Abstract
Patients with locally advanced resectable (LAR) gastric/gastroesophageal junction (G/GEJ) adenocarcinomas have a high recurrence risk despite pre- and post-operative treatment. In the PLAGAST prospective study (NCT02674373), we investigated the ability of circulating tumor DNA (ctDNA) to predict treatment response and improve risk stratification. Plasma samples were prospectively collected before neoadjuvant therapy (NAT), during-NAT, post-NAT, and post-surgery. The primary endpoint was recurrence-free survival (RFS), and the secondary endpoints were overall survival (OS), tumor regression grade (TRG), and pathological tumor stage. ctDNA positivity decreased over these four therapeutic timelines (69.6%, 51.2%, 26.8%, and 20%, respectively). ctDNA-positivity was associated with significantly worse outcomes during-NAT (RFS: HR = 6.17, P = 0.002; OS: HR = 4.71, P = 0.022), post-NAT (RFS: HR = 5.26, P = 0.001; OS: HR = 7.35, P = 0.001) and after surgery (RFS: HR = 12.94, P < 0.0001; OS: HR = 14.54, P < 0.0001). Patients with early ctDNA clearance during NAT had better outcomes compared to those who cleared ctDNA post-NAT, while patients who remained ctDNA-positive pre-, during-, and post-NAT had worse outcomes (RFS: HR = 18.57, P = 0.01; OS: HR = 16.06, P = 0.007). Our data suggests that longitudinal ctDNA monitoring is prognostic of patient outcomes and may guide therapeutic decision-making in patients with LAR G/GEJ adenocarcinoma.
Similar content being viewed by others
Introduction
Gastric and gastroesophageal junction (G/GEJ) adenocarcinomas are associated with a dismal prognosis and often present with advanced disease at diagnosis1. Surgical resection of localized disease is potentially curative; however, most patients relapse following surgery, highlighting the need for combinatory modality therapies and reliable predictors of recurrence, recommended for locally advanced resectable (LAR) G/GEJ adenocarcinoma2,3.
In the pivotal, controlled, phase 2/3 FLOT4 trial, the FLOT regimen (fluorouracil, leucovorin, oxaliplatin, and docetaxel) reported significantly improved overall survival (OS) (median OS: 50 vs. 35 months) compared to the previous standard ECF/ECX (epirubicin, cisplatin, 5-fluorouracil/ epirubicin, cisplatin, capecitabine), supporting the incorporation of FLOT regimen as the new standard perioperative treatment for LAR G/GEJ cancer2. Recently, the ESOPEC trial has compared perioperative FLOT versus neoadjuvant chemoradiation according to the CROSS protocol (41.4 Gy plus carboplatin/paclitaxel) in patients with LAR esophageal adenocarcinoma. In this trial, the perioperative FLOT chemotherapy was associated with a significant improvement of OS (median OS: 66 vs. 37 months) and a higher pathological complete response (pCR) rate (16.8% vs. 10%)4. Furthermore, recently, immune checkpoint inhibitor (ICI) has also emerged as a promising therapeutic approach in gastroesophageal adenocarcinoma. The phase III Checkmate 577 trial demonstrated a significant improvement in disease-free survival with adjuvant nivolumab compared to placebo in patients who had received neoadjuvant chemoradiation for esophageal cancer, including GEJ adenocarcinoma5. In gastric cancer, ongoing randomized clinical trials (DANTE, MATTERHORN) evaluate the benefits of using ICI in the perioperative setting in addition to FLOT6,7. However, despite the advances in multimodal treatment approaches that have improved patient survival, recurrence risk remains high. The clinical incorporation of effective biomarkers to assess treatment efficacy early on, predict recurrence, or monitor response to therapy would benefit clinical decision-making and thus warrants investigation in future similar trials.
Many studies across tumor types have demonstrated the utility of circulating tumor DNA (ctDNA) in assessing molecular residual disease (MRD) to guide treatment decisions and predict recurrence8,9,10,11. Using a personalized, tumor-informed ctDNA assay for patients with esophagogastric cancers, longitudinal assessment of ctDNA allowed for enhanced patient risk stratification and prognostication during curative-intent therapy in several recent studies12,13,14.
Here, we present results from the PLAGAST study, evaluating the association of ctDNA dynamics with prognosis and treatment response in patients with LAR G/GEJ adenocarcinoma.
