Abstract
Although vaccines and treatments have strengthened our ability to combat the COVID-19 pandemic, new variants of SARS-CoV-2 continue to emerge in human populations. Because the evolution of SARS-CoV-2 is driven by mutation, a better understanding of its mutation rate and spectrum could improve our ability to forecast the trajectory of the pandemic. Here, we use circular RNA consensus sequencing (CirSeq) to determine the mutation rate of six SARS-CoV-2 variants and perform a short-term evolution experiment to determine the impact of these mutations on viral fitness. Our analyses indicate that the SARS-CoV-2 genome mutates at a rate of ∼1.5 × 10−6/base per viral passage and that the spectrum is dominated by C → U transitions. Moreover, we find that the mutation rate is significantly reduced in regions that form base-pairing interactions and that mutations that affect these secondary structures are especially harmful to viral fitness. In this work, we show that the biased mutation spectrum of SARS-CoV-2 is likely a result of frequent cytidine deamination and that the secondary structure of the virus plays an important role in this process, providing new insight into the parameters that guide viral evolution and highlighting fundamental weaknesses of the virus that may be exploited for therapeutic purposes.
Similar content being viewed by others
Introduction
COVID-19 is a highly contagious respiratory disease that is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Although multiple vaccines and treatments have been developed to combat the COVID-19 pandemic, new variants of SARS-CoV-2 continue to emerge in human populations. These variants renew the threat of COVID-19 to public health, extend the socio-economic impact of the pandemic, and highlight a growing need to understand the evolution of SARS-CoV-2. Two parameters that are key to viral evolution are the mutation rate and mutation spectrum. Together, these parameters dictate how many variants will emerge in the future and what type of mutations they are likely to carry. Their impact on the overall fitness of the virus will then decide whether these mutations are selected for or against. To understand the parameters that guide the evolution of SARS-CoV-2, we cultured 6 viral strains and used CirSeq to determine their mutation rate and spectrum. In addition, we used these measurements to determine the impact of the 3603 most common mutations on SARS-CoV-2 fitness. These experiments showed that in addition to nonsense and non-synonymous mutations, synonymous mutations have a substantial impact on viral fitness, especially if they disrupt secondary structures present in the viral genome. Remarkably, these structures also display reduced mutation rates, suggesting that the bases that are most essential to viral fitness are protected from mutation. These observations highlight a strong evolutionary link between the structure of the SARS-CoV-2 genome, its mutation rate, and viral fitness, and suggest that this relationship may be exploited to combat both existing and future variants of SARS-CoV-2.
Results
Overall strategy
The mutation rates and spectra of RNA viruses (including SARS-CoV-2) are notoriously difficult to measure. For example, even though more than 8 million SARS-CoV-2 genomes have been documented across the globe (GISAID, https://gisaid.org/), this dataset only contains mutations that were successful enough to become major variants in patients. Accordingly, most studies of within-host genetic diversity are limited to variants with an allele frequency that exceeds 0.5%, while mutations that are detrimental to the virus are missed1. In contrast, the negative correlation between mutation rate and genome size observed in viruses suggests that the spontaneous mutation rate of the >30 kb SARS-CoV-2 genome is <1 × 10−5 per base2, which is significantly lower than the detection threshold of the sequencing methods used on patients3. As a result, RNA-sequencing methods with improved sensitivity are required to determine the mutation rate of SARS-CoV-2. With these considerations in mind, we used an ultra-sensitive and highly accurate rolling-circle RNA consensus sequencing method termed CirSeq4 to determine the mutation rate and spectrum of 6 SARS-CoV-2 variants. This method was previously used to determine the mutational landscape of other RNA viruses, including the polio virus5, the Ebola virus6, the Dengue virus7, and the Zika virus8. The improved accuracy of CirSeq relies on the circularization of short RNA fragments to synthesize long cDNA molecules that carry tandem repeats of the original RNA template. These tandem repeats can then be analyzed to generate a consensus sequence, which eliminates sequencing and reverse-transcription errors from the final sequencing results (Supplementary Fig. 1). Mutation frequencies are then obtained by dividing the number of mutations observed at a given position by the number of molecules that covered this position.
To explore the mutational landscape of SARS-CoV-2, we cultured the virus in VeroE6 cells, a preferred cell line for COVID-19 research because of its susceptibility to infection, efficient viral replication, and permissiveness to mutations9. Accordingly, VeroE6 cells can support a higher degree of viral genetic diversity than other cell lines, which is useful for studies that examine viral evolution during prolonged culture conditions. In total, we cultured 6 major strains of the SARS-CoV-2 virus, including the USA-WA1/2020, Alpha and Delta strains (corresponding to clades 19B, 20I and 21J, respectively), as well as the Beta, Gamma and Omicron strains. Although each strain was cultured in duplicate, the majority of our experiments were performed on the USA-WA1/2020, Alpha, and Delta strains, which we cultured over seven serial passages, while the Beta, Gamma, and Omicron strains were profiled for a single passage (Table 1). For the strains we cultured over seven passages, we initiated each passage at a low multiplicity of infection (MOI = 0.1) to minimize potential complementation effects. This strategy ensures that most cells are infected by a single virion during the initial phase of each passage, significantly reducing the likelihood of co-infections. Co-infections, where multiple viral particles infect the same cell, could allow defective viral genomes to be rescued by functional ones, distorting the mutation spectrum and artificially lowering the observed fitness cost of deleterious mutations. Thus, by maintaining a low MOI across passages, we consistently and repeatedly limit the propagation of defective genomes that may have been rescued transiently during the expansion phase of the prior cycle. Identical approaches were previously used to limit the impact of co-infections on fitness measurements of other viruses5. Finally, because the VeroE6 cells were derived from the kidney of an African green monkey, we wanted to make sure that our measurements were not skewed by this unique biological environment. To do so, we also cultured the Delta strain for 1 passage in Calu-3 cells (a human lung adenocarcinoma cell line) and primary human nasal epithelial cells (HNEC) that were grown in an air–liquid interface (ALI), which more closely mimics human SARS-CoV-2 infections (Table 1). After each passage, we monitored the sequence of the SARS-CoV-2 genome by CirSeq to take a snapshot of its mutational landscape. A schematic of our cell culture and sequencing approach is depicted in Fig. 1. Across all strains and conditions, we sequenced over ~200 billion bases and identified more than three million mutations. Finally, we assigned the most common mutations a fitness value to determine if they are selected for or against by the SARS-CoV-2 virus and mapped these mutations onto the viral genome and proteome to determine the biological basis for selection.
Nasal swabs of SARS-CoV-2 patients were collected, and the virus was cultured in Vero-E6 cells. Positive culture supernatant was serially diluted to an extinction endpoint, and dilutions with less than 33% positive wells were cultured further in Vero E-6 to attain unique cultures. Two of these cultures were propagated per variant. The TCiD50 was determined, and the virus was passed at an MOI of 0.1 for all subsequent passages. Viral supernatant was concentrated, and RNA was extracted from 15 mL of viral culture. After extraction, viral RNA was cleaved into 60–80 bp fragments, which were ligated to themselves to form circular RNA molecules. These circular RNA molecules were then reverse transcribed to generate linear concatemers of the RNA template. If a mutation were present in the template (yellow line), this mutation would be present in every copy of the concatemer. In contrast, sequencing errors (green line) or reverse transcription errors (red line) would be present in only one, thereby allowing true mutations to be discriminated from technical artifacts. This figure was created in BioRender. Nijhuis (2025) https://BioRender.com/mftt8y6.
