Abstract
Adenovirus is an icosahedral, non-enveloped DNA virus that infects humans and other animals. The capsid of adenovirus is mainly assembled by the major capsid protein hexon. Folding and assembly of hexon require the viral encoded chaperone 100K, of which the detailed structure and chaperoning mechanism remain unknown. Here, we report the cryoEM structure of 100K in complex with a pre-mature hexon trimer. The structure shows that 100K dimers bind to the bottom double jelly-roll domains of the pre-mature hexon, mainly through a hook-like domain and a loop extruded from the dimerization domain. Additionally, a groove formed at the dimerization interface of 100K accommodates the N-terminal fragment 49-53 of an adjacent hexon protomer. Mutagenesis studies indicate that the interactions at the jelly-roll domain and the N-terminus of hexon are all essential for the proper folding and assembly of hexon. 100K binds and stabilizes the partially folded hexon, preventing premature aggregation of hexon, promoting the folding of the hexon top insertion loops, and facilitating hexon trimerization.
Similar content being viewed by others
Introduction
Correct folding of newly synthesized polypeptides in cells is critical for generating functional proteins1. Protein folding is a physical process that is driven mainly by hydrophobic interactions between different amino acid residues and may contain multiple stages. Folding of some proteins requires molecular chaperones in stabilizing conformations at the intermediate stages, which usually have large hydrophobic surface exposed. The functions of many protein chaperones remain to be characterized and the corresponding mode of action needs to be investigated, especially for these functioning in the assembly of complex cellular machinery.
Assembly of viral capsids is a process that requires coordinated capsid protein folding and assembly. The viral capsid is built up with an exact number of different capsid proteins, which are synthesized in cytosol of host cells as monomers and have multiple quasi-equivalent areas exposed for oligomerization2. The formation of correct interactions between these exposed surface areas is critical for capsid protein assembly. Otherwise, inappropriate interactions can cause protein aggregation. Viruses utilize chaperones or scaffold proteins for precise control of their capsid assembly. Different types of viral encoded chaperones that function in assisting viral capsid assembly have been described2,3,4,5. Some viruses, such as hepatitis B virus, picornavirus, pseudorabies virus, and polyomavirus, directly use host chaperones for assisting capsid assembly6,7,8,9,10. The major capsid protein hexon of adenovirus and p72 of the African swine fever virus both adopt a jelly-roll fold and assemble into trimers as capsomeres for the assembly of their icosahedral capsids11,12. The correct folding and assembly of hexon and p72 require the viral encoded chaperone 100K and B602L, respectively3,4.
Adenovirus is a non-enveloped dsDNA virus with an icosahedral capsid of 90–100 nm in diameter. The adenovirus capsid is assembled mainly by the hexon capsomeres, each of which is a homotrimer of the hexon protein. The structure of hexon can be divided into the top and the base two distinct parts13. The pseudo-hexagonal base of hexon is formed by the double jelly roll β barrels of the three hexon protomers. The triangular top of hexon is formed by extended insertion loops of the jelly-roll domains and is exposed on the surface of the virus, constituting the major immunological epitopes. Hexon of the human species D adenovirus plays an important role in mediating viral entry via direct binding to the receptor CD4614. Nuclear import of adenovirus DNA involves direct interaction of the hexon protein with the nuclear pore complex15.
In addition to the chaperone function, the late phase non-structural protein 100K of adenovirus also functions in aiding the efficient translation of late viral mRNAs, in blocking host protein synthesis via competitive binding to eIF4G16, in aiding the nucleus transportation of hexon17,18, and in inhibiting the enzyme activity of granzyme B, a cellular protease presents in granules of cytotoxic lymphocytes19. All the evidence shows that 100K is a multi-functional protein critical to the adenovirus life cycle. However, there is still no detailed structural information of 100K.
In this work, we report that cryo-electron microscopy (cryoEM) structural studies of 100K in complex with hexon. The structure shows that 100K forms homodimers and has two functional domains, including the hook-like domain and the dimerization domain. The hook-like domain binds to the hydrophobic pocket of hexon and blocks the premature contact of hexon protomers. Further mutagenesis studies show that the dimeric form of 100K and three major regions in contact with hexon are indispensable for its chaperone function.
Results
Purification and structure determination of the adenovirus 100K-hexon complex
Expression of serotype 3 human adenovirus (ad3) hexon along with an N terminal 3× Flag tag in HEK293F cells resulted in heavily degraded fragments, while co-expression of ad3 hexon and 100K resulted in correctly folded and assembled trimeric hexon particles (Supplementary Fig. 1a). This result is concordant with the previous finding that 100K is required for trimerization and stabilization of the hexon20. Previous studies showed that ad3-hexon and ad2-100K can form a stable complex18. To investigate the molecular basis of the chaperone function of 100K, we co-expressed ad3-hexon, fused with an N terminal 3 × Flag tag, and ad2-100K, fused with a C terminal 2 × Strep tag, in HEK293F cells. The ad3-hexon-ad2-100K complex was purified using sequential affinity chromatography with Strep and Flag tags, respectively (Supplementary Fig. 1b). SDS-PAGE analysis of the purified complex showed that ad3-hexon and ad2-100k are present in a molar ratio of ~1:1 (Supplementary Fig. 1b). For comparison, we also co-expressed ad3-hexon (with an N terminal 3 × Flag tag) and ad3-100K (with a C terminal 2 × Strep tag) in HEK293F cells. Notably, ad3-100K has much higher expression levels than ad2-100K, both when expressed alone and when co-expressed with ad3-hexon (Supplementary Fig. 1c, d). SDS PAGE analysis of the purified ad3-100K-ad3-hexon complex through Strep tag showed a dominant band for ad3-100K, while the ad3-hexon band was significantly weaker (Supplementary Fig. 1e).
In both cases, when the complex was enriched using only the Flag tag on the hexon, size exclusion chromatography revealed a prominent peak corresponding to hexon alone, while the hexon-100K complex peak was barely detectable (Supplementary Fig. 1f). This indicates that the 100K-free hexon predominates, either in the presence of ad2-100K or ad3-100K. The ad3-hexon and ad3-100K complex was less enriched compared to the ad3-hexon-ad2-100K complex. These data suggest that, although ad2-100K and ad3-100K share a highly homologous middle region (Supplementary Fig. 2), they have subtle difference in aiding the folding and assembly of hexon. We selected the ad2-100K-ad3-hexon complex, which has an appropriate molar ratio, for the following studies.
To further stabilize the complex of ad2-100K-ad3-hexon for cryoEM structural analysis, the elution from sequential affinity chromatography was subjected to GraFix21 with a 10–40% glycerol gradient and 0.15% glutaraldehyde (Supplementary Fig. 1b, g). Fractions containing the 100K-hexon complex were collected for cryoEM grid preparation (Supplementary Fig. 1g, h).
