Abstract
Olfactomedin-2 (OLFM2) is a pleiotropic glycoprotein emerging as a regulator of energy homeostasis. We here show the expression of OLFM2 to be adipocyte-specific and inversely associated with obesity. OLFM2 levels increase during adipogenesis and are suppressed in inflamed adipocytes. Functionally, OLFM2 deficiency impairs adipocyte differentiation, while its over-production enhances the adipogenic transformation of fat cell progenitors. Loss and gain of function experiments revealed that OLFM2 modulates key metabolic and structural pathways, including PPAR signaling, citrate cycle, fatty acid degradation, axon guidance and focal adhesion in 3T3 cell lines and primary human adipocytes. On the molecular level, OLFM2 deficiency in differentiated adipocytes predominantly downregulates genes involved in cell cycle. Extending these findings in vivo, both whole-body Olfm2 knockout and adipose-specific Olfm2 depletion in mice resulted in impaired adipose cell cycle gene expression, with the latter also displaying fat mass accretion and metabolic dysfunction. Collectively, our results underscore a critical role for OLFM2 in adipocyte biology, and support a causative link between reduced adipose OLFM2 and the pathophysiology of obesity.
Similar content being viewed by others
Introduction
The olfactomedin family (OLFM1 through 4 in humans) comprises cell adhesion and membrane-associated proteins that appear to be critical mediators for development1. Consequently, disruption of their expressions and/or activity usually results in mild-to-intense physiological perturbations leading to disease2,3. One of the family members, Olfactomedin-2 (OLFM2), a pleiotropic glycoprotein with widespread but most prominent neural expression4, mediates several developmental processes, and is implicated in pathologies ranging from cancer to glaucoma5,6,7. Currently available literature, however, holds a limited number of studies exploring the regulatory mechanisms in which this scaffold protein seems to be implicated. The most recent reports show TGFβ-induced OLFM2 to drive smooth muscle cell proliferation and differentiation8, potentially connecting it to the pathogenesis of a variety of cardiovascular diseases, including atherosclerosis and hypertension9. Our own previous study10, on the other hand, characterized the response of Olfm2-null mice to an obesogenic regime, providing evidence linking this olfactomedin to the hypothalamic regulation of energy homeostasis. Accordingly, mice lacking the Olfm2 gene showed resistance to high-fat diet-induced obesity, mostly due to elevated brown and white adipose tissue catabolic activity and enhanced energy expenditure10. The intriguing phenotype of this mouse model, which also manifests reduced exploration behavior and locomotion, abnormal motor coordination, anxiety-associated behavior patterns, and defective olfactory sensitivity11, is likely related to the loss of Olfm2 in brain, as further suggested through hypothalamus-specific, viral over-expression of this protein, which was found to induce weight gain10. Here, we present a body of in vitro, in vivo, clinical, as well as functional data to support the hypothesis of a previously unknown biological function and clinical relevance for the olfactomedin-domain containing protein OLFM2. Starting from the observation of increased OLFM2 levels in the adipose tissue of subjects with obesity following weight loss, a steady increase during adipogenesis, as well as substantial expression levels in adipocytes within depots of subcutaneous and omental fat, we show that inflammation impairs the expression of OLFM2 in fat cells. Beyond its function as an adipocyte marker, untargeted proteomics and transcriptomics in settings of gain and loss of function assays demonstrated that OLFM2 governs pathways of key relevance in fat cell commitment, including cell cycle, fatty acid metabolism, axon guidance and focal adhesion. Concomitantly, virus-mediated depletion of adipose Olfm2 in healthy mice drove a cluster of molecular changes related to defective adipogenesis and impaired fat cell function, leading to adipocyte hypertrophy and fat mass accretion, which triggered a host of metabolic alterations related to the obese phenotype. Collectively, our findings functionally connect OLFM2 to obesity, and highlight its significance in maintaining adipocyte function to avoid metabolic disease.
Results
Expression of OLFM2 is adipocyte-specific and inversely proportional to obesity
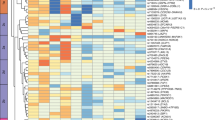
We have previously reported transcriptional variations in the adipose tissue of patients with obesity following weight loss, closely associated with decreased inflammation and improved adipocyte function12. In this context, we noticed the expression of OLFM2 as upregulated (Fig. 1a), and thus inversely related to the obese phenotype. First, we confirmed this hitherto overlooked expression of OLFM2 in adipose tissue in comprehensive public genomic resources. Results collected from the Genotype-Tissue Expression database (http://www.gtexportal.org), for instance, show elevated transcript levels of OLFM2 in human fat, surpassed only by expression levels found in brain, skin and adrenal gland (Fig. S1a). This observation was further corroborated through data from the T2D Knowledge Portal (T2DKP), which integrates much of the available genomic data relevant to type 2 diabetes and related traits (https://t2d.hugeamp.org). Here, the median expression of adipose OLFM2 (n = 2421) of 35.9 transcripts per million (TPM), with 21.3 and 115.4 TPM for the 1st and 3rd quartile, respectively, ranks only below the expression reported in central nervous system (70.04 [29.2–115.4]; n = 3279) and eye (49.4 [33.5–69.7]; n = 453). Additional mining of our microarray data further revealed individual fluctuations in adipose OLFM2 that were concomitant with variations in important adipocyte transcripts such as AACS, ACACA, CIDEC, FASN, GYS1, PNPLA3 and SREBF1 (Table S1), and therefore linked to cellular signaling pathways of relevance in adipocyte commitment (Table S2). Using real time-PCR analysis of OLFM2 gene expression in additional longitudinal and cross-sectional samples, we set out to experimentally corroborate the association with obesity. To this end, we analyzed samples from 24 women without obesity (body mass index (BMI) < 30 kg/m2), as well as 24 age-matched female patients with severe obesity (BMI > 35 kg/m2) followed for an average of ~2 years after bariatric surgery (Table S3)12. In confirmation of our microarray results, expression of OLFM2 in subcutaneous (SC) adipose tissue peaked following weight loss (2.3-fold, p = 1.4E-4), reaching transcript levels even exceeding those observed in female participants with a BMI below 30 kg/m2 (Fig. 1b). Measures of Olfactomedin 1–4 were further retrieved from refs. 13,14. In the former13, biopsies of SC adipose tissue were obtained by core needle aspiration from 50 participants with obesity at the baseline and 2 (n = 49) and 5 years (n = 38) following gastric bypass, and also from a non-operated group of 28 volunteers of healthy weight. On the other hand, Shah et al.14 collected adipose tissue before and 24 h after an intravenous injection bolus infusion of 3 ng/kg lipopolysaccharide (LPS), an endotoxin with proven ability to activate the immune system15 with profound impact on adipose tissue16 and the onset of obesity-related metabolic disturbances17. The study was conducted in 14 healthy volunteers aged 18–40 years and with a BMI of 18–30 kg/m2, and paired fat samples subsequently underwent microarray gene expression profiling18. In line with our results, of the different olfactomedin family protein members, OLFM2 alone presented with increased transcript levels tracking the significant loss of fat upon bariatric surgery (Fig. 1c). It is noteworthy that impaired OLFM2 (Fig. 1d) shadowed dynamic adaptations following the negative impact of LPS on adipocyte biomarkers (SREBF1, ACSL1, LGALS12), and correlated negatively with the expression of lipopolysaccharide-binding protein (LBP) (Fig. S1b, c), which represents a surrogate for obesity-related inflammation and metabolic deterioration19. We further proceeded to analyze the expression of OLFM2 in an independent cohort of 185 individuals (23% males) with a wide BMI range (Table S4), in which paired (n = 82) and unpaired omental (OM) fat samples were collected lateral to umbilicus. Here, OLFM2 was diminished in the SC (Fig. 1e) and OM (Fig. 1f) adipose tissue of subjects with obesity (BMI ≥ 30 kg/m2), and correlated inversely to BMI (Fig. 1g), as well as percent body fat (Table S5). In agreement with our initial microarray analyses, expression of OLFM2 reflected the transcript abundance of a hub of genes related to adipocyte function (Table S5), including ACACA, FASN, CIDEA and IRS1 in both SC and OM fat (Fig. 1h). Overall, expression measures in bulk adipose tissue suggest OLFM2 gene expression as a biomarker of fat cell commitment and function. In a proof of concept, the assessment of ex vivo isolated mature adipocytes (MA) and the stromal vascular cell (SVC) component of human adipose tissues further validated the enrichment of OLFM2 in the former, as shown by our gene expression results (Fig. 1i) and the transcriptomic analysis performed by Vijay et al.20 (Fig. 1j). Taken together, the lines of qualitative as well as quantitative evidence compiled here from human patient cohorts link the expression of adipocyte-specific OLFM2 in fat to the burden of obesity.
a Gene expression dynamics (microarray) of Olfactomedin 1–4 (OLFM1-4) in the SC adipose tissue of 16 women with obesity after weight loss12. b OLFM2 gene expression levels (real time-PCR) in an extended sample of 24 female patients with obesity following bariatric surgery, and 24 age-matched women without obesity (BMI < 30 kg/m2). See in Table S3 for additional details. Boxplots show expression of SC OLFM1-4 in c 50 female patients with obesity at the baseline, and after 2 (n = 49) and 5 (n = 38) years of surgery-induced weight loss (post-WL), and in 28 age-matched women without obesity (NO)13, and in d 14 healthy volunteers before and 24 h after an intravenous bolus of 3 ng/kg LPS18, as retrieved from publicly available microarray datasets. See expression of others biomarkers in Supplementary Fig. S1b, c. OLFM3 gene expression was not detectable in the dataset of ref. 18. RMA, Robust multiarray analysis. Below, bean plots show OLFM2 gene expression (real time-PCR) in e SC and f omental (OM) adipose tissues of an independent sample of subjects with and without obesity, while g scatter dot plots show the association of SC (upper) and OM (lower panel) OLFM2 with BMI (further details in Tables S4 and S5). Individual values for men (blue) and women (red) are provided in gender glyphs. h Additional scatter dot plots associate OLFM2 with biomarkers of adipocyte function. Two-sided Spearman’s rank correlation coefficients (r) and p values are shown for each association. i OLFM2 transcript levels were also measured (real time-PCR) in stroma-vascular cells (SVC) and mature adipocytes (MA) isolated from paired SC and OM samples of adipose tissue, biopsied from 12 women with obesity while being bypassed, and in j the single-cell RNA sequencing of 14 individuals with obesity undergoing bariatric surgery, as explained in ref. 20. RPKM, Reads per kilobase per million reads. The box plots show center line at the median, upper and lower lines bound at 75th and 25th percentiles, respectively, and whiskers at minimum and maximum values. Statistical significance was determined by two-tailed Fisher’s exact t-test (cross-sectional comparisons), or two-tailed paired t-test (longitudinal changes). r.u. stands for relative units. *p < 0.05, **p < 0.01 (two-sided Student’s test). Source data are provided as a Source data file.