The mutation rate and spectrum of the SARS-CoV-2 genome
After we profiled the mutational landscape of the SARS-CoV-2 strains across the length of its genome (Fig. 2A), we used lethal and highly detrimental mutations to estimate their mutation rate. Because these mutations cannot be carried over between passages, they must be produced anew each generation, so that their frequency is equal to the mutation rate5. We used two complementary methods to identify these mutations. First, we considered mutations to be lethal or highly deleterious if they introduce premature stop codons (PTC) in the open reading frame of the RNA-dependent RNA polymerase (RdRP), an essential viral protein required for replication10. This strategy provides the most reliable way to identify lethal mutations and, by extension, to calculate the mutation rate. However, one limitation of this approach is that it cannot capture A → C, U → C, G → C, and A → G mutations, which cannot produce stop codons. To ensure a comprehensive assessment of the mutation rate across all base substitutions, we therefore employed a second, complementary strategy.
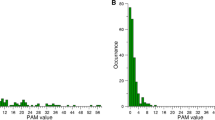
A Circos plot depicting the coverage and mutations detected in SARS-CoV-2 sequencing libraries. Black dots represent the mutations detected, with mutations detected at higher frequencies located further away from the center of the plot. The blue surface represents sequencing coverage, with higher coverage extending further away from the center of the plot. The two outer rings depict the genes present in the SARS-CoV-2 genome, while the numbering outside of these rings indicates the position of these genes along the genome in 1 kb steps. B Mutation rate of six variants of the SARS-CoV-2 virus. Each dot represents a single measurement of a variant at 1 out of 7 passages for 2 replicates (n = 14 for USA, Alpha, and Delta, Mann–Whitney U-test, two-sided alternative, no correction for multiple testing). The mutation rates of the Beta, Gamma, Delta, Delta Calu, and Omicron variants were only measured once, for one replicate and one passage, and are depicted here for comparison. All mutations are presented in the context of the sense strand of the SARS-CoV-2 genome. C The mutation spectrum of the USA, Alpha, Delta, and Omicron strains. Of these strains, USA, Alpha, and Delta were monitored in duplicate across seven passages, while Omicron was monitored once across one passage. Error bars represent the standard deviation of the mean across the seven passages.
For this strategy, we analyzed over eight million SARS-CoV-2 genomes previously aligned by UShER11,12 and Ensembl13 and identified mutations that are absent from these databases. These genomes represent the consensus sequences of the most common viral variants in individual patients, meaning that mutations with severe fitness consequences, including lethal or highly detrimental mutations, are unlikely to be present. Consistent with this idea, we found that the mutations identified through this method were significantly depleted in our experimental dataset (Supplementary Fig. 2), supporting their classification as highly detrimental or lethal. However, we noticed that 68 of these mutations were present in our own dataset at frequencies exceeding 1 × 10⁻⁴ (>10-fold higher than the average mutation frequency), strongly suggesting that they are neither lethal nor highly deleterious. Thus, we excluded them from the list of mutations used to determine mutation rates.
By combining these strategies, we created a comprehensive list of lethal and highly detrimental mutations and used it to calculate the mutation rate across the length of the SARS-CoV-2 genome. This analysis revealed that ~1.5 × 10⁻⁶ mutations occur per nucleotide per viral passage (Fig. 2B), whether the virus was grown in VeroE6 cells, Calu-3 cells, or primary HNEC grown in an ALI (Table 1). To ensure that our “combination strategy” was an appropriate tool to determine mutation rates, we also calculated separate mutation rates, based on either the PTC or ‘absent mutations’ method and found that they yielded nearly identical mutation rate estimates, strongly supporting the idea that these approaches provide appropriate, complementary datasets for determining the mutation rate of the SARS-CoV-2 genome (Supplementary Fig. 3). Interestingly, we found that the Delta strain displayed the highest mutation rate of the three strains that we monitored over seven passages, potentially contributing to the increased virulence it displayed compared to the USA and Alpha strain. For each strain we found that the mutation rate varied greatly between different base substitutions, ranging from ~2 × 10⁻5 for C → U mutations to ~1 × 10⁻6 for G → C mutations, with C → U substitutions being ~4 times more common than any other base substitution (Fig. 2C and Supplementary Fig. 4). The rate with which C → U substitutions arose depended to a significant degree upon the upstream (i.e., 5′ adjacent) and downstream (i.e., 3′ adjacent) nucleotides. For example, we found that C → U mutations occur most commonly in a 5′-UCG-3′ context (Fig. 3A–D), consistent with analyses based on SARS-CoV-2 phylogeny14. When taken together, these observations demonstrate that C → U substitutions add the greatest amount of genetic variation to the SARS-CoV-2 genome and provide the largest substrate for evolution to act upon, a conclusion that is also supported by more indirect observations15. Because our measurements are independent of positive or negative selection, though (which play a key role in published SARS-CoV-2 genome sequences), our analyses provide an unfiltered view of the impact of genetic context on viral mutagenesis.
A–D The mutation rate of the SARS-CoV-2 genome differs depending on the bases that directly flank the focal base. The focal base (the mutated base at the center of a triplet) is listed underneath the graph, while the bases that are on its 5’ or 3’ side are located above and below the focal base. The type of mutation that is analyzed is depicted above the bars. So, for example, the first 4 bars on the left-hand side of (A) correspond to an adenine base that is at the center of the triplet (the focal base), and is flanked on its 5’ base by adenine, and on its 3’ side by one of four possible bases. Each bar then corresponds to the impact of these flanking bases on the mutation of adenine (the focal base) to uracil. For example, the mutation rate of A → C is highest when adenine is flanked by guanine on its 5’ side and 3’ side. In contrast, the mutation rate of A → C substitutions is lowest when adenine is flanked on its 5’ side with uracil, and adenine, cytosine, or guanine on its 3’ side. Note that the y-axes for each panel differ for increased visibility. Average of all passages generated (n = 47), error bars represent standard deviation of the mean.
It’s notable that the mutation rate of SARS-CoV-2 is ~10-fold lower compared to the poliovirus5 and ~5-fold lower than the Dengue virus7, two other RNA-based viruses previously examined by CirSeq. The decreased mutation rate of the SARS-CoV-2 genome is most likely due to the proofreading ability of its RdRp10,16, which is absent in the polio and Dengue virus. In this context, it is important to note that G→A and U→C mutations displayed the largest reduction in mutation rate compared to the polio virus (48-fold and 28-fold, respectively, Fig. S5). When the proofreading activity of eukaryotic RNA polymerases II is compromised17,18,19, these base substitutions increase the most, suggesting the existence of a universal set of rules that govern the proofreading capabilities of RNA polymerases in eukaryotes and viruses.
Selection for nucleotide composition
It is likely that the mutation rate and spectrum of the SARS-CoV-2 genome affect the evolution of the virus in various ways. One of the most fundamental attributes of a genome is its nucleotide composition, which depends on the balance between the mutation spectrum and the intensity of selection for each of the four nucleotides. Using the mutation rate for each of the 12 possible base substitutions, we estimate the equilibrium frequencies for all four nucleotides as: U = 0.42, A = 0.29, G = 0.21, and C = 0.07 (Table 2). This analysis translates into an equilibrium GC content of 28%, which is substantially higher than the 17% previously reported15. However, this previous estimate is based on indirect estimations of mutation rates at 4-fold degenerate sites across lineages sequenced in GISAID, which might be impacted by selection. Regardless, both estimates are significantly lower than the observed 38% GC content of the SARS-CoV-2 genome, indicating that the GC content in the SARS-CoV-2 genome is actively preserved by natural selection, particularly in the case of cytidines. Cytidines were even preserved at 4-fold degenerate sites (Table 2), suggesting that natural selection also preserves cytidines at sites where mutations would not alter amino acid composition. This pattern indicates a broader, possibly structural or regulatory, role for cytidines in the SARS-CoV-2 genome. To gain more insight into the molecular mechanisms that suppress cytidine depletion at 4-fold degenerate sites, and examine the impact of C → U mutations on viral fitness, we calculated fitness values for 3603 C → U mutations that were scattered across the SARS-CoV-2 genome.
