Abstract
Plants rapidly induce strong abscisic acid (ABA) signaling in response to stress, but how they weaken ABA signaling to resume normal growth after stress is unclear. Here, we find that arginine methyltransferase 6b (OsPRMT6b) methylates three arginine residues (R48, R79, R113) in ABA receptor OsPYL/RCAR10 (OsPYR1-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR, R10), thereby enhancing its interaction with Tiller Enhancer (TE) and promoting its ubiquitination and degradation through the 26S-proteasome pathway. OsPRMT6b is induced by ABA at both transcriptional and translational levels. Further, we find that R10 protein accumulates under high temperature stress but declines as temperature drops, whereas OsPRMT6b protein shows an opposite trend. And WT plants display a better growth recovery than osprmt6b mutants after high temperature. These findings suggest that OsPRMT6b acts as a switch to downregulate ABA signaling for growth recovery after high temperature stress.
Similar content being viewed by others
Introduction
Abscisic acid (ABA) is a major plant hormone well known for its role in regulating stress response and plant growth and development1,2,3,4,5. When subjected to environmental stresses, such as extreme temperature, drought, and salinity, plants activate biosynthesis of ABA and ABA signaling, leading to genome-wide reprogramming of the transcriptome. As a result, plant growth is inhibited and adaptation to stress is enhanced. Thus, understanding the balance of ABA-mediated adaptation to stress and the growth recovery is of fundamental importance to plant biology as well as to breeding environmentally resilient crops.
Over the past few decades, the framework of ABA signaling has been extensively studied and is well-established in model plant species Arabidopsis thaliana and rice. PYR/PYL/RCARs receptors, classified as the steroidogenic acute regulatory protein-related lipid transfer (START) domain superfamily, as well as type 2C protein phosphatases (PP2Cs) and SNF1-related kinases 2 (SnRK2s), are the core components of the ABA signaling pathway5,6,7,8. During the ABA signaling, ABA recruits PYR/PYL/RCARs receptors to inhibit the activities of PP2Cs by forming ternary complexes, thus releasing the kinase activity of SnRK2s to phosphorylate downstream ABA-responsive factors for activating ABA responses3,5,9,10,11, and as a result, growth is often inhibited. However, the way by which ABA signaling is downregulated to enable growth recovery after relieving of stress remains largely elusive.
Post-translational modifications including phosphorylation, ubiquitination, acetylation and methylation are involved in multiple biological processes, such as signal transduction, pre-mRNA splicing, DNA repair, and protein interaction12,13,14,15,16,17. As one of the important post-translational modifications, arginine methylation catalyzed by protein arginine methyltransferases (PRMTs) is involved in many vital cellular processes such as mRNA splicing, RNA metabolism, spliceosome assembly and transcriptional regulation15,16,17,18,19,20. PRMTs are evolutionarily conserved in eukaryotes and widely exist in organisms ranging from single-cell yeast to animals and plants. Nine PRMTs in humans15,16,17, nine in Arabidopsis and eight in rice have been identified18,19. Various studies in Arabidopsis have shown that this family controls essential traits including the circadian clock, vegetative growth, flowering time and response to high salinity and ABA21,22,23,24,25,26. For example, AtPRMT3 regulates ribosome biogenesis21; AtPRMT5 plays a significant role in regulating spliceosome assembly, pre-RNA splicing, flowering, iron homeostasis22,23,24,25 and methylates RNA binding factors, U snRNP (uridine-rich small nuclear ribonucleoprotein particles) Sm protein AtSmD1, D3, and AtLSm4 protein to mediate RNA metabolism22; and AtPRMT10 regulates flowering by upregulating expression of flowering repressor FLOWERING LOCUS C (FLC)26. Although the function of PRMT family proteins in animals and Arabidopsis has been studied in depth27,28, their functions in rice remain largely unclear.
The ABA receptor OsPYL/RCAR10 (hereafter, R10) in rice plays a critical role in inhibiting seed germination and seedling growth under adverse environmental conditions29,30,31. ABA stabilizes R10 by weakening the interaction between R10 and Tiller Enhancer (TE), the activator of the E3 ligase APC/CTE complex that targets R10 for ubiquitin-26S proteasome mediated degradation32. In this study, we found that OsPRMT6b is induced by ABA at both the transcriptional and translational levels and that it mediates methylation of R10, thus enhancing the interaction between R10 and TE, leading to destabilization of R10 and growth recovery after relief of high temperature stress. These findings suggest that OsPRMT6b acts as a regulatory knob to fine-tune ABA signaling for growth recovery as the stress fades away.
Results
OsPRMT6b methylates R10
Post-translational modifications such as methylation are key covalent modification processes that occur in almost every protein during or after translation and play important roles in regulating the activity, conformation, stability and subcellular localization of target proteins33,34. To investigate post-translational modifications of ABA receptors, we selected R10, which is highly conserved in maize, wheat and rice (Fig. 1a) for analysis by mass spectrometry. Total proteins from previously generated R10-GFP overexpressing transgenic lines32 were harvested and immunoprecipitated with anti-GFP magnetic beads. Proteins collected from GFP-tag transgenic line were used as the control. The enriched proteins were further examined by mass spectrometry to search for post-translational modification residues in the R10 protein resulting in identification of three monomethylated residues in R10: arginine 48 (R48), arginine 79 (R79) and arginine 113 (R113) (Fig. 1b–d).
a The full-length amino acid sequences and locations of arginine methylation sites in R10 from rice, wheat and maize. Higher energy Collisional Dissociation (HCD) MS/MS spectrum assays identify three arginine methylated peptides of R10: YAVGPGQCSSLLAQR (R48) (b), SCVLR (R79) PDPHHDDNGNDCRPGR (c) and EVSVISGLPASTSTER (R113) (d). ‘R’ represents methylated arginine.
To identify the arginine methyltransferase responsible for methylating R10, an in vitro methyltransferase activity assay using His-tagged R10 protein and all eight GST-tagged OsPRMTs in rice was performed. GST-OsPRMT6a (Os10g0489100) and GST-OsPRMT6b (Os04g0677066), but not the other six GST-OsPRMTs and GST, produced obviously methylated bands in R10, with GST-OsPRMT6b producing stronger methylated bands than GST-OsPRMT6a (Fig. 2a). This indicated that R10 is a substrate of OsPRMT6a and OsPRMT6b, and that OsPRMT6b has higher enzyme activity on R10.
a In vitro methyltransferase activity assay showing methylation levels of R10 catalyzed by eight GST-OsPRMT proteins. Recombinant GST-tagged OsPRMTs were incubated with R10 in the presence of the methyl donor S-adenosyl-L-methionine. R48, R79 and R113 of R10 are key methylation targets of OsPRMT6b. R10 and various demethylation mimicking R10 mutants (48 K, 79 K, 113 K) were incubated with OsPRMT6b (b) or OsPRMT6a (c). Autoradiography indicates methylated R10 proteins. Coomassie blue staining indicates the size and initial amount of each protein in a–c. d In vivo Co-IP assay validating the interaction between R10 and OsPRMT6b in R10-GFP transgenic plants. R10-GFP was precipitated with anti-GFP agarose. OsPRMT6b and R10 proteins were separated by immunoblot and detected with OsPRMT6b and GFP antibodies, respectively (n = three independent experiments). e Bimolecular-fluorescence complementation (BiFC) assay showing the interaction between OsPRMT6b and R10 in the nucleus. mCherry is used as the nuclear marker. The full-length of OsPRMT6b and OsPRMT5 was fused to nYFP. R10 was fused to cYFP. The OsPRMT5 and free R10 fused YFP was used as controls (n = three independent experiments). Merged, merged images of YFP channel, mCherry channel and bright field. Bar, 50 μm. f Luciferase complementation imaging (LCI) assay showing that OsPRMT6b interacts with R10, but the control cannot interact in Nicotiana benthamiana. The full-length of OsPRMT6b and R10 was fused to cLUC and nLUC vectors, respectively (n = three independent experiments). Source data are provided as a Source Data file in a–d.
