Abstract
Macrophages play vital roles in innate and adaptive immunity, and their functions are mediated via phagocytosis and antigen presentation. Despite the effort to identify phagocytic checkpoints and explore their mechanism of action, current checkpoint-scanning strategies cannot provide a complete and systematic list of such immune checkpoints. Here, we perform in vitro phagocytosis assays using primary healthy donor macrophages co-cultured with breast cancer cells followed by ribosome profiling of sorted macrophages, to identify immune system-specific checkpoints. We observe a downregulation of CD37 in phagocytic macrophages and demonstrate that targeting CD37 with a specific antibody promotes the phagocytosis of multiple cancer cells in vitro. Mechanistically, tumorous macrophage migration inhibitory factor (MIF) directly binds to CD37, promoting the phosphorylation of CD37Y13 and activating a transduction cascade that involves the recruitment of SHP1 and inhibition of AKT signaling, ultimately impairing phagocytosis. In vivo, targeting CD37 promotes tumor clearance in multiple preclinical mouse models and synergizes with anti-CD47 therapy. Thus, our study identifies a previously unidentified phagocytic checkpoint and provides new potential for precise therapy.
Similar content being viewed by others
Introduction
Immune checkpoint-based (ICB) therapy has shown unprecedented success in preventing cancer by boosting T cell responses. However, most patients do not benefit from ICB therapy1,2,3,4. Advances in single-cell sequencing have enabled an understanding of the complex interactions between different immune cell subsets and tumor cells in the tumor microenvironment (TME), which is crucial for combating resistance to ICB therapy5. Macrophages are crucial for innate and adaptive immunity6,7. The indispensable function of macrophages in adaptive immunity is to present antigens via phagocytosis, a multistep cellular process involving target cell recognition, cellular engulfment, and lysosomal digestion. The process of phagocytosis is regulated by receptor–ligand interactions between the phagocyte and target cells expressing the so-called “eat me” signals8,9. Intrinsic “eat me” signals may be activated by mediating ITAM and subsequently remodeling the cytoskeleton of phagocytic cells, which sequentially engulfs cancer cells10. Cancer cells evade immune eradication by expressing anti-phagocytosis molecules, including cluster of differentiation 47 (CD47)11, cluster of differentiation 24 (CD24)12, PD-L113, the beta-2 microglobulin subunit of the major histocompatibility class I complex14, stanniocalcin 115, and GD216, which are termed “don’t eat me” signals17. Phagocytosis-associated ICB therapy is mainly achieved by blocking “don’t eat me” signals, such as anti-CD47 and anti-PD-L1 signals.
In this study, we employ a systematic methodology, based on phagocytic assay combined with ribosome profiling, and identify CD37 as a phagocytic checkpoint. Our data elaborate the role of CD37 in suppressing macrophage phagocytosis. Furthermore, we show that inhibition of CD37 effectively promotes the phagocytosis of multiple tumor cells by macrophages and enhances the efficacy of combination therapy with CD47. Thus, our work identified CD47 as a target for phagocytosis checkpoint therapy and tumor immunotherapy.
Results
Identification of potential phagocytosis checkpoints
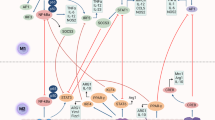
To further understand the potential regulatory mechanism of phagocytosis, we initiated an in vitro phagocytosis assay using the following methodology: peripheral blood from healthy donors was collected and peripheral blood monocytes were isolated. Macrophages were subsequently induced and cultured using ImmunoCult-SF Macrophage Medium. Macrophages were labeled with CellTrace Blue, MDA-MB-231 breast cancer cells were labeled with 5(6)-carboxyfluorescein diacetate N-succinimidyl ester (CFSE). Thereafter, the macrophages were incubated with MDA-MB-231 cells for 2 h and subjected to fluorescence-activated cell sorting (FACS) analysis. In addition to regular FACS, mild trypsinization was performed to avoid cell adhesion. Cells labeled with a blue cell tracker and CFSE were considered phagocytic macrophages (hereafter referred to as phagocytes). Cells stained with only the blue cell tracker were identified as macrophages without phagocytic ability. Equal numbers of phagocytes, macrophages, and MDA-MB-231 cells were harvested and subjected to ribosome profiling (Fig. 1A). Ribosome protection fragments (RPFs) were collected and subjected to transcription. The reads mapped to the transcription products showed a distinct 3-nt periodicity, indicating reliable quality control (Supplementary Fig. 1A). For further confirmation, we utilized the relative FPKM value to scrutinize the well-studied phagocytosis checkpoints. Phagocytosis-inhibiting molecules, such as CLEC-1, Siglec-10, SIRPα, LILRB1 and PD1, exhibited significant upregulation in macrophages compared with phagocytes (Supplementary Fig. 1C, D). The above data showed strong consistency with those of the current relevant studies and demonstrated reliable specificity and sensitivity. Based on identification of known immune checkpoints using our detection method, we employed individual analytical procedures in the subsequent studies. However, the phagocytes identified using FACS were actually two types of cells that clustered together—phagocyte “eating” cancer cells. To guarantee the reliability of determining phagocyte-associated checkpoints, we applied a strict analysis process. First, we identified phagocyte-specific genes by comparing phagocytes with cancer cells (FPKM = 0 in MDA-MB-231 cells). Next, we compared phagocyte-specific genes with those of control macrophages, and differentially expressed genes (DEGs) were identified (p < 0.05, fold change > 1). A total of 1087 DEGs were downregulated, whereas 486 were upregulated in phagocytes compared to those in macrophages (Fig. 1B, Supplementary Fig 1B, Supplementary Dataset 1 and 2). The dysregulated pathways were enriched and detailed; notably, the dysregulated pathways were mostly enriched in receptor activity and receptor-mediated downstream cascade activity, especially immune-related receptor activity (Fig. 1C, D). Thus, we applied GO annotation to identify the differentially expressed receptors and transmembrane proteins in phagocytes. A total of 1663 receptors and transmembrane proteins were identified (Fig. 1E, Supplementary Dataset 3). To avoid false positive results, we further narrowed the dataset (FPKM = 0 in MDA-MB-231 cells and FPKM > 0 in phagocytes) and identified 40 receptors and transmembrane proteins (Fig. 1F, Supplementary Dataset 4). CD37 is a previously unidentified potential checkpoint gene and has thus attracted great interest (FPKM = 0 in MDA-MB-231 cells, FPKM = 0.41 in phagocytes, and FPKM = 298.9 in macrophages).
A Illustration of the ribosome profiling in phagocytes, macrophages, and cancer cells. Peripheral blood monocytes were isolated from healthy donors and differentiated into macrophages. The macrophages were labeled with CellTracker Blue and co-cultured with CFSE-labeled cancer cells for 2 h. Subsequently, flow cytometry was employed to sort the macrophages, phagocytes, and cancer cells as indicated. Cells were then subjected to ribosome profiling analysis. A total of 100 healthy donors were enrolled. B The volcano plot illustrates the identification of dysregulated genes, with phagocyte-specific genes determined by comparing phagocytes to MDA-MB-231 cancer cells (FPKM = 0 in MDA-MB-231 cells). Dysregulated genes were subsequently identified by comparing these phagocyte-specific genes to those in control macrophages. Differentially expressed genes (DEGs) were defined as those meeting the criteria of p < 0.05 and fold change > 2. FC fold change, FDR false detecting rate. C GO analysis of the dysregulated genes, dysregulated genes were identified, and Gene Ontology Analysis was applied to analyze the enriched pathways of dysregulated genes. D The detail of the dysregulated genes enriched pathways, gene ontology analysis was applied to analyze the dys-enriched pathways, the detail of the pathway, including the q value and the number of dysregulated genes, was shown. E The rank of the dysregulated receptor and transmembrane protein, PD1, Siglec-10, and CD37, was indicated, X-axis, the rank of the genes, Y-axis, the log2 FC, FC fold change. F The rank of the dysregulated receptor and transmembrane protein (RPKM > 0 in phagocytes), X-axis, the rank of the genes, Y-axis, the log2 FC, FC fold change.