The structure of the hexon-100K complex was determined through single particle cryoEM at an overall resolution of 3.23 Å with C3 symmetry imposed (Supplementary Figs. 3 and 4). However, the 100K part of the map has a worse resolution of 4.3 Å as indicated by the local resolution map (Supplementary Fig. 4a). The limited resolution of the 100K density map may be due to the dynamic nature and flexibility of the 100K molecules. To improve the reconstruction of 100K, further focused refinements were performed on 100K and significantly improved the resolution to 3.79 Å (Supplementary Figs. 3 and 4b), indicating the rigid-body motion of the bound 100Ks. A model of 100K was built with the Alphafold predicted model as a reference22 (Supplementary Figs. 4b and 5a, b, Supplementary Table 1). The N terminal residues 1–158, loops 217–241, 555–567, 600–608, and the C terminal residues 696–805 of 100K were not built due to the highly disordered densities (Supplementary Figs. 4b and 5b). A model of the 100K-bound hexon was built with the crystal structure of the ad5 hexon as a reference23. Residues 1–47, 173–179, 628–640, 724–730, 740–753, 766–781, 868–891, and 915–944 of the 100K-bound hexon were not built due to the highly disordered densities (Supplementary Figs. 4a and 5c, Supplementary Table 1). Through 3D classifications, we could also obtain a 3.24 Å structure of the hexon that does not have the bound 100K (Supplementary Fig. 3). As the structure of the 100K-free hexon is closely similar to those found in the viral capsid (Supplementary Fig. 5d, e), we refer to this hexon structure as the “mature” hexon. A model of the mature hexon was built (Supplementary Table 1). Except for residues 1–5, 173–179, and 940–944, the model contains the rest residues of hexon. Of note, a conformation of the complex with disordered hexon top domains of the insertion loops was obtained through classifications, which may represent an intermediate state in hexon assembly (Supplementary Fig. 3).
Overall structure of the 100K-hexon complex
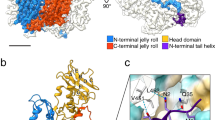
The 100K-hexon complex resembles a “short rocket”, which is assembled by three hexon protomers and three 100K dimers. The hexon constitutes the main body of the rocket, whereas the three 100K symmetric dimers constitute the rocket boosters wrapping around the double jelly-roll bases of the hexon (Fig. 1). The symmetry axis of the 100K dimer is tangential to the base of the rocket-like main body. Each 100K dimer inserts between the double jelly-roll bases of two adjacent hexon protomers, and establishes direct and asymmetric interactions with the two adjacent hexon protomers (Fig. 1). One hexon protomer is clamped by the bound 100K dimer through two contact sites A and B, which are located on the inner and outer surfaces of the jelly-roll barrels, respectively (Fig. 1). Interactions with the adjacent hexon protomer occur at contact site C, which involves a groove at the dimerization interface of 100K and the N-terminal residues 48-60 of the hexon protomer (Fig. 1). The buried surface areas at the three contact sites (A, B and C) are ~2184.5 Å2, 1167.9 Å2 and 633.7 Å2, respectively (Supplementary Fig. 6).
a Color zoned cryo-EM map showing the overall structure of the 100K-hexon complex. The hexon trimer and 100K dimer are colored cornflower blue and hot pink, respectively. The contact sites A, B, and C are indicated with boxes in dash lines. b Overall structure of the 100K-hexon complex. Left: Front view. Right: Top view. The structure is shown as a cartoon with the contact sites A, B, and C highlighted with boxes in dash lines. The hexon top domain, jelly-roll domain, 100K protomer a, and protomer b are colored cyan, purple, lime, and hot pink, respectively.
Structure of 100K
In the complex of 100K-hexon, 100K molecules form symmetric dimers, which have a “χ” shaped structure and are mainly composed of α-helices and loops (Fig. 2a). A Dali search against all the experimental structures revealed no homologous structures of 100K. We further used Foldseek to search against all available libraries, including the Big Fantastic Virus Database and the Alphafold Proteome Database24. With an E value cutoff of 0.001, the search yielded 11 hits, all of which originated from different adenoviruses. These results suggest that 100K may represent a distinct structural fold and is characteristic to adenoviruses. The structure of each 100K protomer can be roughly divided into two domains, including the hook-like domain (residues 555–695) and the dimerization domain (residues 159–554) (Fig. 2a and Supplementary Fig. 7).
a Structure and topology of the 100K dimer. Left: Ribbon diagrams showing the structure of a 100K dimer. Position of the 2-fold symmetry axis is marked by a diamond. The hook-like domain a, dimerization domain a, dimerization domain b, and the disordered region are colored hot pink, cornflower blue, cyan, and light pink, respectively. The N terminal residues Pro159 and Ala 161, the loop Ile285-Gly310, the missing loop Ser600-Pro608, the C terminal residue Pro695, and helices are labeled. Right: Diagrams showing the topology of 100K. b The dimerization interface of the 100K dimer. The helices, loops, and residues at the dimerization interface are labeled. Red and black labels were used to distinguish elements from different protomers. The C atoms of negatively charged, positively charged, polar, and hydrophobic residues are colored dark red, cyan, green, and orange, respectively. The N and O atoms are colored blue and red, respectively. The disulfide bonds are colored deep pink.
Formation of the 100K dimer is solely mediated by the dimerization domains. The dimerization domain contains 17 helices and several short β strands, including α1–α17, β1 and β2 (Fig. 2a). Among these, α4-α8, α10-14, and α17 form an arrowhead-like helix bundle. A long loop between residues 285 and 310 protrudes from the dimerization domain. The two long loops in the dimer are arranged in trans and form the two extended arms of the “χ” shaped structure. The dimerization interface has a buried surface area of 1938.9 Å2. Helix α15 and loop 508-518 are located at the dimerization interface (Fig. 2b). Interactions between the two symmetry-related α15s are mainly through interactions between polar residues Q498, N499, and N502. The two symmetry-related loops (508–518) intertwine and are enriched with hydrophobic residues at the contact interface (Fig. 2b). Furthermore, C515 and C516 and the symmetry related C516’ and C515’ at the distal tips of the loops form two intermolecular disulfide bonds (C515-C516’, C516-C515’), as confirmed by mass spectrometry analysis (Supplementary Fig. 8). These disulfide bonds covalently crosslink the two 100K protomers. Additionally, underneath the helices α15 and α15’, two symmetry-related loops 163-172 interact with each other through mainly hydrophobic interaction to further stabilize the 100K dimer (Fig. 2b).
The hook-like domain contains three helices α18, α19, α20, and a hairpin loop (residues 678–695) (Fig. 2a). The loop 600–608 that connects α18 and α19 is disordered. The hook-like domains are located at the distal ends of the 100K dimer and constitute the other two extended arms of the “χ” shaped structure. The two hook-like domains do not directly interact with each other. In each 100K dimer, one of the hook-like domains has interactions with the hexon and has an ordered conformation, while the other hook-like domain does not interact with the hexon and is highly disordered (Figs. 1 and 2a).
Structure of the pre-mature hexon
Two major conformational states of the hexon were obtained with the ad2-100K-ad3-hexon sample, including the 100K bound and unbound conformations. The 100K unbound conformation of the hexon has a similar structure as that of the hexon capsomere in the viral capsid, referred to here as the “mature” state. In contrast, the structure of the hexon in the 100K-bound complex is significantly different, representing a “pre-mature” state (Figs. 1a and 3a). Structure superimposition of the “pre-mature” and “mature” hexons showed that the top regions of the hexons align well, while the jelly-roll barrels in the bottom region of the “pre-mature” hexon spread out for ~8.5 degrees compared to those of the “mature” hexon (Fig. 3a). Additionally, in the “pre-mature” state, the double jelly-roll barrel of each hexon protomer lacks direct interactions with the jelly-roll barrels of adjacent protomers (Fig. 3a, b). Several hydrophobic loops of the jelly roll barrels, which mediate the interactions between the jelly roll barrels of the mature hexon, are disordered in the “pre-mature” hexon. These include loops 724–730, 740–753, 766–781, and 868–891 (Fig. 3b and Supplementary Fig. 5c). The connecting β sheet (residues 616–640 and 914–939) in between the two jelly-roll domains is also disordered in the “pre-mature” hexon (Fig. 3b and Supplementary Figs. 5c, d and 9a). Moreover, the N-terminal fragment (residues 6–64) of the “mature” hexon, which contains three short helices (residues 7–12, 20–23 and 26–36), interacts with a helix (residues 616–626) of the neighboring N-jelly-roll domain (Fig. 3b). In the “pre-mature” state, however, the N-terminal helices 7–12, 20–23, 26–36 are disordered, and the fragment containing residues 48–60 protrudes outwards without interacting with the neighboring hexon protomers (Fig. 3b). However, when the top insertion loops and the bottom jelly-roll barrels were separated and compared individually, the structures show no significant differences when the disordered regions were excluded, indicating that folding of these domains has already been accomplished in the “pre-mature” conformation (Fig. 3c).
