Abstract
Human Papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSCC) remains a challenging malignancy, with radiotherapy, alone or combined with immune checkpoint inhibitors, often failing to achieve durable disease control. Here, by conducting longitudinal multi-omic analyses of pre- and post-radiation biopsies from patients receiving a pre-operative hypofractionated radiation regimen, we uncover that radiation rapidly depletes a subpopulation of tumor-infiltrating lymphocytes (TIL), characterized by a proliferative, cytotoxic, and tissue-resident gene signature (TProlif_Tox). We provide multi-dimensional evidence for tumor antigen-specificity of TProlif_Tox clonotypes and show that post-radiation tumors are instead repopulated by regulatory and non-specific clones. Finally, TIL depletion correlates with radiorecurrent disease after conventional radiation, emphasizing the potential impact of radiation-induced TIL loss regardless of fractionation. Thus, this study provides key insights into radiotherapy-induced alterations in the immune microenvironment that drive immunologic radioresistance and proposes restoring tumor antigen-specific T cell clonotypes as a strategy to improve radioimmunotherapy responses in HNSCC.
Similar content being viewed by others
Introduction
Human papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSCC), which includes tumors of the oral cavity, pharynx, and larynx, exceeds 65,000 cases per year, resulting in over 14,000 deaths annually in the United States1. It also remains a global health problem and is among the top cancers in South Asia2. Unlike patients with HPV-positive disease3,4, the HPV-negative HNSCC population carries a particularly poor prognosis, and current therapies often lead to profound detriments in quality of life5,6. Surgery and radiation are the mainstays of HPV-negative HNSCC management, but 5-year survival remains under 50% for locoregionally advanced disease using current treatments7. The genomic heterogeneity of HNSCC and the absence of uniform oncogenic drivers have stymied efforts at targeted therapies such as EGFR inhibition3,8,9. Immunotherapeutic approaches, including adoptive T cell therapies and immune checkpoint inhibition (ICI), have demonstrated long-term, durable disease control in a select group of HNSCC patients10,11,12. While these results emphasize the potential power of immuno-oncology for HNSCC, greater than 80% of patients treated with available immunotherapeutic agents do not derive long-term benefit, experiencing eventual disease progression13.
To improve HNSCC outcomes, recent clinical trials have combined radiation-based therapies with ICI. These efforts were largely based on pre-clinical evidence, which had suggested that radiation efficacy depended on immunologic mechanisms, including upregulation of antigen presentation and increased effector T cell function14,15,16,17,18,19. In light of these data, there was hope that such an approach would expand the number of HNSCC patients who benefit from immunotherapy. However, three recent large randomized trials combining radiation with ICI in HNSCC have had disappointing results5,20,21. These data imply that immunologic mechanisms beyond immune checkpoint receptors (ICR) may contribute to disease relapse and immunologic radioresistance in HNSCC.
One possible contributor to these unexpected results is radiotherapy fractionation. While conventionally fractionated radiation (2 Gy per fraction, delivered in 30–35 fractions over 6–7 weeks) has long been considered immunosuppressive, shorter-course high-dose-per-fraction regimens (hypofractionated) have shown immunogenic potential in pre-clinical studies, including improvements in CD8 T cell infiltration, effector function, and tumor antigen presentation14,22,23,24,25. For these reasons, we initiated a Phase I clinical trial testing a Hypofractionated Pre-operative Radiotherapy in HNSCC (HyPR-HN, NCT05538533), which includes three progressively hypofractionated regimens (46 Gy in 10 fractions, 40 Gy in 7 fractions, or 35 Gy in 5 fractions). The design of all dose levels maintains an EQD2 to late-responding tissues identical to conventionally fractionated radiation (70 Gy) while providing an adequate dose to clear microscopic disease (EQD2 α/β = 10 ≥ 50 Gy)26,27,28.
Here, we report translational results from patients treated on the first dose-level (4.6 Gy per fraction, delivered in 10 fractions over 2 weeks), which has completed accrual, including all sample acquisition and immunological correlative analyses. Specifically, we obtain samples of the primary tumor pre-treatment, on the last day of radiation, and 6 weeks later at the time of definitive surgery for multi-omic analysis, including single-cell RNA and T cell receptor (TCR) sequencing. Through this approach, we demonstrate that clinical radiotherapy induces immediate and profound depletion of tumor-infiltrating lymphocytes (TIL), including ablation of TIL sub-populations demonstrating consistent expression of proliferative, exhausted, tissue-resident, and cytotoxic markers (which we have termed TProlif_Tox), indicative of a capacity for tumor-reactivity. Importantly, TProlif_Tox clonotypes are almost entirely depleted by clinical radiation and fail to reappear in the tumor microenvironment post-therapy. In their absence, T cell reintegration into HNSCCs is governed predominantly by non-specific infiltration from the periphery, including regulatory, naïve, and common effector clonotypes. Finally, we analyze an institutional tumor registry of patients treated with conventionally fractionated radiotherapy, including biospecimen analysis pre-treatment and at the time of recurrence in matched patients. We find that TIL reduction is strongly associated with long-term in-field cancer recurrence, emphasizing the potential impact of radiation-induced TIL depletion irrespective of fractionation. Relapsed disease is further characterized by a persistent loss of the ICRhigh TIL populations critical for effective anti-tumor immunity.
Collectively, through multi-omic analyses of serial biopsies obtained across radiotherapy timepoints, we find that clinical radiation profoundly depletes TILs, including a subset of tumor-reactive T cells that do not recover following treatment. This depletion is associated with long-term tumor persistence and a failure of tumor antigen-specific immune reconstitution within the tumor microenvironment. These findings suggest that radiotherapy may inadvertently eliminate the very T cells required for durable immune control. Future strategies should prioritize preserving or replacing tumor antigen-specific T cells to optimize the synergy between radiotherapy and immunotherapy.
Results
HyPR-HN study design and biospecimen acquisition
The Phase I trial HyPR-HN (NCT05538533, MPIs JZ/MA/HH) tests pre-operative hypofractionated radiation in HPV-negative HNSCC. In this study, biopsies of the primary tumor are taken pre-treatment, on the last day of radiation (immediately after the last fraction), and then 6 weeks later at the time of definitive surgical resection of remaining disease. Here, we report translational data from four patients with locoregionally advanced HNSCC enrolled on the first radiation dose level, which included 10 fractions of 4.6 Gy to the primary tumor and involved nodes (PTVhigh, EQD2 = 56 Gy) delivered without chemotherapy (Supplementary Table 1). Elective nodal radiation was also given simultaneously to a dose of 35 Gy in 10 fractions. All enrolled patients had locoregionally advanced oral squamous cell carcinomas. Enrolled patients included 3 females and 1 male patient with ages ranging from 59 to 73 years.
The response to hypofractionated radiation in HNSCC may be constrained by immediate and profound T cell depletion
To dissect the immune alterations occurring in response to hypofractionated radiation and identify mechanisms of immunologic radioresistance, we performed multi-omic analyses on serial tumor biopsy specimens. To maintain consistency, all biopsies were taken from the same anatomical location in the primary tumor for each patient at each timepoint by a single study investigator (JZ, Fig. 1a). Biopsies were completed for all timepoints apart from the final specimen for the third enrolled patient due to a medical complication unrelated to protocol treatment (n = 11 total samples). For each patient, biopsy specimens were divided into pieces for parallel analytic pathways, including paired single-cell RNA and TCR sequencing, multiplex immunofluorescence (mIF), multiplex flow cytometry (mFC), whole-exome sequencing (WES), bulk RNA sequencing, and spatial transcriptomics. Dissociative single-cell sequencing was prioritized and performed immediately on freshly digested biopsy tissues. Additional analytical methods were performed when a sufficient quantity of biopsy material was available, which was determined on a specimen-by-specimen basis (Supplementary Table 2).
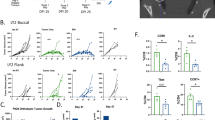
a Schematic of trial design including timepoints and translational correlative analyses. b Combined integrated single-cell datasets including 11 separate HyPR-HN biopsies from four patients over three timepoints. Analysis across timepoints reveals immediate T cell depletion on the last day of radiation both (c) quantitatively as a percent of the single-cell transcriptome and (d) visually across UMAP profiles. e mFC confirms immediate TIL depletion in response to radiation, despite (f) preserved overall immune infiltrate and increased intra-tumoral myeloid cells. g Representative mFC plots of CD8 and CD4 T cell infiltrate and (h) mIF with corresponding clinical MR (pre-treatment and last day of radiation) and CT (6 weeks post-radiation) imaging are shown for HyPR-HN Patient 01 serially across treatment timepoints. Antibody fluorescent conjugates include CD8: light blue, CD4: green, CD20: red, Foxp3: yellow, CD68: orange, Pan-CK: magenta, DAPI: dark blue. Scale bar = 50 µM. T-tests with Holm-Sidak’s multiple comparison correction were used to analyze differences between groups. Flow cytometry gating strategy demonstrated in Supplementary Fig. 1. Graphic created in BioRender. Zenga, J. (2025) https://BioRender.com/zu8pe9n.
Initial processing of single-cell sequencing datasets was performed using unsupervised clustering analyses with cell-type annotation based on differential gene expression and canonical markers (Fig. 1b). Comparative analysis of transcriptomic profiles across timepoints revealed significant and selective lymphocyte depletion on the last day of radiation as a percentage of single cells sequenced, with a relative increase in myeloid cell subsets (Fig. 1c, d). To validate this finding on the protein level, we simultaneously performed mFC (all HyPR-HN patients, Fig. 1e–g, and Supplementary Fig. 1) and mIF (HyPR-HN Patient 01, Fig. 1h), which revealed immediate and profound TIL depletion after radiotherapy with a preserved overall immune infiltrate. Despite this significant T cell depletion, however, tumors exhibited substantial radiographic regression, indicating a radiation-induced direct cytotoxic effect independent of T cell activity (Fig. 1h). Although intra-tumoral T cell populations began to recover by 6 weeks post-treatment, they did not reach pre-treatment levels, most notably in CD8 sub-populations.
