Abstract
PIEZO1, a mechanosensor in endothelial cells, plays a critical role in fetal vascular development during embryogenesis. However, its expression and function in placental trophoblasts remain unexplored. Here, we demonstrate that PIEZO1 is expressed in placental villus trophoblasts, where it is essential for trophoblast fusion and placental development. Mice with trophoblast-specific PIEZO1 knockout exhibit embryonic lethality without obvious vascular defects. Instead, PIEZO1 deficiency disrupts the formation of the syncytiotrophoblast layer in the placenta. Mechanistically, PIEZO1-mediated calcium influx activates TMEM16F lipid scramblase, facilitating the externalization of phosphatidylserine, a key “fuse-me” signal for trophoblast fusion. These findings reveal PIEZO1 as a crucial mechanosensor in trophoblasts and highlight its essential role in regulating trophoblast fusion and placental development, expanding our understanding of PIEZO1’s functions beyond endothelial cells during pregnancy.
Similar content being viewed by others
Introduction
Mechanical forces are essential regulators of embryonic development, shaping processes from gastrulation to organogenesis1,2,3,4. In contrast, mechanobiology of the placenta, a vital yet understudied organ5, remains poorly defined. During pregnancy, the placenta undergoes profound mechanical adaptations, including trophoblast migration, decidual invasion, syncytialization (trophoblast fusion), villous morphogenesis, and spiral artery remodeling1,6,7,8. While mechanical cues drive these processes, the molecular sensors transducing forces into biochemical signals in trophoblasts remain elusive.
PIEZO1, a mechanosensitive ion channel9,10, has emerged as a key force transducer in development. Constitutive PIEZO1 knockout (KO) mice are embryonically lethal, exhibiting severe defects in fetal angiogenesis11,12. Similarly, endothelial-specific deletion of PIEZO1 recapitulates this phenotype, underscoring its essential role in endothelial mechanotransduction. Beyond vascular development, PIEZO1 also regulates neural stem cell lineage specification13,14,15, osteogenesis16,17, and lymphangiogenesis18,19. However, despite trophoblasts being exposed to dynamic shear stress and/or compressive forces during gestation8, whether PIEZO1 mediates force sensing in trophoblasts remains unknown.
Here, we identify PIEZO1 as a mechanosensor in villous trophoblasts and demonstrate its essential role in trophoblast fusion and placental development. By shifting the focus from endothelial cells to trophoblasts, this study reveals a previously unrecognized PIEZO1-dependent mechanotransduction pathway in trophoblasts, providing insights into the mechanical regulation of the placenta development and identifying potential therapeutic targets for pregnancy complications associated with placenta dysfunction.
Results
PIEZO1 is expressed in placental trophoblasts
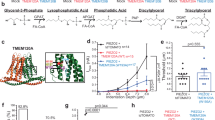
Immunofluorescence analysis revealed that PIEZO1 is expressed in both single-nucleated cytotrophoblasts (CTB) and multi-nucleated syncytiotrophoblasts (STB) in human chorionic villi (Fig. 1A), as well as in the human placental trophoblast cell line, BeWo (Fig. 1B), and the human trophoblast stem cells (hTSCs) (Fig. S2A). Our qRT-PCR analysis further demonstrated that PIEZO1 not PIEZO2 mRNAs are expressed in BeWo cells (Fig. S3A). The Comparative Transcriptomic Placental Model Atlas20, PIEZO1 transcripts are highly expressed in various in vitro models of human trophoblasts, including BeWo, JAR, JEG3 and HTR8 trophoblast cell lines, as well as primary trophoblasts and hTSCs (Fig. S3B), further supporting PIEZO1 expression in human trophoblasts.
A Immunofluorescence staining of PIEZO1 (green) and nuclei (Hoechst, blue) in human first-trimester placenta villi with higher magnification shown on the right. B Immunofluorescence staining of PIEZO1 (green) in BeWo cells. C Representative single channel mechanosensitive currents from BeWo cells. Currents were elicited by −60 mmHg pressure and recorded at voltage steps from −100 mV to −40 mV in 20 mV increments with holding potential at 0 mV. D The current-voltage (I-V) relationship for the currents in (C). The data were fitted with linear regression, obtaining a single-channel conductance of 29.3 ± 1.8 pS (n = 5 individual cells). E Representative cell-attached pressure-clamp recording of control-siRNA and PIEZO1-siRNA treated BeWo cells (left). The macroscopic current was elicited by pressure steps ranging from 0 to −60 mmHg with a holding potential at −80 mV. F The current-pressure relationship for the mechanosensitive current recorded from BeWo cells treated with control-siRNA and PIEZO1-siRNA. G PIEZO1 current amplitudes at –60 mmHg in (F). Unpaired 2-sided Student t-test. ∗∗∗∗: p = 0.0000044338. In F and G, n = 10 and 11 for control siRNA and PIEZO1 siRNA treated cells, respectively. H Representative Ca2+ (red) imaging of control and PIEZO1 siRNA knockdown BeWo cells stimulated with 10 µM Yoda1. I Time course quantification of Ca2+ levels in control (n = 21) and PIEZO1 siRNA (n = 25) knockdown BeWo cells following 10 µM Yoda1 stimulation (indicated by the arrow). Data are presented as mean ± s.e.m from 7 biological replicates. J Immunofluorescence of tdTomato (magenta), MCT1 (a marker for SynT-1 layer, green), MCT4 (a marker for SynT-2 layer, red), and DAPI (blue) for the placenta from Piezo1-td-Tomato transgenic mouse at E13.0. Left: schematic of mouse placental labyrinth showing the maternal-fetal interface. Circular tdTomato+ and MCT1+ signals are red blood cells. n = 4 biological replicates. Source data are provided as a Source Data file.