Results
Patient cohort
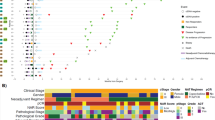
A total of 82 eligible patients with G/GEJ adenocarcinoma were identified. Of these, one presented with two primary cancers, fifteen cases failed QC testing or WES generation, and four patients had metastatic disease at diagnosis and were excluded (Fig. 1). Sixty-two patients had annotated clinical and ctDNA data available (median age 66 years, range: 34−86 years; 63% male) (Table 1). Patients were followed up for a median of 29 (range: 2−93) months. Radiological recurrence was observed in 47% of patients (29/62) and 69% of patients (43/62) were alive at the end of the follow-up. Demographics, clinical characteristics, and genomic profiling data for the entire clinical cohort are listed in Table 1, Supplementary Data 1, and Supplementary Fig. 1.
ctDNA detection rates at various time points for the evaluable patient population (N = 62) are also shown. ACT adjuvant chemotherapy, ctDNA circulating tumor DNA, G/GEJ gastric/gastroesophageal junction, MRD molecular residual disease, NAT neoadjuvant therapy, QC quality control, WES whole-exome sequencing.
ctDNA detection rates, quantification at pre-NAT, and association with outcomes
ctDNA detection rates at pre-NAT, during-, and post-NAT were 69.6% (39/56), 51.2% (21/41), and 26.8% (11/41), respectively, across all stages. In patients analyzed during the MRD window, the overall ctDNA detection rate was 20% (10/50). A significantly higher rate of ctDNA positivity was observed among stage II and III compared to stage I patients (Fig. 1).
At the pre-NAT time point, ctDNA-positive patients showed a trend towards inferior RFS compared to those who tested ctDNA-negative. However, this analysis was not statistically significant (HR = 2.51, 95% CI: 0.94-6.72, P = 0.068; median (m) RFS 22.3 months vs. not reached [NR], respectively). Similar results were observed for OS (HR = 2.09, 95% CI: 0.67-6.48, P = 0.203) (Supplementary Fig. 2A, B). However, when evaluating MTM/mL ctDNA values on ctDNA-positive patients at pre-NAT (N = 35, excluding patients that did not receive NAT), we observed that patients with higher MTM/mL (≥0.71 MTM/mL, based on the AUC value for recurrence), experienced poorer RFS (HR = 4.61, 95% CI: 1.05-20.25; P = 0.043) (Supplementary Fig. 2C).
ctDNA status during- and post-NAT and association with outcomes
The median duration of NAT was 43 days (Q1–Q3: 42–56 days, IQR: 14 days), the median time from NAT initiation to blood draw for ctDNA analysis during NAT was 14 days (Q1–Q3: 14–28 days, IQR: 14 days), and the median time from NAT completion to blood draw for ctDNA analysis post-NAT was 0 days (Q1–Q3: 0–12 days, IQR: 12 days). Of the 41 patients evaluated during NAT, 51% (21/41) were ctDNA-positive and compared to ctDNA-negative patients (49%, 20/41) experienced a significantly inferior RFS (HR = 6.17, 95% CI: 1.99-19.12, P = 0.002; mRFS 13.3 months for ctDNA-positive patients vs. NR in ctDNA-negative patients) and OS (HR = 4.71, 95% CI: 1.24-17.86, P = 0.022; mOS 24.5 months for ctDNA-positive patients vs. NR in ctDNA-negative patients) (Fig. 2A, B). Similarly, when evaluating ctDNA status post-NAT, we observed that patients who were ctDNA-positive exhibited a significant reduction in RFS (RFS: HR = 5.26, 95% CI: 1.96-14.12, P = 0.001; mRFS 7.79 months for ctDNA-positive patients vs. NR in ctDNA-negative patients) and OS (HR = 7.35, 95% CI: 2.35-22.95, P = 0.001; mOS 18.9 months for ctDNA-positive patients vs. NR in ctDNA-negative patients) (Fig. 2C, D). Multivariate analysis post-NAT showed ctDNA status to be the most significant independent risk factor for both RFS and OS when compared with other clinicopathological factors (P < 0.001) (Fig. 2E, F).
Kaplan–Meier estimates of patients with LAR G/GEJ adenocarcinoma representing (A) RFS & (B) OS stratified by ctDNA status during NAT. C RFS & (D) OS stratified by ctDNA status post-NAT. HRs and 95% CIs were calculated using the Cox proportional hazard model; P-values were calculated using the two-sided log-rank test. Forest plot depicting multivariate analysis for RFS (E) and OS (F) in patients with LAR G/GEJ adenocarcinoma post-NAT. P = 0.000467 (E), P = 0.000735 (F). Various prognostic factors and their association with RFS/OS, as indicated by HR, were analyzed across the cohort using the two-sided Wald chi-squared test. The unadjusted HRs (squares) and 95% CIs (horizontal lines) are shown for each prognostic factor; the vertical dotted line represents the null hypothesis (E, F). ctDNA circulating tumor DNA, Cl Confidence interval, GEJ gastroesophageal junction, HR Hazard ratio, NAT neoadjuvant therapy, NR not reached, RFS Recurrence-free survival, OS Overall survival.
