Abstract
Altitude significantly affects both fruit length (FL) and seed germination timing (SGT) in plants. Alpine plants often require prolonged chilling to enhance seed germination rates, yet the molecular mechanisms underlying these adaptations remain largely unexplored. In this study, we have identified the gibberellin biosynthesis gene GA20ox3 as a key regulator of FL and SGT in Arabidopsis thaliana. Our findings demonstrate that DELLA proteins form a negative feedback loop by interacting with the transcription factor DNA-BINDING ONE ZINC FINGER6 (DOF6), which directly binds to the GA20ox3 promoter. Allelic variations in the GA20ox3 promoter of one alpine ecotype influence the binding affinity of DOF6, leading to variations in both FL and SGT. The geographical distribution of the GA20ox3 promoter alleles, corresponding well to different altitudes and temperatures, underscores the significant role of such variation in the A. thaliana’s adaptation to the varied habitats, especially concerning seed germination timing. Our results reveal a cold-mediated gibberellin signaling mechanism that controls both FL and SGT, thereby optimizing reproductive success in varied environments.
Similar content being viewed by others
Introduction
High-altitude environments impose formidable constraints on plant survival, characterized by prolonged winters, abbreviated growing seasons and delayed germination timing1. These pressures select for life-history strategies that balance survival with reproductive success, driving adaptations in phenology, morphology, and physiology. Plants navigate such extremes through three primary strategies—avoidance (e.g., dormancy), tolerance (e.g., cold-acclimated metabolism), and escape (e.g., accelerated flowering)—with their prevalence shaped by genetic diversity and environmental heterogeneity2,3. These adaptive responses not only determine species distributions but also manifest in suites of correlated traits optimized for cold resilience4,5. A hallmark of alpine and Arctic flora is the reduction in fruit size, a strategy hypothesized to minimize energy expenditure during development under resource-limited, low-temperature conditions6,7,8,9. Fruits integrate phytohormonal cues, particularly gibberellin (GA) signaling, which coordinates embryogenesis and maturation10. Smaller fruits may thus represent an energy-saving adaptation, enabling viable seed production within truncated growing seasons7,9,11. Concurrently, precise timing of life-history transitions (for example, annual to biennial or winter to summer)—notably germination—is critical12,13. Alpine plants frequently exhibit physiological seed dormancy, requiring extended cold stratification to break dormancy, thereby synchronizing germination with favorable spring/summer conditions14,15,16,17,18,19,20. This delay avoids premature growth susceptible to frost mortality while ensuring reproductive output aligns with ephemeral summer windows21,22,23,24. The regulation of dormancy and germination hinges on antagonistic interactions between GA and abscisic acid (ABA), hormones that promote and suppress germination, respectively. Prolonged cold exposure not only degrades ABA but also primes GA sensitivity, creating a permissive state for germination25,26. Paradoxically, persistent low temperatures may reactivate ABA pathways, inducing secondary dormancy in stratified seeds27,28,29. Such a duality highlights the multifaceted role of cold stratification as both a terminator of dormancy and an inducer, showcasing the intricate interplay between genetic factors and environmental cues. This dual effect of cold stratification on seed dormancy is particularly evident in diverse Arabidopsis thaliana ecotypes around the world30. Concurrently, with increasing altitude, there is a shift in the life cycle of annual plants from winter to spring blooming13,24, which is accompanied by a decline in reproductive output31. However, the genetic foundations of these adaptations, as well as their underlying molecular relationships and interactions—such as the link between small fruit size and delayed seed germination—remain largely unexplored.
GAs are a group of tetracyclic diterpenoid carboxylic acids, with GA1 and GA4 being the main bioactive forms found in higher plants32. They stimulate growth by promoting cell division and elongation, and they facilitate transitions between different developmental stages33. GAs are synthesized from trans-geranylgeranyl diphosphate in plastids through the methylerythritol phosphate pathway34, involving terpene cyclases, cytochrome P450 monooxygenases, and subsequently dioxygenases35. The isozymes GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) comprise two key elements of these dioxygenases that are regulated by both developmental and environmental signals36. During the fruit (silique) development stage in A. thaliana, GA3ox1 is predominantly expressed in the replum, funiculus, and fruit receptacle, whereas its three homologs (GA3ox2–GA3ox4) are expressed exclusively in developing seeds. Mutants lacking GA3ox1, either alone or in combination with GA3ox2–GA3ox4, exhibit shorter fruits and delayed seed germination10,37. Additionally, A. thaliana possesses five GA20ox genes (GA20ox1–GA20ox5)38,39. Among these, GA20ox3 exhibits redundant functions in fruit development alongside GA20ox1 and GA20ox240,41,42. Furthermore, GA20ox3 plays a crucial role in both embryogenesis and seed germination43,44.
It should be noted that DELLA proteins, including GA INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), and the RGA-LIKE1 (RGL1), RGL2, and RGL3 family members, act as crucial intracellular repressors in the GA signaling pathway45,46. Bioactive GAs are recognized by the receptors known as GID1 (GA INSENSITIVE DWARF1) proteins. This interaction forms a GA-GID1 complex, which then facilitates binding with DELLA proteins. The formation of this complex triggers the ubiquitination and subsequent proteasomal degradation of DELLA proteins. This process relieves the repression, thereby enabling the activation of GA-responsive genes47. DELLA proteins also display both redundant and unique roles in various developmental processes. For example, GAI, RGA, RGL1, and RGL2 collectively inhibit pistil growth and fruit development48,49. While GAI and RGA are involved in seed germination, RGL2 serves as the primary regulator of this GA-mediated process50,51. Although DELLA proteins lack DNA-binding domains, they can participate in transcriptional regulation by forming functional complexes with transcription factors52. For example, GAI interacts with the transcription factor DAG1 to repress the expression of the GA biosynthetic gene GA3ox137. RGL2 also interacts with the NF-YC3, NF-YC4, and NF-YC9 transcription factors to enhance the expression of ABI553. Additionally, RGL2 interacts with the DOF6 transcription factor, which promotes the expression of GATA12, thereby inhibiting the germination of freshly harvested or unstratified seeds54,55.
In this study, we performed genome-wide association studies (GWAS) on a population of recombinant inbred lines (RILs) derived from two A. thaliana ecotypes—one from low altitudes and the other from high altitudes56. These ecotypes exhibit significantly different fruit length (FL) and cold-induced seed germination timing (SGT). We identified the GA biosynthetic gene GA20ox3 as the key regulator controlling these two traits. Significant differences were observed in the expression levels of GA20ox3 during fruit development and SGT between the two ecotypes. Our findings revealed that the DELLA-DOF6 transcriptional regulatory complex directly and negatively regulates GA20ox3 expression during these physiological processes, illustrating a DOF6-dependent negative feedback loop between GA synthesis and signaling. In comparison to the low-elevation ecotype, the GA20ox3 variants due to the allelic changes in the promoter of the high-elevation ecotype exhibit increased binding affinity for the DOF6 protein. This interaction leads to reduced GA20ox3 transcription, resulting in shorter fruits and a longer cold stratification period required for seed germination, which is an adaptive strategy for the colder environment. Finally, we identified four distinct haplotypes of the GA20ox3 promoter from global A. thaliana ecotypes and demonstrated their varying binding affinities for DOF6. These differences likely contribute to the local adaptation of A. thaliana. Our findings underscore the importance of allelic variations in the GA20ox3 promoter in shaping natural differences in FL and SGT, which facilitate A. thaliana’s adaptation to diverse environmental temperatures. Additionally, these results offer insights into the molecular mechanisms underlying morphological and phenological adaptations to cold in this annual herb.
Results
The genetic variations at the GA20ox3 locus are associated with differences in FL and SGT
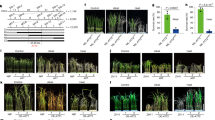
The A. thaliana Tibet (Tib) ecotype originates from an elevation of 4,194 meters, which is much higher than the elevation of the Sichuan (Sic) ecotype at 456 meters (Fig. 1a). The Tib ecotype, characterized by a small and branching statue, germinates in late spring and early summer, flowering and setting fruits during summer and early autumn. In contrast, the Sic ecotype has a taller structure with fewer branches, germinating in winter and procuring flowers and fruits in winter and spring. Various temperature indices show that the Tib habitat is much colder than the Sic habitat and experiences a longer duration of cold weather during the non-growing season (Supplementary Fig. 1). As expected, the Tib ecotype exhibits significantly shorter FL compared to the Sic ecotype, even when both are grown in a greenhouse throughout their entire developmental stages (Fig. 1b, c). Additionally, freshly harvested seeds of the Tib ecotype require more cold stratification to germinate compared to those of the Sic ecotype; however, a prolonged cold stratification period of 31 days does not inhibit the germination rate of either ecotype (Fig. 1c, d). These two specific adaptive traits suggest that they arise from genetic variations between the two ecotypes.