OLFM2 is a marker of adipocyte commitment
To extend the observed expression phenotype of OLFM2 down to the cellular level, we examined primary human preadipocytes (PA) (Fig. 2a) differentiating towards lipid-filled MA under hormonal adipogenic stimulation. We found OLFM2 transcription to be strongly induced during adipocyte differentiation (Fig. 2b), reaching maximum levels in adipocyte progenitors visually differentiated, as evident from microscopically visible lipid droplets in the cytoplasm. A reanalysis of global transcriptomic analyses performed in fat cells following differentiation21 also yielded consistent up-regulation of OLFM2 (but not OLFM1, 3 or 4) during adipogenesis (Fig. 2c). At the protein level, confocal microscopy performed in differentiating adipocytes revealed OLFM2 immunostaining distributed in filamentous structures that were morphologically reminiscent of the appearance of actin stress fibers in PA (Fig. 2d). Starting with day 7 post-differentiation, we observed an increased steady-state OLFM2 redistribution into the perinuclear region, as well as nuclei. OLFM2 signal in fully differentiated human adipocytes presented with comparatively increased intensity in the cytosol (Fig. 2d), and reduced co-localization with DAPI (nuclei). Next, we hypothesized the reduced expression of OLFM2 in the adipose tissue of subjects with obesity to be primarily driven by the influence of a microenvironment rich in macrophage-derived cytokines. To test this, we challenged terminally differentiated adipocytes (Fig. 2a) with 2% macrophage media (MM) or LPS-conditioned macrophage media (MCM). In this setting, we confirmed impaired OLFM2 in inflamed adipocytes (Fig. 2e–g), which was also corroborated by a significant reduction of adipocyte-specific OLFM2 (but not OLFM1, 3 or 4) transcript in the Simpson-Golabi-Behmel syndrome (SGBS) fat cell line responding to 14% MCM22 (Fig. 2h). To further substantiate these results, PA from one female donor without obesity (BMI < 25 kg/m2) and one female donor with obesity (BMI > 30 kg/m2), both of comparable age (35–40 years), were cultured and stimulated with adipogenic conditions. The resulting MA where challenged with recombinant human Transforming Growth Factor β1 (TGFβ1), which has been shown to induce OLFM2 in the context of smooth muscle cells differentiation8, and is linked to the cluster of pathological alterations believed to affect fat cells in obese adipose tissues23. On the other hand, aiming to mimic in vitro the approach of Shah et al.14, adipose cell cultures were exposed to a range of LPS doses. These experiments replicated the increased expression of OLFM2 in MA vs PA and the reduced levels bounded to an obese phenotype, even in cultured adipocytes, while reacting negatively to recombinant TGFβ1 (Fig. 2i, j), as well as to the presence of endotoxins in the media (Fig. 2k). To narrow down underlying mechanisms regulating these gene expression dynamics, we queried the hTFtarget database24 to predict transcription factors (TF) potentially working upstream of OLFM2 gene. This analysis yielded the presence of 64 different overlapping candidate TF-binding sites specific to the OLFM2 locus, as compared to the OLFM1, 3 and 4 loci (Fig. 2l). Amongst these, the occurrence of binding motifs for TF related to the adipogenic program, namely Peroxisome proliferator-activated receptor gamma (PPARγ), CCAAT/enhancer-binding protein beta (CEBPβ), and cAMP-response element binding protein 1 (CREB1) (Fig. 2l, m), can account for the higher expression of OLFM2 in differentiated fat cells, which, together with the expression of well-recognized constituent adipocyte cell-type identity markers (Fig. 2n)25, may cease when adipocytes respond to the pro-inflammatory milieu that usually characterizes large amounts of body fat26. Taken together, these results confirm the expression of OLMF2 as a biomarker linked to adipocyte maintenance and opposite to the obese phenotype.
a Pipeline diagram of in vitro differentiated human preadipocytes (PA) growing into lipid-containing mature adipocytes (MA). DM-2 (first) and AM-1 (second week and thereafter) stand for differentiation and adipocyte media, respectively. b Expression dynamics of OLFM2 during adipogenesis (n = 3 biological replicates for each day of hormonal stimuli), and c dynamic changes with regard to unstimulated precursor cells (n = 4/group/time point) and the expression of the others olfactomedin family protein members21. d OLFM2 (red) immunofluorescent staining captures in human PA (n = 4), differentiating adipocytes (Day 7; n = 4), and MA (n = 4 biological replicates). Nuclei are stained in blue (DAPI). White arrows point at the apparent perinuclear red signal in differentiating adipocytes. e, f MA showed less OLFM2 immunofluorescent staining signal when challenged with macrophage LPS-conditioned media (MCM; n = 4), as confirmed by g measures of gene expression in MA challenged with macrophage media (MM) or MCM (n = 4 biological replicates/group). The scale bars denote 100 μm length. h The scatter plot displays the expression (microarray) of transcripts coding for OLFM1-4, adiponectin (ADIPOQ), and pro-inflammatory interleukins (IL6 and IL1β) in inflamed Simpson Golabi Behmel syndrome (SGBS)-derived adipocytes, as explained in ref. 22. Red ink in labels depicts increased levels, and green labels show transcripts decreased in inflamed adipocytes. Blue labels for transcript with no significant variation. Bar plot shows expression of OLFM2 in human PA and MA from i lean (BMI < 25 kg/m2) and j sex, age and fat depot-matched donors with obesity, and changes in MA when challenged with TGFβ for 24 h (n = 4 biological replicates/group). k Variations in MA responding to LPS in the media (n = 6 replicates/group). In bar plots, results are presented as mean ± S.E.M. The box plots in c show center line at the median, upper and lower lines bound at 75th and 25th percentiles, respectively, and whiskers at minimum and maximum values. r.u. stands for relative units. *p < 0.05, **p < 0.01 (One-way ANOVA and Tukey’s multiple comparisons test). Source data are provided with the article as a Source data file. (l) Number of transcription factors (TF) affecting human olfactomedin coding genes, and m OLFM2 gene region ChIP-seq peaks for human datasets and TF in WashU EpiGenome Browser v40.6113. Only signal values for PPARG, CEBPB, and CREB1 (listed in red in Fig. 2l) are represented. n Mean TPM (transcript per million), detectability (TPM > 0.1%), and heat map of pairwise Spearman correlation coefficients for OLFM2 (ENSG00000105088) and reference genes for different populations of adipose tissue resident cells, in the case of adipocytes (AC), ADIPOQ, LIPE and PLIN1. The lack of values for mesothelial cells (MesoC) and neutrophils (NP) in SC adipose tissue is due to the low representation of these cell types, as explained in ref. 25.
Mapping the protein landscape of fat cells with altered OLFM2
We next investigated OLFM2 dynamics in murine 3T3-L1 cells following the adipogenic course, as well as its response in mouse adipocytes challenged with different stimuli (Fig. 3a). In agreement with our observations in human adipocytes, 3T3-L1 cells displayed a rapid and transient increase of OLFM2 when an adipogenic cocktail was added to the media (Fig. 3b). While acquiring the signet ring appearance of adipose cells and amassing visible lipid droplets in the cytoplasm, 3T3-L1 cells were found to accumulate cytoplasmic OLFM2 proximally to the plasma membrane, primarily located beneath cortical actin, together with β-tubulin (Fig. 3c). On the other hand, pharmacological boosting of adipocyte function (Rosiglitazone)27 increased the levels of Olfm2, while molecules producing pro-fibrotic (recombinant TGFβ1) and inflammatory (LPS) responses compromised the expression of this olfactomedin (Fig. 3d). Subsequently, we engineered two 3T3 mouse embryonic fibroblasts models of gain (GoF) and loss (LoF) of function to study the mechanisms linking the expression of Olfm2 to fat cell differentiation. To that end, plasmids containing sh-RNAs (LoF), or the EX-Mm36031-Lv225 eukaryotic expression vector (GoF) were lentivirally delivered into 3T3-L1 (Fig. 3e) and 3T3-F442A (Fig. S2a) adipocyte-precursor cells. When cell cultures enriched for transfected adipocyte progenitors were allowed to differentiate, the modulation of Olfm2 (Fig. 3f and Fig. S2b) resulted in impaired adipogenesis when diminished, while bolstering the adipogenic transformation when increased, as demonstrated by the expression of a hub of genes related to the adipocyte phenotype (Fig. 3g and Fig. S2c), Oil Red O (Fig. 3h and Fig. S3a in 3T3-L1, and Fig. S2d in 3T3-F442A cells), green fluorescent lipid staining (Fig. 3i and S3b), and western blotting of adipocyte biomarkers (Fig. 3j, k) in protein extracts of differentiated 3T3-L1 adipocytes. Extending these findings, we conducted untargeted in-depth proteome-wide analysis using high-resolution liquid chromatography-tandem mass spectrometry to identify functional variations following the modulation of Olfm2 in 3T3 adipocytes and undifferentiated precursor cells. In 3T3-L1 cells, a total of 7,756 protein groups (≥2 unique peptides; subgroup replication required) were identified and further analyzed (Fig. S4a, b). Hierarchical clustering (Fig. S4c, d) and correlation plots (Fig. S4e, f) of the results obtained yielded clear segregation of biological replicates into their respective subgroups, validating the experiment. In 3T3-F442A cells, spectra identification software yielded 6,255 protein groups (≥2 unique peptides; subgroup replication required) retained for further analysis (Fig. S5a, b). Here, hierarchical clustering (Fig. S5c, d) and correlation plots (Fig. S5e, f) confirmed proper segregation of biological replicates with synthetically altered Olfm2 gene expression levels. Principal component analysis (PCA) also readily distinguished 3T3-L1 MA and PA with experimentally increased (GoF) and diminished (LoF) amounts of Olfm2 gene expression from controls (Fig. 4a). Differentially abundant proteins (DAPs) were determined (FDR adjusted p < 0.05) comparing opposing cell models (GoF vs LoF) in each 3T3-L1 cell type (MA and PA). This yielded 1539 DAPs (946 up and 593 down-regulated) in MA (Fig. 4b), and 1691 DAPs (1,125 up and 566 down-regulated) in PA (Fig. 4c). Remarkably, OLFM2 was not detectable during the proteomic assessment of control nor knocked-down cells, and only passed our identification threshold (≥2 peptide Ids) under Olfm2 overexpression. The inability of MS-based proteomics to detect OLFM2 at native expression levels in mouse adipocytes matches the low amounts revealed by western blot (Fig. 3j, k), and reproduces its absence from cataloging efforts for adipose tissue (see e.g., in https://www.proteomicsdb.org/protein/293869/expression). Next, we proceeded to intersect the DAPs increased in OLFM2 GoF and those down-regulated in LoF cell models, and vice versa. The resulting protein groups were analyzed for enrichment of terms from Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). MetaboAnalyst 5.0 (http://metaboanalyst.ca) was used to visualize the most important biological processes affected by the synthetic modulation of OLFM2 in 3T3-L1 cells. Among others (Table S6), proteins regulated in MA cultures with altered OLFM2 were found enriched for the terms Focal adhesion (46/199 hits, Holm-Bonferroni adjusted p = 2.93E-7; Fig. S6a), Fatty acid degradation (19/50, p = 6.21E-6), Pyruvate metabolism (15/38, p = 1.1E-4), Axon guidance (37/180, p = 3E-4; Fig. S6b), and Citrate cycle (12/32, p = 3E-3) (Fig. 4d). Of these, Focal adhesion (44/199 hits, p = 2.11E-5), Citrate cycle (13/32, p = 1E-3), and Axon guidance (37/180, p = 1.5E-3), were also found enriched in undifferentiated PA with modulated OLFM2 levels, as indicated in Fig. 4e, Supplementary Fig. S6a, b, and in Table S6. Among all these DAPs, only four showed regulations inversely correlated to OLFM2 (Fig. S6c), while 22 were neatly modulated together with it in both MA and PA (Fig. 4f and Fig. S6d). This latter group was related to GO terms ranging from Axon guidance receptor activity to Cell adhesion (Fig. 4g), which further supports a major role in adipocyte and precursor cells for OLFM2, a cell surface glycoprotein primarily known for its involvement in nerve development6, with consequential functions for regulating cell commitment and adhesion to the substratum. Subgroup segregation in PCA validated the experiments synthetically modulating OLFM2 levels in 3T3-F442A-derived fat cells (Fig. S7a). Here, as many as 1082 DAPs (619 up and 463 down-regulated) in MA (Fig. S7b), and 1360 DAPs (858 up and 502 down-regulated) in PA (Fig. S7c), were identified when comparing models of GoF vs LoF. In agreement with results in 3T3-L1, alterations in 3T3-F442A proteomes highlighted, among others (Table S7), an enrichment for the terms Focal adhesion, Fatty acid degradation, Propanoate metabolism, Alzheimer disease, and Citrate cycle in both MA (Fig. S7d) and undifferentiated PA (Fig. S7e). Functional enrichment of proteins conjointly regulated in engineered 3T3-F442A and 3T3-L1-derived adipocytes and precursor cells (Fig. S7f and Table S8) indicated that, in adipocytes, PPAR signaling (11/87 hits, Holm-Bonferroni adjusted p = 8.78E-5) (Fig. S7g) ranked together with Citrate cycle (7/32, p = 2.6E-4), Fatty acid degradation (9/50, p = 4.03E-5), and Focal adhesion (16/199, p = 7.13E-5) among the 10 pathways most prominently modulated in both approaches (Fig. S8a), comprising Citrate cycle (7/32, p = 5E-4), Fatty acid degradation (8/50, p = 1.1E-3), and Focal adhesion (17/199, p = 5.1E-5) also observed in PA (Fig. S8b and Table S8). To further examine the relevance of OLMF2 in adipogenesis, we expanded the characterization of OLFM2 loss (Fig. S9a) and gain (Fig. S9b) of function in differentiating primary human adipocyte progenitors, and found the adipogenic induction to be compromised by the siRNA-mediated reduction of OLFM2 transcript abundance (Fig. S9c), as evident from impaired expression of medium-to-terminal biomarkers (ACSL1, ADIPOQ, LPL, CS, ACLY) of adipocyte differentiation (Fig. S9e), as well as the reduced appearance of lipid droplets in the cytoplasm (Fig. S10a–c). Remarkably, defective OLFM2 in human PA also yielded altered expression of genes related to Citrate cycle (e.g., CS, IDH1), Fatty acid degradation (ACADL, ACADS, ACAT1, ECI1), and Focal adhesion (COL4A1, FLNB, ITGA1, LAMC1) (Fig. S9e). By contrast, OLFM2 GoF in human MA (Fig. S9d) exhibited the overrepresentation of genes related to Focal adhesion (Fig. S9f), and an overall enhancement of the adipogenic conversion, as evident from the amount of lipid droplets accumulated in the cytoplasm of differentiated adipocytes (Fig. S10d–f). Collectively, these results suggest that OLFM2 controls pathways of relevance in the adipogenic transformation of fat precursor cells.
