhibits FXR, activated FXR also converts YAP from a transcriptional activator to a transcriptional repressor. Therefore, in the absence of Fxr, loss of Fxr activation and elevated BA levels may promote YAP activation and accelerate HCC formation. To test this, YAP was activated in hepatocytes of the Fxr KO mice by injecting them with AAV-TBG-YAPS127A (Fig. 8g, Supplementary Fig. 8b). Three weeks after YAP activation, WT control mice showed no change in liver-to-body-weight ratios (Fig. 8g). Strikingly, YAP activation in the Fxr KO mice induced severe hepatomegaly, steatosis, uneven and overcrowded hepatocytes, and increased cell death, fibrosis, and inflammation, with drastically increased serum BA levels and ALT activities, suggesting severe cholestasis (Fig. 8h–j, and Supplementary Fig. 8c–g). Notably, these defects were similar but more severe than those in the Fxr KO mice fed with excessive BAs16,64. Consistently, YAP transcription co-activator activity was increased, as indicated by the expression of multiple YAP target genes in the Fxr KO + YAP* liver (Fig. 8k)65,66, showing Fxr activation inhibits YAP transcription activator function in hepatocytes.
a Luciferase activity assay in Huh-7 cells, tracking the canonical YAP-TEAD transcription activator activity. 5 µM GW4064 treatment was 24 h, ****P < 0.0001. b Immunoprecipitation of HA-FXR in the nuclear-enriched fraction of Huh-7 cells. c Immunoprecipitation of HA-FXR in the nuclear-enriched fraction of Huh-7 cells, 8xGTII and 3xIR1 plasmids were co-transfected with FXR and TEAD4 plasmids, as indicated. d Immunoprecipitation of FXR, YAP and pan-TEAD in the nuclear-enriched fraction of Huh-7 cells. 5 µM GW4064 treatment was 24 h. e ChIP-qPCR analysis of TEAD4 and HDAC1 binding at the 8xGTII TBE sites in the Huh-7 cells. 8xGTII plasmid was co-transfected with GFP or Flag-YAP5SA plasmids, as indicated. 5 µM GW4064 treatment was 24 h, ****P < 0.0001. f Immunoprecipitation of indicated proteins in the nuclear-enriched fraction of liver tissues. g Representative images of liver from the indicated mice. h Liver/body-weight ratios, ****P < 0.0001. i Serum BA concentrations, ****P < 0.0001. j Serum ALT levels, ****P < 0.0001. k–m qRT-PCR analysis of indicated genes in the liver tissue, ****P < 0.0001. **P = 0.0012, ***P = 0.0009 (Ctgf, k); **P = 0.006, ***P = 0.0005 (Cyr61, k); **P = 0.007 (Klf6, k); ***P = 0.0002 (Fos, k); *P = 0.042, ***P = 0.0002, ***P = 0.0005 in sequence (Cyp7a1, i); *P = 0.038, *P = 0.018, ***P = 0.0001, **P = 0.005 in sequence (Cyp8b1, i); **P = 0.004, ***P = 0.0005, **P = 0.0017 in sequence (Cyp7b1, i); **P = 0.0035, ***P = 0.0006 (Cyp27a1, i); *P = 0.016 (Bsep, m). n, o Luciferase activity assay in primary mouse hepatocytes with indicated genotype, tracking the canonical YAP-Tead transcription activity (n), and Fxr transcription activity (o), ****P < 0.0001, ***P = 0.0001 (n). 5 µM GW4064 and 3 µM TDI treatment was 24 h. Scale bars: 1 cm (g). n = 3 independent experiments (a, e, n, o), n = 5 biological replicates in each group (h–j), n = 3 biological replicates in each group (k–m). Data are presented as Mean ± SD, p-values were determined by 1-way ANOVA with Sidak’s test (a, e, h–o). Similar results were obtained from three independent experiments (b–d, f). Source data are provided as a Source Data file.
To further investigate the change of BA metabolism in these mice, we performed qRT-PCR analysis of Fxr target genes and BA-related gene expression (Fig. 8l, m). Indeed, Cyp7a1 and Cyp8b1 levels were upregulated, while expression of the Fxr target genes Shp and Bsep was suppressed in the YAP* and Fxr KO mice at a similar level (Fig. 8l, m). Strikingly, the Fxr KO + YAP* liver exhibited significantly higher expression of Cyp7a1 and Cyp8b1, while Shp and Bsep expression was lower (Fig. 8l, m), suggesting that in addition to directly suppressing Fxr transcriptional activity, YAP may play additional roles in regulating BA synthesis and export. To further understand the changes in the Fxr KO + YAP* livers, we dissect the canonical YAP-Tead activity and the IR-1 motif-dependent Fxr activity using luciferase reporter assay in the Fxr+/− and Fxr−/− primary hepatocytes. Consistently, after YAP activation, YAP activity was higher in the Fxr−/− compared to the Fxr+/− hepatocytes, and GW4064-induced suppression of YAP activity was abolished in the absence of Fxr (Fig. 8n). Similarly, GW4064-induced Fxr transcriptional activity in the Fxr+/− hepatocytes was suppressed by TDI treatment, while the effects of GW4064 and TDI on Fxr reporter activity were completely abolished in the Fxr−/− hepatocytes (Fig. 8o). This data suggests that the suppressive role of YAP on the IR1-dependent transcriptional activity is entirely dependent on the presence of Fxr. Further inhibition of Fxr target gene expression by YAP in vivo is likely mediated by Fxr-independent mechanisms.