To determine whether OsPRMT6a or OsPRMT6b is mainly responsible for methylation of R10 at R48, R79 and R113, we mutated these sites into lysine (K) to mimic the demethylation status35,36,37,38 of R10 (single mutants: R10(R48K), R10(R79K) and R10(R113K); double mutants: R10(R48K/R79K), R10(R48K/R113K) and R10(R79K/R113K); triple mutant: R10(R48K/R79K/R113K)). In vitro methyltransferase activity assays showed that when incubated with OsPRMT6b, the methyl-R10 levels were slightly reduced in single mutants, more reduced in the double mutants, and most reduced in the triple mutant, compared to the non-mutated R10 (Fig. 2b). However, no obvious differences in methyl-R10 levels between the single, double and triple mutants and non-mutated R10 were detected when incubated with OsPRMT6a (Fig. 2c). These results indicated that OsPRMT6b, but not OsPRMT6a, was responsible for methylation at the R48, R79 and R113 sites of R10. To confirm whether OsPRMT6b interacts with R10, in vivo Co-IP assay was performed using R10-GFP transgenic lines. The result showed that OsPRMT6b was pulled down by GFP antibodies from R10-GFP lines but not from GFP lines (Fig. 2d). Bimolecular fluorescence complementation (BiFC) assay and a luciferase complementation imaging (LCI) assay in tobacco (Nicotiana benthamiana) further confirmed the interaction between OsPRMT6b and R10 (Fig. 2e, f). These results demonstrated that OsPRMT6b physically interact with R10 to catalyze R10 methylation in vitro.
OsPRMT6b is a negative regulator of ABA signaling
To explore the function of OsPRMT6b in rice, the expression pattern of OsPRMT6b and its protein subcellular localization were investigated. Reverse transcriptase-quantitative PCR (RT-qPCR) analysis indicated that OsPRMT6b was expressed ubiquitously in all tissues examined, including roots, leaves and panicles (Supplementary Fig. 1a). Subcellular localization analysis showed that like R1032, the OsPRMT6b protein was mainly localized in the nucleus (Supplementary Fig. 1b).
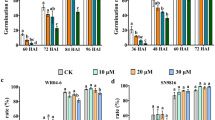
Two osprmt6b mutants, osprmt6b-1 and osprmt6b-2, were generated using CRISPR-Cas9 technology, to further explore the biological function of OsPRMT6b. Western blot analysis confirmed the lack of OsPRMT6b protein in the two mutants (Fig. 3a, b). Both mutants had reduced plant height and increased tiller numbers compared to wild-type (WT) plants under natural long-day conditions in the field (Supplementary Fig. 2). In addition, the osprmt6b-1 and osprmt6b-2 mutants exhibited lower seed germination rates and slower seedling growth in the absence of ABA treatment, and these changes became more evident compared to the WT under treatments with increasing concentrations of ABA (Fig. 3c–h). These results indicate that OsPRMT6b is a negative regulator of ABA signaling.
a OsPRMT6b gene structure and mutation sites in two osprmt6b mutants. The gene structure of OsPRMT6b is from NCBI. Blue boxes, exons; lines, introns. Sequence alignment indicates a “T or A” insertion in the fifth exon close to PAM site recognized by Cas9. The sequencing chromatograms identifying two osprmt6b mutants were attached. b Western blot showing OsPRMT6b protein levels in WT, and osprmt6b-1 and osprmt6b-2 mutants. Germination time course of WT, and osprmt6b-1 and osprmt6b-2 mutant seeds on ½ MS medium containing 0 μM ABA (c), 1 μM ABA (d) or 2 μM ABA (e). n = 21 seedlings. f Phenotypes of 5-day-old WT, and osprmt6b-1 and osprmt6b-2 mutant seedlings in the presence or absence of exogenously applied ABA. Bars, 1 cm. Seedling height (g) and root length (h) of WT, and osprmt6b-1 and osprmt6b-2 seedlings in f. Data are means ± SD in c–e, g and h (n = 16 seedlings). Three biological replicates were used for each treatment. All above P values were calculated by two-tailed t-test. All descriptive statistical values for all box plots and source data are provided as a Source Data file in b–h.
To investigate the molecular basis of the mutant phenotype, we performed RNA-sequencing of osprmt6b-1 mutant and WT seedlings under mock (represented by CK) and ABA (represented by ABA) treatments. Results showed that 2,226 genes in the WT and 2,996 genes in the osprmt6b-1 mutant were differentially expressed (fold change ≥ 1.5, p-value ≤ 0.05) between the mock and ABA treatments (Fig. 4a, b, and Supplementary Data 2). In addition, we identified 903 DEGs (Differentially Expressed Genes) between the osprmt6b-1 mutant and WT under mock conditions, and 1,043 DEGs between the osprmt6b-1 mutant and WT in the presence of ABA (Fig. 4b and Supplementary Data 2). Venn diagram analysis showed that 236 DEGs were up-regulated by ABA (ARUGs, fold change ≥ 1.5, p-value ≤ 0.05) and 936 DEGs that were down-regulated by ABA (ARDGs, fold change ≤ 1.5, p-value ≤ 0.05) in both the WT and osprmt6b-1 mutant (Fig. 4c, d and Supplementary Data 3). We identified that 154 ARUG genes (65.25% of ARUGs) and 592 ARDG genes (63.25%) showed larger expression changes in mutant than in WT (Supplementary Data 4, 5). Gene ontology (GO) analysis showed that the ARUG genes were significantly enriched in response to stimulus and stress, and many of them were related to ABA biosynthesis, ABA signaling and ABA regulation, whereas several ARDG genes were related to GA (Gibberellin) biosynthesis and GA signaling (Fig. 4e, f and Supplementary Data 4–6). We confirmed the expression changes of some representative stress responsive genes related to ABA such as OsDREB1D39, LEA3 and LEA1840, and ABI341 in osprmt6b-1-CK, osprmt6b-1-ABA, WT-CK and WT-ABA using RT-qPCR (Supplementary Fig. 3). All these results demonstrated that OsPRMT6b globally down-regulated expression of ABA-related genes, supporting OsPRMT6b as a negative regulator of ABA signaling.
a RNA-sequencing analysis showing global transcriptional changes between WT and osprmt6b-1 seedlings under mock and ABA treatments. Orange represents genes without significant change; blue indicates genes with significant changes (fold change ≥ 1.5, p-value ≤ 0.05). Numbers of up-regulated and down-regulated genes are given for each sample (n = two independent experiments). b Comparisons of the transcriptome data between samples (straight lines) identified different numbers of DEGs. c Venn diagram displaying shared and up-regulated genes in WT and osprmt6b-1 mutant between mock and ABA treatments. Orange, up-regulated genes between WT-ABA and WT-CK; blue, up-regulated genes between osprmt6b-1-ABA and osprmt6b-1-CK. d Venn diagram displaying shared and down-regulated genes in WT and the osprmt6b mutant between mock and ABA treatments. Orange, down-regulated genes between WT-ABA and WT-CK; blue, down-regulated genes between osprmt6b-1-ABA and osprmt6b-1-CK. e GO analyses show significant functional enrichment of genes in the biological process related to “Response to stimulus and stress”. These genes were from Fig. 4c and d. Gene count represents the number of genes. FDR, false discovery rate. f Heat map showing expression levels of genes related to ABA regulation, ABA biosynthesis, ABA signaling, GA biosynthesis, and GA signaling in the WT (CK) and osprmt6b-1 mutant with or without ABA treatments. “1” and “-1” represent the maximum and minimum value of the relative expression, respectively.