a Pipeline diagram of differentiating 3T3-L1 cells, and b changes affecting OLFM2 protein and transcript levels during adipogenic conversion (n = 4 biological replicates/time point/experiment). c IF images show OLFM2 distribution in 3T3-L1 PA when transforming into lipid-containing MA. The scale bars denote 50 μm length. d Different treatments applied to 3T3-L1-derived MA underscored significant variations affecting Olfm2 gene expression levels (n = 4/group). e Engineered 3T3-L1 PA and MA cell cultures were obtained by means of lentivirus-mediated plasmid transfections, and expressed higher (GoF) or lower (LoF) amounts of OLFM2 coding transcripts, as shown in f (n = 6/group). These modifications increased the adipogenic transformation of 3T3-L1 Olfm2_GoF cells, while OLFM2_LoF preadipocytes differentiated less that control, as demonstrated by g the expression levels of a hub of genes related to the adipocyte phenotype (n = 4/group), measures of h Oil Red O (n = 8/group) and i fluorescent lipid staining (n = 4 wells/group; the scale bar denotes 100 μm length), and j, k protein amounts (n = 4/group) of different adipogenic markers and OLFM2 (western blot). The box plots show center line at the median, upper and lower lines bound at 75th and 25th percentiles, respectively, and whiskers at minimum and maximum values. In bar plots, results are presented as mean ± S.E.M. r.u. stands for relative units. *p < 0.05, **p < 0.01 (One-way ANOVA (plus Tukey’s test p values), and Two-way ANOVA after controlling for FDR in multiple comparisons). Source data are provided as a Source data file.
Mass spectrometry-based quantitative proteomics were conducted in engineered 3T3-L1-derived MA and PA obtained as illustrated in Fig. 3e. a Multivariable analysis (PCA) shows the clustering of non-targeting controls (Ctrl) and models of gain (GoF) and loss of function (LoF) in MA and PA. Volcano plots of up (red) and down-regulated (blue) proteins in b MA and c PA with synthetically altered OLFM2 levels. Dots in gray show proteins meeting our exclusion criteria in a Bayesian moderated t-test (nominal p ≥ 0.05), when assuming equal variance in comparisons of n = 5 biological replicates/group. Horizontal p value thresholds show the number of DAPs (adjusted p < 0.05) after correcting for multiple comparisons with the false discovery rate (fdr) and Bonferroni (bon). Labels show some key gene symbols. Metabolic pathway analysis bubble plots created by applying MetaboAnalyst 5.0 to DAPs identified in d MA and e PA (additional details provided in Table S6). Statistical significance was acknowledged for Holm-Bonferroni adjusted p < 0.05 (dashed blue line). f The Heat map shows variations in 26 proteins (genes) significantly (Bayesian moderated t-test adj. FDR p < 0.05) modulated in both MA and PA, strikingly opposed to (4), or directly compelled by (22) OLFM2 levels (see also in Fig. S6c, d). Color represents row z-scores calculated for each cell type and replicate by subtracting the mean and then divide by the standard deviation of each column. g Gene Ontology (GO) enrichment analysis applied to this subset of 26 DAPs.
Defective OLFM2 alters adipocyte transcriptomes
Next, we explored the functional contribution of defective OLFM2 to adipocyte function, as changes occurring in inflamed fat cells correlated with the reduced expression of this factor observed in subjects with obesity. To that end, we reduced OLFM2 transcript levels in terminally differentiated human adipocytes using si-OLFM2 for 3 days, as well as lentiviral-mediated transfection of short-hairpin (sh-)RNA particles for a more stable 6-day down-regulation of OLFM2 (sh-OLFM2) in an independent experiment (Fig. 5a). First, measures of gene expression (Fig. 5b) and protein levels (Fig. 5c) confirmed the knock-down (KD) of OLFM2 in both assays. We next proceeded to query the resulting cell models by transcriptomics using Affymetrix oligonucleotide microarrays, comprising a total of 20,893 transcripts. Stringent threshold criteria (adjusted false discovery rate (FDR) p < 0.05 and fold-change ≥ [1.5]) yielded 243 differentially expressed transcripts (82% decreased) between si-OLFM2 and non-targeting control (NTC) adipocytes (Fig. 5d). For the harsher lentiviral-mediated KD (Fig. 5b, c), the analysis produced as many as 1226 differentially expressed transcripts (54% decreased) (Fig. 5e). The experimental approach was validated by unsupervised PCA (Fig. 5f, g) and hierarchical clustering (Fig. 5h, i), neatly splitting results according to treatment. Gene Set Enrichment Analysis (GSEA)28 applied to the landscape of genes regulated by the loss of OLFM2 revealed striking changes in both assays (Fig. 5j, k), as for example, impaired expression of genes related to the cell cycle (Fig. 5l and Fig. S11a), or the enrichment of factors related to the Tnfα signaling canonically driven through the transcriptional activity of nuclear factor kappaB (NF-κB) (Fig. 5m and Fig. S11b, c), known to play a critical role in obesity-related oxidative stress and metabolic inflammation29. Combining results from these two complementary model systems, we shortlisted 41 genes up and 27 down-regulated concomitantly with defective OLFM2 in human adipocytes (Fig. 5n). Found among the up-regulated gene candidates was adipocyte-derived angiopoietin-like protein 2 (ANGPTL2), known to be abundantly expressed in adipose tissue in close association with obesity-related inflammatory cues, leading to impaired metabolic homeostasis30. Also increased in both approaches were calcium regulated heat stable protein 1 (CARHSP1), which enhances the stability of single-stranded RNAs such as those coding for TNFα31, and β8 integrin (ITGB8), participating in the activation of TGFβ32, which in fibroblasts may led to increased fibrosis and pathologic inflammation33. On the other hand, the set of down-regulated genes comprised NDC80, PBK, STIL, PLK4, KIF20B, SMC4, KPNA2 and GEN1, prominently involved in cell cycle regulation (i.e., E2F targets, mitotic spindle, and G2M checkpoint), potentially partaking of the adipocyte response to a pro-inflammatory microenvironment, enabling further adaptation to the expansion of adipose tissue and metabolic alterations in subjects with obesity34. Global transcriptomics performed in two independent experimental settings underscored, in conclusion, an apparent defect in cell cycle characterizing human adipocytes with impaired OLFM2 levels, and indicated that, in addition (or simultaneously) to its contribution to the adipogenic conversion of fat precursor cells, this specific isoform of olfactomedin may act to maintain proper adipocyte function, thus contributing to the common hallmark of the obesity-related metabolic imbalance, when defective.
a Scheme illustrating the treatments applied to accomplish our 3 (si-OLFM2) and 6 (sh-OLFM2) days-lasting OLFM2 loss of function assay in human MA. Defective OLFM2 was first checked at b gene expression (n = 4/group; two-sided Student’s test p value when comparing each modification vs. their respective control) and c protein (n = 3/group; One-way ANOVA) levels. Results are presented as mean ± S.E.M, and source data are provided as a Source data file. For transcriptome profiling (multiarrays), 3 biological replicates of treated cells and 4 controls were used in each experiment. d, e Volcano plots, f, g PCA, and h, i hierarchical clustering show the impact of impaired OLFM2 in MA transcriptomes. Inked dots in Volcano plots depict genes with adjusted p < 0.05 (Bayes moderated t-statistics after correcting for FDR multiple comparisons) and fold-change (FC) ≥ 1.2 (red) and FC ≤ −1.2 (blue). Green dots show genes related to the G2M checkpoint hallmark. Interpretation of genes differentially expressed in j si-OLFM2 and k sh-OLFM2 adipocytes when compared to their respective controls (si-NTC and sh-NS) was carried out by means of Gene set enrichment analysis (GSEA), which highlighted significant variations happening in both experiments, such as those affecting l G2M checkpoint (decreased) and m Tnf signaling via NFκB (increased). n Heat maps and Venn diagrams resume significant changes in a list of 68 common genes, 41 up and (including OLFM2) 27 down-regulated in each approach.