Discussion
In this study, we showed that YAP directly controls a unique and key metabolic function of the liver—BA production and exportation—by inhibiting Fxr. We further identified a transcriptional repressor function of YAP that is responsible for liver tumorigenesis. Furthermore, transcriptional activation by YAP-Tead binding to TBE can also be reprogrammed by activated Fxr in hepatocytes. All these findings show that YAP-induced liver tumor formation can be alleviated by pharmacological Fxr activation, or suppression of HDAC1-mediated Fxr inhibition, or genetically restoring Fxr-controlled BA export or reduction of serum BAs by feeding the mice with BA-absorbing resin. Our results show BA metabolism is critically controlled by a reciprocal and repressive epigenetic reprogramming of YAP and Fxr. Therefore, BA metabolism can be therapeutically targeted for various liver diseases caused by YAP abnormality.
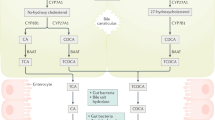
Significant risk factors for HCC include cholestasis in CLD and cirrhosis, in which persistent liver injury and inflammation are observed3,5,67. A recently published study demonstrated that resin feeding drastically reduced HCC formation in the Fxr; Shp DKO mice40. We showed previously that similar to cholestatic liver defects, YAP activation in hepatocytes induces severe liver fibrosis and inflammation, which contributes to liver tumor formation18,19,20. Our findings of YAP-regulated BA metabolism further reveal a direct regulation of liver injury and inflammation by YAP-induced BA accumulation in hepatocytes due to Fxr inhibition. Epigenetic Fxr inhibition by YAP is mediated by Tead-dependent protein binding, not Tead-dependent DNA binding as previously shown for YAP-regulated cell proliferation, differentiation and survival52,53. BAs are known to promote both liver regeneration and carcinogenesis15,68,79,70, and BAs can promote YAP activation through IQGAP1 mediated cytoskeleton remodeling or via S1PR2 mediated G-protein-coupled receptor signaling15,29. Moreover, YAP activation is also required for cholestasis-induced HCC, suggesting YAP activation is both a key inducer, and a key responder of cholestasis in HCC induction. Our studies here show that defects in BA export due to Fxr inhibition, are key mediators of YAP activation in HCC induction. YAP, Fxr, and BAs comprise a dynamic regulatory network (Fig. 9a), the breakdown of which mediates and amplifies liver disorders, including HCC. Indeed, curtailing BA production and promoting BA export by activating Fxr and/or relieving YAP inhibition of Fxr via Hdac1/2, or restoring expression of Bsep, an Fxr target gene that promotes BA export from hepatocytes, alleviated tumor formation caused by YAP activation, further demonstrating the key function of BA intracellular accumulation in YAP-induced HCC.
a YAP regulation of Fxr activity and BA metabolism. A proposed model for YAP, Fxr, and BAs forms a dynamic regulatory network. YAP activation inhibits Fxr transcriptional activity, causing BA accumulation in hepatocytes by downregulating Bsep expression. Pharmacological activation of Fxr by GW4064 not only curtails BA production and promotes BA exportation but also directly inhibits YAP activity, alleviating tumor formation caused by YAP activation. b Dynamic regulation of YAP and FXR transcriptional activity by P300 and HDAC1. Pharmacological activation of FXR by GW4064 relieves YAP inhibition of FXR by recruiting P300 and inhibiting HDAC1 binding to FXR. Meanwhile, it promotes the repressor function of the DNA-binding YAP-TEAD complex, converting it from a transcriptional activator to a transcriptional repressor.
Since YAP and BA metabolism can be regulated by many factors, breaking the positive feedback loop between YAP activation and BA accumulation may represent an effective strategy to reduce liver fibrosis, inflammation, and tumorigenesis in many different contexts. Notably, in cholangiocytes, YAP is necessary for bile duct maintenance65,71 and previous research with YAP deletion in both hepatocytes and cholangiocytes showed induction of cholestasis due to bile duct disruption71, making it difficult to pinpoint the role of hepatic YAP in BA metabolism. This study used hepatocyte-specific AAV-TBG-Cre to manipulate YAP activity, which allowed us to identify the central role of hepatic YAP in BA metabolism. It is important to note that YAP activity is normally suppressed in hepatocytes but activated in cholangiocytes65, while Fxr is only functional in hepatocytes. The strong inhibitory role of Yap activation imposed on Fxr explains the need to keep YAP activity low in hepatocytes under normal conditions, to allow Fxr to be promptly activated to regulate BA metabolism.
The inhibitory role of Fxr in HCC mainly depends on the hepatic modulation of BA homeostasis16. The liver-specific Fxr knockout mice are extremely susceptible to BA-feeding-induced liver injury and liver carcinogenesis64, indicating that the liver requires Fxr to clear excessive BAs. Interestingly, our Fxr−/− mice were also extremely susceptible to HCC induction by YAP activation, with more severe cholestasis, inflammation, liver damage, and faster tumor growth. Importantly, YAP in these mice may play additional Fxr-independent roles in regulating BA metabolism. Previous reports also showed that YAP can induce epigenetic reprogramming both as a transcriptional co-activator and co-repressor in the TBE-dependent YAP-TEAD transcription52,53,72,73, which drives liver inflammation and overgrowth18,19,49,74. The DNA binding YAP-TEAD complex can act as a transcriptional repressor in various contexts, including tumors, by recruiting the NuRD complex52,53,54 or directly competing with other transcription-activating factors75,76. In these contexts, TEAD DNA binding is required for transcriptional repression functions. In this study, FXR activation also promoted the repressor function of DNA-bound YAP-TEAD. FXR activation by GW4064, while relieving YAP inhibition of FXR, reprogrammed the DNA-binding YAP-TEAD complex from transcriptional activator to transcriptional repressor. Importantly, we also identified a TEAD-DNA-binding-independent mechanism of YAP-TEAD repression of FXR. Interestingly, specific mutations that abolished TEAD4 DNA binding, further enhanced its repression of FXR transcriptional activity, suggesting competition between FXR and TBE (protein-DNA interaction) for TEAD-mediated transcription inhibition (Fig. 9a, b). Such competition may be an underlying mechanism for enhanced YAP transcription activity and therefore tumor formation in the Fxr−/− mutant liver15. Interestingly, the Tead4/Fxr interaction is weak in normal mouse hepatocytes but drastically enhanced by hepatic YAP activation. This change in the Tead4 binding pattern may be due to increased YAP-Tead4 complex binding to Fxr. Taken together, Fxr activation by GW4064 treatment in mice reduced YAP-induced HCC by restoring both BA metabolism and YAP inhibition, therefore modulating Fxr activation by BAs and/or HDAC1/2/3 inhibitors may represent novel strategies to reduce HCC formation. A previous study showed the tumor-suppressive role of the Fgf15-Fgfr4-Mst1/2 pathway44. The direct, Mst1/2-independent role of YAP in regulating BA metabolism by reprogramming Fxr identified here suggests BA metabolism is regulated in multiple ways.