R10 is essential for OsPRMT6b-mediated ABA response
To investigate the genetic relationship between OsPRMT6b and R10, we constructed an r10 mutant using CRISPR/Cas9 and selected an r10/osprmt6b-1 double mutant from a cross between the r10 and osprmt6b-1 mutants, and western blot confirmed the lack of R10 protein in the r10 mutant (Supplementary Fig. 4a, b). Seed germination assays revealed that like the r10 mutant, the r10/osprmt6b-1 double mutant was hyposensitive to ABA in germination and early seedling growth compared with the WT and osprmt6b-1 single mutant (Fig. 5), suggesting that R10 was epistatic to OsPRMT6b and necessary for OsPRMT6b-mediated ABA response.
a Phenotypes of WT, and osprmt6b-1, r10, and r10/osprmt6b-1 mutant seedlings treated with 0 μM, 1 μM or 2 μM ABA. Photographed on the fifth day. Bar, 1 cm. Germination rates of WT, and osprmt6b-1, r10, and r10/osprmt6b-1 mutant seedling on ½ MS medium treated with 0 μM (b), 1 μM (c), or 2 μM (d) ABA. n = 18 seedlings. Heights (e) and root lengths (f) of 5-day-old WT, and osprmt6b-1, r10, and r10/osprmt6b-1 mutant seedlings grown on ½ MS medium. Data are means ± SD (n = 14 seedlings) in b–f. Three biological replicates were used for each treatment in a–d. All above P values were calculated by two-tailed t-test. All descriptive statistical values for all box plots and source data are provided as a Source Data file in (a-f).
We constructed an RNAi vector using the coding sequence of R10 obtained from Lin et al.32 and transformed it into the osprmt6b-1 mutant to create a stable R10RNAi/osprmt6b-1 (abbreviated as R10i/osprmt6-1) transformant with decreased expression of R10 compared to the WT (Supplementary Fig. 4c). We found that compared with WT and the osprmt6b-1 mutant, the R10i and R10i/osprmt6b-1 mutant lines were also ABA-hyposensitive in germination and early seedling growth (Supplementary Fig. 5), further confirming that R10 was necessary for OsPRMT6b-mediated ABA response.
Methylated R10 is vulnerable to ubiquitination and degradation
To test the biological function of arginine methylation of R10 in planta, we substituted the arginine residues R48, R79 and R113 with phenylalanine (F) or lysine (K) to mimic methylated (R103F) or demethylated (R103K) R10 based on previous studies35,36,37,38 and fused them with the GFP-tag in the binary vector pCAMBIA1390 to create transgenic overexpressing lines. We selected overexpressing lines with similar transcriptional expression levels for each R10-GFP variant for further study (Supplementary Fig. 4d). Seed germination assays on media containing 0 μM, 1 μM, or 2 μM ABA showed that all three overexpressing lines (R10-GFP, R103F-GFP, and R103K-GFP) were hypersensitive to ABA compared to WT. Among them, the R103K-GFP lines were more hypersensitive and the R103F-GFP lines less hypersensitive to ABA than the R10-GFP line (Fig. 6). To further confirm this result, we overexpressed R103K and R103F in the r10 mutant background (R103K/r10 and R103F/r10). Further seed germination assays indicated that both R103K/r10 and R103F/r10 overexpressing lines restored the ABA-insensitive phenotype of r10 mutant lines, while R103K/r10 lines showed a more ABA sensitive phenotype than R103F/r10 lines (Supplementary Fig. 6). These observations verified that methylation of R10 attenuates ABA response.
a Germination phenotypes of WT and R10-GFP, R103K-GFP, and R103F-GFP overexpressing lines treated with different concentrations of ABA. Photographed on the seventh day. Bars, 1 cm. Germination time course for WT and R10-GFP, R103K-GFP, and R103F-GFP overexpressing lines on ½ MS medium supplemented with 0 μM (b), 1 μM (c), or 2 μM (d) ABA. Data are means ± SD (n = 18 seedlings). Heights (e) and root lengths (f) of 5-day-old WT, and R10-GFP, R103K-GFP, and R103F-GFP overexpressing lines grown on ½ MS. Data are means ± SD (n = 17 seedlings). Three biological replicates were used for each treatment in a–d. All above P values were calculated by a two-tailed t-test. All descriptive statistical values for all box plots and source data are provided as a Source Data file in a–f.
To explore the mechanisms of R10 methylation attenuating ABA response, we examined the protein levels of R10 in WT and osprmt6b-1 plants treated with or without ABA. Western blot analysis showed that R10 protein accumulated in the osprmt6b-1 mutant, and there was further accumulation with ABA treatment (Fig. 7a). RT-qPCR assays of R10 in the WT and osprmt6b-1 mutant showed that the R10 mRNA level was not affected by ABA treatment (Supplementary Fig. 7), which excludes the possibility that R10 accumulation in the osprmt6b-1 mutant was caused by upregulating R10 expression. Notably, the R10 level had obviously declined in WT but not in osprmt6b-1 mutant just when the expression of OsPRMT6b is induced in WT at 6 hours after ABA treatment (Fig. 7a–c), suggesting that the stability of R10 protein is likely affected by OsPRMT6b-mediated methylation. Supporting this, a time-course western blot assay showed that the R103K-GFP protein was quite stable, whereas R103F-GFP was unstable compared to R10-GFP (Fig. 7d), indicating that OsPRMT6b-mediated methylation of R10 clearly weakens its stability.
a Protein levels of R10 in WT and osprmt6b-1 mutant treated with 0 μM or 10 μM ABA for 3 and 6 hours. HSP82, loading control. The WT and osprmt6b-1 seedings were simultaneously treated with 1 mM CHX (n = three independent experiments). b The mRNA levels of OsPRMT6b in rice seedlings treated with 0 μM or 1 μM ABA. Samples were harvested at 0, 1, 3, 6, 12, and 24 h after treatments. CK served as the control (n = two independent experiments). Error bars, standard deviation (All above P values were calculated by two-tailed t-test. c Levels of OsPRMT6b protein in WT and osprmt6b-1 mutant seedlings treated with 0 μM or 1 μM ABA. The membrane was probed with OsPRMT6b and HSP82 antibodies (n = three independent experiments). (d) Western blot assay showing the protein levels of R10-GFP, R103K-GFP, and R103F-GFP in 14-d-old R10-GFP, R103K-GFP, and R103F-GFP overexpressing lines. Samples for western blot analysis occurred at the indicated time points. HSP82, loading control. e Ubiquitination assays of R10, R103K, and R103F proteins incubated with WT and osprmt6b-1 plant total protein extractions. The ubiquitination of R10, R103K or R103F was detected by Anti-Ub antibody (n = three independent experiments). Anti-His, the protein loading control. Ub, Ubiquitination. f Ubiquitination and methylation of R10-GFP protein in rice protoplasts of WT and osprmt6b-1 mutant. Total protein of protoplasts was extracted, proteins separated using SDS-PAGE, and levels of ubiquitination and methylation detected using respective antibodies. Anti-GFP was used as a loading control (n = three independent experiments). g In vivo co-immunoprecipitation assays of TE with R10, R103F, and R103K in rice. The R10, R103F, and R103K proteins were enriched by GFP beads, and an equal amount of these enriched proteins was incubated with the total protein of His-TE overexpressing transgenic line (n = three independent experiments). Numbers below the top band show relative protein levels in a, c–g. Source data are provided as a Source Data file in a–g.
Since degradation of R10 relies on ubiquitination mediated by the APC/CTE complex32, we predicted that OsPRMT6b-mediated methylation would enhance ubiquitination of R10. Supporting this, in vitro ubiquitination assays showed that the methylation mimic R103F underwent higher ubiquitination whereas the demethylation mimic R103K showed lower ubiquitination than R10 in WT total proteins, and the ubiquitination levels of R10 were clearly reduced while R103F still was effectively ubiquitinated in osprmt6b-1 mutant total protein extracts compared to in WT total proteins (Fig. 7e and Supplementary Fig. 8). Furthermore, levels of both methylation and ubiquitination of R10 were higher in protoplasts from WT than in osprmt6b-1 mutant protoplasts (Fig. 7f). These findings support the idea that the increased ubiquitination level of R10 is due to the methylation of R10 by OsPRMT6b, which in turn affects the stability of R10. To verify whether OsPRMT6b affects the interaction between TE and R10, we conducted in vivo co-immunoprecipitation assays using plants overexpressing R10-GFP, R103F-GFP, R103K-GFP, and His-TE. We incubated the same amounts of R10-GFP, R103F-GFP and R103K-GFP proteins with His-TE overexpressing plant total proteins and discovered that R103F-GFP interacted more strongly with TE than R103K-GFP or R10-GFP (Fig. 7g and Supplementary Fig. 9), supporting the contention that OsPRMT6b affects the stability of R10 by enhancing interaction between TE and R10, thus destabilizing R10 through the ubiquitination-proteasome pathway.