ctDNA dynamics pre-, during-, and post-NAT and association with outcomes
Among 22 patients with ctDNA status available at pre-NAT, during-, and post-NAT time points, 86% (6/7) with persistent positive ctDNA status, experienced a clinical relapse. Overall, patients who remained ctDNA-positive at all time points showed a significantly higher risk of recurrence compared to patients who converted negative at subsequent time points (mRFS 6.2 m, HR = 18.57, 95% CI: 1.92-179.22, P = 0.012 and OS: HR = 16.06, 95% CI: 1.87-2102, P = 0.007, respectively), (Fig. 3A, B).
Kaplan–Meier estimates of patients with LAR G/GEJ adenocarcinoma representing (A) RFS & (B) OS stratified by ctDNA dynamics pre-, during- (on-NAT), and post-NAT. HRs and 95% CIs were calculated using the Cox proportional hazard model; P-values were calculated using the two-sided log-rank test. For (B) the HR value was based on Firth’s penalized maximum likelihood. C Violin plot showing pre-NAT MTM/mL levels for ctDNA dynamics pre-, during-, and post-NAT categories. N denotes the number of patients within each subgroup of ctDNA dynamics (ctDNA + /−/−, ctDNA + /+/−, ctDNA + /+/+). Center line: median MTM/ml; bottom of box: 25% quantile; top of box: 75% quantile; data points represent outliers. P-values were calculated using the two-sided Wilcoxon Rank Sum test. D The contingency plot shows the association of ctDNA dynamics pre-, during-, and post-NAT with the TRG score. The p-value for the bar plot was calculated based on the two-sided chi-square test. The TRG score was unavailable for one patient in the ctDNA + /−/− subgroup. ctDNA circulating tumor DNA, Cl Confidence interval, HR Hazard ratio, MTM mean tumor molecules, NAT neoadjuvant therapy, NR not reached, RFS Recurrence-free survival, OS Overall survival.
Of note, we observed that mMTM/mL levels were significantly higher in patients who remained persistently positive at all the time points (mMTM/mL: 34.63) compared to patients who subsequently converted negative at the post-NAT time point (mMTM/mL: 0.96) (P = 0.0015) (Fig. 3C). All of these patients with persistent ctDNA-positivity (7/7) had poor pathological response with a higher TRG 4/5 grade disease compared to TRG 1/2/3 (P = 0.035). In contrast, all patients with early ctDNA clearance (during-NAT timepoint; N = 4/4) correlated with TRG 1/2/3 (Fig. 3D).
Postoperative ctDNA status during MRD window and association with outcomes
For survival analysis, 47 and 50 patients within the MRD window were included with ctDNA data for RFS and OS analysis, respectively. The median time from surgery to blood draw at the MRD time point was 41 days (Q1 – Q3: 30 – 47 days, IQR: 17 days). ctDNA-positivity was associated with a significantly shorter RFS (HR: 12.94, 95% CI: 4.23−39.59; P < 0.0001; mRFS of 3.57 months for ctDNA-positive vs. NR for ctDNA-negative patients), and OS (HR: 14.54, 95% CI: 4.54-46.6, P < 0.0001, mOS of 8.59 months for ctDNA-positive vs. NR for ctDNA-negative patients) (Fig. 4A, B). Of the 10 ctDNA-negative patients that recurred, four patients had their last ctDNA test 12 months (median: 10.51 months; range: 15.8-38.54 months) from radiologic evidence of recurrence. We also conducted a sensitivity analysis for the prognostic value of ctDNA within the MRD window for RFS and OS in patients who received NAT (Supplementary Fig. 3A, B) and adjuvant treatment (Supplementary Fig. 3C, D). ctDNA positivity was observed to be highly prognostic for RFS (NAT-treated: HR = 9.34, 95% CI: 2.8−31.08, P < 0.0001; adjuvant treatment: HR = 11.88, 95% CI: 3.76−37.57, P < 0.0001) and OS (NAT-treated: HR = 10.86, 95% CI: 3.21-36.78, P < 0.0001; adjuvant treatment: HR = 10.88, 95% CI: 3.03-39.08, P < 0.0001) in both these settings.