Pressure clamp electrophysiology recording elicited a mechanosensitive current from BeWo cells with a unitary conductance of 29.3 ± 1.8 pS (Fig. 1C–G), which aligns with the typical single channel conductance of PIEZO110,21. Yoda1, a PIEZO1 agonist, robustly increased intracellular Ca2+ ([Ca2+]i), which can be attenuated by the PIEZO1 inhibitor, GsMTx4 (Fig. S1). Silencing PIEZO1 using siRNA not only markedly attenuated this mechanosensitive current from BeWo cells (Fig. 1E–G) and hTSCs (Fig. S2D–F) but also reduced Yoda1-induced [Ca2+]i increase in these cells (Fig. 1H, I for BeWo and S2B, C for hTSCs). Taken together, these results support that PIEZO1 is functionally expressed in human trophoblasts.
We previously reported that the TRPV4 ion channel is specifically expressed in human but not mouse trophoblasts, indicating differential expression profiles in trophoblasts from different species22. To determine whether Piezo1 is also expressed in mouse trophoblasts, we examined its protein expression in the mouse placenta utilizing the Piezo1-tdTomato reporter mice (Piezo1P1-tdT) in which tdTomato is covalently tagged to endogenous Piezo112. The td-Tomato signal was detected in the placental labyrinth where SynT-1 and SynT-2, two layers of STBs, are located23 (Fig. 1J). We found strong td-Tomato signal in SynT-2 and weaker signal in SynT-1 using antibodies for MCT1 and 4, which are two monocarboxylate transporters specific to SynT-1 and -222,24,25. Consistently, single-cell RNA sequencing of the mouse placenta from the Spatiotemporal Transcriptomic Atlas of Mouse Placenta (STAMP)26 also shows that Piezo1 (Fig. S4A, B) is highly expressed in SynT-2 STBs that specifically express the fusogenic gene Synb, but weakly expressed in SynT-1 STBs that specifically express another fusogenic gene Syna23 (Fig. S4C, D). Taken together, our qRT-PCR, immunostaining and functional analysis confirm that, unlike TRPV4, PIEZO1 is expressed in both human and mouse placental trophoblasts and predominantly localizes to SynT-2 STBs in the mouse labyrinth.
Trophoblast-specific deletion of Piezo1 results in embryonic lethality
Piezo1 constitutive KO mice are embryonically lethal, a phenotype previously attributed to endothelial dysfunction and defective angiogenesis due to PIEZO1 deficiency11,12. To dissect the role of PIEZO1 in placental trophoblasts, we generated a trophoblast-specific knockout (cKO) of Piezo1 mice by breeding Piezo1f/f27 with Elf5-Cre (Fig. 2A), a transgenic mouse line that specifically drives Cre recombinase expression in the trophectoderm as early as embryonic day 4.5 (E4.5)28. The heterozygous mice (HET) were born at near-Mendelian ratios (Fig. 2B) and exhibited no signs of embryonic lethality or developmental abnormalities, indicating that this breeding scheme did to lead to haploinsufficiency. On the other hand, nearly all cKO embryos died during pregnancy, with only 1 out of 105 (<1%) cKO mice surviving after birth (Fig. 2B, right). This drastically deviates from the anticipated Mendelian ratio of 25% for cKO mice (Fig. 2B, left). Morphologically, the cKO embryos and placentas were markedly smaller compared to their litter mates; some even showed signs of degradation and resorption (Fig. 2C and S5), suggesting that cKO mice suffer from intrauterine growth restriction (IUGR) and stillbirth. Embryonic death of Piezo1 trophoblast cKO mice starts as early as E11.5 (Fig. S5A).
A Breeding scheme for the generation of Piezo1 conditional knockout (cKO) mice. B Genotype distribution of wild-type (WT), heterozygous (HET), and cKO pups compared to the expected Mendelian inheritance ratio. The numbers of animals with specific genotypes over 105 pups were shown in the parentheses. C Representative embryos and placentas from Piezo1 WT, HET, and cKO mice at embryonic day 13.0 (E13.0). Images of the complete litter can be found in Fig. S5B.
Piezo1 deficiency disrupts trophoblast fusion
We conducted histological analysis to identify the defects in the cKO placenta. H&E staining demonstrated that E13.0 cKO placentas had markedly enlarged maternal blood sinuses in the labyrinth compared to those in WT placentas from the same litter (Fig. S6, red asterisks). Interestingly, the numbers of fetal capillaries, which were identified by nucleated fetal red blood cells (RBCs), did not show an obvious difference between WT and cKO placentas (Fig. S6, blue stars). Our CD31 staining of WT and cKO placentas further supports that trophoblast-specific deletion of Piezo1 does not significantly impact fetoplacental blood vessel development (Fig. 3A, B). This is distinct from the severe angiogenesis defects observed in the constitutive Piezo1 KO and the endothelial-specific Piezo1 cKO placentas11,12. Instead, our histological analysis suggests that defective trophoblast differentiation, rather than impaired angiogenesis, is the primary cause of embryonic lethality in trophoblast-specific Piezo1 cKO mice.
CD31 immunohistochemical staining of PIEZO1 WT (A) and cKO (B) placentas at E13.0 with enlarged areas shown on the right. n = 3 biological replicates for WT and 3 biological replicates for cKO. MCT1 and MCT4 immunofluorescence staining of the PIEZO1 WT (C) and cKO (D) placentas (upper panels) with enlargement (lower panels) at E12.5. n = 5 biological replicates for WT and 6 biological replicates for cKO.
Defects in trophoblast fusion, a crucial step of trophoblast differentiation, are a major contributor to placental insufficiency, IUGR, and in severe cases, stillbirth29,30,31. To evaluate STB formation in the cKO labyrinth, we stained MCT1 and MCT4, markers of the SynT-1 and SynT-2 STBs, respectively22,24,25. In stark contrast to the smooth, uniform, and colocalized MCT1 and MCT4 staining in WT labyrinth, MCT4 expression was dramatically reduced, and in some regions, entirely absent in the Piezo1 cKO labyrinth. Although MCT1 staining was largely preserved, it exhibited a more irregular, zigzag pattern (Fig. 3C, D). These findings indicate that trophoblast-specific deletion of Piezo1 severely disrupts the formation of the SynT-2 STB layer while having a milder effect on the SynT-1 STB layer, consistent with Piezo1’s expression profile in these STB layers (Figs. 1J and S4).