CD37 was widely expressed in macrophages
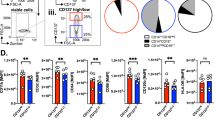
CD37 is specifically expressed on B cells18. Before further investigating the biological function and underlying mechanism of CD37 in phagocytosis, we first derived macrophages from the tumor species and detected the proportion of CD37+ macrophages (the data were normalized to CD14+CD86+ cells and CD14+MRC1+ cells). The proportion of CD37+ cells ranges from 0 to 80% and tend to be enriched in MRC1+ macrophages in multiple cancers, including breast cancer, colon cancer, rectal cancer, glioma, and PDAC (Fig. 2A, Supplementary Fig. 2A). For further confirmation, we analyzed single-cell sequence data from breast cancer (BRCA, GSE150660)19, glioblastoma (GBM, GSE163108)20, colon-rectum cancer (CRC, GSE139555)21, lung cancer (LC, GSE739555)21 and pancreatic ductal adenocarcinoma patients (PDAC, GSE148673)22 and analyzed the proportion of CD37+ macrophages. These results suggest that CD37 is widely expressed in macrophages from different cancers and is preferentially expressed in a certain cell subpopulation (Fig. 2B). A violin plot of CD37 and other critical phagocytosis checkpoints, PDCD1, SIRPα, and SLAMF7, was generated for the macrophages from the above datasets. As expected, CD37 was detected in tumor-derived macrophages (TDMs), and compared to other checkpoint molecules, CD37 had the highest expression level, suggesting its biological importance of CD37 (Fig. 2C). Next, we analyzed the distribution of CD37+ macrophages in the BRCA dataset. The results indicated that CD37 was expressed in almost all macrophages, specifically enriched in MRC1+ macrophages CD37 and barely expressed in CD86+ macrophages (Fig. 2D). MRC1 and CD86 are two classical markers of widely used antitumor/proto-tumor (AT/PT) macrophages23,24. Analysis of the biological information of CD37−/+ macrophages from the scRNA-seq dataset (GSE150660) was performed. In total, 1408 DEGs were identified (Supplementary Fig. 3A, Supplementary Dataset 5), and the top 20 DEGs are shown (Supplementary Fig. 3B). Dysregulated genes were enriched in phagocytosis-associated pathways such as phagosome, antigen processing, cell adhesion, and autoimmune disease pathways. These data further confirm the biological importance of CD37 in phagocytosis (Supplementary Fig. 3C–E).
A The proportion of CD37+ macrophages derived from different cancer species measured by FACS, BRCA breast cancer, GBM glioblastoma, CRC colon-rectum cancer, LC lung cancer, PDAC pancreatic ductal adenocarcinoma, n = 38 for each cancer, data were presented as individual dots. B The proportion of CD37+ macrophages derived from different single-cell sequence of different cancer species, BRCA breast cancer, GBM glioblastoma, CRC colon-rectum cancer, LC lung cancer, PDAC pancreatic ductal adenocarcinoma, n = 5 for each cancer, data were presented as box plot, the mean value, maximum and minimum values, and the quartile values were presented. C The violin plot of CD37 and three other checkpoints (PDCD1, SIRPA, and SLAMF7) in macrophages from different cancer species, BRCA breast cancer, GBM glioblastoma, CRC colon-rectum cancer, LC lung cancer, PDAC pancreatic ductal adenocarcinoma, n = 5 for each cancer, each white dot within the violin plot represents the median. The thick black bar in the center indicates the interquartile range, with the upper end marking the third quartile and the lower end marking the first quartile. The thin black line extending from the bar represents the data range, with the upper end indicating the maximum value and the lower end the minimum value. The width of the violin reflects the relative density of the data, with wider regions indicating a higher probability of observed values. D The single-cell sequence of breast cancer (GSE150660) was analyzed, the UMAP of macrophages was obtained, and the expression distribution of CD37, MRC1, and CD86 was demonstrated. All the experiments were repeated at least three times with similar results.
Targeting CD37 promotes phagocytosis of multiple cancer cells
Macrophages and phagocytes were collected from in vitro phagocytosis assays and stained with CD37. As expected, CD37 was enriched in macrophages and barely detectable in phagocytes (Fig. 3A, B, Supplementary Fig. 2A, C). Donor-derived macrophages (DDM) were collected and divided into the CD37-negative and -positive subgroups. An in vitro phagocytosis assay was performed, and CD37-negative macrophages exhibited enhanced phagocytosis of multiple cancer cells, especially BRCA and GBM cells (Fig. 3C, Supplementary Fig. 4A). This could be attributed to the discrepant significance of CD37 in suppressing phagocytosis in diverse tumors. Some tumor cells predominantly depend on the CD37 pathway to realize immune escape, whereas in other tumors, the role played by CD37 is not predominant. Thirteen separate DDMs were collected and treated with CD37-specific antibody naratuximab. DDMs were then incubated with MDA-MB-231 cells, or the glioblastoma cell lines U251, DDM1, and DDM2 were picked for presentation. These results suggest that targeting CD37 promotes phagocytosis to eliminate cancer cells (Fig. 3D, E, Supplementary Fig. 2C). CD37 was knocked out in the THP1 and U937 cell lines using the CRISPR-Cas9 system, and macrophages were obtained after PMA application. Macrophages were incubated with MDA-MB-231 and U251 cells. CD37 knockout promoted the phagocytosis of MDA-MB-231 and U251 cells (Fig. 3F, G). Antigen presentation is another critical function that promotes T cell recognition and cytotoxicity. Thus, we used T cell proliferation and chemokine detection assays to validate the antigen presentation ability of these cells. T cell function was strongly enhanced, as measured by increased proliferation rate and increased production of IFNγ (Supplementary Fig. 4B–E, Supplementary Fig. 2B). This was further confirmed using in vivo experiments in immunocompromised (BALB/c Nude) and C57BL/6 mice. Cancer cells were transplanted into the kidney capsules of mice, and naratuximab and control IgG were injected intraperitoneally. Naratuximab treatment resulted in tumor eradication, as measured by the decreased flux intensity and prolonged survival time (Fig. 3H, I). For further confirmation, we established a conditional CD37 knockout model in C57BL/6 mice (CD37fl/fl-CD11b Cre). Cancer cells were engrafted into the kidney capsule, and flux intensity and overall survival time were measured. Consistent with previous results, knocking out CD37 in macrophages promoted the elimination of multiple cancer cells and prolonged the overall survival time (Supplementary Fig. 4 F–H).