Fitness landscape of SARS-CoV-2
Because fitness analyses require large amounts of data gathered from a single strain over an extended period of time, we selected one replicate of the SARS-CoV-2 Delta variant and tracked it over the course of seven passages. After each passage, we monitored its genome by Cirseq, ultimately sequencing 155 billion bases and covering each base 1.7 million times on average. This sequencing effort allowed us to identify 64,967 unique mutations across all passages, with each mutation being observed 42 times on average, for a total of 2.7 million mutation observations. Because the SARS-CoV-2 genome is ∼30,000 bases in length, and each base can be mutated into 3 different nucleotides, a total of ∼90 K base substitutions is theoretically possible, meaning that we identified 66% of all possible mutations in the SARS-CoV-2 genome. We then used this dataset to determine the consequences of C → U mutations on viral evolution by characterizing their impact on the fitness of the SARS-CoV-2 virus with a strategy previously employed for the polio virus5. Due to technical considerations, we did not determine fitness values for other base substitutions (see “Methods” section). Briefly, the fitness of a mutation is related to its change in frequency between consecutive passages as described in the following equation:
With fn and fn−1 being the observed frequency of the mutation at passages n and n − 1, w the relative fitness, and µ the rate of C → U substitutions. (Fig. 4A and Supplementary Data 1). These fitness values were calculated as a weighted average of the fitness values derived at each of the seven passages, so that values with higher coverage (and thus higher precision) contribute more to the final estimates. We performed three tests to determine the veracity of these fitness values. First, we separated the C → U mutations into three groups and found that, as expected, synonymous C → U mutations were less deleterious than non-synonymous C → U mutations (0.78 vs 0.70, P = 2.0 × 10-6, Mann–Whitney U-test), and non-synonymous mutations were less deleterious than non-sense mutations (0.70 vs 0.62, P = 0.006, Mann–Whitney U-test). In a second test, we examined the fitness values of mutations that were predicted to be either lethal or highly detrimental because they produce a PTC, or because they were absent from the 8 million genomes alignment. We found that these mutations displayed significantly lower fitness values compared to all the other mutations we detected (mean fitness: 0.38 vs 0.73, P = 3.4 × 10-4, Mann–Whitney U-test). Moreover, mutations with similar fitness values frequently clustered together, as expected of mutations that affect similar regions of the genome or the proteome (Fig. 4B). Finally, we compared our fitness estimates to studies that used changes in mutation frequency throughout the SARS-CoV-2 phylogeny to infer fitness values20,21. When we compared our values to the only other study to provide fitness estimates for both synonymous and nonsynonymous mutations21, we found a moderate but significant correlation between our data and this independent dataset (r = 0.47, P < 2 × 10−16, Fig. S6). Together, these analyses strongly support the idea that our algorithms provide predictive information about the impact of mutations on viral fitness.
A Fitness distribution of the 3603 C → U mutations which we calculated fitness values. B Mutations with similar fitness values tend to cluster together. Blue bases indicate locations where C → U mutations are positively selected for, and orange bases indicate locations where C → U mutations are selected against. The intensity of the color indicates the intensity of selection. Five clusters are highlighted. C, D Fitness distribution of all synonymous C → U mutations for which we calculated fitness values, split up into paired or unpaired bases. E, F Fitness distribution of all non-synonymous C → U mutations for which we calculated fitness values, split up into paired or unpaired bases. G, H Fitness distribution of all non-sense C → U mutations that we calculated fitness values for, split up into paired or unpaired bases I. Mutation rate of all mutations, split up into paired or unpaired bases (P = 0.0006 for C-to-U, Mann–Whitney U-test, n = 7). J Presence of paired and unpaired bases in hot spots or cold spots of mutation. K Mutation rate of cytosine in CpG vs non-CpG islands (P = 0.002, Mann–Whitney U-test, n = 7). All analyses were done by the Mann–Whitney U-test. **P < 0.01; ***P < 0.001. All error bars represent the standard deviation of the mean across the seven passages of the Delta (replicate B) virus.
Paired bases contribute disproportionately to SARS-CoV-2 fitness
Next, we used our fitness values to investigate why synonymous C → U mutations are selected against in the SARS-CoV-2 genome, even if they occur at 4-fold degenerate sites. Potentially, this phenomenon could be explained by stronger, more frequent purifying selection against synonymous mutations in SARS-CoV-2 compared to other viruses, such as the polio virus5. It was recently shown that the SARS-CoV-2 genome adopts a highly specific secondary structure22 and that bases that pair with each other to form these structures tend to display lower nucleotide diversity23. Interestingly, we observed a similar specificity for secondary structures in our CirSeq dataset. The enzyme used to fragment viral RNA (RNAse III) prefers to cleave RNA at specific double-stranded structures, causing strong peaks and valleys in genome coverage that reflect the secondary structure of the SARS-CoV-2 genome. We found that these coverage peaks are identical between all the variants we tested, indicating that the secondary structure of the genome is highly conserved across the SARS-CoV-2 phylogeny (Fig. S7). Based on these observations, we hypothesized that the need to preserve this secondary structure could be a significant factor driving purifying selection against synonymous mutations. To test this hypothesis, we split synonymous C → U mutations into two groups: those that form base-pair interactions (henceforth referred to as “paired” sites) and those that do not (henceforth referred to as “unpaired” sites). This classification is based on a study that used DMS MapSeq to determine whether nucleotides are paired or not22.
Consistent with the idea that there is strong purifying selection against synonymous mutations that affect secondary structures in the SARS-CoV-2 genome, we found that the average fitness value of synonymous C→U mutations was lower at paired sites compared to unpaired sites (0.60 vs 0.93, P < 2 × 10−16, Mann–Whitney U-test, Fig. 4C, D). We observed a similar pattern for nonsynonymous mutations (average fitness: 0.50 vs 0.81 for paired and unpaired sites, respectively, P < 2 × 10−16, Mann–Whitney U-test, Fig. 4E, F) and non-sense mutations (average fitness 0.29 vs 0.71 for paired and unpaired sites, respectively, P < 2 × 10−16, Mann–Whitney U-test, Fig. 4G, H). To support this idea further, we re-examined our fitness values with the help of an independent assessment of secondary structures based on SHAPE scores24 and found a weak but significant positive correlation between the shape reactivity score and our fitness estimates for synonymous C → U mutations (r = 0.28, P < 2 × 10−16, Fig. S8). Taken together, these results suggest that mutations that disrupt base-pairing interactions are more likely to be deleterious to SARS-CoV-2 fitness than those that don’t.
Because our fitness estimates are limited to C → U mutations, we used the fitness estimates previously published by Bloom and Neher21 to investigate if purifying selection for synonymous mutations at paired sites was present for all types of base-substitutions. Restricting our analysis to base-substitutions with enough observations in both paired and unpaired categories, we found that synonymous mutations are significantly more deleterious at paired vs unpaired sites for U → C, G → U, G → C, C → U, C → A, and A → U base substitutions (P < 0.01 for all, Mann–Whitney U-test, Fig. S9). U → A, U → G, C → G, and A → C substitutions did not yield enough observations to calculate fitness values, while two types of base-substitutions (G → A and A → G) showed no significant difference. Interestingly, though, it was previously shown that A:C and G:U base pairs (which would arise from G → A and A → G mutations, respectively) allow wobble base pairing in RNA molecules25. Therefore, it is possible that these mutations do not significantly alter the secondary structure of the SARS-CoV-2 genome, even when they occur at paired sites, allowing them to escape purifying selection.