a Top Left: Ribbon diagrams showing domain organization of the mature hexon. The N terminal fragment (residues 1–55), N-jelly roll, C-jelly roll, loops DE1, DE2, FG1, FG2 and the connecting β sheet are colored firebrick, spring green, orange, yellow, red, deep sky blue, lime and cornflower blue, respectively. Bottom Left: Structural comparisons of the pre-mature and mature hexons. Middle: Top (Left) and front views (Right) of the superimposition of pre-mature and mature hexons. The black triangle depicts the position of the 3-fold symmetry axis. Right: Relative positions of the double jelly-roll domains in the pre-mature and mature hexons. The centroids were calculated with the top insertion and bottom jelly-roll domains, respectively. Centroids of the pre-mature and mature hexons are colored red and blue, respectively. b Structural comparisons of the jelly-roll domains in mature and pre-mature hexons. The N terminal fragment (residues 1–55), N-jelly roll, C-jelly roll, loop DE2 and the connecting β sheet of one hexon protomer are colored firebrick, spring green, orange, red and cornflower blue, respectively. The positions of the disordered regions (1–47, 724–730, 740–753, 766–781, 868–891, and 915–944) in the pre-mature hexon are represented with dash lines. The N- and C-termini of the colored premature hexon protomer are highlighted with red balls. c.Structural superimposition of the top insertion loops and the bottom double jelly-roll domains of the pre-mature and mature hexons. The top insertion loops and the bottom double jelly-roll domains of the mature hexon are colored the same as in the left top panel of (a). The pre-mature hexon are colored gray.
Interactions between 100K and hexon at the contact sites A, B, and C
The two extended arms in cis of the 100K dimer, consisting of the hook-like domain of one 100K protomer and the loop 285-310 of the dimerization domain from the other 100K protomer, clamp onto the double jelly-roll barrels of a single pre-mature hexon protomer. These arms make direct interactions with the pre-mature hexon protomer at contact sites A and B, respectively (Fig. 1). Contact site A is located on the inner surface of the jelly-roll bases, where helix α20 and loop 678–695 of the 100K are partially buried in a hydrophobic pocket formed by the N terminal jelly-roll domain and α-helix 616–626 of the pre-mature hexon protomer (Fig. 4a). Helix 20 and loop 678–695 of the 100K hook domain are connected to the 100K dimerization domain via helices α18 and α19, which trans pass the pre-mature hexon protomer at the bottom, between the N and C-jelly-roll domains (Supplementary Fig. 9a). These helices occupy the position where the connecting β sheet is located in the mature hexon protomer (Supplementary Fig. 9a). The loop 590–613 in between α18 and α19 contains a disordered segment (residues 600–608) and a short β strand (residues 594–597). The short β strand further stabilizes the interactions at site A by forming a parallel β-sheet with residues 564–566 of the pre-mature hexon protomer (Fig. 4a). Helix α20 and loop 678–695 of 100K are rich in hydrophobic residues, including Ile659, Leu663, Ile666, Phe673, Leu674, Leu675, Val681, Tyr682, Leu683, and Pro685. These hydrophobic residues are all buried in the pocket and are in close contact with residues Leu622, Leu541, Phe612, Ala97, Leu367, Leu370, Pro555, Phe326, Ile552, Pro549, and Phe377 of the pre-mature hexon protomer within the pocket (Fig. 4a). Of note, the C-terminal region (696–805 residues) of the 100K hook-like domain, which is in proximity to the loop 678–695, is highly disordered and not visible in the structure. At the contact site A, the hook-like domain inserts between two pre-mature hexon protomers, obstructing the approach of the jelly-roll barrels from neighboring protomers.
The top and bottom jelly-roll domains of one hexon protomer are colored cyan and purple, respectively. Other hexon protomers are colored gray. The 100K protomer a and protomer b in one dimer are colored lime and pink, respectively. a Interactions between 100K and hexon at contact site A. Top Left: Ribbon diagrams showing the interactions between the 100K hook-like domain and the inner surface of the hexon jelly-roll domain. Top Right: The hexon jelly-roll domain shown in a surface-rendered representation and the 100K hook-like domain shown in ribbons. The surface is colored according to the hydrophobicity of the residues with yellow, white, and blue representing high, mediate, and low hydrophobicity, respectively. Residues involved in forming hydrophobic interaction between α20 of hook-like domain and hexon are labeled in red (100K) and black (hexon). Bottom: Ribbon diagrams showing the parallel β-sheet formed by residues 594–597 from 100K and 564-566 from hexon. The contact site A is indicated with a dashed frame. Residues of 100K and hexon are labeled red and black, respectively. b Interactions between 100K and hexon at contact site B. Left: Ribbon diagrams showing the interactions between the loop 285–310 of 100K and the outer surface of the hexon jelly-roll domain. The contact site B is highlighted with a dashed frame. Right: The C terminal jelly-roll domain of hexon is shown in a representation of the electrostatic potential surface. The loop 285–310 of 100K is shown in ribbon and sticks. Residues of hexon are labeled black. Negatively charged, positively charged and hydrophobic residues of 100K are in red, blue, and yellow, respectively. c Interactions between 100K and hexon at contact site C. Left: Ribbon diagrams showing the interaction of the hexon N-terminal fragment and the groove at the dimerization interface of 100K. The contact site C is highlighted with a dashed frame. Right: a zoom-in view of the contact site C. The hydrophobic residues are colored orange. The residues of hexon, 100K protomer a and protomer b are labeled purple, lime and pink, respectively.
In the mature hexon, the hydrophobic pocket is occupied by loop 740-785 and the N terminal residues 6-52 from two neighboring hexon protomers (Supplementary Fig. 9b). Loop 740–785 replaces loop 678–695 of 100K and has ten hydrophobic residues (Ala749, Phe758, Leu759, Met762, Leu763, Tyr770, Gly772, Phe773, Tyr774 and Ile775) buried in the pocket. Additionally, the α-helix (residues 26–36) in the N-terminal region has residues Leu28 and Phe31 buried in the pocket (Supplementary Fig. 9b). Apart from hydrophobic interactions, residues 740–754 of the neighboring hexon protomer also establish electrostatic interactions with nearby residues (Supplementary Fig. 9b). The position of 100K helices α18 and α19 is taken by the newly formed connecting β-sheet between the N- and C-jelly-roll domains (Supplementary Fig. 9a).
Contact site B is located on the exterior surface of the jelly-roll bases, where loop 285–310 of the 100K dimerization domain attaches to the C-terminal jelly-roll barrel of the pre-mature hexon protomer. The loop 285–310 extends ~28 Å from its connection to the 100K dimerization domain and is aligned vertically to the β-sheet of the C-terminal jelly-roll barrel. Loop 285–310 is stabilized by interactions with the pre-mature hexon protomer (Figs. 1 and 4b). Electrostatic interactions are formed between residues Asp756 and Asp894 from the pre-mature hexon protomer and Lys178 and Arg288 from 100K (Fig. 4b). Local clusters of hydrophobic residues formed by residues Val286, Ala289, Pro291, Leu292, Ala296, Leu298, Pro307, Ala308, Val309 of 100K and Leu645, Tyr646, Pro656, Ile657, Pro660, Thr677 of the pre-mature hexon protomer are also observed. In proximity to the contact site B, the dimerization domain of the 100K protomer (protomer b in Fig. 4b) blocks the approach of the adjacent hexon protomer to form the mature hexon (Figs. 1 and 4b).
