Abstract
Azole effectiveness against candidiasis can be compromised by Candida albicans resistance and tolerance, and unfortunately, few clinically useful compounds can enhance azole antifungal activities. We find that the amino acids V234, F235 and L238 of Erg11 are critical for its interaction with Ncp1, and the Ncp1-Erg11 association is important in azole response. Ellipticine and its analog phiKan 083 block this Erg11-Ncp1 interaction by targeting Ncp1, and boost antifungal effects of fluconazole in vitro and in vivo. A series of steps influencing this process—an initial elevation in reactive oxygen species, leading to protein oxidation and misfolding in the endoplasmic reticulum (ER) that causes ER stress. This stress leads to Ca2+ release from the ER, mitochondrial Ca2+ accumulation and dysfunction, increased ROS production, and apoptosis of C. albicans cells. Overall, disrupting the Erg11-Ncp1 interaction in C. albicans can serve as a useful approach to enhancing the antifungal properties of azoles.
Similar content being viewed by others
Introduction
Candida albicans, a common fungus that can cause serious infections, may lead to fatal disseminated candidemia in patients with weakened immune systems1,2. Azoles, which inhibit lanosterol 14-α-demethylase (Erg11), are key antifungal agents for treating and preventing candidiasis3,4. However, long-term use can lead to C. albicans resistance, raising the minimum inhibitory concentration (MIC) of azoles needed to inhibit growth compared to susceptible strains5. As well, azoles may not effectively treat infections from non-resistant C. albicans due to the organism’s tolerance, which is linked to the fungistatic effect of azoles6. This characteristic allows tolerant strains to grow beyond the MIC7, leading to reduced azole efficacy against candidiasis, increased resistance, and persistent candidemia3,8,9. Therefore, the development of resistance and tolerance to azoles presents a significant clinical challenge in treating candidiasis.
Combination therapy represents a promising strategy for augmenting the antifungal activities of azoles10. However, translating in vitro efficacy enhancements to clinical use faces challenges6,11. Some compounds show target protein conservation, limited fungal selectivity, and human toxicity. Calcium ion (Ca2+) homeostasis maintains C. albicans azole tolerance, and calcineurin inhibition by cyclosporin A could enhance azole effects, but cyclosporin A’s immune suppression limits its use6,11. Poor pharmacokinetic parameters of azole adjuvants also pose issues. For example, CZ66 enhances azole efficacy by inhibiting Erg251 but has a short half-life in mice12. Overcoming these challenges requires exploring different mechanisms to improve azole antifungal efficacy.
Erg11, a cytochrome P450 enzyme, catalyzes the 14-methylation of lanosterol in C. albicans through three cycles13,14, each requiring two electrons from nicotinamide adenine dinucleotide phosphate (NADPH)-dependent Ncp1 reductase15,16. This electron transfer forms a high-affinity complex between the proteins17. In coupled P450 systems, electrons from NADPH are fully used for substrate hydroxylation, but leakage can produce reactive oxygen species (ROS). The ER produces ROS, oxidizing proteins like thioredoxins and causing ER stress18. Tunicamycin enhances C. albicans’ susceptibility to FLC by inhibiting protein N-glycosylation and inducing ER stress3. Carvacrol also boosts FLC’s anti-C. albicans activity via ER stress19. In response, C. albicans activates the unfolded protein response (UPR) pathway, involving the protein kinase Ire1 and the transcription factor Hac120. Deleting the IRE1 gene in Saccharomyces cerevisiae or C. albicans increases sensitivity to FLC and impedes ergosterol synthesis gene expression, respectively20,21. Removing the HAC1 gene enhances C. albicans’ susceptibility to azole drugs22. Thus, inhibiting the Erg11-Ncp1 interaction may suppress Erg11, elevate ER-derived ROS, augment ER stress, and enhance azole efficacy, but further research is needed to elucidate the mechanism.
This study investigated the role of amino acids V234, F235, and L238 of Erg11 in the Erg11-Ncp1 interaction. Disrupting this interaction increases C. albicans’ susceptibility to azoles by promoting misfolded protein production, leading to ER stress, Ca2+ buildup in mitochondria, dysfunction, and apoptosis. Ellipticine, targeting Ncp1, inhibits the Erg11-Ncp1 interaction, hyphal growth, and enhances antifungal activity of FLC in vivo. This study shows that impairing the Erg11-Ncp1 interaction can be a useful strategy for enhancing the antifungal activity of azoles.
Results
The Erg11200-251 region and the Ncp11-63 region are important for the interaction between Erg11 and Ncp1
Previous studies have provided evidence of the physical interaction between Erg11 and Ncp1 in both C. albicans and S. cerevisiae23,24. However, the specific mechanism of this interaction remains elusive. To investigate this, we employed a membrane yeast two-hybrid (MYTH) system. In this system, Erg11 was fused with the C-terminal half of ubiquitin (Cub) and LexA-VP16 (an artificial transcription factor), while Ncp1 was fused with the mutated N-terminal half of ubiquitin (NubG). The interaction between Erg11 and Ncp1 resulted in the proximity of Cub and NubG, thereby facilitating the reconstitution of split-ubiquitin and the activation of two auxotrophic genes, HIS3 and ADE2, allowing the yeast’s survival on a medium devoid of histidine and adenine (Fig. 1a)25. To prevent self-activation in the MYTH system, it is necessary to add the HIS3 gene product inhibitor 3-aminotriazole (3-AT) to the SD-Leu-Trp-His-Ade medium. Spotting assays demonstrated that co-expression of Erg11 and Ncp1 allowed yeast to grow on the selective medium, providing evidence for the interaction between Erg11 and Ncp1 (Fig. 1b and Supplementary Fig. 1a). To further validate this interaction, fusion proteins Erg11-13Myc and 3HA-Ncp1, were generated with a 13×Myc tag at the C-terminal of Erg11 (Supplementary Fig. 1b, d) and a 3×HA tag at the N-terminal of Ncp1 (Supplementary Fig. 1c, d). Co-immunoprecipitation (co-IP) assays demonstrated that Erg11 was immunoprecipitated using an anti-Myc antibody, and the co-IP of Ncp1 was detected through an anti-HA antibody. Reciprocally, when Ncp1 was immunoprecipitated employing an anti-HA antibody, the presence of Erg11 was detected using an anti-Myc antibody (Fig. 1c).
a The schematic diagram of a membrane yeast two-hybrid (MYTH) system. b MYTH analyzed the interaction between Erg11 and Ncp1. “Erg11” refers to yeast cells transformed with the bait Erg11 and the empty prey vector. “Erg11 + Ncp1” denotes the co-transformation of both the bait Erg11 and the prey Ncp1 into yeast cells. “Positive control” denotes the co-transformation of both the bait APP and the prey Fe65. “Negative control” refers to yeast cells that have been transformed with the bait APP and the prey vector without any protein insertions. c Co-IP assay analyzed the interaction between Erg11 and Ncp1. “Input” refers to lysates without any other manipulations used for immunoblotting. “IP” stands for immunoprecipitate, and the Protein A columns are used to collect the antibody-protein complex. “+“ indicates co-incubation of lysates with anti-Myc or anti-HA antibody, and “−“ represents lysates not incubated with antibodies. d Spotting assays of the yeast cells co-transformed to Ncp1 and C-terminal truncated fragments of Erg11. “Erg111-x “ represents the retained amino acids of Erg11. The schematic diagram depicts the interception strategy utilized to investigate the crucial interaction regions of Erg11. e Co-IP assay analyzed the interaction between Ncp1 and Erg111-251 or Erg111-200. The upper panel precipitates Erg111-251 with anti-Myc antibody and Ncp1 with anti-HA antibody. The lower panel precipitates Erg111-200 with anti-Myc antibody and Ncp1 with anti-HA antibody. f Spotting assays of the yeast cells containing Erg11 and N-terminal truncated fragments of Ncp1. “Ncp1y-680” represents the retained amino acids of Ncp1. g Co-IP assay analyzed the interaction between Erg11 and Ncp163-680 or Ncp191-680. The experiment was repeated three times with similar results. Source data, including uncropped and unprocessed scans with molecular weight markers are provided as Source Data file (c, e, g).
It has been proposed that the interaction between Erg11 and Ncp1 occurs via the electrostatic interaction between the C-terminal heme region of Erg11 and the N-terminal flavin mononucleotide (FMN) region of Ncp126,27. Thus, we utilized the MYTH system to examine the interaction between Ncp1 and a series of C-terminal truncated fragments of Erg11, including Erg111-490, Erg111-458, Erg111-428, Erg111-378, Erg111-293, Erg111-279, Erg111-251, and Erg111-200. Although the interaction between the full length of Erg11 and Ncp1 gives the strongest signal, most of the C-terminal truncated fragments of Erg11 are still capable of interacting with Ncp1. However, the fragment Erg111-200 is unable to interact with Ncp1 (Fig. 1d and Supplementary Fig. 1e), suggesting the importance of the Erg11200-251 region in facilitating the interaction between Erg11 and Ncp1. We further employed a C. albicans mutant in which the N-terminus of Ncp1 was tagged with 3×HA, and subsequently truncated Erg11 from the C-terminus and labeled it with 13×Myc, generating two combinations, Erg111-251-13Myc/3HA-Ncp1 and Erg111-200-13Myc/3HA-Ncp1 (Supplementary Fig. 1d). Co-IP findings revealed that the Erg111-251 fragment exhibited an interaction with Ncp1, whereas the Erg111-200 fragment did not (Fig. 1e). This supports the significance of the Erg11200-251 region in the Erg11-Ncp1 interaction.
Similarly, we employed the MYTH system to assess the interaction between Erg11 and various N-terminal truncated fragments of Ncp1, including Ncp163-680, Ncp191-680, Ncp1134-680, and Ncp1171-680. Our findings demonstrate that only full-length Ncp1 exhibits binding affinity towards Erg11, while all N-terminal truncated fragments of Ncp1 fail to interact with Erg11 (Fig. 1f). We further tagged the C-terminus of Erg11 with 13×Myc, and truncated Ncp1 from the N-terminus and tagged it with 3×HA, creating two mutants, 3HA-Ncp163-680 and 3HA-Ncp191-680 (Supplementary Fig. 1d). Co-IP results indicated that neither the Ncp163-680 fragment nor the Ncp191-680 fragment exhibited any interaction with Erg11 (Fig. 1g). In addition, we also truncated the C-terminus of Ncp1 and tagged it with 3HA, generating three mutants, Ncp1-3HA, Ncp11-600-3HA and Ncp11-500-3HA (Supplementary Fig. 1d). The Co-IP results establish that both the Ncp11-600 fragment and the Ncp11-500 fragment interacted with Erg11 (Supplementary Fig. 1f); full-length Ncp1 is not required. These findings demonstrated that the Ncp11-63 region plays a key role in the Erg11-Ncp1 interaction.