Multi-omic evaluation identifies persistent loss of ICRhigh CD8 TILs post-radiotherapy
Given the absence of synergy between radiotherapy and ICI for HNSCC in multiple recent Phase II and III clinical trials5,20,21, we evaluated the immediate effects of radiation on TIL ICR expression using several orthogonal approaches. We first performed integrated single-cell RNA sequencing analyses of TILs across treatment timepoints. T cells identified by both transcriptional clustering and expression of CD3 complex genes were subsetted and re-clustered again (Supplementary Fig. 2a) through unsupervised analysis with immunophenotypic sub-cluster annotation based on canonical markers (Supplementary Fig. 2b) and differentially expressed genes (Supplementary Fig. 2c) which were globally conserved across treatment timepoints (Supplementary Fig. 3). These analyses confirmed TIL depletion across sub-clusters immediately upon completion of radiation. Surprisingly, the phenotypic pattern of TIL repopulation was selective, with early infiltration of regulatory, naïve, and pre-exhausted TILs and persistent near-complete loss of the terminally exhausted ICRhigh CD8 sub-cluster (ExCD8) (Fig. 2a–c).
a Integrated single-cell datasets of 11 separate HyPR-HN biopsies with T cells extracted and re-clustered from four patients over three timepoints with UMAP profiling demonstrating global loss of TILs post-radiation and revealing persistent loss of exhausted CD8 T cells. b Analysis across timepoints confirms immediate T cell depletion on the last day of radiation quantitatively as a percent of the T cell transcriptome. c Characterization of T cells co-expressing multiple ICRs (PDCD1, HAVCR2, TIGIT, and LAG3) demonstrates immediate and persistent loss of ICR expression in response to radiotherapy. d mFC confirms immediate loss of ICRhigh TILs as a percentage of viable cells in response to radiation, despite (e) preserved expression as a percentage of CD8 T cells. Representative mFC plots of CD8 T cell infiltration and ICR expression are shown for HyPR-HN Patient 02 (f) pre-treatment and (g) on the last day of radiation. Gating is provided as a percentage of total live cells. Pre-ex Pre-exhausted T cells, Reg Regulatory T cells, TRM Tissue-resident memory T cells, Prolif Proliferative T cells, ExCD8 Exhausted CD8 T cells, ExCD4 Exhausted CD4 T cells, IFN Type I interferon-responsive T cells. T-tests with Holm-Sidak’s multiple comparison correction were used to analyze differences between groups. Flow cytometry gating strategy demonstrated in Supplementary Fig. 4.
To confirm these findings at the protein level, we simultaneously performed mFC on the same biopsy samples, which revealed a significant numerical loss in viable ICRhigh (PD-1, TIM3, LAG3, and TIGIT) CD8 TILs immediately post-radiotherapy (Fig. 2d–g and Supplementary Fig. 4). We further confirmed these findings using spatial analysis from one enrolled patient for whom we had pre-treatment and 6-week post-radiotherapy samples of sufficient quality for spatial transcriptomics (HyPR-HN Patient 02). This analysis demonstrated spatial depletion of post-radiotherapy TILs throughout the tumor microenvironment with minimal ICR expression despite preserved myeloid infiltrate in both pre- and post-radiation samples (Supplementary Fig. 5).
These findings raised critical questions about the immunological impact of radiation on TIL functionality. The loss of the highly expanded ICRhigh ExCD8 population, a key target of ICIs, may have detrimental consequences if this sub-cluster contains tumor antigen-specific TILs in HNSCC. Furthermore, it remained uncertain whether the re-infiltrating non-exhausted T cell landscape supports or impairs anti-tumor immunity. To address these issues, we investigated whether radiation preferentially depletes tumor antigen-specific T cells and whether repopulating TILs retain a tumor-reactive immunophenotype or are instead more likely dominated by non-specific TCRs.
Post-radiation T cell clonal repopulation patterns suggest conserved mechanisms of T cell infiltration in HNSCC
The serial biopsy design of the HyPR-HN trial, combined with paired single-cell RNA and TCR sequencing, enabled precise tracking of T cell clonotypes across treatment timepoints. First, using cell-specific barcode pairing, clonotype frequency data were projected onto the matching TIL transcriptomic states. These projections demonstrated consistency with TIL sub-cluster immunophenotypic classifications, with naïve-like and regulatory clusters largely composed of singleton and low-frequency clonotypes while proliferative, pre-exhausted, and exhausted clusters showed extensive clonal expansion (Fig. 3a). T cells repopulating the tumor post-radiation largely mirrored the clonal architecture of pre-treatment TILs. The clonal and transcriptional overlap observed among TIL subsets associated with less differentiated states, such as naïve-like and pre-exhausted clusters, suggested that the same conserved chemotactic forces driving TIL infiltration pre-treatment remained active post-radiation. However, the notable absence of the expanded ICRhigh ExCD8 sub-cluster post-radiation raised the possibility of selective tumor-reactive clonotype depletion or functional alteration, which may have significant implications for anti-tumor immunity.
a Clonal frequency projections onto T cell UMAP profiles from all 11 HyPR-HN datasets demonstrate a diverse T cell landscape of both singletons and hyper-expanded clones. Repopulating T cells largely recapitulate the pre-treatment clonal architecture, particularly among less differentiated states including naïve-like and pre-exhausted populations. b While exhausted CD8 T cell clonotypes have overlap with several sub-clusters, they demonstrate the greatest clonal overlap with proliferative TILs, which are subsequently lost post-radiotherapy and do not return to the tumor microenvironment. c Overlapping exhausted and proliferative TIL clonotypes contain a conserved and unique gene transcriptional profile suggesting tumor antigen-reactivity. Pre-ex Pre-exhausted T cells, Reg Regulatory T cells, TRM Tissue-resident memory T cells, Prolif Proliferative T cells, ExCD8 Exhausted CD8 T cells, ExCD4 Exhausted CD4 T cells, IFN Type I interferon-responsive T cells.
T cell clonal tracking demonstrates clonal and transcriptional overlap between pre-treatment ICRhigh and proliferative TILs, suggestive of anti-tumor immunogenicity
To further investigate the immuno-oncologic relevance of ExCD8 T cell loss, we tracked pre-treatment ExCD8 T cell clonotypes within tumors across timepoints for each patient to determine if these clones were fully lost or instead reprogrammed into a new phenotypic sub-cluster. Pre-treatment ExCD8 clonotypes overlapped with several sub-clusters, including regulatory T cells, suggesting that an exhaustion gene signature alone is insufficient to reliably identify tumor antigen-specific and reactive TILs (Supplementary Fig. 6a). Surprisingly, the most prominent clonotype overlap was observed between ExCD8 and proliferative T cell clusters in all pre-treatment samples (Fig. 3b and Supplementary Fig. 6b–d). This suggested that a subset of ExCD8 TILs is not terminally exhausted but rather exists in dynamic equilibrium between over-stimulated anergic and activated proliferating states. Through differential gene expression analyses, we found that this overlapping TIL sub-population within pre-treatment proliferative and ExCD8 not only shares TCR clonotypes but has a remarkably conserved gene expression profile unique among pre-treatment TILs and distinct from post-radiation proliferating TILs (Fig. 3c). This conserved gene expression profile was enriched for CD8 markers (CD8A, CD8B), proliferative genes (MKI67, STMN1, TYMS), immune checkpoint molecules (PDCD1, HAVCR2, LAG3, TIGIT, ENTPD1), indicators of cytotoxicity (GZMA, GZMB, GZMH, IFNG), a marker of tissue-residence (ITGAE), and CXCL13, which has been previously associated with tumor antigen load, development of tertiary lymphoid structures, and anti-tumor immune response29,30. This constellation of gene expression programs, to include proliferative, ICRhigh, cytotoxic, tissue-residence, and CXCL13+ (hereafter referred to as the TProlif_Tox expression signature), combined with TCR clonotype overlap between exhausted and proliferating TILs, suggested the possibility of tumor reactivity and, therefore, tumor antigen-specificity within this sub-population. Although these TProlif_Tox clonotypes were conserved within each patient across exhausted and proliferative clusters, we found no overlap in TCR sequences between patients, suggesting each patient carried unique antigenic targets.
The TProlif_Tox gene signature is present across HNSCCs but is absent in normal oral mucosa
To further investigate the potential of the TProlif_Tox gene expression profile to predict tumor antigen-specificity, we next asked whether this signature was unique to malignant tissues and whether it was present across other HNSCCs, external to the HyPR-HN dataset. To address this, we integrated our pre-treatment single-cell RNA sequencing data with publicly available single-cell datasets, including other HPV-negative HNSCCs and single-cell sequencing of normal oral mucosal biopsies31,32. As above, unsupervised clustering was performed and T cells were subsetted, re-clustered, annotated, and compared across studies and tissue types (Fig. 4a, b). T cell gene expression profiling revealed that the TProlif_Tox gene signature was present across HNSCCs but fully absent from normal tissue single-cell sequencing profiles (Fig. 4c), demonstrating the tumor-specificity of TProlif_Tox.
a Integrated analyses of multiple internal and external datasets with T cells subsetted demonstrates (b) conserved clustering and T cell functional groups. c Despite globally conserved transcriptional T cell sub-classifications, the TProlif_Tox gene signature is tumor-specific without any expression in normal oral mucosa. Pre-ex Pre-exhausted T cells, Reg Regulatory T cells, Prolif Proliferative T cells, Ex Exhausted T cells.
The TProlif_Tox gene signature and clonotypes are conserved across metastatic sites in HNSCC
We next evaluated whether the TProlif_Tox gene signature and associated clonotypes were conserved across metastatic sites. If TProlif_Tox TCRs have tumor antigen-specificity and similar tumor antigens are shared across metastatic sites, the same TProlif_Tox clonotypes should also be present in both the primary tumor and metastatic deposits. To investigate this, we performed single-cell RNA and TCR sequencing of a previously untreated HNSCC patient (P120, not enrolled on HyPR-HN), including samples of the primary tumor, a metastatic regional lymph node, and circulating T cells from peripheral blood (Fig. 5a). We first confirmed that TILs from P120 harbored a subset of T cells that could objectively recognize tumor antigen and kill patient-matched tumor cells. To test this, we derived in vitro TIL cultures and a malignant cell line from separate biopsy pieces of P120’s primary tumor, with isolated cancer-associated fibroblasts (CAF) as a patient-matched control line. To ensure translational validity of this system, we performed whole-exome sequencing (WES) and bulk RNA sequencing of P120’s primary tumor, involved regional node, and the tumor-derived malignant cell line. Through tumor mutational analyses and a neo-antigen prediction pipeline, we identified substantial neo-antigenic overlap between all three samples (Fig. 5b). This finding supports the existence of shared neo-antigens across both primary tumor and metastatic sites, which are also present in the tumor-derived malignant line. Through co-culture cytotoxicity assays using CD8-selected TILs and patient-matched malignant cells, we found concentration-dependent tumor-specific CD8 TIL cytotoxicity (>80% kill at 10 TIL:1 tumor cell), suggesting that a subset of CD8 TILs can recognize tumor antigen and induce a relevant immunologic response (Fig. 5c). Next, we analyzed single-cell sequencing data from additional pieces of P120 biopsy samples, which were processed at the time of surgical resection. We performed integration, clustering, T cell subsetting, and clonal frequency projections (Fig. 5d and Supplementary Fig. 7). Gene expression profiling identified the TProlif_Tox gene signature in both tumor and regional node (Fig. 5e). We additionally found that several identical TProlif_Tox clonotypes were present in both the primary tumor and metastatic node despite being undetectable in blood (Fig. 5f). Taken together, these results suggest that a subset of TILs in HNSCC have tumor-killing capability and that several of these clonotypes may target similar antigens across primary tumor and metastatic disease.
a Schematic of biospecimens and tissue analyses for patient P120. b Exomic and transcriptional mutational analyses demonstrates conserved coding mutations across tumor, involved node, and patient-derived malignant line. c Co-culture between a patient-derived malignant line and autologous extracted TILs demonstrates concentration-depended tumor cell killing, while cultures of autologous cancer-associated fibroblasts (CAF) enriched for patient-matched non-malignant cells are largely preserved. Ratios represent target-to-TIL concentration. Single-cell sequencing of tumor, involved regional node, and circulating T cells from a HNSCC patient (P120) reveals (d) similar clustering and clonal architecture to HyPR-HN samples, and (e) the conserved presence of TProlif_Tox signature expression in both tumor and node, without expression in blood. f Several TProlif_Tox clonotypes demonstrate identical overlap across primary tumor and involved node but are undetectable in blood. Pre-ex Pre-exhausted T cells, Reg Regulatory T cells, Prolif Proliferative T cells, ExCD8 Exhausted CD8 T cells, ExCD4 Exhausted CD4 T cells, IFN Type I interferon responsive T cells, HSP Heat-shock protein-enriched T cells, SCM stem-cell memory T cells, EM effector memory T cells, CM central memory T cells. Graphic created in BioRender. Zenga, J. (2025) https://BioRender.com/qj8bqqo.