TCA and TβMCA were the major BAs that accumulated in large quantities in the liver and serum after YAP hepatic activation. TCA and TβMCA play distinct roles in the BA-YAP feedback loop. TCA is oncogenic in hepatic Fxr knockout mice16, and promotes tumor progression by activating S1PR2-YAP signaling29,69,70. In our study, TCA induced YAP activation in MHOs. Unlike TCA, which is an FXR agonist, TβMCA is a natural Fxr antagonist31 that in our studies did indeed suppress Fxr activity. High serum levels of TβMCA might further suppress Fxr in hepatocytes and in gut epithelial cells, to worsen liver cholestasis. Of note, TβMCA is a mouse-specific BA, although human patients may have other BAs with similar functions. Considering that human HCC is more complex, further translational and clinical studies using human HCC samples are needed to understand BA metabolism in YAP-activated human livers or organoids. A recently published paper identified a subset of human hepatoblastoma cells that are hypersensitive to HDAC1/2 inhibition77, suggesting that HDAC1/2 represents a promising target for treating poorly differentiated liver tumors. Given that YAP-activated HCC is also poorly differentiated20, a combination of Fxr activation and HDAC1/2 inhibition may be a potent therapeutic strategy for this subtype of HCC.
Methods
Mice
Animal protocols and procedures were approved by the Harvard Medical School Institutional Animal Care and Use Committee (HMS-IACUC). Mice were housed in a pathogen-free facility in a 12 h light/dark cycle. All mice were fed ad libitum with LabDiet® 5053. In the in vivo experiments, the maximal tumour burden was permitted by HMS-IACUC and the maximal tumour burden in the experiments was not exceeded the standard. The timeline of liver tumor formation and liver enlargement has been described previously20. In this study, the endpoints chosen for the current mouse models represent the minimum duration required to observe moderate liver enlargement and tumor formation. No mice developed severe visible illness that met the veterinarian’s criteria for euthanasia at the endpoint. All mice were sacrificed between Zeitgeber Time (ZT) 5–8. Both male and female mice were used in the experiments. The source of the mice is provided: Rosa26lox-stop-lox-rtTA/+; Col1a1Teto-YapS127A/+ (TetO-YAP) mice were obtained from Dr. Fernando Camargo’s lab23. Other mice were procured from the Jackson Laboratory: The Fxr−/− mice (JAX: 004144), B6J mice (JAX: 000664), Mst1fl/fl; Mst2fl/fl mice (JAX: 017635), Sox9fl/fl (JAX: 013106) and Yapfl/fl (JAX: 027929)
AAV injection
AAV viruses were diluted in sterile PBS, 1 × 1011 genome copies (GC) of each AAV were administered to 3-week old mice via retro-orbital injection at a volume of 30 μl. pAAV-TBG-Cre (Addgene, #107787) and pAAV-TBG-Null (Addgene, #105536) were purchased from Addgene. The pAAV-TBG-YAPS127A and pAAV-TBG-Bsep plasmids were generated in our lab. All AAVs were produced in the lab. For YAPS127A overexpression in the TetO-YAP mice, 1 x 1011 GC of AAV-TBG-Cre were administered to 8-week old TetO-YAP mice, two weeks later, YAPS127A expression was induced in the mice by Dox water (0.2 g/L) feeding.
RNA isolation, reverse transcription, and quantitative PCR
RNA from liver tissue was isolated using the TRI Reagent (Sigma-Aldrich) according to manufacturers’ instruction, cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Cwbio) subsequently. RNA was isolated from organoid pellets using the PureLink RNA mini kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNA was synthesized from RNA using the SuperScript IV VILO Master Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. qPCR was performed using UltraSYBR Mixture (Cwbio) on the One-Step Plus qPCR System (Thermo Fisher Scientific). The gene expression data were normalized to Gapdh (mouse) or GAPDH (Human) and presented as mRNA fold change using the 2 − ΔΔCt method. Primers is provided in the Supplementary Data 2.
Immunohistochemistry
Mouse liver was perfused with ice-cold PBS and cut into small pieces in ice-cold PBS. For frozen sections, the liver tissue was fixed in 4% paraformaldehyde for 12 h, and then processed according to standard procedures. 10 µm Sections were collected on slides, and immunohistochemistry was performed according to standard procedures. Briefly, Sections were washed in 0.05% Triton/PBS (PBST), followed by incubation with the blocking buffer (5% Donkey serum/PBST) for 1 h at room temperature and then incubated overnight at 4 °C with diluted primary antibodies in the blocking buffer. Then the sections washed with PBST and incubated with diluted secondary antibodies in the blocking buffer for 1 h at room temperature. After incubation, the sections were washed with PBST and mounted with Fluoroshield™ with DAPI (Sigma-Aldrich) mounting medium. The following antibodies were used for immunostaining: F4/80 (MCA497, AbD Serotec). ACTA2 (A17910, ABclonal), donkey anti-rat IgG Alexa Fluor 647 (A48272, Invitrogen) and donkey anti-rabbit IgG Alexa Fluor 568 (A10042, Invitrogen) Images were captured on a Leica DM6 confocal microscope.