Contact site C involves a hydrophobic groove formed by helix α3 and α3’ at the dimerization interface of the 100K dimer, where the N terminal fragment (residues 48-60) of the pre-mature hexon protomer binds and is partially buried (Fig. 4c). The groove at the dimerization interface is largely constituted by hydrophobic residues (Supplementary Figs. 7 and 10). The groove accommodates hydrophobic residues Val50, Ala51, and Pro52 of the hexon N-terminal fragment (Fig. 4c).
Functional verification of the contacts at sites A, B, and C
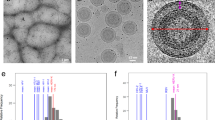
To further pinpoint the key contacts required for the chaperone function of 100K, we mutated residues in 100K and hexon that are involved in the interactions. Specifically, we deleted the loops and helices at contact site A, generating 100K deletion mutants Δ657–805, Δ680–805, and Δ693–805. Correct assembly of hexon was indicated by SEC analysis. The SEC results showed that deletion of either helix α20 and loop 678–695 (Δ657–805) or loop 678–695 alone (Δ680–805) completely abolished the chaperone function of 100K (Fig. 5a). However, the mutant Δ693–805 has only marginal influence on the chaperone function of 100K (Fig. 5a), indicating that the disordered C-terminal region (residues 696–805) of 100K is not essential for its chaperone function (Fig. 5a). Replacement of the disordered fragment 600–608 in the loop that connects α18 and α19 with a linker SGGSGGSGG had no significant influence on the formation of mature hexon (hexon trimer in Fig. 5a).
a SEC analysis of hexon co-expressed with ad2-100K, △657–805, △680–805, △693–805, △600–608(GS),△285–310(GS), △1–38, △1–93, and △1–135, respectively. SEC analysis of hexon mutant V50L-A51F-T53W co-expressed with ad2-100K mutant A251F-A254W-V255L, hexon mutant V50L-A51F-T53W co-expressed with ad2-100K, and hexon co-expressed with ad2-100K mutant A251F-A254W-V255L. The peaks of soluble aggregation, hexon trimer, and monomer are indicated by red, orange, and blue dash lines, respectively. b Representative negative staining images of the mature hexon (hexon trimer) peak fractions collected from SEC analysis of hexon co-expressed with ad2−100K, soluble aggregation of 100K and hexon, mature hexon (hexon trimer) peak fractions collected from SEC analysis of hexon mutant V50L-A51F-T53W co-expressed with ad2−100K, and hexon monomer peak fractions collected from SEC analysis of hexon co-expressed with ad2-100K mutant A251F-A254W-V255L. c Top Left: SEC analysis of hexon mutant V50L-A51F-T53W co-expressed with ad2-100K. Top Right: SDS-PAGE analysis of the sample for SEC and the peak fractions collected form SEC analysis of hexon mutant V50L-A51F-T53W co-expressed with ad2-100K. Bottom Left: SEC analysis of hexon co-expressed with ad2-100K mutant A251F-A254W-V255L. Bottom Right: Western blot (WB) analysis of the peak fractions collected form SEC analysis of hexon co-expressed with ad2-100K mutant A251F-A254W-V255L.
Replacement of the protruding loop 285–310 with a linker SGGSGGSGGSGG at the contact site B led to the absence of a mature hexon peak (Fig. 5a). This indicates that the interaction between hexon and 100K at site B is critical for correct folding and assembly of mature hexon. Furthermore, we made the 100K mutant A251F-A254W-V255L, in which the bulky side chains of the mutated residues are predicted to occupy the groove at the dimerization interface. This prevents binding of the hexon N-terminal fragment and disrupts interactions at contact site C. SEC analysis showed that this mutant significantly impairs the formation of mature hexon (Fig. 5a). Similarly, we made the hexon mutant V50L-A51F-T53W, where the bulky side chains of the mutated residues cannot be accommodated by the groove, thus preventing interactions at contact site C. SEC results showed a dramatic decrease in the mature hexon peak (Fig. 5a, b and c). With the 100K mutant A251F-A254W-V255L, we observed an increase in the hexon monomer peak accompanied by a significant decrease in the mature hexon peak (Fig. 5a, c). The 100K dimer recruits additional hexon protomers through contact site C, which could further promote the formation of the hexon trimer. Therefore, disruption of contact site C likely leads to the increase in the hexon monomer peak. With the hexon mutant V50L-A51F-T53W, we observed the presence of a soluble aggregation peak, likely caused by hexon-100K aggregates as shown by SEC, SDS-PAGE, and negative staining microscopy analysis (Fig. 5a, b and c). These data combined indicate that the interactions at site C are indispensable for the chaperone function of 100K.
Sequence comparisons show that the middle segments of ad2 and ad3 100Ks are homologous, while the N- and C-terminal fragments differ (Supplementary Figs. 2, 7, 10). Since the C-terminal fragment is not essential for the chaperoning function of 100K, we further explored the possible role of the N-terminal fragment in its chaperone activity. The N-terminal region of 100K (residues 1–158) is highly disordered and not visible in the structure. Residue159, the first visible N-terminal residue in the structure, is located in proximity to the dimerization interface of 100K. We removed the N-terminal region and generated deletion mutants Δ1–38, Δ1–93, and Δ1–135. SEC analysis showed that the peak of mature hexon was significantly reduced, suggesting that the chaperone function of 100K is affected but not abolished (Fig. 5a). However, further analysis showed that the N-terminal deletions led to a significant decrease in the expression level of 100K (Supplementary Fig. 11a). Thus, the reduced chaperone activity in those mutants may be attributed to the lower expression levels. Furthermore, we replaced the N-terminal region of ad2 100K with that of ad3 100K to generate the chimeric mutant ad3N1-121-ad2-100K. Co-expression of ad3N1-121-ad2-100K and ad3-hexon yielded results similar to those obtained with ad2-100K and ad3-hexon, suggesting that the differences between ad2 and ad3 100Ks are unlikely caused by the N-terminal region (Supplementary Fig. 11b).
To investigate whether the mutations affect the folding of 100K, all 100K mutants were purified using Strep-Tactin resin. SDS-PAGE and western blot analyses showed that those mutants are soluble and have the expected size (Supplementary Fig. 11a–d). Analysis with GraFix showed that those mutations have a similar oligomeric state as that of the wild type 100K, which exists mainly as a dimer in solution (Supplementary Fig. 11e–h). These data suggest that the mutations unlikely impact the proper folding of 100K. ELISA analysis showed fractions of the soluble aggregation peak, the mature hexon peak, the hexon monomer peak with the 100K mutant A251F-A252W-V255L and the elution of hexon when expressed without 100K can react with the rabbit polyclonal adenovirus antibody, suggesting that some epitopes of hexon have been formed even with the dis-functional 100K or in the absence of 100K (Supplementary Fig. 11i).