The TProlif_Tox gene signature is conserved across other malignant histologies and can predict tumor antigen-specific TCRs
We next analyzed publicly available single-cell datasets to evaluate whether the TProlif_Tox gene signature may represent a tumor antigen-specific population, not only in HNSCC but also across other solid tumors. We first examined a breast cancer dataset covering both the primary tumor and a matched metastatic regional lymph node (Supplementary Fig. 8a)33. We again identified a similar clustering (Supplementary Fig. 8b) and clonal architecture (Extended Data Fig. 3c) to HNSCC, including expanded Prolif and ExCD8 sub-clusters. We identified the TProlif_Tox gene signature in both tumor and regional node (Supplementary Fig. 8d) with shared TProlif_Tox clonotypes across primary and metastatic sites (Supplementary Fig. 8e). Next, we evaluated the effects of ICI therapy on TProlif_Tox clonotypes through a secondary analysis of an ICI-treated renal cell carcinoma (RCC) single-cell dataset (Supplementary Fig. 9a)34. If TProlif_Tox harbors tumor antigen-specific TCRs, release from immune checkpoint blockade by ICI therapy may lead to TProlif_Tox clonal expansion. Consistent with this hypothesis, in ICI-treated RCC we identified the TProlif_Tox gene signature among clonally expanded sub-clusters (Supplementary Fig. 9b, c). Hyper-expanded TProlif_Tox clonotypes were shared multi-spatially across tumor biopsies, but were absent in peripheral blood (Supplementary Fig. 9d, e). Finally, we performed a secondary analysis of a single-cell pancreatic adenocarcinoma dataset for which tumor antigen-specific TCRs had been externally validated through in vitro studies (Supplementary Fig. 10a)35. We again identified the TProlif_Tox gene signature and clonotypes across proliferative and exhausted sub-clusters (Supplementary Fig. 10b, c). We found extensive overlap between in vitro validated tumor antigen-specific TCRs and TProlif_Tox clonotypes predicting tumor-reactivity (Supplementary Fig. 10d).
In silico predicted binding of TCRs to neo-antigen-HLA complexes supports tumor antigen-specificity of TProlif_Tox clonotypes in HNSCC
To assess the potential of TProlif_Tox clonotypes to bind tumor neo-antigen in HNSCC, we used a custom prediction pipeline to evaluate the binding affinity of patient-matched neo-antigen peptide-HLA pairs with TProlif_Tox clonotypes. Based on the availability of samples, we performed WES and bulk RNA sequencing of HyPR-HN Patient 02’s tumor biopsy pre-treatment and at 6 weeks post-radiation. These analyses confirmed a substantial overlap in identical coding mutations between pre-treatment and post-radiotherapy samples (Fig. 6a), suggesting the existence of similar neo-antigens across treatment timepoints. To evaluate the in silico potential of TProlif_Tox clonotypes to bind patient-specific neo-peptide-loaded HLA (pHLA), we employed TCRdock36. This package uses structural modeling and TCR:pHLA docking algorithms to simulate the binding interface between a given TCR and its corresponding pHLA. To develop inputs for this model, we first predicted neo-antigens from HyPR-HN Patient 02 using a custom script creating 8-11mer neo-peptides sliding across all conserved coding mutations identified from WES and validated in RNA sequencing. These were filtered for patient-specific HLA-binding through netMHCpan 4.1 and prioritized based on normalized expression in bulk RNA sequencing37. From this filtered set of neo-peptides, we then identified the pHLA complexes with the closest predicted docking geometry to TProlif_Tox TCRs, indicating high binding affinity. These pHLAs demonstrated significantly greater predicted binding to select TProlif_Tox TCRs compared to a set of patient-matched control TCRs extracted from patient-matched naïve intra-tumoral T cells (Fig. 6b and Supplementary Table 3). We then repeated the TCRdock pipeline using predicted neo-antigens from the WES and bulk RNA sequencing data from patient P120’s tumor, described above. Binding prediction through TCRdock again demonstrated greater predicted pHLA complex binding affinity to TProlif_Tox clonotypes when compared to control patient-matched intra-tumoral naïve T cells (Supplementary Fig. 11a), which were found with the TProlif_Tox phenotype in both tumor and node but were absent from peripheral blood (Supplementary Fig. 11b). In particular, one clonotype (CAVIILQSQGNLIF_CAWASTGELFF) was predicted to strongly bind several neo-antigens, consistent with the known degeneracy of TCR specificity38. An ability to target multiple neo-antigens may further explain the observed expansion of this clonotype in P120 across both the primary tumor and metastatic node, and its high expression of the T cell activation marker TNFRSF9.
a Whole exome and RNA sequencing mutational analyses were performed pre-treatment and at 6 weeks post-radiation for the primary tumor of HyPR Patient 02, demonstrating substantial overlap in transcribed coding mutations, which translation into candidate neo-antigens. b Predicted binding between TCRs and neo-antigen-HLA complexes for HyPR-HN Patient 02. Binding scores were zero-shifted and normalized. Binding prediction for each neo-peptide-HLA complex was compared between TProlif_Tox TCRs and patient-matched irrelevant naïve TCRs. Within each pHLA complex, we performed 10,000 bootstrap replicates by sampling with replacement from the control group and, for each replicate, recording the lowest PAE_SD scores. This bootstrap procedure generated an empirical null distribution of extreme binding scores. The 95% confidence interval for the minimum PAE_SD was calculated using the T distribution. The empirical p-value was then calculated using this same T distribution. For each neo-peptide-HLA complex, a TProlif_Tox clonotype demonstrates the greatest predicted binding affinity. Among all HyPR-HN patients, (c) the TProlif_Tox expression profile and (d) the associated clonotypes that express it are lost after radiation and do not return to the tumor. Pre-ex Pre-exhausted T cells, Reg Regulatory T cells, TRM Tissue-resident memory T cells, Prolif Proliferative T cells, ExCD8 Exhausted CD8 T cells, ExCD4 Exhausted CD4 T cells, IFN Type I interferon-responsive T cells. The box plot represents the 25th to 75th percentile, and the whiskers represent 1.5x the interquartile range.
In vitro patient-matched tumor-TIL co-culture demonstrates tumor-reactive potential of TProlif_Tox
To more directly test the tumor-reactive potential of TProlif_Tox clonotypes, we leveraged the availability of a patient-derived malignant cell line and a large number of patient-matched TILs expanded from P120s primary tumor biopsy. With these samples, we performed co-culture, flow sorting by activation marker expression (CD137), single-cell sequencing, clonotype validation and enrichment by high activation marker gene expression (TNFRSF9), and subsequent clonotype projection onto the initial P120 primary tumor single-cell dataset (Supplementary Fig. 12a). Malignant cell-TIL co-culture demonstrated approximately 1% CD137-positive TILs (Supplementary Fig. 12b), which were then flow sorted for single-cell RNA and TCR sequencing. A CD137-negative TIL population was also flow-sorted and sequenced as a control. Clustering and clonality demonstrated several proliferative and activated sub-clusters enriched in the CD137-positive sorted population (Supplementary Fig. 12c, d). Given the low CD137 expression in most sorted TILs, we leveraged single-cell transcriptional profiling to ensure tumor-specific reactivity by isolating T cells with high TNFRSF9 expression, minimizing the inclusion of weak or non-specific TCR interactions (Supplementary Fig. 12e). Of these, only four TIL clonotypes exhibited high TNFRSF9 expression across multiple clones (Supplementary Fig. 12f). Although two of these clonotypes were not found in the original P120 single-cell dataset, one mapped to TProlif_Tox cluster (Supplementary Fig. 12f). The final clonotype was identified within the interferon-responsive sub-cluster, suggesting the presence of additional intra-tumoral T cell clonotypes with tumor-antigen specificity that may either require further priming in vivo for efficacy or are advancing along an earlier pseudo-time trajectory towards exhaustion.
TProlif_Tox clonotypes are lost after radiotherapy and do not return to the tumor microenvironment
With these data supporting the potential tumor antigen-specificity and immunological relevance of the TProlif_Tox sub-population, we next evaluated the effects of radiation on TProlif_Tox clonotypes across treatment timepoints. Remarkably, across all enrolled patients, the TProlif_Tox gene expression profile (Fig. 6c) and the clonotypes (Fig. 6d) that express these markers were both lost post-radiotherapy and did not return to the tumor in any patient studied (Supplementary Fig. 13). Together, these data suggested that pre-treatment tumor antigen-specific TILs are ablated by radiotherapy and do not re-infiltrate the tumor, at least within 6 weeks of treatment.
T cell repopulation of TIL-depleted tumors is driven by non-specific re-infiltration, including high-frequency circulating clonotypes and low-frequency regulatory and naïve T cells
In light of the loss of TProlif_Tox clonotypes in response to radiation, we sought to understand the immuno-oncologic relevance of TIL repopulation through the evaluation of clonotype dynamics. Unsurprisingly, the number of unique clonotypes was profoundly reduced immediately after radiation-induced TIL depletion but began to recover by the 6-week timepoint (Fig. 7a, b). Despite extensive TIL ablation and subsequent repopulation, however, amino acid-level entropy at each position along the CDR3 region remained unchanged across all timepoints, indicating that neither overall clonotype depletion nor repopulation was specific to TCR sequencing pattern (Fig. 7c). This aligns with evidence that tumor antigen-specific TILs constitute only a small fraction of the total repertoire, such that their loss or recruitment would have minimal impact on overall TCR diversity dynamics. Clonotype alluvial plotting revealed that approximately 30% of clones return to the tumor microenvironment post-radiation (Fig. 7d), with the majority of these representing the most frequent TCR sequences within each pre-treatment tumor (Fig. 7e).
a Radiation-induced TIL depletion is associated with a major reduction in overall TCR clonotype diversity, which begins to return by 6 weeks post-radiation in all patients. b Clonotype frequency mapping reveals broad immediate TIL depletion with return of high-frequency and intermediate-frequency clonotypes by 6 weeks post-radiation. c Despite alterations in overall number of clonotypes at each timepoint, diversity is conserved on the amino acid-level at the CDR3 region. d Among all repopulating clonotypes approximately 30% are returning clones while the remaining are newly infiltrating. e Clonal tracking of the 30 most frequent clonotypes within pre-treatment tumors reveals that returning clonotypes are predominantly composed of pre-treatment expanded populations.