PIEZO1 is essential for trophoblast fusion in vitro
To further confirm the essential role of PIEZO1 in trophoblast fusion, we pharmacologically and genetically manipulated PIEZO1 in BeWo cells and quantified its effects on forskolin-induced BeWo cell fusion in vitro. Inhibition of PIEZO1 with GsMTx4 abolished BeWo cell fusion (Fig. 4A, B). Similarly, PIEZO1 knockdown via siRNA significantly reduced the fusion index (Fig. 4C, D). Conversely, PIEZO1 overexpression significantly increased PIEZO1 current (Fig. S7) and enhanced BeWo cell fusion index from 0.42 ± 0.03 to 0.65 ± 0.02 (Fig. 4E, F). Human chorionic gonadotropin (hCG) is specifically secreted by STBs32. Using hCG as a biomarker of trophoblast fusion22,33,34, we found that GsMTx4 treatment significantly reduced hCG secretion from hTSCs after inducing differentiation (Fig. S8), further supporting that PIEZO1 plays an important role in regulating trophoblast fusion. Taken together, our in vitro experiments in human trophoblast models thus support our in vivo findings in the mouse placenta, confirming that PIEZO1 is a critical regulator for trophoblast fusion.
Representative images (A) and fusion index quantification (B) of untreated control (n = 6) and 5 μM GsMTx4 treated (n = 6) BeWo cells after 48-h forskolin treatment. Unpaired 2-sided Student t-test. ****: p = 0.000000576. Representative images (C) and fusion index quantification (D) of control (n = 8) and PIEZO1 siRNA knockdown (n = 8) BeWo cells after forskolin treatment for 48-h to induce fusion. Unpaired 2-sided Student t-test. ****: p = 0.00000384529. Representative images (E) and fusion index quantification (F) of BeWo cells that were overexpressed with empty vector (n = 8) and PIEZO1 (n = 8) after 48-h forskolin treatment. Unpaired 2-sided Student t-test. ****: p = 0.00002456791. Each dot represents fusion indexes averaged from six random fields of one coverslip. Data are presented as mean ± s.e.m. Source data are provided as a Source Data file.
PIEZO1-TMEM16F coupling regulates trophoblast fusion
We previously reported that TMEM16F, a Ca2+-activated phospholipid scramblase (CaPLSase), plays a critical role in trophoblast fusion and placental development by mediating the externalization of phosphatidylserine (PS), an essential ‘fuse-me’ signal for cell fusion24,35. Subsequently, we demonstrated that Ca2+ influx through Ca2+ permeable channels such as TRPV4 can activate TMEM16F, and the functional coupling between TRPV4 and TMEM16F modulates trophoblast fusion22. In our recent investigations of RBCs, we found that PIEZO1-mediated Ca2+ influx activates TMEM16F and enhanced PIEZO1-TMEM16F coupling contributes to the pathophysiology of RBC disorders such as hereditary xerocytosis36 and sickle cell disease37. Interestingly, Piezo1 and Tmem16f transcripts are both highly expressed in the SynT-2 layer (Fig. S4B–E), and the deficiency of either protein results in embryonic or perinatal lethality24 accompanied by severe defects in SynT-2 STB layer development (Figs. 2 and 3C, D). Based on these findings, we hypothesized that PIEZO1 and TMEM16F could be functionally coupled in trophoblasts and this coupling plays a key role in trophoblast fusion by regulating PS exposure.
To test this hypothesis, we employed a fluorescence imaging-based CaPLSase assay38 to investigate PIEZO1-TMEM16F coupling in BeWo cells. Treatment with 10 μM Yoda1 induced robust Ca²⁺ influx in both TMEM16F WT and KO BeWo cells (Fig. 5A). However, Yoda1-induced PS exposure was observed only in WT but not KO cells (Fig. 5A, B), indicating that Ca2+ influx through PIEZO1 can mediate TMEM16F-dependent PS exposure. On the other hand, gene silencing or GsMTx4 inhibition of PIEZO1 abolished both Yoda1-induced Ca2+ elevation and PS exposure in BeWo cells (Fig. 5C, D and S9). These findings demonstrate that PIEZO1 and TMEM16F are functionally coupled to regulate PS exposure in trophoblasts (Fig. 5G).
A Representative images of Ca2+ and PS exposure in WT (left) and TMEM16F KO (right) BeWo cells stimulated with 10 μM Yoda1. B Time course quantifications of Yoda1-induced PS exposure in WT (n = 25) and TMEM16F KO (n = 18) BeWo cells. C Representative images of Ca2+ and PS exposure in control (left) and PIEZO1 siRNA knockdown (right) BeWo cells stimulated with 10 μM Yoda1. D Time course quantifications of Yoda1-induced PS exposure control (n = 21) and PIEZO1 siRNA (n = 25) knockdown BeWo cells. E Representative images of TMEM16F KO BeWo overexpressed with empty vector and PIEZO1 after forskolin treatment for 48 h. F Fusion index quantification in WT (the same data in Fig. 4F) and TMEM16F KO BeWo cells, which were overexpressed with an empty vector or PIEZO1 (n = 8 each) after forskolin treatment for 48 hrs. Data are presented as mean ± s.e.m. n.s. non-significant; ****P < 0.0001 (BeWo WT+ empty vs. BeWo WT+ Piezo1 WT: P = 0.000000028606201; BeWo WT+ empty vs. BeWo 16FKO+ empty, P = 0.000000000000011; BeWo WT+ empty vs. BeWo 16FKO+ Piezo1 WT: P = 0.000000000000011), one-way ANOVA with Tukey’s multiple comparisons test). G Cartoon illustration (Created in BioRender. Shan, Z. (2025) https://BioRender.com/jkftvy7) of PIEZO1-TMEM16F coupling in regulating trophoblast fusion. Source data are provided as a Source Data file.