A Phagocytes and macrophages were sorted from the in vitro phagocytosis assay, and CD37 was detected in phagocytes and macrophages using FACS. B The statistical analysis of CD37+ cells in macrophages and phagocytes as described, n = 5, data were presented as mean ± SD, two-tailed paired T-test, p < 0.0001. C Donor-derived macrophages were collected and divided into CD37-negative and -positive groups. The CD37-negative and -positive macrophages were then subjected to an in vitro phagocytosis assay to detect the phagocytic ability. D Donor-derived macrophages were labeled with cell tracker-Blue and treated with IgG or Naratuximab and then subjected to phagocytosis assay. E The statistical analysis of the percentage of phagocytosis of cells with indicated modifications, n = 3, data were presented as mean ± SD, two-tailed paired T-test, ***p < 0.0001 for all four statistical analyses. F THP1 and U937 cells were transfected with guide RNA targeting CD37 and were then induced to macrophages. Cells were collected, labeled with cell tracker-Blue, and incubated with cancer cells; the representative image of the in vitro phagocytosis assay is illustrated. G The statistical analysis of the percentage of phagocytosis of cells with indicated modifications, n = 3, data were presented as mean ± SD, paired T-test, ***p < 0.001. H Mice bearing MDA-MB-231 cancer cells were treated with IgG or Naratuximab, respectively. The representative image of the flux intensity was obtained, the overall survival time was calculated, n = 5 for each group, unpaired T-test for flux intensity, two-tailed Mantel–Cox test for overall survival analysis, p = 0.0008 for flux intensity and p = 0.0006 for overall survival analysis. I Mice bearing U251 cancer cells were treated with IgG and Naratuximab, respectively. The representative image of the flux intensity was obtained, the overall survival time was calculated, n = 5 for each group, unpaired T-test for flux intensity, two-tailed Mantel–Cox test for overall survival analysis, p = 0.0008 for flux intensity and p = 0.0007 for overall survival analysis. All the experiments were repeated at least three times with similar results.
Phosphorylation of CD37Y13 inhibits phagocytosis
Different ligands activate CD37 signaling at different phosphorylation sites25. To further understand the role of CD37 in phagocytosis, we generated specific antibodies against p-CD37Y13 and p-CD37Y274. The kinase-dead alleles Y13A/Y274A and constitutively active Y13E/Y274E were transfected into CD37 null cells, and p-CD37Y13 and p-CD37Y274 were detected for confirmation (Fig. 4A, Supplementary Fig. 5D). We collected macrophages from different tumor species and detected CD37 phosphorylation using immunoblotting and flow cytometry. While the p-CD37Y13+ proportion ranged from 40 to 90%, the p-CD37Y274+ proportion was only ~10% in different cancers (Fig. 4B–E). As previously indicated, CD37 is enriched in macrophages, but not in phagocytes. To investigate the phosphorylation of CD37 in phagocytes, phagocytes were collected using FACS. Because CD37 was downregulated in phagocytes, only a small number of phagocytes were collected. Notably, CD37 tended to be phosphorylated at Y13 rather than at Y274 as measured using immunoblotting and flow cytometry (Fig. 4F–H, supplementary Fig. 2A). These data suggest that CD37 on TDMs is activated at Y13 rather than at Y274. To further understand the biological importance of CD37 in phagocytosis, we divided THP1 and U937 cells into CD37-negative and -positive subgroups and treated CD37+ macrophages with naratuximab or activated CD37Y274E, which resulted in the inhibition of CD37Y13 and activation of CD37Y274. We treated the CD37- cells with CD37Y13E, which activates CD37Y13 signaling. Cells were collected and subjected to an in vitro phagocytosis assay. Narratuximab and CD37Y274E increased the proportion of phagocytes, whereas CD37Y13E inhibited phagocytosis (Fig. 4I, J). The downstream signaling cascade was detected using immunoblotting. CD37Y13E activated immunoreceptor tyrosine-inhibiting motifs (ITIM) and inhibited the PI3K-AKT pathway, whereas CD37Y274E activated ITAM and promoted AKT phosphorylation (Fig. 4K). This was further confirmed in TDMs, which were divided into CD37-negative and -positive cells, and the phosphorylation of CD37 and AKT was determined. As expected, in CD37+ macrophages, p-CD37Y13 was elevated, whereas the AKT pathway was dramatically inhibited; in CD37- macrophages, SHP1 was attenuated, and the AKT pathway was activated (Fig. 4L). Taken together, these findings suggest that CD37 tends to be phosphorylated at Y13 in macrophages and that this phosphorylation inhibits phagocytosis.
A CD37 null cells (MCF7) were transfected with kinase-dead (Y13A, Y274A) and constitutively active (Y13E, Y274E) CD37 alleles, and cells were collected and subjected to Immunoblot detecting p-CD37Y13 and p-CD37Y274. B Macrophages derived from different cancer species were collected and subjected to immunoblot detecting p-CD37Y13 and p-CD37Y274. CD37 null cell (MCF7) transfected with constitutively active (Y13E, Y274E) CD37 alleles were applied as a positive control. C Macrophages derived from different cancer species were collected and subjected to cytometry to detect the proportion of p-CD37Y13 and p-CD37Y274 positive cells. D The statistical analysis of p-CD37Y13+ cells in CD37+ macrophages, BRCA breast cancer, GBM glioblastoma, CRC colon-rectum cancer, LC lung cancer, PDAC pancreatic ductal adenocarcinoma, n = 30 for each cancer species, data were shown as mean and individual dots. E The statistical analysis of p-CD37Y274+ cells in CD37+ macrophages, BRCA breast cancer, GBM glioblastoma, CRC colon-rectum cancer, LC lung cancer, PDAC pancreatic ductal adenocarcinoma, n = 30 for each cancer species, data were shown as mean and individual dots. F CD37-positive phagocytes were sorted from the in vitro phagocytosis assay, and immunoblotting detecting p-CD37Y13 and p-CD37Y274 in CD37+ phagocytes was applied, CD37 null cell (MCF7) transfected with constitutively active (Y13E, Y274E) CD37 alleles were applied as positive control. G Cytometry detecting the proportion of p-CD37Y13 and p-CD37Y274 positive cells in CD37+ phagocytes as described. H The statistical analysis of (G), n = 3, data were presented as mean ± SD, two-tailed unpaired T-test, ***p < 0.0001. I CD37-negative and -positive THP1 and U937 cells were sorted. CD37-positive cells were treated with Naratuximab, or transfected with CD37Y274E, CD37-negative cells were transfected with CD37Y13E, in vitro phagocytosis assay was applied. J The statistical analysis of the percentage of phagocytosis in cells with modifications, n = 3, data were presented as mean ± SD, two-tailed unpaired T-test, p < 0.0001 for all the statistical analyses. K CD37-negative and -positive THP1 and U937 cells were sorted and subjected to immunoblot detecting p-CD37Y13 and p-CD37Y274. L Macrophages derived from different cancer species were divided into CD37-negative and -positive subgroups, immunoblot detecting p-CD37Y13 and p-CD37Y274 was applied. All the experiments were repeated at least three times with similar results.