H&E staining and Sirius red staining
Fixed liver tissues were dehydrated, embedded in paraffin, and sectioned at 6μm for Hematoxylin and Eosin (H&E) staining and Sirius red staining. Paraffin sections were rehydrated before staining. For H&E staining, tissue sections were stained with Mayer’s Hematoxylin solution (G-Biosciences) and Eosin staining solution (Thermo Fisher Scientific) subsequently. For Sirius red staining, Sections were incubated in Sirius staining solution (0.1% Direct Red 80 and 1.3% picric acid in distilled water) for 1 hour then washed in two changes of acidified water (0.5% acetic acid in distilled water). The stained sections were dehydrated and mounted for imaging. Images were captured using a Keyence BZ-X710 microscope.
Biochemical assays in the serum and cell culture medium
Total bile acid in the serum, tissue, and media was measured using the Total Bile Acids Assay kit (Diazyme) according to the manufacturer’s instructions. ALT Activity in the serum was measured using the Assay kit (Sigma-Aldrich), according to the manufacturer’s instructions. BAs in the media were concentrated by mixing 100 µL of media with 1 mL of chloroform: methanol (1:1). The mixture was vortexed for 15 s, sonicated for 10 min, and then centrifuged at 8000 g for 10 min. The supernatant was dried and resolved in 10 µL of methanol. To extract BAs in the HOs and liver tissues, the samples were weighed and digested with proteinase K (1 mg/mL) for 3 h at 50 °C. After digestion, the samples were centrifuged at 8000 g for 10 min. The BAs in the supernatant were extracted and partitioned by adding 3.3 volumes of chloroform: methanol (1:1) and resolved in 10-100 µL of methanol.
Bile acid analysis by LC-MS
The sample preparation and BA analysis by LC-MS were performed by the McGuire VA Medical Center LC-MS Core Lab. At least three biological replicates were used in each experiment group. Serum and liver tissues were processed for bile acid analysis as described previously78. High-throughput profiling of BAs was conducted using a Shimadzu LC–MS 8600 system. BA metabolite data in the original scale in terms of raw area counts were normalized by median centering. Missing values were imputed with the lower limit of detection for a given metabolite. When ≥ 30% of the values were missing, data were excluded from analyses. The peak and area under the curve for individual analytes were related to validated library standards to compute BA levels, which are read out as related to these library standards. To construct the calibration curves, 10 μL of each standard solution was mixed with 20 μL of IS solution (a mixture of 10 stable isotope-labeled BAs: d4-CA, d4-GCA, d4-TCA, d4-CDCA, d4-DCA, d4-GCDCA, d4-TCDCA, d4-LCA, d4-GLCA, d4-TLCA at 0.5 µM/each BA in acetonitrile/methanol/water (25/25/50, v/v/v)) and diluted to 400 µL with acetonitrile/methanol/water (25/25/50, v/v/v). A 2 μL aliquot was injected into the LC/MS/MS system. For quantification of bile acids in the liver, 20 mg of liver tissue was homogenized with 500 µL of acetonitrile/methanol/water (25/25/50, v/v/v) using a issue Homogenizer. After centrifugation, 10 µL supernatant was spiked with IS solution (20 µL, 10 pmol). The mixture was diluted to 400 µL with methanol/acetonitrile/water (25/25/50, v/v/v). For the serum specimen, 10 µL was spiked with IS solution (20 µL, 10 pmol), and the mixture was diluted with 250 µl of acetonitrile/methanol (1:1, v/v). After centrifugation at 12,000 g for 2 min at room temperature, the supernatant (200 µL) was diluted with water (200 µL). All samples were filtered through 0.2 µm PTFE membrane, and 2 μL aliquots were injected into the LC/MS/MS system. The Shimadzu LCMS-8600 CL liquid chromatography triple-quadrupole tandem mass spectrometer equipped with a dual ion source (DUIS) interface was used. Data were collected and processed using Lab Solutions software. Normalized data is provided in the source data file.
TUNEL assay
The TUNEL assay was performed using the Click-iT™ Plus TUNEL Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Mouse Hepatocyte isolation
Two-step liver perfusion was performed. Briefly, the mouse heart and liver were surgically exposed after anesthesia, a 25 G infusion set (Terumo) was immediately inserted into the right atrium, and the portal vein was cut with scissors at the same time. Mouse livers were then perfused sequentially with pre-warmed (37 °C) liver perfusion medium (Thermo Fisher Scientific) at a flow rate of 3 ml/min for 2 min, and pre-warmed (37 °C) digestion solution (0.75 mg/ml collagenase type I in DMEM; Thermo Fisher Scientific) was perfused at the same flow rate for 3 min. During the perfusion, the portal vein was manually occluded by cotton tips every 30 s for 10 s. After perfusion, the liver was surgically removed and cut into small pieces in ice-cold DMEM, and hepatocytes from the digested tissue were released into the medium by gentle pipetting. Next, hepatocytes were passed through a 70μm cell strainer (Corning) and centrifuged at 50 g for 3 min. The viable hepatocytes were purified using 40% Percoll in PBS (Cytiva) and centrifuged at 150 g for 5 min.