Paired bases display a reduced mutation rate
Our data suggests that the secondary structure of the SARS-CoV-2 genome is critical for viral fitness and that SARS-CoV-2 conserves these structures by strong purifying selection. However, the pace of evolution is also controlled by the mutation rate. Accordingly, we wanted to test the impact of the secondary structure on the mutation rate. To do so, we compared the rate of mutation between paired and unpaired bases and found that C → U mutations (but not other base substitutions) are ~3 times more frequent at unpaired bases compared to paired bases in all strains (P = < 0.01, Figs. 4G and S10). Other base substitutions are not increased at paired bases (Fig. 4G), indicating that the mechanism responsible for this observation is highly specific. A similar discrepancy is seen at mutational hot spots and cold spots, which are defined by locations where the mutation frequency either increases or decreases 10-fold. In hot spots, only 14.2% of nucleotides are predicted to be paired (vs 47.3% overall, P < 2 × 10−16, chi-square test), while they make up 81.1% of bases in cold spots (P < 2 × 10−16, chi-square test, Fig. 4H). One potential explanation for this observation is spontaneous cytidine deamination, as unpaired cytidines are 140 times more prone to spontaneous deamination into uracil than paired bases26. In support of this idea, we found that C → U mutations are elevated at CpG sites (Fig. 4I). The electron density of guanine slightly alters a cytosine’s electron distribution, particularly around the amino group at the 4-position, thereby increasing the likelihood of spontaneous hydrolytic deamination27,28. Another possibility is that the reduced rate of cytidine deamination at paired sites reflects the preference of APOBEC proteins for ssRNA. For example, APOBEC3A has a strong preference for unpaired cytidines that are flanked by a 5’ uracil and a 3’ guanosine29,30,31, the exact conditions that show the highest C → U mutation rate in our dataset (Fig. 3C). Regardless of the mechanism though, these observations suggest that the secondary structure of the SARS-CoV-2 genome is not only preserved by strong purifying selection, but also by local changes in the mutation rate that spare paired bases. Since paired bases display lower C → U mutation rates, we hypothesized that selection should result in an excess of essential components of proteins in paired regions of the genome. In support of this hypothesis, we found that the more detrimental a mutation is for a protein, the greater the chance it is located in a paired region (Fig. 4D, F, H). For example, bases in which C → U mutations would result in a premature stop codon with a fitness value of 0 have a 60% chance of being placed in a paired region, compared to 40% for non-synonymous mutations and 30% for synonymous mutations. Moreover, when we compiled a short-list of 120 amino acids that have been proposed to undergo mutational scanning, conservation between coronaviruses and patient information, that together, these observations together suggest a synergistic relationship between the secondary structure of the SARS-CoV-2 genome and its mutation rate, which reinforces each other to promote viral fitness.
Fitness values, viral evolution, and potential weaknesses of SARS-CoV-2
Because fitness values reflect the forces of natural selection, we wondered whether they could predict the evolution of the SARS-CoV-2 virus. Since the emergence of the Delta variant, we sequenced for our fitness analysis, multiple variants have evolved that swept the globe. Each of these strains contains defining mutations that were positively selected for during the evolution of SARS-CoV-2 in human populations (Fig. 5A, B). Interestingly, some of these mutations were also detected in our short-term evolution experiment. When we examined the fitness values of the mutations that define strain 23H (the most advanced strain at the time of writing), we found that these mutations displayed significantly higher fitness values compared to all other mutations detected in our evolution experiment (0.97 vs 0.72, P = 5 × 10−5, Wilcoxon rank sum test). Thus, the fitness landscape obtained from our dataset could help predict the mutations that arise in future variants. Accordingly, mutations with high fitness values that have not been observed in known variants so far could be of interest to researchers trying to predict the evolutionary trajectory of SARS-CoV-232,33. Although mutations with high fitness values tend to be dispersed across the SARS-CoV-2 genome (Fig. 5C), regions where they cluster together might be of particular interest for this purpose (Fig. 5D).
A Distribution of fitness values for all defining C → U mutations in SARS-CoV-2 clade 23H compared to all other mutations detected during our short-term evolution experiment (P = 5 × 10−5, Wilcoxon rank sum test, two-sided alternative). B Details of all defining C → U mutations in SARS-CoV-2 clade 23H. C Although mutations with similar fitness values tend to cluster together, clusters that are positively or negatively selected for tend to be semi-randomly distributed across the genome. Orange bases indicate locations where C → U mutations are selected against, and blue indicates positive selection. The intensity of the color indicates the intensity of selection. D–F However, some regions are strongly enriched in either positive or negative selected cytosines. G–L We plotted the mutations that are most detrimental to viral fitness (ω < 0.5) on the structure of the SARS-CoV-2 spike protein (G–I) and the replisome (J–L) in various orientations. The most interesting mutations are those that occur in clusters, highlighting regions of the viral proteome, or the underlying structure of the genome, that are especially vital to fitness. Three subunits of the spike protein are depicted in pink, blue, and green. Mutations that are predicted to be detrimental are depicted in red (all mutations detected were detrimental) or purple (a subset of mutations detected were detrimental). In J–L, individual units of the replisome are depicted in various colors, including the exonuclease, polymerase, and the viral genome itself.
Conversely, we also identified clusters of mutations with fitness values below 0.5, suggesting that these mutations are targets for negative selection. Our data suggests that one mechanism by which these clusters affect viral fitness is by disrupting secondary structures in the genome. Consistent with this idea, we identified multiple secondary structures in which mutations on either side of the structure lead to large fitness defects, even if they are hundreds of bases apart in the primary sequence of the genome (Fig. 5E, F). Figure S11 contains a comprehensive 2D map of the SARS-CoV-2 genome containing all the C → U mutations for which we established fitness values. In addition to the secondary structure of the genome, C → U mutations may also affect critical amino acids in protein structures. To visualize the potential impact of mutations on the proteome, we mapped clusters of mutations that are subject to negative selection onto the spike protein and the viral replisome (Fig. 5G–L and see also Supplementary Movie 1–4). Because the clusters of mutations that negatively affect the structure of the genome or the proteome highlight immutable components of the SARS-CoV-2 virus, they could be valuable targets for vaccines and treatments. Frequently, viruses develop resistance to vaccines and treatments by mutation, leading to variants that lack the targets that treatments or vaccines were developed against; however, because mutation of these essential components significantly lowers viral fitness, targeting these clusters could limit the number of escapees that emerge.
Discussion
Single-stranded RNA viruses are often considered to be fast-evolving organisms, with some of the highest known mutation rates. Indeed, a recent study estimated the mutation rate of SARS-CoV-2 to be as high as 1 × 10−4 per nucleotide per round of infection34. Our results indicate, though, that the mutation rate of SARS-CoV-2 is substantially lower than that of other ssRNA viruses, allowing ~1.5 × 10−6 base-substitutions per nucleotide per viral passage. Despite this relatively low mutation rate, our measurements are in line with expectations based on genome size2 and indirect estimates relying on alternative methods35.
An important contributor to the enhanced fidelity of the SARS-CoV-2 replisome is its proofreading domain, which reduces the error rate of multiple base substitutions10,16, but is especially effective against transitions. This aligns with the evolutionary role of proofreading domains in correcting the most frequent errors made by RNA polymerases. RNA polymerases, including the SARS-CoV-2 RdRp, tend to generate more transitions than transversions because transitions better resemble Watson-Crick base pair geometries, and have lower steric constraints. Consistent with this hypothesis, we found that G → A and U → C transitions are suppressed >20-fold in SARS-CoV-2 compared to the polio virus, which does not have a proofreading domain. Genetic or pharmacological disruption of nsp14, the gene encoding the proofreading domain, could provide direct insight into this phenomenon in the future.