As Yoda1 is not a physiological activator of PIEZO1, we next examined whether mechanical forces could directly trigger PIEZO1-TMEM16F coupling in BeWo cells. We leveraged TMEM16F’s moonlighting function as a Ca2+-activated ion channel39 and applied a −50 mmHg suction to a cell-attached membrane patch using a pressure clamp device. This mechanical stimulation elicited a time- and voltage-dependent, outward-rectifying current, a biophysical hallmark of TMEM16F current (Fig. S10A, B). Notably, this force-induced TMEM16F current depends on extracellular Ca2+ and requires the expression of both PIEZO1 and TMEM16F in BeWo cells (Fig. S10C), providing direct evidence that mechanical forces can directly activate PIEZO1-TMEM16F coupling in trophoblasts (Fig. 5G).
To examine whether PIEZO1-TMEM16F functional coupling directly regulates trophoblast fusion, we overexpressed PIEZO1 in TMEM16F KO BeWo cells and quantified forskolin-induced fusion. While PIEZO1 overexpression robustly enhanced fusion in WT BeWo cells (Fig. 4E, F), it failed to rescue the fusion deficit in TMEM16F KO cells (Fig. 5E, F). Our pressure clamp electrophysiology confirmed comparable PIEZO1 channel activity in WT and TMEM16F KO cells (Fig. S11), ruling out differential PIEZO1 expression or function as confounding factors. These results establish that PIEZO1’s pro-fusogenic role depends on TMEM16F, likely through TMEM16F-mediated PS exposure, a “fuse-me” signal required for trophoblast fusion22,24. This functional coupling defines a mechanochemical axis where PIEZO1 senses mechanical cues and TMEM16F executes downstream biochemical signaling via PS externalization (Fig. 5G).
Discussion
Our identification of PIEZO1 as a key regulator of trophoblast fusion expands its known functions in mechanobiology and development. Complementary to previous studies showing PIEZO1’s critical role in fetoplacental angiogenesis and embryonic development11,12,40,41,42, our findings reveal that PIEZO1 in trophoblasts is also essential for embryonic survival. By manipulating PIEZO1 activity, our experiments demonstrate that mechanical forces play a pivotal role in driving trophoblast fusion during placental development.
Unlike endothelial PIEZO1, which responds to shear stress from blood flow43,44, CTB-to-STB fusion mainly occurs at the basolateral side of STBs (Fig. 5G), distant from maternal and fetal circulation. This suggests that blood flow-induced shear forces are less likely to activate PIEZO1 at the fusogenic CTB-STB interface. Instead, trophoblasts may rely on local compressive forces at the fusogenic synapses45, similar to those observed in myoblast fusion46,47, to trigger PIEZO1 activation. Future investigation of these intrinsic mechanical cues will be crucial for understanding how PIEZO1 regulates trophoblast fusion.
Our in vitro experiments further demonstrate that PIEZO1-mediated Ca²⁺ influx activates TMEM16F, leading to surface exposure of PS, an essential ‘fuse-me’ signal required for trophoblast fusion24. Notably, both PIEZO1 and TMEM16F are highly expressed in the SynT-2 layer (Fig. S4), and loss of either protein in trophoblasts severely disrupts the development of this STB layer, further supporting the importance of PIEZO1-TMEM16F coupling in trophoblast fusion (Fig. 5G). Beyond promoting TMEM16F-mediated PS externalization, PIEZO1-mediated Ca²⁺ influx may also activate additional signaling pathways that influence trophoblast gene expression, migration, and proliferation, processes that collectively impact trophoblast fusion, placental maturation, and overall embryonic development. All of these require future systematic investigations, both in vitro and in vivo using various approaches, such as live cell imaging and histological analysis including electron microscopy, at different placental developmental stages.
By identifying PIEZO1 as a crucial mechanosensitive ion channel in trophoblasts, our study lays a foundation for further exploration of trophoblast mechanobiology in placental physiology and pregnancy complications. Given the central role of trophoblast fusion in placental functions, dysregulation of PIEZO1 may contribute to pregnancy disorders such as preeclampsia, fetal growth restriction, preterm birth, and placental abruption. Future studies targeting PIEZO1 and its downstream pathways may offer therapeutic opportunities for managing these pregnancy-related complications.
It is noteworthy that PIEZO1 is broadly expressed in mouse and human placentas, not only in trophoblasts but also in endothelial cells, fibroblasts, smooth muscle cells, and various immune cell types (Figs. S4 and 12). This widespread expression suggests that the embryonic lethality observed in global Piezo1 knockout mice, which occurs between E9.5 and E14.511,12, likely stems from cumulative dysfunction across multiple cell lineages. Supporting this idea, our data show that trophoblast-specific Piezo1 cKO embryos exhibit lethality as early as E11.5, closely phenocopying the global Piezo1 knockout mice. Interestingly, while endothelial-specific Piezo1 cKO mice display similar vascular defects as global Piezo1knockout mice, there are discrepancies in the timing of lethality, with both midgestation and perinatal lethality reported11,12,19. This further supports the notion that the embryonic lethality observed in global Piezo1 knockout mice likely results from the combined loss of Piezo1 function across multiple cell types. In addition, the role of PIEZO1 in various placental cell types in the context of pregnancy complications, such as preeclampsia, preterm birth, intrauterine growth restriction (IUGR), premature rupture of membranes (PROM), and placental abruption, remains an important area for future investigation. In summary, our findings on PIEZO1’s role in trophoblast fusion and placental development provide new insights into placental mechanobiology and lay the groundwork for developing novel therapeutic strategies to treat pregnancy complications.
Methods
Human placenta tissue
Human placental tissues were obtained under institutional review board approval (IRB# PRO00014627 and XHEC-C-2018-089), with a waiver of consent, to procure de-identified samples exclusively for research purposes. Placental specimens were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, #15710) at 4 °C for 48 h. Following fixation, the tissues were preserved in 70% ethanol for 3 to 5 days and subsequently processed using a 4-h protocol on the Leica ASP6025 tissue processor (Leica Biosystems). Processed tissues were paraffin-embedded using the HistoCore Arcadia system (Leica Biosystems) and sectioned at 5 μm thickness with a Leica RM2255 microtome (Leica Biosystems, Buffalo Grove, IL) for histological examination.