OsPRMT6b promotes the decline of the ABA signal after high temperature stress
To further investigate the physiological significance of OsPRMT6b, we performed the following analyses. The above results had shown that both the mRNA and protein levels of OsPRMT6b were obviously induced after 6 h ABA treatment (Fig. 7b, c) whereas previous studies had shown that the mRNA expression of R10 was repressed after 6 h of ABA treatment31,32. Thus, the expression of OsPRMT6b and R10 appears to be oppositely regulated by ABA. Examination of the protein levels of OsPRMT6b and R10 in field-grown WT and osprmt6b-1 mutant plants at three time points during the day indicated that the R10 protein level increased with rising temperature and decreased with falling temperature whereas levels of OsPRMT6b showed the opposite trend (Fig. 8a). However, the R10 protein level in the osprmt6b mutant did not decline with falling temperature (Fig. 8a). The R10 protein levels over a 24 h period in WT plants grown in normal and high temperature conditions increased with rising temperature and decreased upon with falling temperature while the OsPRMT6b protein exhibited an opposite pattern (Supplementary Fig. 10a, b). In addition, we detected the concentration of ABA in the field. The ABA content in rice at 2:00 p.m. was higher than at 8:00 a.m. (Supplementary Fig. 10c), which is consistent with the higher levels of R10 protein accompanying with higher temperature levels at 2:00 p.m. (Fig. 8a) and previous studies42,43. At the same time, we transferred 14-day-old WT and osprmt6b-1 mutant seedlings into a simulated high-temperature climate chamber with continuous light to exclude the effect of other environmental factors in the field. In the WT, higher temperature induced the accumulation of R10 but repressed the protein level of OsPRMT6b while lower temperature repressed the protein level of R10 but induced the accumulation of OsPRMT6b, and these opposite expression patterns were disrupted in the osprmt6b-1 mutant (Fig. 8b). To further eliminate the influence of circadian rhythms and other factors, we have measured the ABA content of WT at 8:00 a.m. and 2:00 p.m. under the same temperature conditions (30 °C). The ABA content of WT at 8:00 a.m. was similar to that at 2:00 p.m. (Supplementary Fig. 10d), indicating that the accumulation of ABA at 2:00 p.m. (38 °C) is indeed caused by the high temperature, not related to circadian. To eliminate the influence of water Vapor Pressure Deficit (VPD), we have further conducted temperature treatment assays again by transfer 3-day-old WT and osprmt6b-1 seedlings on agar plates with long shoots always in close contact with the ½ MS medium to the simulated high-temperature climate chamber with continuous light. Consistent with the above results, The R10 protein was accumulated under high-temperature conditions and reduced under normal temperature conditions, while OsPRMT6b protein displayed an opposite change trend in WT. However, the expression pattern of R10 protein was disrupted in osprmt6b-1 mutant (Supplementary Fig. 10e). All of these results indicated that the levels of OsPRMT6b and R10 proteins undergo opposing temperature-dependent expression patterns, suggesting that OsPRMT6b plays a positive role in ABA signal decline and recovery of growth following high temperature stress. To confirm this negative regulator role of OsPRMT6 for R10 in genetics, a growth chamber experiment of WT and osprmt6b simulating a typical day and a day with high temperature in the field was conducted. One-week-old WT and osprmt6b-1 seedlings grown in a simulated natural long-day growth chamber were transferred into a simulated natural high-temperature growth chamber for 5 days. The plant heights at different stages of growth were recorded, and the growth ratio was calculated. We found that no difference in the relative growth of the WT and osprmt6b-1 mutant under typical conditions, whereas under high temperature simulation, the relative growth of the WT was significantly higher than that of the osprmt6b-1 mutant (Fig. 8c, d). Similarly, the phenotypes of the r10 and r10/osprmt6b-1 mutants were examined in growth chambers with simulated high-temperature field conditions. It was found that the r10 and r10/osprmt6b-1 mutants exhibited a phenotype of faster growth and development than WT in the high-temperature stress conditions (Supplementary Fig. 10f). These biochemical and genetic findings indicate that OsPRMT6b functions in the recovery of growth following high temperature stress by reducing the level of R10, and that OsPRMT6b functions in fine-tuning plant growth and stress balance through feedback inhibition of ABA signaling.
a OsPRMT6b and R10 protein levels in WT and osprmt6b-1 mutant seedlings grown in the field. 60-day-old seedlings were collected for protein extractions at the indicated time points during the day. Numbers below the bands show the relative protein levels of OsPRMT6b/HSP82 or R10/HSP82 (n = three independent experiments). The hour numbers represent daytime hours, and the corresponding temperatures refer to the temperature achieved at that moment. b OsPRMT6b and R10 protein levels in WT and osprmt6b-1 mutant seedlings grown in the climate chamber. WT and osprmt6b-1 seedlings were grown in a normal growth chamber (10 h darkness, 25 °C/14 h light, 30 °C conditions, light intensity of 500–800 μmol·m−2·s−1 and relative humidity of 70%) for 14 days and then placed in a simulated high-temperature growth chamber with continuous light for 2 days (n = three independent experiments). Samples were taken at the indicated times. Numbers below the bands show the relative protein levels of OsPRMT6b/HSP82 or R10/HSP82. The hour numbers represent daytime hours, and the corresponding temperatures refer to the temperature achieved at that moment. c Plant heights of WT and osprmt6b-1 seedlings grown in mock and after high temperature conditions. One-week-old WT and osprmt6b-1 seedlings grown in a simulated natural long-day growth chamber were transferred into a simulated high-temperature growth chamber for 5 days prior to recording plant height. Three biological replicates were used for each treatment. Data are means ± SD (n = 17 plants). Regression analysis was conducted on the data of plant heights. The slope represents the change in value. All descriptive statistical values for all box plots are provided in the Source Data files. d Relative changes in plant height for c. Data are means ± SD (n = 17 plants). All above P values were calculated by a two-tailed t-test. Source data are provided as a Source Data file in a–c.
Discussion
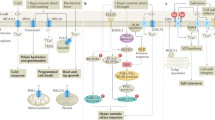
Plants have evolved a variety of mechanisms, including RNA processing, post-transcriptional and post-translational modifications, and reprogramming of gene expression, to coordinate growth and respond to stress. Under stress conditions, plant growth is inhibited. During the recovery phase after subsiding of the stresses, plant growth is restored44. Understanding the balance between stress resistance and growth recovery following stress has significance for improving crop yield. Despite reports of several critical molecular components regulating stress response and plant growth, including SnRK2s, ABA receptors, and TOR45,46, there are few reports on the regulatory mechanisms of plant recovery after stress. Here, we identified a mechanism that OsPRMT6b acts as a valve to tune down ABA signaling for the plant growth recovery after stresses: under stress conditions like high temperature, ABA amount was rapidly induced to stimulate strong ABA responses for the stress tolerance. At the same time, the strong ABA responses also promote the expression and accumulation of OsPRMT6b, which could methylate R10 to tune down ABA signaling by accelerating its ubiquitination and degradation for plant growth recovery when the ambient temperature drops (Fig. 9). Our study demonstrated that OsPRMT6b-R10-mediated ABA signal decline balances the ABA signal pathway to coordinate resumption of plant growth with the decline of stress.
Ambient stresses like high temperature around mid-day promote ABA biosynthesis. This increased ABA binds to ABA receptors such as R10 to activate the ABA signaling pathway for adaptation to high temperature stress. ABA signaling in the WT induces expression of OsPRMT6b to methylate ABA receptors such as R10, rendering their ubiquitination and degradation. Decrease of ABA signaling to recover normal plant growth occurs when the ambient temperature falls to a normal level for plant growth. R10 cannot be methylated in the osprmt6b mutant, and the ABA signaling pathway remains activated, thus inhibiting plant growth and seed germination.