Amino acid residues V234, F235, and L238 in Erg11 contribute to the Erg11-Ncp1 interaction
We further identify the key amino acid residues in the Erg11200-251 region for the Erg11-Ncp1 interaction using molecular dynamics simulations. After 50 ns of kinetic simulation, the Erg11-Ncp1 complex system reached an equilibrium state with a root mean square deviation value of about 8.5 Å (Supplementary Fig. 2a). The binding free energy (ΔGbind) of this complex system was −127.21 kcal/mol. To explore key residues at the interface of this interaction, we modeled the contribution of each residue to the binding energy (Supplementary Table 1). Amino acid residues with lower ΔGbind are considered more critical in the Erg11-Ncp1 interaction. The three residues with the lowest ΔGbind and largest energy contributions in the Erg11200-251 region were V234, F235 and L238, with ΔGbind of −0.85 kcal/mol, −1.95 kcal/mol, and −2.04 kcal/mol, respectively (Fig. 2a). Therefore, we infer that these residues in the Erg11200-251 region play a key role in mediating this interaction.
a The energy decomposition diagram for the Erg11200-251 region revealed that the residues V234, F235, and L238 exhibited the highest energy contributions, with ΔGbind values of −0.85 kcal/mol, −1.95 kcal/mol, and −2.04 kcal/mol, respectively. b The MYTH system analyzed the interaction between Erg11m and Ncp1. “Erg11m” represents the yeast cells with the bait Erg11m and the prey vector (no inserting Ncp1); “Erg11m + Ncp1” is the co-transformation of the bait Erg11 and the prey Ncp1 to the yeast cells. c Co-IP assay analyzed the interaction between Erg11m and Ncp1. An anti-Myc antibody to precipitate Erg11m or Erg11 and utilizes an anti-HA antibody to precipitate Ncp1. The protein content of 3HA-Ncp1 was quantified relative to Erg11m-13Myc or Erg11-13Myc. “Input” refers to lysates without any other manipulations used for immunoblotting. “IP” stands for immunoprecipitate, and the Protein A columns are used to collect the antibody-protein complex. “+“ indicates co-incubation of lysates with anti-Myc antibody, and “−“ represents lysates not incubated with antibodies. d Co-IP assay analyzed the interaction between Erg11m and Ncp1. An anti-HA antibody to precipitate Ncp1. The protein content of Erg11m-13Myc or Erg11-13Myc was quantified relative to 3HA-Ncp1. “+“ indicates co-incubation of lysates with anti-HA antibody, and “−“ represents lysates not incubated with antibodies. Three biological replicates were carried out. One of the representative images is shown (c, d). Data were presented as mean ± standardized variance (SD) (n = 3). Two-tailed unpaired t-test (c, d). Source data including uncropped and unprocessed scans with molecular weight markers are provided as Source Data file (c, d).
To validate these predictions, we synthesized the ERG11 gene de novo, substituting the amino acid residues V234, F235, and L238 with alanine residues A234, A235, and A238, respectively. The co-expression of the mutated Erg11V234A, F235A, L238A (Erg11m) and Ncp11 showed no association in the MYTH system, indicating the need for these amino acid residues in the Erg11-Ncp1 interaction (Fig. 2b and Supplementary Fig. 2b). We further synthesized a nucleic acid sequence of Erg11m-13Myc and recombined it at the ADE2 gene locus in the 3HA-Ncp1 mutant, generating the Erg11m-13Myc/3HA-Ncp1 mutant. The co-IP results indicated that the interaction of Erg11m with Ncp1 was lost (Fig. 2c, d). We also built the single mutant constructs Erg11V234A-13Myc/3HA-Ncp1, Erg11F235A-13Myc/3HA-Ncp1, and Erg11L238A-13Myc/3HA-Ncp1; the co-IP results showed that all individual amino acid mutations did not affect the interaction between Erg11 and Ncp1 (Supplementary Fig. 2c–f). We additionally selected and mutated three amino acid residues (D214, D223, and D225) within the Erg11200-251 region to alanine, based on minimal involvement in the Erg11-Ncp1 interaction suggested by molecular docking results. Subsequently, the nucleic acid sequence of Erg11D214A, D223A, D225A (Erg11m’)-13Myc was integrated into the ADE2 gene locus of the 3HA-Ncp1 mutant, yielding the Erg11m’-13Myc/3HA-Ncp1 mutant. Co-IP experiments confirmed that the interaction between Erg11m’ and Ncp1 remained intact (Supplementary Fig. 2g, h). Overall, amino acid residues V234, F235, and L238 in Erg11 play a pivotal role in facilitating the Erg11-Ncp1 interaction.
Impairing the Erg11-Ncp1 interaction increases the susceptibility of C. albicans to azoles
To evaluate the importance of the Erg11-Ncp1 interaction for the susceptibility of C. albicans to azoles, we synthesized a nucleic acid sequence for Erg11m and then inserted it at the ADE2 gene locus in the heterozygous deleted mutant ERG11/erg11Δ. Subsequently, the remaining allele of the ERG11 gene was eliminated, generating an erg11m/erg11Δ, in which Erg11m and Ncp1 are unable to interact (Fig. 3a). To determine whether mutations in the amino acid residues V234, F235, and L238 of Erg11 affect protein expression and localization, we performed western blotting and confocal microscopy. Results showed no difference in Erg11 protein expression between the wild-type (Erg11-13Myc) and mutant (Erg11m-13Myc) strains (Supplementary Fig. 3a). Additionally, both GFP-tagged Erg11 and Erg11m localized to the ER (Supplementary Fig. 3b).
a The schematic diagram illustrating the generation of the erg11m/erg11Δ mutant involved the insertion of synthesized Erg11m at the ADE2 gene locus in the heterozygous deleted mutant ERG11/erg11Δ. Subsequently, the ARG4 gene was employed to replace the remaining allele of the ERG11 gene, resulting in the creation of the erg11m/erg11Δ mutant, where Erg11m and Ncp1 were found to be incapable of interacting. b Spotting assays were conducted to assess the hypersensitivity of the erg11m/erg11Δ mutant to fluconazole (FLC) in comparison to the wild-type strain SN152 and the ERG11/erg11Δ, NCP1/ncp1Δ, erg11Δ/erg11Δ, and ncp1Δ/ncp1Δ mutants on YPD plates with or without 4 μg/mL FLC. Images of the cultured cells were captured after 48 h of incubation at 30 °C. c The FLC disk diffusion assays were conducted on the erg11m/erg11Δ mutant, which exhibited a larger and cleaner inhibition zone on plates compared to the wild-type strain SN152 and the mutants of ERG11/erg11Δ, NCP1/ncp1Δ, erg11Δ/erg11Δ, and ncp1Δ/ncp1Δ. d The MIC assays were conducted to evaluate the hypersensitivity of the erg11m/erg11Δ mutant to FLC contrasted with the wild-type strain SN152 and the mutants of ERG11/erg11Δ, NCP1/ncp1Δ, erg11Δ/erg11Δ, and ncp1Δ/ncp1Δ. “NA” stands for “not applicable”. e The MIC assays demonstrated that ergosterol was ineffective in completely mitigating the antifungal effects of FLC in the erg11m/erg11Δ mutant. “NA” stands for “not applicable”. Source data are provided as a Source Data file.
Furthermore, we have created mutants of NCP1/ncp1Δ, erg11Δ/erg11Δ and ncp1Δ/ncp1Δ (Supplementary Fig. 3c–e). In comparison to the wild-type strain SN152 and the mutants ERG11/erg11Δ, NCP1/ncp1Δ, erg11Δ/erg11Δ, and ncp1Δ/ncp1Δ, the erg11m/erg11Δ mutant exhibited increased susceptibility to azoles such as FLC, miconazole, voriconazole, ketoconazole, and itraconazole (Fig. 3b and Supplementary Fig. 3f). Additionally, given C. albicans’ tolerance to FLC, disk diffusion assays revealed significant growth of C. albicans SN152 cells within the inhibition zone of 25 μg FLC when cultured on YPD plates and incubated at 30 °C for 48 h3,12. FLC paper disk displayed a larger and cleaner inhibition zone on the plate spread with the erg11m/erg11Δ mutant when compared with the wild-type strain SN152 (Fig. 3c). Furthermore, relative to these strains, the values of MIC and SMG of FLC were both lower against the erg11m/erg11Δ mutant (Fig. 3d). Time-growth curve assays also indicated that the erg11m/erg11Δ mutant exhibited increased susceptibility to FLC (Supplementary Fig. 3g). Notably, ergosterol effectively nullified the antifungal activity of FLC against the above strains, except for the erg11m/erg11Δ mutant, as shown in MIC (Fig. 3e) and time-growth curve assays (Supplementary Fig. 3h). These findings suggest that unlike the depletion of ergosterol resulting from Erg11 or Ncp1 deficiency, the blockade of the Erg11-Ncp1 interaction induces additional detrimental effects beyond ergosterol depletion.
Impairing the Erg11-Ncp1 interaction increased misfolded proteins and induced ER stress
The catalytic function of Erg11 is dependent on the presence of Ncp1 to facilitate electron transfer16. The efficient exchange of electrons between these redox partners is contingent upon the formation of a specific high-affinity 1:1 complex between Erg11 and Ncp117. In cases where the interaction between Erg11 and Ncp1 is disrupted, electrons may escape from the hydroxylation process and instead be directed towards O2, leading to the production of ROS. We conducted RNA-seq analysis to compare the transcriptome differences between the erg11m/erg11Δ mutant and the ERG11/erg11Δ mutant following 16 h of treatment with 4 μg/mL FLC. Pathway enrichment analysis identified notable regulatory distinctions in response to ROS, apoptotic processes, oxidative stress, and chaperone-mediated protein complex assembly between the ERG11/erg11Δ and erg11m/erg11Δ mutants (Fig. 4a). We further employed the flow cytometer to detect ROS production with the fluorescent dye 2’,7’-Dichlorodihydrofluorescein diacetate (DCFH-DA) and observed that ROS levels elevated 35 ± 0.5% (p < 0.0001) in the erg11m/erg11Δ mutant compared with the ERG11/erg11Δ mutant (Fig. 4b). Additionally, we discovered that more ROS was produced in the erg11m/erg11Δ mutant treated with 8 μg/mL menadione, an oxidative stress inducer28, with an increase of 60 ± 7.8% (p < 0.001) relative to the ERG11/erg11Δ mutant (Fig. 4b). Similarly, MIC and time-growth curve assays demonstrated that the erg11m/erg11Δ mutant exhibited greater susceptibility to menadione compared to the ERG11/erg11Δ mutant (Supplementary Fig. 4a, b). These findings suggested that blocking the Erg11-Ncp1 interaction enhances ROS generation.
a Go pathway enrichment analysis using RNA-seq to compare the transcriptome data of the ERG11/erg11Δ and erg11m/erg11Δ mutants after 16 h at 30 °C. The Q-value, which is the P-value adjusted for multiple hypothesis testing, ranges from 0 to 1. A lower Q-value indicates a higher level of significance in terms of enrichment. The Rich Ratio is calculated as the ratio of differentially expressed genes within a pathway term to all annotated genes within that pathway term. b Cellular levels of ROS were measured in the absence of drug treatment and following treatment with 8 μg/mL menadione for 4 h. c Quantification of cellular misfolded proteins was conducted by treating mutants ERG11/erg11Δ and erg11m/erg11Δ without or with 4 μg/mL FLC. d The mRNA expression of the HAC1 and IRE1 genes in the absence or presence of 4 μg/mL FLC for 4 h was quantified. The mutant ERG11/erg11Δ was treated with 5 mM DTT for 2 h as the positive control. The ACT1 gene served as the control. Three biological replicates were carried out. Data were presented as mean ± SD (n = 3). Statistical analysis was performed using an unpaired two-tailed t test (b, c, d). Source data are provided as a Source Data file.
Increased ROS can oxidize proteins within the ER, resulting in misfolded proteins29. Tetraphenylethene maleimide (TPE-MI) shows aggregation-induced emission, becoming fluorescent when molecules aggregate, which limits non-radiative decay. It binds to thiol groups in misfolded proteins, causing fluorescence30. TPE-MI can identify many misfolded proteins because thiol exposure is linked to protein misfolding, with cysteine residues being exposed during unfolding31. Dithiothreitol (DTT) was selected as a positive control due to its ability to disrupt disulfide bonds in proteins, resulting in the aggregation of misfolded proteins32. The misfolded protein content in the erg11m/erg11Δ mutant has a 29 ± 1.4% (p < 0.0001) increase compared to the ERG11/erg11Δ mutant. Additionally, the misfolded protein content increases 73 ± 5.3% (p < 0.0001) in the erg11m/erg11Δ mutant after treatment with 4 μg/mL FLC, compared to the ERG11/erg11Δ mutant (Fig. 4c).