To further investigate the functional relevance and potential antigenic targets of these repopulating TILs, we identified the most common circulating T cell clones through single-cell RNA and TCR sequencing of blood samples, which were available from two enrolled patients prior to radiation (HyPR-HN Patient 02 and 04) and one patient at 6 weeks post-radiotherapy (HyPR-HN Patient 02). Since very high-frequency circulating clonotypes carry predominantly viral-epitope-specific TCRs, this approach allowed us to track clonotype dynamics among a population of T cells irrelevant to the tumor antigen response39,40,41,42. As expected, this analysis revealed that the frequency of these abundant circulating clones remained unchanged in the blood pool pre- and post-radiation (Fig. 8a). Importantly, we observed that many of these clones were also present in the pre-exhausted niche of the pre-treatment tumor and reappeared post-radiation with nearly identical transcriptional and clonal distributions (Fig. 8b–d). Conversely, no TProlif_Tox clonotypes from HyPR-HN Patients 02 or 04 were found in their respective tested circulating T cell pools, consistent with TProlif_Tox tumor-specificity. Additionally, we found that the remaining repopulating TILs were largely composed of newly infiltrating regulatory and naïve clonotypes that were absent in the pre-treatment tumor (Fig. 8e, f). Together, these findings suggest that the migration of diverse repopulating TILs is driven by non-specific and generalized mechanisms, such as chemotactic gradients, rather than selective pressures from tumor antigens.
Integrated single-cell sequencing across treatment timepoints demonstrates extensive clonal overlap between high-frequency circulating clonotypes and the pre-exhausted sub-cluster within tumors, showing that T cell re-infiltration after radiation-induced TIL depletion is driven in part by common circulating clonotypes in both (a, b) HyPR-HN Patient 02 and (c, d) HyPR-HN Patient 04. Clonal mapping of post-radiation regulatory and naïve clonotypes further demonstrates that these sub-clusters are composed largely of new clonotypes not seen in the pre-treatment tumor in both (e) HyPR-HN Patient 02 and (f) HyPR-HN Patient 04. Pre-ex Pre-exhausted T cells, Reg Regulatory T cells, TRM Tissue-resident memory T cells, Prolif Proliferative T cells, ExCD8 Exhausted CD8 T cells, ExCD4 Exhausted CD4 T cells, IFN Type I interferon-responsive T cells.
Recurrent HNSCCs after conventionally fractionated radiotherapy are relatively CD8 T cell-depleted
To determine if the post-radiation TIL depletion observed in hypofractionated radiotherapy may also play a role in immunologic radioresistance and HNSCC recurrence after conventionally fractionated standard of care radiation, we compared immune infiltration in both stromal and cancer compartments between previously untreated (n = 51 patients) and radiation recurrent (n = 16 patients) HNSCCs enrolled on a prospective tumor registry. All patients had HPV-negative HNSCC involving a mucosal subsite of the upper aerodigestive tract (Supplementary Table 4). Through mIF analysis, we observed that previously untreated HNSCCs demonstrated a wide range of T cell infiltration from heavily infiltrated to immune-cold, and overall, pre-treatment TIL infiltrate had no clear association with disease control. However, in-field radiation recurrent tumors were comparatively T cell-depleted (Fig. 9a) with the most significant effects on CD8 TIL populations, including both stromal and cancer compartments (Fig. 9b). To assess for potential confounders, we performed a multivariable regression to identify clinical variables associated with CD8 TIL density. This analysis revealed that only radiation recurrence significantly impacted TIL infiltration (Supplementary Table 4). For a subset of patients (n = 20 previously untreated, n = 11 radiation recurrent), we also performed concurrent immunophenotyping of fresh preparations from the same biopsy specimens using mFC. These orthogonal analyses confirmed T cell depletion in recurrent HNSCCs with the greatest effect on CD8 T cells, both as a percentage of total viable cells and of the intra-tumoral immune fraction (Supplementary Fig. 14a–d).
a 67 samples from 61 HNSCC patients included in a prospective tumor registry underwent multiplex immunofluorescence (mIF) demonstrating lower CD8 and CD4 T cell populations in radiation-recurrent compared to previously untreated disease. Six patients are represented twice due to analysis of matched tumor samples obtained both pre-treatment and at the time of radiation recurrence from the same patients in the same anatomic location. Therefore, each sample represents an independent biological replicate. b The finding of decreased T cells in radio-recurrent samples was consistent across both stromal and cancer compartments. c Analysis of patient-matched biopsies obtained pre-treatment and at the time of in-field radiation recurrence confirms relative CD8 TIL depletion of radiation recurrent tumors compared to matched pre-treatment levels, d an effect not seen other immune cell subsets. Representative PET/CT, mIF, and multiplex flow cytometry (mFC) images are shown for (e) a patient-matched previously untreated and (f) radiorecurrent HNSCC. g mFC with a T cell exhastion panel was also performed from adjacent tissue fragements in 32 of the samples, which also underwent mIF. For these patients, exhausted T cell populations were particularly diminished in radiorecurrent disease as measued by mFC. Gate frequencies are provided as percentage of live cells. Antibody fluorescent conjugates include CD8: light blue, CD4: green, CD20: red, Foxp3: yellow, CD68: orange, Pan-CK: magenta, DAPI: dark blue. Scale bar = 50 µM. T-tests with Holm-Sidak’s multiple comparison correction were used to analyze differences between groups. Flow cytometry gating strategy demonstrated in Supplementary Fig. 4.
We next asked whether the relatively low CD8 T cell infiltrate seen in recurrent tumors was a pre-treatment feature of resistant HNSCCs or, instead, whether dynamic changes in T cell populations during therapy, as seen in hypofractionated radiation, may contribute to cancer recurrence. To investigate this, we analyzed a subset of patients for whom we were able to obtain HNSCC biopsies pre-treatment and then at the time of local recurrence after conventionally fractionated radiotherapy, from the same anatomical location in the same patients (n = 6 patients). T cell infiltration was assessed through both mIF and mFC of freshly digested HNSCC surgical biopsies. In each case, we found reduced CD8 T cell infiltration in radiation-recurrent HNSCCs, despite high pre-treatment CD8 TIL populations in several cases (Fig. 9c–f).
ICRhigh CD8 TILs in radiation recurrent HNSCC are markedly reduced
Finally, to evaluate whether limited repopulation of tumor-reactive TILs may contribute to cancer recurrence, we tested whether the relative T cell depletion observed in radiation recurrent HNSCCs also affected ICRhigh CD8 TILs. To address this, we measured PD-1 and TIM3 expression among HNSCC TILs by mFC from freshly digested surgical biopsies. We observed that viable ICRhigh CD8 TILs were profoundly depleted in recurrent HNSCCs. We confirmed this effect in both unmatched cohorts comparing previously untreated and radiorecurrent disease (Fig. 9g) as well as in matched biopsies obtained pre-treatment and at the time of in-field radiation recurrence from the same HNSCC patient in the same anatomical location of the primary tumor (Supplementary Fig. 14e, f). Together, these data suggest that radiation-induced TIL depletion and loss of pre-treatment tumor antigen-specific clonotypes with limited repopulation by new tumor-reactive TILs may be a barrier to cancer control for both hypofractionated and conventionally fractionated radiation.
Discussion
Through paired single-cell RNA and TCR sequencing analyses on serial biopsies at multiple timepoints across the treatment period for individual patients, this study provides a unique window into the real-time effect of radiation on the T cell landscape within HNSCCs. Remarkably, we find that radiotherapy leads to profound T cell depletion in every patient studied immediately upon completion of treatment. Importantly, T cell clonotypes with transcriptional features highly suggestive of tumor-reactivity and neo-antigen specificity are persistently lost from the tumor microenvironment and do not return post-radiation. Instead, T cell re-population of the tumor is primarily driven by regulatory, naïve, and high-frequency circulating effector-memory clones, which are unlikely to exert beneficial immuno-oncologic effects.
These findings help to clarify the transcriptional and functional states of TILs in HNSCC with direct implications for treatment. It is clear that TILs in HNSCC are a heterogeneous group, and overall TIL infiltrate has proven an inconsistent predictor of prognosis43,44,45,46,47,48. Similarly, while we show that radiorecurrent disease is associated with low TIL levels, pre-treatment TILs range widely in HNSCC and have no clear association with disease control. Yet despite the limited predictive power of overall TIL infiltrate, the objective clinical success of ICI therapy in metastatic HNSCC has established that immunogenic and oncologically relevant TILs exist and can be leveraged for therapy11. Further, recent data have demonstrated and visualized objective killing of patient-derived tumor cells by individual patient-matched TILs in HNSCC49. It appears, however, that such TILs are rare within clinical samples, and we show that the majority of intra-tumoral T cells are either naïve, regulatory, or high-frequency non-tumor-specific clones prevalent in the circulation. Taken together, this evidence suggests that a small subset of HNSCC TILs are tumor antigen-specific, carrying T cell receptors that can recognize cancer as a foreign entity and induce a relevant immunologic response. Although directly and efficiently characterizing these tumor antigen-specific TILs has remained elusive, such efforts could facilitate the development of targeted therapies, including TCR engineering.
Our present study provides deeper insight into this tumor antigen-specific TIL population in HNSCC, and we develop a rational gene expression signature to identify these TILs across patients and metastatic sites. We find that a subset of TILs exist across HNSCCs as expanded clonotypes found in both proliferating and exhausted sub-clusters. The existence of such a TIL population, which has also been identified in other malignancies50,51, suggests that a subset of ICRhigh TILs is not terminally exhausted but instead dynamically fluctuates between activation-induced proliferation and temporary anergy. We then use overlapping transcriptional programs of these clonotypes shared between proliferative and ICRhigh subsets to develop a predictive transcriptional signature associated with tumor-reactivity, including multi-dimensional supporting evidence of tumor antigen-specificity. The TProlif_Tox signature is found in proliferative and ICRhigh sub-clusters in every pre-treatment HNSCC patient we have analyzed, including internal and external datasets, but it is completely absent from normal oral mucosa. It is found across both primary and metastatic sites, including identical clonotype overlap, suggesting shared antigen specificity at both primary tumor and regional nodes. The TProlif_Tox signature can be found in other malignancies and is consistent with in vitro-confirmed tumor antigen-specific TCRs. Finally, we further validate tumor antigen-specificity of TProlif_Tox clonotypes in HNSCC through patient tumor- and HLA-specific in silico binding prediction and in vitro testing.