Human and mouse MHO culture
Mouse and human MHO culture was performed as previously described in ref. 31. Briefly, freshly isolated mouse hepatocytes were embedded in the Cultrex BME, Type R1 (R&D Systems for mouse HO generation. Mouse HOs were maintained in the +GF medium. The +GF medium for mouse HO culture contained Advanced DMEM/F12 (Thermo Fisher Scientific) with B27 minus vitamin A (Thermo Fisher Scientific), 10 mM HEPES (Thermo Fisher Scientific), GlutaMAX (Thermo Fisher Scientific), 50 μg/ml Primocin (InvivoGen), 15% RSPO1-conditioned medium (made in the lab using Cultrex HA-R-Spondin1-Fc 293 T cells (Bio-Techne) according to the manufacturer’s instructions), 3 μM CHIR-99021 (MCE), 50 ng/ml EGF (Biolegend), 25 ng/ml HGF (Biolegend), 50 ng/ml FGF7 (Peprotech), 50 ng/ml FGF10 (Biolegend), 1 μM A83-01 (Sigma), 5 μM ROCK inhibitor Y-27632 (MCE), 10 nM gastrin (Sigma), 10 mM nicotinamide (Sigma), and 1 mM N-acetylcysteine (Sigma). The cryopreserved human fetal HOs were retrieved, embedded in the Cultrex BME, Type R1 and maintained in the +GF medium. The +GF medium for human HO culture contained Advanced DMEM/F12 (Thermo Fisher Scientific) with B27 minus vitamin A (Thermo Fisher Scientific), 10 mM HEPES (Thermo Fisher Scientific), GlutaMAX (Thermo Fisher Scientific), 50 μg/ml Primocin (InvivoGen), 15% RSPO1-conditioned medium (home-made), 3 μM CHIR-99021 (MCE), 50 ng/mlEGF (Biolegend), 50 ng/ml HGF (Biolegend), 50 ng/ml FGF7 (Peprotech), 50 ng/ml FGF10 (Biolegend), 20 ng/ml TGFα (BioLegend), 1 μM A83-01(Sigma), 5 μM ROCK inhibitor Y-27632 (MCE), 10 nM gastrin (Sigma), 2.5 mM nicotinamide (Sigma) and 1.25 mM N-acetylcysteine (Sigma). Before the induction of MHOs, the HO cultures underwent one passage to enrich well-formed HOs. HOs were retrieved from the BME using Cultrex Organoid Harvesting Solution (R&D Systems). Well-formed organoids were picked from the bottom of a well and further washed in Advanced DMEM-F12 and re-embedded into the BME for subsequent culture. The cystic bile duct organoids were hand-picked or further removed by 36% Percoll (Cytiva) by centrifugation at 150 g for 5 min during passage. The +GF medium was switched to the maturation medium (RGF medium) for the induction of MHOs. The basal medium for human and mouse HO maturation contained William’s E Medium (Thermo Fisher Scientific) with B27 minus vitamin A (Thermo Fisher Scientific), 10 mM HEPES (Thermo FisherScientific), GlutaMAX (Thermo Fisher Scientific), penicillin-streptomycin (Thermo Fisher Scientific), 50 μg/ml Primocin (InvivoGen), 15% RSPO1-conditioned medium (home-made), 3 μM CHIR-99021 (MCE), 1 μM A83-01 (MCE), 5 μMROCK inhibitor Y-27632 (MCE), 10 nM gastrin (Sigma), 2.5 mM nicotinamide (Sigma) and 1.25 mM N-acetylcysteine (Sigma). For maturation of mouse HOs, the medium used was RGF with 10 nM T3 (Sigma).
The fetal human HOs (fhHOs) used in this study were cryo-recovered from previously generated fhHOs derived from human fetal hepatoblasts, as described in our previous publication30. The human fetal hepatoblasts used for fhHO generation were obtained from Dr. Lishan Su’s lab at the University of Maryland School of Medicine. The project was reviewed by the University of Maryland’s Office of Human Research Ethics, which determined that the submission does not constitute human subjects research, as defined under federal regulations [45 CFR 46.102(d or f) and 21 CFR 56.102(c), (e), and (l)]. For maturation fhHOs, the media used was RGF with 20 nM T3 and 500 nM DEX. For both HO maintenance and maturation, the media were changed every 3 days.
AAV-TBG-Cre was mixed with hepatocytes in BME at a concentration of 1 x 1010 genome copies (GC)/ml before seeding to induced conditional knockout in the HOs. To induce YAPS127A expression in the TetO-YAP cells, doxycycline (Sigma) was added to the culture medium at a concentration of 200 ng/ml.
Cell culture and transfection
The Huh-7 cells were obtained from JCRB cell bank (Cat: JCRB0403) and maintained in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS, L-glutamine (Thermo Fisher Scientific), and 100 U/ml penicillin-streptomycin (Thermo Fisher Scientific). Plasmid transfection was performed using the PEI MAX Reagent (Polysciences) according to the manufacturer’s instructions.
Western blotting
RIPA buffer containing Protease Inhibitor Cocktail (PIC) and PhosSTOP (Roche) was used to harvest whole tissue and cell lysates. Protein concentrations were quantified by the BCA protein assay kit (Thermo Fisher Scientific). Equal amounts of protein were resolved by SDS-PAGE and further transferred to the Nitrocellulose Blotting Membrane (Cytiva). Target proteins were detected with the SuperSignal West Pico Plus Chemiluminescent Substrate (Thermo Fisher Scientific) using the PXi4 Chemiluminescent Imaging System (Syngene).