Kaplan–Meier estimates of patients with LAR G/GEJ adenocarcinoma representing (A) RFS & B) OS stratified by ctDNA status within the MRD window. P = 7.17e-06 (A), P = 6.64e-06 (B). C RFS & (D) OS stratified by ctDNA status within the MRD window and ypT stage. P = 9.62e-05 (blue vs. green, C), P = 2.89e-05 (blue vs. red, C), P = 1.1e-06 (blue vs. red, D). E RFS & F OS stratified by ctDNA status within the MRD window and ypN stage. P = 0.000114 (blue vs. green, E), P = 1.38e-06 (blue vs. red, E), P = 2.01e-05 (blue vs. green, F), P = 9.53e-05 (blue vs. red, F). G RFS & (H) OS stratified by ctDNA status within the MRD window and TRG score. P = 0.000647 (blue vs. red, G), P = 1.93e-05 (blue vs. red, H). HRs and 95% CIs were calculated using the Cox proportional hazard model. P-values were calculated using the two-sided log-rank test. The RFS and OS analyses in the MRD window were landmarked from the date of the MRD time point (12 weeks post-surgery). Median RFS/OS and percentage RFS and OS were estimated from the landmark timepoint. I, J Forest plot depicting multivariate analysis for RFS (I) and OS (J) in patients with LAR G/GEJ within the MRD window. Various prognostic factors and their association with RFS/OS, as indicated by HR, were analyzed across the cohort using the two-sided Wald chi-squared test. The unadjusted HRs (squares) and 95% CIs (horizontal lines) are shown for each prognostic factor; the vertical dotted line represents the null hypothesis (I, J). Abbreviations: ctDNA circulating tumor DNA, Cl Confidence interval, NAT neoadjuvant therapy, GEJ gastroesophageal junction, HR Hazard ratio, MRD molecular residual disease, NR not reached, RFS Recurrence-free survival, OS Overall survival.
On evaluating both tumor pathological stage (ypT or ypN) and ctDNA status within the MRD window, we observed that patients who tested ctDNA-positive had worse outcomes irrespective of ypT or ypN stage. Specifically, patients who tested ctDNA-positive and had ypT4 disease had the worst prognosis (RFS: HR = 32.90, 95% CI: 6.4-169.13, P < 0.0001; OS: 90.76, 95% CI: 14.80-556.40, P < 0.0001) followed by ctDNA-positive patients with ypT1-T3 disease (RFS: HR = 14.61, 95% CI: 3.8-56.23, P < 0.0001; OS: HR = 12.57, 95% CI: 2.38-66.49, P = 0.0029). Notably, patients with ypT4 disease who were ctDNA-negative had better outcomes than those with ypT1-T3 disease who were ctDNA-positive, but poorer outcomes compared to the ypT1-3/ ctDNA negative group (Fig. 4C, D). ctDNA positivity was also associated with worse outcomes in both ypN0 and ypN+ patients, and similarly, patients who were ctDNA negative/ ypN+ had better outcomes than ctDNA positive patients regardless of their ypN status, but worse than patients who were ctDNA negative/ ypN0 (Fig. 4E, F). Consistently, within the MRD window, patients who were ctDNA-negative and ypT1-T3N0 had favorable RFS and OS compared to ctDNA-positive, or ypT4, or ypN+ patients (Supplementary Fig. 4). In addition, we evaluated the timing of molecular recurrence relative to anatomic recurrence (i.e. ctDNA turning positive relative to radiographic evidence of recurrence post-MRD until the last clinical follow-up), and we observed that ctDNA detected recurrence with a median of 184 (range 2–323) days ahead of imaging.
When utilizing ctDNA status as an adjunct to the TRG score within the MRD window, we observed that patients who were ctDNA-positive with TRG scores 4/5 had higher rates of recurrence (100% [3/3]) and inferior RFS and OS when compared to those who were TRG 1/2/3 and ctDNA-negative (RFS: HR = 20.47, 95% CI: 3.61-116, P < 0.001; OS: HR = 23.62, 95% CI: 5.54-100.75, P < 0.0001). ctDNA-negative TRG 4/5 patients had a non-statistically significant inferior RFS and OS compared to ctDNA-negative TRG 1/2/3 (Fig. 4G, H). Multivariate analysis during the MRD window also showed ctDNA status to be a significant independent risk factor for recurrence (P = 0.002, P = 0.006) (Fig. 4I, J).
Discussion
In this initial analysis of the PLAGAST observational study, we utilized blood samples from 62 patients with LAR G/GEJ adenocarcinomas for retrospective ctDNA analysis at pre-NAT, during-NAT, post-NAT, and post-surgical MRD window. We report that ctDNA-positivity at these time points was highly prognostic of inferior RFS and OS. Additionally, patients who showed early ctDNA clearance during NAT had better outcomes than those who cleared only at the post-NAT time point or remained persistently positive during- and post-NAT. Overall, ctDNA positivity emerged as the most significant independent risk factor associated with worse clinical outcomes in a multivariate analysis at post-NAT, and within the MRD window (RFS/OS post-NAT: P < 0.001) (MRD: RFS, P < 0.001; OS, P = 0.006).