Images of Sichuan and Tibet ecotypes at the reproductive growth stage in their natural habitat (a) and a controlled growth chamber (b). c Fruit development stages in A. thaliana from the Sichuan and Tibet ecotypes are depicted from the first day after pollination (1 DAP) through to seed shedding (21 DAP), with a scale bar representing 2 mm. Freshly harvested seeds were subjected to cold stratification for 0, 3, 10 or 31 days, and germination phenotypes were assessed after 4 days. d Statistical analysis of fruit length and germination rate in (c). e Manhattan plot of the GWAS analysis for fruit length (FL). f Manhattan plot of the GWAS analysis for T50. g Manhattan plot of the bivariate GWAS analysis for FL and T50. The dashed red line indicates the 5% Bonferroni-corrected genome-wide significance threshold. h Regional association landscape and linkage disequilibrium (LD) heatmap of FL around Chr5:1,701-2,700 kb. i Regional association landscape and LD heatmap of T50 around Chr5:1,701-2,700 kb. j Beta estimates for SNPs within the intersection peak interval of both traits. Red dots indicate SNPs with P values from the bivariate GWAS that exceed the Bonferroni-corrected significance threshold. k, l RT-qPCR analysis of the expression of nine candidate genes in fruits and freshly harvested seeds from the Sichuan and Tibet ecotypes. ND denotes genes not detected. m Quantification of bioactive gibberellin GA1 and GA4 levels in 9-DAP fruits, imbibed seeds, and stratified seeds from the Sichuan and Tibet ecotypes. In d (left), l, m data are means ± s.d. (n = 21/26 biological replicates in d; n = 3 biological replicates in l and m), P values are determined using unpaired two-tailed Student’s t-test. In d (right), data are presented as means ±s.d. (n = 4 biological replicates), with different letters indicating statistically significant differences (P < 0.05) determined by one-way ANOVA with Tukey’s multiple comparisons test. Source data are provided as a Source Data file.
To identify the major gene, whose allelic variations are responsible for FL and SGT in the two distinct ecotypes, we first conducted a GWAS based on the FL data of 123 recombinant inbred lines (RILs, F7 generation) derived from the Sic and Tib ecotypes (Supplementary Fig. 2b, d). This analysis revealed the strongest associated signals in the proximal region of chromosome 5 (Fig. 1e). Quantile-quantile plot (QQ plot) analysis exhibited the expected distribution of observed P-values (Supplementary Fig. 3). In addition, the time required for seeds to reach 50% germination (T50) after 3 days of cold stratification from 206 RILs served as the basis for another GWAS. Interestingly, the results once again highlighted a peak on chromosome 5 at a similar position as that identified for FL (Fig. 1f, Supplementary Fig. 3b). There were 1844 single nucleotide polymorphisms (SNPs) around the intersection interval Chr5:2,079,298–2,378,363 bp that were significantly associated with both traits, explaining 44.71% of the phenotypic variation in FL and 37.08% in the time required for seeds to reach 50% germination (T50), the SGT trait (Fig. 1h, i). Moreover, we conducted a bivariate GWAS for the two traits, and 1580 of the 1844 SNPs passed the significance threshold. The beta estimates for these SNPs showed a highly significant linear correlation (P < 2.2e−16, Fig. 1g, j). These findings suggest that the two adaptive traits are likely governed by the same genetic locus.
To identify candidate genes for two traits, we scanned the genes within the potential interval. We focused on 14 candidate genes related to FL or SGT (Supplementary Data 1). Additionally, we examined their expression patterns during fruit development and seed germination using RT-qPCR analysis. The results showed that the expression of At5g07190 and At5g07200 continually increased over 9 days after pollination (DAP) in both Sic and Tib fruits. However, the amplitude of expression in the Sic ecotype was significantly greater than that in the Tib ecotype (Fig. 1k and Supplementary Fig. 4a). Meanwhile, the relative expression of these two genes, particularly At5g07200, was lower in the Tib ecotype compared to the Sic ecotype during the initial stages of seed germination (Fig. 1l and Supplementary Fig. 4b). The gene At5g07200, located within the peak of our GWAS results, encodes the key enzyme GA20ox3, which is crucial for catalyzing GA biosynthesis41,57. It exhibits tissue-specific expression patterns in seeds and fruits compared to its four homologs (Supplementary Fig. 4c). We further quantified two bioactive forms of gibberellins, GA1 and GA4, in both ecotypes and found that the contents were lower in the developing fruits, imbibed seeds, and cold-stratified seeds of the Tib ecotype compared to those of the Sic ecotype (Fig. 1m). These results collectively suggest that GA20ox3 (At5g07200) is the most likely candidate gene, with its allelic variations potentially accounting for the differences in both FL and SGT between the two ecotypes.
Allelic variations in GA20ox3 influence both FL and SGT
To identify the allelic variations in the GA20ox3 responsible for functional differences between the two ecotypes, we analyzed all SNPs within the approximately 2.1 kb promoter region upstream of the start codon and the coding sequence of GA20ox3 in the Sic and Tib ecotypes (Fig. 2a, Supplementary Fig. 5a). We identified five substitutions and a 3-bp insertion/deletion (indel) in the promoter region of GA20ox3. In the coding region, only two SNPs were found, leading to a single amino acid substitution (Supplementary Fig. 5b). Given that amino acid variations can potentially affect gene function, we overexpressed GA20ox3 from both the Sic and Tib ecotypes in the Col-0 background using a 35S promoter. The resulting transgenic lines, GA20ox3Sic-OE4 and GA20ox3Tib-OE6, which had comparable expression levels (Supplementary Fig. 6a, b), exhibited significantly longer fruits compared to Col-0. However, no notable differences were observed between GA20ox3Sic-OE4 and GA20ox3Tib-OE6 (Supplementary Fig. 6c, d). Furthermore, both transgenic lines exhibited the same germination rate, which was higher than that of Col-0 when treated with paclobutrazol (PAC), a GA biosynthesis inhibitor (Supplementary Fig. 6e, f). This indicates that overexpression of both GA20ox3Sic and GA20ox3Tib increases GA content. These findings suggest that the GA20ox3Sic and GA20ox3Tib enzymes have identical activity in fruit development and seed germination, implying that the amino acid variation does not impact their enzymatic function.
a Gene structure and allelic variation of GA20ox3 between the Sichuan and Tibet ecotypes. b Representative GUS staining of fruits and seeds carrying the pGA20ox3Sic:GUS and pGA20ox3Tib:GUS constructs in a Col-0 genetic background. Seeds were grown on 1/2 MS medium and subjected to either 3 days of cold stratification (+cold) or no cold stratification (-cold) at 4 °C. c Quantitative analysis of GUS activity in pGA20ox3Sic:GUS and pGA20ox3Tib:GUS is shown in (b). Each biological replicate involved testing more than 20 fruits or 300 seeds from the same batch. d Fruit images of Col-0, ga20ox3-1, ga20ox3-1 pGA20ox3Sic:GA20ox3, and ga20ox3-1 pGA20ox3Tib:GA20ox3 at stage 17-B. Scale bar, 2 mm. e Statistic analysis of fruit length in (d). f Germination phenotypes of Col-0, ga20ox3-1, ga20ox3-1 pGA20ox3Sic:GA20ox3, and ga20ox3-1 pGA20ox3Tib:GA20ox3 freshly harvested seeds. Seeds were cultivated on 1/2 MS medium with various treatments. g Statistic analysis of germination rate in (f). PAC denotes paclobutrazol. h Quantitation of bioactive gibberellin GA1 and GA4 levels in 9-DAP fruit, imbibed seeds, and stratified seeds (blue background) from ga20ox3-1 pGA20ox3Sic:GA20ox3, and ga20ox3-1 pGA20ox3Tib:GA20ox3. In c (top) data are presented as means ±s.d. (n = 9 biological replicates) P values determined using unpaired two-tailed Student’s t-test. In c (bottom), e, g, h data are shown as means ± s.d. (n = 5 biological replicates in (c); n = 23/22/20/16/23/21 biological replicates in (e); n = 3/4/3 biological replicates in (g); n = 3/4 biological replicates in h), with different letters indicating significant differences (P < 0.05) based on one-way ANOVA followed by Tukey’s multiple comparisons test. Source data are provided as a Source Data file.