Tumorous MIF activates CD37Y13 and inhibits phagocytosis
We hypothesized that the activation of CD37Y13 was due to tumor ligation. To exclude somatic differences between species, we collected TDMs and blood-derived macrophages (BDMs) from the same patient with BRCA or GBM. The phosphorylation of CD37 was detected using immunoblotting and cytometry. TDMs from different tumors tended to activate the p-CD37Y13-SHP1 pathway, whereas in BDMs, the p-CD37Y13-SHP1 pathway and p-CD37Y274-PI3K/AKT pathway were activated at low levels (Fig. 5A, B). For further confirmation, we co-cultured the two BDMs with MDA-MB-231 (BRCA cells) or U251 (GBM cells). B-cell lymphoma cell lines (TMD8, DB, and SC11) without co-cultured with BDMs were used as positive control, and BDMs without co-culture were used as blank control. After co-culture, phosphorylation of CD37 was detected; the p-CD37Y13 level increased, whereas the p-CD37Y274 level dramatically decreased. This was further confirmed using flow cytometry (Fig. 5C, D). We hypothesized that different ligands induced CD37Y13/ Y274 phosphorylation, tumorous ligation promotes the activation of CD37Y13 and inhibits phagocytosis (Fig. 5E). To identify potential interacting ligands, we generated purified CD37 and added it to the MDA-MB-231/U251 cell medium. Co-IP was performed using a CD37 antibody, and the protein complexes were subsequently subjected to an LC-MS assay. A total of 329 potentially interacting proteins were identified in U251 cells, 163 interacting proteins were identified in MDA-MB-231 cells, and 30 proteins were detected in both MDA-MB-231 and U251 cells (Fig. 5E). Protein interaction network analysis was performed, and of note, proteins were enriched in the chromosome stability pathway. Only three transmembrane proteins were identified (CD44, CD74, and macrophage migration inhibitory factor, MIF) (Fig. 5F). Next, we generated purified CD44, CD74, and MIF and performed an in vitro pull-down assay after incubation with purified CD37. MIF directly interacted with CD37 (Supplementary Fig. 5C) and was upregulated in multiple cancers in both the TCGA database and our in-house cohort (Supplementary Fig. 5A, B). A multi-fluorescence immunohistochemical assay was used to detect potential interactions in breast cancer samples (CK was stained to track tumor cells, and MIF and CD37 were detected), suggesting that MIF colocalizes with CD37 (Fig. 5G). Next, we established stable MIF-knockout MDA-MB-231 and U251 cell lines using the CRISPR-Cas9 system. BDMs were co-cultured with MIF WT or KO cells and subjected to immunoblotting and flow cytometry. Coculturing with MIF WT cells induced p-CD37Y13, and co-culture with MIF K.O. abrogated this phosphorylation (Supplementary Fig. 5E). This was further confirmed using flow cytometry (Fig. 5H, I). Next, we treated the cells with the MIF-specific antagonist-MIF098, and detected CD37 phosphorylation. As expected, MIF098 inhibited the phosphorylation of CD37Y13 and promoted the activation of CD37Y274 (Fig. 5J, K, Supplementary Fig. 5F). The DDMs were incubated with MDA-MB-231 or U251 cells and treated with MIF098. The results suggested that targeting MIF promotes phagocytosis to eliminate cancer cells (Supplementary Fig. 6A, B). This was further confirmed using in vivo experiments in both immunocompromised and C57BL/6 mice (Supplementary Fig. 6C, D).
A Tumor-derived macrophages (TDMs) and blood-derived macrophages (BDMs) were isolated from the patient., Immunoblot was performed to detect the phosphorylation of CD37, SHP1, and AKT signaling, BRCA breast cancer, GBM glioblastoma. B Cytometry detecting p-CD37Y13 and p-CD37Y274 positive cells in tumor-derived macrophages (TDMs) and blood-derived macrophages (BDMs), BRCA breast cancer, GBM glioblastoma. C Blood-derived macrophages (BDMs) were incubated with the indicated cancer cells and subjected to immunoblot. TMD8, DB, and SC1 without co-culture were applied as positive control, BDM blood-derived macrophages. D Cytometry detecting p-CD37Y13 and p-CD37Y274 positive cells in BDMs before/after incubation with indicated cancer cells, BDM blood-derived macrophages. E Left, different ligands induced the phosphorylation of CD37 and the downstream cascade. Right, Co-IP assay was applied using CD37 antibody in MDA-MB-231 and 251 cells, LC-MS was applied to detect the potential interacting protein, and the venn plot of the potential interacting proteins was generated. F The protein interacting network of the 30 proteins in (E). G The multi-fluorescence immunohistochemical assay was applied to detect CK, CD37, and MIF in breast cancer species, scale, 200 μm. H BDMs were incubated with MIF WT/K.O. cancer cells, cytometry was applied detecting p-CD37Y13 and p-CD37Y274 positive cells. I The statistical analysis of p-CD37Y13 and p-CD37Y274 positive cells described in (H), n = 3, data was presented as mean ± SD, two-tailed unpaired T-test, p < 0.0001. J BDMs were treated with DMSO and MIF098, phagocytosis assay detecting p-CD37Y13 and p-CD37Y274 positive cells was applied. K The statistical analysis of p-CD37Y13 and p-CD37Y274 positive cells described in (J), n = 3, data was presented as mean ± SD, two-tailed unpaired T-test, p < 0.0001. All the experiments were repeated at least three times with similar results.
Targeting CD37 enhances the efficacy of anti-CD47 therapy both in vitro and in vivo
CD47 is one of the most important phagocytic immune checkpoints, and the specific blockade of CD47 by an antibody promotes phagocytosis26. To verify the clinical application of narratuximab in terms of phagocytosis. We treated DDMs with naratuximab, urabrelimab (anti-CD47 antibody), or a combination of both. Cells were subjected to a phagocytosis assay, and the results indicated that naratuximab and ubrelimab promoted phagocytosis as expected, and urabrelimab plus naratuximab exerted a synthesized effect (Fig. 6A, B). Next, we established para-orthotopic (engrafted into the kidney capsule) xenograft models and treated the mice with naratuximab, Urabrelimab, or both agents in combination. The fluorescence intensity was measured, and the overall survival time was calculated. Both naratuximab and urabrelimab promoted the elimination of cancer cells and prolonged the overall survival time (Fig. 6C–G). These findings were further confirmed using a C57 model. Naratuximab and urabrelimab were synthesized and inhibited tumor progression (Supplementary Fig. 7A–F).
A, B DDMs were treated with Urabrelimab, Naratuximab, or in combination, phagocytosis assay was applied as indicated. DDM donor-derived macrophages. C Mice were treated with Urabrelimab, Naratuximab, or in combination. Live imaging of mice with indicated modifications is presented. D The flux intensity of mice bearing MDA-MB-231 and U251 cells, n = 5 for each group, was shown as a single dot. E The statistical analysis of (D), two-tailed unpaired T-test. F The overall survival of mice with indicated modifications, n = 5 for each group. G The survival analysis of (F), two-tailed Mantel–Cox test. All the experiments were repeated at least three times with similar results.
Discussion
Macrophages and phagocytes play key roles in adaptive immunity, and the identification of phagocytosis-associated checkpoints is one of the most popular fields of scientific research worldwide27. With these advances, researchers have selected potential target sites based on experience and further narrowed the range of available targets according to tumor specificity; these include CD47, Siglec7 and CD2412,28,29,30,31. Currently, systematic scanning strategies are far from satisfactory, possibly because of the lack of phagocyte induction methods. However, further studies have attempted to identify the potential checkpoints in mice using CRISPR-Cas928. This approach helps to identify the “don’t eat me” signaling molecule expressed on the cancer cell surface. The mechanisms of phagocytosis in mice and humans are not entirely identical, and the phagocytosis of heterotypic and homotypic tumor cells is completely different. In the present study, we collected macrophages from healthy donors and performed an in vitro phagocytosis assay. The phagocytes were sorted and subjected to ribosomal profiling. First, we identified phagocyte-specific genes by comparing phagocytes with cancer cells (FPKM = 0 for MDA-MB-231 cells). We compared phagocyte-specific genes with those of macrophages and identified putative phagocytosis-associated dysregulated genes. Interestingly, in phagocytes, most DEGs were downregulated, which is consistent with the expression features of the checkpoints. GO analysis was applied to uncover enriched biological functions and associated diseases. The results indicated that the DEGs were enriched in receptor signaling activity, chemokine receptor activity, and phagocyte-associated diseases. Therefore, we selected transmembrane DEGs for further investigation. Ribosome profiling can only detect RPFs and not full-length RPFs. We further narrowed down the dysregulated transmembrane DEGs (FPKM > 0 in phagocytes) and identified 139 transmembrane DEGs. Thus, we selected CD37 for further investigation. in vivo and in vitro experiments confirmed the function of CD37 in inhibiting phagocytosis, indicating that our ribosome profiling and scanning strategies are highly reliable.