Recent reports have demonstrated the prognostic and predictive value of ctDNA assessment for MRD in esophagogastric cancers. Findings from the phase 2 PANDA trial, in which patients with LAR G/GEJ adenocarcinoma received one cycle of monotherapy atezolizumab followed by four cycles of atezolizumab combined with docetaxel, oxaliplatin, and capecitabine, highlight the potential clinical utility of presurgical ctDNA status and its association with pathological response and recurrence risk14. ctDNA levels were significantly higher in patients who did not respond to NAT when compared to responders (P = 0.0065). Furthermore, none of the ctDNA-positive patients after NAT obtained pCR and had a higher recurrence risk than ctDNA-negative patients. Lastly, MRD-positivity after NAT and surgical resection was associated with 100% rapid recurrence <6 months from surgery. These data indicate a clear correlation between ctDNA status and pathologic response to NAT.
In an exploratory analysis of the phase 2 ICONIC trial (NCT03399071), ctDNA monitoring was used to predict recurrence and determine the efficacy of FLOT plus avelumab both before and after surgery in LAR G/GEJ adenocarcinoma15. Post-neoadjuvant chemotherapy (NAC)/pre-surgical ctDNA positivity correlated with worse pathological response. Post-surgical persistent ctDNA positivity after NAC and surgery was the strongest predictor of recurrence (HR = 27; P < 0.0001) with 83% (5/6) of the patients showing clinical recurrence, compared to parameters such as lymph node status or TRG in the resection specimen. Moreover, postoperative adjuvant therapy failed to clear ctDNA in any ctDNA-positive patients, indicating the need to test new adjuvant therapies in these patients who remain ctDNA-positive after surgery. ctDNA-based MRD assessment in the post-NAC and post-surgical setting in esophageal squamous cell carcinoma was evaluated by Takei et al. where ctDNA-positivity post-NAC and post-surgery was associated with a significantly shorter RFS (post-NAC: HR = 3.3; P = 0.05; post-surgery: HR = 14; P < 0.001), underscoring the potential clinical utility of using ctDNA to direct post-surgical management in these patients16. The prognostic value of longitudinal ctDNA monitoring postoperatively has also previously been explored by Huffman et al. in 212 patients with stage I-III esophagogastric cancers12. ctDNA detection post-surgery within the MRD window (HR = 10.7; P < 0.0001) was associated with shorter RFS. Our findings here are consistent and build upon previous evidence, showing that early ctDNA clearance in the neoadjuvant setting is the best predictor of RFS and OS, while patients who failed to clear their ctDNA during- and post-NAT showed a significantly higher risk of recurrence and death (RFS: HR = 18.57; P = 0.012 and OS: HR = 16.06; P = 0.007). Furthermore, all these patients with persistent ctDNA-positivity had poor pathological responses and higher TRG 4/5 grade disease.
Furthermore, our data also demonstrate that ctDNA positivity within the MRD window is associated with a significantly shorter RFS (mRFS 3.57 m, HR: 12.94; P < 0.0001) and OS (mOS 8.59 m, HR: 14.54; P < 0.0001). Our data also showed that postoperative ctDNA status can further refine the prognostic value of pathological tumor staging. As noted above, ypT4 who were MRD-negative had superior 2-year RFS rate compared to ypT1-3 MRD-positive patients. Importantly, the 6-month lead time of molecular recurrence ahead of clinical recurrence provides a window of opportunity to intensify imaging surveillance to capture oligometastatic recurrence, wherein several studies have shown improved survival with local therapy in addition to systemic therapy compared to systemic therapy alone17,18,19.
As noted in other trials, all patients who remained ctDNA-positive after surgery had a 100% recurrence rate with a mRFS of ~3.6 m and mOS of ~8.6 m. These data support the clinical utility of ctDNA in the MRD setting, where ctDNA positivity predicts the futility of adjuvant systemic therapy when using the same pre-operative regimen and highlights the need for a therapy switch approach in those patients. Our findings also support incorporating ctDNA-guided treatment escalation and de-escalation options in future prospective studies and clinical trials in the perioperative setting. Two such trials were recently launched, the DECIPHER20 and TRINITY21 trials, investigating trastuzumab-deruxtecan (T-DXd) either alone or in combination with capecitabine/5-fluorouracil as an adjuvant therapy regimen for patients with Her-2 positive LAR G/GEJ adenocarcinoma and positive post-operative ctDNA status. Conversely, a de-escalation approach could be evaluated for patients with postoperative ctDNA negativity, particularly those with localized disease.