The dimeric form of 100K is essential for its chaperone function
As described above, 100K dimer clamped hexon through the hook-like domain from one 100K and the dimerization domain from the other 100K. Moreover, a groove formed at the dimerization interface constitutes the contact site C. We determined the oligomerization state of 100K in solution by GraFix. In consistence with the structural analysis, 100K exists as a dimer in solution (Supplementary Fig. 11e). These structural and biochemical information suggested the dimeric form could be essential for the chaperone function of 100K. To verify the importance of the dimeric form, we performed systematic mutagenesis studies on the residues at the dimerization interface of 100K. Intriguingly, the disulfide bonds between cys515 and cys516’ are not essential for the dimerization of 100K, which was proved by the mutant C515S-C516S (Fig. 6a, b). This mutant also has normal chaperone function as indicated by SEC result (Fig. 6a). Furthermore, sequence alignments show that C515 and C516 of 100K are not conserved among 100Ks from different adenoviruses (Supplementary Figs. 7 and 10). Mutagenesis studies at the α15-α15’ interface showed that the double mutation Q498A-N499A resulted in trace amounts of recombinant 100K, which could be detected only by western blot (WB) analysis (Fig. 6c). However, the double mutation Q498A-N499A together with the C-terminal deletion of 100K (Δ693–805 + Q498A-N499A) has detectable expression (Fig. 6d). SEC data showed that the C-terminal fragment 693-805 of 100K is not important for its chaperone function (Fig. 5a). Further biochemistry studies with a 10–30% (w/v) glycerol gradient and GraFix showed that the double mutation Q498A and N499A in Δ693-805 disrupted the dimeric form of 100K and caused the formation of soluble aggregations (Fig. 6e). Co-expression of hexon and the 100K mutant Δ693-805 + Q498A-N499A led to the absence of the mature hexon peak (Fig. 6f), confirming that the dimeric form of 100K is essential for its chaperone function.
a Left: SEC analysis of hexon co-expressed with ad2−100K mutant C515S-C516S. Middle: SDS-PAGE analysis of the SEC peak fractions. Right: negative staining analysis of the purified recombinant hexon coproduced with 100K mutant C515S-C516S. Class averaged images from representative 2D classes are shown at the bottom. b Grafix and WB analysis of ad2-100K mutant C515S-C516S-strepII. Elution fractions from Strep-Tactin resin were analyzed. c SDS-PAGE and WB analysis of ad2-100K-Q498A-N499A-strepII. Elution fractions from Strep-Tactin resin were analyzed. d SDS-PAGE analysis of ad2-100K Δ693-805 + Q498A-N499A-strepII. Elution fractions from Strep-Tactin resin were analyzed. e Grafix analysis of ad2-100K Δ693-805 + Q498A-N499A-strepII. Elution fractions from Strep-Tactin resin were analyzed. f SEC analysis of hexon co-expressed with wild type ad2-100K (left) and ad2-100K mutant Δ693-805 + Q498A-N499A (right), respectively. The peak positions of the hexon trimer and monomer are indicated by an orange arrow/dashed line and a blue arrow/dashed line, respectively.
Discussion
The intricate folding and remarkable stability of the hexon trimer have been extensively described11,23. Evidence has been provided suggesting that the nascent hexon polypeptide chain can form a complex with 100K while hexon is still interacting with ribosomes25. In addition, the trimeric hexon is transported into the cell nucleus in the presence of the 100K protein and independently of any other adenovirus proteins18. 100K was found to interact with both hexon monomers and trimers within the cytoplasm, whereas it interacts predominantly with hexon trimers in the nucleus26,27. There is still limited information about the function of hexon nucleus transport during adenovirus assembly. Based on these prior observations and our research, we have come up with a model of the 100K-aided hexon trimerization (Fig. 7). 100K pre-exist as dimers before engaging with hexon in the cytoplasm. 100K dimers immediately interact with the newly synthesized N-terminus of hexon polypeptides during ribosome systhesis. Meanwhile, the hexon-bound 100K dimers would bind the hydrophobic area of hexon monomer to prevent aggregation (Figs. 1 and 3). In addition, hydrophobic interaction is one of the major forces to drive protein folding. Helix α20 of the hook-like domain may provide the platform in aiding the folding of hexon jelly-roll domain. Still, we can’t confirm the exact chronological order of 100K bind to the N-terminus or double jelly-roll domain hydrophobic area of hexon. Two hexon monomers are indirectly linked by one 100K dimer, which results in the trimerization of three hexon protomers. In this procedure, the top insertion loops of hexon can fold into correct structures and further trimerize by interacting with neighboring hexon molecules. The observation of a class of 100K-hexon complexes with disordered hexon top domains of the insertion loops in 3D classifications could provide supporting evidence for the above point (Supplementary Fig. 3).
100K pre-exists as dimers before engaging with hexon in the cytoplasm. 100K dimers immediately interact with the nascent hexon polypeptides during ribosome synthesis. The 100K dimer forms a complex with the partially folded hexon monomer. This primary 100K-hexon complex serves as a platform for further recruiting additional 100K-bound hexon subunits, ultimately assembling into the immature hexon with 100K. The pre-mature hexon undergoes further conformational changes to release the bound 100K, resulting in the formation of the stable mature hexon. The annotations of the components involved in the cartoon are showed in the bottom.
Although 100K predominantly exists as a dimer in solution, various oligomerization states of 100K were observed by GraFix (Supplementary Fig. 11e). Notably, the disulfide bonds between the two 100K protomers are not essential for maintaining its chaperone function or its dimeric form. Additionally, residues involved in the formation of the disulfide bonds are not conserved among adenoviruses (Supplementary Figs. 7 and 10). Since 100K functions in the cytoplasm, where the environment is reducing. The disulfide bonds observed in the structure might be an artifact of the purification process. The role of these disulfide bonds in 100K’s function remains to be further investigated. Direct interactions between the 100K dimers were not observed in the 100K-hexon map with C3 symmetry. The correct assembly of hexon bottom jelly-roll domains is dependent on the extrusion of 100K dimers between the domains, which requires the synergistic action of adjacent hexon structures to compete with the bound 100K (Fig. 7 and Supplementary Fig. 9). This process may not occur spontaneously after the formation of the hexon-100K trimer. We prepared the hexon-100K complex by co-expressing 100K of ad2 and hexon of ad3. However, our results showed that a small portion of proteins is in the form of hexon-100K complex even when co-expressing the 100K of ad3 and hexon of ad3 (Supplementary Fig. 1e). Thus, the hexon-100K complex may stably exist in the cytoplasm and await nucleus transportation. Hexon transport to the nucleus may function as a checkpoint for ensuring hexon trimerization and release of 100K.
Multiple sequence alignments of the 100Ks from different adenoviruses (Supplementary Figs. 7 and 10) showed that residues of 100K involved in hexon binding, especially at contact sites A and C, are conserved. Sequences of the disordered N- and C-terminal fragments, which are not essential for the chaperone function of 100K, are not conserved. Multiple sequence alignments of hexons from different adenoviruses showed that residues at the contact sites with 100K are highly conserved (Supplementary Figs. 12, 13). The conservation at the contact regions between hexon and 100K could be a result of coevolution, which is worth further investigation.