Olfm2-null mice show impaired adipose cell cycle
The notion that olfactory modulation influences obesity outcomes beyond food choice and nutritional balance has recently gained traction in the literature35,36,37. We contributed to the onset of that conversation by studying diet-induced obesity in an Olfm2 knockout mouse model10, which is characterized by defective olfactory function, as well as a cluster of mild-to-intense motor and behavioral defects11. In that study we concluded OLFM2 to act as a major regulator of whole-body energy homeostasis, as expenditure and distribution of energy surplus in Olfm2-null mice was improved10. In the present work, we took advantage of the available mouse model to try to corroborate those findings in adipocyte cell systems. The appearance of brown adipose tissue-like gene expression signatures in the adipose tissue of Olfm2-null mice (Fig. S12a–d), for instance, paralleled these of human adipocytes with impaired OLFM2, which showed a trend towards the overrepresentation of thermogenic genes (Fig. S12e–h). Accordingly, when the responsiveness to a β3-agonists was explored in vivo, the higher thermogenic tone and gene expression patterns at the basal level of white fat pads in Olfm2-knockout mice resembled those of wild-type mice under β3-agonists activation (Fig. S13a–e). On the other hand, since OLFM2 in mouse adipocytes appears to be involved in the control of genes related to axon guidance (Fig. 4d, e), and as sympathetic innervation is critical for adipose tissue function38,39, we resolved to study the abundance of somatosensory neurons innervating fat pads in Olfm2-null mice. By using tyrosine hydroxylase (TH) immunolabelling40 and Adipo-Clear (adipose tissue delipidation and clearing protocol to facilitate the observation of its architecture41), our data indicated the impairment in length and abundance of sympathetic fibers innervating fat pads in Olfm2-null mice (Fig. S13f–h). Together, these experiments further defined the impact of whole-body Olfm2 ablation on the thermogenic tone of white adipose tissue (enhanced), local response to adrenergic activation (blunted), and abundance of adipose-innervating sympathetic fibers (impaired). Finally, to investigate molecular programs associated with the loss of Olfm2 in mouse adipose tissues, paired OM (perigonadal) and SC (inguinal) fat samples were prepared from five control and five Olfm2-null animals and subjected to bulk microarray analysis (Fig. 6a). As the latter are protected against diet-induced obesity, and thus exhibited reduced fat mass when compared to controls10, we expected adipose tissue transcriptomes grouping according to genotype (Fig. 6b, c). Accordingly, scrutiny of our microarray results revealed differentially expressed genes (Fig. 6d, e), highlighting enriched pathways and transcriptional networks delineating each genotype (Fig. 6f, g and Table S9). Some of the hallmarks underscored in SC and OM adipose tissues were related to cell cycle entry and progression (Fig. 6h), which agreed with our results in vitro, when impaired cell cycle-specific gene expression patterns in Olfm2-null mice (Fig. 6i) mirrored to some extent the results assessed in human adipocytes with defective OLFM2 (Fig. 6j). As a matter of fact, 21 out of 31 genes with a fold-change ≤ −1.2 and FDR p < 0.05 in mouse adipose tissues and human adipocytes were associated with cell cycle (Fig. 6k), including CCNB1, CDK1, NDC80, PRC1, STIL, KIF18A, and BUB1; genes coding for centrosome-associated proteins such as CENPE, CENPF, CENPI, and CENPN; and the proliferation marker MKI67. With regard to genes with expression patterns opposing OLFM2, the eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1), primarily related to PI3K-Akt signaling, adipogenesis42 and adipose tissue accretion43, as well as adipsin (CFD), were of highest consistency amongst the 12 biomarkers shortlisted by integrated transcriptomics (Fig. 6l). Remarkably, by deconvolving the cellular landscape of bulk adipose tissue transcriptomes, we observed the reduced representation of interneurons in Olfm2-null SC fat (−33%, adj. p = 1.9E-2), together with gene signatures further indicative of impaired innervation concomitant with defective OLFM2 (Fig. S14a–c). Taken together, in vitro and in vivo evidence connected defective OLFM2 signal to molecular changes suggestive of impaired cell cycle dynamics and compromised adipose tissue function in the context of obesity, including altered adipocyte-nerve fiber crosstalk and depleted adipose tissue neurite density in whole-body Olfm2-knockout mice.
a We ran the transcriptomic analysis of epigonadal omental (OM) and inguinal subcutaneous (SC) adipose tissues of 10 diet-induced obese (DIO) Olfm2-null (Olfm2_KO) and wild-type (Wt) mice. Principal component analysis (PCA) shows b OM and c SC sample distribution in two-dimensional charts. Correlation plots picture transcript changes in d OM and e SC adipose tissues. Inked dots depict genes with FDR p < 0.05 and FC ≥ 1.2 (red) or FC ≤ −1.2 (blue), and green dots show genes related to the G2M checkpoint hallmark. GSEA summarizes the hallmarks affected in f OM and g SC fat. h Genes ascribed to G2M checkpoint and/or being E2F targets were broadly compromised. Below, heat maps show changes affecting a hub of 50 transcripts related to cell cycle and compromised in i Olfm2-null adipose tissues, as well as in our j human adipocyte cultures with impaired OLFM2 (si-OLFM2 and sh-OLFM2 cells vs controls). Heat maps represent z-scores based on gene-counts for each sample using the package Bioinfokit (2.0.8) for Python. Gene symbols inked in blue depict transcripts with FDR p < 0.05 in at least one fat depots and/or experimental approach in vitro. Venn diagrams summarize coincidences within genes significantly k down (31) and l up-regulated (12 transcripts) in Olfm2-null mice and human adipocytes with defective OLFM2 signal.
Adipose-specific Olfm2 knock-down triggers an obese phenotype
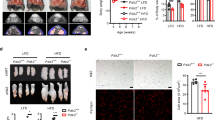
Following our findings in Olfm2-null mice, and in human as well as mouse adipocytes, we sought to further confirm the relevance of OLFM2 to the physiology of adipose tissue. To survey the specific contribution of defective adipose OLFM2, a transient, tissue-centric KD was achieved by direct lentiviral transfection of depots of OM (perigonadal) and SC (inguinal) adipose tissue during surgical exposure, as explained in ref. 44 and ref. 45. Ten 12-week-old C57BL/6J female mice were treated with vectors coding for short hairpin constructs targeting Olfm2 (Olfm2_KD), and ten age-matched specimens were injected with non-silencing (NS) controls, followed by sacrifice of five individuals each after two and 5 weeks, respectively (Fig. 7a). This design was chosen to monitor transient changes caused by reduced adipose OLFM2 levels in animals on a standard diet, as well as any potential physiological adaptation to this modification. Down-regulation of Olfm2 transcript levels was confirmed in OM and SC adipose tissues, parallel to increased Impa2, Itgb8 and Col16a1 gene expression and interference with transcript levels of cell cycle-related genes in OM fat (Fig. 7b). On the other hand, depots of OM (Fig. 7c) and, to a greater extent, SC (Fig. 7d) adipose tissue, were characterized by altered expression of other proteins identified along this research, including Lpl (triglyceride metabolism), Ephb2 and Slit2 (axon guidance), and Pdgfra and Pdgfrb (focal adhesion, also considered early adipocyte progenitor markers, together with Zfp42346, Klf5 and Klf1447). This treatment failed, however, to modify white adipose tissue gene expression signatures towards a brown fat phenotype (Fig. S15a–d), as well as to compromise sympathetic activation of nerves innervating fat depots, as deduced from a lack of changes in neurotransmitter concentrations (Fig. S15e). At the physiological level, Olfm2_KD mice were characterized by faster body weight acquisition (Fig. 7e and Fig. S15f–g) to a maximum exceeding that of control animals (Fig. 7f), even in the absence of significant differences in terms of food-intake (Fig. 7g). Olfm2_KD mice also exhibited decreased adiponectin (Fig. 7h) and higher fasting blood glucose than their control counterparts (Fig. 7i), as well as impaired response to sugar, as revealed by glucose tolerance tests (GTT) performed in both groups (Fig. 7j, k). Concurrently, histological assessment of fat samples collected at the end of the experiment indicated a substantial enlargement of existing SC adipocytes in Olfm2_KD mice (Fig. 7l), which may be indicative of alterations affecting fat cell turnover, as suggested by expression levels of genes related to early adipocyte commitment at 2 weeks (Fig. 7d). Integrating these findings with evidence compiled from adipocytes, human patient cohorts and mice models reinforces the notion of a functional relevance of diminished OLFM2 in adipocytes, aggravating impaired fat cell function and hypertrophy in the context of obesity, also displaying metabolic disturbances of an obese phenotype, including impaired glucose tolerance even in healthy animals under a standard diet.
a The largest depots of white adipose tissue were surgically exposed and injected with our lentiviruses carrying sh-Olfm2 (Olfm2_KD) or a non-silencing (NS) plasmid control. For the molecular assessment of changes affecting these adipose tissues, half of the specimens were terminated 2 weeks after treatment (n = 10; 1:1). Here, b impaired Olfm2 transcript levels were confirmed, paralleling the expression of a hub of gene markers for inflammation and fibrosis (increased), as well as cell cycle (decreased) in omental (OM) fat pads. c Also in OM, but primarily in d subcutaneous (SC) adipose tissue, impaired expression of gene markers for triglyceride metabolism (Lpl), axon guidance (Ephb2, Slit2), and focal adhesion (Pdgfra, Pdgfrb) ran together with the down-regulation of early adipocyte progenitor markers (Zfp423, Klf5, Klf14) in Olfm2_KD mice. To evaluate the physiological consequences of impaired OLFM2 in adipose tissues, the remaining animals (n = 10; 1:1) were maintained under a standard laboratory chow (NC) and monitored for 5 weeks, before being terminated. Notably, e the Olfm2_KD group depicted a higher growth, leading to f increased (p = 0.17) final body weight, although g not significant differences were observed in terms of food intake. Accompanying theses variations, h impaired adiponectin and i increased basal glucose levels were suggestive of obesity-related impaired metabolic homeostasis affecting our adipose-specific Olfm2 knocked-down mice, as further indicated by j glucose tolerance tests (GTT) and k area under the curve (AUC). l Representative images of the hematoxylin–eosin staining of SC and OM adipose tissues administered with either sh-Olfm2 or NS lentiviral particles in animals fed a normal chow. The scale bars denote 50 μm length. The morphometric analysis of adipocytes indicated higher adipocyte area in the Olfm2_KD group. Data in error bars are expressed as mean ± S.E.M. The box and violin plots show center line at the median, upper and lower lines bound at 75th and 25th percentiles, respectively, and whiskers at minimum and maximum values. *p < 0.05, **p < 0.01 (Two-way ANOVA for multiple comparisons by controlling the FDR (Benjamini, Krieger & Yekutieli) q value, or two-sided Student’s test p value). Source data are provided as a Source data file.
Connecting OLFM2 to obesity
A number of chromosomal loci associated with the timing of pubertal initiation in women appear to be located in or near genes associated with BMI and/or implicated in energy homeostasis48. Our intriguing data from different in vitro and in vivo models suggesting a causative involvement of defective OLFM2 in metabolic outcomes prompted us to test for further translational value of these observations in humans. We used the T2DKP database (type2diabetesgenetics.org, RRID: SCR_003743: OLFM2 Gene page. 2025 Jan 15; https://hugeamp.org/gene.html?gene=OLFM2) to explore Genome-Wide Association Studies (GWAS) for any relationship between OLFM2 gene variants and traits accompanying an obese phenotype. As shown in Fig. 8a, significant associations with anthropometric values were detected. Further corroboration came from PhenoScanner (https://phenoscanner.medschl.cam.ac.uk), indicating the significant correlation of a number of polymorphisms affecting OLFM2 and anthropometric traits related to obesity (Fig. 8b), as well as a connection to the average onset age of menstrual period in female adolescents, reported a number of times in the literature48,49,50,51. Among the associations retrieved from GWAS, the potential clinical implications of intergenic variants affecting OLFM2 gene location (Fig. 8c) included reference numbers rs80076900 and rs8108147, which also highlight the apparent relationship between OLFM2 gene variants and anthropometric and physiological traits related to the risk of being obese (e.g., waist-hip ratio with52 or without51 adjustments for BMI, and blood pressure53). Remarkably, these two regulatory variants are in linkage disequilibrium (LD) with the SNPs with reference numbers rs11668085, rs55732232 and rs56243392, which have been associated with significant differences among individuals with regard to OLFM2 protein levels in blood54,55,56,57. However, only the missense variant with reference number rs2303100, and the variant with reference number rs80076900 reached the maximum FORGEdb score of 10 (Fig. 8b), and thus are most likely to be regulatory variants, as calculated by using various human DNA datasets58. As a matter of fact, the SNP with reference number rs2303100 may, as a predicted missense R[Arg] > Q[Gln] (106) and loss of function mutation, be of particular relevance. Accordingly, this polymorphic variation has been suggested as a disease-causing factor for open-angle glaucoma7, as well as associated with the age at menarche59. In line with this, PhenoScanner (inclusion criteria p-value < 1E-5 and r2 ≥ 0.8) found a connection between this specific mutation (Fig. S16a) and the age at menarche (P = 4.7E − 17, β = 2.09 × 10−2), whole body impedance (a surrogate for body fat percentage) (P = 4.34E − 15, β = 1.67 × 10−2), as well as comparative body size at age 10 (P = 8.08E − 6, β = 0.75 × 10−2). In silico analysis of OLFM2 protein structure conformation (Fig. S16b) underscored potential steric effects in the amino acid sequence towards the N-Terminal end lacking homology with the structurally resolved olfactomedin-like domains in other olfactomedin family protein members. At least two sequences are predicted to form coiled-coil motifs (58–85 and 136–193) (Fig. S16c). When comparing in silico generated structures, helixes 96-112 and 116-125 (helixes W-X), and 143-162 and 173-187 (helixes Y-Z in Fig. S16d) differed markedly for the polymorphic OLFM2-Q106 protein, and a drop in the confidence of supercoiled alpha helix formation was observed between residues 109 and 116. These observations suggest structural divergence between OLFM2-R106 and OLFM2-Q106 potentially translating into phenotypic variations even though an intermediate degree of conservation (21/100) was found in SuSPect (Fig. S16e), suggesting no expected relationship with disease (based on sequence, structure and systems biology-based features including known and predicted active sites, post-translational modifications and sequence conservation at the mapped position60). In conclusion, exploration of GWAS datasets further connects OLFM2 to obesity, which is in line with evidence compiled in our previous publication10, as well as the study presented here. A diagram summarizing our framework is presented in Fig. 9.