For analysis, tissue sections were de-paraffinized at 60 °C, followed by two washes in xylene (Fisher Chemical, #X5S-4), and then rehydrated through a graded ethanol series (100%, 95%, and 70%). Sections were rinsed in MilliQ water for 5 min. Antigen retrieval was performed, after which tissue slices were permeabilized with 0.2% Triton X-100 and blocked with 10% goat serum prior to antibody incubation. PIEZO1 antibody (Proteintech, #82625-4-RR) was applied at a 1:200 dilution and incubated overnight. Fluorescent labeling was achieved using Alexa Fluor 488 fluorescence systems (Molecular Probes, #35552).
Mice
PIEZO1-floxed27 mice (B6.Cg-Piezo1tm2.1Apat/J, Strain #:029213) and Piezo1P1-tdT12 mice (B6;129-Piezo1tm1.1Apat/J, Strain #:029214) were obtained from the Jackson Laboratory or a kind gift from Bailong Xiao. To generate trophoblast-specific PIEZO1 knockout mice, PIEZO1flox/flox mice were crossed with Elf5-Cre mice28. Genotyping was performed via PCR using DNA extracted from tail samples. Genomic DNA was extracted using an alkaline lysis method and PCR performed with GoTaq® Master Mix (Promega, #M7122). Amplified products were analyzed on a 2% agarose gel. All mouse handling and experimental procedures were conducted in strict accordance with the protocol approved by the Institutional Animal Care and Use Committee at Duke University (#A057-24-02) and complied with National Institutes of Health guidelines. Animals were housed at a constant 24 °C in a 12 h light–dark cycle (lights on at 7:00 AM) with ad libitum access to food and water.
Cell lines
The wildtype BeWo cell line was authenticated by the Duke University Cell Culture Facility and the TMEM16F knockout (KO) BeWo cell line was generated by sgRNAs targeting exon 2, as previously reported22,24. BeWo cells were cultured in Dulbecco’s modified Eagle’s medium/Hams F12 (DMEM/F12) medium (Gibco, #11320-033) in a 5% CO2-95% air incubator at 37 °C. The media were supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, #F2442) and 1% penicillin/streptomycin (Gibco, #15-140-122). The human trophoblast stem cell (hTSC) line CT29 was a generous gift from Dr. Danny Schust. Cells were maintained in DMEM/F12 supplemented with 0.1 mM 2-mercaptoethanol, 0.2% FBS, 0.5% Penicillin-Streptomycin, 0.3% BSA, 1% ITS-X supplement, 1.5 μg/ml L-ascorbic acid, 50 ng/ml EGF, 2 μM CHIR99021, 0.5 μM A83-01, 1 μM SB431542, 0.8 mM VPA, and 5 μM Y2763248. Cells were seeded on tissue culture plates precoated with 5 μg/ml Collagen IV (Corning, #354233) for 2 h at 37 °C. Culture medium was replaced every two days and cells were passaged at ~80% confluence using TrypLE Express Enzyme (Gibco, #12604-021).
siRNA transfection
BeWo cells were transfected with PIEZO1 SMARTPool siRNAs (Horizon, # M-020870-00-0005) using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, #13778075) following the manufacturer’s instructions. One day after siRNA transfection, the cells were switched to fresh medium and further cultured for another 24 h before confocal imaging or fusion experiments.
qRT-PCR
Total RNA was extracted using RNA isolator Reagent (Vazyme, #R401), quantified via NanoDrop Spectrophotometer (Thermo Scientific), and reverse-transcribed (500 ng input) with HiScript IV All-in-One Ultra RT SuperMix (Vazyme, #R433-01) to generate cDNA. qRT-PCR was performed using Taq Pro Universal SYBR Master Mix (Vazyme, #Q712-03) on a QuantStudio 6 Pro system (Thermo Scientific) under the following conditions: 95 °C for 30 s (initial denaturation); 40 cycles of 95 °C for 10 s and 60 °C for 30 s, with fluorescence acquisition during extension. Reactions were run in duplicate, and gene expression was normalized to GAPDH. All primer sequences are provided below. Human GAPDH primers: Forward 5’- ACAACTTTGGTATCGTGGAAGG −3’. Reverse 5’- GCCATCACGCCACAGTTTC −3’; Human Piezo1 primers: Forward 5’- GGACTCTCGCTGGTCTACCT −3’. Reverse 5’- GGGCACAATATGCAGGCAGA −3’;Human Piezo2 primers : Forward 5’- GACGGACACAACTTTGAGCCTG −3’. Reverse 5’- CTGGCTTTGTTGGGCACTCATTG −3’; Human TMEM16F primers: Forward 5’- GGCCTCCATCATCAGCTTTAT −3’. Reverse 5’- GTCCTTGGGAGTTCGAAGTTAG −3’.
Fluorescence imaging of calcium and PS exposure
The procedures were done as previously described22,24. Briefly, to detect calcium dynamics, BeWo cells were treated with 1 μM Calbryte 590 AM (AAT Bioquest, #20700) for 10 min at 37 °C and 5% CO2. PS exposure was detected using 500 ng/ml CF 488-tagged AnV (Biotium, #29005). 10 µM Yoda1 (Cayman Chemical, #21904) and 5 µM GsMTx4 (MedChemExpress, #HY-P1410) were used to stimulate and inhibit PIEZO1 activity, respectively. Time-lapse imaging of BeWo cells was conducted before and after Yoda1 stimulation at room temperature using a Zeiss 780 inverted confocal microscope or an Olympus IX83 inverted epi-fluorescent microscope. ImageJ and a custom MATLAB code were used to quantify cytosolic calcium and AnV binding24.