Elevated levels of misfolded proteins lead to ER stress, as evidenced by the increased susceptibility of the erg11m/erg11Δ mutant to tunicamycin, an inducer of ER stress33, compared to the ERG11/erg11Δ mutant (Supplementary Fig. 4c). The UPR pathway in C. albicans, activated by increased misfolded proteins, involves Ire1 and Hac1. Ire1, sensing ER stress, initiates a cascade by splicing Hac1 mRNA to upregulate genes related to protein folding, degradation, and ER-associated degradation34. We further found that the mRNA expression levels of IRE1 and HAC1 increased to 1.9 ± 0.1 fold (p < 0.001) and 1.8 ± 0.1 fold (p < 0.001) in the ERG11m/erg11Δ mutant compared with the ERG11/erg11Δ mutant. Under 4 μg/mL FLC treatment for 8 h, the mRNA expression levels of IRE1 and HAC1 elevated to 1.7 ± 0.2 fold (p < 0.01) and 2.2 ± 0.1 fold (p < 0.001) in the erg11m/erg11Δ mutant compared to the ERG11/erg11Δ mutant. Disruption of the Erg11-Ncp1 interaction results in the activation of the UPR signaling pathway, akin to the effects observed with the ER stress inducer DTT (Fig. 4d).
ER stress leads to the release of Ca2+ from the ER and accumulation in the mitochondria
Under ER stress, Ca2+ in the ER may leak into the cytoplasm, potentially resulting in an increase in the availability of Ca2+ within the mitochondria35,36. This suggests that the inhibition of the Erg11-Ncp1 interaction will result in the buildup of Ca2+ within mitochondria. Laser confocal assays revealed that there was no difference in the Ca2+ content within the vacuoles in both the ERG11/erg11Δ and erg11m/erg11Δ mutants (Supplementary Fig. 5a, b). However, in comparison to the ERG11/erg11Δ mutant, the erg11m/erg11Δ mutant cells exhibited an enhanced green fluorescence signal (representing Ca²⁺ by Fluo-3 AM) that did not overlap with the red fluorescence (indicative of the ER by ER-Tracker Red) (Fig. 5a and Supplementary Fig. 5c). Quantitative analysis the integrated green fluorescence intensity that remained green after merging with red fluorescence within individual cells and found that in the erg11m/erg11Δ mutant, the integrated green fluorescence intensity (post-merge) was 3.7 ± 0.2 times higher than that in the ERG11/erg11Δ mutant (p < 0.0001) (Fig. 5b). This suggests that disrupting the Erg11-Ncp1 interaction leads to the leakage of Ca2+ into the cytoplasm.
a Representative images depicting the localization of Ca2+ localization (green) in relation to ER membranes (red) were obtained from ERG11/erg11Δ and erg11m/erg11Δ mutants. Scale bar = 5 µm. b The integrated green fluorescence intensity (post-merge) was quantitatively analyzed between the ERG11/erg11Δ and erg11m/erg11Δ mutants using Image J software. The integrated fluorescence intensity was calculated as the product of the fluorescence density and the fluorescence area. c An assay was conducted to observe for Ca2+ activation (red) in the mitochondria using confocal microscopy. In the diagram provided, the large black ellipse symbolizes the C. albicans cell, the small orange ellipse represents the mitochondria, and the red dot signifies Ca2+. Scale bar = 5 µm. d The mitochondrial Ca2+ levels in confocal microscopy images were quantitatively analyzed between the ERG11/erg11Δ and erg11m/erg11Δ mutants using Image J software. The integrated fluorescence intensity was calculated as the product of the fluorescence density and the fluorescence area. Three biological replicates were performed, and a representative image is shown (a, c). Violin plots display the distribution density (width of the violin shape), median (black points), interquartile range (white points), and range (minimum and maximum values) (b, d). Two-tailed unpaired t-test (b, d). Source data are provided as a Source Data file.
We further used a Ca2+ fluorescent probe, Rhod-2 AM, which specifically accumulates in mitochondria and emits red fluorescence, to measure the concentration of Ca2+ within the mitochondria. Laser confocal assays demonstrated a significant increase in mitochondrial Ca2+ levels in the erg11m/erg11Δ mutant compared to the ERG11/erg11Δ mutant (Fig. 5c and Supplementary Fig. 5d). The integrated fluorescence intensity in the erg11m/erg11Δ mutant was found to be 6.9 ± 1.1 times (p < 0.01) higher than in the ERG11/erg11Δ mutant (Fig. 5d). Collectively, the induction of ER stress through inhibition of the Erg11-Ncp1 interaction leads to the release of Ca2+ from the ER, followed by its accumulation in mitochondria.
Mitochondrial Ca2+ accumulation results in mitochondrial dysfunction and apoptosis of C. albicans cells
Elevated intracellular Ca²⁺ levels can lead to mitochondrial overload, disrupting the mitochondrial membrane potential and impairing ATP synthesis. This disruption can trigger the opening of the mitochondrial permeability transition pore, leading to the release of pro-apoptotic factors and ultimately resulting in cell death37, and thus, the buildup of Ca2+ within mitochondria serves as an important contributor to mitochondrial dysfunction. Depolarization of the mitochondrial membrane is an important characteristic of mitochondrial dysfunction38. Rhodamine 123, a cationic dye, assesses mitochondrial function by accumulating in active mitochondria with a negative membrane potential. A drop in its fluorescence signifies mitochondrial dysfunction due to potential loss39,40. The results indicated a significant decrease of 26 ± 2.7% (p < 0.001) in the mitochondrial membrane potential of the erg11m/erg11Δ mutant, similar to the effects induced by the ER stressor DTT (Fig. 6a).
a Rhodamine 123 staining was utilized to observe alterations in mitochondrial membrane potential. A positive control group that the ERG11/erg11Δ mutant treated with 5 mM DTT for 2 h was included. b A quantitative analysis was conducted on mitochondrial ROS levels in the ERG11/erg11Δ and erg11m/erg11Δ mutants. A positive control group was included, consisting of the ERG11/e rg11Δ mutant incubated with 5 mM DTT for 2 h. c The fluorescence intensity of misfolded proteins within mitochondria was assessed using flow cytometry. A positive control group that the ERG11/erg11Δ mutant was treated with DTT was included. d Density plots were used to visualize the fluorescence patterns of the ERG11/erg11Δ and erg11m/erg11Δ mutants for 8 h. Data were obtained from three biological repeats, and representative images are shown. e Quantification of Annexin V/PI double staining assay. Data are mean ± SD of n = 3 biological replicates (a, c, e). Two-tailed unpaired t-tests were used (a, c, e). Source data are provided as a Source Data file.
Mitochondrial dysfunction leads to an increase in ROS production within the mitochondria. Mitochondrial ROS levels were quantified using MitoSOX Red, a dye that oxidizes in mitochondria and binds to nucleic acids, producing red fluorescence41. The results indicated a significant 20 ± 0.7% (p < 0.0001) increase in mitochondrial ROS levels in the erg11m/erg11Δ mutant (Fig. 6b). EGTA is a chelator of Ca²⁺ that is widely utilized due to its capacity to form stable complexes with Ca²+42. As well, in the erg11m/erg11Δ mutant, exposure to 200 μM EGTA led to a 21 ± 1.8% decrease in mitochondrial ROS levels (p < 0.0001) (Fig. 6b). These results indicated a correlation between elevated mitochondrial ROS levels and heightened mitochondrial Ca2+ content. Increased ROS caused mitochondrial oxidized proteins to be misfolded. We quantified misfolded protein mass in the mitochondrial matrix using MitoTracker Green, a dye that selectively targets this area and binds to exposed cysteine residues on misfolded proteins43. The data indicated a significant increase in the content of mitochondrial misfolded proteins by 40 ± 5.9% (p < 0.001) in the erg11m/erg11Δ mutant compared to the ERG11/erg11Δ mutant (Fig. 6c). Moreover, the erg11m/erg11Δ mutant exhibited a reduction in mitochondrial misfolded protein mass by 15 ± 0.6% (p < 0.0001) following treatment with 200 μM EGTA, suggesting a relationship between increased mitochondrial misfolded proteins and elevated mitochondrial Ca2+ levels (Fig. 6c).
Mitochondrial dysfunction and increased ROS produced by mitochondria will lead to apoptosis of C. albicans cells. In the Annexin V/Propidium Iodide (PI) assay, Annexin V, a protein that binds to phosphatidylserine on apoptotic cells, is conjugated to a fluorescent marker (FITC). Cells positive for Annexin V indicate apoptosis44. Our results indicate that inhibition of the Erg11-Ncp1 interaction resulted in enhanced apoptosis (Fig. 6d), with an apoptosis induction rate of 10.5 ± 0.3% (p < 0.0001) in the erg11m/erg11Δ mutant compared to 3.4 ± 0.5% in the ERG11/erg11Δ mutant (Fig. 6e).
Ellipticine acts as an inhibitor of the Erg11-Ncp1 interaction by targeting Ncp1
Given that inhibiting the interaction between Erg11 and Ncp1 enhances the susceptibility of C. albicans to FLC, we employed the MYTH system to characterize potential inhibitors that could augment the antifungal effects of azoles. If a compound obstructs the Erg11-Ncp1 interaction, the reconstitution of the split ubiquitin is prevented, preventing activation of the two auxotrophic genes, HIS3 and ADE2. Furthermore, the candidate inhibitor of the Erg11-Ncp1 interaction must not exert any influence on the interaction between APP (amyloid A4 precursor protein) and Fe65 (amyloid beta A4 precursor protein-binding family B member 1), two other proteins giving a positive signal in the MYTH assay, thus ensuring the candidate compound has no general effect on the assay. The extent of yeast growth in the SD-Leu-Trp-His-Ade medium compared to that in the SD-Leu-Trp medium is indicative of the inhibition of the Erg11-Ncp1 interaction by the compound (Fig. 7a).
a Schematic of Erg11-Ncp1 inhibitor screening using MYTH. b Relative growth rates of six Erg11-Ncp1 interaction candidates measured by MYTH. c Dose-matrix assays of albicans cmp1-aidΔ/cmp1-aidΔ with ellipticine and FLC; DTT as positive control. d Growth inhibition curve assays were conducted to assess the combined effects of FLC and ellipticine. Data represent mean ± SD (n = 3). e Co-IP analysis shows ellipticine blocked the Erg11-Ncp1 interaction. “Input” is the Erg11-13Myc/3HA-Ncp1 lysate used for immunoblotting. “NC” is the wild-type SN152 lysate as a negative control. The binding rate of 3HA-Ncp1 to Erg11-13Myc was quantified. f Intracellular ROS was quantified after treatment with 16 μg/mL ellipticine and 4 μg/mL FLC. g Misfolded proteins in ERG11/erg11Δ mutants were analyzed after treatment with 16 μg/mL ellipticine and 4 μg/mL FLC. h Mitochondrial misfolded proteins were determined by MitoTracker Green. i Intracellular Ca2+ levels were quantified following treatment with 16 μg/mL ellipticine and 4 μg/mL FLC in the ERG11/erg11Δ mutant. j Mitochondrial membrane potential changes were assessed by Rhodamine 123 staining. k CYC3 gene expression was quantified after treatment with 16 μg/mL ellipticine and 4 μg/mL FLC. l Annexin V/PI double staining density plots were generated. m Quantification of Annexin V/PI double staining assay. n Ellipticine protects Ncp1 from pronase degradation in DARST assays. Uncropped, unprocessed scans with molecular weight markers are provided in the Source Data file. The quantification shows 3HA-Ncp1 protein levels in the control group relative to the ellipticine group. The representative images shown are from 3 biological repeats (e, I, n). Data were presented as mean ± SD of n = 3 biological replicates (b, e–k, m, n). Two-tailed unpaired t-test (b, e–k, m, n). Source data are provided as a Source Data file.