Despite the efforts of the proofreading domain, C → U transitions remain the most prevalent mutation in the SARS-CoV-2 genome, suggesting that these transitions may arise through a mechanism that is outside of the purview of the proofreading domain. Interestingly, C → U transitions are increased threefold in regions of the genome that are not involved in base-pairing interactions. This increase is unlikely to be caused by a change in the fidelity of the viral RdRP, as there is no reason to think that this enzyme would be less accurate in paired vs unpaired regions of the genome. Consistent with this hypothesis, we found that every other base substitution displays identical mutation rates in paired and unpaired regions (Fig. 4G). Thus, it is likely that a substantial number of C → U mutations are generated by other mutational mechanisms, including cytosine deamination. Cytosine deamination converts cytosine to uracil, and can either occur spontaneously, or in an APOBEC-mediated fashion. Importantly, both of these mechanisms are specific to C → U mutations and occur preferentially in unpaired regions of RNA molecules. Additionally, we found that C → U mutations are enriched in UCG triplets—a sequence targeted by APOBEC3A—suggesting that APOBEC3A may play a key role in the mutagenesis and evolution of the SARS-CoV-2 virus. Disruption of the endogenous APOBEC3A gene could provide direct evidence for this mechanism in the future.
Our fitness measurements suggest that the reduced rate of C → U mutations in paired regions of the genome is further amplified by stronger selection against these mutations in paired vs unpaired regions. The dual role of RNA secondary structures in facilitating essential viral processes and protecting sensitive regions from mutations highlights a synergistic relationship that may have shaped the evolution of the SARS-CoV-2 genome. Viruses like SARS-CoV-2 may prefer to encode the most essential features of their genome in intricate secondary structures because these structures have the added advantage that they are protected from mutation. In support of this idea, we found that 30% of synonymous C → U mutations are lethal when they occur in paired regions, but only 10% when they occur in unpaired regions (Fig. 4D). Because this trend increases for non-synonymous (40% vs 10%, Fig. 4F) and non-sense mutations (60% vs 10%, Fig. 4H), our results suggest that a similar form of protection may apply to cytidines that encode critical amino acid sequences. Together, these observations suggest that the secondary structure of the RNA-encoded genome of SARS-CoV-2 modulates its local mutation rate and imposes significant constraints on its evolution.
Similar to SARS-CoV-2, synonymous mutations affect the fitness of other RNA viruses as well36,37, either by destabilizing secondary structures36,38,39, or potentially by altering codon usage40,41, although the latter is still debated41. Compared to other viruses, though, the percentage of synonymous mutations that affect SARS-CoV-2 fitness is relatively high. However, it has been reported that the fraction of lethal mutations among synonymous mutations increases with genome size. For example, 3% of synonymous mutations are lethal for the 4.2 kb Qβ genome36,42, 10% for the 7.5 kb polio virus5, 9% for the 9.5 kb TEV genome42,43, and 13% for the 11 kb VSV genome36. Our study now suggests that this fraction may be as high as 20% for the 30 kb SARS-CoV-2 genome. Note, though, that in contrast to these other studies, our measurements exclusively report on the percentage of synonymous C → U mutations that are detrimental to viral fitness. There are two main reasons why the percentage of C → U mutations that are detrimental for viral fitness may be higher than other base substitutions. First, because C → U mutations dominate the mutation spectrum of the SARS-CoV-2 genome, it is likely that most non-essential cytidines have been replaced by uracil over the course of viral evolution. The observation that cytidines are markedly diminished at 4-fold degenerative sites provides strong evidence for this idea (Table 2). It is reasonable to assume, then, that the cytidines that remain in the genome are somehow important for viral fitness, skewing the fitness values of C → U mutations towards 0. Second, C → U mutations could have an outsized impact on the stability of secondary structures because C:G base pairs are held together by three hydrogen bonds, while A:U base pairs are held together by 2. Thus, loss of a C:G base pair could have a greater destabilizing effect on secondary structures and viral fitness than other base substitutions.
Our focus on the impact of C → U mutations on vital fitness is a limitation that is the direct result of the low mutation rate of the SARS-CoV-2 genome. Because mutations in the viral genome tend to be rare, they can be lost between passages if they are absent from the subset of viral particles that infect the next culture, creating the appearance of lethal or highly detrimental effects. If ignored, this sampling issue could artificially increase the number of mutations with a fitness value of 0. For this reason, we restricted our analysis to C → U mutations, the most common mutation in the viral genome. In most cases, these mutations exceed the frequency threshold at which this sampling bias is likely to occur. Thus, future studies with greater sequencing power could significantly expand upon the fitness values reported here by including the contribution of other base substitutions to the fitness landscape.
If possible, we recommend that these studies consider long-read sequencing technology for genome analysis. Due to the short-read sequencing technology employed here, we were unable to resolve viral haplotypes and account for epistatic interactions between mutations within the same viral genome. Thus, while our fitness measurements provide valuable insights into the impact of individual mutations on viral fitness, they may not fully capture the effects of interactions between multiple mutations. Such epistatic interactions could amplify or mitigate the fitness effects of individual mutations, complicating the interpretation of their evolutionary significance. We expect, though, that the impact of these epistatic interactions in our experiment was limited by the relatively low mutation rate of SARS-CoV-2. Given a mutation rate of 1.5 × 10−6 per viral passage and a genome that is 30,000 bases long, 0.3 mutations are likely to arise in each genome over the course of seven passages, limiting the number of genomes that acquired multiple mutations over the course of our short-term evolution experiment.
Despite these limitations, though, our findings were robust enough to demonstrate a clear evolutionary trade-off between genome size, secondary structure, and viral fitness. The observation that synonymous mutations can strongly affect viral fitness also has important implications for our understanding of the selective pressures acting on the evolution of SARS-CoV-2. The ratio of the rates of non-synonymous to synonymous substitutions (dN/dS) is one of the most widely used tests to detect the types of selection acting on coding sequences. This test relies on the assumption that synonymous mutations are mostly neutral and elevated dN/dS ratios are often interpreted as evidence of positive selection. Recent studies comparing the sequences of different variants of SARS-CoV-2 have reported dN/dS ratios in the range of ~0.5–1.0(45), which is substantially higher than the ratios typically observed in prokaryotic and eukaryotic protein-coding genes. The elevated dN/dS ratios for SARS-CoV-2 are usually interpreted as evidence for positive selection acting on the sequence of viral proteins, driven by an arms race between the virus and the host immune system. However, our results now show that these elevated dN/dS ratios may also stem from widespread purifying selection on synonymous mutations rather than positive selection for rapid evolution.
Notably, similar conclusions were previously drawn for other RNA viruses. For example, it was previously shown that synonymous mutations in the HIV-1 genome can disrupt conserved RNA structures that are important for replication efficiency, sometimes more so than non-synonymous mutations44,45. Synonymous mutations were also reported to impact the fitness of the poliovirus46, the hepatitis C virus47, and the Zika virus48 by disrupting secondary structures important for protein translation and interactions with the host cell. Our results now indicate that the same is true in the case of SARS-CoV-2. Intriguingly, multiple strategies have been proposed to suppress viruses that rely on the secondary structure of their genome for fitness. Thus, our results suggest that these therapies may aid treatments targeting SARS-CoV-2 as well. For example, methods based on genome editing by CRISPR Cas13b or RNA translation interference (RNAi) could be targeted against regions that are important for the secondary structure of the genome. If these regions are essential for viral fitness, this strategy should reduce the likelihood of facing mutations that escape treatment49,50. Alternatively, it may be possible to exploit the importance of secondary structures for viral fitness with treatments that are aimed at the viral proteome. It is tempting to target amino acids that code for essential components of proteins, because those amino acids are unlikely to escape treatment by mutation. However, our data now suggests that even amino acids that play a relatively unimportant role for viral fitness could be targets for antibody-based treatments and therapies because the bases that encode them are essential to maintain immutable secondary structures. Because these structures can be quite large, multiple bases with low fitness values cluster together in defined areas of the proteome (Fig. 5C–G), which could make them valuable targets for antibody-based approaches.