a Meta-analyzed associations between genetic variants of OLFM2 and anthropometric traits mapped on PhenoScanner (https://phenoscanner.medschl.cam.ac.uk). The illustrated SNPs (x axis) are lead SNPs for the presented anthropometric traits (y axis). Different shades of blue are used to distinguish between the traits. b Heatmap matrix of pairwise linkage disequilibrium statistics of lead SNPs for anthropometric traits identified in c the OLFM2 locus (chr19: 9,853,718–9,936,515). An apparent LD block, consisting of rs2303100, rs8112411, rs889122, rs1862471, and rs12979274, and led by rs2303100 missense variant, shows the strongest associations with BF% (P = 8.3E-05, 2.0E-04, 1.3E-03, 2.3E-04 and 1.9E-04) and Trunk fat ratio (P = 3.2E-05, 4.7E-04, 6.2E-05, 1.6E-04 and 1.8E-04, respectively). Publicly available reference haplotypes from the 1000 Genomes Project were used by LDlink (https://ldlink.nih.gov/) to calculate population-specific measures of linkage disequilibrium (LD). FORGEdb scores, ranging between 0 and 10, are used for predicting regulatory genetic variants, and are calculated using different regulatory DNA datasets, including data for transcription factor (TF) binding and chromatin accessibility (https://forgedb.cancer.gov). eQTL data were derived from Open Targets Genetics (https://genetics.opentargets.org/). Source data are provided as a Source data file.
Framework proposal laid out during this research, which consequentially connects obesity to changes in OLFM2 levels during adipogenesis and upon the inflammatory activation of adipocytes. In turn, impaired OLFM2 may compromise adipocyte phenotype in obesity through the disruption of paramount mechanisms in adipocyte function, as differentiation, cell cycle and focal adhesion, to the point of exacerbate the pathogenesis of adipocyte hypertrophy and impaired metabolism in mice. Coincidentally, scrutiny of OLFM2 gene variants (GWAS) and the metabolic phenotype of our Olfm2-null mouse model further suggests physiological implications connecting OLFM2 to obesity-related traits. Created in BioRender. Gómez Serrano, M. (2025) https://BioRender.com/0l2biim.
Discussion
Members of the olfactomedin family of proteins, the founding member of which was first described in frog olfactory neuroepithelium61,62, are characterized by a homology domain of ~250 amino acids. Despite being overrepresented in neural tissues, many olfactomedin domain-containing proteins show specific peripheral tissue expression patterns3, and may be found in species ranging from nematodes to humans3,4. Overall, they appear to act as transmembrane receptors, facilitating cell cycle regulation, intercellular interactions, and cell-cell adhesion. They may further serve as modulators of critical signaling pathways5. Notwithstanding this assumed broad range of biological mechanisms they are involved in, individual functional relevance remains elusive for a majority of olfactomedin protein family members. This study was aimed at remedying that for OLFM2, a scaffold protein mostly related to brain development6,11, as well as implicated in vascular remodeling8,9, and assigns it molecular, cellular and physiological significance in adipose tissue. We present a plethora of evidence showing that this member of the olfactomedin family is highly expressed in adipocytes and relevant for the homeostatic balance of adipose tissues. Effects of altered expressions in human adipose tissue are shown to be related to the inflammatory component of obesity, as well as to play a key role in adipocyte differentiation and fat mass expansion. Our results demonstrate that expression modulation impacts the commitment of fat cells, modifying the transcriptional program and proteomic landscapes of adipocytes and fat cell progenitors. Concomitantly, while the loss of whole-body Olfm2 protects against diet-induced obesity10, its transient depletion in adipose tissues disrupts the gene expression program of fat cells, fostering adipocyte malfunction, cell hypertrophy, fat mass accretion, and impaired metabolism in mice. Altogether, our study demonstrates that adipose OLFM2 is important for adipocyte biology, and suggests that its association with obesity is not only correlative but also causative.
In functional depots of adipose tissue, hormonally activated mesenchymal stem cells adapt gene expression to further the phenotypic transition to MA63. The differentiation cascade leading to the final fat-cell phenotype (adipogenesis) is tightly controlled, and involves coordination of the thousands of genes involved in terminal differentiation64. During this process, the amount of OLFM2 (but none of the others olfactomedin family proteins) gradually increases, first in the nucleus, then in the cytoplasm, and even at the plasma membrane of differentiating adipocytes, reaching peak levels at full differentiation. The presence of binding motifs for transcription factors orchestrating adipocyte differentiation in the OLFM2 locus, such as PPARγ65, CEBPβ66,67,68, and CREB169, both present in the promoter region of OLFM224, but not located upstream the genes coding for OLFM1, 3 or 4, likely underlies this differentiation-correlated expression pattern. The occurrence of these binding motifs may further explain impaired expression of OLFM2 in adipocytes under the influence of inflammatory cytokines, capable of compromising fat cell function through the corresponding transcription programs70,71,72,73. Concomitant to the fact that this unique olfactomedin may serve as a biomarker of adipocyte cell conversion and function, experimental boosting of its abundance in fat cell precursors promoted adipogenic capacity, while reduction yielded a partial compromise of the transition to adipocytes in mouse and human cell progenitors. Based on the apparent relevance to modulate human adipocyte transcriptomes, as well as proteomic molecular phenotypes of both adipocytes and precursor cells, our results point at OLFM2 as a novel contributor to the phenotypic maintenance of fat cells, and thus the physiopathology of obesity.
Considerable progress has been made in identifying the signaling pathways that govern alterations of adipocyte gene expression patters linked to obesity74,75,76. Recent advances in single cell RNA sequencing, in particular, yielded increased appreciation for a transcriptomic profile suggestive of systematic cell cycle activation in human adipocytes, intrinsically linked to the metabolic dysregulation found in subjects with obesity77,78,79. As a matter of fact, although adipocytes are largely assumed to no longer be capable of entering mitosis to replicate, a study by Li et al.34 demonstrated that these cells may reenter cell cycle to accommodate large volume increases, when required. Consequently, cell cycle exit in adipocytes was noted as associated with impaired metabolism in obesity34. These findings are given further credibility by related mechanisms other olfactomedin-domain containing proteins are involved in, namely OLFM480,81,82,83,84 and olfactomedin-like 2A (OLFML2A)85, which appear to influence cell cycle to the extent of controlling cell proliferation. Our results for OLFM2 mirror such activity in MA and bulk adipose tissue, where it appears to act as a key regulator in coordinating signaling-induced transcriptional modifications leading to alterations in cell cycle. Thus, impaired adipocyte-specific OLFM2 may contribute to fat cell hypertrophy and adipose tissue expansion, as reflected by the increased weight of mice challenged with the virogenetic knock-down of adipose Olfm2 and maintained under a non-obesogenic dietary regime.
In turn, experimentally altered OLFM2 levels in 3T3 cell lines and primary human preadipocytes modulated differentiation and aspects of utmost importance in adipocyte biology, also linked to the physiopathology of obesity. These included pathways related to lipid homeostasis and tissue consistency, ranging from fatty acid degradation, pyruvate and citrate metabolism (energy handling) to axon guidance (contributors to neurite outgrowth and axon elongation that can also control cytoskeletal and plasma membrane rearrangements in other cells86) and focal adhesion (protein structures connecting dynamic intracellular protein complexes and actin bundles to the extracellular substrate87 that may also influence axon guidance88). In our previous work89, we demonstrated that impaired expression levels of a component of desmosomes (macromolecular cell structures specialized in cell-to-cell adhesion) can break cell cycle dynamics to yield impaired metabolism and premature senescence in adipocytes. Intriguingly, the findings reported for Plakophilin-2 (PKP2)89 and these of the current study are largely congruent. Expression of PKP2 and OLFM2, both, rise during adipocyte differentiation, and are inversely associated with obesity and inflammation, while normalized upon weight loss. Defective PKP289 or OLFM2 in human adipocytes resulted in large transcriptional alterations suggestive of an impaired cell cycle program. Taken together, our functional and mechanistic data imply possible new avenues of research in the context of obesity, in which alterations affecting these scaffolding proteins, relevant in the maintenance of anchoring junctions, either interconnecting fat cells (PKP2), or attaching adipocytes to the extracellular substrate (OLFM2), may compromise adipocyte function to the extent that their ability to response to nutritional and hormonal inputs is impaired, aggravating the disrupted metabolic homeostasis characterizing obesity. As a matter of fact, adhesion structures are necessary when cells progress through cell cycle90, while modulation of the cytoarchitecture to achieve dynamic adaptations in cell shape implies the existence of fundamental mechanisms that link cell cycle machinery to cell adhesion90,91. In the light of a paucity of literature reporting on cell cycle and cytoplasmic proteins mediating focal adhesion in adipocytes, our findings may represent a first step toward better understanding of how the pathogenesis of adipocyte malfunction in patients with obesity relies on the successive disruption of these pathways.
In relation to the phenotype that characterized adipose-specific Olfm2-deficient mice, the sharp contrast to what was observed for Olfm2-null mice requires a closer look. Another instance of this apparent discrepancy was found in animal models and cell systems with impaired AMP-activated protein kinase (AMPK)92 signaling. In brain (hypothalamus), the genetic ablation and pharmacological blockage of brain-borne AMPK function protects against an obese phenotype93,94,95, which is in line with what was hypothesized when investigating the metabolic phenotype of the Olfm2-null mouse model10. In adipocytes (adipose tissue), however, activation of AMPK inhibits acetyl-CoA carboxylase and promotes mitochondrial β-oxidation, while simultaneously suppressing fatty acid biosynthesis96, thus controlling part of the obese phenotype and its sequelae97. Therefore, in adipose tissue, defective AMPK may aggravate obesity, as would seem to be the case also for OLFM2. Another possible explanation may rely in compensatory adaptations during the development of brain, adipose tissue, or any other organ directly or indirectly influenced by the lack of OLFM2, causing the metabolic phenotype observed in our Olfm2-null mouse. In line with this hypothesis, available data connecting OLFM2 to axon guidance and sympathetic innervation was based on the morpholino experiments conducted in zebrafish6. These approaches showed that the transient antisense oligonucleotides-mediated knock-down of Olfm2 produced an even more pronounced defect in nervous system development than the whole-body knockout in mice11. There again, intriguing differences related to target location, strength, and the timing of treatments challenging OLFM2 signaling were observed. Notwithstanding this, the results reported here show, collectively, expression patterns of OLFM2 in adipose tissue to be functionally related to obesity and adipocyte function, underpinning that this particular olfactomedin not only represents an important regulator of adipogenesis, but also that its partial loss in inflamed adipocytes acts as a critical driver of defective cell anchorage and impaired fat cell cycle, resulting in exacerbated adipocyte hypertrophy, fat mass accretion, disturbed adipose tissue function, and impaired metabolism in obesity.