BeWo cell fusion and quantification of fusion index
After BeWo cells were seeded on poly-L-lysine (Sigma-Aldrich)-coated coverslip for one day, cells were treated with 30 μM forskolin (Cell Signaling Technology, #3828 s) for 48 h to induce cell fusion. The forskolin-containing media was changed every 24 h. After forskolin treatment, cells were stained with Hoechst (Invitrogen, #H3570, 1:2000) and Wheat Germ Agglutinin Alexa Fluor-488 Conjugate (Invitrogen, #W11261, 1:1000) for 15 min to visualize nuclei and cell membranes, respectively. Six random fields of view were acquired using a Zeiss 780 inverted confocal microscope for each group. Cell fusion was quantified by calculating the fusion index as previously described22,24. To evaluate the effect of PIEZO1 inhibition on BeWo fusion, 5 µM GsMTx4 was co-applied with forskolin at the onset of fusion induction.
hTSC differentiation and hCG Measurement
For hTSC differentiation into STB(2D)48, 1.5 × 10⁴ hTSCs were seeded into 48-well plates precoated with 2.5 μg/ml Collagen IV. Cells were cultured in DMEM/F12 supplemented with 0.1 mM 2-mercaptoethanol, 0.5% Penicillin-Streptomycin, 0.3% BSA, 1% ITS-X supplement, 2.5 μM Y27632, 2 μM forskolin, and 4% KSR. After 24 h, culture supernatants were collected and analyzed for human chorionic gonadotropin (hCG) levels using the Human hCG (intact) ELISA Kit (Sigma, #RAB0092), according to the manufacturer’s instructions.
PIEZO1 lentiviral overexpression
To overexpress PIEZO1, we utilize the lentiviral overexpression system as described previously49. Briefly, we designed lentiviral vectors based on Addgene plasmid #62554 where the puromycin resistance gene was replaced with a hygromycin resistance gene from the pSBtet-GH plasmid (Addgene #60498) and the TMEM16F/Ano6 gene was replaced with the hPIEZO1 sequence (Genebank AGH27891.1) followed by an IRES sequence. For empty vector controls, hPiezo1-IRES was removed. All subcloning was done using an In-Fusion Snap Assembly (Takara #638947). Lentiviral constructs of PIEZO1 were packaged into lentivirus by transfecting HEK-293T cells with lentiviral vectors, pMD2.G, and psPAX2 (Addgene #12259 and #12260) using Lipofectamine 2000. WT and TMEM16F KO BeWo cells were then transduced with the filtered lentiviral supernatant and polybrene (10 µg/ml) and selected with hygromycin.
Immunofluorescence staining
Cells were fixed with 1% PFA in phosphate-buffered saline (PBS) for 10 min, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 5% goat serum in PBS for an hour. Coverslips were incubated in anti-PIEZO1 antibody (Proteintech, 15939-1-AP, 1:400) at 4 °C overnight. A secondary antibody, Alexa Fluor 488 fluorescence system (Molecular Probes, #35552, 1:1000), was used for fluorescent staining. After staining nuclei with 4′,6-diamidino-2-phenylindole (DAPI), coverslips were mounted using ProLong Diamond Antifade Mountant (Invitrogen, #P36961) and imaged with a Zeiss 780 inverted confocal microscope.
Mouse placenta histological analysis
Mice were deeply anesthetized using isoflurane. Placentas and embryos were freshly collected and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, #15710) for 2 days at 4 °C and then transferred to 70% alcohol for 3 to 5 days before being processed by using a 4-h tissue processing setting with Leica ASP6025 (Leica Biosystems). Placentas were embedded in paraffin (HistoCore Arcadia H and Arcadia C, Leica Biosystems) immediately after the processing and then sectioned at 5 μm by using Leica RM2255 (Leica Biosystems, Buffalo Grove, IL) for histological staining.
H&E staining was performed through deparaffinization at 60 °C followed by two washes with xylene (Fisher Chemical, #X5S-4) and a graded alcohol series (100%, 95%, 70%). The tissue was rinsed in MilliQ water for 5 min and then incubated in Hematoxylin 560 MX (Leica Biosystems, c#3801575) for 2 min, followed by rinsing with running water for 1 min. Tissue was dipped seven times in 0.3% ammonium hydroxide (Fisher Chemical, #A669S-500) and then rinsed in running water for 1 min. The tissue was incubated in 70% ethanol for 1 min followed by 10 dips in Alcoholic Eosin Y 515 (Leica Biosystems, #3801615). Dehydration was performed through an alcohol gradient of 95%, 100%, and twice in xylene, 1 min each. All slides were mounted using DPX Mounting Medium (Electron Microscopy Sciences, #13512). CD31 immunohistochemistry (IHC) staining was performed by using rabbit polyclonal to CD31 (Abcam, #ab28364) with the Avidin/Biotin Blocking Kit (Vector Laboratories Inc., #SP-2001), VECTASTAIN Elite ABC HRP Kit (Peroxidase, Standard) (Vector Laboratories Inc., #PK-6100), and VECTOR VIP Peroxidase (HRP) Substrate Kit (Vector Laboratories Inc., #SK-4600). Nuclei were counterstained with VECTOR Methyl Green (Vector Laboratories Inc., #H-3402-500). Microscopic imaging of histology samples was taken at 20X by using an Olympus microscope (BX63l, Olympus, Center Valley, PA) with cellSens Dimension software.
Immunofluorescence staining against MCT1 and MCT4 was performed on TMEM16F WT and KO placenta by using MCT1 antibody (Millipore-Sigma, #AB1286-I), which stains the fused SynT-1 layer facing the maternal blood sinuses, and MCT4 antibody (Santa Cruz Biotechnology Inc., #sc-376140), which stains the fused SynT-2 layer facing the fetal blood vessels. Immunofluorescence staining were imaged with a Zeiss 780 inverted confocal microscope.
Mouse placenta transcriptomic analysis
The reference clustering and individual single-cell RNAseq UMAP results were obtained from the Spatiaotemporal Transcriptomic Atlas of Mouse Placenta (STAMP, https://db.cngb.org/stomics/stamp/)26.