Methods
Nasal swabs used for virus generation were obtained from a study involving health care workers, approved by the Institutional Review Board of the University Medical Center Utrecht (ABR NL73903.041.20). For ALI cultures, nasal brushings were collected from individuals who provided written informed consent for the use and storage of their cells. This procedure was approved by TcBIO (Toetsingscommissie Biobanks), the institutional Medical Research Ethics Committee for biobanked materials at the University Medical Center Utrecht (protocol ID: 21/265).
Cell lines
Vero C1008 African green monkey kidney Cell Line, Clone E6, (Kindly provided by Assoc. Prof. Bosch Veterinary Medicine, University Utrecht, The Netherlands) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with L-Glutamine (Lonza) supplemented with 10% fetal bovine serum (FBS (FBS), 100 U/mL Penecilin/Streptomycin (Gibco), 1% Ultraglutamine (250 mM in 0.85% NaCl) (Lonza) (cell line culture media) in a humidified incubator at 37 °C 5% CO2. Calu E-3 cells (kindly provided by Prof. Beekman, Hubrecht Institute, the Netherlands) were cultured in cell line culture media and maintained in a humidified incubator at 37 °C 5% CO2.
HNEC culturing
Nasal brushing-derived HNECs (n = 1 healthy donors) were collected, isolated, and stored with informed consent of all participants and were approved by a specific ethical board for the use of biobanked materials TcBIO (Toetsingscommissie Biobanks), an institutional Medical Research Ethics Committee of the University Medical Center Utrecht (protocol ID: 16/586). HNEC were cultured as previously described in ref. 7. In brief, cells (passage 5) were first expanded in 6-well culture plates coated with 50 µg/mL collagen IV (Sigma-Aldrich), using a defined expansion medium composed of: 50% (v/v) Bronchial epithelial cell medium-basal (BEpiCM-b; Sciencell), 23.5 % (v/v) Advanced DMEM F12 (Thermo Fisher), 2% (v/v) B-27 (Thermo Fisher), 1% (v/v) GlutaMAX (Thermo Fisher), 10 mM HEPES (Thermo Fisher), 0.5 μg/mL (±)-Epinephrine hydrochloride (Sigma-Aldrich), 0.5 μg/mL Hydrocortisone (Sigma-Aldrich), 100 nM 3,3′,5-Triiodo-L-thyronine (Sigma-Aldrich), 1.25 mM N-Acetyl-L-cysteine (Sigma-Aldrich), 5 mM Nicotinamide (Sigma-Aldrich), 500 nM SB 202190 (Sigma-Aldrich), 1 μM DMH-1 (Selleck Chemicals), 1 μM A83-01 (Tocris), 5 μM Y-27632 (ROCKi) (Selleck Chemicals), 5 μM DAPT (Fisher Scientific), 25 ng/mL recombinant human FGF-7, 100 ng/mL recombinant human FGF-10, 5 ng/mL recombinant human EGF, 25 ng/mL recombinant human HGF (all Peprotech), 20% (v/v) Rspondin 1 conditioned medium (from Rspo1 cells Cultrex®), 1% (v/v) Penicillin-Streptomycin (Thermo Fisher), and 100 μg/mL Primocin (Invivogen). After reaching confluency, HNEC (0.2 × 106 cells) were seeded on PureCol (Advanced BioMatrix) coated 24-well transwell inserts (0.4 μm pore size polyester membrane, Corning). Cells were first cultured in submerged conditions in expansion medium. Next, confluent monolayers were differentiated as ALI-cultures in differentiation medium, consisting of: 98.5% (v/v) Advanced DMEM F12, 0.5 μg/mL (±)-Epinephrine hydrochloride, 0.5 μg/mL Hydrocortisone, 100 nM 3,3′,5-Triiodo-L-thyronine, 1% (v/v Penicillin-Streptomycin, 50 nM A83-01, 100 nM TTNPB (Cayman), 5 μM DAPT, and 0.5 ng/mL recombinant human EGF. ALI-HNEC cultures were differentiated for at least 18 days before use in experiments.
Viral culture
SARS-CoV-2/human/USA/WA-CDC-WA1/2020 was obtained from the BEI resources. Other SARS-CoV-2 virus strains were cultured from nasal swabs derived from patients from the University Medical Center Utrecht, the Netherlands. SARS-CoV-2/B1.1.7/Alpha/2021, SARS-CoV-2/ B 1.617.2/Delta/2021, SARS-CoV-2/ B.1.351/Beta/2021, SARS-CoV-2/ P.1/Gamma/2021 and SARS-CoV-2/ B.1.1.529/Omicron/2021 were obtained by culturing in Vero E6 cells. 1.67 million Vero E6 cells were cultured in a 25 cm2 filtered tissue flask (T25) in cell line culture medium one day prior to infection. At the day of infection Vero E6 cells were washed in culture flask with PBS (Lonza) and 1 mL infected with viral sample in universal transport medium (UTM) (Copan) was supplemented with 4 mL cell line culture media adapted to 2.5% FCS and 0.1 mg/mL Normicin (Invivogen) and passed through a 0.22 µm Stericup Durapore (Millipore) for 2 h at 37 C 5% CO2. Subsequently, cells were washed with PBS, and 5 mL culture media (2.5% FCS, 0.1 mg/mL Normicin) was added, and cells were incubated at 37 °C 5% CO2 until cytopathic effect (CPE) was observed.
Viral clone generation
Virus clones obtained from the first passage were serially diluted in Vero E6 cells in cell line culture medium by infecting 10,000 cells in a flat-bottom 96-well cell culture plate. The virus was harvested after 5 days when CPE was observed in less than 1/3 of a serial dilution series. This increases the likelihood of clonality. We harvested two clones of SARS-CoV-2/human/USA/WA-CDC-WA1/2020, SARS-CoV-2/B1.1.7/Alpha/2021 and SARS-CoV-2/B1.617.2/Delta/2021 and one clone of SARS-CoV-2/B.1.351/Beta/2021, SARS-CoV-2/P.1/Gamma/2021 and SARS-CoV-2/B.1.1.529/Omicron/2021. Viral load for serial passaging was increased by infecting a T25 tissue culture flask with 1.67 million Vero E6 cells. The virus was harvested when CPE appeared throughout the culture.
Viral passage
From the first passage, 50% tissue culture infectious dose (TCID50) was determined by limiting dilution in Vero E6 cells. One day prior to infection, 10,000 VeroE6 cells/well were seeded in a flat-bottom tissue culture 96-well plate (Costar) in cell line culture media (2.5% FBS). Cells were infected in a limiting dilution in 5-fold dilutions in quadruplicate. Four days post-infection, CPE was scored, and TCID50 was determined.
Serial passaging of the clones of SARS-CoV-2/human/USA/WA-CDC-WA1/2020, SARS-CoV-2/B1.1.7/Alpha/2021 and SARS-CoV-2/B1.617.2/Delta/2021 was performed by infecting 5 million Vero E6 cells in a T75 filtered culture flask at a MOI of 0.1 in 18 mL of cell line culture media (2.5% FBS). Cells were subsequently cultured for 5 days at 37 °C 5% CO2. TCID50 was determined as described above to allow for subsequent infection with an MOI of 0.1. The remainder of the virus was concentrated using 30 kDa centrifugal filter (Amicon) tubes, and viral RNA of the concentrated virus was isolated using a total RNA purification kit (Norgenbiotek) according to the manufacturer's protocol. SARS-CoV-2/human/USA/WA-CDC-WA1/2020, SARS-CoV-2/B1.1.7/Alpha/2021, and SARS-CoV-2/B1.617.2/Delta/2021 were passed seven times.