Immunoprecipitation
To prepare the whole cell lysates, immunoprecipitation buffer (20 mM Tris-HCl pH 8137 mM NaCl, 1% Nonidet P-40 (NP-40), and 2 mM EDTA) containing PIC and PhosSTOP (Roche) was used to harvest the cells. After a brief sonication at a low power setting (one second x 4), cell debris was removed from the lysates by centrifugation at 4 °C. To prepare the nuclear lysates, briefly, cells were incubated in 5x volume buffer CE (10 mM HEPES, 60 mM KCl, 1 mM EDTA, 0.075% NP-40, 1 mM DTT, with PIC and PhosSTOP) on ice for 3 min, then centrifuged at 600 g for 5 min. The cytoplasmic extract was removed, and the pellet was washed twice with buffer CE without NP-40 by centrifugation at 600 g for 5 min. The pellet was then resuspended in 2x volume buffer NE (20 mM Tris-HCl pH 8, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA with PIC and PhosSTOP) and incubated on ice for 10 min, then slowly diluted three-fold with buffer D (20 mM Tris-HCl, 25% glycerol, 1% NP-40 with 1x PIC and PhosSTOP). The lysate was cleared by centrifugation at max speed for 10 min at 4 °C, and the supernatant (nuclear extract) was used for IPs. Dynabeads™ Protein G (Thermo Fisher Scientific) was used for immunoprecipitation. The antibody-beads complex was prepared according to the manufacturer’s instructions. Whole cell lysates or nuclear lysates were incubated with the antibody-beads complex on a rotator at 4 °C for 2 h. The magnetic beads were washed on a magnetic rank six times using the immunoprecipitation buffer. Bead-bound protein was retrieved by incubating with sample loading buffer at 70 °C for 10 min. Antibody information is provided in the Supplementary Data 3.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed using the ChIP-Grade™ Protein G Enzymatic Kit (BioLegend, 699904) according to the manufacturer’s instructions. Briefly, 0.1–0.2 g of frozen liver tissue was pulverized with a pre-cooled hammer on a pre-cooled metallic block in dry ice. Tissue powder was cross-linked with 1% formaldehyde in PBS containing protease inhibitor (Roche) for 15 min and stopped by adding 2.5 M glycine (final concentration 0.125 M), followed by 5 min incubation. The tissue was pelleted by centrifugation at 500 g for 5 min, resuspended in the hypotonic buffer provided in the kit, and passed through a 20-gauge needle several times until the slurry was uniform. Chromatin was enzymatically digested for 5 min at 37 °C. Antibodies used in the ChIP experiment: Fxr (Santa Cruz), Tead4 (Santa Cruz), IgG antibody (BioLegend). The sequence information of qRT-PCR primers is provided in the Supplementary Data 2.
Luciferase assay
Reporter plasmids of Bsep enhancer (3xIR1) were generated by subcloning the genomic DNA PCR products into the pGL4.10 basic vector (Promega). 8xGTIIC-luciferase plasmid was from Addgene (#34615). Cells were transfected with pGL4.10 reporter plasmids, with pTK-Renilla (Promega) and effector plasmids. The luciferase activity was measured with a dual-luciferase reporter assay kit (Promega) according to the manufacturer’s instruction 48 h after transfection. The Renilla luciferase activity was used to normalize the luciferase reporter activity.
RNA-seq library preparation and sequencing
DNA was removed from RNA using a RapidOut DNA Removal Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. 400–1000 ng RNA per sample reaction was used for mRNA isolation using a NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). Libraries were prepared using an xGen RNA Library Prep Kit (IDT) according to the manufacturer’s instructions. Barcoded libraries were sequenced on an Illumina NextSeq 550/1000 at the Genomics Technology Laboratory of NCI. Paired-end sequencing mode (2x45 bp) was used in the experiments.
Computational analysis of bulk RNA-seq data
Raw counts were normalized, and differential expression analysis was performed using the edgeR package (Robinson and Oshlack, 2010). Heatmaps were generated using the Morpheus web tool (https://software.broadinstitute.org/morpheus/). Gene ontology analysis was performed using the ‘ClusterProfiler’ in the RStudio79. GSEA was performed using the GSEA 4.0 software or ‘ClusterProfiler’. KEGG functional analysis was performed using ClusterProfiler’.
Computational analysis for ICGC RNA-seq cohort
For transcriptomic analysis of human cohort data (LIRI-JP)50, a normalized count matrix was extracted from the ICGC data portal (https://dcc.icgc.org/). A ‘YAP liver tumor signature’ was reported previously, and a ‘hepatic FXR signature’ was generated in this paper (Supplementary Data 1). Predictions of these gene signatures in human samples were performed using the nearest template prediction (NTP) module80 in Gene Pattern81 (https://www.genepattern.org/) with the default setting. Overexpressed or under-expressed genes in a signature set were assigned a value of 1 or −1, respectively. Samples predicted as having a signature with False Discovery Rate (FDR) of less than 0.05 were compared with the rest of the samples in the cohort.
Computational analysis of ChIP-seq data
The ChIP-seq tracks were presented by the Integrative Genomics Viewer (IGV) software82. Heatmaps and plots of the ChIP-seq peaks were generated using DeepTools83. Peak analysis and motif analysis were performed using the HOMER software84.
Statistical analysis
Data were presented as mean ± standard deviation (SD). Data quantification and analyses were plotted using Prism 8. Unless mentioned otherwise, P-values were determined by 1-way ANOVA with Sidak’s test for comparison of more than two sample groups, and presented as ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. Processed LC-MS data for bile acids are included in the source data (Supplementary Fig. 2a-d, Supplementary Fig. S7d). RNA-seq data were deposited in GEO under accession GSE272343. The published RNA-seq datasets re-analyzed in this study can be found in GEO under accession number GSE16444743 and GSE14658920. The published ChIP-seq datasets re-analyzed in this study can be found in GEO under accession number GSE13819155 and GSE8786642. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
References
Chiang, J. Y. L. & Ferrell, J. M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu Rev. Nutr. 39, 175–200 (2019).