Our TProlif_Tox approach is distinct from prior algorithms predicting tumor antigen specificity as it relies largely on a concerted expression of transcriptional programs, rather than requiring full expression of an exact geneset35,50,51,52,53,54,55,56,57,58,59,60,61,62. While our signature has overlap with prior prediction models including expression of CXCL1335,50,51,52,53,55,57,58,59,60,62, PDCD150,53,55,61,62, IFNG50,54,55,58, MKI6762, and ITGAE 50,62, approaches requiring full expression of precise genesets are limited in applicability across different tumors and disease sites given the low transcriptomic coverage, technical dropout, temporal-dependent transcription, and stochasticity inherent to single-cell sequencing data. For example, while TNFRSF9 is commonly used as an indicator of T cell activation50,62, and we find it highly correlated to the TProlif_Tox gene signature, its expression is temporally restricted and may only be captured in a short window after TCR ligation. Further, many algorithms developed to predict tumor antigen-specific TILs are based only on TCRs reacting to synthesized neo-antigens in vitro. While valuable, such studies of in vitro peptide presentation may include false positives or negatives due to differential peptide processing or HLA-loading between tumor and cell culture. Further, in vitro studies may miss key neo-antigens undergoing post-translational modification, RNA editing, or non-canonical expression63,64,65. Instead, we leverage clonotype overlap in clustering, transcriptional program expression biologically associated with tumor-reactivity, and changes in TIL states at multiple timepoints across the treatment period.
Importantly, we find that radiotherapy persistently ablates TProlif_Tox TILs, which may help to explain the failure of combination immunoradiotherapy to improve disease control outcomes in multiple recent large clinical trials in HNSCC5,20,21. These data are unique given our ability to perform serial single-cell RNA and TCR sequencing of tumor samples across timepoints, including both the last day of radiation and 6 weeks post-therapy, enabling us to track individual clonotypes across time within the same patients. While the response to ICI therapy is multi-factorial66, without a sufficient presence of tumor antigen-specific TILs, agents designed to activate or potentiate T cell function will be ineffective. These findings are surprising given the wealth of pre-clinical data emphasizing the immunogenicity of radiotherapy and supporting its concurrent use with ICI14,15,16,17. Such discordance between pre-clinical models and clinical findings demonstrates the importance of multi-omic investigation of on-trial longitudinal clinical biopsies.
Despite profound radiation-induced TIL depletion and loss of TProlif_Tox clonotypes, T cells do reinfiltrate the tumor microenvironment. However, this is driven by new regulatory and naïve TILs as well as common circulating clonotypes, which appear at similar frequencies in both pre-treatment and post-therapy blood samples. Since the most common circulating clonotypes are likely viral-epitope specific from prior infectious agent exposures, these are unlikely to induce an anti-tumor immune response as they reinfiltrate post-radiation. Similarly, regulatory T cells will suppress post-radiation anti-tumor immunity, while new naïve T cells, in the absence of tumor antigen specificity and intra-tumoral priming, are unlikely to mount an effective anti-tumor response. However, these findings demonstrate that a TIL-depleted tumor in the immediate post-radiation setting, temporarily devoid of regulatory T cells, could be receptive to prevalent circulating clonotypes. If a tumor antigen-specific T cell therapy product can be delivered at this time point, or if new tumor antigen-specific T cells can be primed and released systemically, existing chemotactic gradients are likely to lead to T cell infiltration from the circulation. Recent data in other malignancies supports this possibility, suggesting that tumor antigen-specificity may be key to the efficacy and infiltration of adoptive cell therapies67. Further, given the mutational and neo-antigenic overlap identified between pre-treatment and post-radiation tumors, a TCR-engineered T cell product designed based on pre-treatment TProlif_Tox clonotypes may be efficacious in the post-radiation setting.
While these data provide a valuable window into the real-time effects of radiation on the tumor microenvironment with implications for current and future therapeutics, there are several limitations. Due to the small study sample size and available biopsy tissue, the generalizability of our conclusions remains constrained. While we provide extensive supporting data that TProlif_Tox is tumor antigen-specific, including in vitro testing, this will require further confirmation in larger datasets. Additionally, the prognostic significance of the baseline clonal diversity of TProlif_Tox remains uncertain, and it is unclear whether these TILs can be directly reinvigorated for in vivo expansion or if TCR engineering will be required to effectively leverage their tumor antigen-specificity. Further, while post-radiation T cell reinfiltration appears largely non-specific, we cannot exclude the possibility that non-exhausted T cell populations may also include newly infiltrating tumor antigen-specific T cells targeting different epitopes in the post-radiation environment. Identification and expansion of such populations may improve the efficacy of radiation in future studies.
Additionally, while the clinical radiation dosage delivered in this study is iso-equivalent to the standard of care, it was delivered using a hypofractionated regimen. Whether intra-tumoral T cell responses are fractionation-dependent remains uncertain and will be investigated in future cohorts from the HyPR-HN trial. Further, the radiation prescription included elective doses to regional nodes to treat micrometastatic disease. How this might affect TIL repopulation of tumors is unclear. There is increased interest in neo-adjuvant radiotherapy approaches that spare draining lymphatics, hypothesizing that radiotherapy will be immunogenic in this setting68,69. Omission of radiation to elective nodal regions may be a viable strategy to increase the priming of new tumor antigen-specific T cells and should be evaluated in future studies. Finally, while tumor antigen-specific TILs must be at the foundation of any T cell-based immuno-oncologic therapy, there are numerous other factors that influence clinical cancer outcomes, including immune editing, immune exclusion, nutrient deprivation, and other soluble and cellular mechanisms of intra-tumoral immunosuppression which must be addressed. In particular, we demonstrate that recurrent HNSCCs are commonly CD8 TIL-depleted. Whether delivery or activation of tumor antigen-specific TILs in the post-radiation setting can improve TIL infiltration, expansion, and anti-tumor functionality is not yet established.
Methods
HNSCC specimen acquisition: prospective institutional tumor registry
Patients who had HPV-negative HNSCC undergoing standard of care treatment of their tumor at the Medical College of Wisconsin (MCW) were eligible for inclusion. Patients have consented to additional research-specific biopsies, which were performed in the clinic setting or during a standard of care surgical procedure. The study size was planned as a fixed available sample of HNSCCs obtained over approximately a one-year period. Patients were consented consecutively. Biopsy samples were triaged for translational studies depending on specimen size. Samples were first preserved for subsequent multiplex immunofluorescence (mIF). In select cases with sufficient tissue, additional pieces were freshly digested for single-cell sequencing, immunophenotyping by multiplex flow cytometry (mFC), and tissue preservation for whole-exome and bulk RNA sequencing.
HNSCC specimen acquisition: HyPR-HN Study
The clinical trial Hypofractionated Pre-operative Radiation for HNSCC (HyPR-HN, NCT05538533) is a single-institution Phase I study of pre-operative neo-adjuvant hypofractionated radiation in HPV-negative HNSCC. Eligible patients have advanced-stage disease (T3-4 and/or clinical node-positive) without radiographic extracapsular extension. Enrolled patients undergo biopsies pre-treatment and immediately after the last fraction of radiotherapy in the clinic setting. Approximately 6 weeks after completion of radiation, patients are taken for definitive resection of residual disease with an additional research-specific biopsy at the time of surgery. All enrolled patients included in this report were treated with 46 Gy in 10 fractions to PTVhigh regions (to encompass gross disease) and 35 Gy in 10 fractions to PTVlow regions (to encompass microscopic spread, including nodal regions at risk).
As above, biopsy samples were triaged for translational studies. However, for the HyPR-HN study, samples were prioritized first for single-cell sequencing. After single-cell suspensions were processed for sequencing, the remaining cells from the sample underwent mFC. In cases with additional sufficient tissue samples, separate pieces were preserved for mIF, bulk RNA sequencing, whole-exome sequencing, and spatial transcriptomics.
Multiplex immunofluorescence
Biopsy pieces planned for multiplex immunofluoresence (mIF) were first fixed in 10% neutral-buffered formalin for 24 h. The tissues were subsequently dehydrated, cleared in xylene, and embedded in paraffin wax. The paraffin-embedded tissue blocks were then sectioned at 5 µm and mounted on glass slides. Tumor-immune spatial heterogeneity was subsequently quantified through histologic analysis as previously described in ref. 49. Briefly, multiplex immunostaining was performed using an Opal Polaris 7-Color Automation IHC Kit (Akoya Biosciences, NEL871001KT), as recommended by the manufacturer. All multiplex immunofluorescence slides were scanned on an Akoya Vectra Polaris (RRID:SCR_025508) at 20X using MOTiF™ protocol. Whole slide images were then loaded into InForm image analysis software for automated cell type density quantification.
Biopsy processing for single-cell analyses
To facilitate analysis by single-cell sequencing and flow cytometry, the biopsy specimen was freshly digested according to the Miltenyi Tumor dissociation kit protocol (130-095-92) in a Miltenyi gentleMACS C tube using the Miltenyi gentleMACS tissue dissociator (RRID:SCR_020267). Cellular debris and dead cells were removed from the dissociated cell suspension when necessary, using Debris Removal Solution (Miltenyi, 130-090-101) and the Dead Cell Removal Kit (Miltenyi, 130-090-101), respectively.
Multiplex flow cytometry
At least 1 × 105 cells per sample were used for immunostaining with conjugated monoclonal antibodies. 7-AAD (BioLegend, 420404) was used to exclude dead cells. Single color tubes were employed to set up a compensation matrix, and a fluorescence minus one control tube was included to ensure specific staining. Antibody panels are listed in Supplementary Table 5. Cells were washed and suspended in 0.2 mL staining buffer (PBS with 2% FBS). Staining was done at 4 °C for 30 min. The samples were washed again and then acquired on a MACSQuant 10 Analyzer Flow Cytometer (Miltenyi RRID:SCR_020268). Data was analyzed using FlowJo software version 10.10 (BD Life Sciences). To verify gating and purity, all populations were routinely backgated. Cell-type-specific immunophenotyping was performed on live single-cell populations and gated using canonical surface marker expression.
Whole exome sequencing
DNA was extracted and exome libraries prepared. All tumor WES was complimented by patient-matched germline WES obtained from peripheral blood mononuclear cells (PBMC) to validate mutational calls. Library preparation and sequencing was completed using the Illumina DNA prep with enrichment that utilizes on-bead transposase activity and panel capture by hybridization. Processing followed the Illumina prep protocol, and WES was completed on the NovaSeq6000 (RRID:SCR_016387), obtaining 150 bp paired-end reads and aiming for >100x depth of coverage across all samples (Novogene)70. DNA reads were cleaned using TrimGalore. Illumina short reads were mapped to the current reference genome, GRCh38, using BWA-mem71. Somatic mutations, including single-nucleotide variants and indels, were evaluated by Mutect2, SomaticSniper, and Varscan2 with comparison to patient-matched germline WES obtained from isolated peripheral blood mononuclear cells72,73,74.