Methods
Human adipose tissue
Samples of SC adipose tissue were retrieved at the mesogastric level during and ~2 years after bariatric surgery, as described elsewhere12. SC fat samples were also obtained during elective surgical procedures (i.e., cholecystectomy and surgery of abdominal hernia) on 24 age-matched women with BMI < 30 kg/m2. Subjects’ characteristics are provided in Table S3. Additionally, biopsies of SC and OM adipose tissue were assessed in 70 men and 130 women with BMI ranking between 18 and 60 kg/m2. BMI was calculated by dividing weight in kilograms by the square of the height in meters (kg/m2), and obesity was set up at BMI > 30 kg/m2. For the analysis of the SVC fraction and ex vivo isolated adipocytes, ~5 g of paired SC and OM adipose tissue of 12 women with obesity were removed of fibrous material, finely minced, and disaggregated enzymatically in a collagenase buffer (Sigma, C0130); then centrifuged, washed, and filtrated according to the Bunnell method98 to separate the infranatant of SVC from floating adipocytes. All participating subjects were White, gave written informed consent, and reported that their body weight had been stable for at least 3 months before entering the study. In all cases, sex was self-reported by the study participants. Samples and subject data were given by the FATBANK platform, promoted by the CIBEROBN, and coordinated by the IDIBGI Biobank (Biobanc IDIBGI, B.0000872). All samples were processed following standard operating procedures with the appropriate approval of the Ethics, External Scientific and FATBANK Internal Scientific Committees.
Olfm2-null mice
For this study, the transcript landscape of OM (perigonadal) and SC (inguinal) adipose tissue of 25 weeks-old, diet-induced obese male Olfm2-null and wild-type C57BL/6 littermate mice (n = 10)10 was analyzed by means of microarrays. These mice were allowed free access to water and a high-fat dietary regime (Research-Diets Inc., D12451) for 10 weeks before being terminated (Fig. 6a), as explained in ref. 10. On the other hand, 15-to-25 weeks-old, standard weight Olfm2-null and age-matched wild-type C57BL/6 control male mice were used to test the response to beta-agonists. Animals were housed under a 12-h artificial light cycle (08:00–20:00), maintaining a controlled room temperature (22–24 °C) and humidity (55%). They were provided with free access to standard laboratory chow diet and tap water. Mice were subjected to intraperitoneal injections of CL-316,243 (1 mg/kg BW of a solution at 0.2 mg/ml diluted in saline) or an equivalent volume of saline. Skin temperature surrounding brown adipose tissue was recorded with in an infrared camera (B335: Compact-Infrared-Thermal-Imaging-Camera, FLIR) and analyzed with a specific software package (FLIR-Tools-Software). Measurements were taken immediately just before and 1 h after CL-316,243 administration. All animals were terminated by cervical dislocation and decapitation. The experiments were performed in agreement with the International Law on Animal Experimentation and were approved by the USC Ethical Committee (15010/14/006).
TH-immunostaining
Teen Olfm2-null and wild-type (1:1) C57BL/6 male mice were also terminated by anesthesia overdose with ketamine/xylazine and transcardially perfused with saline. Then, gonadal adipose tissue was dissected, collected and fixed with 4% PFA overnight, rotating at 4 °C. Next, fixed samples were washed in 20%, 40%, 60%, 80%, and twice in 100% methanol for 30 min each at RT. The samples were bleached with 6% H2O2 diluted in 100% methanol at 4 °C overnight with shaking. The samples were then rehydrated with 80%, 60%, 40%, and 20% methanol for 30 min each at RT. Next, samples were incubated overnight in permeabilizing solution (20% DMSO, 0.2% Triton X-100, 0.3 M glycine, 0.02% sodium azide in PBS) at 37 °C and subsequently blocked using blocking buffer (10% DMSO, 2% Triton X-100, 0.02% sodium azide and 5% goat serum in PBS). For immunolabelling, samples were incubated in primary antibody dilution (5% DMSO, 5% goat serum, 0.2% Tween-20, 10 μg/ ml heparin, 0.02% sodium azide in PBS) for 5 days, with shaking at 37 °C. In this Study, an antibody (1:500; AB152, Millipore) against Tyrosine Hydroxylase (TH) was used to label TH-immunoreactive cells (nervous system). Samples were washed in PTxwH 6 times (1 h each) and left washing overnight at RT. Samples were then incubated with secondary antibody conjugated with Alexa-647 (1:500; Invitrogen) for a further 5 days at 37 °C, with shaking. After immunolabelling, samples were first washed 6 times with PTxwH (1 h each) and then embedded in 1% agarose. Embedded samples were dehydrated in 20%, 40%, 60%, 80%, and 100% methanol for 1 h each, and then in 100% methanol overnight at RT. Following dehydration, samples were washed with 100% dichloromethane (Sigma Aldrich) for 2 h twice, followed by overnight shaking at RT. Samples were then incubated in 100% dibenzyl ether (Sigma Aldrich) overnight. Samples were stored at RT in 100% ethyl cinnamate (Sigma Aldrich) in the dark until imaging. All whole-cleared tissue samples were imaged using a Miltenyi Biotec Ultramicroscope II light-sheet microscope equipped with x4 (used for whole -tissue imaging at low magnification) and ×12 objective lenses (used for high-magnification images). For 4× magnification, step size was 14 μm, thickness of the light sheet was 3.9 μm, horizontal dynamic focusing was set to 10 steps, and exposure time was 37.9 ms. For 12× magnification, step size was 6 μm, thickness of the light sheet was 3.9 μm, horizontal dynamic focusing was set to 3 steps, and exposure time was 38.6 ms. Samples were illuminated using a bidirectional light sheet and scanned with a 680/30 filter for AF647. All whole tissue images were generated by Imarisx64 software (version 8.0.1, Bitplane). Images were quantified by means of ImageJ99 v.1.54k (https://imagej.net/software/imagej/) by adjusting threshold and measuring the stained area in 5 biological replicates.
Adipose Olfm2 knock-down
The assay was performed using 12-week-old C57BL/6JOlaHsd female mice (n = 20) purchased from Envigo. Animals were housed under standard SPF conditions, in groups of up to 3 mice per cage, and kept in ventilated isolators. Mice had full access to a standard laboratory diet (SAFE-A40) and sterilized drinking water during the whole experiment. The environmental conditions were 22 °C (±2 °C) of temperature, 50% (±20%) of relative humidity and light/dark cycle (12:12). The animals were randomized, weighed, and identified by ear tags, generating the different groups (Fig. 7a), either mice treated with lentiviral particles harboring sh-Olfm2 or the non-silencing (NS) control (n = 10/each). The lentiviral particles were kept frozen at −80 °C until administration, time at which they were slowly thawed on ice and administered at room temperature. The lentiviral particles were administered into the adipose tissue (both depots of OM gonadal and SC inguinal adipose tissue) following the surgical procedure detailed in ref. 44. First, lentivirals were applied with a Hamilton pipette the surgically exposed gonadal fat pads. Four injections of 12.5 μl were made along the fat pad of each flank. Once the injection was finished, both fat pads were placed back in the abdominal cavity of the mouse, and the abdominal wall was sutured. Once the abdominal wall was sutured, the skin of the abdominal wall was separated by blunt dissection in the direction of the hind legs to expose the inguinal fat pads. A tissue retractor was placed to expose the SC adipose tissue on each flank, and 2 injections (12.5 μl/each) were made throughout the fat pad with the Hamilton pipette. Finally, the skin is sutured with U-stitches, using 5/0 braided non-absorbable silk suture. Once sutured, povidone-iodine is applied with a swab over the entire surgical field. Two weeks after the surgery, the endpoint of the assay was carried out in half of specimens, which were terminated (3% isoflurane-induced deep anesthesia and exsanguination) to collect both gonadal and inguinal white adipose tissues. The remaining specimens were kept alive for 3 additional weeks to assess the physiological impact of defective Olfm2 at the long term. In both groups, animals’ welfare was monitored (weight, appearance, behavior), and body weight and food intake were recorded three times per week. Circulating adiponectin was measured by means of the Mouse Adiponectin/Acrp30 Quantikine ELISA Kit (R&D, MRP300). A glucose tolerance test (GTT) was performed at week 4 after lentiviral injections. There, blood glucose was measured at 0, 15, 30, 60, 90, 120, and 150 min after an intraperitoneal injection of a 20% glucose solution. Animals terminated (3% isoflurane-induced deep anesthesia and exsanguination) at week 5 were processed equally, and sample collection included serum and inguinal and perigonadal white adipose tissues. Fat pads were fixed in formalin and embedded in paraffin, minced and stained with H&E for histological assessment. Adipocyte area was determined by ImageJ software99. The research was conducted in accordance with the European Guidelines for the Care and Use of Laboratory Animals (directive 2010/63/EU) and was approved by the Ethical Committee for Animal Experimentation of Barcelona Science Park (PCB).
HPLC
Small pieces of frozen adipose tissue were homogenized, sonicated, and centrifuged (14,000 × g for 20 min at 4 °C). The supernatant fraction was filtered and injected (20 μl/injection) into a high-performance liquid chromatography (HPLC) system (Shimadzu LC Prominence; Kyoto, Japan). Norepinephrine (NE), dopamine (DA) and serotonin (5-HT) were separated using a reverse phase analytical column (Waters Symmetry 300C18; Milford, MA, USA). The mobile phase, consisted of a 10% MeOH solution (pH = 4) containing 70 mM KH2PO4, 1 mM octanesulfonic acid, and 1 mM EDTA, was delivered at a rate of 1 ml/min. Detection was performed with a coulometric electrochemical detector ESA Coulochem III (Thermo Scientific; Waltham, MA, USA). The first and the second electrode of the analytical cell were set at +50 mV and +350 mV, respectively, and the guard cell was set at −100 mV. Data were acquired and processed with the Shimadzu LC solution software (Kyoto, Japan). The concentrations were expressed as pg/mg of wet tissue.
Human adipocytes
Commercially available cryopreserved preadipocytes of White women aged 30–50 years and BMI of 25-30 kg/m2 (SP-F-2), or above 35 kg/m2 (SP-F-3), were purchased (Zen-Bio, Inc.). To induce the adipogenic conversion, adipocyte progenitors were led to grow until reaching confluence in preadipocyte medium (PM-1), then incubated with adipocyte differentiation medium (DM-2) for 7 days. This media is composed of DMEM/Ham’s F-12 (1:1), HEPES, FBS, biotin, pantothenate, insulin, dexamethasone, IBMX, PPARγ agonist, penicillin, streptomycin and amphotericin B. Thereafter, differentiating adipocytes were maintained in adipocyte maintenance (AM-1) media (i.e., DM-2 without IBMX and PPARγ agonists) for 7 days. During this process, the shape of preadipocytes evolves from the flattened form to rounded cells containing abundant lipid droplets, and are considered terminally differentiated MA (~12th day and thereafter). The human monocyte cell line THP-1 (ATCC, TIB-202) was cultured in RPMI 1640 medium (Gibco, 21875-034) containing 10% FBS, 5 mM glucose (Sigma, G8644), 2 mM L-glutamine, 50 mg/ml Gentamicin (Gibco, 15710-064), and 20 mM HEPES. Floating monocytes were incubated with 0.162 mM phorbol 12-myristate 13-acetate (Merck, P1585) to induce the adherent pro-inflammatory type 1 macrophage-like state (M1). Then, M1 macrophages were washed with PBS and incubated 24 h with fresh medium containing 10 ng/ml LPS (Sigma, L4516). The resulting macrophage LPS-conditioned medium (MCM) was collected and centrifuged at 400 g for 5 min, filtered, diluted in adipocyte media, and applied to adipocyte cultures. Human adipocyte cultures were also challenged during 24 h with AM-1 containing 5 ng/ml of recombinant human TGFβ (R&D Diagnostics, 240-B), or different dosages of LPS from Escherichia coli O26:B6 (Sigma-Aldrich, L8274).