Gene transcriptional regulation depends on the interaction between transcription factors and their corresponding cis-acting elements. To assess the differences in GA20ox3 expression patterns between the two ecotypes, we introduced pGA20ox3Sic:GUS and pGA20ox3Tib:GUS constructs into the same Col-0 background. We used a GUS reporter system to assess the transcriptional activity of the two promoters. The results indicated significantly higher GUS activity driven by the GA20ox3Sic promoter in developing fruits and germinating seeds compared to the GA20ox3Tib promoter. Additionally, the transcriptional activity of the GA20ox3Tib promoter increased markedly after cold stratification (Fig. 2b, c). These results, which align with our RT-qPCR findings, suggest that the differences in GA20ox3 expression patterns are primarily due to variations in the promoter sequences rather than specific transcription factors. We therefore hypothesize that allelic SNPs in the GA20ox3 promoter are responsible for the observed variations in transcription, ultimately affecting FL and SGT. To explore this possibility, we introduced the coding sequence of Col-0 GA20ox3, driven by the respective promoters of GA20ox3Sic and GA20ox3Tib, into Col-0 wild-type background (Supplementary Fig. 7). Two stable expression lines were selected and crossed with the loss-of-function mutant ga20ox3-1. This resulted in the generation of transgenic lines: ga20ox3-1 pGA20ox3Sic:GA20ox3 (#4 and #6) and ga20ox3-1 pGA20ox3Tib:GA20ox3 (#3 and #4). We found that while both transgenes could rescue the shorter fruit phenotype of the ga20ox3-1 mutant, the degree of restoration was more effective with the GA20ox3Sic promoter compared to the GA20ox3Tib promoter (Fig. 2d, e). Seed germination analysis revealed that the ga20ox3-1 pGA20ox3Sic:GA20ox3 lines had a significantly higher germination rate than the ga20ox3-1 pGA20ox3Tib:GA20ox3 lines, although both lines rescued the low germination rate of the ga20ox3-1 mutant (Fig. 2f, g). Additionally, cold stratification treatment alleviated germination inhibition caused by PAC in all transgenic lines, suggesting that both GA20ox3Sic and GA20ox3Tib promoters are up-regulated in response to cold treatment. To determine whether these observed phenotypes were linked to increased GA content, we measured endogenous GA1 and GA4 in developing fruits, imbibed seeds, and cold-stratified seeds of these lines. The results showed that the ga20ox3-1 pGA20ox3Tib:GA20ox3 line had significantly lower levels of both GAs in fruits and cold-stratified seeds (Fig. 2h). These findings collectively indicate that natural variations in the GA20ox3 promoter region affect both FL and SGT by modulating GA20ox3 transcript abundance.
The SNP-418 impacts the affinity of the GA20ox3 promoter for the DOF6 transcription factor, which negatively regulates GA20ox3 expression during fruit elongation and seed germination
To understand how promoter variations affect GA20ox3 expression, we hypothesized that variations within or near cis-acting elements could influence their affinity for transcription factors. We analyzed the SNPs and indel between the GA20ox3Sic and GA20ox3Tib promoters and discovered that SNP-1,211 and SNP-418 are located in the flanking regions of two DNA-BINDING ONE ZINC FINGER (DOF) protein binding elements58. However, the core sequence (5’-AAAG-3’) is highly conserved across the Sic, Tib, and Col-0 ecotypes (Fig. 3a). Additionally, no typical elements were identified surrounding the remaining SNPs. These findings suggest the involvement of DOF transcription factors in directly regulating GA20ox3 expression. To identify the candidate DOF transcription factors, we examined the expression data for 33 DOF genes (out of a total of 37 members in this family59) obtained from the ePlant database. Interestingly, we observed that DOF6 gene expression decreased progressively during fruit development, showing an inverse pattern compared to GA20ox3 expression (Fig. 3b). This finding was confirmed by RT-qPCR analysis (Fig. 3c). DOF6, known as a negative regulator of seed germination, is expressed throughout embryo maturation, peaks in dry seeds, and then declines during stratification and dry storage55. Therefore, we hypothesized that DOF6 negatively regulates GA20ox3 expression during fruit development and seed germination. To test this hypothesis, we generated DOF6 overexpression lines (DOF6-OE) and a CRISPR/Cas9-mediated mutant in the Col-0 background (Supplementary Fig. 8). The fruits of the DOF6 overexpression line were shorter compared to Col-0, while the dof6 mutant fruits were slightly longer, though the difference was not significant. Additionally, the germination rate was reduced in the DOF6 overexpression line, whereas the dof6 mutant showed an increased rate compared to Col-0 (Fig. 3d–g). Furthermore, crossing the ga20ox3-1 mutant with the dof6 mutant produced a ga20ox3-1 dof6 double mutant, which exhibited similar FL and SGT to the ga20ox3-1 mutant. These results indicate that DOF6 negatively regulates FL and SGT through its impact on the GA20ox3 gene.
a DOF proteins recognize the core sequence AAAG near the -418 SNP and -1,211 SNP. b A heatmap visualizes DOF family genes and GA20ox3 expression during fruit development. c RT-qPCR analysis of GA20ox3 (left) and DOF6 (right) across various stages of fruit development in the Col-0. Data are shown as means ± s.d., n = 3 independent experiments. d Fruit images of Col-0, DOF6-OE1, ga20ox3-1, dof6 and ga20ox3-1 dof6 at stage 17-B. Scale bar: 2 mm. e Statistic analysis of FL in (d). f Germination assays of different lines using freshly harvested seeds on 1/2 MS medium. g Statistical analysis of the germination rate in (f). h RT-qPCR analysis of GA20ox3 in freshly harvested seeds from Col-0 and DOF6-OE1. i Dual-luciferase assays of GA20ox3Col-0 promoter activity modulated by DOF6. j The schematic diagram illustrates the A/TAAAG-box regions used for ChIP in the GA20ox3 promoter. Fragments P1-P9 represent the regions amplified for ChIP-qPCR analysis. k The EMSA results demonstrate that DOF6 directly binds to the AAAAG-box of P8 in the GA20ox3Col-0 promoter. l The mutant site is indicated, where Sic-mutant and Tib-mutant denote the substitution of T for A and A for T, respectively, at nucleotide position -418 bp of the GA20ox3 promoter. m Dual-luciferase assays were conducted to assess the promoter activity of GA20ox3Sic, GA20ox3Tib, GA20ox3Sic-mut, and GA20ox3Tib-mut in response to modulation by DOF6. n, o EMSAs reveal variations in the binding affinity of DOF6 to the GA20ox3Sic, GA20ox3Tib, GA20ox3Sic-mut, and GA20ox3Tib-mut promoters. In e, g, data are presented as means ±s.d. (n = 24/18/10/15/15 biological replicates in e; n = 3 biological replicates in g), with different letters indicating statistically significant differences (P < 0.05) determined by one-way ANOVA with Tukey’s multiple comparisons test. In h, i, j, m, data are shown as means ± s.d. (n = 3 biological replicates), with P values obtained using an unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
To investigate the effect of DOF6 on GA20ox3 expression, we performed RT-qPCR analysis on seeds from the DOF6 overexpression line. The results showed a significant down-regulation of GA20ox3 compared to Col-0 (Fig. 3h), indicating that DOF6 represses GA20ox3 expression. To pinpoint the specific region of the GA20ox3 promoter regulated by DOF6, we conducted dual-luciferase (LUC) assays. These assays revealed that DOF6 inhibits GA20ox3 transcription when using 2143 bp and 614 bp fragments of the GA20ox3 promoter to drive the LUC gene. However, this repressive effect was lost when the promoter lacked the region from −397 to −614 (Fig. 3i). Additionally, we conducted ChIP-qPCR analysis to identify the critical binding site of DOF6 within the GA20ox3 promoter. The results indicated that only the P8 region was significantly enriched (Fig. 3j). Within this region, we identified a single DOF binding motif, located near SNP-418, which was further confirmed to be bound by DOF6 using an electrophoretic mobility shift assay (EMSA) (Fig. 3k). To investigate how SNP-418 affects the differential affinities of the two haplotypes (Sic and Tib) in the GA20ox3 promoter for DOF6 protein, we performed a dual-LUC transactivation assay. We found that DOF6 from Col-0 significantly suppressed the transcriptional activation of the GA20ox3Tib promoter compared to the GA20ox3Sic promoter (Fig. 3l, m). However, when SNP-418 was swapped, the LUC signals of the mutated GA20ox3Sic promoter decreased to levels similar to those of the GA20ox3Tib promoter (Fig. 3l, m). Additionally, EMSAs demonstrated that DOF6 from Col-0 preferentially bound to the GA20ox3Tib probe over the GA20ox3Sic probe, with this preference reversing when SNP-418 was exchanged (Fig. 3n, o). These results indicate that SNP-418 is a functional site that determines the differential binding affinity of DOF6, leading to the distinct expression patterns of GA20ox3Sic and GA20ox3Tib.