Bulk RNA sequencing
RNA was extracted, quantified, and integrity assessed using RIN values from Agilent Fragment Analyzer 5200 (RRID:SCR_019417). RNA libraries were prepared according to manufacturer’s protocols utilizing Illumina’s TruSeq stranded mRNA library kit before sequencing on the Illumina NovaSeq6000 (RRID:SCR_016387) with paired-end 100 base pair reads targeting >100 million reads per sample (Novogene).
Single-cell RNA and TCR sequencing
Biopsy specimens designated for immediate single-cell sequencing underwent tumor digestion, as above. After dissociation, single cells were resuspended, counted, and viability assessed. Library preparation was then completed at the MCW Mellowes Center (RRID:SCR_022926) with 10x Genomics Chromium Next GEM Single Cell 5’ Reagent Kits (10x Genomics, sample dual indexes with cellular and molecular barcodes with VDJ amplification). Target capture of 10,000 cells per sample was used. Prior to sequencing, libraries were quantified and pooled by qPCR (Kapa Library Quantification Kit, Kapa Biosystems). Sequencing was completed on the NovaSeq6000 (RRID:SCR_016387), targeting 5000 reads for VDJ libraries and 50,000 transcript reads per cell per condition for expression. For all patients included in this study, apart from HyPR-HN Patient 01, single-cell RNA and TCR sequencing were obtained simultaneously from the same samples to enable T cell clonotype tracking across transcriptomic profiles. Samples from HyPR-HN Patient 01 underwent single-cell RNA sequencing only.
Single-cell data pre-processing
CellRanger (v8.0.1) was used to demultiplex raw reads, align to GRCh38, and quantify unique molecular identifiers (UMI). Sequencing quality was assessed based on the number of genes detected per cell and the proportion of mitochondrial genes expressed. Cells with <200 features or >5000 features were filtered out to exclude low-quality cells or doublets, respectively. Cells were also filtered out when the proportion of mitochondrial genes exceeded 15% to exclude dead or dying cells (Supplementary Fig. 15). Finally, T cell receptor variable genes (TRAV, TRBV, TRDV, TRGV) were excluded from the analysis to avoid clonotype bias in clustering. We did not perform regression of cell cycle or type I interferon genes, as has been previously described58, to avoid loss of relevant biological information in the context of radiation response.
Single-cell data integration and clustering
Seurat (5.1.0) was used to normalize the raw count data, identify highly variable features, scale features, and integrate samples75. Canonical Correlation Analysis (CCA) was used to mitigate batch effects between samples. The filtered log-transformed UMI matrix was used to perform truncated singular value decomposition with k = 50. Principal component analysis was performed based on the 4000 most variable features identified using the vst method implemented in Seurat. Cell types were then annotated based on expression of known marker genes visualized on the Uniform Manifold Approximation and Projection (UMAP) plot and by performing unbiased gene marker analysis. For the latter, a Wilcoxon Rank-Sum Test was used to perform differential gene expression (DEG) by comparing cells in each cluster to the rest of the cell profiles. Genes with FDR < 0.01 and log-fold change ≥ 1 were selected as candidate cell-type markers. Sub-cluster DEGs and canonical cell-type annotation markers were used to define cell subsets as described previously31,76,77. To evaluate changes in cell-type composition within single-cell sequencing profiles across treatment, the number of cells within each immunophenotypic subset was extracted. Cell subset numbers were normalized to total number of cells sequenced within each sample to enable proportional comparison across samples and timepoints.
To further analyze T cell subsets, T cell-containing clusters identified through DEGs and known marker analyses were extracted, and cells within this set expressing at least one CD3 complex subunit (CD3D | CD3E | CD3G | CD247) were retained. These cells were re-clustered and T cell subset immunophenotyping was performed through canonical marker expression and unbiased gene marker analysis, as above78,79.
Single-T cell clonotype tracking
scRepertoire (v2.0.3) was used to link T cell clonotype with single-cell gene expression profiling80. Barcode-linked TCR data were extracted from the filtered contig annotation output derived from the CellRanger pipeline. Individual TCRα and TCRβ chains were paired into full clonotypes through barcode matching. T cell clonotypes were then projected onto the T cell UMAP profiles again through barcode linkage. Within this framework, T cell clonotype tracking was then performed within individual patients between T cell sub-clusters, across primary and metastatic tumor sites, and across treatment timepoints.
Differential expression tests for development of the TProlif_Tox signature
To identify transcriptional programs enriched in T cells that demonstrate clonotype overlap between classical terminally exhausted and proliferating T cell sub-clusters, DEG analyses were performed on pre-treatment samples from the HyPR-HN trial. Cells present in exhausted or proliferative T cell sub-clusters with TCRs overlapping across both sub-clusters were identified. Transcriptional profiles of this cell subset were compared to all other T cells within the pre-treatment HyPR-HN samples and to post-treatment proliferative T cells using DEG analysis through a Wilcoxon Rank-Sum Test. DEG results were confirmed orthogonally with Model-based Analysis of Single-cell Transcriptomics (MAST)81. Of the top differentially expressed genes, we selected transcriptional programs rationally associated with T cell-mediated tumor-specific reactivity. These included non-regulatory (FOXP3-negative), CD8 markers (CD8A, CD8B), proliferative genes (MKI67, STMN1, TYMS), immune checkpoint molecules (PDCD1, HAVCR2, LAG3, TIGIT, ENTPD1), indicators of cytotoxicity (GZMA, GZMB, GZMH, IFNG), a marker of tissue-residence (ITGAE), and CXCL13. Due to the limitations in sequencing depth of single-cell approaches, which can lead to stochastic technical dropout on a cell-by-cell basis, we set the requirement that at least one gene from each program exhibit scaled expression greater than 1 within any given cell. For ITGAE and CXCL13, which were represented by single genes, we only required a scaled expression greater than 0.
Predicted TCR binding to tumor neo-antigens
To evaluate whether TProlif_Tox clonotypes within each patient may target tumor neo-antigens, we evaluated in silico prediction of patient-specific TCR:neo-peptide-HLA (TCR:pHLA) binding pairs involving mutated neo-peptides. To accomplish this, we first performed neo-antigen prediction based on mutations identified through WES data, as described above. Bam files obtained from WES from tumor and matched germline, bam files obtained from bulk RNA sequencing of tumor, and variant call files (VCF) files obtained from all three variant callers were imported into custom R software with use of the following libraries: VariantAnnotation1.48.1, stringr1.5.1, AnnotationHub3.12.0, ensembldb2.26.0, Rsamtools2.20.0, GenomicAlignments1.40.0. Coding sequence (CDS) transcripts annotated in Ensemble Genome Browser 111 that overlap regions of somatic mutations called by any of the three variant callers were extracted from tumor RNA bam files. Germline RNA transcripts (including up to a 36-nucleotide flank) overlapping the somatic mutation locus were extracted from the tumor bulk RNA sequencing (which come from normal cells within the tumor and/or unmutated tumor haplotypes), with the two most common transcripts expressing the germline sequence at the locus representing the expressed germline alleles. Tumor RNA transcripts containing the known mutation and up to 36 nucleotides on each end of the somatic mutation were extracted to determine up to 11 adjacent amino acids to each end of the mutation. The most common RNA transcript in the tumor that identically matched one of the germline alleles except at the site of the mutation, was used as the ground truth tumor mutation. Any mutation identified in WES that did not exist in tumor bulk RNA sequencing was discarded. Using the known CDS site from EnsemblDB to obtain the reference coding frame, both germline and tumor transcripts were translated using VariantAnnotation. Custom R software was then used to determine 8, 9, 10, and 11-mer peptides around all mutated amino acids from these translated sequences to select all neo-peptides derived from the translated tumor transcript that were not present in the translated germline transcript. Neo-peptides were designed to slide across the mutation to form all possible combinations. From this set of neo-peptides, we then normalized mutated transcript expression to total reads to adjust for depth of sequencing. This scaled expression value was then used to prioritize neo-peptides for import into netMHCpan4.1 to assess for HLA binding to any of the 6 major type I HLA types for that patient37. Neo-peptides predicted to be strong binders to any HLA (predicted binding affinity IC50 < 50 nM) were carried forward for testing against patient-matched TCRs for in silico TCR:pHLA binding prediction using TCRdock36.
For each confirmed TCR and pHLA complex, a pdb file was generated for AlphaFold using TCRdock and then imported into the TCRdock-specific version of AlphaFold. Docking geometries were determined with AlphaFold TCR pipeline simulations using sequence homology, as described previously36. Fifty naïve TCRs from patient-matched blood identified through single-cell RNA sequencing data were used to control for variations attributable to diverse pHLA complexes. We then calculated the residue-residue TCR:pHLA predicted aligned error (PAE) for TProlif_Tox TCRs. Low residue-residue PAE scores indicate close TCR:pHLA geometric approximation, suggesting favorable molecular interactions and high binding affinity. We corrected for TCR-specific variability by subtracting the mean binding of naïve TCRs to each pHLA complex from the PAE of each TProlif_Tox TCR:pHLA complex for every tested pHLA. We then zero-shifted and normalized these TCR-corrected binding scores for a given TCR by subtracting the mean binding score of each pHLA complex to a specific TCR and dividing by the standard deviation (SD) of each TCR. pHLA complexes with predicted binding to TProlif_Tox TCRs demonstrating extremely low residue-residue PAE scores ( > 3 SD below mean) were selected for further interrogation. For each patient, binding of these pHLA complexes was tested against both TProlif_Tox TCRs and an additional set of 25 separate patient-matched intra-tumoral naïve TCRs.
Spatial transcriptomics
Slides were processed through the 10x Genomics gene expression workflow, with pre-sequencing H&E imaging completed on the Keyence Microscope. Next, transcripts were de-crosslinked, hybridized to a probe panel, ligation completed, and probes released for capture on the gridded, barcoded surface of the Visium slide (CytAssist, 10x Genomics, RRID:SCR_024570). After barcoding, the samples underwent library preparation, amplification, and clean-up with quality assessment, ensuring that fragments were ~240 bp. Libraries were sequenced at the Mellowes Center Facility on the Illumina NovaSeq6000 with paired-end reads per the 10x Genomics protocol, and generating 25 million read pairs per spot covered by tissue. The quality control of raw reads was processed by FastQC. Raw reads were mapped to the human GRCh38 reference genome with Visium Probe_Set_v2.0 by Space Ranger. Fiducial detection, barcode/UMI counting, zero-count spot filtering, and normalization were also performed by Space Ranger. Visualization was performed with 10x Loupe Browser. Co-expression of canonical markers (CD3E, CD8A) was used to identify transcriptomic spots containing CD8 T cells with subsequent evaluation for immune checkpoint receptor (ICR) co-expression (PDCD1, HAVCR2, LAG3, TIGIT). The spatial distribution of these co-expression networks was evaluated before and after radiation treatment for HyPR-HN Patient 02.