While the PLAGAST study was conducted prospectively, the ctDNA analysis performed was retrospective, which is a limitation. Although the median follow-up of 29 months is long enough for OS analysis, continued follow-up is necessary to determine the long-term longitudinal impact of ctDNA status on OS. Tumor tissue samples used for WES were from surgical resection or biopsy if surgical tissue was unavailable. The most common reason for WES QC failures in this cohort was insufficient tumor volume; however, no significant differences were found between any of the clinicopathological factors, recurrence rates, and median follow-up between the subgroup of patients included and those excluded. Furthermore, although we report a median lead time of 184 days, we acknowledge that this is specific to the imaging frequency and clinical follow-up as outlined in the protocol. Another potential shortcoming is that this study is observational; future interventional trials are warranted to support actionability on ctDNA results for clinical decisions and adopt ctDNA-guided treatment intervention to improve outcomes for this patient population. There is a need for larger, potentially multi-institutional cohorts and international validation studies to confirm the findings of this study.
In summary, our data highlight the prognostic and predictive value of tumor-informed ctDNA results for NAT response and its correlation with long-term outcomes. Early ctDNA clearance was associated with superior survival outcomes; meanwhile, persistent ctDNA positivity post-NAT/pre-surgery/ at MRD correlated with inferior outcomes and poor pathological response. This initial analysis of the PLAGAST study demonstrates the clinical utility of ctDNA in prognosticating outcomes, supporting its potential use to inform treatment decisions or prompt early radiographic imaging. Given its minimally invasive approach, ctDNA-based testing offers an advantage over conventional methodologies for monitoring therapeutic response. Risk stratification using MRD-based ctDNA assay has the potential to truly personalize precision medicine by facilitating informed decision-making for patients with LAR G/GEJ adenocarcinoma.
Methods
Patients and study design
PLAGAST is a prospective, observational biobanking study in patients with LAR (≥cT2 and/or cN + ) G/GEJ adenocarcinoma. Eligible patients included those aged over 18 years with histologically proven adenocarcinoma of G/GEJ and receiving treatment for localized tumor stage (surgical resection associated with perioperative chemotherapy, adjuvant chemotherapy, or preoperative chemoradiotherapy) at the European Georges Pompidou Hospital (Paris, France).
Patients were enrolled over 2 years and were excluded for one or more of the following reasons: if they were unable to undergo medical monitoring for geographical, social, or psychic reasons, patients under guardianship or unable to read, understand, and sign the information sheet and consent form, and any non-affiliated to the French social security institution. This study was sponsored by the Association des Gastro-Entérologues Oncologues (AGEO) - Gastroenterologists Oncologists Association and was conducted per the Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice. All participants provided written informed consent. This study is registered at ClinicalTrials.gov (NCT03079427).
Biospecimen collection and processing, and personalized ctDNA assay
Tumor DNA was extracted from formalin-fixed paraffin-embedded (FFPE) specimens, and plasma samples were prospectively collected. For germline analysis, genomic DNA was extracted from whole blood collected in EDTA tubes. Plasma samples (n = 280) were collected between 07/2015 and 12/2022 from 62 patients with LAR G/GEJ adenocarcinoma at baseline (anytime post-diagnostic biopsy/enrollment and before NAT or before surgery for NAT-naive), during-NAT, post-NAT, and post-surgery at the molecular residual disease (MRD) window (within 2−12 weeks, before adjuvant treatment). Retrospective ctDNA analysis was performed using a personalized, tumor-informed 16-plex mPCR-NGS assay (SignateraTM, Natera, Inc.)11. Briefly, whole-exome sequencing (WES) was performed on DNA extracted from FFPE tumor tissue from a core needle biopsy or surgical resection of primary tumor tissue along with matched normal blood samples from each patient. Based on WES results, up to 16 patient-specific, somatic, single nucleotide variants (SNVs) were selected for mPCR-NGS testing in the plasma cell-free DNA (cfDNA) of the respective patient. cfDNA was extracted from a median of 3.2 mL (range: 1.8−5.2 mL) of plasma. Detection of 2 or more SNVs above a predefined statistical algorithm confidence threshold was considered ctDNA-positive. ctDNA concentration (levels) was reported as mean tumor molecules per mL of plasma (MTM/mL).