CD37 is a member of the tetraspanin family and plays key roles in B-cell malignancies32,33. CD37 is present on the cell surface, endosomes, and exosomes, and contributes to intracellular transport and immune regulation34. The biological functions of CD37 have not been fully elucidated; however, recent advances suggest that this protein promotes both survival and apoptotic signaling through the PI3K/AKT pathway. Additionally, it controls IL-6 receptor signaling by interacting with SOCS335. CD37 can mediate the clustering of α4β1 integrin, subsequently activating PI3K/AKT signaling and promoting cell survival36. However, CD37 can also form complexes with SHP1, LYN, SYK, and PI3Kγ, leading to AKT inactivation and recruitment of SOCS3 to restrict IL-6/STAT3 signaling36. Research has indicated that CD37 is restricted to the immune system, especially in B cells, and is barely expressed in NK cells or granulocytes35,37. The conclusions of previous studies regarding the role of CD37 in phagocytosis are not consistent. This may be due to the complexity of the TME. This study is an advancement of previous research: our research methods are more advanced. We used techniques such as ribosome profiling to screen phagocytic checkpoints and explored the specific mechanisms of CD37 expression using single-cell sequencing technology. Our research methods were more systematic, as we combined in vivo and in vitro experiments and constructed transgenic mice to elucidate the functions of CD37. Thus, a mechanistic study is more accurate. We not only discovered different ligands for CD37 but also explained the role of phosphorylation at different sites in phagocytosis. The prospects for translation are clear. We identified the specific function of CD37 in phagocytosis, and showed that targeted antibodies can effectively enhance phagocytosis efficiency. Our ribosome profiling data suggest that CD37 is expressed in macrophages and potentially inhibits phagocytosis, which conflicts with previous conclusions. We detected CD37 expression in TDMs from multiple cancers in our in-house cohort. The percentage of CD37+ macrophages ranged from 60 to 90%. Single-cell RNA-Seq was performed for further confirmation. Macrophages were identified according to their annotations, and CD37 expression was analyzed. As expected, CD37 was widely expressed in the macrophages. Compared with other phagocytosis checkpoints, the expression of CD37 was markedly greater. The RNA sequence was applied to CD37−/+ macrophages, and pathway enrichment was analyzed. The pathways associated with phagosomes and related diseases were also enriched. These data support the biological importance of CD37 in phagocytosis. Our data suggested that CD37 is widely expressed in macrophages and is critical for phagocytosis.
However, the potential mechanisms underlying CD37 ligation and activation are not yet fully understood. Different ligands mediate phosphorylation at different sites and downstream cascades. Consistent with these results, our results suggest that CD37Y13 promotes the recruitment of SHP1 and that Y274 activates the PI3K-AKT pathway. To determine the potential underlying mechanism, we collected TDMs and BDMs and subjected them to immunoblotting. In TDMs, CD37Y13 was activated and SHP1 was recruited, whereas in BDMs, both CD37Y13 and CD37Y274 were activated. Next, we co-cultured BDMs with cancer cells, after which CD37Y13 was strongly activated and CD37Y274 was inhibited. Thus, we performed a pull-down assay using purified CD37 protein and identified a potential interacting protein complex (CD44-CD74-MIF). An in vitro pull-down assay was performed using purified CD37, CD44, CD74, and MIF. MIF has been identified as a direct ligand for CD37. Our data demonstrate that MIF is a potential ligand of CD37 that causes the phosphorylation of CD37Y13 and ultimately inhibits phagocytosis.
Macrophage MIF is a pro-inflammatory pleiotropic cytokine that was first identified in 196638,39,40,41. MIF is upregulated in multiple cancer types, including breast cancer and GBMs42,43. MIF promotes cancer progression through multiple mechanisms MIF binds to the CD44-CD74 complex and activates the downstream MAPK and AKT pathways44,45. MIF was recently reported to be a nuclease that enzymatically catalyzes the tautomerization of l-dopachrome to 5,6-dihydroxyindole-2-carboxylic acid46. Moreover, MIF was subsequently found to promote the recruitment and function of tumor-associated macrophages (TAM)47; however, the underlying mechanism is not fully understood. In the present study, we identified MIF as a direct ligand of CD37 and determined that it causes the phosphorylation of CD37Y13. This finding provides practical insights into the mechanism of MIF in TAM recruitment; however, the biological function and mechanism of TAM heterogeneity require further investigation.
“Eat me” or “don’t eat me” signals are mediated through ligand‒receptor specific activation of immunoreceptor tyrosine activating motifs (ITAM) or ITIM, respectively48,49. In our study, tumor MIF directly bound to CD37 on macrophages and inhibited the phagocytosis of multiple cancer cells. Naratuximab is a CD37-specific antibody used to treat B-cell malignancies. We treated macrophages with naratuximab and performed an in vitro phagocytosis assay. Naratuximab dramatically promoted phagocytosis. To test the biological importance of MIF in regulating CD37 expression and phagocytosis, we generated a specific antibody that blocked the interaction between MIF and CD37. Moreover, anti-MIF therapy promotes phagocytosis, which further supports our findings. In addition, anti-CD37 had synergistic effects with anti-CD47 on phagocytosis both in vivo and in vitro. Naratuximab enhances the efficacy of ubrelimab in eradicating cancer cells.
CD37 is an identified and characterized phagocytosis-related immune checkpoint protein. This markedly differs from known checkpoints such as CD47. These differences are not merely manifested in cell distribution and receptor–ligand interactions. Upon activation by different ligands, CD37 is phosphorylated at different sites to exert diverse functions. Activation of the Y13 site inhibited phagocytosis, whereas activation of the Y274 site promoted phagocytosis. This approach offers a novel option for the development of targeted therapeutic strategies. When targeting CD37 with antibodies is challenging, specific agonists of the Y274 site can be used to transform the original phagocytosis-inhibiting CD37 into a phagocytosis-promoting one. This is an advantage that other checkpoints do not have.
Although our study has elucidated the crucial role of CD37 in macrophage endocytosis, targeting CD37 still faces inevitable problems. First, the targeting of CD37 may result in foreseeable off-target effects. Because CD37 is predominantly expressed on B cells, in addition to acting on macrophages to facilitate endocytosis, targeting CD37 can also influence B cells and affect their biological functions. However, the effects of CD37 antibodies on B cells and their role in the tumor remain unclear. To solve this problem, macrophage-specific targeting systems could be developed to deliver CD37 antibodies only to CD37-positive macrophages to exert their functions. This not only avoids off-target effects but also effectively inhibits tumor progression. The second issue is drug resistance. T cells play a vital role in tumor clearance and immune surveillance. The inhibitory effect of tumor cells on T cells was not mitigated by the enhanced phagocytic function of macrophages. Therefore, combining immune checkpoint inhibitors such as anti-PD1/PDL1 can effectively enhance therapeutic efficacy while preventing the emergence of drug resistance.
In summary, CD37 was identified as a phagocytosis checkpoint through ribosome profiling using an in-house in vitro phagocytosis assay. Tumor-related MIF directly binds to CD37, activates CD37Y13, and promotes SHP1 recruitment. Anti-CD37 promotes phagocytosis of multiple cancer cells and enhances the efficacy of anti-CD47 therapy (Fig. 7). Our study identified a phagocytic checkpoint and precise therapeutic targets, and we will undertake clinical experiments to clarify its efficacy and safety in the near future.
Methods
Healthy donors’ recruitment, tumor specimens, and ethical approval
Healthy donors were enrolled in Guangzhou Women Medical Center, 20 ml peripheral blood was drawn from each donor for an in vitro phagocytosis assay. Tumor species were obtained from each department. Informed and signed consent were obtained from each healthy donor and patient, and the study was approved by the Institutional Review Board of the Guangzhou Women and Children’s Medical Center (Reference Number: 060A01, 2024-168B00). The study was compliant with all relevant ethical regulations regarding research involving human participants.