Generation of patient-derived malignant HNSCC lines
Cell lines were directly derived from fresh patient HNSCC biopsies. Tumor specimens were minced and cultured such that a broad surface of tumor was directly adherent to the culture plate. Culture media included DMEM/F12 with 1.2 g/L sodium bicarbonate and 2.5mM L-glutamine (Life Technologies, 11320033), supplemented with 400 ng/mL hydrocortisone (Sigma Aldrich, H0135), 10 µg/mL insulin (Sigma Aldrich 16634), 5 ng/mL epidermal growth factor (Millipore Sigma, 01-107), and 10% FBS (Life Technologies, A5256701). 2% antibiotic-antimycotic (Gibco, 15240062) was used to limit contamination. Bulk tumor pieces were removed from the plate once adherent cells were established. All cell lines were cultured in an incubator at 37 °C with 5% CO2. Malignant populations were separated from cancer-associated fibroblasts (CAF) through serial differential trypsinization, given the absence of fully reliable surface markers for malignant cells in HNSCC. Mixed cell lines were incubated for approximately 2 min in the presence of TrypLE (Gibco, 12604021), which preferentially harvests the less adherent fibroblastic population. Since malignant HNSCC cells can lose epithelial surface markers and express fibroblastic and mesenchymal signatures82,83, serial differential trypsinization remains the most reliable method to enrich for malignant lines while maximally preserving tumor clonal heterogeneity.
TIL isolation
TILs were extracted from separate pieces of the same biopsy specimens used to generate malignant lines. For TIL generation, fresh HNSCC specimens were finely minced (1 mm sections) and cultured. TIL culture media included ImmunoCult XF Expansion media (StemCell Technologies, 10981), 20 ng/mL IL7 (Peprotech, 200-07-100UG) and 20 ng/mL IL15 (Peprotech, 200-15-100UG), and 2% antibiotic-antimycotic (Gibco, 15240062). Cultures were inoculated on Day 0 with soluble CD3/CD28 activator (StemCell Technologies, 10991). Once expanding T cell colonies were established, T cells were enriched with CD3 magnetic column selection (Miltenyi, 130-097-043). Once fully expanded, TILs were then cryopreserved (CryoStor, StemCell, 07930) until patient-matched malignant lines had matured.
In vitro evaluation of the tumor-reactive potential of TProlif_Tox
To assess the tumor-reactive potential of TProlif_Tox clonotypes, we utilized a patient-derived malignant cell line and autologous TILs expanded from the P120 primary tumor biopsy. Malignant cell and TIL isolation procedures are described above. Since expanded TILs retain a persistently positive CD137 fraction (~5%) following non-specific CD3/CD28 stimulation, CD137-negative TILs were first flow sorted (BD FACSAria III) prior to co-culture. The resulting CD137-negative TIL population was then co-cultured with patient-matched malignant cells for 24 h at approximately 2:1 TIL-to-tumor ratio, without exogenous cytokines. Following co-culture, TILs were stained with CD137 antibody (Miltenyi) and flow-sorted into CD137-positive and CD137-negative populations. Both fractions underwent single-cell RNA and TCR sequencing using the 10x Genomics platform. The resulting sequencing datasets were processed using Cell Ranger, followed by integration and dimensionality reduction in Seurat, as described above. To ensure tumor-specificity and exclude TILs with weak or non-specific TCR interactions, we identified CD137-positive clonotypes that also exhibited high TNFRSF9 expression across multiple clones. These clonotypes were then projected back onto the initial P120 primary tumor single-cell dataset to assess their in vivo transcriptional states and tumor-reactive potential.
Circulating T cell isolation
For HNSCC patients who underwent peripheral blood sampling concurrent with biopsy, ~45 mL of blood was taken. Ficoll-gradient separation was used to isolate the buffy coat, which was aspirated. For flow cytometry, circulating T cells were immediately processed fresh. The remaining T cells were then cryopreserved (Cryostor) for later analyses.
xCELLigence
Autologous adherent tumor cells derived from a HNSCC biopsy were plated in a 96-well xCELLigence E-Plate (300601010) at 10,000 cells/well. The plate was incubated at room temperature for 30 min to facilitate uniform immobilization of the target cells on plate bottom and then placed in the xCELLigence instrument. Data acquisition was initiated, and the plate was incubated at 37 °C. After 24 h, TILs were plated in triplicate at several effector-to-target ratios. Appropriate negative (TIL or tumor cells only) and positive (addition of Agilent cytolysis reagent) controls were performed. After addition of the effector cells, the plate was incubated at room temperature for 30 min to facilitate uniform distribution of the effectors. The plate was placed back onto the xCELLigence instrument and incubated for 24 h at 37 °C with continuous data acquisition. Data were analyzed using RTCA Software Pro v2.3.2 and Graphpad Prism v10.0.3.
Statistics
Statistical analysis was performed using GraphPad Prism v10.0.3, R v4.3.3, and SPSS v28.0.0. For analysis of flow cytometry and immunofluorescence data multiple T-tests with Holm-Sidak’s multiple comparison correction were used to analyze differences between groups. Differential gene expression analyses between single-cell clusters were performed in Seurat using the Wilcoxon Rank-Sum Test and MAST, as described.
To evaluate the influence of clinical and tumor characteristics on CD8 T cell density (cells/mm²), a multivariable linear regression analysis was conducted. Independent variables included gender, smoking history, ACE27 comorbidity score, prior treatment history (de novo vs radiation recurrence), tumor site category, T stage, and N stage. To assess potential collinearity, variance inflation factors were examined. Model significance was determined using an F-test, while individual predictor significance was assessed via t-tests, with p < 0.05 considered statistically significant. Residual plots and collinearity diagnostics were reviewed to confirm the appropriateness of the model.
For evaluation of the strength of the predicted binding affinity between identified TProlif_Tox pHLA:TCR pairs, we first isolated pHLAs where a TProlif_Tox TCR exhibited high predicted binding affinity, defined as a PAE_SD score more than 3 standard deviations below the mean (<−3 SD). For each such pHLA complex, the candidate TCR was defined by the lowest PAE_SD score among the TProlif_Tox TCRs. To assess whether such an extreme binding score was an outlier, we employed a bootstrap resampling approach. Specifically, for each pHLA complex, we constructed a control group comprising 25 intra-tumoral naive-like TCRs together with all other TProlif_Tox TCRs for that complex (i.e., excluding the candidate extreme binder). Within each pHLA complex, we performed 10,000 bootstrap replicates by sampling with replacement from the control group and, for each replicate, recording the lowest PAE_SD scores. This bootstrap procedure generated an empirical null distribution of extreme binding scores. The 95% confidence interval for the minimum PAE_SD was calculated using the T distribution. The empirical p-value was then calculated using this same T distribution.
Study approval
Prospective tumor registry activities were approved by the MCW Institutional Review Board (approval PRO00040992) and the Institutional Biosafety Committee (approval IBC20210026). The HyPR-HN clinical trial (NCT05538533), including all tissue biopsies and handling, was approved by the MCW Institutional Review Board (approval PRO00044863). Specifically, the approved protocol allows for publication of translational data from initial dose levels, prior to full study completion and endpoint reporting. All recruited volunteers provided written informed consent to participate in biospecimen sampling and analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All bulk and single-cell sequencing datasets generated in this study have been deposited in the Gene Expression Omnibus database under the GSE280982, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE280982. Details of external datasets re-analyzed in this study are listed in Supplementary Table 6. Values for all data points in plots and gene expression profiling are reported in the Supporting Data Values file. Raw flow cytometry and immunohistochemistry files have been deposited in the Zenodo repository (10.5281/zenodo.15258024 and 10.5281/zenodo.15265351). All unique biological materials in this study are available upon request from academic researchers, provided appropriate data transfer agreements are established. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
Code availability
Custom R code used for analyses of these datasets has been deposited in the Zenodo repository (https://doi.org/10.5281/zenodo.15258024).
References
American Cancer Society. Cancer Statistics: Analysis Tool. Accessed September 26, 2024. https://cancerstatisticscenter.cancer.org.
Cunha, A. R. D. et al. The global, regional, and national burden of adult lip, oral, and pharyngeal cancer in 204 countries and territories: a systematic analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 9, 1401–1416 (2023).
Gillison, M. L. et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet 393, 40–50 (2019).
Ferris, R. L. et al. Phase II randomized trial of transoral surgery and low-dose intensity modulated radiation therapy in resectable p16+ locally advanced oropharynx cancer: an ECOG-ACRIN cancer research group trial (E3311). J. Clin. Oncol. 40, 138–149 (2022).
Lee, N. Y. et al. Avelumab plus standard-of-care chemoradiotherapy versus chemoradiotherapy alone in patients with locally advanced squamous cell carcinoma of the head and neck: a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol. 22, 450–462 (2021).
Rathod, S., Livergant, J., Klein, J., Witterick, I. & Ringash, J. A systematic review of quality of life in head and neck cancer treated with surgery with or without adjuvant treatment. Oral. Oncol. 51, 888–900 (2015).
Rosenthal, D. I. et al. Final report of a prospective randomized trial to evaluate the dose-response relationship for postoperative radiation therapy and pathologic risk groups in patients with head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 98, 1002–1011 (2017).
Ang, K. K. et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J. Clin. Oncol. 32, 2940–2950 (2014).
Leemans, C. R., Snijders, P. J. F. & Brakenhoff, R. H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 18, 269–282 (2018).
Ohtani, T. et al. Activated cytotoxic T-lymphocyte immunotherapy is effective for advanced oral and maxillofacial cancers. Int. J. Oncol. 45, 2051–2057 (2014).
Cohen, E. E. W. et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet 393, 156–167 (2019).
Burtness, B. et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 394, 1915–1928 (2019).
Cramer, J. D., Burtness, B. & Ferris, R. L. Immunotherapy for head and neck cancer: recent advances and future directions. Oral. Oncol. 99, 104460 (2019).
Morisada, M. et al. PD-1 blockade reverses adaptive immune resistance induced by high-dose hypofractionated but not low-dose daily fractionated radiation. Oncoimmunology 7, e1395996 (2018).
Sharabi, A. B. et al. Stereotactic radiation therapy augments antigen-specific PD-1-mediated antitumor immune responses via cross-presentation of tumor antigen. Cancer Immunol. Res. 3, 345–355 (2015).
Burnette, B. C. et al. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 71, 2488–2496 (2011).
Lim, J. Y., Gerber, S. A., Murphy, S. P. & Lord, E. M. Type I interferons induced by radiation therapy mediate recruitment and effector function of CD8(+) T cells. Cancer Immunol. Immunother. 63, 259–271 (2014).
Twyman-Saint Victor, C. et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520, 373–377 (2015).