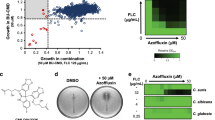
In theory, the inhibition of the Erg11-Ncp1 interaction may be achieved by the binding of a small molecule compound to either Erg11, Ncp1, or both simultaneously. We searched three categories of compounds through a literature review: (1) azoles targeting Erg11 (n = 5); (2) compounds potentially targeting Ncp1 (n = 12); (3) compounds preventing electron transfer between Ncp1 and Erg11 (n = 5) (Supplementary Table 2). Six compounds were identified as inhibitors of the Erg11-Ncp1 interaction: arachidonic acid (57 ± 3.1% inhibition), carbonyl cyanide m-chlorophenylhydrazone (75 ± 3.6% inhibition), ellipticine (57 ± 2.5% inhibition), menadione (45 ± 3.3% inhibition), piperine (37.9 ± 9.9% inhibition), and tamoxifen (68 ± 2.1% inhibition) (Fig. 7b).
Considering that inhibiting the Erg11-Ncp1 interaction leads to heightened susceptibility of C. albicans to FLC, we tested if compounds targeting the Erg11-Ncp1 interaction could augment the antifungal efficacy of FLC. The dose-matrix titration assays showed that only ellipticine can enhance the efficacy of FLC (Fig. 7c and Supplementary Fig. 6a–e). We further observed that ellipticine markedly decreased both the MIC and SMG values of FLC in FLC-resistant and -tolerant clinical strains of C. albicans (n = 13). However, in strains that were only FLC-tolerant (n = 4), ellipticine did not further reduce the MIC values, although it did decrease the SMG values, thereby lowering the tolerance level of C. albicans to FLC. Conversely, in non-FLC-resistant and -tolerant clinical C. albicans strains (n = 8), the addition of ellipticine did not result in further reductions in either MIC or SMG values for FLC. For FLC-resistant and -tolerant Nakaseomyces glabratus (formerly Candida glabrata) strains (n = 9), ellipticine significantly reduced both the MIC and SMG values of FLC. However, ellipticine did not enhance the efficacy of FLC against FLC-resistant and -tolerant clinical Candidozyma auris (formerly Candida auris) strains (n = 9) (Supplementary Table 3). Given the phylogenetic divergence between C. auris and C. albicans, which leads to substantial differences in their carbon utilization, lipid composition, and protein content45, this divergence may account for the observed enhancement of the antifungal activity of FLC by ellipticine against C. albicans, but not against C. auris. In addition, considering that the disruption of the Erg11-Ncp1 interaction causes additional detrimental effects beyond ergosterol depletion, ellipticine, a potential inhibitor of Erg11-Ncp1 interaction, enhances the antifungal activity of FLC against the ERG11/erg11Δ mutant in the presence of ergosterol, as demonstrated through time-growth curve assays (Fig. 7d).
We further used the co-IP assay to evaluate the inhibitory effect of ellipticine on the Erg11-Ncp1 interaction. Cell lysates from the Erg11-13Myc/3HA-Ncp1 mutant were exposed to 25 μg/mL ellipticine for 1 h. The relative protein content of Ncp1 and Erg11 in the absence of drug treatment was established as 100%. Upon exposure to concentrations of ellipticine as above, the ratios of Ncp1 to Erg11 protein content decreased to 64 ± 8.3% (p < 0.05) (Fig. 7e). Taken together, this suggests ellipticine acts as an inhibitor of the Erg11-Ncp1 interaction.
The combination of FLC and ellipticine resulted in a significant increase in intracellular ROS levels of 2.6 ± 0.1 times (p < 0.0001) in the ERG11/erg11Δ mutant, as detected using the dye DCFH-DA (Fig. 7f). Additionally, this combination elevated misfolded protein levels by 2.3 ± 0.01 times (p < 0.0001) in the ER, detected using the dye TPE-MI (Fig. 7g) and in mitochondria by 20.3 ± 2.3% (p < 0.01), detected using the dye MitoTracker Green (Fig. 7h). Intracellular Ca2+ concentration increased by 29.0 ± 0.7% (p < 0.0001), as measured by Fluo-3AM (Fig. 7i). Concurrently, a significant decrease of 41.2 ± 8.8% (p < 0.01) in mitochondrial membrane potential was observed using Rhodamine 123 assays (Fig. 7j). The CYC3 gene encodes cytochrome c heme lyase, crucial for cytochrome c maturation, electron transport, and mitochondrial respiration. Its expression indicates mitochondrial health and functionality46. As anticipated, the expression of the CYC3 gene exhibited a significant 1.9 ± 0.1-fold (p < 0.001) upregulation, specifically after 8 h of co-treatment with 16 μg/mL ellipticine and 4 μg/mL FLC (Fig. 7k). Subsequently, we utilized the Annexin V/PI double staining assay to demonstrate the induction of apoptosis. A notable 23.9 ± 1.1% (p < 0.0001) of cells underwent apoptosis when exposed to ellipticine in conjunction with 4 μg/mL FLC (Fig. 7l, m). In summary, ellipticine disrupts the interaction between Erg11 and Ncp1 and induces ER stress and mitochondrial dysfunction.
We used drug affinity responsive target stability (DARTS) assays to reveal the inhibitory mechanism of ellipticine on the Erg11-Ncp1 interaction. The theory of the method is that a protein becomes stable upon binding to a small molecule compound, contributing to the protection of the target protein from degradation by proteases47. The mutant Erg11-13Myc/3HA-Ncp1 was employed, and its lysates were digested with different concentrations of pronases. It was discovered that Ncp1 was progressively degraded in the absence of ellipticine. When no pronase was added, the relative protein content was 100 ± 1%. Nevertheless, without ellipticine protection, it was reduced to 78.0 ± 4.6% (p < 0.01) and 62.0 ± 7.0% (p < 0.0001) in the presence of 8 μg/mL and 16 μg/mL pronases, respectively (Fig. 7n). However, we did not observe this protective function of ellipticine on Erg11 (Supplementary Fig. 6f, g). Hence, ellipticine appears to act as an inhibitor of the Erg11-Ncp1 interaction by targeting Ncp1.
Ellipticine and its analog phiKan 083 enhance the antifungal activity of FLC in vivo
The therapeutic efficacy of ellipticine against the wild-type C. albicans strain SN152, was tested in Galleria mellonella infections. The larvae were randomly assigned to one of four groups: (1) control group receiving PBS treatment, (2) group receiving 1 mg/kg FLC alone, (3) group receiving 4 mg/kg ellipticine alone, and (4) group receiving a combination of 1 mg/kg FLC and 4 mg/kg ellipticine. Treatment was administered 2 h post-infection and was administered as a single dose. The administration of 4 mg/kg ellipticine significantly enhanced the antifungal effect of FLC (1 mg/kg) over a 15-day experimental period (Fig. 8a). The survival rate of the ellipticine-FLC combination group was notably higher compared to the other three groups (p = 0.0003, compared to the control group; p = 0.0021, compared to the FLC alone group; p < 0.0001, compared to the ellipticine-treated group; assessed using the log-rank test). These in vivo data suggest that ellipticine can synergize with FLC to create potential antifungal effects.
a An investigation was conducted on the survival of animals exposed to ellipticine against the wild-type SN152 in Galleria mellonella (n = 10 per group). b An investigation was conducted on the survival of animals exposed to ellipticine in a highly FLC-tolerant C. albicans strain, cmp1-aidΔ/cmp1-aidΔ in Galleria mellonella (n = 10 per group). c The fungal load in the kidneys of phiKan 083 was conducted in C. albicans SN152 was assessed. n = 12 mice. d The CFUs analysis of phiKan 083 was conducted in the C. albicans strain cmp1-aidΔ/cmp1-aidΔ. n = 12 mice. e The histopathological examination of phiKan 083 in the C. albicans SN152 involved staining the kidneys of mice with PAS, followed by the display of representative images from 3 biological repeats. f The histopathological examination of phiKan 083 in the C. albicans strain cmp1-aidΔ/cmp1-aidΔ involved staining the kidneys of mice with PAS, followed by the display of representative images from 3 biological repeats. g An investigation into the survival of Galleria mellonella larvae (n = 10 per group) infected with the C. albicans SN152, specifically focusing on the impact of phiKan 083 treatment. h An investigation into the survival of Galleria mellonella larvae (n = 10 per group) infected with the highly FLC-tolerant C. albicans strain cmp1-aidΔ/cmp1-aidΔ, specifically focusing on the impact of phiKan 083 treatment. Data were presented as mean ± SD (c, d). Log-rank (Mantel-Cox) test (a, b, g, h), two-tailed unpaired t-test (c, d). Source data are provided as a Source Data file.
The CMP1 gene encodes the catalytic subunit of calcineurin, crucial for FLC tolerance in C. albicans. Deletion of the C-terminal autoinhibitory domain of Cmp1 results in a constitutively active calcineurin complex48,49,50. Consequently, the cmp1-aidΔ/cmp1-aidΔ mutant, with the C-terminal 110 residues of Cmp1 deleted, exhibits high FLC tolerance51. In addition, we validated that cmp1-aidΔ/cmp1-aidΔ mutant did not cause the ROS generation or the elevation of misfolded proteins and Ca2+ content compared with wild-type SN152 (Supplementary Fig. 7a–c). We further constructed Galleria mellonella infection model used the FLC-tolerant cmp1-aidΔ/cmp1-aidΔ strain to evaluate the therapeutic efficacy of ellipticine. The survival rate of the ellipticine-FLC combination group was notably higher compared to the other three groups (Fig. 8b). Regrettably, the enhancement effect of ellipticine on the antifungal activity of FLC in a murine model of systemic C. albicans infection was suboptimal (Supplementary Fig. 8a, b). This outcome may be attributed to the inadequate pharmacokinetic parameters of ellipticine52, which likely hinder its efficacy in enhancing the antifungal action of FLC in this context.
Analogues of ellipticine were pursued through an examination of its core structure, which is composed of two benzene rings surrounding a pyrrole ring. Three distinct series of ellipticine analogues were studied, each maintaining the fundamental structure of the parent nucleus: the a-series, which investigated substitutions on the benzene ring on the left side of the parent nucleus structure; the b-series, which concentrated on alterations to a nitrogen atom of the pyrrole ring; and the c-series, which explored modifications to the connecting substituents on the benzene ring on the right side of the parent nucleus structure (Supplementary Fig. 9). Dose-matrix titration assays and spotting assays identified three compounds, namely 7H-Dibenzo[c,g]carbazole (Supplementary Fig. 10a), 10-Bromo-7H-benzo[c]carbazole (Supplementary Fig. 10b), and phiKan 083 (Supplementary Fig. 10c), that demonstrated promising antifungal properties through the inhibition of FLC tolerance. Subsequently, we conducted further evaluation of the antifungal activity of these compounds through time-growth curves, leading to the discovery that only phiKan 083 demonstrated dose-dependent inhibition of fungal growth when combined with FLC (Supplementary Fig. 10d–f). Similar to ellipticine, phiKan 083 can also impair mitochondrial function (Supplementary Fig. 10g, h).