OsPRMT6a and OsPRMT6b are homologs of PRMT6 in rice20. We found that both OsPRMT6a and OsPRMT6b could methylate R10 in vitro, but only OsPRMT6b methylated the three methylation sites identified in planta (Fig. 2). We speculate that other methylation sites in R10 might be specifically methylated by OsPRMT6a in planta or that OsPRMT6b is the major enzyme to methylate R10 in planta. Phylogenetic tree analysis showed that PRMT6 divided into subtypes OsPRMT6a and OsPRMT6b at an early evolutionary stage in monocots (Supplementary Fig. 11). As sequence alignment showed that OsPRMT6a shared 57.3% amino acid similarity with OsPRMT6b (Supplementary Fig. 12), it is possible that the functions of OsPRMT6a and OsPRMT6b diverged during evolution. On the other hand, phylogenetic analysis revealed the presence of putative orthologs of OsPRMT6b in numerous monocotyledonous and dicotyledonous plant species (Supplementary Fig. 11) with AtPRMT6 sharing high homology with OsPRMT6 (Supplementary Fig. 12). We thus speculate that the ABA-OsPRMT6b-R10 regulatory module likely has a conserved role in controlling plant growth and stress response across multiple species.
We showed that OsPRMT6b-mediated methylation of R10 renders strong interaction between R10 and the activator subunit TE in E3 ubiquitin ligase APC/CTE (anaphase-promoting complex/cyclosomeTiller Enhancer), thereby influencing the stability of R10 (Fig. 7d, e, g), consistent with a previous report that arginine methylation impacts protein-protein interactions, protein-DNA or protein-RNA interactions, protein stability, subcellular localization, and enzymic activity. Arginine methyltransferase mediates different fates of substrates by recognizing different hydrogen-bond donors12,13,14,15,16,17. Given the properties of arginine, with five potential hydrogen bond donors, the addition of methyl groups due to spatial effects will affect interacting partners with hydrogen bond acceptors without altering the charge47. The most common type of amino acid sequences favored by PRMTs are arginine- and glycine-rich motifs, termed RGG/RG motifs, which function in nuclear acid binding to mediate protein-protein interactions48. Motifs with arginine adjacent to glycine are predicted to enhance conformational flexibility17. PRMT6 is mainly localized in the nucleus and can simultaneously methylate RGG/RG motifs and arginine adjacent to charged residues in humans49. We speculate that specific changes in the structure and conformation of R10 affected its interaction with TE.
In addition, we also found ABA receptors OsRCAR1, OsRCAR2, OsRCAR3, OsRCAR4, OsRCAR5, OsRCAR6, OsRCAR7, OsRCAR8, OsRCAR9 can also interact with OsPRMT6b (Supplementary Fig. 13), indicating that OsPRMT6b could also regulate ABA signal through modifying another ABA receptors. Besides, mass spectrometry analysis of OsRCAR2-GFP, OsRCAR7-GFP, OsRCAR8-GFP and OsRCAR9-GFP transgenic overexpressing plants showed that the methylation sites were detected in those overexpressing plants (Supplementary Data 7, 8, 9, 10), further indicating that other ABA receptors may also be methylated by OsPRMTs. However, the specific mechanisms by which OsPRMTs methylates other ABA receptors require further investigation. PP2Cs interact with SnRK2 protein kinases to inhibit the kinase activity of the latter, thereby negatively regulating ABA signaling3,50,51,52. To investigate whether methylation affects the interaction between PP2Cs and ABA receptors, we selected four PP2Cs (PP2C8, PP2C30, PP2C51, and PP2C53) to perform pull-down assays with R103F and R103K. The results showed that the interaction between GST-PP2C8, GST-PP2C30, GST-PP2C51, GST-PP2C53, and R103F was slightly stronger than that with R103K (Supplementary Fig. 14), indicating that the methylated ABA receptors also enhanced their interaction with PP2Cs. However, our study indicated that the methylated R103F is more prone to be ubiquitinated and degraded, leading to a significant decline of R103F to affect ABA response (Fig. 7d, e). Therefore, we think there are other mechanisms to affect ABA signaling via the activities of PP2Cs regulated by methylated R10, which still need further study.
With global warming, the rising high temperatures will have negative effects on crop growth and development. Understanding how rice maintains normal growth following high temperature stress and whether it is conserved among various crop species will provide insights on how to produce more stable crop yields in response to climate change. Thus, how to maintain stable plant growth and development amidst fluctuating temperatures and how to balance stress tolerance and plant growth will be key research areas in crop improvement. Our results show that OsPRMT6b can mediate the balance of stress response and plant growth during or following high temperature stress, thus providing a valuable strategy for achieving tolerance to high temperatures in rice and also for enhancing its regional adaptation.
Methods
Accession numbers, vector construction, and growth conditions
The accession number mentioned in this article: TE (Os03g0123300), R10 (Os10g0573400), OsPRMT1 (Os09g0359800), OsPRMT3 (Os07g0640000), OsPRMT4 (Os07g0671700), OsPRMT5 (Os02g0139200), OsPRMT6a (Os10g0489100), OsPRMT6b (Os04g0677066), OsPRMT7 (Os06g0105500), OsPRMT10 (Os06g0142800), OsPP2C8 (Os01g0656200), OsPP2C30 (Os03g0268600), OsPP2C51 (Os05g0572700) and OsPP2C53 (Os05g0592800). An R10-GFP overexpressing transgenic line was obtained from Lin et al.32. To generate R10, R10(48K), R10(79K), R10(113K), R10(48K/79K), R10(48K/113K), R10(79K/113K) and R10(48K/79K/113K) proteins, the coding sequences of seven R10 variants involving three sites were cloned into the pET-28a vector and expressed in the E. coli BL21 (DE3) strain. To obtain R103K-GFP and R103F-GFP overexpressing plants, full-length cDNAs of R103K(48K/79K/113K) (R103k) and R103F(48F/79F/113F) (R103F) were subcloned into the transformed binary vector pCAMBIA1390 driven by Ubi promoter with an attached GFP tag and transformed into wild type plants. To create R103K/r10 and R103F/r10 overexpressing plants, full-length cDNAs of R103k K and R103F were subcloned into the pCAMBIA2300 vector driven by the CaMV35S promoter with no tag and transformed into r10 mutant lines. To create R10-cYFPN, R10-nYFP, PRMT5-nYFP, PRMT6b-nYFP, and nYFP, the full-length of R10, OsPRMT5, and OsPRMT6b genes were fused to the split N- and C-terminal fragment of the Venus vector obtained from Nagai et al.53. The osprmt6b-1 and osprmt6b-2 mutants were produced by CRISPR-Cas9. The full-length CDS of OsPRMT6b driven by the cauliflower mosaic virus (CaMV35S) promoter was inserted into the pAN580 vector to produce a Pro35S::OsPRMT6b-GFP plasmid. Except for specific experimental requirements, plants were grown in the field at Shunyi, Beijing, during the natural growing season. Plants used for germination and seeding growth assays, ubiquitination assays, degradation, RT-qPCR, methyltransferase activity assays, subcellular localization and mass spectrometry were grown in growth chambers with 10 h darkness, 25 °C/14 h light, 30 °C conditions, light intensity of 500–800 μmol·m−2·s−1 and relative humidity of 70%.
Germination assays and ABA treatments
Seed germination and ABA treatment assays were conducted according to Lin et al.32. WT cultivar Kitaake, various mutants, and transgenic rice lines were sterilized in 70% ethanol for 1 min and washed twice with sterilized water. The seeds were submerged in NaClO for 40–50 min, and subsequently washed 5-7 times with sterilized water before planting on half-strength MS (½ MS) medium and various concentrations of ABA. The seeds were placed in a climate chamber, and germination was considered complete when the coleoptile was 5 mm. For ABA treatment, WT and osprmt6b-1 plants were grown in a small chamber with water. After growth for 2 weeks, 10 μM ABA was applied for 24 h; sampling occurred at 0, 1, 3, 6, 12, and 24 h after treatment.