DOF6 negatively regulates GA20ox3 expression through its interaction with DELLA proteins
DOF6 regulates the expression of downstream genes by forming a complex with RGL2, a member of the DELLA protein family54,60. To determine if the DELLA-DOF6 interaction influences GA20ox3 transcription during fruit development and cold-induced seed germination, we measured the expression levels of five DELLA genes (GAI, RGA, RGL1, RGL2, and RGL3) in fruits at various developmental stages and in seeds undergoing cold stratification, using RT-qPCR. The results indicated that all five DELLA genes were expressed in developing fruits, with their expression levels decreasing in seeds as the cold stratification period increased (Fig. 4a). To identify which DELLA protein interacts with DOF6, we conducted a yeast two-hybrid assay. This assay revealed strong interactions between DOF6 and four DELLA proteins—GAI, RGA, RGL1, and RGL2—while RGL3 did not show significant interaction (Fig. 4b). These interactions were further confirmed by a bimolecular fluorescence complementation (BiFC) assay (Fig. 4c). To further explore the genetic relationship between DELLAs and GA20ox3, we selected RGL2 as the representative DELLA protein due to its significant role in seed germination. We created a ga20ox3-1 rgl2 double mutant to evaluate its phenotype. The double mutant showed longer fruits and a higher seed germination rate compared to the ga20ox3-1 mutant (Supplementary Fig. 9). However, the RGL2 single mutant did not fully restore the phenotypes of FL and SGT to those of Col-0, likely due to functional redundancy among DELLA proteins48,61. Therefore, we crossed the ga20ox3-1 mutant with the della-q (rga gai rgl1 rgl2) mutant (Supplementary Fig. 10) and observed a complete rescue of the phenotypic defects in FL and SGT caused by ga20ox3-1 (Fig. 4d–f). These findings indicate that these four DELLA genes, acting downstream in GA signal transduction, are epistatic to GA20ox3. To explore the influence of DELLAs on DOF6-mediated GA20ox3 expression, we performed dual-LUC assays. The results showed that all four DELLAs could significantly enhance DOF6’s transcriptional repression of GA20ox3 (Fig. 4g). Subsequent in vivo ChIP-qPCR analysis further validated that RGL2 was specifically enriched in the P8 region of the GA20ox3 promoter (Fig. 4h), consistent with the binding of DOF6 and corroborating that RGL2 binding to the GA20ox3 promoter is mediated by DOF6. Additionally, in vitro EMSA results revealed that RGL2 alone did not bind to the DNA probe of the GA20ox3 promoter but could significantly enhance DOF6’s DNA binding affinity (Fig. 4i). Moreover, the GA20ox3Tib probe showed a much greater affinity for the RGL2-DOF6 complex compared to the GA20ox3Sic probe (Fig. 4j). Collectively, these findings demonstrate that the DELLAs-DOF6 complexes repress GA20ox3 expression to regulate FL and SGT, revealing a conserved mechanism across multiple A. thaliana ecotypes.
a RT-qPCR analysis of DELLAs was performed on fruits from 1 to 17 days after pollination (DAP) and on cold-stratified seeds from Col-0. b Yeast two-hybrid assays show the interactions between DOF6 and DELLAs. c BiFC assays demonstrate that DOF6-nGFP interacts with GAI-cGFP, RGA-cGFP, RGL1-cGFP, and RGL2-cGFP in N. benthamiana mesophyll protoplasts. Scale bar: 20 μm. d Images of fruits at stage 17-B for Col-0, ga20ox3-1, della-q, and ga20ox3-1 della-q are shown. Scale bar: 2 mm. e Germination phenotypes of Col-0, ga20ox3-1, della-q, and ga20ox3-1 della-q freshly harvested seeds. Seeds were grown on 1/2 MS medium, with and without PAC. f Statistical analysis of fruit length and germination rate is presented in (d, e). g Dual-luciferase expression assays of GA20ox3Col-0 promoter activity modulated by DOF6 and DELLAs (GAI, RGA, RGL1, and RGL2). REN serves as an internal control. h ChIP-qPCR analysis was performed to assess the enrichment of the GA20ox3 promoter using the p35S:FLAG-RGL2 transgenic lines. Col-0 wild-type plants served as a negative control, and EIF4A was used for data normalization. i, j EMSA demonstrates that RGL2 protein enhances the binding of DOF6 to the GA20ox3Col-0/Sic/Tib promoter fragment. In f data are presented as means ± s.d. (n = 15/15/13/13, 3/3/3/3, 4/4/4/4 biological replicates), with different letters indicating statistically significant differences (P < 0.05) determined by one-way ANOVA with Tukey’s multiple comparisons test. In g, h, data are presented as means ± s.d. (n = 3 biological replicates), with P values determined using an unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
Haplotype distribution of GA20ox3
To explore the natural variation of the GA20ox3 promoter across the total distribution of A. thaliana, we retrieved the 600 bp promoter sequences preceding the start codon for 576 globally distributed ecotypes (accessions) with clearly identified orthologous sequences (Supplementary Data 2 and Supplementary Data 3). After sequence alignment and haplotype analysis, our findings pinpointed a cluster of polymorphisms predominantly within the −412 to −434 bp interval of the GA20ox3 promoter region (Supplementary Fig. 11), with particular emphasis on SNP-417, SNP-418, and SNP-419. These SNPs serve as the flanking sequences of a DOF protein binding core and play a crucial role in influencing the affinity for DOF binding58. Based on the nucleotides in these three sites, we categorized the 576 accessions into four distinct haplotypes: Hap1 (AAA), Hap2 (ATA), Hap3 (TTA), and Hap4 (ATG) (Fig. 5a and Supplementary Fig. 11). Subsequent EMSA and dual-LUC assays showed that the DOF6Sic protein exhibited the highest binding efficiency with the Hap2-specific probe containing the ATA sequence, which corresponded to the lowest transcriptional activity. This was followed by Hap3, Hap4, and Hap1 in turn (Fig. 5b, c). These findings indicate that this differential binding activity directly results in variations in promoter activity, with stronger binding leading to lower expression activity.
a Haplotype network of the GA20ox3 promoter region created using 576 accessions, revealing four clades: Hap1, Hap2, Hap3, and Hap4. Each circle represents one haplotype, with the size corresponding to the haplotype frequency. b EMSAs showing the differences in DOF6-binding affinity with four haplotypes containing the GA20ox3 promoter region. c Dual-luciferase expression assays of GA20ox3Hap1/Hap2/Hap3/Hap4 promoters activity modulated by DOF6Sic. Data are presented as means ± s.d., with different letters indicating statistically significant differences (P < 0.05) determined by one-way ANOVA with Tukey’s multiple comparisons test. All experiments were conducted three times with consistent results. d, e Geographic distribution of 576 accessions. The heat map displayed on the map shows the minimum temperature of December (d) and altitude (e). f Comparison of the minimum temperature in December between Hap2 and other haplotypes. g Comparison of the altitude between Hap2 and other haplotypes. h Comparison of DSDS50 (the days of seed dry storage required to reach 50% of germination) between Hap1/4 and Hap2/3 is shown. i Comparison of the germination rate between Hap1/4 and Hap2/3. In f–i, the number of plants (n) is shown. Center lines represent the medians, edges of the box indicate the 25th and 75th percentiles, and whiskers indicate the minima and maxima. P values are determined using unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
Further exploration of the geographical distribution of the 576 ecotypes and their associated temperature and altitude data revealed no significant difference in the annual average temperature between Hap2 and Hap4 (Supplementary Fig. 12a, b). It appears that the high temperatures in summer offset the low temperatures in winter, resulting in an insignificant difference in average annual temperatures. Annual herbaceous plants face the decision of whether to endure the coldest seasons in the form of seeds or seedlings. Therefore, monthly minimum temperatures in winter (December, January, and February in the Northern Hemisphere) may exert a more pronounced impact on plant life history. Our findings indicate that Hap2 exhibited significantly lower minimum temperatures in December, January, and February compared to other haplotypes, as well as lower minimum temperatures of the coldest month (Fig. 5d, f and Supplementary Fig. 12c–e). Temperature gradually decreases with increasing elevation and latitude. Analysis of latitudes and elevations indicates that Hap1 is predominantly located in low-latitude and low-elevation regions, whereas Hap2 and Hap3 are primarily distributed in mid-latitude high-elevation regions. Moreover, Hap4 shows a preference for low-elevation regions in mid-to-high latitudes (Fig. 5e, g and Supplementary Fig. 13a). Furthermore, the minimum temperature in the Hap2 region seems to be primarily influenced by high elevation, while in the Hap1, Hap3, and Hap4 regions, it is predominantly determined by latitude (Fig. 5g and Supplementary Fig. 13b-e).