To compare the mutation rate of SARS-CoV-2/B1.617.2/Delta/2021 between different host cells and culture conditions, clone b from passage 3 was used to infect Calu-3 cells, and SARS-CoV-2/B1.617.2/Delta/2021 clone b from passage 2 was used to infect HNECs. Six ALI filters of HNECs were infected using an MOI of 0.1 for 2 h at the apical level. The virus was washed after 2 h of infection, and the supernatant was pooled and harvested 5 days post-infection. RNA was extracted using a total RNA purification kit (Norgenbiotek). Clones of SARS-CoV-2/B.1.351/Beta/2021, SARS-CoV-2/P.1/Gamma/2021, and SARS-CoV-2/B.1.1.529/Omicron/2021 were used to infect T75 of Vero E6 at an MOI of 0.01, as an MOI of 0.1 was not obtained. This was done for one passage. Virus and viral RNA were harvested as described above.
Library preparation and sequencing
We followed a refined protocol of CirSeq to prepare sequencing libraries3. First, RNA was DNase-treated for 30 min at 37 °C, followed by DNase inactivation (AM1926, Ambion). DNase-treated RNA was quantified with a Qubit RNA HS (High Sensitivity) Assay Kit (Q32852, Invitrogen) and a Qubit 4 Fluorometer (Q33238, Invitrogen). 500 ng RNA was then fragmented using RNase III (AM2290, Ambion) for 9 min at 37 °C. After a clean-up with an Oligo Clean & Concentrator kit (D4061, Zymo Research), RNA fragments were circularized with T4 RNA ligase 1 (M0204S, New England Biolabs) for 2 h at 25 °C. Circular RNA was purified with an Oligo Clean & Concentrator kit and used to generate cDNA with tandem repeats by rolling-circle reverse transcription. For cDNA synthesis, circular RNA was first primed by incubation with random hexamers (N8080127, Thermo Fisher) for 10 min at 25 °C. Then, the reaction was shifted to 42 °C for 20 min to allow for primer extension and cDNA synthesis (18080044, Invitrogen). cDNA was purified with an Oligo Clean & Concentrator kit, and second-strand synthesis was performed with the NEBNext mRNA Second Strand Synthesis Module (E6111S, New England Biolabs). After a clean-up with an Oligo Clean & Concentrator kit, the remaining steps for library preparation were then performed with the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530L, New England Biolabs) and NEBNext Multiplex Oligos for Illumina (New England Biolabs) according to the manufacturer’s guidelines. Size selection and clean-up during sequencing library preparation were performed with AMPure XP Beads (A63881, Beckman Coulter). An Agilent TapeStation (G2991AA, Agilent) with appropriate High Sensitivity ScreenTapes and a Qubit 4 Fluorometer with HS Assay Kits were used for precise sizing and quantification of nucleic acids during different steps of library preparation. Paired-end reads (250nt) were then generated using the Illumina NovaSeq 6000 System (SP Reagent Kit v1.5).
Initial data analysis
Rolling-circle RNA sequencing reads were processed as described in ref. 3, using the pipeline available at https://github.com/jfgout/tr-errors-pipeline-v1/tree/master. Briefly, reads are first trimmed using fastp8 with default parameters except for trimming poly-G ends (minimum size of 6) and requesting a minimum read size of 120 nucleotides after trimming. Trimmed reads are inspected to find repeats and generate a consensus sequence from the stack of repeats. Only positions in the consensus supported by at least three repeats, with all repeats giving the same base call and with a cumulative quality score of at least 100, are considered reliable and are included in the analysis. Consensus sequences are mapped to the reference genome with Kallisto9 followed by a local optimization of the alignment with the seqan2 library10. For every reliable consensus sequence mapped to the genome, we record the genotype supported by the consensus sequence (= a call) to generate a table of all the reliable calls made at every position in the genome.
Exclusion of subgenomic RNA
While our sequencing protocol is not strand-specific, we took significant precautions to ensure that only full-length genomic RNA molecules were collected for analysis. The virions collected after shedding from infected cells are not expected to contain negative-sense RNA strands, as these are replication intermediates that are not typically packaged into virions. To confirm that this is the case, we analyzed the sequencing coverage of the SARS-CoV-2 genome. Importantly, our coverage is evenly distributed between the 5′ and 3′ ends of the genome (see Fig. 2A). If subgenomic RNAs had contaminated our data, we would expect a significant skew in coverage toward the 3′ end of the genome, as ORF1ab (located at the 5′ end) produces very few subgenomic RNAs, while the genes located near the 3′ end generate abundant subgenomic RNAs (see https://doi.org/10.1101/2020.03.05.976167). The absence of such a bias strongly indicates that our sequencing data primarily represent full-length genomic RNA molecules.
As a second test, we examined the mutation spectrum for evidence of contamination by negative-sense RNA strands. If both positive- and negative-sense RNA molecules were present in our dataset, we would expect to observe a symmetrical elevation of complementary mutations. For example, the elevated C → U mutation frequency on the positive-sense strand should be mirrored by an elevated G → A mutation rate on the negative strand, as cytosine-to-uracil (C → U) mutations in the positive strand would correspond to guanine-to-adenine (G → A) mutations on the complementary strand. However, we did not observe this symmetry in our mutation spectrum, further confirming that our data originate exclusively from positive-sense RNA molecules. Thus, all mutation data is presented in the context of the sense strand.
Mutation rate calculations
We downloaded the list of variants published from UShER (8) based on the alignment of about eight million SARS-CoV-2 genomes on November 15th, 2024. Mutations that were never observed among the eight million genome alignment were considered lethal and retained for the calculation of the mutation rate. To prevent a small subset of non-lethal mutations missed by the alignment from biasing the result, we also excluded mutations that occurred at a frequency of more than 0.01% and therefore restricted our analysis to genomic positions covered by at least 1000 reliable consensus sequences. Mutation rates were then calculated, using the genomic sites that passed these criteria, for each of the twelve possible base-substitutions by dividing the number of calls supporting the base-substitution considered by the total number of calls at the genomic position with the nucleotide considered.
Calculation of equilibrium for base frequencies
Equilibrium frequencies were calculated by simulating the genome evolution for 1,000,000 generations using the mutation rate of each possible base substitution. Equilibrium was confirmed by the lack of changes in nucleotide composition in subsequent generations of the simulation.
Selection of mutations for fitness value calculations
Rare mutations may be absent from the subset of viral particles used to seed the next passage. Under this scenario, the bottleneck between each passage would reset the frequency of these mutations to zero, so that only new mutations generated in the current passage could be detected, mimicking the pattern expected for lethal mutations. This would result in a systematic underestimation of the fitness values for these mutations occurring at low frequency. Therefore, we transfer approximately half a million viral particles between each passage. We only included C → U mutations in our fitness analysis that occur at a rate greater than 1 × 10−5. At this frequency, at least five mutations should be transferred on average from one passage to the next.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RAW sequencing reads have been deposited at the NCBI Short Read Archive database under project ID: PRJNA1135749. A pre-compiled table of all mutation frequencies is available for download from GitHub at https://github.com/jfgout/SARS-CoV-2/tree/main/data.
Code availability
All the code required to reproduce these analyses is available on GitHub at: https://github.com/jfgout/SARS-CoV-2.
References
Tonkin-Hill, G. et al. Patterns of within-host genetic diversity in SARS-CoV-2. Elife. https://doi.org/10.7554/eLife.66857 (2021).
Peck, K. M. & Lauring, A. S. Complexities of viral mutation rates. J. Virol. https://doi.org/10.1128/JVI.01031-17 (2018).
Minoche, A. E., Dohm, J. C. & Himmelbauer, H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 12, R112, (2011).
Acevedo, A. & Andino, R. Library preparation for highly accurate population sequencing of RNA viruses. Nat. Protoc. 9, 1760–1769 (2014).
Acevedo, A., Brodsky, L. & Andino, R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505, 686–690 (2014).
Whitfield, Z. J. et al. Species-specific evolution of ebola virus during replication in human and bat cells. Cell Rep. 32, 108028 (2020).
Dolan, P. T. et al. Principles of dengue virus evolvability derived from genotype-fitness maps in human and mosquito cells. Elife. https://doi.org/10.7554/eLife.61921 (2021).