High temperature assays
For the field, we collected samples from a high-temperature day in July in the field at Shunyi, Beijing. For climate chamber, the detailed information on the temperature time course was supplied in Supplementary Fig. 15. Normal growth of 14-day-old (or 3-day old on ½ MS medium) WT and osprmt6b-1 seeds were placed in the high-temperature chamber under continuous light conditions for 2 days firstly, and then the samples were taken at specific time points after high temperature treatment. For ½ MS medium treatment, the seedlings on agar plates are in good contact with the agar medium (the long shoots are always in close contact with the agar) to avoid the effect of VPD.
The information of antibodies
For production of R10-specific, OsPRMT6b-specific and methylation antibodies (Dilute 1000-fold for use): Anti-R10 was obtained from Lin et al.32, and R10 antibody specifically detected endogenous R10 (Supplementary Fig. 4b). Synthetic peptides corresponding to amino acid residues 229 to 331 of the OsPRMT6b protein were used to produce polyclonal rabbit antibodies (GenScript). OsPRMT6b antibody specifically detected endogenous OsPRMT6b (Fig. 3b). The “LDLLDDA” amino acid residues were used to produce the methylation antibody. For commercial antibodies (Dilute 5000-fold for use): Anti-GFP (Roche, Code No: 11814460001, http://www.bio-equip.com/show1equip.asp?equipid=2971069);
Anti-lgG (H + L chain) Rabbit pAb-HRP (MBL, Code No: 458, http://www.mbl-chinawide.cn/search012?keyword=458), is a secondary antibody and universal for all rabbit applications that require secondary antibodies;
Antibody-lgG (H + L chain) Mouse pAb-HRP (MBL, Code No: 330, http://www.mbl-chinawide.cn/search012?keyword=330), is a secondary antibody and universal for all mouse applications that require secondary antibodies;
Antibody-GST (pAb-HRP-Direct Rabbit, WBL, Code No: PM013-7, http://www.mbl-chinawide.cn/search012?keyword=PM013-7);
Anti-His (pAb-HRP-DirecT, MBL, Code No: D291-7, http://www.bio-republic.cn/product/4024.html);
Anti-HSP82 (BPI, Code No: AbM51099-31-PU, http://www.proteomics.org.cn/product/202.html);
Anti-Ub (Ubiquitin, P4D1 Mouse mAb, Code No: sc-8017, https://www.scbt.com/zh/p/ubiquitin-antibody-p4d1).
Protein expression and purification
Constructed vectors were expressed in BL21 (DE3). Bacteria pre-cultured in 1 mL Luria–Bertani (LB) liquid at 37°C were transferred into 100 ml LB and cultured to a concentration of OD600 = 0.6, before adding 0.1–1 mM isopropyl b-D-1-thiogalactopyranoside (IPTG) for 12-16 h at 16°C. The bacteria were resuspended with PBS buffer (1× PBS, 1× protease inhibitor cocktail tablets (PI, supplied from Roche), and 1 mM dithiothreitol (DTT)) and harvested by centrifugation at 2000 x g for 10 min. Intermittent sonication was used to disrupt bacteria by using 3 s pulses and 3 s breaks on ice. Subsequently, the samples were centrifuged twice at 4 °C for 10 min at 6000–14000 x g and the supernatants were incubated with GST beads for 2-4 h. The incubation beads were collected and washed three times with PBS buffer, and the proteins were eluted with the corresponding eluents.
Methyltransferase activity assay
The full-length cDNAs of OsPRMT1, OsPRMT3, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, OsPRMT7, and OsPRMT10 were inserted into pGEX-4T-,1 and R10 cDNA was inserted into pET28a for protein expression assays. 1-2.5 μg of GST-OsPRMT1, GST-OsPRMT3, GST-OsPRMT4, GST-OsPRMT5, GST-OsPRMT6a, GST-OsPRMT6b, GST-OsPRMT7, GST-OsPRMT10 and GST were separately incubated in the HMT buffer (20 mM Tris–HCl (pH 8.0), 4 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), 4 μM S-(methyl-3H) adenosyl-L-methionine (NET155)) with 2.5 μg of His-R10. After incubation at 30°C for 1.5 h, reactions were stopped with an SDS loading buffer. For in vitro methyltransferase assays, 5 μg of enzyme OsPRMT6b and 5 μg of substrate GST-OsPRMT1, GST-OsPRMT3, GST-OsPRMT4, GST-OsPRMT5, GST-OsPRMT6a, GST-OsPRMT6b, GST-OsPRMT7, GST-OsPRMT10, GST were mixed initially, followed by the addition of 5 μL of 10X PBS solution (a Master Mix containing [³H] SAM) and 2 μCi of [methyl ³H]-SAM (78 Ci/mmole). The reaction was then brought to a final volume of 50 μL with water and incubated at 30 °C for 1 hour. Subsequently, 25 μL of 4X SDS Dye and 2 μCi of SAM were added, and the samples were heated at 95 °C for 5 minutes. 10 to 15 μL aliquot of the reaction was then subjected to electrophoresis on a 10% to 15% Acryl/Bis Gel. The protein was separated by 12% SDS-PAGE, the lower section of the gel containing the dye front and [³H] SAM was removed. The gel was stained with Coomassie Brilliant Blue R 250 and imaged. The gel was treated with an amplifier (Amersham Biosciences), dried, and fluorographed on X-ray film. One biological replicate was used for the assay.
In vitro mass spectrometry assays
Seeds of the WT and GFP-R10 overexpressing lines were surface sterilized and germinated in water in the dark at 30 °C for two days. The germinated seedlings were then transferred to soil and grown for about 30 days under normal conditions. 3-5 g seedling tissues were collected and were ground in liquid nitrogen and precipitated overnight with 45 mL TCA/acetone (1:9, 65 mM DTT) at −20 °C. After centrifugation at 5000 x g for 20 minutes, the supernatant was discarded. The pellet was washed with 40 mL cold acetone and centrifuged at 5000 x g for 15 minutes, repeated three times, and then dried at room temperature. The dried powder was resuspended in urea lysis buffer, sonicated, and centrifuged at 16000 x g for 20 minutes. 6 mg of each sample was treated with 10 mM DTT at 37 °C for 2.5 hours, followed by 50 mM IAA in the dark for 30 minutes. The samples were diluted to 1.5 M UA with water and digested with trypsin at 37 °C for 18 hours. The digested samples were desalted using SPE C18 columns and freeze-dried. Methylated peptides were enriched by incubation with Anti-Ac-K antibody beads (PTMScan PTMScan Methyl-R Motif (Methyl-R) Kit, Cell Signal Technology)54. Enriched peptides were separated using an EASY-nLC1000 system. The separated peptides were analyzed by Q-Exactive mass spectrometry. Raw LCMS/MS files were analyzed using Maxquant software (v1.3.0.5). The experiment concluded three control groups and three experimental groups with three biological replicates.
Bimolecular fluorescence complementation (BiFC) assays
The full-length coding regions of R10, OsPRMT5, and OsPRMT6b were fused with cYFP and nYFP of P2YC vector. The plasmids containing R10-cYFP, OsPRMT5-nYFP, OsPRMT6b-nYFP, and nYFP with strain p19 and mCherry for BiFC were co-infiltrated into tobacco leaves for 72 h. R10-cYFP and the empty vectors nYFP, OsPRMT5-nYFP were used as negative controls. Fluorescence signals were observed with a ZESS LSM880 confocal microscope.
Reverse transcription-quantitative PCR (RT-qPCR)
The total RNA was extracted and reverse transcribed it into cDNA. An SYBR Premix Ex Taq TM kit (TaKaRa) and an ABI prism 7500 Real-Time PCR System were used for RT-qPCR analysis. For each assay, RT-qPCR was performed with three technical and two biological repeats. Primers are listed in Supplementary Data 1. Rice ubiquitin (UBQ) was used as the internal control.