In addition to 100K’s functions in hexon trimerization and hexon transport to the nucleus, biochemical evidence suggests that 100K plays a critical role in the scaffolding process of assembly-intermediate capsid. This function appeared to correspond with the region between amino acid 300-400 from the N terminus of 100K27,28. This region mapped to our structure is well exposed and may serve as a platform to interact with other scaffold proteins. Intriguingly, the key residues RRK (residues 329–331) response for eIF4G binding to drive ribosome shunting are also mapped in this region29. The inhibition of Granzyme B-mediated cell death is absolutely dependent on D4819. The nucleus transport of 100K relies on the arginine methylation of RGG motif (residues 741–743)30. D48 and RGG motif are located at the disordered N-terminus, and the C-terminus in our structure, respectively, which means 100K has multi-functional domains to fulfill the requirements of different biological processes. Our biochemical results show that C-terminus (693–805) and the N-terminus (1-135) are not essential for the chaperone function of 100K (Fig. 5a). Sequence alignments showed that residues of 100K involved in the contact sites A, B and C are highly conserved in human adenoviruses, which explains why ad2 100K can help ad3-hexon correct-folding (Supplementary Fig. 7). The results of the chimera mutant ad3N1-121-ad2-100K and the C-terminal deletion mutants (Fig. 5a and Supplementary Fig. 11b) revealed that the non-conserved N- and C-terminal regions are not responsible for the different behaviors of ad2-100K and ad3-100K in aiding the folding and assembly of ad3-hexon (Supplementary Fig. 11b). Therefore, the differences between ad2-100K and ad3-100K are likely related to other non-conserved sites beyond the N-and C-terminal regions. Of note, the N-terminal disordered region of 100K affects the expression level of 100K, which needs further investigation to elucidate the underlying mechanisms. In consideration of the multi-function, the structure of the disordered portion of 100K may become ordered while binding target proteins or ligands. 100K may adopt various conformations to execute different functions. We attempted to solve the structure of 100K without hexon. Although the cryoEM micrographs of 100K seemed homogeneous, we couldn’t get the ordered feature from 2D classifications or model-guided 3D classifications. This suggested the ordered conformations of 100K may rely on the interactions with other proteins.
Methods
Recombinant protein purification and sample preparation
The hexon and 100K genes of type2 and type3 adenovirus were synthesized from the Qinglan company (Wuxi, China). The synthesized genes were cloned into the vector pCMV. An N-terminal 3 × Flag tag was added to the recombinant hexon. A C-terminal 2×Strep was added to the recombinant 100K. Sequences of the primers are listed in the Supplementary Table 2. HEK293F cells (Thermo Fisher, Cat# R79007) were cultured in suspension at 37 °C with the 293-SIM medium (Sino Biological company, Product M293II) and 5% CO2. For one-liter cell culture, the cells were transfected by a plasmid mixture containing 2 mg hexon plasmid, 2 mg 100K plasmid, and 12 mg PEI at a cell density of 2 × 106 cells/ml. The transfected cells were harvested 48 h post-transfection by centrifugation at 1000 × g for 20 min. The cell pellet was resuspended by a buffer containing 20 mM HEPES at pH 7.4, 300 mM NaCl, and cocktail protease inhibitors (Thermo Scientific, Product A32965). The suspended cells were sonicated for 3 min, and the cell lysate was centrifuged for 20 min at 30,966 × g (JA 25.50 rotor, Beckman). The recombinant hexon-100K complex protein in the supernatant was collected and applied to the Strep-Tactin resin (IBA, Product 2-1201-010) (Supplementary Fig. 1b). The bound protein was washed with the resuspension buffer twice and then was eluted from the resin with a buffer containing 5 mM desthiobiotin, 20 mM HEPES at pH 7.4, and 300 mM NaCl (Supplementary Fig. 1b). The eluted sample was applied to the anti-Flag resin (GenScript, Product L00432). The bound protein was washed with the resuspension buffer twice and then was eluted from the resin with a buffer containing 0.1 mg/mL Flag peptide, 20 mM HEPES at pH 7.4, and 300 mM NaCl. The eluted sample then was concentrated and applied to a 10–40% w/v linear glycerol gradient supplemented with 0.15% glutaraldehyde for gradient fixation (GraFix) (Supplementary Fig. 1g). The gradient was run for 13 h at 240,000 × g. Fractions containing hexon-100K were collected, and the glycerol was removed by buffer exchange through centrifugation with 20 mM HEPES at pH 7.4 and 150 mM NaCl.
Aliquots of 3.5 μl cross-linked hexon-100K protein at a concentration of 0.3 mg/ml were applied to glow-discharged holey carbon grids (Quantifoil, Cu 400 mesh, R1.2/1.3). The grids were blotted for 5.5 s in 100% humanity at 8 °C and were then immediately plunged into liquid ethane by using a Vitrobot Mark VI (Thermo Fisher).
Antibodies
The following antibodies were used in western blot and ELISA. The anti-Flag-Tag Mouse Monoclonal Antibody (Cat. No. CW0287M, 1:5000) was obtained from CWBIO. The anti-Strep-Tag II Monoclonal Antibody (8C12) (Cat. No. A02230, 1:5000) was obtained from Abbkine. The HRP conjugated goat Anti-Mouse IgG (CWBIO, Cat. No. CW0102S, 1:10000) was obtained from CWBIO. The GAPDH Mouse mAb (High Dilution) (No. AC033,1:10000) was obtained from ABclonal. The anti-Adenovirus Type 5 antibody (No. ab6982, 1:5000) was obtained from Abcam. The VHH anti-Rabbit IgG (Lot#L1904, 1:5000) was obtained from AlpalifeBio.
Comparative studies of ad2 and ad3 100K
The synthesized gene of ad3-100K was cloned into the vector pCMV. A C-terminal 2 × Strep was added to the recombinant 100K. We transfected HEK293F cells with ad3-hexon and ad2-100K, and ad3-hexon and ad3-100K, respectively. For both cases, the volumes and concentrations of the cells were kept the same. The expression of hexon and 100K were analyzed by western blot (Supplementary Fig. 1c). GAPDH was used as an internal control to normalize the loading of proteins (Supplementary Fig. 1c). We also transfected HEK293F cells with ad2-100K and ad3-100K, respectively. The expression of ad2-100K and ad3-100K were analyzed by western blot with GAPDH as an internal control (Supplementary Fig. 1d).
The cells co-expressed ad2-100K and ad3-hexon were divided into two equal parts and purified with anti-Flag and Strep-Tactin resin, respectively. The cells co-expressed ad3-100K and hexon were also divided into two equal parts and purified with anti-Flag and Strep-Tactin resin, respectively. The eluted samples from anti-Flag and Strep-Tactin resin were analyzed by SDS-PAGE (Supplementary Fig. 1e, f). The eluted samples from anti-Flag resin were concentrated under 500 µL and subjected to SEC analysis with an AKTA pure system and a Superose 6 increase 10/300 GL column (Supplementary Fig. 1f).
To investigate the differences between ad2-100K and ad3-100K, we replaced the N-terminal amino acids 1-93 of ad2-100K with the N-terminal amino acids 1-121 of ad3-100K based on the sequence alignment (Supplementary Fig. 2) to obtain the mutant ad3N1-121-ad2-100K. We co-expressed hexon and ad3N1-121-ad2-100K in HEK293F cells and purified the complex with Strep-Tactin resin. The eluted fractions from the Strep-Tactin resin were analyzed by SDS-PAGE (Supplementary Fig. 11b).
Data acquisition, image processing, model building, and structure refinement
CryoEM micrographs of hexon-100K were collected at a nominal magnification of 22,500 (which yields a calibrated pixel size of 1.25 Å) on a 300 kV Titan Krios equipped with a Gatan K3 camera. Images were recorded as movie stacks under the counting mode in a defocus range of −1.0 μm to −3.5 μm. A total dose of ~50 electrons per Å2 was used. AutoEMation2 was used for the data collection. A total of 10,560 movie stacks were collected. The 32 frames in each movie stack were aligned, summed and 2 × binned by utilizing the program MotionCor231. The CTF parameters of the micrographs were determined by using the program Gctf, which takes local defocus variations into consideration32.