Grass, V. et al. Adaptation to host cell environment during experimental evolution of Zika virus. Commun. Biol. 5, 1115 (2022).
Jefferson, T., Spencer, E. A., Brassey, J. & Heneghan, C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin. Infect. Dis. 73, e3884–e3899 (2021).
Denison, M. R., Graham, R. L., Donaldson, E. F., Eckerle, L. D. & Baric, R. S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 8, 270–279 (2011).
McBroome, J. et al. A daily-updated database and tools for comprehensive SARS-CoV-2 mutation-annotated trees. Mol. Biol. Evol. 38, 5819–5824 (2021).
Turakhia, Y. et al. Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat. Genet. 53, 809–816 (2021).
Harrison, P. W. et al. Ensembl 2024. Nucleic Acids Res. 52, D891–D899 (2024).
Sung, W. et al. Asymmetric context-dependent mutation patterns revealed through mutation-accumulation experiments. Mol. Biol. Evol. 32, 1672–1683 (2015).
Rice, A. M. et al. Evidence for strong mutation bias toward, and selection against, U content in SARS-CoV-2: implications for vaccine design. Mol. Biol. Evol. 38, 67–83 (2021).
Moeller, N. H. et al. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. USA. https://doi.org/10.1073/pnas.2106379119 (2022).
Chung, C. et al. Evolutionary conservation of the fidelity of transcription. Nat. Commun. 14, 1547 (2023).
Fritsch, C. et al. Genome-wide surveillance of transcription errors in response to genotoxic stress. Proc. Natl. Acad. Sci. USA. https://doi.org/10.1073/pnas.2004077118 (2021).
Gout, J. F. et al. The landscape of transcription errors in eukaryotic cells. Sci. Adv. 3, e1701484 (2017).
Obermeyer, F. et al. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science 376, 1327–1332 (2022).
Bloom, J. D. & Neher, R. A. Fitness effects of mutations to SARS-CoV-2 proteins. Virus Evol. 9, vead055 (2023).
Lan, T. C. T. et al. Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. Nat. Commun. 13, 1128 (2022).
Simmonds, P. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses. mBio. https://doi.org/10.1128/mBio.01661-20 (2020).
Sun, L. et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell 184, 1865–1883.e1820 (2021).
Garg, A. & Heinemann, U. A novel form of RNA double helix based on G.U and C.A(+) wobble base pairing. RNA 24, 209–218 (2018).
Frederico, L. A., Kunkel, T. A. & Shaw, B. R. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 29, 2532–2537 (1990).
Lindahl, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993).
Fryxell, K. J. & Zuckerkandl, E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol. Biol. Evol. 17, 1371–1383 (2000).
Stavrou, S. & Ross, S. R. APOBEC3 proteins in viral immunity. J. Immunol. 195, 4565–4570 (2015).
Smith, H. C. APOBEC3G: a double agent in defense. Trends Biochem. Sci. 36, 239–244 (2011).
Sharma, S. & Baysal, B. E. Stem-loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. PeerJ 5, e4136 (2017).
Rodriguez-Rivas, J., Croce, G., Muscat, M. & Weigt, M. Epistatic models predict mutable sites in SARS-CoV-2 proteins and epitopes. Proc. Natl. Acad. Sci. USA. https://doi.org/10.1073/pnas.2113118119 (2022).
Han, W. et al. Predicting the antigenic evolution of SARS-COV-2 with deep learning. Nat. Commun. 14, 3478 (2023).
Bradley, C. C. et al. Targeted accurate RNA consensus sequencing (tARC-seq) reveals mechanisms of replication error affecting SARS-CoV-2 divergence. Nat. Microbiol. 9, 1382–1392 (2024).
Amicone, M. et al. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol. Med. Public Health 10, 142–155 (2022).
Cuevas, J. M., Domingo-Calap, P. & Sanjuan, R. The fitness effects of synonymous mutations in DNA and RNA viruses. Mol. Biol. Evol. 29, 17–20 (2012).
Lauring, A. S., Acevedo, A., Cooper, S. B. & Andino, R. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe 12, 623–632 (2012).
Tubiana, L., Bozic, A. L., Micheletti, C. & Podgornik, R. Synonymous mutations reduce genome compactness in icosahedral ssRNA viruses. Biophys. J. 108, 194–202 (2015).
Zanini, F., Puller, V., Brodin, J., Albert, J. & Neher, R. A. In vivo mutation rates and the landscape of fitness costs of HIV-1. Virus Evol. 3, vex003 (2017).
Le Nouen, C. et al. Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc. Natl. Acad. Sci. USA 111, 13169–13174 (2014).
Tulloch, F., Atkinson, N. J., Evans, D. J., Ryan, M. D. & Simmonds, P. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. Elife 3, e04531 (2014).
Domingo-Calap, P., Cuevas, J. M. & Sanjuan, R. The fitness effects of random mutations in single-stranded DNA and RNA bacteriophages. PLoS Genet. 5, e1000742 (2009).
Carrasco, P., de la Iglesia, F. & Elena, S. F. Distribution of fitness and virulence effects caused by single-nucleotide substitutions in Tobacco etch virus. J. Virol. 81, 12979–12984 (2007).
Watts, J. M. et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716 (2009).
Pollom, E. et al. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 9, e1003294 (2013).
Burrill, C. P. et al. Global RNA structure analysis of poliovirus identifies a conserved RNA structure involved in viral replication and infectivity. J. Virol. 87, 11670–11683 (2013).
Romero-Lopez, C. & Berzal-Herranz, A. A long-range RNA-RNA interaction between the 5’ and 3’ ends of the HCV genome. RNA 15, 1740–1752 (2009).
Nicolas Calderon, K., Fabian Galindo, J. & Bermudez-Santana, C. I. Evaluation of Conserved RNA Secondary Structures within and between Geographic Lineages of Zika Virus. Life. https://doi.org/10.3390/life11040344 (2021).
Fareh, M. et al. Reprogrammed CRISPR-Cas13b suppresses SARS-CoV-2 replication and circumvents its mutational escape through mismatch tolerance. Nat. Commun. 12, 4270 (2021).
Becker, J. et al. Ex vivo and in vivo suppression of SARS-CoV-2 with combinatorial AAV/RNAi expression vectors. Mol. Ther. 30, 2005–2023 (2022).
Acknowledgements
We thank Prof. J.M. Beekman (Hubrecht Institute, Utrecht, the Netherlands) for providing Calu E-3 cells. We thank Dr. Jeffrey Jensen for his comments on an earlier draft of this manuscript. This work was supported by a National Science Foundation award 2032784, RAPID: Probing SARS-CoV-2 evolution and vulnerabilities through its mutation and fitness landscape (M.V. and J.-F.G.) and a Health Holland LSHM20058 Clear Covid-19 project, ZonMw 114025009 (J.S. and M.N.).
Author information
Authors and Affiliations
Contributions
Conceptualization: J.-F.G., M.V., J.S. and M.N. Formal analysis: J.-F.G. Software: J.-F.G. Methodology: J.-F.G., M.V., J.S. and M.N. Investigation: J.-F.G., J.S., C.C., B.M.V., S.S., D.d.J., G.A., M.N. and M.V. Visualization: J.-F.G. and M.V. Funding acquisition: J.-F.G., M.V. and M.N. Project administration: J.-F.G., M.V., J.S. and M.N. Supervision: J.-F.G., M.V. and M.N. Writing—original draft: J.-F.G., M.V., J.S. and M.N. Writing—review and editing: J.-F.G., M.V., J.S., and M.N.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Raul Andin, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Symons, J., Chung, C., Verheijen, B.M. et al. The mutational landscape of SARS-CoV-2 provides new insight into viral evolution and fitness. Nat Commun 16, 6425 (2025). https://doi.org/10.1038/s41467-025-61555-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61555-x