Subcellular localization
The full-length OsPRMT6b coding sequence was ligated into the pAN580 vector with GFP at the C terminal. D53 protein was fused to mCherry as a nuclear marker. The GFP-OsPRMT6b vector and D53-mCherry were transiently co-expressed in rice leaf protoplasts. The method for rice leaf protoplast transformation was described previously55. Fluorescence signals were detected by a ZESS LSM880 confocal microscope. Primers are listed in Supplementary Data 1.
In vitro ubiquitination assays
The fusion proteins His-R10, His-R103K, and His-R103F were expressed in E. coli strain BL21 (DE3). Total proteins were extracted from the leaves of 8-day-old rice plants. Equal amounts of His-R10, His-R103K, or His-R103F protein beads and total rice protein extracts were each incubated in ubiquitination buffer (25 mM Tris-HCl (PH 7.5), 10 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1× PI, 10 mM ATP, 40 μM MG132) at 28 °C for 1 h. The beads were collected and thrice washed with ubiquitination buffer before 10% SDS-PAGE and protein detection with anti-His (D291-7, Bio-REPUBLIC) and anti-polyubiquitin (3936T, Gene Lab) antibodies.
RNA sequencing
WT and osprmt6b-1 mutant seedlings were grown on ½ MS media supplemented with 0 μM (mock) or 1 μM ABA under a 14 h light/10 h darkness at 30/25°C for five days. RNAs were extracted using a RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The constructed libraries were sequenced on an Illumina Hiseq X platform. Qualified samples mRNA was sheared into small fragments and subsequently used as a template for the synthesis of double-stranded cDNA in conjunction with primers, deoxynucleotide triphosphates (dNTPs), and DNA polymerase I. The cDNA was purified and size-selected using AMPure XP beads, followed by PCR amplification to generate the final cDNA library. After quality assessment and confirmation of library integrity, sequencing was performed on the HiSeq X platform. HISAT (v.2.2.1) and DESeq (1.18.0) software were used for alignment and DEGs analysis, respectively. Each sample comprised two biological replicates. Genes with p-value ≤ 0.05 and fold change ≥ 1.5 were identified as DEGs. The experiment concluded two control groups and two experimental groups.
In vivo Co-immunoprecipitation (Co-IP) Assay
The full-length coding regions fused a GFP tag or His tag of R10 or TE were transferred into vector pCAMBIA1390 to produce R10-GFP and His-TE transgenic overexpressing plants. The R10, R103F, and R103K proteins were enriched by GFP beads. For each assay, equal amounts of these enriched proteins were added and incubated with His-TE protein. The detailed method of immunoprecipitation is described in previous report32. 10% SDS-PAGE gels were used to separate proteins which were detected by GFP, OsPRMT6b and His antibodies.
Luciferase complementation imaging (LCI) assay
The full-length coding regions of OsRCAR1(R1), OsRCAR2 (R2), OsRCAR3 (R3), OsRCAR4 (R4), OsRCAR5 (R5), OsRCAR6 (R6), OsRCAR7 (R7), OsRCAR8 (R8), OsRCAR9 (R9), R10 and OsPRMT6b were amplified and introduced to pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vector, respectively. Co-transfect the constructed vectors into N. benthamiana leaves via Agrobacterium-mediated transformation. Spray the transformed leaves with 10 mM beetle luciferin (Promega, E1602). Capture the luminescence image using imaging apparatus (Berthold, LB985).
CRISPR-Cas9 method
The CRISPR-P 2.0 website (http://cbi.hzau.edu.cn/cgi-bin/CRISPR) is used to identify the target sites for primer design. Selected primers with high scores that are located within the coding sequence close to the ATG start codon were annealed. The annealed sequences were subsequently cloned into the Cas9 (pOs-cas9) vector. The correct plasmids were transformed into Agrobacterium, which were used to infect rice callus tissue.
Pull-down assay in vitro
The full-length CDS of PP2C8, PP2C30, PP2C51 and PP2C53 were inserted into pGEX-4T-1 vector. GST-PP2C8, GST-PP2C30, GST-PP2C51, GST-PP2C53, R103F-His and R103K-His plasmids were transformed into BL21 (DE3). The protein purification was extracted from BL21 (DE3) as the above method of protein expression and purification. For pull-down assay, GST-PP2C8, GST-PP2C30, GST-PP2C51, GST-PP2C53 and R103F-His, R103K-His fusion proteins were co-incubated with GST beads at 4°C for 1 hour in Tris buffer (25 mM Tris (PH 7.5), 50 mM NaCl, 5 mM DTT, 1 × PI). GST, R103F-His, R103K-His, and GST beads were co-incubated under the same conditions as control. The beads were collected and washed three times with Tris buffer (containing 1% Triton X 100 and 0.1% SDS) and examined by 10% SDS-PAGE. Western blot was conducted with anti-GST and anti-His antibodies.
Measurement of endogenous ABA
60-day-old seedlings in the field were collected for ABA measurement at the indicated time points during the day. WT seedlings were grown in a normal growth chamber for 7 days and then placed in the growth chamber for 2 days with a constant temperature (30 °C), and were collected at 8:00 a.m. and 2:00 p.m. for endogenous ABA measurement. The measurement of endogenous ABA was commissioned to the National Centre for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China. Measurement of endogenous ABA. 200–300 mg rice leaves were ground in liquid nitrogen, weighed, and extracted with methanol and 2H6-ABA for 24 h. The endogenous ABA was subsequently purified and quantified following the methods outlined by Fu et al.56 and Chu et al.57, with slight alterations to the detection parameters. The LC-MS/MS analysis was executed on an ultra-performance liquid chromatography (UPLC) system from Waters, interfaced with the 5500 Qtrap mass spectrometer from AB SCIEX. Chromatographic separation was achieved using a BEH C18 column (1.7 μm particle size, 100 × 2.1 mm dimensions; Waters), with a binary mobile phase consisting of A, 0.05% (v/v) acetic acid in water, and B, 0.05% (v/v) acetic acid in acetonitrile. The gradient program initiated at 20% phase B and ramped up to 70% phase B over a period of 6 minutes. ABA was monitored using multiple reaction monitoring (MRM) with specific transition pairs: 263.0 > 153.1 for ABA and 269.2 > 159.2 for [2H6]-ABA. Each sample was assessed in triplicate for biological consistency. Three biological replicates were used for the assay.
Statistics and reproducibility
Two-tailed student’s t-tests were performed for two two-group comparisons. The statistical methods used and the sample sizes (n) for each experiment are both detailed in the figure legends. Three separate plants per tissue sample were combined to form three biological replicates in RT-qPCR analysis. Each experiment was performed a minimum of three times to ensure consistency.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this work are available in the article and its Supplementary Information files. Plant materials generated in this study are available from the corresponding author upon request. RT-PCR data applied ABI Prism 7500 Real-Time PCR System. Subcellular localization and BiFC assay were applied ZESS LSM880 confocal microscope. LCI assay applied imaging apparatus (Berthold, LB985). Image J was used for the quantitative analysis of all western blots. The software R 4.3.1 was used for data analysis of RNA-seq and plotting of germination assays. MEGA 7.0 software is used for protein sequence alignment and phylogenetic tree construction. The gene structures of OsPRMT6b and R10 are from NCBI; Homologs were obtained from NCBI BLAST. The target sites of primer design were obtained from the CRISPR-P 2.0 website (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). Kitaake reference genome is available at the Gramene website (http://www.gramene.org). The raw data of RNA-seq generated in this study have been deposited in the NCBI database under the BioProject ID: PRJNA1063164. The raw data of mass spectrometry are not available due to the deletion of raw data by the sequencing institution. The processed data of mass spectrometry are available as Source Data. Source data are provided with this paper.
References
Gonzalez-Guzman, M. et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24, 2483–2496 (2012).
Miyakawa, T., Fujita, Y., Yamaguchi-Shinozaki, K. & Tanokura, M. Structure and function of abscisic acid receptors. Trends Plant Sci. 18, 259–266 (2013).
Fujii, H. et al. In vitro reconstitution of an abscisic acid signaling pathway. Nature 462, 660–664 (2009).