Altered OLFM2 in adipocytes
Human preadipocytes and MA were transduced with 100 nM of short-interfering (si-)RNAs directed against OLFM2 mRNA, or with a non-targeting control (NTC). Synthetic si-RNAs (SASI_HS01_00053740; Sigma), with sequences 5′-GUCAUGCGGUCCUGGGACA[dT][dT]-3′ and 5′-UGUCCCAGGACCGCAUGAC[dT][dT]-3′, and the MISSION® siRNA Universal NTC #1 (Sigma, SIC001), were used together with Lipofectamine RNAiMAX (Invitrogen, 13778-075), and merged 1:3 in the respective culture media. The short-hairpin (sh-)RNA-mediated knock-down of OLFM2 was achieved in differentiated human adipocytes by means of lentiviral particles (sc-97434-V). Treatment control consisted of scrambled non-silencing (NS) lentiviruses (sc-108080), also purchased from Santa Cruz Biotechnology, Inc. Lentiviral particles were used together with 10 μg/ml polybrene (Sigma, TR-1003) merged in AM-1 culture media. Forty-eight hours’ post-infection, positive adipocytes harboring sh-RNA constructs were enriched by means of 3 µg/ml puromycin dihydrochloride (Sigma, P8833) added to the media. The expression-ready human OLFM2 cDNA ORF clone (#EX-A6251-M90), and the negative empty control vector for pReceiver-M90 (#EX-NEG-M90), were obtained from Genecopoeia (Tebu-bio, Spain) and transfected (0.25 µg/cm2) into primary human preadipocytes. The pReceiver-M90 vector incorporates the human cytomegalovirus (CMV) promoter and SV40-eGFP-IRES-Puromycin Tag. Successful transfection of human adipocytes was achieved by Fugene® Transfection Reagent (Promega, WI, USA) at ratio 1:3, following manufacturer’s instructions. Efficiency was checked by measuring OLFM2 levels in harvested cells.
Microarrays
Microarray expression profiling (Genechip® Affymetrix) was performed in human adipocytes with depleted OLFM2 (sh-OLFM2 and si-OLFM2 cells versus their respective controls), and in the white adipose tissue (both SC and OM) of the Olfm2-null mouse model. The purified RNA was processed according to the protocols GeneChip WT PLUS Reagent Kit (P/N 703174 2017) and “Expression Wash, Stain and Scan” User Manual (P/N 702731 2017), from Affymetrix. RNA integrity was checked by means of an Agilent 2100 Bioanalyzer (Agilent Technologies). For statistical evaluation, the R programming environment (R Core Team (2018) version 3.4.4) was used together with different R analysis packages. The interpretation of genes with significant changes in groups was performed by means of the GSEA computational method, based on the GO set, and evaluated according to Hallmark and C5 gene sets (versions 7.4), which display coherent gene expression patterns to summarize specific and well-defined biological processes, including online pathway databases and the biomedical literature100. The adjusted FDR q value score of 0.05 was set as threshold. Over-representation analysis (ORA) implemented in clusterProfiler101 package version 3.18.0 was used in order to determine whether known biological functions or processes are over-represented in adipocytes. The gene list was obtained from differential expression analysis assessed previously and genes were separated depending on the logFC value (down-regulated and up-regulated). Functional annotations were obtained based on the enrichment of gene sets belonging to gene set collections in Molecular Signatures Database (MSigDB).
Cell deconvolution
Trying to assess the presence of neurite fibers embedded in Olfm2-null mice, we used bulk adipose tissue transcriptomes to calculate the abundance of different cell types. To this end, we collected 6,160 gene markers from multiple single-cell sequencing datasets encompassing 142 cell types from the PanglaoDB (https://panglaodb.se/) database102. Deconvolution analysis was performed using R version 4.2.1 (https://www.r-project.org/) and TBSignatureProfiler package (https://github.com/wejlab/TBSignatureProfiler). Normalized Robust Multiarray Analysis (RMA) data was log2 transformed and entered into the ssGSEA algorithm, resulting in individual enrichment scores for each sample-cell type pairing. Then, a t-test was used to compare differences in cell types between the Olfm2 KO and WT groups.
3T3 cells
The embryonic fibroblast mouse cell line 3T3-L1 (ATCC, CL-173) was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 4.5 g/l glucose (Gibco, 21013-024), 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Three days’ post-confluence, differentiation was induced with 0.5 mM isobutylmethyl-xanthine (Sigma, I7018), 0.25 µM dexamethasone (Sigma, D1756) and 1 μg/ml insulin (Sigma, I3536) in DMEM 10% FBS. At 4th day and thereafter, isobutylmethyl-xanthine and dexamethasone were removed, and insulin was maintained until day 8th. To assess Olfm2 gene expression dynamics, 3T3-L1 cells were harvested at different stages of adipogenic conversion. Upon differentiated, cultures of 3T3-L1-derived adipocytes were challenged for 24 h with either 0.5 µM rosiglitazone (Sigma, R2408), 10 ng/ml LPS, or 10 ng/ml of recombinant mouse TGFβ (R&D Diagnostics, 7666-MB). The embryonic fibroblast mouse cell line 3T3-F442A (Kerafast, EF3002) was cultured in DMEM containing 4.5 g/l glucose, 10% HyClone Iron-Supplemented Calf Serum (Cytiva, SH30072.03), 100 U/ml penicillin, and 100 μg/ml streptomycin. After reaching confluence, differentiation was induced during 8 days with 0.5 µg/ml insulin in DMEM 10% FBS.
Engineered 3T3 cells
To modify OLFM2 levels in 3T3-L1 and 3T3-F442A cell cultures, a sh-Olfm2 plasmid (#217MSH098533-LVRU6GP; clone set against mouse Olfm2, NM_173777.4), and an open reading frame vector plasmid (#217EX-Mm36031-Lv225-GS; ORF expression clone for Olfm2, NM_173777.3), were purchased (Tebu-Bio Spain S.L.). Lentiviruses were obtained by co-transfection into human embryonic kidney HEK-293T epithelial-like cells (ATCC, CRL-3216) of these plasmids with a combination of packaging (pCMV-VSV-G) and envelope (pCMV-dR8.2 dvpr) plasmids from Addgene, used together with LipoD293TM Transfection Reagent (SignaGen, #SL100668). All virus preparations were purified by two cesium chloride density gradients, dialyzed against Hepes-buffered saline, and heated at 56 °C for 45 min to ensure purity and stability of the vector. Empty vectors were synthetized and used as control, and transduction efficiency was optically verified and monitored in all assays by means of green fluorescent protein (GFP) signal, used as a reporter gene.
BODIPY
BODIPY (C9H7BN2F2) staining was used to determine intracellular lipid droplets content. Cells grown and differentiated on coverslips were fixed with 4% para-formaldehyde for 10 min at RT, and stained with 5 µM BODIPY 493/503 (Invitrogen, #D3922) for 3 h. Then, cells were stained with 0.1 µg/ml DAPI solution (Thermo Fisher Scientific, #62247). Coverslips were mounted on slides, and microscopic images were obtained and analyzed using a fluorescence microscope (Nikon eclipse 50i). Images were quantified by means of ImageJ v.1.54k (https://imagej.net/software/imagej/)99 by splitting channels, adjusting threshold and measuring the selected area in 4 biological replicates.
Oil-Red O
Oil-Red O staining (Sigma, O0625) was used to determine changes in the lipid content of 3T3-L1 adipocytes with impaired Olfm2. Differentiated 3T3-L1 cell cultures were washed twice with PBS and fixed with 7% formaldehyde solution (Sigma, #47608) for 1 h. After removing the paraformaldehyde, 60% isopropanol containing filtered Oil-Red O was applied for 10 min at RT. The solution was then discarded, wells were rinsed four times with distilled water, and pictures were taken using a Leica DM IL LED inverted microscope. Then, distilled water was removed, cells were left to air dry, and 200 µl of 100% isopropanol was poured to each well. After 10 min at RT, the isopropanol merged with Oil-Red O stained lipids was collected, and optical density was measured in each well at 500 nm using a PowerWave™ XS spectrophotometer (BioTek Instruments). When Oil Red O staining was not available (as in Fig. S10a–f), lipid droplet area was quantified by means of the Fiji 2.15.1 software (https://imagej.net/software/fiji/), a platform for custom biological image analysis103. Image processing approaches were applied to the area took by lipid droplets, which was quantified using a threshold plugin after sharpening pixel intensity. Values obtained were expressed as the percentage (%) of total microscopy image filled by highly refringent vacuoles of lipid within the cytoplasm of adipocytes observed under bright-field illumination.
Immunofluorescence
Preadipocytes were plated on collagen-coated glass coverslips and differentiated as per normal. Cells were rinsed with PBS and fixed for 1 h in 4% para-formaldehyde at room temperature (RT) in 0.01 M, pH 7.4 PBS buffer. Then, fixed cells were rinsed with PBS, incubated 3 min with 0.1% Triton X-100 (Sigma, X100), and blocked with 10% Gibco® goat serum (#16210064) and 1% Pierce™ Bovine Serum Albumin Standard (#23210) for 1 h. Human and mouse cells were then incubated with a 1:200 dilution of anti-human OLFM2 rabbit polyclonal (Proteintech, #11997-1-AP), or recombinant anti-mouse OLFM2 [EPR10605(B)] (Abcam, ab154196) rabbit monoclonal antibodies for 2 h, respectively. Mouse monoclonal anti-alpha Tubulin antibodies [DM1A] (ab7291), also purchase from Abcam, were used in 3T3-L1 adipocytes. Next, 1:200 dilution of Alexa Fluor 546 Goat anti-rabbit (Invitrogen, A-11035) and Alexa Fluor 488 Goat anti-mouse (A-11001) were applied for 1 h. Finally, fixed cells were treated with 1 μg/mL DAPI solution for nucleic acid staining (D9542) for 5 min at RT. Slides were washed in PBS and mounted with dako fluorescent mounting medium (DakoCytomation, #237-S302380). Images were captured on a Zeiss Axiovert 200 inverted microscope equipped with an AxioCam camera system (ZEISS).
Real time-PCR
Total RNA was obtained from cell cultures and mouse and human samples using the RNeasy Mini Kit (QIAgen, 74104). Integrity was checked by means of a Bioanalyzer 2100 (Agilent Technologies) and RNA quantified in a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). RNA was retro-transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4374967) using the following cycle parameters: 10 min at 25 °C, 120 min at 37 °C, and a final inactivation step of 5 min at 85 °C. All cDNA samples were diluted to 10 ng/µl to obtain results within the range of the standards. Commercially available TaqMan hydrolysis probes (Applied Biosystems) and SYBR Green I, with forward/reverse paired primers (KiCqStart® SYBR® Green Primers; Sigma, KSPQ12012), were used to assess the expression of gene candidates in a Light Cycler 480 II sequence detection system (Roche Diagnostics). The following thermocycler conditions were used: 2 min at 50 °C and 10 min at 95 °C, followed by 40 cycles for 30 s at 95 °C, 45 s at 60 °C and 72 °C for 30 s. The respective human and mouse reference gene transcript cyclophilin A was used to normalize gene expression levels using the 2−ΔΔCt method. Thus, results are expressed as the normalized fold expression for each gene as compared to untreated/reference control cells. Replicates and positive and negative controls were included in all reactions. TaqMan hydrolysis assays and paired SYBR Green primer sequences are listed in Table S10.