To determine if these promoter variations are linked to dormancy or germination, we combined published phenotypic data and performed a correlation analysis30. The analysis revealed that the primary dormancy of Hap1/4 seeds was significantly lower than that of Hap2/3 seeds, and the basal germination rate of Hap1/4 seeds was significantly higher than that of Hap2/3 seeds (Fig. 5h, i). These observations align with the geographical distribution patterns, where Hap2 and Hap3 are predominantly found in high-altitude regions, whereas Hap1 and Hap4 are more prevalent in low-altitude areas. Collectively, these findings suggest that genetic diversity in the GA20ox3 promoter serves as a beneficial strategy for A. thaliana to optimize germination timing, thereby contributing to local adaptation.
Discussion
Across different altitudinal gradients, plants exhibit phenological adaptations to germinate and complete their reproductive cycles during the most favorable growing seasons13,24. Local plants have undergone regional selection to choose genetic variations that confer adaptive changes in freezing resistance, morphology, and phenology6. In this study, the A. thaliana ecotypes from Tibet (Tib) and Sichuan (Sic) experience markedly different minimum temperatures during both their growth and dormancy stages (Supplementary Fig. 1). Compared to the low-altitude Sic ecotype, the high-altitude Tib ecotype has shorter fruits (Fig. 1a–d) and fewer seeds per fruit (Supplementary Fig. 14c) as found for most alpine plants, a morphological change that reduces convective heat loss and helps retain warmth during seed development6. Additionally, the Tib ecotype requires more cold stratification for seed germination (Fig. 1c, d), a phenology that helps safeguard seedlings against unpredictable growth conditions13,62. These cold-adaptive traits in morphology and phenology ensure that the Tib ecotype can survive the extremely low temperatures in the winter and complete successful sexual reproduction in the temperature-oscillated summer in the high-elevation habitats. Through GWAS and gene expression analyses, we identified the GA20ox3 gene, which encodes a crucial enzyme in GA biosynthesis, as a major regulator affecting fruit length (FL) and cold-indued seed germination timing (SGT). Allelic variations at nucleotides −417, −418, and −419 bp in the GA20ox3 promoter significantly impact its binding affinity to the DELLA-DOF6 transcriptional repression complex. This altered affinity modulates GA20ox3 expression levels without completely changing its spatiotemporal expression pattern. Such precise regulation of GA20ox3 expression fine-tunes both FL and SGT, facilitating adaptation of A. thaliana to varying altitudinal gradients across global habitats (Fig. 6). These findings highlight the critical role of the DELLA-DOF6-GA20ox3 module in regulating FL and SGT via GA signaling in plants, revealing a molecular link between these seemingly unrelated traits and offering insights into their evolutionary adaptation to local climates. Additionally, high-altitude adaptation traits, such as reduced stature and increased silique density, are also achieved through the suppression of GA signaling63,64, suggesting that GA deficiency may be a potential evolutionary driver in high-altitude environments.
The transcript of the gibberellin biosynthetic gene GA20ox3 is negatively feedback-regulated by DELLA proteins in a DOF6-dependent manner. The DOF6-GA20ox3-mediated GA signaling pathway is influenced by cold. SNPs in the GA20ox3 promoter region affect the recognition efficiency of DOF6, causing differences in fruit length and seed germination timing. Hap1/4 and Hap2/3 have developed distinct adaptive mechanisms to cope with variations in cold exposure duration resulting from differences in high and low altitudes.
The five GA20ox genes in A. thaliana show distinct expression patterns (Supplementary Fig. 4c). Compared to GA20ox3, the knockout of GA20ox1 or GA20ox242 has a minimal effect on FL and SGT (Fig. 3d–g). This suggests that the GA20ox genes have evolved differentiated functional roles within specific tissues, with GA20ox3 being the primary regulator of fruit development and seed germination. Additionally, GA3ox1, another enzyme involved in GA biosynthesis, converts the products of GA20ox into bioactive GA4, which also plays a crucial role in promoting fruit growth and seed germination10,65. These findings confirm that the bioactive GAs involved in both fruit development and seed germination are synthesized through the same metabolic pathway, which includes the GA20ox3 and GA3ox1 enzymes. Our further analysis of GA20ox3 expression patterns indicates that it is specifically active during certain stages of fruit development and seed germination, rather than being constitutively expressed (Figs. 2b, c and 3c). Thus, the regulation of its expression initiation is crucial for these biological processes. Our study reveals that GA20ox3 expression is negatively regulated by a transcriptional repression complex composed of DELLAs and DOF6. In non-stratified seeds, this DELLA-DOF6 complex maintains seed dormancy by inhibiting GA20ox3 transcription (Figs. 1d and 2b, c). After cold treatment, the expression of DOF6 decreases55, and DELLA proteins are degraded by GA48,66, leading to the activation of GA20ox3 transcription and the subsequent initiation of seed germination. Given the delayed effect of transcriptional regulation and the need for a sufficient amount of GA to degrade DELLA proteins, we speculate that an unidentified protein degradation mechanism targeting DOF6 might act as the initial trigger for the cold-induced activation of GA20ox3. Similarly, we observed that DOF6 expression gradually decreased while GA20ox3 expression increased during fruit development (Fig. 3c). We infer that GA20ox3 is repressed by the DELLA-DOF6 complex and remains at low levels in the unfertilized pistil. Following fertilization, growth signals released by the ovule likely downregulate DOF6 expression, thereby promoting fruit growth and development, although further evidence is needed to support this hypothesis. Additionally, previous studies have shown that RGL2-DOF6 protein complex promotes the expression of GATA12, which inhibits the germination of freshly harvested or unstratified seeds54. These results indicate that the DELLA-DOF6 complex exerts bidirectional effects on downstream genes involved in germination, though the underlying mechanisms require further investigation. Although previous models have proposed that DELLA proteins interact with specific DOF proteins to regulate gene expression37,54, our results provide substantial evidence that the four DELLA genes (RGA, GAI, RGL1, RGL2) have redundant functions mediated by DOF6 in fruit development and SGT (Fig. 4). This finding underscores the multifaceted role of DELLA-DOF6 protein complexes in the feedback inhibition of GA20ox3 and sheds light on the efficacy of such pleiotropic regulatory pathways in modulating multiple traits.
Globally distributed natural ecotypes of A. thaliana exhibit local adaptation to varied environments, demonstrating the diverse strategies employed by various ecotypes to optimize their life cycles in response to natural selections67. Ecotypes exhibiting deep primary dormancy require extensive cold stratification to break dormancy, while those lacking primary dormancy may enter secondary dormancy when subjected to prolonged low temperatures28,30,68. This differential response highlights the varied dormancy mechanisms among ecotypes, which are critical for their adaptation to specific environments. Winter cold not only regulates germination but also induces vernalization, a process where prolonged low-temperature exposure enables spring flowering69, regulated by the FLC and FRI networks70,71. In contrast, ecotypes that do not require vernalization may optimize germination timing based on factors such as spring or summer rainfall, or the winter cold cycle. For instance, FLOE1 regulate seed germination in challenging environments through reversible hydration-dependent phase separation72. Under unpredictable natural conditions, seeds may employ two germination strategies: “best bet” and “bet hedging”. The “best bet” strategy involves germinating immediately when conditions seem favorable, while the “bet hedging” strategy involves delaying germination to create a seed bank in the soil, thereby increasing the chances of survival through varied seasonal conditions73,74. Similar to many alpine plants1,24, the Tib ecotype of A. thaliana relies on longer cold stratification to overcome seed dormancy and successfully germinate compared to the low-altitude Sic ecotype (Fig. 1c, d). However, it remains unclear whether the seeds of Tib ecotype enter the secondary dormancy after longer winter cold than 31-day conducted here and/or other factors, as suggested before27,28,29. Otherwise, after the initial enough but longer cold stratification than Sic ecotype, it is temperature that determines the germination of the Tib ecotype in the Qinghai-Tibet Plateau. Additionally, the GA impact on plant stature raises further questions. We do not yet understand how allelic variations of GA20ox3 contribute to the small stature of the Tibet ecotype (Fig. 1a), particularly in light of the presence of four other GA20ox genes and additional regulators that influence GA syntheses38,39. Overall, the shorter FL and changed SGT observed in the Tib haplotype, driven by GA20ox3 allelic variation, indicate a “bet hedging” strategy designed to enhance seedling survival in later spring and total summer. This approach, while improving survival rates, may compromise FL and reproductive success within a shorter growing season (Supplementary Fig. 14). These findings suggest that A. thaliana accessions have developed varied germination strategies to adapt to their specific environmental conditions. The Tib ecotype seems to prioritize seedling survival over reproductive success, reflecting an adaptation to the harsh alpine climate (Supplementary Fig. 15). This trade-off between survival and fecundity illustrates the complex evolutionary pressures that influence local adaptation in this species.