Segovia-Miranda, F. et al. Three-dimensional spatially resolved geometrical and functional models of human liver tissue reveal new aspects of NAFLD progression. Nat. Med. 25, 1885–1893 (2019).
Jansen, P. L. et al. The ascending pathophysiology of cholestatic liver disease. Hepatology 65, 722–738 (2017).
Davit-Spraul, A., Gonzales, E., Baussan, C. & Jacquemin, E. Progressive familial intrahepatic cholestasis. Orphanet. J. Rare Dis. 4, 1. https://doi.org/10.1186/1750-1172-4-1. (2009)
Jungst, C. et al. Intrahepatic cholestasis in common chronic liver diseases. Eur. J. Clin. Invest 43, 1069–1083 (2013).
Hirschfield, G. M., Heathcote, E. J. & Gershwin, M. E. Pathogenesis of cholestatic liver disease and therapeutic approaches. Gastroenterology 139, 1481–1496 (2010).
Qin, L. X. & Tang, Z. Y. Hepatocellular carcinoma with obstructive jaundice: diagnosis, treatment and prognosis. World J. Gastroenterol. 9, 385–391 (2003).
Sun, L., Cai, J. & Gonzalez, F. J. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 18, 335–347 (2021).
Goodwin, B. et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 (2000).
Lu, T. T. et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6, 507–515 (2000).
Ananthanarayanan, M., Balasubramanian, N., Makishima, M., Mangelsdorf, D. J. & Suchy, F. J. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276, 28857–28865 (2001).
Sinal, C. J. et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 (2000).
Yang, F. et al. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 67, 863–867 (2007).
Kim, I. et al. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 28, 940–946 (2007).
Anakk, S. et al. Bile acids activate YAP to promote liver carcinogenesis. Cell Rep. 5, 1060–1069 (2013).
Takahashi, S. et al. Role of Farnesoid X Receptor and Bile Acids in Hepatic Tumor Development. Hepatol. Commun. 2, 1567–1582 (2018).
Zheng, Y. & Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell 50, 264–282 (2019).
Kim, W. et al. Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. J. Clin. Invest 127, 137–152 (2017).
Kim, W. et al. Hepatic Hippo signaling inhibits protumoural microenvironment to suppress hepatocellular carcinoma. Gut 67, 1692–1703 (2018).
Liu, Y. et al. Yap-Sox9 signaling determines hepatocyte plasticity and lineage-specific hepatocarcinogenesis. J. Hepatol. 76, 652–664 (2022).
Russell, J. O. & Camargo, F. D. Hippo signalling in the liver: role in development, regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 19, 297–312 (2022).
Zeng, J., Fan, J. & Zhou, H. Bile acid-mediated signaling in cholestatic liver diseases. Cell Biosci. 13, 77 (2023).
Yimlamai, D. et al. Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 (2014).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Camargo, F. D. et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 (2007).
Zollner, G. et al. Expression of bile acid synthesis and detoxification enzymes and the alternative bile acid efflux pump MRP4 in patients with primary biliary cirrhosis. Liver Int. 27, 920–929 (2007).
Schaap, F. G., van der Gaag, N. A., Gouma, D. J. & Jansen, P. L. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology 49, 1228–1235 (2009).
Schneider, K. M. et al. Gut microbiota depletion exacerbates cholestatic liver injury via loss of FXR signalling. Nat. Metab. 3, 1228–1241 (2021).
Liu, R., Li, X., Hylemon, P. B. & Zhou, H. Conjugated Bile Acids Promote Invasive Growth of Esophageal Adenocarcinoma Cells and Cancer Stem Cell Expansion via Sphingosine 1-Phosphate Receptor 2-Mediated Yes-Associated Protein Activation. Am. J. Pathol. 188, 2042–2058 (2018).
Liu, Y. et al. Generation and characterization of mature hepatocyte organoids for liver metabolic studies. J Cell Sci. https://doi.org/10.1242/jcs.261961. (2024)
Sayin, S. I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Maloney, P. R. et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med Chem. 43, 2971–2974 (2000).
Gerloff, T. et al. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J. Biol. Chem. 273, 10046–10050 (1998).
Knisely, A. S. et al. Hepatocellular carcinoma in ten children under five years of age with bile salt export pump deficiency. Hepatology 44, 478–486 (2006).
Jansen, P. L. et al. Hepatocanalicular bile salt export pump deficiency in patients with progressive familial intrahepatic cholestasis. Gastroenterology 117, 1370–1379 (1999).
Strautnieks, S. S. et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat. Genet 20, 233–238 (1998).
Wang, R. et al. Targeted inactivation of sister of P-glycoprotein gene (spgp) in mice results in nonprogressive but persistent intrahepatic cholestasis. Proc. Natl Acad. Sci. USA 98, 2011–2016 (2001).
In LiverTox: Clinical and Research Information on Drug-Induced Liver Injury (2012).
Schumacher, J. D. et al. Direct and Indirect Effects of Fibroblast Growth Factor (FGF) 15 and FGF19 on Liver Fibrosis Development. Hepatology 71, 670–685 (2020).
Patton, M. E. et al. (eLife Sciences Publications, Ltd, 2024).
Pellicciari, R. et al. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 45, 3569–3572 (2002).
Lien, F. et al. Metformin interferes with bile acid homeostasis through AMPK-FXR crosstalk. J. Clin. Invest 124, 1037–1051 (2014).