Skinner, H. D. et al. Integrative analysis identifies a novel AXL-PI3 kinase-PD-L1 signaling axis associated with radiation resistance in head and neck cancer. Clin. Cancer Res. 23, 2713–2722 (2017).
Bourhis, J. et al. LBA35 Avelumab-cetuximab-radiotherapy versus standards of care in patients with locally advanced squamous cell carcinoma of head and neck (LA-SCCHN): randomized phase III GORTEC-REACH trial. Ann. Oncol. 32, S1310 (2021).
Bourhis, J. et al. LBA38 Pembrolizumab versus cetuximab, concomitant with radiotherapy (RT) in locally advanced head and neck squamous cell carcinoma (LA-HNSCC): results of the GORTEC 2015-01 “PembroRad” randomized trial. Ann. Oncol. 31, S1168 (2020).
Morisada, M. et al. Dose-dependent enhancement of T-lymphocyte priming and CTL lysis following ionizing radiation in an engineered model of oral cancer. Oral. Oncol. 71, 87–94 (2017).
Qian, J. M. & Schoenfeld, J. D. Radiotherapy and immunotherapy for head and neck cancer: current evidence and challenges. Front. Oncol. 10, 608772 (2020).
Demaria, S. et al. Radiation dose and fraction in immunotherapy: one-size regimen does not fit all settings, so how does one choose?. J. Immunother. Cancer 9, e002038 (2021).
Runnels, J. et al. Combining radiotherapy and immunotherapy in head and neck cancer. Biomedicines 11, e2097 (2023).
Stevens, G. et al. Locally advanced melanoma: results of postoperative hypofractionated radiation therapy. Cancer 88, 88–94 (2000).
Kumar, A. M. S. et al. Postoperative hypofractionated stereotactic brain radiation (HSRT) for resected brain metastases: improved local control with higher BED. J. Neurooncol. 139, 449–454 (2018).
Murray Brunt, A. et al. Hypofractionated breast radiotherapy for 1 week versus 3 weeks (FAST-Forward): 5-year efficacy and late normal tissue effects results from a multicentre, non-inferiority, randomised, phase 3 trial. Lancet 395, 1613–1626 (2020).
Workel, H. H. et al. A transcriptionally distinct CXCL13+CD103+CD8+ T-cell population is associated with B-cell recruitment and neoantigen load in human cancer. Cancer Immunol. Res. 7, 784–796 (2019).
Veatch, J. R. et al. Endogenous CD4+ T cells recognize neoantigens in lung cancer patients, including recurrent oncogenic KRAS and ERBB2 (Her2) driver mutations. Cancer Immunol. Res. 7, 910–922 (2019).
Kürten, C. H. L. et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 12, 7338 (2021).
Williams, D. W. et al. Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 184, 4090–4104.e15 (2021).
Liu, T. et al. Single cell profiling of primary and paired metastatic lymph node tumors in breast cancer patients. Nat. Commun. 13, 6823 (2022).
Krishna, C. et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell 39, 662–677.e6 (2021).
Meng, Z. et al. Transcriptome-based identification of tumor-reactive and bystander CD8. Sci. Transl. Med. 15, eadh9562 (2023).
Bradley, P. Structure-based prediction of T cell receptor:peptide-MHC interactions. Elife 12, e82813 (2023).
Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 48, W449–W454 (2020).
Wooldridge, L. et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 287, 1168–1177 (2012).
Cohen, C. J. et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Investig. 125, 3981–3991 (2015).
Gros, A. et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22, 433–438 (2016).
Cafri, G. et al. Memory T cells targeting oncogenic mutations detected in peripheral blood of epithelial cancer patients. Nat. Commun. 10, 449 (2019).
Gros, A. et al. Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. J. Clin. Investig. 129, 4992–5004 (2019).
Spector, M. E. et al. Prognostic value of tumor-infiltrating lymphocytes in head and neck squamous cell carcinoma. JAMA Otolaryngol. Head. Neck Surg. 145, 1012–1019 (2019).
Rodrigo, J. P. et al. Tumor-infiltrating lymphocytes in the tumor microenvironment of laryngeal squamous cell carcinoma: systematic review and meta-analysis. Biomedicines 9, e0486 (2021).
Borsetto, D. et al. Prognostic significance of CD4+ and CD8+ tumor-infiltrating lymphocytes in head and neck squamous cell carcinoma: a meta-analysis. Cancers13, e0781 (2021).
Soopanit T., Laokulrath N., Chayopasakul V., Pongsapich W. Prognostic value and clinicopathological status of PD-L1 expression and CD8+ TILs in oral squamous cell cancer patients with or without traditional risk factors. Head Neck https://doi.org/10.1002/hed.27330 (2023).
de Ruiter, E. J. et al. Digital pathology-aided assessment of tumor-infiltrating T lymphocytes in advanced stage, HPV-negative head and neck tumors. Cancer Immunol. Immunother. 69, 581–591 (2020).
Koukourakis, I. M. et al. Prognostic and predictive relevance of tumor-infiltrating lymphocytes in squamous cell head-neck cancer patients treated with radical radiotherapy/chemo-radiotherapy. Curr. Oncol. 29, 4274–4284 (2022).
Zenga, J. et al. Tumor-specific T cells in head and neck cancer have rescuable functionality and can be identified through single-cell co-culture. Transl. Oncol. 42, 101899 (2024).
Li, H. et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 176, 775–789.e18 (2019).
He, J. et al. Defined tumor antigen-specific T cells potentiate personalized TCR-T cell therapy and prediction of immunotherapy response. Cell Res. 32, 530–542 (2022).
Tan, C. L. et al. Prediction of tumor-reactive T cell receptors from scRNA-seq data for personalized T cell therapy. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02161-y (2024).
Oliveira, G. et al. Phenotype, specificity and avidity of antitumour CD8. Nature 596, 119–125 (2021).
Zhang, C. et al. Single-cell sequencing of tumour infiltrating T cells efficiently identifies tumour-specific T cell receptors based on the T cell activation score. Cancer Immunol. Immunother. 73, 123 (2024).
Lowery, F. J. et al. Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science 375, 877–884 (2022).
Pétremand, R. et al. Identification of clinically relevant T cell receptors for personalized T cell therapy using combinatorial algorithms. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02232-0 (2024).
Veatch, J. R. et al. Neoantigen-specific CD4+ T cells in human melanoma have diverse differentiation states and correlate with CD8+ T cell, macrophage, and B cell function. Cancer Cell 40, 393–409.e9 (2022).
Caushi, J. X. et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 596, 126–132 (2021).
Zheng, C. et al. Transcriptomic profiles of neoantigen-reactive T cells in human gastrointestinal cancers. Cancer Cell 40, 410–423.e7 (2022).
Hanada, K. I. et al. A phenotypic signature that identifies neoantigen-reactive T cells in fresh human lung cancers. Cancer Cell 40, 479–493.e6 (2022).
Chatani, P. D. et al. Cell surface marker-based capture of neoantigen-reactive CD8. J. Immunother. Cancer 11, e006264 (2023).
van der Leun, A. M., Thommen, D. S. & Schumacher, T. N. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer 20, 218–232 (2020).
Li, J. et al. The screening, identification, design and clinical application of tumor-specific neoantigens for TCR-T cells. Mol. Cancer 22, 141 (2023).
Schwarz, S. et al. T cells of colorectal cancer patients stimulated by neoantigenic and cryptic peptides better recognize autologous tumor cells. J. Immunother. Cancer 10, e005651 (2022).
Yamaguchi, S. et al. TCR function analysis using a novel system reveals the multiple unconventional tumor-reactive T cells in human breast cancer-infiltrating lymphocytes. Eur. J. Immunol. 51, 2306–2316 (2021).
Chang, T. G. et al. LORIS robustly predicts patient outcomes with immune checkpoint blockade therapy using common clinical, pathologic and genomic features. Nat. Cancer https://doi.org/10.1038/s43018-024-00772-7 (2024).
Chiffelle, J. et al. Tumor-reactive T cell clonotype dynamics underlying clinical response to TIL therapy in melanoma. Immunity https://doi.org/10.1016/j.immuni.2024.08.014 (2024).
Darragh, L. B. et al. A phase I/Ib trial and biological correlate analysis of neoadjuvant SBRT with single-dose durvalumab in HPV-unrelated locally advanced HNSCC. Nat. Cancer 3, 1300–1317 (2022).
Saddawi-Konefka, R. et al. Lymphatic-preserving treatment sequencing with immune checkpoint inhibition unleashes cDC1-dependent antitumor immunity in HNSCC. Nat. Commun. 13, 4298 (2022).
Kong, S. W., Lee, I. H., Liu, X., Hirschhorn, J. N. & Mandl, K. D. Measuring coverage and accuracy of whole-exome sequencing in clinical context. Genet. Med. 20, 1617–1626 (2018).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Quah, H. S. et al. Single cell analysis in head and neck cancer reveals potential immune evasion mechanisms during early metastasis. Nat. Commun. 14, 1680 (2023).
Osumi-Sutherland, D. et al. Cell type ontologies of the human cell atlas. Nat. Cell Biol. 23, 1129–1135 (2021).
Guo, X. et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018).
Andreatta, M. et al. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 12, 2965 (2021).
Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47 (2020).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Puram, S. V., Parikh, A. S. & Tirosh, I. Single cell RNA-seq highlights a role for a partial EMT in head and neck cancer. Mol. Cell Oncol. 5, e1448244 (2018).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck Cancer. Cell 171, 1611–1624.e24 (2017).
Acknowledgements
This work was supported by funding from the National Institutes of Health (NIH) through grant R21CA279935 (JZ) and R01DE034360 (J.Z. and T.K.). Funding was also provided by the Medical College of Wisconsin Department of Radiation Oncology (HAH) and in large part by the Medical College of Wisconsin Cancer Center (J.Z., M.J.A., and H.A.H.). This project was also funded through the Benjamin Garmer Family Gift Fund (J.Z.) and the Don and Sharyn Blatnik Endowment (J.Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders or NIH.
Author information
Authors and Affiliations
Contributions
Manuscript drafting was performed by J.Z., H.A.H., and M.J.A. All three authors were involved in study design, conducting experiments, analyzing data, providing reagents, and bioinformatic processing. The remaining authors contributed through study design, conducting experiments, analyzing data, interpretation of data analysis, manuscript editing, and final approval (A.F., J.F., R.K., J.K., B.M., J.B., K.A., M.S., S.J.W., A.J.M., J.W.A., B.H., A.P., V.X.J., A.A.M., O.V.E., F.C., M.S., T.K., and P.H.).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Özcan Met, N. Gopalakrishna Iyer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zenga, J., Awan, M.J., Frei, A. et al. Radiation therapy results in preferential tumor antigen-specific lymphodepletion in head and neck cancer. Nat Commun 16, 5660 (2025). https://doi.org/10.1038/s41467-025-60827-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60827-w