Methods
Plant materials and growth conditions
The Sichuan ecotype seeds (ABRC stock no.: CS79063) were sourced from Yilong County, Nanchong City, Sichuan Province. The Tibet ecotype seeds (ABRC stock no.: CS79062) were collected from Chengguan District, Lhasa City, Tibet Autonomous Region. To create a recombinant inbred line (RIL) population, an initial cross was made between the Sichuan and Tibet ecotypes, followed by seven generations of self-pollination. All A. thaliana mutants and transgenic plants utilized in this study were in the Columbia-0 (Col-0, ABRC stock no.: CS76778) background. The ga20ox3-1 mutant (CS86016) was acquired from the Nottingham Arabidopsis Stock Center (NASC). The rgl2 mutant (SALK_124231) and the della-q mutant have been described in previous reports61. dof6 mutants were generated using the CRISPR/Cas9 system, following a previously established protocol75. Adult plants were grown in a controlled growth chamber with consistent conditions: long-day (LD) conditions (16 h light, 22 °C; 8 h dark, 19 °C) and humidity levels between 60%. The soil was regularly irrigated, and seeds were harvested simultaneously for each batch to ensure experimental uniformity.
Plasmid construction and plant transformation
For the dual-LUC assay, three upstream fragments (2143 bp, 614 bp, and 397 bp) of the GA20ox3 translational start codon from the Col-0 ecotype were cloned into the pGreen-0800-LUC vector as reporter constructs. Additionally, an upstream fragment (614 bp) of the GA20ox3 translational start codon from the Sichuan and Tibet ecotypes were also cloned into the pGreen-0800-LUC vector as reporter constructs. For the construction of pGA20ox3Sic:GUS and pGA20ox3Tib:GUS, a 2143 bp fragment upstream of the GA20ox3 translational start codon was PCR-amplified and inserted into the pCXGUS-P vector using XcmI restriction sites. The p35S:GA20ox3Sic, p35S:GA20ox3Tib, and p35S:GA20ox3Col-0 constructs were created by inserting the coding sequences of GA20ox3 from the Sichuan, Tibet, and Col-0 ecotypes into the pCAMBIA1300-EGFP vector. Both the pGA20ox3Sic:GA20ox3Col-0 and pGA20ox3Tib:GA20ox3Col-0 constructs were generated by replacing the CaMV35S promoter in the pCAMBIA1300-GA20ox3Col-0-EGFP vector with a 2,143-bp upstream fragment of GA20ox3 from the Sichuan and Tibet ecotypes. p35S:FLAG-RGL2 and p35S:FLAG-DOF6 constructs were created by inserting the coding sequences of RGL2 and DOF6 into the pCAMBIA1300-FLAG vector. The primers used for plasmid construction are detailed in Supplementary Data 4.
The binary constructs were introduced into the Agrobacterium tumefaciens strain GV3101 and used to infect Col-0 wild-type plants through the floral dip method76. Transgenic plants were screened on 1/2 MS plates containing 50 mg/l hygromycin. Double mutant and transgenic lines were generated through genetic crossbreeding, and homozygous plants were selected for further investigations.
Germination assay
Seed germination assays were conducted on 0.65% agar-solidified plates containing half-strength Murashige and Skoog (MS) medium (Sigma, USA) and 1% sucrose (pH 5.7) supplemented with PAC (Sigma-Aldrich) upon the experiment requirement. Freshly harvested seeds from mature wild-type and transgenic plants, grown under controlled conditions in the same tray, were used for these assays unless otherwise specified. Seeds were dried over silica beads for 5 days before the experiments. For cold stratification, plates with seeds were kept in darkness at 4 °C for 3 days, then transferred to a tissue culture room under long-day (LD) conditions (16 h light at 22 °C; 8 h dark at 19 °C) with fluorescent white light at 7000 lux (Philips F17T8/TL841 17 W). Each assay included a minimum of 50 seeds per genotype and was performed in triplicate to ensure rigorous statistical analysis. All experiments were conducted at least three times, and the phenotypic photographs present one representative result.
Genome-wide association analysis
The FLs of 123 RILs grown under controlled conditions were meticulously measured, with at least 10 fruits assessed per RIL. Additionally, the seed germination process was recorded for 206 RILs grown on 1/2 MS medium, with a minimum of 50 seeds analyzed per RIL (Supplementary Data 5). Separate GWAS analyses were performed for FL and T50. SNP identification involved aligning re-sequencing reads of the RILs to the Sichuan ecotype HiFi reference using BWA software (version 0.7.17). Duplicate reads were removed using Sambamba (version 0.7.1), followed by SNP and indel calling with the Genome Analysis Toolkit (GATK version 4.1.1.7). Vcftools (version 0.1.15) was then used to retain biallelic SNPs with a minor allele frequency of at least 0.05 and genotypes called in at least 80% of the samples. A total of 1,048,575 SNPs were used for the subsequent GWAS analysis.
The GWAS analysis was conducted using GEMMA software (version 0.98.1) with a standard linear mixed model to account for population structure, and a Bonferroni correction was applied for multiple testing. For the bivariate GWAS on fruit length and T50, a multivariate linear mixed model was utilized in GEMMA. Significant SNPs were identified by comparing P-values to a threshold of 0.05 divided by the total number of SNPs. Linkage disequilibrium (LD) analysis was performed using LDBlockShow (version 1.40) to define LD blocks around significant peaks based on confidence intervals. To estimate the phenotypic variation explained (PVE) by the interval SNPs, a mixed linear model was employed:
\(y\) is the phenotypic value. \(\mu\) is the intercept of the model. \({Z}_{1}\) and \({Z}_{2}\) represent the corresponding design matrix that satisfies \({Z}_{1}{{{{{\rm{Z}}}}}_{1}}^{T}={G}_{1}\) and \({Z}_{2}{{{{{\rm{Z}}}}}_{2}}^{T}={G}_{2}\), where \({G}_{1}\) denotes the relatedness matrix for the interval SNPs within the population, while \({G}_{2}\) refers to the relatedness matrices for the remaining SNPs. \({u}_{1}\) represents the effect of \({Z}_{1}\) follows \(N\left(0,I{\sigma }_{g1}^{2}\right).\,{u}_{2}\) represents the effect of \({Z}_{2}\) follows \(N\left(0,I{\sigma }_{g2}^{2}\right)\). \(e\) is the residual with \(e \sim N\left(0,I{\sigma }_{e}^{2}\right)\). The PVE was calculated using the formula \(\frac{{\sigma }_{g1}^{2}}{{\sigma }_{g1}^{2}+{\sigma }_{g1}^{2}+{\sigma }_{e}^{2}}\) in the variance component estimation function of GEMMA.
Gene expression analysis
Total RNA was extracted from various plant tissues, including leaves, seeds, seedlings, and fruits, using TRIzol reagent (Bio Fit, USA). For cDNA synthesis, 1 μg of RNA was reverse transcribed with the PrimeScript RT reagent kit (Takara, Japan). Relative gene expression was analyzed using real-time quantitative PCR (RT-qPCR). The reaction mixtures were prepared with SYBR Green PCR Master Mix (Yeasen, China) and analyzed using a Bio-Rad CFX96 Real-Time System. ACTIN2 (At3g18780) was used as the internal control for RT-qPCR in all tissues except seeds and fruits, for which EIF4A (At3g13920) served as the control. A comprehensive list of primers used in the study is provided in Supplementary Data 4.
Quantitation of gibberellins
To measure endogenous levels of GA1 and GA4, fruits at 9 days after pollination (DAP), seeds imbibed for 24 h, and seeds cold-stratified for 3 days were collected. These samples were promptly frozen in liquid nitrogen and then ground into a fine powder. A modified extraction procedure was used, involving a solvent mixture of methanol, water, and formic acid in a 15:4:1 ratio by volume. The mixture was thoroughly agitated and then centrifuged at 12,000 × g for 10 min at 4 °C to separate the phases. The supernatant was evaporated under nitrogen gas at room temperature, and the resulting residue was dissolved in 100 μl of methanol for analysis. The extracts were analyzed using an LC-ESI-MS/MS system, which included a Shimadzu LC30AD liquid chromatograph, a QTRAP 6500+ mass spectrometer, and a Thermo Hypersil Gold column.