Electrophysiology
All ionic currents were recorded in the cell-attach configuration using an Axopatch 200B amplifier (Molecular Devices) and the pClamp software package (Molecular Devices). Glass pipettes were pulled from borosilicate capillaries (Sutter Instruments) and fire-polished using a microforge (Narishge) with a resistance of 2−3 MΩ for macroscopic current and 5−7 MΩ for single channel recordings. The bath solution consisted of the following (in mM): 140 CsCl, 10 HEPES, and 5 EGTA, with the pH adjusted to 7.4 using CsOH. For PIEZO1 experiments, the pipette solution contained (in mM): 140 CsCl, 10 HEPES, and 1 MgCl₂, with the pH adjusted to 7.4 using CsOH. PIEZO1 activation was achieved by applying pressure using a high-speed pressure clamp (ALA Scientific Instruments, #HSPC-2-SB). Pressure was applied using a step protocol starting from 0 mmHg to −60 mmHg with −10 mmHg increments. In the PIEZO1-TMEM16F coupling experiments, the pipette solution contained (in mM): 140 CsCl, 10 HEPES, 1 MgCl₂, and either 0 or 2.5 mM Ca²⁺, as indicated. The pH was adjusted to 7.4 using CsOH. The pressure was maintained at a constant value of −50 mmHg during the recordings to activate PIEZO1 and enable Ca2+ influx. A voltage step protocol from −100 mV to +160 mV in 20 mV increments was used to elicit TMEM16F channel activation. Cs⁺-based solutions were used to suppress contaminating K⁺ current in BeWo cells.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software. Unpaired two-tailed Student’s t-tests were used for single comparisons between two groups, while one-way ANOVA (with Tukey’s multiple comparisons test) was used for multiple comparisons. Sample number (n) values are provided in the figure legends. All data are presented as the mean ± standard error of the mean (s.e.m.).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The original immunohistochemistry images in this study have been deposited with a https://doi.org/10.7924/r4fj2qc4x. All remaining data are available in the main text or the supplementary materials. Source data are provided with this paper.
References
Huang, H., Gao, S. & Bao, M. Exploring mechanical forces shaping self-organization and morphogenesis during early embryo development. Annu. Rev. Cell Dev. Biol. 40, 75–96 (2024).
Gomez-Galvez, P., Anbari, S., Escudero, L. M. & Buceta, J. Mechanics and self-organization in tissue development. Semin. Cell Dev. Biol. 120, 147–159 (2021).
Goodwin, K. & Nelson, C. M. Mechanics of development. Dev. Cell 56, 240–250 (2021).
Burton, G. J., Charnock-Jones, D. S. & Jauniaux, E. Regulation of vascular growth and function in the human placenta. Reproduction 138, 895–902 (2009).
Khorami-Sarvestani, S. et al. Placenta: an old organ with new functions. Front. Immunol. 15, 1385762 (2024).
Ma, Z., Sagrillo-Fagundes, L., Mok, S., Vaillancourt, C. & Moraes, C. Mechanobiological regulation of placental trophoblast fusion and function through extracellular matrix rigidity. Sci. Rep. 10, 5837 (2020).
Xie, B. et al. Mechanical forces on trophoblast motility and its potential role in spiral artery remodeling during pregnancy. Placenta 123, 46–53 (2022).
Saw, S. N., Dai, Y. & Yap, C. H. A review of biomechanics analysis of the umbilical-placenta system with regards to diseases. Front. Physiol. 12, 587635 (2021).
Coste, B. et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010).
Coste, B. et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483, 176–181 (2012).
Li, J. et al. Piezol integration of vascular architecture with physiological force. Nature 515, 279–U308 (2014).
Ranade, S. S. et al. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc. Natl. Acad. Sci. USA 111, 10347–10352 (2014).
Pathak, M. M. et al. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl. Acad. Sci. USA 111, 16148–16153 (2014).
Del Marmol, J. I., Touhara, K. K., Croft, G. & MacKinnon, R. Piezo1 forms a slowly-inactivating mechanosensory channel in mouse embryonic stem cells. Elife 7, e33149 (2018).
Nourse, J. L. et al. Piezo1 regulates cholesterol biosynthesis to influence neural stem cell fate during brain development. J. Gen. Physiol. 154, e202213084 (2022).
Sugimoto, A. et al. Piezo type mechanosensitive ion channel component 1 functions as a regulator of the cell fate determination of mesenchymal stem cells. Sci. Rep. 7, 17696 (2017).
Nie, X., Abbasi, Y. & Chung, M. K. Piezo1 and Piezo2 collectively regulate jawbone development. Development 151, e202213084. https://doi.org/10.1242/dev.202386 (2024).
Du, J. et al. The mechanosensory channel PIEZO1 functions upstream of angiopoietin/TIE/FOXO1 signaling in lymphatic development. J. Clin. Investig. 134. https://doi.org/10.1172/JCI176577 (2024).
Nonomura, K. et al. Mechanically activated ion channel PIEZO1 is required for lymphatic valve formation. Proc. Natl. Acad. Sci. USA 115, 12817–12822 (2018).
Lapehn, S. et al. A transcriptomic comparison of in vitro models of the human placenta. Placenta 159, 52–61 (2025).
Xiao, B. Mechanisms of mechanotransduction and physiological roles of PIEZO channels. Nat. Rev. Mol. Cell Biol. 25, 886–903 (2024).
Zhang, Y. et al. Functional coupling between TRPV4 channel and TMEM16F modulates human trophoblast fusion. eLife 11, e78840 (2022).
Dupressoir, A. et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. USA 108, E1164–E1173 (2011).
Zhang, Y. et al. TMEM16F phospholipid scramblase mediates trophoblast fusion and placental development. Sci. Adv. 6, 10 (2020).