Statistical analysis
The primary endpoint was recurrence-free survival (RFS), defined as the time from surgery until radiological or clinical recurrence. Recurrence was determined based on diagnostic imaging or any other diagnostic procedure if imaging was not confirmative (e.g., gastroscopy to diagnose local recurrence). The secondary endpoints were OS, tumor regression grade (TRG), and pathological tumor stage. OS was defined as the time between the date of surgery and the date of death due to any cause or latest clinical follow-up. TRG was defined according to the Mandard classification22. To account for potential immortal time bias, analyses for the prognostic value of ctDNA at the MRD Window were landmarked at 12 weeks post-surgery. Patient characteristics were summarized using descriptive statistics, and statistical significance was evaluated using Fisher’s exact test or Chi-square test for categorical variables and Wilcoxon rank sum two-sided test for continuous variables, as indicated in corresponding figure legends. Survival analyses were conducted using R software v4.4.0 using packages survminer (v0.4.9) and survival (v3.2.13). Survival curves were compared using the Kaplan-Meier method. Hazard ratios (HR) associated 95% confidence intervals (CI), and p-values were calculated using Cox regression analysis (R packages survminer v0.4.9, coxphf, and survival v3.2.13). The log-rank test was used to compare two survival distributions. Overall, P-values < 0.05 were considered statistically significant.
A multivariable Cox proportional hazards model assessed prognostic factors associated with RFS and OS. The proportional hazard assumption was tested using a global test of the Schoenfeld residuals. The receiver operating characteristic (ROC) analysis was utilized to determine an optimal ctDNA threshold at baseline by maximizing sensitivity and specificity for RFS events.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all relevant data used to conduct the analyses are available within the article. Complete clinical information and ctDNA results are presented in the heat map and overview plot (Supplementary Fig. 1). Patient characteristics and demographic information are available in Table 1 and Supplementary Data 1. Supplementary Data 2 includes the de-identified Raw data of patient characteristics, outcomes (RFS and OS), and complete ctDNA results. In order to be compliant with the ethics committee and to protect the privacy and confidentiality of patients in this study, the sequencing data and source data supporting the findings of this study are not made publicly available but can be requested from the corresponding author for academic use only, within the limitations of the provided informed consent. Data will not be made available for commercial use. Any request will be reviewed within a timeframe of 2−4 weeks to verify whether the request is subject to any intellectual property or confidentiality obligations. All data shared will be de-identified and will be provided to researchers with access limited for scientific verification purposes and with strict prohibitions on secondary use. Applying researchers will be required to sign a data usage agreement.
Code availability
The fully documented code for the R statistical computing environment for analyses related to this manuscript is deposited in the GitHub repository and can be accessed at https://github.com/Natera-TMED/Zaanan-et-al_AGEO-PLAGAST.git.
References
Mantziari, S. et al. A comprehensive review of prognostic factors in patients with gastric adenocarcinoma. Cancers 15, 1628 (2023).
Al-Batran, S. E. et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet 393, 1948–1957 (2019).
Shapiro, J. et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol. 16, 1090–1098 (2015).
Hoeppner, J. B. T. et al. Prospective randomized multicenter phase III trial comparing perioperative chemotherapy (FLOT protocol) to neoadjuvant chemoradiation (CROSS protocol) in patients with adenocarcinoma of the esophagus (ESOPEC trial). J. Clin. Oncol. https://doi.org/10.1200/JCO.2024.42.17_suppl.LBA1 (2024).
Lin, Y., Liang, H. W., Liu, Y. & Pan, X. B. Nivolumab adjuvant therapy for esophageal cancer: a review based on subgroup analysis of CheckMate 577 trial. Front. Immunol. 14, 1264912 (2023).
Janjigian, Y. Y. et al. Pathological complete response (pCR) to durvalumab plus 5f luorouracil, leucovorin, oxaliplatin and docetaxel (FLOT) in resectable gastric and gastroesophageal junction cancer (GC/GEJC): interim results of the global, phase III MATTERHORN study. ESMO Ann. Oncol. 34, S1315–S1316 (2023).
Lorenzen, S. et al. Perioperative atezolizumab plus fluorouracil, leucovorin, oxaliplatin, and docetaxel for resectable esophagogastric cancer: interim results from the randomized, multicenter, phase II/III DANTE/IKF-s633 trial. J. Clin. Oncol. 42, 410–420 (2024).
Kotani, D. et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat. Med. 29, 127–134 (2023).
Loupakis, F. et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis. Oncol. 5, PO.21.00101 (2021).
Powles, T. et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 595, 432–437 (2021).
Reinert, T. et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131 (2019).
Huffman, B. M. et al. Analysis of circulating tumor DNA to predict risk of recurrence in patients with esophageal and gastric cancers. JCO Precis. Oncol. 6, e2200420 (2022).
Ococks, E. et al. Serial circulating tumor DNA detection using a personalized, tumor-informed assay in esophageal adenocarcinoma patients following resection. Gastroenterology 161, 1705–1708.e2 (2021).
Verschoor, Y. L. et al. Neoadjuvant atezolizumab plus chemotherapy in gastric and gastroesophageal junction adenocarcinoma: the phase 2 PANDA trial. Nat. Med. 30, 519–530 (2024).