A total of 5,856,543 particles were boxed by using Gautomatch. All the extracted particles were binned by 4 pixels × 4 pixels and subjected to reference-free 2D classifications by using RELION3.033. The particles from classes with apparent features were selected for further 3D classifications with C3 symmetry imposed (Supplementary Figs. 3 and 4d). All the classes from the 3D classifications were processed for further 3D auto-refinements. The resultant maps from the three classes show clear structural details and better resolutions than other classes. Then the class of hexon with extra densities was used as a reference model to guide 3D classifications. After two rounds of 3D classifications, the selected particles were combined and used for 3D refinements that resulted in a 3.23 Å density map. To improve the reconstruction of 100K, the densities of hexon were subtracted from the complex to allow focused 3D refinements of 100K. The refined map was used as reference model to guide multi-rounds of 3D classifications. The selected classes were used for final refinements, resulting in a 3.79 Å map of 100K. The local resolution of the cryoEM density map was calculated by using ResMap34. Combined with alphafold predicted model, this 3.79 Å map was used for model building. The atomic model was built and adjusted by using COOT35. The model was refined by using the PHENIX cryoEM Real-space Refinement tool36. Some of the figures to show the density maps and models were prepared by using the UCSF chimera37 and chimeraX38.
Functional verification of the contacts at sites A, B, and C
Six truncation mutants of ad2-100K were made with a C terminal 2 × Strep tag, including mutants Δ1–38, Δ1–93, Δ1–135, Δ657–805, Δ680–805, and Δ693–805, which contain residues 39–805, 94–805, 136–805, 1–656, 1–679 and 1–692 of ad2-100K, respectively (Supplementary Fig. 11a, c). Two additional mutants of ad2-100K, ad2-100K-Δ285-310 (GS) and ad2-100K-Δ600-608 (GS), were made with a C terminal 2 × Strep tag. In these two mutants, the loops 285–310 and 600–608 were replaced by SGGSGGSGG and SGGSGGSGGSGG, respectively. These 100K mutants were co-expressed with 3 × Flag-ad3 hexon, respectively, and the recombinant hexon was purified by anti-Flag resin following a similar procedure as mentioned before. For comparisons, we also co-expressed the ad2-100K with a C terminal 2xStrep tag, and 3×Flag-Ad3 hexon. The eluted fractions from anti-Flag resin were concentrated and subjected to SEC analysis with an AKTA pure system and a Superose 6 increase 10/300 GL column. Peak fractions were collected and further analyzed by using SDS-PAGE and negative staining microscopy (Fig. 5a).
To verify the function of contacts at site C, we made the 3×Flag tagged ad3 hexon mutant V50L-A51F-T53W (hexon-V50L-A51F-T53W), 2×Strep tagged ad2-100K mutants A251F-A254W-V255L and ad2-100K mutant A251F-A254W-V255L without any tag (100K-251F-A254W-V255L). The C-terminal 2×Strep tagged ad2-100K mutant A251F-A254W-V255L was used to test the expression level of 100K (Supplementary Fig. 11d). We co-expressed Flag-hexon-V50L-A51F-T53W with 100K-notag, hexon with ad2-100K-A251F-A254W-V255L, and the Flag-hexon-V50L-A51F-T53W with ad2-100K-A251F-A254W-V255L, respectively. The recombinant hexon proteins were purified by anti-Flag resin following a similar procedure as mentioned before. The eluted fractions from the anti-Flag resin were concentrated and subjected to SEC analysis with an AKTA pure system and a Superose 6 increase 10/300 GL column. Peak fractions were collected and further analyzed by using SDS-PAGE, negative staining microscopy, and ELISA (Fig. 5a–c and Supplementary Fig. 11i).
All 100K mutants were purified using Strep-Tactin resin and the elution fractions were analyzed by SDS-PAGE and western blot (Supplementary Fig. 11a–d). The eluted samples of mutants ad2-100K Δ680–805, Δ693–805, Δ285–310 (GS) were concentrated and applied to a 10–30% w/v linear glycerol gradient supplemented with 0.15% glutaraldehyde for gradient fixation (GraFix). The gradient was run for 13 h at 200,000 × g (Supplementary Fig. 11e–h)
Mutagenesis studies of the dimeric 100K
Ad2-100K mutant ad2-100K C515S-C516S were made at the dimerization interface. The 100K mutants were co-expressed with 3 × Flag-ad3-hexon. The hexon was purified by anti-Flag resin following a similar procedure as mentioned before. The eluted fractions from the anti-Flag resin were concentrated to a volume of approximately 500 µL and subjected to SEC analysis with an AKTA pure system and a Superose 6 increase 10/300 GL column. Peak fractions were collected and further analyzed by using SDS-PAGE gels and negative staining microscopy (Fig. 6a). Additionally, we made the mutant ad2-100K-Q498A-N499A-strep and tested the expression of this mutant. By following a similar procedure as mentioned above, we did not detect any band of 100K on the SDS-PAGE gel (Fig. 6c).
The oligomerization state and chaperone function of the 100K mutant Δ693–805 is similar to those of the wild type protein (Fig. 5a and Supplementary Fig. 11f), we then made the ad2 100K mutants ad2-100KΔ 693-805 + Q498A-N499A-strepII and ad2-100K Δ693–805 + Q498A-N499A (no tag). Expression yield of the mutant ad2-100K Δ693-805 + Q498A-N499A-strepII is normal (Fig. 6d). To determine the oligomerization state of 100K, the eluted 100K from the Strep resin was concentrated and applied to a 10–30% w/v linear glycerol gradient supplemented with 0.15% glutaraldehyde for gradient fixation (GraFix). The gradient centrifugation was run for 13 h at 200,000 × g. We also co-expressed 3 × Flag-ad3-hexon with ad2-100K Δ693-805 + Q498A-N499A. The recombinant hexon was purified by following a similar procedure as having been described above. The eluted fractions from the anti-Flag resin were concentrated and subjected to SEC analysis with an AKTA pure system and a Superose 6 increase 10/300 GL column (Fig. 6f). The oligomerization state of ad2-100K-Δ694-805 + Q498A-N499A was also determined by GraFix following a similar procedure as that for the wild type 100K (Fig. 6e).
ELISA
High-binding 96 well ELISA plates (Corning #9018) were coated overnight at 4 °C with 100 µL 1 µg/mL fractions of the trimer peak in SEC with samples prepared by co-expressing ad3-hexon and ad2 100K, and fractions of the soluble aggregation peak, trimer peak and protomer peak in SEC with samples prepared by co-expressing ad3-hexon and ad2-100K-A251F-A254W-V255L. Samples prepared by expressing hexon alone and by treating the SEC fractions at 96 °C for 20 min were used as controls. The wells were washed with 1×PBS and 0.05% Tween20 (washing buffer) and were blocked for 1 h at 37 °C with 200 µL of 1×PBS, 3% BSA, and 0.05% Tween20 (blocking buffer). The rabbit polyclonal adenovirus type 5 antibody (catalog no. ab6982; Abcam) was diluted 1:5000 in the blocking buffer. Then 100 µL diluted antibody was added to each well, and incubated for 1 h at 37 °C. The plates were washed 3 times with washing buffer and then was added with the 1:5000 diluted horseradish peroxidase (HRP) conjugated VHH anti-Rabbit IgG (AlpalifeBio) antibody. The plate was incubated for 1 h at 37 °C. Following five additional washes, 100 µL of TMB was added to each well and the absorbance at 450 nm was read after 20 min (BioTek 800 TS).
Sample preparation for mass spectrometry
The 100K-hexon complex sample collected from the sequential affinity purification was prepared without DDT and β-mercaptoethanol and was subjected to SDS-PAGE analysis. After electrophoresis, the gel was stained with Coomassie Brilliant Blue to visualize protein bands. The protein gel bands were cut and excised for in-gel digestion, and proteins were identified by mass spectrometry. In-gel digestion was performed using sequencing grade-modified trypsin in 50 mM ammonium bicarbonate at 37 °C overnight. The peptides were extracted twice with 1% trifluoroacetic acid in 50% acetonitrile aqueous solution for 30 min. The peptide extracts were then centrifuged in a SpeedVac to reduce the volume.