Nagai, A., Takebe, K., Nio-Kobayashi, J., Takahashi-Iwanaga, H. & Iwanaga, T. Cellular expression of the monocarboxylate transporter (MCT) family in the placenta of mice. Placenta 31, 126–133 (2010).
Wu, Y. et al. A spatiotemporal transcriptomic atlas of mouse placentation. Cell Discov. 10, 110 (2024).
Cahalan, S. M. et al. Piezo1 links mechanical forces to red blood cell volume. Elife 4 https://doi.org/10.7554/eLife.07370 (2015).
Kong, S. et al. Generation of Elf5-Cre knockin mouse strain for trophoblast-specific gene manipulation. Genesis. 56, e23101 (2018).
Zhou, H. et al. Regulators involved in trophoblast syncytialization in the placenta of intrauterine growth restriction. Front. Endocrinol. 14, 1107182 (2023).
Kosinska-Kaczynska, K. Placental syndromes-a new paradigm in perinatology. Int .J. Environ. Res. Public Health 19. https://doi.org/10.3390/ijerph19127392 (2022).
Redman, C. W. G., Staff, A. C. & Roberts, J. M. Syncytiotrophoblast stress in preeclampsia: the convergence point for multiple pathways. Am. J. Obstet. Gynecol. 226, S907–S927 (2022).
Yabe, S. et al. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc. Natl Acad. Sci. USA 113, E2598–E2607 (2016).
Gaspard, U. J., Hustin, J., Reuter, A. M., Lambotte, R. & Franchimont, P. Immunofluorescent localization of placental lactogen, chorionic gonadotrophin and its alpha and beta subunits in organ cultures of human placenta. Placenta 1, 135–144 (1980).
Deglincerti, A. et al. Self-organization of the in vitro attached human embryo. Nature 533, 251–254 (2016).
Whitlock, J. M. & Chernomordik, L. V. Flagging fusion: Phosphatidylserine signaling in cell-cell fusion. J. Biol. Chem. 296, 100411 (2021).
Liang, P. F. et al. Deciphering and disrupting PIEZO1-TMEM16F interplay in hereditary xerocytosis. Blood 143, 357–369 (2024).
Liang, P. et al. Targeting PIEZO1-TMEM16F coupling to mitigate sickle cell disease complications. bioRxiv, 2025.2005.2027.656389. https://doi.org/10.1101/2025.05.27.656389 (2025).
Le, T. et al. An inner activation gate controls TMEM16F phospholipid scrambling. Nat. Commun. 10, 13 (2019).
Yang, H. et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell 151, 111–122 (2012).
Allerkamp, H. H. et al. In vitro examination of Piezo1-TRPV4 dynamics: implications for placental endothelial function in normal and preeclamptic pregnancies. Am. J. Physiol. Cell Physiol. 328, C227–C244 (2025).
Morley, L. C., Debant, M., Gaunt, H. J., Simpson, N. A. B. & Beech, D. J. Nitric oxide synthase phosphorylation in fetoplacental endothelium is enhanced by agonism of Piezo1 mechanosensor in small for gestational age babies. Reprod. Fertil. 4, https://doi.org/10.1530/RAF-22-0100 (2023).
Morley, L. C. et al. Piezo1 channels are mechanosensors in human fetoplacental endothelial cells. Mol. Hum. Reprod. 24, 510–520 (2018).
Morley, L. C., Beech, D. J., Walker, J. J. & Simpson, N. A. B. Emerging concepts of shear stress in placental development and function. Mol. Hum. Reprod. 25, 329–339 (2019).
Morley, L. C., Debant, M., Walker, J. J., Beech, D. J. & Simpson, N. A. B. Placental blood flow sensing and regulation in fetal growth restriction. Placenta 113, 23–28 (2021).
Parameshwar, P. K. et al. Directed biomechanical compressive forces enhance fusion efficiency in model placental trophoblast cultures. Sci. Rep. 14, 11312 (2024).
Kim, J. H. & Chen, E. H. The fusogenic synapse at a glance. J. Cell. Sci. 132. https://doi.org/10.1242/jcs.213124 (2019).
Tsuchiya, M. et al. Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nat. Commun. 9, 2049 (2018).
Okae, H. et al. Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50 (2018).
Shan, K. Z. et al. TMEM16F scramblase regulates angiogenesis via endothelial intracellular signaling. J. Cell Sci, 137, https://doi.org/10.1242/jcs.261566 (2024).
Acknowledgements
We sincerely appreciate the valuable insights and discussions provided by Drs. Carolyn Coyne and Liheng Yang. Many thanks to Drs. Danny Schust, Jie Zhou and Namya Mellouk for helping with the human trophoblast stem cells, Dr. Shuang Liang for qRT-PCR, and Dr. Hua Pan for assistance with H&E and CD31 staining. We are also grateful to Drs. Haibin Wang and Bailong Xiao for generously providing the Elf5-Cre and Piezo1P1-tdT mouse lines, respectively. This work was supported by National Institutes of Health grants R35GM153196 (HY) and DP2GM126898 (HY). We acknowledge the use of ChatGPT (https://chatgpt.com/) to assist with grammar checking and writing refinement in the Introduction and Discussion. Prompts used were [help check grammatical errors] or [analyze the following and suggest how it can be more concise without losing precision]. All final edits were reviewed manually to ensure accuracy and originality. BioRender was used to create the summary illustration.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.Y. and Y.Z. Methodology: Y.Z., K.Z.S., P.L., A.J.L., H.Y. and L.F. Investigation: Y.Z., K.Z.S. and P.L. Visualization: Y.Z., K.Z.S., P.L. and H.Y. Funding acquisition: H.Y. Project administration: H.Y. Supervision: H.Y. Writing – original draft: H.Y., K.Z.S., P.L. and Y.Z. Writing – review & editing: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks In-Jeoung Baek, Leonid Chernomordik and Olivier Schwartz for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Y., Shan, K.Z., Liang, P. et al. PIEZO1 drives trophoblast fusion and placental development. Nat Commun 16, 6895 (2025). https://doi.org/10.1038/s41467-025-62254-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62254-3