In vitro phagocytosis assay
Briefly, peripheral blood was extracted from each donor, and monocyte was isolated using the EasySep Direct Human Monocyte Isolation Kit (Stem cell technology, Catalog # 19669RF). Macrophage was induced by PMA (100 ng/ml) (MedChemExpress, Catalog # HY-18739). Macrophages were then labeled with CellTracker CMF2HC (Thermo Fisher Scientific, Catalog # C12881), and breast cancer cells MDA-MB-231 were labeled with CellTrace CFSE (Thermo Fisher Scientific, Catalog # C34554). Cells were co-cultured for 2 h and then subjected to FACS. Cells labeled with CFSE and CMF2HC were identified as phagocytes, cells only labeled with CMF2HC were macrophage, while CFSE-labeled cells were MDA-MB-231. An equal amount of phagocyte, macrophage, and MDA-MB-231 cells were collected and subjected to ribosome profiling.
Monocyte isolation
Peripheral blood from donors was collected, and monocytes were isolated using the EasySep Direct Human Monocyte Isolation Kit (STEMCELL Technologies, Catalog #19669RF) following the manufacturer’s protocol. Briefly, peripheral blood cells, except monocytes, were washed through binding with the beads. Monocytes were harvested and subjected to in vitro inductions.
Single-cell RNA sequence
Single-cell RNA sequence datasets were obtained and subjected to sequence analysis. For each sample, 33–95M RNA-seq clean reads were obtained that mapped to the genome using HISAT2 (hierarchical indexing for spliced alignment of transcripts) v2.0.477. Sequencing read counts were calculated using Stringtie78,79 (v.1.3.0). Then, expression levels from different samples were normalized by the Trimmed Mean of M values method. The normalized expression levels of different samples were converted to FPKM (Fragments Per Kilobase of transcript per Million mapped fragments). The edgeR package of R was used to analyze the difference between intergroup gene expression, the P values were calculated, and a multiple hypothesis test was performed. The P value threshold was determined by controlling the FDR (False Discovery Rate) with the Benjamini algorithm. The corrected P value is called the q value. DEGs were defined as transcripts with a fold change in expression level (according to the FPKM value) greater than 2.0 and a q value less than 0.05. GO enrichment analysis was performed with the clusterProfiler package of R and the enrichment criteria including a q value < 0.05. Heatmaps of specific genes were generated using the pheatmap package of R. PCA analysis was visualized using the scatterplot3d package of R.
Flow cytometry
Samples were prepared and resuspended in staining buffer after incubating with fluorescence-labeled primary antibody. Samples were subjected to BD FACSymphony A3 for flow cytometry analysis. Cells are initially gated on forward scatter (FSC) and side scatter to define the overall population. Doublets are excluded by comparing FSC-A and FSC-H. Positive cell populations are then determined using appropriate isotype controls and unstained samples to set the threshold. Both unstained samples and isotype control antibodies were used to ensure accurate gating and to validate the specificity of the antibody staining. The expression of CD37 on different cells is examined as follows: After collecting the corresponding cells, they are stained with CD37 or p-CD37Y274, p-CD37Y13 antibodies (1:100) at room temperature for 30 min, washed twice, stained with Alexa Fluor488 secondary antibody (1:100) at room temperature in the dark for 30 min, washed twice, and resuspended in cell staining solution. The GFP positive rate is detected by BD LSRFortesa (×20) flow cytometer. For the T cell proliferation experiment, donor-derived somatic cells are sorted using magnetic beads with the EasySepDirect Human CD8+ T cell isolation kit (STEMCELL Technologies, Catalog # 19663) to obtain CD8+ T cells from donor peripheral blood. T cells are labeled with CFSE and co-cultured with macrophages and cancer cells. Cells are treated with IgG and naratuximab (1 mg/ml). T cells are collected and stained according to the cell surface and intracellular marker staining protocol with CD8-PE (20ul/test) and IFN-γ (BV421) antibodies. T cell proliferation is detected by BD LSRFortesa (×20) flow cytometer. Cells that are double positive for CD8 and INF-γ are the target cell population. For the in vitro phagocytosis experiment, macrophages are labeled with CellTrace Blue (Thermo Fisher Scientific, Catalog # C34568) and co-cultured with CFSE-labeled tumor cells for 2 h. Cells are collected and analyzed by flow cytometry. Cells that are double positive for CellTrace Blue and CFSE are phagocytic cells. The antibody information was detailed in Supplementary Table1.
Ribosome profiling
The ribosomal profiling technique was carried out following the protocols. After obtaining ribosome footprints, ribosomal profiling libraries were constructed using a NEBNext Multiple Small RNA Library Preset for Illumina (E7300S, E7300L). Briefly, adapters were added to both ends of ribosome frames (RF), followed by reverse transcription and PCR amplification. The 140–160-bp-sized PCR products were enriched to generate a complementary DNA library and sequenced using Illumina HiSeq X10.
Metagene analysis and identification of differentially expressed genes
Low-quality reads were filtered using FASTP. The short reads alignment tool Bowtie2 was used for mapping reads to a ribosome RNA and transfer RNA database. After removing the reads mapped to ribosome RNA and transfer RNA. The retained reads from each sample were mapped to the reference genome using Bowtie2, and no mismatches were allowed. To visualize the RFs surrounding the start and stop codons of metagenes, reads were counted at each position of each gene, then these counts were summed across all genes. Metagene plots were generated using R, taking the read counts and the CDS (coding sequence) boundaries in the transcript coordinates as input. Phagocytes were actually two cells together—phagocyte “eating” cancer cells. To identify the DEGs between phagocyte and macrophage, we first identify phagocyte-specific genes by comparing the phagocyte with cancer cells (FPKM = 0 in MDA-MB-231 cells, FPKM > 1 in phagocytes). We next compared the phagocyte-specific genes with macrophage and finally DEGs between phagocyte and macrophage were identified (Fold change > 1, p < 0.05).
Animal experiments
Four-week-old female mice were used for the in vivo experiments. For the para-orthotopic experiments, 1 × 106 MDA-MB-231-Luc or 1 × 106 U251-Luc tumor cells were implanted into the renal capsule as previously described. Mice bearing tumor cells were treated with anti-CD37 (MedChemExpress, Catalog # HY-P99533) or anti-CD47 (MedChemExpress, Catalog # HY-P99374). A total of 400 μg of anti-CD37 and anti-CD47 was injected into mice intraperitoneally every other day. The growth of tumor xenografts was detected using bioluminescence imaging with an IVIS machine (PhotoSound PAFT/256), and relative flux intensity was automatically measured. We recorded the relative fluorescence intensity every 3 days. When a mouse needed to be euthanized immediately to terminate the experiment, we also made a record on that day. The overall survival time was measured and analyzed. Immunocompromised mice (BALB/c Nude, Ruiye model animal, stock#4482270043435,44822700042968), C57BL/6 (Ruiye model animal, stock#4482270045179,44822700051162), and CD37 knockout mice (C57BL/6, Shanghai Model Organisms Center, NM-CKO-230164) were used for in vivo experiments. Mice were reared in an SPF environment. The mice of the control group and the experimental group were housed separately, in their own cages, in the same environment.
Animal ethics
The studies involving animals were approved by the Ruiye model animal (Guangzhou) Biotechnology Co., Ltd Experimental Animal Ethics Committee (RYEth-20230615261). They complied with the WMA Statement on animal use in biomedical research and the EU recommendations (Directive 2010/63/EU) for experimental design.