For LC-MS/MS analysis, peptides were separated by a 60 min gradient elution at a flow rate 0.300 μL/min with a Thermo-Dionex Ultimate 3000 HPLC system, which was directly interfaced with the Thermo Orbitrap Fusion mass spectrometer. The analytical column was a homemade fused silica capillary column (75 μm ID, 150 mm length; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 A, 5 μm; Varian, Lexington, MA). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid. The Orbitrap Fusion mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur3.0 and there is a single full-scan mass spectrum in the Orbitrap (350–1550 m/z, 120,000 resolution) followed by 3 seconds data-dependent MS/MS scans in an Ion Routing Multipole at 30% normalized collision energy (HCD).
The MS/MS spectra from each LC-MS/MS run were searched using the software pLink 239. pLink search parameters: linkers SS, precursor mass tolerance 20 parts per million (ppm), fragment mass tolerance 0.02 Da, peptide length minimum 6 amino acids and maximum 60 amino acids per chain, peptide mass minimum 600 and maximum 6000 Da per chain, variable modification M 15.994915, enzyme Trypsin-specific up to 3 missed cleavages. The protein sequences were downloaded from Uniprot (P24932).
Structure prediction
The structure of 100K was predicted by using the AlphaFoldColab40. The predicted structure of 100K was used to refine the 100K model based on the cryo-EM map. The refined 100K model have been deposited into the Protein Data Bank (http://www.pdb.org) under ID 9IVW.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and EM maps have been deposited into the Protein Data Bank (http://www.pdb.org) and the EM Data Bank (http://www.emdataresource.org), respectively, with the accession numbers EMD-60935 (100K dimer), EMD-60936 (hexon-100K complex), EMD-60937 (mature hexon), 9IVW (atomic models of 100K dimer), 9IVX (atomic models of hexon-100K complex), 9IW0 (atomic models of mature hexon). Source data are provided as a Source Data file. Source data are provided with this paper.
References
Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 14, 630–642 (2013).
Cobbold, C., Windsor, M. & Wileman, T. A virally encoded chaperone specialized for folding of the major capsid protein of African swine fever virus. J. Virol. 75, 7221–7229 (2001).
Cepko, C. L. & Sharp, P. A. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31, 407–415 (1982).
Epifano, C., Krijnse-Locker, J., Salas, M. L., Rodríguez, J. M. & Salas, J. The African swine fever virus nonstructural protein pB602L is required for formation of the icosahedral capsid of the virus particle. J. Virol. 80, 12260–12270 (2006).
van der Vies, S. M., Gatenby, A. A. & Georgopoulos, C. Bacteriophage T4 encodes a co-chaperonin that can substitute for Escherichia coli GroES in protein folding. Nature 368, 654–656 (1994).
Seo, H. W., Seo, J. P. & Jung, G. Heat shock protein 70 and heat shock protein 90 synergistically increase hepatitis B viral capsid assembly. Biochem. Biophys. Res. Commun. 503, 2892–2898 (2018).
Chen, C., Wang, J. C. & Zlotnick, A. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog. 7, e1002388 (2011).
Chromy, L. R., Pipas, J. M. & Garcea, R. L. Chaperone-mediated in vitro assembly of Polyomavirus capsids. Proc. Natl. Acad. Sci. USA 100, 10477–10482 (2003).
Zhang, W. J. et al. Hsp90 is involved in pseudorabies virus virion assembly via stabilizing major capsid protein VP5. Virology 553, 70–80 (2021).
Geller, R., Vignuzzi, M., Andino, R. & Frydman, J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 21, 195–205 (2007).
Roberts, M. M., White, J. L., Grutter, M. G. & Burnett, R. M. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232, 1148–1151 (1986).
Liu, Q. et al. Structure of the African swine fever virus major capsid protein p72. Cell Res 29, 953–955 (2019).
Burnett, R. M. The structure of the adenovirus capsid: II. The packing symmetry of hexon and its implications for viral architecture. J. Mol. Biol. 185, 125–143 (1985).
Persson, B. D. et al. Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46. Proc. Natl. Acad. Sci. USA 118, e2020732118 (2021).
Cassany, A. et al. Nuclear import of adenovirus DNA involves direct interaction of hexon with an N-terminal domain of the nucleoporin Nup214. J. Virol. 89, 1719–1730 (2015).
Xi, Q., Cuesta, R. & Schneider, R. J. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J. Virol. 79, 5676–5683 (2005).
Makadiya, N., Gaba, A. & Tikoo, S. K. Cleavage of bovine adenovirus type 3 non-structural 100K protein by protease is required for nuclear localization in infected cells but is not essential for virus replication. J. Gen. Virol. 96, 2749–2763 (2015).
Hong, S. S. et al. The 100K-chaperone protein from adenovirus serotype 2 (Subgroup C) assists in trimerization and nuclear localization of hexons from subgroups C and B adenoviruses. J. Mol. Biol. 352, 125–138 (2005).
Andrade, F. et al. Adenovirus L4-100K assembly protein is a granzyme B substrate that potently inhibits granzyme B-mediated cell death. Immunity 14, 751–761 (2001).
Zhu, R. et al. Molecular Mechanism of Adenovirus Late Protein L4-100K Chaperones the Trimerization of Hexon. J. Virol. 97, e0146722 (2023).
Stark, H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol 481, 109–126 (2010).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Rux, J. J., Kuser, P. R. & Burnett, R. M. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77, 9553–9566 (2003).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Yan, J. et al. Interaction between hexon and L4-100K determines virus rescue and growth of hexon-chimeric recombinant Ad5 vectors. Sci. Rep. 6, 22464 (2016).
Yu, B. et al. Chimeric hexon HVRs protein reflects partial function of adenovirus. Biochem. Biophys. Res. Commun. 421, 170–176 (2012).
Morin, N. & Boulanger, P. Hexon trimerization occurring in an assembly-defective, 100k temperature-sensitive mutant of adenovirus-2. Virology 152, 11–31 (1986).
Morin, N. & Boulanger, P. Morphogenesis of human adenovirus type 2: sequence of entry of proteins into previral and viral particles. Virology 136, 153–167 (1984).
Xi, Q., Cuesta, R. & Schneider, R. J. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 18, 1997–2009 (2004).
Iacovides, D. C., O’Shea, C. C., Oses-Prieto, J., Burlingame, A. & McCormick, F. Critical role for arginine methylation in adenovirus-infected cells. J. Virol. 81, 13209–13217 (2007).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Acknowledgements
We thank the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for providing the facility support. We thank Dr. Jianlin Lei, Xiaomin Li, and Fan Yang for cryo-EM data collection. We thank Dr. Haiteng Deng and Xianbin Meng in Proteinomics Facility at Technology Center for Protein Sciences, Tsinghua University, for MS analysis. Y.X. is supported by the National Key R&D Program of China (grants: 2023YFC2306300 and 2021YFA1300204), the National Natural Science Foundation of China (NSFC, grants: 31925023, 21827810, 31861143027), the Beijing Frontier Research Center for Biological Structure, the SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine and the Tsinghua-Peking Center for Life Sciences.
Author information
Authors and Affiliations
Contributions
Q.L., H.N.L., L.Y.S., and Z.L. planned and performed the biochemical experiments. Q.L. and H.N.L. analyzed data, prepared the figures, and wrote the manuscript together with Y.X. Y.X. planned and supervised the experiments, analyzed the data, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Pablo Guardado-Calvo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, H., Shao, L., Liu, Z. et al. Molecular mechanisms of the viral encoded chaperone 100K in capsid folding and assembly of adenovirus. Nat Commun 16, 4013 (2025). https://doi.org/10.1038/s41467-025-59301-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59301-4