Several studies have demonstrated that mitochondrial dysfunction hinders the hyphal growth of C. albicans53,54. Therefore, we hypothesize that ellipticine and phiKan 083, known for impairing mitochondrial function, may inhibit the hyphal growth of C. albicans. The C. albicans SN152 and cmp1-aidΔ/cmp1-aidΔ mutants were induced to form hyphae in RPMI 1640 medium. Interestingly, 4 μg/mL FLC did not prevent this morphological change. However, the combination of FLC with 8 μg/mL ellipticine or 32 μg/mL phiKan 083 significantly inhibited the hyphal growth of C. albicans. The average length of hyphae in the C. albicans SN152 strain and the cmp1-aidΔ/cmp1-aidΔ mutants was 44.5 ± 4.42 μm and 50.3 ± 4.49 μm, respectively, while under FLC treatment, the average lengths were 28.1 ± 4.77 μm and 30.3 ± 4.23 μm, respectively. When FLC was combined with 8 μg/mL ellipticine or 32 μg/mL phiKan 083, the average hyphal lengths were 11.4 ± 1.50 μm and 11.0 ± 2.08 μm in the C. albicans SN152 strain (Supplementary Fig. 11a, b), and 3.1 ± 0.60 μm and 10.8 ± 1.74 μm in the cmp1-aidΔ/cmp1-aidΔ mutants (Supplementary Fig. 11c, d and Supplementary Table 4). The data presented indicates that disrupting the Erg11-Ncp1 interaction results in mitochondrial dysfunction, thereby impeding the morphological transition of C. albicans from yeast to hyphae.
To assess the antifungal efficacy of phiKan 083 in vivo, C. albicans wild-type SN152 and highly FLC-tolerant cmp1-aidΔ/cmp1-aidΔ strains were utilized to establish a model of systemic candidiasis in C57BL/6 mice via tail vein injection. Following infection, all mice were randomly assigned to one of four groups: (1) control group, (2) FLC alone group at a dosage of 0.5 mg/kg, (3) phiKan 083 alone group at a dosage of 6 mg/kg, and (4) combination group receiving FLC (0.5 mg/kg) in conjunction with phiKan 083 (6 mg/kg). The initial intraperitoneal injection was administered 2 h post-infection, with subsequent administrations given at one-day intervals for a total of three consecutive days. On the seventh day post-infection, six mice from each group were euthanized, and the fungal load in their kidneys, livers, and spleens was evaluated. Given that there was no significant interaction between drug treatment and sex, we decided to pool the data from male and female mice together for further analysis. The results indicated a clear decrease in fungal load in the kidneys of mice treated with the drug combination (Fig. 8c, d). However, the impact of the combination therapy on fungal burden in the liver and spleen has no physiological decrease observed compared to FLC monotherapy (Supplementary Fig. 11e, f). Similar to the inhibitory effect observed in vitro with the combination of FLC and phiKan 083 on the formation of C. albicans hyphae, histopathological analysis utilizing Periodic Acid Schiff (PAS) staining revealed the absence of hyphae in the kidneys of the group treated with FLC in combination with phiKan 083. In contrast, a significant presence of hyphae was observed in the kidneys of both the C. albicans infected group, the FLC alone group and phiKan 083 alone group (Fig. 8e, f). These findings demonstrated that phiKan 083 enhanced the antifungal efficacy of FLC in vivo, especially in the highly FLC-tolerant C. albicans cmp1-aidΔ/cmp1-aidΔ.
To further evaluate the therapeutic efficacy of phiKan 083 against the wild-type C. albicans strain SN152 and the highly FLC-tolerant C. albicans cmp1-aidΔ/cmp1-aidΔ, systemic fungal infections were simulated in Galleria mellonella. The larvae were randomly assigned to one of four groups: (1) the control group receiving PBS treatment, (2) the group treated with 1 mg/kg FLC alone, (3) the group treated with 4 mg/kg phiKan alone, and (4) the group treated with a combination of 1 mg/kg FLC and 4 mg/kg phiKan 083. Each group consisted of ten larvae, which were treated 2 h post-infection with a single dose of the respective drug. Our observations indicate that the combination of phiKan 083 with FLC resulted in a 60% survival rate. In contrast, the remaining three groups exhibited a 100% mortality rate over the 20-day observation period (Fig. 8g, h). Taken together, phiKan083 significantly enhance the antifungal activity of azoles.
Discussion
In this study, we have identified the Erg11200-251 region and the Ncp11-63 region as crucial for the interaction between Erg11 and Ncp1. Our findings also highlight the significance of amino acid residues V234, F235, and L238 in Erg11 for this interaction. Mutating these three amino acids effectively disrupts the Erg11-Ncp1 interaction, resulting in heightened susceptibility of C. albicans to azoles. This disruption of the Erg11-Ncp1 interaction results in elevated ROS levels, protein oxidation, and misfolding in the ER, culminating in ER stress. This stress triggers the release of Ca2+ from the ER and its accumulation in mitochondria, leading to mitochondrial dysfunction, heightened ROS production, and, ultimately, apoptosis in C. albicans cells. Additionally, our investigation revealed that ellipticine functions as an inhibitor of the Erg11-Ncp1 interaction by specifically targeting Ncp1, thereby enhancing the efficacy of FLC in the Galleria mellonella infection model caused by the FLC-tolerant C. albicans strain.
Our research has revealed that the cascade amplification of ROS production can impair mitochondrial function and augment the antifungal efficacy of azoles. By disrupting the Erg11-Ncp1 interaction, electrons may escape from the hydroxylation process and be redirected to oxygen, leading to the generation of ROS within the ER. ROS originating from the ER may act as a “trigger,” initiating ER stress. This stress induces the release of Ca2+ from the ER. The flow of Ca2+ may act as a “fuse,” causing an accumulation of Ca2+ in the mitochondria and subsequent mitochondrial dysfunction. The dysfunction of the mitochondria further increases ROS generation within mitochondria. This heightened mitochondrial ROS production may serve as an “explosive package,” ultimately leading to apoptosis in C. albicans cells and demonstrating potential for enhancing the antifungal activities of azoles (Fig. 9). It is important to note that other mechanisms might underlie these phenomena. For instance, the potential disruption of the Erg11-Ncp1 interaction could change the content and composition ratio of sterols in the membrane, thereby affecting mitochondrial functions. Furthermore, the direct impact of ROS on mitochondria can lead to apoptosis, independent of Ca2+ homeostasis. Additionally, there may be intricate crosstalk between the ER and mitochondria involving various signaling pathways that are not solely reliant on Ca2+, such as the UPR.
Ellipticine acts on Ncp1 to inhibit the Erg11-Ncp1 interaction within the ER, disrupting the electron transport chain between the two proteins. This disruption results in electron leakage, leading to the accumulation of excess electrons that react with oxygen to produce elevated levels of ROS. The surplus ROS oxidizes proteins synthesized in the ER, resulting in the production of misfolded proteins and the induction of ER stress. During ER stress, a substantial quantity of Ca2+ is transported from the ER to the cytoplasm, resulting in a notable elevation in cytoplasmic Ca2+ levels. The presence of a mitochondria-associated membrane (MAM) facilitates the transfer of Ca2+ from the ER to mitochondria. The buildup of Ca2+ in the mitochondria contributes to mitochondrial dysfunction and the advancement of cell apoptosis in C. albicans, thereby augmenting the efficacy of azoles.
The elucidation of the interaction between Erg11 and Ncp1 offers a promising approach for the advancement of antifungal drugs inhibiting the activity of Erg11. The ergosterol biosynthesis pathway has been identified as a traditional therapeutic target for addressing invasive candidiasis55. Azoles function by inhibiting the activity of Erg11 through binding to the heme region of Erg1114, making them a preferred choice for primary treatment options due to their favorable bioavailability and safety profile56. However, the emerging issue of azole resistance and tolerance poses a substantial obstacle7 and underscores the necessity for alternative approaches to impede the function of Erg11. The formation of a complex between Erg11 and Ncp1 is essential for the catalytic activity of Erg1157. Thus, disrupting the Erg11-Ncp1 interaction has the potential to inhibit Erg11 activity. Nevertheless, the precise mechanism of the Erg11-Ncp1 interaction has remained elusive57. This study showcases the physical interaction between Erg11 and Ncp1 using the MYTH system and co-IP. Furthermore, we elucidate the crucial domain of the Erg11-Ncp1 interaction through fragment analysis, emphasizing the significance of the Erg11200-251 region in facilitating this interaction. Additionally, we pinpoint key amino acid residues V234, F235, and L238 in Erg11 that are required for the Erg11-Ncp1 interaction through molecular dynamic simulations, the MYTH system, and co-IP. Our study further revealed that the disruption of the Erg11-Ncp1 interaction led to increased susceptibility of C. albicans to azoles as demonstrated by in vitro antifungal drug susceptibility tests.
Our research introduces a potential approach to impair mitochondrial function and trigger apoptosis in C. albicans cells by inhibiting the interaction between Erg11 and Ncp1. This results in inducing ER stress, causing Ca2+ leakage from the ER and its accumulation in mitochondria, ultimately leading to mitochondrial dysfunction. Our previous investigations have demonstrated that baicalin, berberine, and osthole can elevate ROS levels in C. albicans, thereby bolstering the antifungal potency of azoles58,59,60. However, there is substantial conservation in the proteins of the mitochondrial respiratory chain between C. albicans and mammalian species. As a result, although the dysfunction of the mitochondrial respiratory chain induced by these compounds can lead to elevated ROS production in C. albicans cells, they can also cause an increase in ROS levels in mammalian cells, ultimately resulting in oxidative damage to host cells. In response to this issue, we suggest an alternative approach to inhibit the fungal-specific interaction between Erg11 and Ncp1. In this approach, elevated levels of ROS within the ER serve as a stimulus, inducing the release of Ca2+ to the mitochondria akin to a detonation fuse. Ultimately, the escalation of ROS levels within the mitochondria leads to the death of C. albicans cells (Fig. 9).
Ellipticine is a naturally occurring alkaloid with significant antineoplastic properties, originally isolated from Ochrosia elliptica61. An earlier Phase I clinical study conducted in cancer patients has shown that the half-life of ellipticine is ~23.7 h62. In this study, ellipticine has been identified as an inhibitor of the Erg11-Ncp1 interaction, specifically targeting Ncp1 to enhance the antifungal efficacy of azoles. These findings were achieved through various methods, including the MYTH system, in vitro antifungal drug susceptibility tests, co-IP, and DARTS. Ellipticine’s inhibition of the Erg11-Ncp1 interaction resulted in increased misfolded protein levels, intracellular Ca2+ concentration, ROS generation, mitochondrial dysfunction, and, ultimately, apoptosis, thereby augmenting the antifungal effects of FLC. After analyzing the molecular structure of ellipticine, it was discovered that its derivative, phiKan083, exhibits a notable ability to augment the efficacy of FLC in both in vivo and in vitro settings against fungal infections. The results suggest that focusing on the Erg11-Ncp1 interaction for the discovery of azole enhancers is a viable approach.
As Erg11 and Ncp1 are membrane-anchored proteins, challenges arise in expressing and purifying these proteins, as well as in obtaining their crystalline structures. Consequently, this study did not focus on structure-activity relationships when seeking analogues of ellipticine to enhance its antifungal efficacy. Future investigations should prioritize obtaining the crystal structure of the Erg11-Ncp1 complex to develop more effective antifungal inhibitors targeting the Erg11-Ncp1 interaction.
Methods
Ethics statement
All animal experiments comply with relevant ethical regulations. The study protocol was approved by the Animal Ethics Committee of Tongji University.