Dual-luciferase assay
Dual-LUC activity assays were conducted using young Nicotiana benthamiana leaves. Expression vectors were co-transformed into Agrobacterium strain GV3101 and subsequently infiltrated into 4-week-old N. benthamiana leaves. The Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was employed to measure luciferase and Renilla luciferase (REN) activities after 36 hours of dark incubation followed by 36 h of normal light growth. The Renilla luciferase gene served as an internal control to assess transfection efficiency.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were conducted using a previously established protocol77. DNA-protein complexes were extracted from rosette leaves of 3-week-old wild-type Col-0, p35S:FLAG-DOF6 and p35S:FLAG-RGL2 transgenic plants. These complexes were precipitated using an anti-FLAG antibody (Abcam, ab205606, 1:600 dilution) and protein A agarose beads. RT-qPCR analysis was then performed to evaluate the enrichment of specific promoter fragments within the eluted chromatin from the ChIP experiment. Enrichment levels were determined by normalizing against input DNA, with ACTIN2 and EIF4A used as internal controls for data normalization.
Electrophoresis mobility shift assay
Recombinant proteins GST-DOF6Col-0, GST-DOF6Sic, and GST-RGL2Col were purified from E. coli Rosetta2 (DE3) using glutathione affinity chromatography. Oligonucleotide probes were synthesized and biotin-labeled at either the 3’ or 5’ end by Sangon Biotech (Shanghai, China). For competitive assays, unlabeled oligonucleotides were used at concentrations 100-fold or 500-fold higher than the labeled probes. Electrophoretic mobility shift assays (EMSAs) were conducted using the LightShift Chemiluminescent EMSA kit from Thermo Fisher (Waltham, MA, USA). Probe sequences are detailed in Supplementary Data 4.
GUS staining and activity analysis
During the GUS staining assays, 9 DAP fruits and germinating seeds were immersed in a GUS staining solution containing 50 mM Tris-Cl (pH 7.5), 0.2% Triton X-100, and 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-gluc). The samples were subjected to vacuum infiltration for 10 min to ensure thorough penetration of the staining solution into the tissues. After infiltration, the tissues were incubated at 37 °C in the dark for 60 min to allow enzymatic reactions to proceed. Following incubation, the tissues were washed three times with 70% ethanol, and images were captured using an Olympus digital camera.
GUS activity was evaluated using a fluorimetry assay with 4-methylumbelliferyl-β-D-glucuronide (4-MUG) as the substrate. Fluorescence measurements were taken at excitation and emission wavelengths of 365 nm and 455 nm, respectively. The data were normalized to a reference range for 4-methylumbelliferone (4-MU). GUS activity was then expressed as nmol/min/mg based on the fluorescence generation of 4-MU.
Yeast two-hybrid assay
The Matchmaker Gold Yeast Two-Hybrid system was employed to perform yeast two-hybrid (Y2H) screening. To assess the physical interactions between DELLAs (GAI, RGA, RGL1, RGL2, and RGL3) and DOF6, the coding sequences (CDS) of these proteins were extracted from wild-type (Col-0) plants. Subsequently, the DELLAs were cloned into the pGADT7 vector, while DOF6 was inserted into the pGBKT7 vector. Each combination—AD-GAI/BD-DOF6, AD-RGA/BD-DOF6, AD-RGL1/BD-DOF6, AD-RGL2/BD-DOF6, and AD-RGL3/BD-DOF6—was individually transformed into the yeast strain Y2H Gold. The transformed yeast strains were initially grown on SD/-Leu/-Trp medium. Positive interactions were then confirmed by screening the transformants on SD/-Leu/-Trp/-His/-Ade medium.
BiFC assay
For the BiFC assays, the CDS of DOF6, GAI, RGA, RGL1, and RGL2 were fused to either the N-terminal or C-terminal half of green fluorescent protein (GFP), resulting in constructs DOF6-nGFP, GAI-cGFP, RGA-cGFP, RGL1-cGFP, and RGL2-cGFP. These plasmids were co-transformed into N. benthamiana mesophyll protoplasts via PEG-mediated transient transformation. After incubating the samples in the dark for 16 h, fluorescence was observed using a confocal laser scanning microscope (Leica TCS SP5).
Haplotype network construction
The −412 to −434 bp segment of the GA20ox3 promoter region from 576 accessions was analyzed to construct a statistical parsimony network. This haplotype network was generated using Network 10.2 software. Detailed sequences can be found in Supplementary Data 3. Only haplotypes observed in two or more accessions were included in the analysis.
Statistics and reproducibility
Significant differences were analyzed using unpaired two-tailed Student’s t-test or one-way ANOVA with Tukey’s multiple comparisons test with GraphPad Prism 9.0. All experiments were repeated independently at least three times.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing dataset of the GA20ox3 promoters for 506 accessions was retrieved from the Arabidopsis 1001 Genomes project (http://signal.salk.edu/atg1001/3.0/gebrowser.php), with an additional 70 accessions obtained from studies78,79 (Supplementary Data 3). The origin of each sequence is detailed in Supplementary Data 2. Data on mean annual temperature, winter monthly minimum temperature, and the minimum temperature of the coldest month were obtained from the WorldClim database80 (https://www.worldclim.org/, Supplementary Data 6). Elevation data were obtained from WorldClim database80 (https://www.worldclim.org/) and meteoblue database (https://www.meteoblue.com/, Supplementary Data 7). The values for each accession were extracted using ArcMap software (version 10.7) based on their latitude and longitude coordinates, as detailed in Supplementary Data 2. SNP data have been deposited in the figshare public repository (https://doi.org/10.6084/m9.figshare.27188217.v1). The genetic materials used in this study are available from the corresponding authors upon request. Source Data are provided with this paper.
References
Fernandez-Pascual, E. et al. The seed germination spectrum of alpine plants: a global meta-analysis. N. Phytol. 229, 3573–3586 (2021).
Satyakam Zinta, G., Singh, R. K. & Kumar, R. Cold adaptation strategies in plants-an emerging role of epigenetics and antifreeze proteins to engineer cold resilient plants. Front. Genet. 13, 909007 (2022).
Preston, J. C. & Sandve, S. R. Adaptation to seasonality and the winter freeze. Front. Plant Sci. 4, 167 (2013).
Bykova, O., Chuine, I., Morin, X. & Higgins, S. I. Temperature dependence of the reproduction niche and its relevance for plant species distributions. J. Biogeogr. 39, 2191–2200 (2012).
Fernández-Pascual, E., Pérez-Arcoiza, A., Prieto, J. A. & Díaz, T. E. Environmental filtering drives the shape and breadth of the seed germination niche in coastal plant communities. Ann. Bot. 119, 1169–1177 (2017).
Korner, C. Plant adaptation to cold climates. F1000Res 5, F1000 (2016).
McFadden, I. R. et al. Global plant-frugivore trait matching is shaped by climate and biogeographic history. Ecol. Lett. 25, 686–696 (2022).
Ulrich, W., Kusumoto, B., Shiono, T., Fuji, A. & Kubota, Y. Latitudinal gradients of reproductive traits in Japanese woody plants. Ecol. Res. 38, 188–199 (2022).
Singh, A. & Roy, S. High altitude population of Arabidopsis thaliana is more plastic and adaptive under common garden than controlled condition. BMC Ecol. 17, 39 (2017).
Hu, J. et al. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20, 320–336 (2008).
de Villemereuil, P., Mouterde, M., Gaggiotti, O. E. & Till-Bottraud, I. Patterns of phenotypic plasticity and local adaptation in the wide elevation range of the alpine plant Arabis alpina. J. Ecol. 106, 1952–1971 (2018).
Montesinos-Navarro, A., Picó, F. X. & Tonsor, S. J. Clinal variation in seed traits influencing life cycle timing in Arabidopsis thaliana. Evolution 66, 3417–3431 (2012).
Picó, F. X. Demographic fate of Arabidopsis thaliana cohorts of autumn- and spring-germinated plants along an altitudinal gradient. J. Ecol. 100, 1009–1018 (2012).
Gremer, J. R. & Venable, D. L. Bet hedging in desert winter annual plants: optimal germination strategies in a variable environment. Ecol. Lett. 17, 380–387 (2014).
Burghardt, L. T., Edwards, B. R. & Donohue, K. Multiple paths to similar germination behavior in Arabidopsis thaliana. N. Phytol. 209, 1301–1312 (2016).
Childs, D. Z., Metcalf, C. J. E. & Rees, M. Evolutionary bet-hedging in the real world: empirical evidence and challenges revealed by plants. Proc. Biol. Sci. 277, 3055–3064 (2010).
Zacchello, G., Vinyeta, M. & Ågren, J. Strong stabilizing selection on timing of germination in a Mediterranean population of Arabidopsis thaliana. Am. J. Bot. 107, 1518–1526 (2020).
Gremer, J. R., Wilcox, C. J., Chiono, A., Suglia, E. & Schmitt, J. Germination timing and chilling exposure create contingency in life history and influence fitness in the native wildflower Streptanthus tortuosus. J. Ecol. 108, 239–255 (2019).