Gerlinger M. G. A. & Barber L. J. Circulating tumor DNA for recurrence prediction and efficacy analysis in the ICONIC trial of peri-operative FLOT and avelumab (PD-L1) in localized esophago-gastric adenocarcinoma. Medicine (Baltimore). 102, e36228 (2023).
Takei S. K. D. & Sato K. Longitudinal circulating tumor DNA monitoring in patients with esophageal squamous cell carcinoma. JCO. https://doi.org/10.1200/JCO.2023.41.16_suppl.3041 (2023)
Al-Batran, S. E. et al. Effect of neoadjuvant chemotherapy followed by surgical resection on survival in patients with limited metastatic gastric or gastroesophageal junction cancer: the AIO-FLOT3 trial. JAMA Oncol. 3, 1237–1244 (2017).
Kroese, T. E. et al. Stereotactic radiotherapy or metastasectomy for oligometastatic esophagogastric cancer: a nationwide population-based cohort study. Clin. Transl. Radiat. Oncol. 37, 109–115 (2022).
Mizrak Kaya, D. et al. 101 long-term survivors who had metastatic gastroesophageal cancer and received local consolidative therapy. Oncology 93, 243–248 (2017).
ClinicalTrials. Bethesda (MD): National Library of Medicine (US). Identifier NCT05965479 DcGATfGCD-c. https://clinicaltrials.gov/ (2025).
ClinicalTrials. Bethesda (MD): National Library of Medicine (US). Identifier NCT06253650 ATDfH-pGCWPomRD. https://clinicaltrials.gov/ (2025).
Mandard, A. M. et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathol. Correlations. Cancer 73, 2680–2686 (1994).
Acknowledgements
The ROCHE company (donation to the UMR-S1138 laboratory unit for research on biomarkers in gastrointestinal cancers), and the French National Society of Gastroenterology (grant "FARE" from the "SNFGE, Société Nationale Française de Gastro-Entérologie"). Funding support by The Association des Gastro-Entérologues Oncologues (AGEO)—Gastroenterologists Oncologists Association, sponsors the PLAGAST study.
Author information
Authors and Affiliations
Contributions
A.Z. and P.L.P. conceived and designed the study. A.Z. and P.L.P. developed the methodology. A.Z., A.D., C.B., A.S.-F., F.S., M.P., A.M., and W.L. recruited patients and collected the data. E.S., H.S., G.L., S.S., P.D., and MM provided administrative, technical, or material support. G.L. and S.S. managed the data. A.Z., G.L., S.S., P.D., M.M., A.J., M.C.L., and P.L.P. analyzed and interpreted the data. G.L. and S.S. conducted the statistical analysis. All authors drafted and revised the manuscript. All authors critically revised the manuscript for intellectual content. All authors approved the final version of the submitted report and agreed to be accountable for all aspects. All authors verify that this study was done per protocol and vouch for data accuracy and completeness.
Corresponding author
Ethics declarations
Competing interests
All authors have disclosed relevant conflicts of interest. G.L., E.S., P.D., M.M., S.S., H.S., A.J., and M.C.L. are employees of Natera, Inc. and have stock/option to hold stock in the company. Additional COIs for M.C.L.: Grants/Contracts: Funding to Institution (Mayo) from: Eisai, Exact Sciences, Genentech, Genomic Health, GRAIL, Menarini Silicon Biosystems, Merck, Novartis, Seattle Genetics, Tesaro; Travel Support Reimbursement from AstraZeneca, Genomic Health, Ionis; Ad hoc advisory board meetings. All funds to Mayo Clinic. No personal compensation from AstraZeneca, Celgene, Roche/Genentech, Genomic Health, GRAIL, Ionis, Merck, Pfizer, Seattle Genetics, Syndax. A.Z. has participated in consulting and/or advisory boards for Amgen, Astellas, Merck, Roche, Servier, MSD, BMS, Pierre Fabre, Daiichi Sankyo, Astra Zeneca, Bayer, BeiGene, Astra Zeneca, Gilead, Abbvie. P.L.P. is the founder and shareholder of MethysDx, and a member of advisory board of Biocartis, Amgen, BMS, Pierre Fabre.
Peer review
Peer review information
Nature Communications thanks Pashtoon Kasi, Genki Usui and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zaanan, A., Didelot, A., Broudin, C. et al. Longitudinal circulating tumor DNA analysis during treatment of locally advanced resectable gastric or gastroesophageal junction adenocarcinoma: the PLAGAST prospective biomarker study. Nat Commun 16, 6815 (2025). https://doi.org/10.1038/s41467-025-62056-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62056-7
This article is cited by
-
Redefining gastroesophageal junction cancer care with perioperative immunotherapy, minimal residual disease monitoring and new targets
Nature Reviews Gastroenterology & Hepatology (2026)