Strains, primers, agents, and cultural conditions
All strains, plasmids, and primers utilized in this study are presented in Supplementary Tables 5, 6, and 7. The C. albicans strains were cultured in YPD medium containing 2% dextrose (Sangon Biotech, A610219-0500), 1% yeast extract (Oxoid, LP0021B), and 2% peptone (Oxoid, LP0137) at 30°C. The SD medium containing 0.67% yeast nitrogen-based (Sangon Biotech, A610507-0100), required amino acids, and 2% dextrose was used to mutate target genes. To conduct the dual membrane functional assay, we employed SD-Leu-Trp (Takara, Cat#630316) and SD-Leu-Trp-His-Ade (Takara, Cat#630322) selective media. FLC (E129360), tamoxifen (T137974), arachidonic acid (A424359), piperine (P107402), menadione (M408896), terbinafine (T129208), amphotericin B (A408132), 5-fluorocytosine (F123460), and myriocin (M102400) are from Aladdin in Shanghai, China. Congo red (A427695-0100), dimethyl sulfoxide (DMSO) (A610163-0250) and 10% sodium dodecyl sulfate (SDS) solution (B548118-0100) were from Sangon Biotech in Shanghai, China. Carbonyl cyanide m-chlorophenylhydrazone (T7081) and ellipticine (T1166) were from TargetMol in Boston, USA; phiKan 083 (HY-108637) is from MedChemExpress in Shanghai, China. Ergosterol (Sangon Biotech, A351890-0100) is prepared into a 10 mM solution using Tween 80 and ethanol (1:1).
MYTH assays
The MYTH system was operated in accordance with the user manual for the dual membrane starter kits (P01201-P01229) user manual from Dualsystems Biotech, Schlieren, Switzerland63. The yeast strains obtained were subjected to serial dilution and spotted onto selective plates lacking either two amino acids (leucine and tryptophan) or four amino acids (leucine, tryptophan, histidine, and adenine). The bait plasmid pTSU2-APP, expressing the type I integral membrane protein APP (amyloid beta A4 precursor protein), and the prey plasmid pNubG-Fe65, expressing the cytosolic protein Fe65 (amyloid beta A4 precursor protein-binding family B member 1), were used as positive controls. The co-transformation with the control plasmids pTSU2-APP and the pPR3-N prey vector served as the negative controls. 3-AT (Aladdin, A107202) acts as a competitive inhibitor of the HIS3 gene product, and increasing concentrations of 3-AT enhance the stringency of HIS3 selection. The selective plates were then sealed and incubated at 30 °C for 4–6 days.
To identification of inhibitors of the Erg11-Ncp1 interaction using the MYTH assays. The yeasts were subjected in the 96-well plates using fluid selective medium. 4 mM 3-AT was added in the SD-Leu-Trp-His-Ade selective medium to eliminate the self-activation of Erg11. The pTSU2-APP and pNubG-Fe65 were chosen to determine whether the compounds can specifically inhibit Erg11-Ncp1 interaction. The yeasts were sealed and cultured for 16 h at 30 °C and adjusted to 1 × 103 cells/mL. The compounds were initiated at a concentration of 64 μg/mL and subsequently diluted in duplicate using SD-Leu-Trp and SD-Leu-Trp-His-Ade (containing 4 mM 3-AT) selective media. After incubating at 30 °C for 48 h, the 96-well plates were tested for OD600 at the full-wavelength microplate instrument (Thermo Fisher Scientific, Massachusetts, USA). The degree to which a compound disrupts the interaction between Erg11 and Ncp1 can be quantified by evaluating yeast growth in SD-Leu-Trp-His-Ade (containing 4 mM 3-AT) medium compared to its growth in SD-Leu-Trp medium. The inhibition rate of compounds is determined by subtracting the result of the subtraction in yeast growth rates in these two media. Compounds exhibiting ≥30% inhibition are considered potential inhibitors of the Erg11-Ncp1 interaction.
Spotting assay
The spotting assay was performed as follows: Overnight cultured strains were diluted ten-fold with sterilized phosphate-buffered saline (PBS) (Sangon Biotech, B040100-0005) in five gradients. Subsequently, 5 μL of each gradient tube was aliquoted onto spotting plates, with various compounds added to the solid plates as required by the experimental design. The plates were then incubated at 30 °C for 48 h before being photographed using an automatic chemiluminescence image analyzer (Tanon, Shanghai, China).
Co-IP assay
The co-IP assay was conducted as follows: Overnight C. albicans cultures of 10 mL were incubated at 30 °C. The collected C. albicans cells were washed twice with ice-cold sterilized water and then suspended in detergent-free lysis buffer [(PBS containing: 5 mM EDTA (Sangon Biotech, B540625-0500), 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sangon Biotech, A610425-0025), and protease inhibitor cocktail (TargetMol, C0001)]. The cells were lysed by adding 3 volumes of ice-cold lysis buffer and 3 volumes of glass beads per volume of pelleted yeast cells. Then, the C. albicans cell suspension was vigorously vortexed with 425–600 μm glass beads (Sigma-Aldrich, G9268) for four 1-min intervals, with the cells being kept on ice for 1 min between each period. Cell lysates totaling 800 μL were incubated with 8 μL of either anti-Myc (Santa Cruz, sc-47694) or anti-HA (Santa Cruz, sc-7392) antibody for 20 min at room temperature. The sample, pre-incubated with the antibody, was then applied to Protein A columns as per a Co-IP Kit’s (Clontech, 635721) instructions, which are designed to capture antibody-bound target protein complexes. In our investigation of the potential inhibitory effects of ellipticine on the Erg11-Ncp1 interaction, we encountered difficulties with compound binding to Protein A columns. To address this issue, we utilized an anti-Myc (20587ES03) or anti-HA agarose affinity gel (20586ES03) from Yeasen Biotechnology in Shanghai, China. The resulting 1000 μL cell lysates were then individually exposed to 10 μL of either anti-Myc or anti-HA affinity gel for 2 h at room temperature. Finally, the agarose beads containing the protein complexes were retrieved, subjected to three washes with TBS buffer (Sangon Biotech, B040126-0005), and subsequently analyzed through SDS-PAGE and western blot analysis.
Western blot
In the western blot procedure, proteins were initially separated by molecular weight using SDS-PAGE with an 8–20% precast PAGE gel (Beyotime, P0471M). Subsequently, the separated proteins were electroblotted onto a 0.45 μm polyvinylidene fluoride (PVDF) membrane (Millipore, IPVH00010). Following blocking, the primary antibodies were used: c-Myc (1:1000, Santa Cruz, sc-47694), HA (1:1000, Santa Cruz, sc-7392), β-Tubulin (1:1000, Abbkine, ABL1030). After incubating with an anti-mouse IgG secondary antibody m-IgGκ BP-HRP (Santa Cruz, sc-516102) at a dilution of 1:10,000, the Omni-ECL™Femto Light Chemiluminescence Kit (Epizyme Biotech, SQ201) was utilized to reveal the chemiluminescence signal, which was captured by an automatic chemiluminescence image analyzer (Tanon, Shanghai, China)28.
Molecular dynamics simulations
The crystal structure of Erg11 was obtained from the Protein Data Bank (PDB) (ID: 5V5Z) at a resolution of 2.9 Å. Due to the unavailability of the crystal structure of Ncp1 in the existing protein database, the predicted protein structure of Ncp1 generated by AlphaFold was used for molecular docking. The docking procedure was carried out using ZDOCK 3.0.2 software (University of California, San Francisco, USA)64, with default Hdock parameters. The analysis of the 10 docking results yielded the selection of the highest-scoring outcomes for subsequent molecular dynamics simulations. The docked complex structure was enclosed within a 20 Å cubic water box utilizing the TIP3P explicit solvent model. To maintain electrical neutrality, 24 Na+ ions were incorporated based on the volume of the box and the electronegativity of the protein. This process completed the construction of the molecular dynamics model of the complex, facilitating the commencement of traditional molecular dynamics simulation. A conventional molecular dynamics simulation was conducted for a total duration of 50 ns using the AMBER 18 software (University of California, San Francisco, USA), employing the assisted model building with energy refinement method.
Disk diffusion assays
The C. albicans cultures were incubated at 30 °C for 16 h and subsequently diluted 100-fold with sterilized PBS. The diluted cultures were then evenly spread onto drug-free or drug-containing YPD plates using a cotton swab. Following the drying of the plates, a 25 μg FLC (Liofilchem, 9166) paper disk was positioned at the center of each plate. The plates were then placed in an incubator at 30 °C and observed and photographed after 48 h.
Antifungal susceptibility testing
Antifungal susceptibility testing was performed using the following methods: Compounds were diluted in a 96-well plate through a two-fold dilution from column 2 to 11, with 100 μL discarded from column 11. Subsequently, overnight cultures of C. albicans were diluted to a concentration of 1 × 103 cells/mL in the YPD medium. Following this, 100 μL of the diluted C. albicans cell suspension was added to each well of the 96-well plate. Incubate the microdilution plates at 30 °C for 24 h and observe for the presence or absence of visible growth. Use a reading mirror to score the microdilution wells. Compare the turbidity in each well with that of the drug-free growth control well. Assign a numerical score to each well based on the following scale: 0, optically clear; 1, slightly hazy; 2, prominent decrease in turbidity (approximately 50% as determined visibly); 3, slight reduction in turbidity; and 4, no reduction of turbidity. In this study, the MIC is defined as the lowest concentration at which a score of 2 (prominent decrease in turbidity) is observed. MIC values of >64 μg/mL was defined as 128 μg/mL65.
The dose-matrix titration assay was utilized to assess the efficacy of a drug combination with C. albicans culture dilution methods consistent with those employed in MIC assays. The two drugs were diluted at a 2-fold ratio in separate 96-well plates, with one drug diluted horizontally and the other vertically. Subsequently, the contents of one plate were transferred to the other, followed by the addition of 100 μL of 1 × 103 cells/mL cell suspension to each well. The 96-well plate was incubated for 24 h to visually read the MIC for the calculation of the FICI and subsequently incubation was extended to 48 h for OD600 measurement using a full-wavelength microplate instrument (Thermo Fisher Scientific, Waltham, USA). FICI is calculated as follows: FICI = MICAB/MICA + MICBA/MICB. Here, MICAB refers to the MIC of drug A in the presence of drug B; MICA stands for the MIC of drug A alone. Similarly, MICBA refers to the MIC of drug B in the presence of drug A; MICB stands for the MIC of drug B alone. The interaction was categorized as synergic if the FICI was ≤0.5, additive if the FICI was >0.5–1.0, indifferent if the FICI was >1.0–4.0 and antagonistic if the FICI was >4.066. The tolerance, characterized by incomplete growth inhibition over an extended range of antifungal concentrations, was recorded after 48 h of incubation67. We quantified SMG at 48 h (average growth per well above the MIC, normalized to growth in the absence of the drug)3.
Growth inhibition curve assay
C. albicans colonies were introduced into the YPD medium, and the concentration of fungal liquid was standardized to an optical density of approximately 0.2 at a wavelength of 600 nm. Subsequently, compounds were diluted in a 96-well plate as required for the experimental procedure. A volume of 75 μL was then introduced into the 96-well plate. The completed plate was subsequently placed into the Infinite 200 PRO Multifunctional Microplate Reade (Tecan, California, USA) for incubation and analysis. The temperature was set at 30 °C, with OD600 measurements taken and recorded at 2-h intervals for 48 h.
RNA sequencing
The cultures of ERG11/erg11Δ and erg11m/erg11Δ mutants were incubated for 16 h with 4 μg/mL FLC in YPD at 30 °C. RNA was isolated and a DNA probe was utilized for rRNA hybridization. Subsequently, DNase I (Takara, 2270A) digestion of the DNA probe and selective digestion of the DNA/RNA hybridization chain by RNase H (Takara, 2641Q) facilitated the isolation of the desired RNA. The isolated RNA was then reverse-transcribed into the cDNA, which was further synthesized into double-stranded DNA with the use of primers to amplify the ligation products. The purified double-stranded DNA was mixed with an end-repair reaction mix containing DNA polymerase, ATP, and dNTPs to repair the ends of the DNA fragments. Then, the 3’ ends of the DNA fragments were added to an adenine (A) nucleotide via incubation with a dATP and terminal transferase enzyme mix. The DNA fragments were thermally denatured into a single strand, and then a bridge primer was used to cyclize the single-strand DNA to obtain a single-strand circular DNA library and then sequenced (BGI Genomics, Shenzhen, China).