Mathur, B. et al. Nuclear receptors FXR and SHP regulate protein N-glycan modifications in the liver. Sci. Adv. 7. https://doi.org/10.1126/sciadv.abf4865. (2021)
Ji, S. et al. FGF15 Activates Hippo Signaling to Suppress Bile Acid Metabolism and Liver Tumorigenesis. Dev. Cell 48, 460–474 e469 (2019).
Song, H. et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl Acad. Sci. USA 107, 1431–1436 (2010).
Zhou, D. et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 16, 425–438 (2009).
Lu, L. et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl Acad. Sci. USA 107, 1437–1442 (2010).
Kastan, N. R. et al. Development of an improved inhibitor of Lats kinases to promote regeneration of mammalian organs. Proc. Natl Acad. Sci. USA 119, e2206113119 (2022).
Liu-Chittenden, Y. et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300–1305 (2012).
Fujimoto, A. et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet 48, 500–509 (2016).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Beyer, T. A. et al. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5, 1611–1624 (2013).
Kim, M., Kim, T., Johnson, R. L. & Lim, D. S. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 11, 270–282 (2015).
He, L. et al. A Regulation Loop between YAP and NR4A1 Balances Cell Proliferation and Apoptosis. Cell Rep. 33, 108284 (2020).
Biagioni, F. et al. Decoding YAP dependent transcription in the liver. Nucleic Acids Res. 50, 7959–7971 (2022).
Ramos Pittol, J. M. et al. FXR Isoforms Control Different Metabolic Functions in Liver Cells via Binding to Specific DNA Motifs. Gastroenterology 159, 1853–1865 e1810 (2020).
Thakore, P. I. et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015).
Jiao, S. et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180 (2014).
Shi, Z. et al. DNA-binding mechanism of the Hippo pathway transcription factor TEAD4. Oncogene 36, 4362–4369 (2017).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Fang, S. et al. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J. Biol. Chem. 283, 35086–35095 (2008).
Lasko, L. M. et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017).
Falkenberg, K. J. & Johnstone, R. W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 13, 673–691 (2014).
Kong, B. et al. Mice with hepatocyte-specific FXR deficiency are resistant to spontaneous but susceptible to cholic acid-induced hepatocarcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G295–G302 (2016).
Pepe-Mooney, B. J. et al. Single-Cell Analysis of the Liver Epithelium Reveals Dynamic Heterogeneity and an Essential Role for YAP in Homeostasis and Regeneration. Cell Stem Cell 25, 23–38 e28 (2019).
Koo, J. H. et al. Induction of AP-1 by YAP/TAZ contributes to cell proliferation and organ growth. Genes Dev. 34, 72–86 (2020).
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7, 6 (2021).
Huang, W. et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 312, 233–236 (2006).
Liu, R. et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 60, 908–918 (2014).
Liu, R. et al. Taurocholate Induces Cyclooxygenase-2 Expression via the Sphingosine 1-phosphate Receptor 2 in a Human Cholangiocarcinoma Cell Line. J. Biol. Chem. 290, 30988–31002 (2015).
Verboven, E. et al. Regeneration Defects in Yap and Taz Mutant Mouse Livers Are Caused by Bile Duct Disruption and Cholestasis. Gastroenterology 160, 847–862 (2021).
Galli, G. G. et al. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol. Cell 60, 328–337 (2015).
Wu, B. K., Mei, S. C., Chen, E. H., Zheng, Y. & Pan, D. YAP induces an oncogenic transcriptional program through TET1-mediated epigenetic remodeling in liver growth and tumorigenesis. Nat. Genet 54, 1202–1213 (2022).
Guo, X. et al. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 31, 247–259 (2017).
Ma, S. et al. Transcriptional repression of estrogen receptor alpha by YAP reveals the Hippo pathway as therapeutic target for ER(+) breast cancer. Nat. Commun. 13, 1061 (2022).
Li, X. et al. YAP inhibits ERalpha and ER(+) breast cancer growth by disrupting a TEAD-ERalpha signaling axis. Nat. Commun. 13, 3075 (2022).
Kluiver, T. A. et al. Divergent WNT signaling and drug sensitivity profiles within hepatoblastoma tumors and organoids. Nat. Commun. 15, 8576 (2024).
Wang, Y. et al. Berberine Prevents Disease Progression of Nonalcoholic Steatohepatitis through Modulating Multiple Pathways. Cells 10. https://doi.org/10.3390/cells10020210. (2021)
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Hoshida, Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One 5, e15543 (2010).
Reich, M. et al. GenePattern 2.0. Nat. Genet 38, 500–501 (2006).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Acknowledgements
We thank the Yang lab members for stimulating discussion. This work was supported by National Institutes of Health grants R01CA222571 and R01AR070877 to Y.Y., Y.L., and Y.J.; supported by VA Merit Award 5I01BX005730, the VA ShEEP grant (1 IS1 BX004777-01) and the Research Career Scientist Award from the Department of Veterans Affairs (IK6BX004477) to H.Z.; We thank the LC-MS Core of Virginia Commonwealth University for the technical support, and thank Praju Vikas Anekal and Paula Montero-Llopis of the Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School for helpful discussions and training in confocal imaging.
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.Y. and Y.L. Methodology: Y.L., Y.Y., Y.J., and H.Z. Investigation: Y.L., J.Z., Y.J., and Z.S. Visualization: Y.L., and Y.J. Resources: X.W., and H.Z. Supervision: Y.Y. Writing—original draft: Y.L., and Y.Y. Writing—review and editing: Y.Y., and Y.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Zhu, J., Jin, Y. et al. Disrupting bile acid metabolism by suppressing Fxr causes hepatocellular carcinoma induced by YAP activation. Nat Commun 16, 3583 (2025). https://doi.org/10.1038/s41467-025-58809-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58809-z