During the rearing of experimental animals and the experimentation process, we strictly abide by the principles of experimental animal welfare to ensure that the experimental animals can enjoy the following benefits. They have the freedom from hunger and thirst, the freedom to live comfortably, the freedom from pain, injury, and disease, the freedom from fear and distress, and the freedom to express their natural instincts. Meanwhile, during the experiments, the experimenters will anesthetize the experimental animals to minimize their pain as much as possible and complete the experiments as soon as possible while meeting the expected goals of the experiments. The experimenters will perform euthanasia on the animals in the terminal stage to reduce unnecessary pain and suffering for the animals. When the animals present the following conditions, carbon dioxide euthanasia will be immediately carried out:
Rapid weight loss (a 15%–20% reduction in body weight); Presentation of characteristics of terminal animals, such as persistent listlessness, inability to rise, convulsions, limb paralysis, breathing difficulties, etc.; Excessive tumor burden, such as a tumor long diameter reaching 20 mm in mice or a tumor volume reaching 2000 mm³, or tumor ulceration, infection, necrosis, interfering with feeding or hindering walking.
Generation of the CD37 knockout mouse model
CD37 knockout mouse model used in this study was designed and developed by Shanghai Model Organisms Center, Inc. (Shanghai, China). Briefly, Cas9 mRNA was in vitro transcribed with mMESSAGE mMACHINE T7 Ultra Kit (Ambion, TX, USA) according to the manufacturer’s instructions. Two sgRNAs targeted to delete exons 4–5 were in vitro transcribed using the MEGAshortscript Kit (ThermoFisher, USA). One sgRNA targeted to the 5′UTR of the CD37 gene was 5′CTGGAACTCTGTACTCTAGGTGG-3′; the other sgRNA targeted to the 3′UTR of the CD37 gene was 5′-agatctccatgaggtcagatagg-3′. In vitro-transcribed Cas9 mRNA and sgRNAs were injected into zygotes of C57BL/6J mice and transferred to pseudopregnant recipients. Obtained F0 mice were validated by PCR and sequencing using primer pairs: F-5′-GGCTGTCTGGGTGAAAAGGT-3′; R-5′-GTCTCCACCAGTGAACGAGG-3′. The positive F0 mice were chosen and crossed with C57BL/6J mice to obtain F1 heterozygous Cd37 knockout mice. The genotype of F1 mice was identified by PCR and confirmed by sequencing. Male and female F1 heterozygous mice were intercrossed to produce the homozygous CD37 knockout mice.
Co-culture assay
BDMs were co-cultured with cancer cells. BDMs were labeled with CFSE and incubated with cancer cells for about 2 h. BDMs were then sorted using FACS and subjected to flow cytometry and immunoblot.
Pull-down assay
Purified CD44, CD74, and MIF were obtained and incubated with purified CD37, respectively. Pull-down assay was applied using anti-CD37 antibody. The complex was then subjected to an immunoblot to detect the direct interaction.
Protein interacting analysis
CD37 putative interacting proteins were identified after co-IP and LC-MS analysis in MDA-MB-231 cells. The list of potential interacting proteins was submitted to the STRING database (https://www.string-db.org), and the protein interacting network was automatically obtained.
T cell proliferation assay
CD8+ T cells were obtained from donors’ peripheral blood using the EasySep™ Direct Human CD8+ T cells Isolation Kit following the manufacturer’s protocol. T cells were labeled with CFSE and incubated with macrophages and cancer cells. T cells were then harvested and subjected to flow cytometry, and the proliferation of T cells was analyzed.
Cell lines and culture
U251(ATCC Cat#: HTB-17), MDA-MB-231 (ATCC Cat#: HTB-26), TMD8 (ATCC Cat#: CRL-3211), DB (ATCC Cat#: CRL-2289) and SC1 (ATCC Cat#: CRL-3671) cancer cells were directly obtained from ATCC, and U251, MDA-MB-231 were cultured in DMEM (Thermo Fisher Scientific, Gibco Catalog # C11995500BT) while TMD8, DB, and SC1 were cultured in RPMI-1640 (Thermo Fisher Scientific, Gibco Catalog # C11875500BT) supplemented with 10 % FBS (MIKX Catalog # MK1124SS-500c) and 1 % penicillin/streptomycin (P/S) (NCM Biotech, Catalog # C125C5). Macrophages were cultured in the immunoCult-SF Macrophage Medium (Stem cell technology, Catalog # 10961).
Western blot
Lysates from cultured cells or patient tissues were loaded onto a 12% SDS-PAGE gel, followed by electrophoresis. Transfer to PVDF membrane (Millipore, Massachusetts, USA, Catalog # HVLP14250) was then performed. The membrane was first blocked with 5% milk (Biofroxx, Germany, Catalog # 1172), and then incubated with the indicated primary antibody, followed by the corresponding secondary antibody tagged with horseradish peroxidase. Signals were visualized using enhanced chemical luminescence, and the specificity of anti-p-CD37Y13 and p-CD37Y274 was detected using a dot plot. Chemically synthesized polypeptide with a phosphate group added at the Y13 and Y274 sites. Different dosages of peptide were added to each dot, and the antibody was applied to detect the specificity. The antibody information was detailed in Supplementary Table1.
LC-MS analysis
The immunoprecipitates were harvested and subjected to digestion. The resulting peptide was analyzed using a QExactive mass spectrometer coupled to a nano-LC (AdvanceLC). The acquired spectra were analyzed using the SEQUEST HT algorithm.
Plasmid construction and lentivirus production
The CD37 and MIF KO plasmid were generated through CRISP-Cas9 system. Plasmids were co-transfected with the packaging vectors psPAX2 (Addgene) and pMD2G (Addgene) into HEK293T cells for lentivirus production using Lipofectamine 3000 (Thermo Fisher Scientific, Catalog # L3000015) according to the manufacturer’s instructions. Isolated macrophages were transduced using the lentivirus with polybrene (8 mg ml−1, Sigma, Catalog # H9268). Stable cell lines were constructed.
Isolation of tumor-infiltrated macrophages
Tumors were collected, minced with a syringe plunger, washed with PBS, and the cell suspension was collected. Tumor-infiltrated macrophages were isolated by negative depletion using the EasySep Direct Human Monocyte Isolation Kit (Stem cell technology, Catalog # 19669RF) and cultured in the immunoCult-SF Macrophage Medium (Stem cell technology, Catalog # 10961).
Statistical tests
For in vitro analyses, each experiment was performed at least three times. Both the investigators and the analysts were blinded to the conditions. Statistical analysis was conducted using SPSS 13.0 and GraphPad 10.0 software. Experimental data are expressed as the mean ± standard deviation (SD). Statistical significance was determined using Student’s two-tailed paired and unpaired t-test, the details of each statistical test are detailed in the figure legend. The specific details of each statistical analysis are provided in the figure legends, and a p value of less than 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The ribosome profiling data have been deposited in the Genome Sequence Archive under the accession code HRA006465. This dataset was generated from this study. The LC-MS data have been deposited in the ProteomeXchange Consortium under the accession code PXD061433 (https://proteomecentral.proteomexchange.org). This dataset was generated from this study. The single-cell RNA sequence data have been deposited in NCBI under the (scRNA-seq) BRCA, GEO, GSE150660; (scRNA-seq) GBM, GEO, GSE163108; (scRNA-seq) CRC, GEO, GSE139555; (scRNA-seq) LC, GEO, GSE139555; and (scRNA-seq) PDAC, GEO, GSE148673 (https://www.ncbi.nlm.nih.gov). All the single-cell RNA sequence datasets were publicly available and were not generated from this study. The raw data of FACS have been deposited in the Science Data Bank under the 31253.11.sciencedb.25020 (Data CSTR), https://doi.org/10.57760/sciencedb.25020 (Data DOI) (https://www.scidb.cn/en/detail?dataSetId=e454e205797e4dcebe58bdb35981e3ee&version=V1). This dataset was generated from this study. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
References
Ribas, A. & Wolchok, J. D. Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018).