Determination of misfolded protein levels
To determination of intracellular and mitochondrial misfolded protein levels, overnight cultures of C. albicans were inoculated at a 1:100 dilution and grown in the presence of respective compounds (or DMSO equivalent) for 4 h at 30 °C. Exposure to 5 mM DTT (Sangon Biotech, A620058-0025) for 2 h as the positive control. Cell suspensions of 1 × 107 cells/mL were incubated with 50 μM TPE-MI (MedChemExpress, HY-143218) at 30 °C or 0.1 μM MitoTracker Green (MedChemExpress, HY-135056) at 37 °C for 30 min. The excitation and emission wavelength of TPE-MI were 350 nm and 470 nm, respectively, while the MitoTracker Green fluorescence intensities were measured with excitation at 490 nm and emission at 523 nm. Fluorescence intensity measurements were obtained using a BD FACSverse flow cytometer (BD Biosciences, New York, USA). Forward Scatter (FSC) and Side Scatter (SSC) were adjusted at 419 V and 150 V, respectively to encircle the C. albicans cells to exclude cell debris. The gating process was concluded upon reaching a stopping criterion of 10,000 events.
Measurement of ROS generation
The levels of intracellular and intra-mitochondrial ROS were quantified using a BD FACSVerse flow cytometer (BD Biosciences, New York, USA). The overnight cultures of C. albicans were inoculated at a 1:100 dilution and grown in the presence of respective compounds (or DMSO equivalent) for 4 h at 30 °C. Exposure to 5 mM DTT for 2 h as the positive control. Subsequently, Cell suspensions of 1 × 107 cells/mL were incubated with either 10 μM DCFH-DA (Sigma-Aldrich, D6883) at 37 °C or 5 μM MitoSOX Red (MedChemExpress, HY-D1055) at 30 °C for 30 min. The DCFH-DA fluorescence intensities were detected with an excitation wavelength of 492 nm and an emission wavelength of 517 nm, while the MitoSOX Red fluorescence intensities were detected with an excitation wavelength of 510 nm and an emission wavelength of 580 nm. FSC and SSC were adjusted at 415 V and 150 V, respectively to encircle the C. albicans cells to exclude cell debris. The gating process was concluded upon reaching a stopping criterion of 10,000 events.
Determination of the mitochondrial membrane potential
The mitochondrial membrane potential of C. albicans cells was assessed using Rhodamine 123 (Aladdin, R299309) staining. Cells cultured overnight were diluted 1:100 and treated with drugs for 8 h. A volume of 10 mL of cell suspensions was stained with 10 μM Rhodamine 123 at 30 °C for 30 min in the absence of light. Fluorescence intensity was measured with excitation at 507 nm and emission at 529 nm using a BD FACSVerse flow cytometer (BD Biosciences, New York, USA).
Apoptosis assay
The C. albicans cells required protoplast preparation prior to Annexin V and PI staining. Overnight cultured C. albicans were diluted 1:100 and further incubated in the presence or absence of compounds (or DMSO equivalent) for 8 h at 30 °C. The purified C. albicans cells were then exposed to a solution containing 80 μL of 2.4 M sorbitol (comprising 0.1 M PBS, 5 mM EDTA, 50 mM DTT, 50 mM Na2HPO4, 50 mM NaH2PO4, and 40 mM 2-mercaptoethanol) and 5 μL Zymolyase (ZYMO RESEARCH, R1002). After incubating at 37 °C for 30 min, 160 μL of 0.1 M sodium citrate containing 0.1% Triton X-100 was added to the processed protoplasts. Subsequent procedures were carried out in accordance with the FITC Annexin V Apoptosis Detection Kit I (BD Pharmingen, 556547). FITC fluorescence density was measured using 488 nm excitation and 525 nm emission settings, while PI fluorescence density was measured with an excitation wavelength of 535 nm and an emission wavelength of 615 nm. FSC and SSC were adjusted at 380 V and 200 V, respectively to locate the C. albicans cells using the unstained tube. Then proceeding with the samples stained singly with FITC and singly with PI and adjusting the compensation so that the cell population is positioned in the lower left corner of 104. Control the flow rate of the cells at 1000 cells per second.
Measurement of the content of Ca2+
To assess the levels of the cytosolic Ca2+, the overnight cultures of C. albicans were diluted 1:100 and exposed to 4 μg/mL FLC and 16 μg/mL ellipticine (or DMSO equivalent) for 4 h. Subsequently, the cells were stained with Fluo-3 AM (5 μM) (HY-D0716) from MedChemExpress, Shanghai, China, and incubated in darkness for 30 min. The fluorescence intensities of Fluo-3 AM were measured using an excitation wavelength of 488 nm and an emission wavelength of 526 nm. Fluorescence intensity measurements were obtained using a BD FACSverse flow cytometer (BD Biosciences, New York, USA). FSC and SSC were adjusted at 419 V and 150 V, respectively to encircle the C. albicans cells to exclude cell debris. The gating process was concluded upon reaching a stopping criterion of 10,000 events.
Confocal microscope analysis
The co-localization of ER or vacuole with Ca2+ and localization of mitochondrial Ca2+ were observed using the Nikon Laser Scanning Confocal Microscope System (Nikon, Tokyo, Japan). Prior to imaging, C. albicans cells were pre-stained with ER-Tracker Red (2 μM, 30 min) (MedChemExpress, HY-D1431), or Cell Tracker Blue (25 uM, 30 min) (MedChemExpress, HY-D1462), and co-stained with Fluo-3 AM (5 μM, 30 min) at 37 °C. Bright red fluorescence indicated ER, blue fluorescence indicated vacuole, while green fluorescence indicated Ca2+. To detect the level of Ca2+ in mitochondria, C. albicans cells were stained with Rhod-2 AM (5 μM, 30 min) (MedChemExpress, HY-D0989) at 37 °C. The images were acquired utilizing a confocal microscope (Nikon, Tokyo, Japan). The excitation and emission wavelengths for the fluorescent probes used in this study are as follows: ER-Tracker Red (587 nm/615 nm), Rhod-2AM (549 nm/578 nm), Fluo-3 AM (488 nm/526 nm), and Cell Tracker Blue (372 nm/470 nm).
DARTS assay
The DARTS assay was operated according to the reported protocols47. The 10 mL overnight C. albicans cultures were harvested. Following two washes with ice-cold sterilized water, C. albicans cells were resuspended in detergent-free lysis buffer. The resuspended cells were transferred a tube containing 0.5-mm glass beads and vortexed at maximum speed for 1 min, followed by a 1-min rest on ice. Perform four cycles of 1-min vertexing and 1-min resting. The samples underwent four cycles of 1-min vertexing and 1-min resting. Subsequently, the tube containing lysates were centrifuged at 20,000 × g for 10 min at 4 °C. Following centrifugation, 1000 μL of cell lysates with either 5 μL of DMSO or 5 μL of ellipticine (5 mg/mL) separately for 1 h at room temperature. The protein samples were then divided into six aliquots, each containing 50 μL, and subject to a 30-min digestion with an appropriate concentration of pronase. The digestion process was halted by adding SDS loading buffer to each sample, which were then heated at 100 °C for 10 min. Finally, the samples were analyzed using SDS-PAGE and Western blot analysis.
Systemic candidiasis model
Mice were maintained in a controlled environment with a 12-h light/dark cycle, temperature set between 18 and 25 °C, and relative humidity of 30–70%. Mice had unrestricted access to food and water throughout the experiment. C57BL/6 mice (specific-pathogen-free, 6–8 weeks) (Slaccas, Shanghai, China) weighing 17–19 g were chosen for the establishment of a systemic candidiasis model. For each group, a total of 12 mice (half male and half female) were systemically infected through the tail vein injection of 2 × 105 C. albicans cells. In the treatment group, the initial intraperitoneal administration occurred 2 h post-infection and was administered for three consecutive days with one-day intervals between doses. Following completion of dosing, all mice were euthanized at 3-day intervals. Subsequently, the liver, kidney, and spleen of each mouse were extracted and processed in PBS, ground, and plated onto SDA (4% dextrose, 1% peptone, and 2% agar) solid plates at varying dilutions. The resulting colonies on each SDA plate were enumerated, and the colony-forming units were calculated68.
Histopathology
Following the systemic candidiasis model, the left kidney of each mouse was excised and placed in 4% paraformaldehyde for 3 days for histological examination. Subsequently, the finished fixed kidney tissue was sectioned thinly, stained with PAS, and subjected to photographic documentation and analysis.
Statistics and reproducibility
All experiments (except those described otherwise in the legend) were performed independently at least twice with similar results. The number of independent experiments is reported in the figure legend. Statistical analysis was conducted using two-tailed unpaired t-tests. The Log-rank (Mantel–Cox) test was used to manifest differences between survival curves. For all statistical analyses, p < 0.05 was considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq raw data can be acquired from the sequence read archive (SRA) section of the National Center for Biotechnology Information (NCBI) repository under the accession code PRJNA1083027 [http://submit.ncbi.nlm.nih.gov]. Source data are provided with this paper.
References
Lu, H., Hong, T., Jiang, Y., Whiteway, M. & Zhang, S. Candidiasis: From cutaneous to systemic, new perspectives of potential targets and therapeutic strategies. Adv. Drug Deliv. Rev. 199, 114960 (2023).
Denning, D. W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 24, e428–e438 (2024).
Rosenberg, A. et al. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat. Commun. 9, 2470 (2018).
Shafiei, M., Peyton, L., Hashemzadeh, M. & Foroumadi, A. History of the development of antifungal azoles: a review on structures, SAR, and mechanism of action. Bioorg. Chem. 104, 104240 (2020).
Pristov, K. E. & Ghannoum, M. A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 25, 792–798 (2019).
Feng, Y., Lu, H., Whiteway, M. & Jiang, Y. Understanding fluconazole tolerance in Candida albicans: implications for effective treatment of candidiasis and combating invasive fungal infections. J. Glob. Antimicrob. Resist. 35, 314–321 (2023).
Berman, J. & Krysan, D. J. Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 18, 319–331 (2020).
Levinson, T., Dahan, A., Novikov, A., Paran, Y., Berman, J. & Ben-Ami, R. Impact of tolerance to fluconazole on treatment response in Candida albicans bloodstream infection. Mycoses 64, 78–85 (2021).
Cowen, L. E. & Lindquist, S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309, 2185–2189 (2005).
Spitzer, M., Robbins, N. & Wright, G. D. Combinatorial strategies for combating invasive fungal infections. Virulence 8, 169–185 (2017).
Li, W., Shrivastava, M., Lu, H. & Jiang, Y. Calcium-calcineurin signaling pathway in Candida albicans: a potential drug target. Microbiol. Res. 249, 126786 (2021).
Lu, H. et al. A small molecule inhibitor of Erg251 makes fluconazole fungicidal by inhibiting the synthesis of the 14α-methylsterols. mBio 14, e0263922 (2023).
Martel, C. M. et al. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54, 4527–4533 (2010).
Sagatova, A. A., Keniya, M. V., Wilson, R. K., Monk, B. C. & Tyndall, J. D. Structural insights into binding of the antifungal drug fluconazole to saccharomyces cerevisiae lanosterol 14α-demethylase. Antimicrob. Agents Chemother. 59, 4982–4989 (2015).
Cuccioloni, M., Sparapani, L., Amici, M., Lupidi, G., Eleuteri, A. M. & Angeletti, M. Kinetic and equilibrium characterization of the interaction between bovine trypsin and I-ovalbumin. Biochim. Biophys. Acta 1702, 199–207 (2004).