Pan, W. B. et al. The UBC27–AIRP3 ubiquitination complex modulates ABA signaling by promoting the degradation of ABI1 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 117, 27694–27702 (2020).
Park, S. Y. et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1107 (2009).
Kong, L. Y. et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6, 8630 (2015).
Belda-Palazon, B. et al. PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms. Proc. Natl. Acad. Sci. USA. 115, E11857–E11863 (2018).
Zhao, Y. et al. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 23, 3340–3351 (2018).
Kim, H. et al. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024 (2012).
Ye, Y. J. et al. A novel chemical inhibitor of ABA signaling targets all ABA receptors. Plant Physiol. 173, 2356–2369 (2017).
Sun, Y. F. et al. A ligand-independent origin of abscisic acid perception. Proc. Natl. Acad. Sci. USA. 116, 24892–24899 (2019).
Bedford, M. T. & Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 (2005).
Zakrzewicz, D. et al. Protein arginine methyltransferases (PRMTs): Promising targets for the treatment of pulmonary disorders. Int. J. Mol. Sci. 13, 12383–12400 (2012).
Hu, J. L. et al. Nitric oxide regulates protein methylation during stress responses in plants. Mol. Cell 67, 702–710 (2017).
Morales, Y., Cáceres, T., May, K. & Hevel, J. M. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch. Biochem. Biophys. 590, 138–152 (2016).
Wesche, J. et al. Protein arginine methylation: a prominent modification and its demethylation. Cell. Mol. Life. Sci. 74, 3305–3315 (2017).
Blanc, R. S. & Richard, S. Arginine methylation: The coming of age. Mol. Cell 65, 8–24 (2017).
Wang, S. C. M., Dowhan, D. H. & Muscat, G. E. O. Epigenetic arginine methylation in breast cancer: emerging therapeutic strategies. J. Mol. Endocrinol. 62, 223–237 (2019).
Ahmad, A. & Cao, X. F. Plant PRMTs broaden the scope of arginine methylation. J. Genet. Genom. 39, 195–208 (2012).
Ahmad, A., Dong, Y. Z. & Cao, X. F. Characterization of the PRMT gene family in rice reveals conservation of arginine methylation. PLoS One 6, e22664 (2011).
Hang, R. L. et al. Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc. Natl. Acad. Sci. Usa. 111, 16190–16195 (2014).
Deng, X. et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA. 107, 19114–19119 (2010).
Deng, X. et al. Recruitment of the NineTeen complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. USA. 113, 5447–5452 (2016).
Fan, H. J. et al. SKB1/PRMT5-mediated histone H4R3 dimethylation of Ib subgroup bHLH genes negatively regulates iron homeostasis in Arabidopsis thaliana. Plant J. 77, 209–221 (2014).
Schmitz, R. J., Sung, S. & Amasino, R. M. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 105, 411–416 (2008).
Cheng, Y. et al. Crystal structure of the plant epigenetic protein arginine methyltransferase 10. J. Mol. Biol. 414, 106–122 (2011).
Branscombe, T. L. et al. PRMT5 (Janus Kinase-binding Protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276, 32971–32976 (2001).
Kanishk, J. G. C. Steven, PRMT7 as a unique member of the protein arginine methyltransferase family: A review. Arch. Biochem. Biophys. 665, 36–45 (2019).
Tian, X. J. et al. Characterization and functional analysis of Pyrabactin Resistance-Like Abscisic Acid Receptor family in rice. Rice 8, 28 (2015).
Miao, C. B. et al. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA. 115, 6058–6063 (2018).
Wang, Y. P. et al. A regulatory loop establishes the link between the circadian clock and abscisic acid signaling in rice. Plant Physiol. 191, 1857–1870 (2023).
Lin, Q. B. et al. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 6, 7981 (2015).
Chen, H. et al. EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Mol. Plant 11, 706–719 (2018).
Cindy, C. & Michaela, U. G. Post-translational control of intracellular pathogen sensing pathways. Trends Immunol. 38, 39–52 (2017).
Wang, Y. P. et al. Arginine methylation of MDH1 by CARM1 Inhibits glutamine metabolism and suppresses pancreatic cancer. Mol. Cell. 64, 673–687 (2016).
Dong, K. et al. OsPRMT6a-mediated arginine methylation of OsJAZ1 regulates jasmonate signaling and spikelet development in rice. Mol. Plant. 17, 900–919 (2024).
McBride, A. E. et al. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 280, 30888–30898 (2005).
Campbell, M. et al. Protein arginine methyltransferase 1-directed methylation of Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Biol. Chem. 287, 5806–5818 (2012).
Zhang, Y. et al. Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep. 42, 486–492 (2009).
Xu, D. et al. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257 (1996).
Hobo, T., Kowyama, Y. & Hattori, T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA. 96, 15348–15353 (1999).
Liu, J. et al. High temperature and drought stress cause abscisic acid and reactive oxygen species accumulation and suppress seed germination growth in rice. Protoplasma 256, 1217–1227 (2019).
Zhu, Y. et al. A separation defect of tapetum cells and microspore mother cells results in male sterility in Brassica napus: The role of abscisic acid in early anther development. Plant Mol. Biol. 72, 111–123 (2010).
Xiong, J. W. et al. The deubiquitinating enzymes UBP12 and UBP13 positively regulate recovery after carbon starvation by modulating BES1 stability in Arabidopsis thaliana. Plant Cell 34, 4516–4530 (2022).
Wang, P. C. et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69, 100–112 (2017).
Zhang, H. et al. Thriving under stress: How plants balance growth and the stress response. Mol. Cell 55, 529–543 (2020).
Fuhrmann, J. et al. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 115, 5413–5461 (2015).
Thandapani, P. et al. Defining the RGG/RG motif. Mol. Cell 50, 613–623 (2013).
Boulanger, M. C. et al. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79, 124–131 (2005).
Fuchs, S. et al. Type 2C protein phosphatases in plants. FEBS J. 280, 681–693 (2013).
Soon, F. F. et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88 (2012).
Nishimura, N. et al. ABA-Hypersitive Germination 1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant. J. 50, 935–949 (2007).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Thompson, J. W. et al. Quantitative Lys-Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J. Biol. Chem. 289, 28942–28955 (2014).
Zhang, Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30 (2011).
Fu, J. et al. Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28, 1081–1087 (2012).
J. Chu et al., Quantitative analysis of plant hormones based on LC-MS/MS. In book: Hormone Metabolism and Signaling in Plants. London: Elsevier 471–537 (2017).
Acknowledgements
We thank Dr. Xiaofeng Cao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for help with the methylation assay; and Dr. Haiteng Deng (Protein Chemistry Facility, School of Biological Sciences, Tsinghua University) for LC-MS/MS analysis of R10. We appreciate the expertise of Miss Shujing Cheng and Dr Jinfang Chu (National Centre for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) in determining ABA. This work was supported by the National Key Research and Development Program of China (2022YFD1201504, Shanshan Zhu), the National Natural Science Foundation of China (31771886, Fu-Qing Wu), the Innovation Program of the Chinese Academy of Agricultural Sciences, and the earmarked fund for CARS-01-05 (Jianmin Wan).
Author information
Authors and Affiliations
Contributions
J. M. W. supervised the study. F. Q. W., S. S. Z., X. J., and Q. B. L. conceived the study and designed the experiments. X. Y. Z., S. Z., W. W. M., P. K. S., L. M. Z., M. X. W., X. D. Z., B. J. L., J. C. W., Y. P. W., K. D., and M. Y. participated in partial biochemical experiments. Z. W. L. analyzed data. Y. L. R., Z. J. C., C. L. L., X. P. G., and X. W. participated in rice transformation. X. J., S. S. Z., F. Q. W., Q. B. L., and H. Y. W. wrote and revised the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jorge Lozano-Juste, Alexander Christmann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jin, X., Lin, Q., Zhang, X. et al. OsPRMT6b balances plant growth and high temperature stress by feedback inhibition of abscisic acid signaling. Nat Commun 16, 5173 (2025). https://doi.org/10.1038/s41467-025-60350-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60350-y