Topalian, S. L., Drake, C. G. & Pardoll, D. M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015).
Wolchok, J. D. et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 377, 1345–1356 (2017).
Motzer, R. J. et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 378, 1277–1290 (2018).
Luo, Y. & Liang, H. Single-cell dissection of tumor microenvironmental response and resistance to cancer therapy. Trends Genet. TIG 39, 758–772 (2023).
Iwasaki, A. & Medzhitov, R. Regulation of adaptive immunity by the innate immune system. Science 327, 291–295 (2010).
Dranoff, G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer 4, 11–22 (2004).
Jutras, I. & Desjardins, M. Phagocytosis: at the crossroads of innate and adaptive immunity. Annu. Rev. Cell Dev. Biol. 21, 511–527 (2005).
Oldenborg, P. A. et al. Role of CD47 as a marker of self on red blood cells. Science 288, 2051–2054 (2000).
Chen, J. et al. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature 544, 493–497 (2017).
Majeti, R. et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299 (2009).
Barkal, A. A. et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572, 392–396 (2019).
Gordon, S. R. et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545, 495–499 (2017).
Barkal, A. A. et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 19, 76–84 (2018).
Zhang, Y. et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 39, 1578–1593.e1578 (2021).
Theruvath, J. et al. Anti-GD2 synergizes with CD47 blockade to mediate tumor eradication. Nat. Med. 28, 333–344 (2022).
Liu, Y. et al. Emerging phagocytosis checkpoints in cancer immunotherapy. Signal Transduct. Target. Ther. 8, 104 (2023).
Press, O. W. et al. Treatment of refractory non-Hodgkin’s lymphoma with radiolabeled MB-1 (anti-CD37) antibody. J. Clin. Oncol. 7, 1027–1038 (1989).
Smalley, I. et al. Single-cell characterization of the immune microenvironment of melanoma brain and leptomeningeal metastases. Clin. Cancer Res. 27, 4109–4125 (2021).
Mathewson, N. D. et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 184, 1281–1298.e1226 (2021).
Wu, T. D. et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278 (2020).
Gao, R. et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 39, 599–608 (2021).
Mazzieri, R. et al. Targeting the ANG2/TIE2 axis inhibits tumor growth and metastasis by impairing angiogenesis and disabling rebounds of proangiogenic myeloid cells. Cancer Cell 19, 512–526 (2011).
Patel, S. S. et al. The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4-positive T cells that are PD-1-negative. Blood 134, 2059–2069 (2019).
Lapalombella, R. et al. Tetraspanin CD37 directly mediates transduction of survival and apoptotic signals. Cancer Cell 21, 694–708 (2012).
Logtenberg, M. E. W., Scheeren, F. A. & Schumacher, T. N. The CD47-SIRPalpha immune checkpoint. Immunity 52, 742–752 (2020).
Veillette, A. & Chen, J. SIRPalpha-CD47 immune checkpoint blockade in anticancer therapy. Trends Immunol. 39, 173–184 (2018).
Kamber, R. A. et al. Inter-cellular CRISPR screens reveal regulators of cancer cell phagocytosis. Nature 597, 549–554 (2021).
Bradley, C. A. CD24 - a novel ‘don’t eat me’ signal. Nat. Rev. Drug Discov. 18, 747 (2019).
Rodriguez, E. et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 12, 1270 (2021).
Li, Y. CD47 Blockade and Rituximab in Non-Hodgkin’s Lymphoma. N. Engl. J. Med. 380, 497 (2019).
Xu-Monette, Z. Y. et al. Assessment of CD37 B-cell antigen and cell of origin significantly improves risk prediction in diffuse large B-cell lymphoma. Blood 128, 3083–3100 (2016).
Deckert, J. et al. A novel anti-CD37 antibody-drug conjugate with multiple anti-tumor mechanisms for the treatment of B-cell malignancies. Blood 122, 3500–3510 (2013).
Peeters, R. et al. Fatty acid metabolism in aggressive B-cell lymphoma is inhibited by tetraspanin CD37. Nat. Commun. 13, 5371 (2022).
de Winde, C. M. et al. Tetraspanin CD37 protects against the development of B cell lymphoma. J. Clin. Investig. 126, 653–666 (2016).
van Spriel, A. B. et al. The tetraspanin CD37 orchestrates the alpha(4)beta(1) integrin-Akt signaling axis and supports long-lived plasma cell survival. Sci. Signal. 5, ra82 (2012).
Yu, B. et al. Targeted drug delivery and cross-linking induced apoptosis with anti-CD37 based dual-ligand immunoliposomes in B chronic lymphocytic leukemia cells. Biomaterials 34, 6185–6193 (2013).
Bernhagen, J. et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 13, 587–596 (2007).
Bloom, B. R. & Bennett, B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science 153, 80–82 (1966).
O’Reilly, C., Doroudian, M., Mawhinney, L. & Donnelly, S. C. Targeting MIF in cancer: therapeutic strategies, current developments, and future opportunities. Med. Res. Rev. 36, 440–460 (2016).
Bernhagen, J. et al. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature 365, 756–759 (1993).
Mittelbronn, M. et al. Macrophage migration inhibitory factor (MIF) expression in human malignant gliomas contributes to immune escape and tumour progression. Acta Neuropathol. 122, 353–365 (2011).
Wu, M. Y., Fu, J., Xu, J., O’Malley, B. W. & Wu, R. C. Steroid receptor coactivator 3 regulates autophagy in breast cancer cells through macrophage migration inhibitory factor. Cell Res. 22, 1003–1021 (2012).
Bifulco, C., McDaniel, K., Leng, L. & Bucala, R. Tumor growth-promoting properties of macrophage migration inhibitory factor. Curr. Pharm. Des. 14, 3790–3801 (2008).
Mitchell, R. A. & Bucala, R. Tumor growth-promoting properties of macrophage migration inhibitory factor (MIF). Semin. Cancer Biol. 10, 359–366 (2000).
Park, H. et al. PAAN/MIF nuclease inhibition prevents neurodegeneration in Parkinson’s disease. Cell 185, 1943–1959.e1921 (2022).
Klasen, C. et al. MIF promotes B cell chemotaxis through the receptors CXCR4 and CD74 and ZAP-70 signaling. J. Immunol. 192, 5273–5284 (2014).
Bruhns, P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood 119, 5640–5649 (2012).
Barclay, A. N. & Van den Berg, T. K. The interaction between signal regulatory protein alpha (SIRPalpha) and CD47: structure, function, and therapeutic target. Annu. Rev. Immunol. 32, 25–50 (2014).
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2021YFA1302000 to X.G.), National Natural Science Foundation of China (82472834 to X.G., 82172917 to L.J.), and National Natural Science Youth Foundation of China (82203525 to X.G.). The authors also thank TosanBio for their help with pattern diagram making.
Author information
Authors and Affiliations
Contributions
J.L., H.M., and X.G. designed the project. X.G., J.Z., H.Z. (Hui Zhang), and X.L. carried out the experiments. B.Z., H.W., and H.Z. (Hanbing Zhang) performed statistical analysis. X.L. and H.W. consented to the patients and collected clinical samples. X.G. designed the figures for this manuscript. X.G. wrote the original manuscript. W.L., X.H., H.M., and J.L. revised the manuscript. All authors have read, commented, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Matthias Trost, Tomohide Tsukahara, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, X., Zhang, J., Zhang, H. et al. Targeting CD37 promotes macrophage-dependent phagocytosis of multiple cancer cell types and facilitates tumor clearance in mice. Nat Commun 16, 6610 (2025). https://doi.org/10.1038/s41467-025-61348-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61348-2
This article is cited by
-
Targeting phagocytosis checkpoints for cancer immunotherapy
Nature Reviews Cancer (2025)