Bradshaw PC. Cytoplasmic and mitochondrial NADPH-coupled redox systems in the regulation of aging. Nutrients 11, 504 (2019).
Hanukoglu, I. Antioxidant protective mechanisms against reactive oxygen species (ROS) generated by mitochondrial P450 systems in steroidogenic cells. Drug Metab. Rev. 38, 171–196 (2006).
Kritsiligkou, P., Rand, J. D., Weids, A. J., Wang, X., Kershaw, C. J. & Grant, C. M. Endoplasmic reticulum (ER) stress-induced reactive oxygen species (ROS) are detrimental for the fitness of a thioredoxin reductase mutant. J. Biol. Chem. 293, 11984–11995 (2018).
Chaillot, J. et al. The monoterpene carvacrol generates endoplasmic reticulum stress in the pathogenic fungus Candida albicans. Antimicrob. Agents Chemother. 59, 4584–4592 (2015).
Sircaik, S. et al. The protein kinase Ire1 impacts pathogenicity of Candida albicans by regulating homeostatic adaptation to endoplasmic reticulum stress. Cell Microbiol. 23, e13307 (2021).
Hoepfner, D. et al. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol Res. 169, 107–120 (2014).
Iracane, E., Donovan, P. D., Ola, M., Butler, G., Holland, L.M. Identification of an exceptionally long intron in the HAC1 gene of Candida parapsilosis. mSphere 3, e00532-18 (2018).
Lamb, D. C. et al. Generation of a complete, soluble, and catalytically active sterol 14 alpha-demethylase-reductase complex. Biochemistry 38, 8733–8738 (1999).
Tarassov, K. et al. An in vivo map of the yeast protein interactome. Science 320, 1465–1470 (2008).
Thaminy, S., Auerbach, D., Arnoldo, A. & Stagljar, I. Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res. 13, 1744–1753 (2003).
Huang, R., Zhang, M., Rwere, F., Waskell, L. & Ramamoorthy, A. Kinetic and structural characterization of the interaction between the FMN binding domain of cytochrome P450 reductase and cytochrome c. J. Biol. Chem. 290, 4843–4855 (2015).
Khosla, N., Thayil, S. M., Kaur, R. & Kesavan, A. K. MSMEG_3955 from Mycobacterium smegmatis is a FMN bounded homotrimeric NAD(P)H:Flavin mononucleotide (FMN) oxidoreductase. BMC Microbiol. 21, 319 (2021).
Xiong, J. et al. Halofantrine hydrochloride acts as an antioxidant ability inhibitor that enhances oxidative stress damage to Candida albicans. Antioxidants 13, 223 (2024).
Yu, Q., Zhang, B., Li, J., Zhang, B., Wang, H. & Li, M. Endoplasmic reticulum-derived reactive oxygen species (ROS) is involved in toxicity of cell wall stress to Candida albicans. Free Radic. Biol. Med 99, 572–583 (2016).
Shi, Y. et al. Helical sulfono-γ-AApeptides with aggregation-induced emission and circularly polarized luminescence. J. Am. Chem. Soc. 141, 12697–12706 (2019).
Zhang, S. & Hong, Y. Measuring cysteine exposure in unfolded proteins with tetraphenylethene maleimide and its analogs. Methods Mol. Biol. 2378, 3–18 (2022).
Wang, L. T. et al. WLS/wntless is essential in controlling dendritic cell homeostasis via a WNT signaling-independent mechanism. Autophagy 17, 4202–4217 (2021).
Yang, F. et al. Tunicamycin potentiates antifungal drug tolerance via aneuploidy in Candida albicans. mBio 12, e0227221 (2021).
Bernales, S., McDonald, K. L. & Walter, P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4, e423 (2006).
Pittis, A. A., Goh, V., Cebrian-Serrano, A., Wettmarshausen, J., Perocchi, F. & Gabaldón, T. Discovery of EMRE in fungi resolves the true evolutionary history of the mitochondrial calcium uniporter. Nat. Commun. 11, 4031 (2020).
Hong, M. P., Vu, K., Bautos, J. & Gelli, A. Cch1 restores intracellular Ca2+ in fungal cells during endoplasmic reticulum stress. J. Biol. Chem. 285, 10951–10958 (2010).
Bround, M. J. et al. MCU-independent Ca(2+) uptake mediates mitochondrial Ca(2+) overload and necrotic cell death in a mouse model of Duchenne muscular dystrophy. Sci. Rep. 14, 6751 (2024).
Kwun, M. S. & Lee, D. G. Quercetin-induced yeast apoptosis through mitochondrial dysfunction under the accumulation of magnesium in Candida albicans. Fungal Biol. 124, 83–90 (2020).
Kato, M., Tanabe, K., Miki, A., Ichimori, K. & Waki, S. Membrane potential of Plasmodium falciparum gametocytes monitored with rhodamine 123. FEMS Microbiol. Lett. 57, 283–288 (1990).
Divo, A. A., Patton, C. L. & Sartorelli, A. C. Evaluation of rhodamine 123 as a probe for monitoring mitochondrial function in Trypanosoma brucei spp. J. Eukaryot. Microbiol 40, 329–335 (1993).
Shah, A. & Dobrovolskaia, M. A. Detection of induction of mitochondrial oxidative stress by nanoparticles in T cells using MitoSOX red dye. Methods Mol. Biol. 2789, 145–151 (2024).
Peng, L. et al. The V-ATPase regulates localization of the TRP Ca(2+) channel Yvc1 in response to oxidative stress in Candida albicans. Int J. Med. Microbiol. 310, 151466 (2020).
Gautam, N., Sankaran, S., Yason, J. A., Tan, K. S. W. & Gascoigne, N. R. J. A high content imaging flow cytometry approach to study mitochondria in T cells: MitoTracker green FM dye concentration optimization. Methods 134-135, 11–19 (2018).
Kabakov, A. E. & Gabai, V. L. Cell death and survival assays. Methods Mol. Biol. 1709, 107–127 (2018).
Zamith-Miranda, D. et al. Multi-omics signature of Candida auris, an emerging and multidrug-resistant pathogen. mSystems 4, e00257-19 (2019).
Cervera, A. M., Gozalbo, D., McCreath, K. J., Gow, N. A., Martínez, J. P. & Casanova, M. Molecular cloning and characterization of a Candida albicans gene coding for cytochrome c haem lyase and a cell wall-related protein. Mol. Microbiol. 30, 67–81 (1998).
Lomenick, B., Jung, G., Wohlschlegel, J. A. & Huang, J. Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 3, 163–180 (2011).
Mendoza, I., Quintero, F. J., Bressan, R. A., Hasegawa, P. M. & Pardo, J. M. Activated calcineurin confers high tolerance to ion stress and alters the budding pattern and cell morphology of yeast cells. J. Biol. Chem. 271, 23061–23067 (1996).
Sanglard, D., Ischer, F., Marchetti, O., Entenza, J. & Bille, J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48, 959–976 (2003).
Chaudhuri, B., Hämmerle, M. & Fürst, P. The interaction between the catalytic A subunit of calcineurin and its autoinhibitory domain, in the yeast two-hybrid system, is disrupted by cyclosporin A and FK506. FEBS Lett. 357, 221–226 (1995).
Li, W. et al. Pitavastatin calcium confers fungicidal properties to fluconazole by inhibiting ubiquinone biosynthesis and generating reactive oxygen species. Antioxidants 13, 667 (2024).
Garbett, N. C. & Graves, D. E. Extending nature’s leads: the anticancer agent ellipticine. Curr. Med. Chem. Anticancer Agents 4, 149–172 (2004).
Zeng, L. et al. QCR7 affects the virulence of Candida albicans and the uptake of multiple carbon sources present in different host niches. Front. Cell Infect. Microbiol. 13, 1136698 (2023).
Huang, X. et al. Mitochondrial complex I bridges a connection between regulation of carbon flexibility and gastrointestinal commensalism in the human fungal pathogen Candida albicans. PLoS Pathog. 13, e1006414 (2017).
Rodrigues, M.L. The multifunctional fungal ergosterol. mBio 9, e01755-18 (2018).
Chen, S. C. & Sorrell, T. C. Antifungal agents. Med J. Aust. 187, 404–409 (2007).
Wang, M., Roberts, D. L., Paschke, R., Shea, T. M., Masters, B. S. & Kim, J. J. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. USA 94, 8411–8416 (1997).
Xu, Y. et al. Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: endogenous ROS augmentation. J. Proteome Res. 8, 5296–5304 (2009).
Fu, Z., Lu, H., Zhu, Z., Yan, L., Jiang, Y. & Cao, Y. Combination of baicalein and Amphotericin B accelerates Candida albicans apoptosis. Biol. Pharm. Bull. 34, 214–218 (2011).
Li, D.D. et al. Potent in vitro synergism of fluconazole and osthole against fluconazole-resistant Candida albicans. Antimicrob. Agents Chemother. 61, e00436-17 (2017).
Froelich-Ammon, S. J., Patchan, M. W., Osheroff, N. & Thompson, R. B. Topoisomerase II binds to ellipticine in the absence or presence of DNA. Charact. Enzym.Drug Interact. Fluoresc. Spectrosc. J. Biol. Chem. 270, 14998–15004 (1995).
Gouyette, A., Huertas, D., Droz, J. P., Rouesse, J. & Amiel, J. L. Pharmacokinetics of 2-methyl-9-hydroxyellipticinium acetate (NSC-264137) in cancer patients (phase I study). Eur. J. Cancer Clin. Oncol. 18, 1285–1292 (1982).
Hershko, A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 12, 1191–1197 (2005).
Pierce, B. G., Hourai, Y. & Weng, Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One 6, e24657 (2011).
Epp, E. et al. Reverse genetics in Candida albicans predicts ARF cycling is essential for drug resistance and virulence. PLoS Pathog. 6, e1000753 (2010).
Johnson, M. D., MacDougall, C., Ostrosky-Zeichner, L., Perfect, J. R. & Rex, J. H. Combination antifungal therapy. Antimicrob. Agents Chemother. 48, 693–715 (2004).
Zomorodian, K., Bandegani, A., Mirhendi, H., Pakshir, K., Alinejhad, N. & Poostforoush Fard, A. In vitro susceptibility and trailing growth effect of clinical isolates of candida species to azole drugs. Jundishapur J. Microbiol. 9, e28666 (2016).
Wang, H. et al. Myriocin enhances the antifungal activity of fluconazole by blocking the membrane localization of the efflux pump Cdr1. Front. Pharmacol. 13, 1101553 (2022).
Acknowledgements
This study received financial support from the National Natural Science Foundation of China (No. 82020108032 to Y.J.), the Innovation Program of Shanghai Municipal Education Commission (202101070007-E00094 to Y.J.) and the National Key Research and Development Program of China (No.2022YFC2303004 to Y.J.).
Author information
Authors and Affiliations
Contributions
W.L. conducted most of the experiments and performed data analysis. W.L., H. L., M.W., and Y.J. wrote the manuscript draft and revised the manuscript. W.L., S.H., and J.Y. constructed mutant strains. W.L., S.H., and J.Y. evaluated the antifungal activities and data collection. W.L., H. L., M. W., and Y. J. discussed and analyzed the data. H.L. and Y.J. conceived the idea. H.L. and Y. J. directed the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Patrick Van Dijck, who co-reviewed with Hans Carolus, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, W., Whiteway, M., Hang, S. et al. Impairing the interaction between Erg11 and cytochrome P450 reductase Ncp1 enhances azoles’ antifungal activities. Nat Commun 16, 6821 (2025). https://doi.org/10.1038/s41467-025-62131-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62131-z