Donohue, K., Rubio de Casas, R., Burghardt, L., Kovach, K. & Willis, C. G. Germination, postgermination adaptation, and species ecological ranges. Ann. Rev. Ecol. Evol. S. 41, 293–319 (2010).
Rajjou, L. et al. Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533 (2012).
Cavieres, L. A. & Sierra-Almeida, A. Assessing the importance of cold-stratification for seed germination in alpine plant species of the High-Andes of central Chile. Perspect. Plant Ecol. 30, 125–131 (2018).
Fernández-Pascual, E., Jiménez-Alfaro, B. & Bueno, Á Comparative seed germination traits in alpine and subalpine grasslands: higher elevations are associated with warmer germination temperatures. Plant Biol. 19, 32–40 (2017).
García-Fernández, A., Escudero, A., Lara-Romero, C. & Iriondo, J. M. Effects of the duration of cold stratification on early life stages of the Mediterranean alpine plant Silene ciliata. Plant Biol. 17, 344–350 (2015).
Gremer, J. R. et al. Variation in the seasonal germination niche across an elevational gradient: the role of germination cueing in current and future climates. Am. J. Bot. 107, 350–363 (2020).
Finch-Savage, W. E. & Footitt, S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 68, 843–856 (2017).
Sajeev, N., Koornneef, M. & Bentsink, L. A commitment for life: decades of unraveling the molecular mechanisms behind seed dormancy and germination. Plant Cell 36, 1358–1376 (2024).
Finch-Savage, W. E., Cadman, C. S., Toorop, P. E., Lynn, J. R. & Hilhorst, H. W. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51, 60–78 (2007).
Penfield, S. & Springthorpe, V. Understanding chilling responses in Arabidopsis seeds and their contribution to life history. Philos. T. R. Soc. B. 367, 291–297 (2012).
Footitt, S., Hambidge, A. J. & Finch-Savage, W. E. Changes in phenological events in response to a global warming scenario reveal greater adaptability of winter annual compared with summer annual Arabidopsis ecotypes. Ann. Bot. 127, 111–122 (2021).
Martinez-Berdeja, A. et al. Functional variants of DOG1 control seed chilling responses and variation in seasonal life-history strategies in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 117, 2526–2534 (2020).
Gonzalo-Turpin, H. & Hazard, L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J. Ecol. 97, 742–751 (2009).
Hedden, P. & Sponsel, V. A century of gibberellin research. J. Plant Growth Regul. 34, 740–760 (2015).
Colebrook, E. H., Thomas, S. G., Phillips, A. L. & Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75 (2014).
Kasahara, H. et al. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 277, 45188–45194 (2002).
Hedden, P. & Thomas, S. G. Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25 (2012).
O’Neill, D. P. et al. Regulation of the gibberellin pathway by auxin and DELLA proteins. Planta 232, 1141–1149 (2010).
Boccaccini, A. et al. The DOF protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. Mol. Plant 7, 1486–1489 (2014).
Hedden, P. & Kamiya, Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu. Rev. Plant Biol. 48, 431–460 (1997).
Sun, T. P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6, e0103 (2008).
Ogawa, M. et al. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604 (2003).
Plackett, A. R. et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24, 941–960 (2012).
Rieu, I. et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53, 488–504 (2008).
Yan, C., Yan, Z., Wang, Y., Yan, X. & Han, Y. Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J. Exp. Bot. 65, 5933–5944 (2014).
Barboza, L. et al. Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proc. Natl. Acad. Sci. USA 110, 15818–15823 (2013).
Dill, A. & Sun, T. P. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159, 777–785 (2001).
Wen, C. K. & Chang, C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100 (2002).
Daviere, J. M. & Achard, P. Gibberellin signaling in plants. Development 140, 1147–1151 (2013).
Tyler, L. et al. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019 (2004).
Fuentes, S. et al. Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 24, 3982–3996 (2012).
Oh, E. et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208 (2007).
Lee, B. D. et al. CONSTITUTIVE PHOTOMORPHOGENIC 1 promotes seed germination by destabilizing RGA-LIKE 2 in Arabidopsis. Plant Physiol. 189, 1662–1676 (2022).
la Rosa, N. M. D. et al. Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166, 1022–1032 (2014).
Liu, X. et al. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 7, 12768 (2016).
Ravindran, P., Verma, V., Stamm, P. & Kumar, P. P. A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 10, 1307–1320 (2017).
Rueda-Romero, P., Barrero-Sicilia, C., Gomez-Cadenas, A., Carbonero, P. & Onate-Sanchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63, 1937–1949 (2012).
Lou, S. et al. Allelic shift in cis-elements of the transcription factor RAP2.12 underlies adaptation associated with humidity in Arabidopsis thaliana. Sci. Adv. 8, eabn8281 (2022).
Kim, Y. C., Nakajima, M., Nakayama, A. & Yamaguchi, I. Contribution of gibberellins to the formation of Arabidopsis seed coat through starch degradation. Plant Cell Physiol. 46, 1317–1325 (2005).
Yanagisawa, S. & Schmidt, R. J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214 (1999).
Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560 (2002).
Chong, S. N., Ravindran, P. & Kumar, P. P. Regulation of primary seed dormancy by MAJOR LATEX PROTEIN-LIKE PROTEIN329 in Arabidopsis is dependent on DNA-BINDING ONE ZINC FINGER6. J. Exp. Bot. 73, 6838–6852 (2022).
Hu, Y. et al. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat. Plants 4, 289–298 (2018).
Penfield, S. Seed dormancy and germination. Curr. Biol. 27, R874–R878 (2017).
Luo, Y. et al. A single nucleotide deletion in gibberellin20-oxidase1 causes alpine dwarfism in Arabidopsis. Plant Physiol. 168, 930–937 (2015).
Hou X. H. et al. Green revolution gene drives adaptation of Arabidopsis to the extremely high altitude. Sci. China Life Sci. https://doi.org/10.1007/s11427-024-2769-x (2025).
Yamauchi, Y. et al. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16, 367–378 (2004).
Murase, K., Hirano, Y., Sun, T. P. & Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 (2008).
Larran, A. S., Pajoro, A. & Questa, J. I. Is winter coming? Impact of the changing climate on plant responses to cold temperature. Plant Cell Environ. 46, 3175–3193 (2023).
Footitt, S., Huang, Z., Clay, H. A., Mead, A. & Finch-Savage, W. E. Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant J. 74, 1003–1015 (2013).
Bloomer, R. H. & Dean, C. Fine-tuning timing: natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 68, 5439–5452 (2017).
Zhu, P., Lister, C. & Dean, C. Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 599, 657–661 (2021).
Hepworth, J. et al. Natural variation in autumn expression is the major adaptive determinant distinguishing Arabidopsis FLC haplotypes. Elife 9, e57671 (2020).
Dorone, Y. et al. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell 184, 4284–4298.e4227 (2021).
Philippi, T. & Seger, J. Hedging one’s evolutionary bets, revisited. Trends Ecol. Evol. 4, 41–44 (1989).
Pausas, J. G., Lamont, B. B., Keeley, J. E. & Bond, W. J. Bet-hedging and best-bet strategies shape seed dormancy. N. Phytol. 236, 1232–1236 (2022).
Lowder, L. G. et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985 (2015).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Bowler, C. et al. Chromatin techniques for plant cells. Plant J. 39, 776–789 (2004).
Zou, Y. P. et al. Adaptation of Arabidopsis thaliana to the Yangtze River basin. Genome Biol. 18, 239 (2017).
Kang, M. H. et al. The pan-genome and local adaptation of Arabidopsis thaliana. Nat. Commun. 14, 6259 (2023).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
Acknowledgements
The work was equally supported by the Natural Science Foundation of China (32030006) and Science and Technology Projects of Xizang Autonomous Region, China (XZ202402ZD0005).
Author information
Authors and Affiliations
Contributions
J.L. led the research. Y.J. and H.L. co-directed the program. X.G. finished most of the analyses and experiments. X.G. and Y.J. wrote the manuscript. J.L. revised the manuscript. Y.H., Y.Z., X.F., L.F., D.M., X.J. and P.S. performed parts of the bioinformatics analysis. S.L., H.Z., X.L., J.H., Y.Y., Y.S., M.L., S.G., R.L. and L.T. performed parts of the biochemical experiments. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Xingliang Hou, Steven Penfield and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, X., Lou, S., Han, Y. et al. Allelic variations in GA20ox3 regulate fruit length and seed germination timing for high-altitude adaptation in Arabidopsis thaliana. Nat Commun 16, 5053 (2025). https://doi.org/10.1038/s41467-025-60436-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60436-7