Proteomics
Sample preparation started from protein lysates of PA and MA monolayers. Cells were collected in 4% sodium lauroyl sarcosinate (SLS) in 100 mM Tris-HCl (pH ~ 7.5) solution and boiled for 10 min at 90 °C in a ThermoMixer (1800 rpm). Supernatants were collected by 13,000 × g centrifugation at 4 °C for 15 min and protein concentration was estimated by Lowry Assay. Prior to protein digestion, DNA was removed by two cycles of incubation at 90 °C in a ThermoMixer (10 min, 1800 rpm) followed by 1 min sonication. Approximately 50 μg of protein (~50 μl of solution) were taken for further processing. Samples were reduced (addition of 2 μl of 400 mM dithiothreitol to a final concentration of 10 mM and incubation at 95 °C for 10 min), followed by alkylation (additions of 2 μl 550 mM iodoacetamide to a final concentration of 13 mM and incubation for 30 min at 25 °C). All samples were diluted 7x with 50 mM TEAB buffer to allow trypsin digestion. Trypsin was added to a final amount of 1 μg. Digestion was carried for 16 h at 37 °C and was stopped by addition of TFA to a final concentration of 1.5%. Precipitating SLS was removed by 10 min centrifugation at maximum speed in a benchtop centrifuge. Peptides were purified using solid phase extraction on C18 microspin columns according to the manufacturer’s instructions (Macherey-Nagel, Germany). Purified peptides were first dried, then resuspended in 50 μL of 0.1% TFA. Peptide concentration was estimated using the Pierce Fluorimetric Peptide Assay, and sample volumes were adjusted to achieve equal concentrations. Peptides from 3T3-L1 (n = 5 biological replicates/group) cell cultures were analyzed by liquid chromatography-tandem mass spectrometry (MS) carried out on a Thermo Orbitrap Eclipse instrument connected to a Thermo Scientific™ UltiMate™ 3000 RSLCnano UHPLC System. Approximately 800 ng of peptides were loaded onto a µPAC™ -Trapping-column (2 mm diameter, 10 mm length) and eluted with a gradient from 98% solvent A (0.1% formic acid) and 2% solvent B (99.9% acetonitrile and 0.1% formic acid) to 7% solvent B over 15 min, continued from 7 to 38% of solvent B for another 90 min, then from 38 to 55% of solvent B for 15 min over a reverse-phase high-performance liquid chromatography (HPLC) separation column (Thermo Scientific™ µPAC™ HPLC Columns, 50 cm) with a flow rate of 300 nL/min. Peptides were infused via an Advion TriVersa NanoMate (Advion BioSciences, Inc.) into an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Scientific). The mass spectrometer was operating in positive-ionization mode with a spray voltage of the NanoMate system set to 1.7 kV and source temperature at 275 °C. Data were acquired by means of a data-independent acquisition (DIA) paradigm. In short, spectra were acquired with a MS1 resolution of 60,000 and mass range from 375 to 1500 m/z for the precursor ion spectra, maximum injection time was set to 50 ms, AGC target of 400k and normalized AGC target of 100%. This was followed by DIA scans with 40 fixed DIA windows of 15 m/z width and 1 m/z of overlap, ranging from 375 to 975 m/z. MS2 spectrum was acquired with the resolution of 30,000, collision energy of 30%, maximum injection time of 54 ms and AGC target of 2000%. On the other hand, purified peptides from 3T3-F442A (n = 4 biological replicates/group) cell cultures were analyzed by liquid chromatography-tandem MS carried out on a Bruker Daltonics timsTOF Ultra instrument connected to a Bruker Daltonics nanoElute instrument. Approximately 50 ng of peptides were loaded onto a C18 precolumn (Thermo Trap Cartridge 5 mm, µ-Precolum TM Cartridge/PepMap TM C18, Thermo Scientific) and then eluted in the backflush mode with a gradient from 98% solvent A (0.15% formic acid) and 2% solvent B (99.85% acetonitrile and 0.15% formic acid) to 17% solvent B over 36 min, continued from 17 to 25% of solvent B for another 18, then from 25 to 35% of solvent B for another 6 min over a reverse-phase high-performance liquid chromatography (HPLC) separation column (PepSep Ultra, C18, 1.5 μm, 75 μm × 25 cm, Bruker Daltonics) with a flow rate of 300 nL/min. The outlet of the analytical column was coupled to the MS instrument by CaptiveSpray 20 μm Emitter. Data were acquired using a DIA paradigm using a default method provided by Bruker. In short, spectra were acquired with fixed resolution of 45,000 and mass range from 100 to 1700 m/z for the precursor ion spectra and a1/k0 range from 0.64 to 1.45 V s/cm2 with 100 ms ramp time for ion mobility, followed by DIA scans with 24 fixed DIA windows of 25 m/z width, ranging from 400 to 1000 m/z. In both experiments, peptide spectrum matching and label-free quantitation were subsequently performed using DIA-NN (ref. 90) and a library-free search against the Mouse Uniprot.org database (17,136 reviewed Swiss-Prot entries; April 2023). In brief, output was filtered to a 1% FDR on precursor level. Deep learning was used to generate an in silico spectral library for library-free search. Fragment m/z was set to a minimum of 200 and a maximum of 1800. In silico peptide generation allowed for N-terminal methionine excision, tryptic cleavage following K*, R*, a maximum of one missed cleavage, as well as a peptide length requirement of seven amino acid minimum and a maximum of 30. Cysteine carbamidomethylation was included as a fixed modification and methionine oxidation (maximum of two) as a variable modification. Precursor masses from 300 to 1550 m/z and charge states one to four were considered. DIA-NN was instructed to optimize mass accuracy separately for each acquisition analyzed and protein sample matrices were filtered using a run-specific protein q value (“--matrix-spec-q” option). Downstream data processing and statistical analysis were carried out by the Autonomics package developed in-house (version autonomics 1.13.19). Proteins with a q value of <0.01 were included for further analysis. MaxLFQ104 values were used for quantitation and missing values imputed. All intensities and maxLFQ values containing only 1 precursor (Np) per sample were exchanged by NA for that particular sample. Differential abundance of protein groups was evaluated by Autonomics (10.18129/B9.bioc.autonomics) employing Bayesian moderated t-test as implemented by limma105. Functional enrichment analysis of DAPs was performed using MetaboAnalyst 5.0106 and g:Profiler (version e109_eg56_p17_1d3191d), with g:SCS multiple testing correction method applying significance threshold of 0.05107. Detailed settings for data acquisition, spectrum matching and statistical analysis can be found along with the mass spectrometric raw data at the ProteomeXchange Consortium with dataset identifier: PXD065388, via the MassIVE partner repository (https://massive.ucsd.edu/, MassIVE ID: MSV000098300; https://doi.org/10.25345/C5Z31P19P).
Western blot
Tissues were homogenized using a IKA T10 basic ULTRA-TURRAX (IKA-Werke GmbH) and embedded in Radioimmunoprecipitation (RIPA) lysis buffer supplemented with phenylmethylsulfonyl fluoride (PMSF), protease inhibitors and Sodium Orthovanadate (Santa Cruz Biotechnologies, sc-24948). Cell monolayers were first washed with cold sterile phosphate buffered saline (Gibco, #20012027), then disaggregated in cell lysis buffer (Cell Signaling, #9803) supplemented with 1 mM PMSF and cOmplete™ Protease Inhibitor Cocktail (Roche, #11697498001), and collected by scraping on ice. Lysates were kept in agitation for 30 min at 4 °C, then centrifuged at 10.000 × g for 10 min at 4 °C to remove any cell debris. Protein concentration was assessed using the PierceTM BCA Protein Assay Kit (Thermo, #23225). Protein extracts were separated using 4–20% Mini-PROTEAN® TGX Stain-Free™ Protein Gels (Bio-Rad, #4568096) and transferred to polyvinylidene fluoride (PVDF) membranes using the Trans-Blot Turbo RTA LF PVDF Transfer Kit (#1704275) also from Bio-Rad. To ensure equal loading of protein we took advantage of the Stain-Free imaging technology, which utilizes a trihalo compound to detect proteins fluorescent with a short photoactivation. To do so, both Criterion Stain-Free gels (Bio-Rad) and PVDF membranes were placed into the transilluminator and exposed 1 min to ultraviolet light to activate fluorescence signal. Membranes were blocked with 5% BSA in TBS buffer with 0.1% Tween 20 (Sigma-aldrich, P2287) and incubated with rabbit monoclonal anti-OLFM2 antibody [EPR10605(B)] (Abcam, #ab154196) or sheep polyclonal anti-human Olfactomedin-2/Noelin-2 (R&D Systems, AF5614). Primary antibodies from Cell Signaling Technology were used to detect PPARG (#2443S), FAS (#3180S), and PLIN1 (#9349S), and S14 (SCBT, # sc-67299). As housekeeping proteins, 1:2000 anti-β-actin (SCBT, sc-47778, clone C4) mouse monoclonal antibody and 1:1000 hFABTM Rhodamine Anti-Tubulin Primary Antibody (BioRad, #12004166). All primary antibodies were prepared at 1/1000 in 5% BSA in 0.1% TBS-Tween 20. Then, incubation with an anti-rabbit IgG HRP-linked antibody (Cell Signaling Technology, #7074), or a HRP-conjugated anti-sheep IgG secondary antibody (R&D Systems, HAF016), followed. Protein signal was detected by chemiluminescence with a ChemidocTM MP Imaging System (Bio-Rad). Optical densities of the immunoreactive bands and lanes in UV activated membranes were quantified by means of the ImageJ analysis software (https://imagej.net/software/imagej/)99.
Conformational protein analysis
As crystallographic or spectroscopic structures are not available yet, we used in silico tools to recreate the human OLFM2 full-chain molecular model, combining homology modeling and ab initio protein structure predictions. Then, we set out to assess structural variations likely related to the rs2303100 missense polymorphism. To this end, prediction of solvent accessibility was obtained by introducing the corresponding FASTA sequence of OLFM2-wild type (WT) NM_058164 and OLFM2-R106Q (rs2303100) into I-TASSER108. Next, tertiary structures were analyzed by ROBETTA109. DeepView/Swiss-PDB Viewer 4.1.0 software110 was used to perform three-dimensional alignments for OLFM2-R106 (ancestral) and OLFM2-Q106 (polymorphic protein), taking the former as the reference isoform. The quality of alignments (1.82 Å) was evaluated according to Root Mean Squared (RMS) value (structures that are related show an RMS value of 0–2 Å110). N and C-terminal-side chains were colored per atom, as follows: Carbon-white, Oxygen-red, Nitrogen-dark Blue, Sulfur-yellow, and Hydrogen-light Blue (Fig. S16b). Spatial conservation of OLFM2 regions was shown by coloring the structure with the Alignment Diversity option (Fig. S16c, d). Hydrophobicity analysis and active domain prediction of PROSITE111 was run as well. All captures were rendered with POV-Ray v3.7 software to maximize the quality of the images. SuSPect (https://www.sbg.bio.ic.ac.uk/suspect/), which uses sequence, structure and systems biology-based features to predict the phenotypic effects of missense mutations60, was queried to investigate whether OLFM2-R106Q variant may be considered disease-causing or neutral (Fig. S16e).
Statistical analyses
For cell studies, data is expressed as mean ± standard error of means (S.E.M.). For human study analyses, results are presented in bean plots, and provided in Supplementary Tables as mean ± standard deviation. Statistical significance between groups was determined using two-tailed paired and Fisher’s exact t-test (2 groups), or one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference post-hoc test when comparing multiple groups (3 and above). Spearman’s correlation analysis was performed between OM and SC OLFM2 and other quantitative variables, and by stepwise multiple linear regression analysis after adjusting for potential confounders, when necessary. Statistical analyses were performed with the SPSS statistical software (IBM Analytics Inc., Chicago, IL), GraphPad Prism Version 5.0 (GraphPad Software Inc., San Diego, CA), and R Statistical Software (http://www.r-project.org/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Complete gene expression profiles have been deposited following MIAME compliant guidelines in the community-endorsed repository Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/)112, with accession numbers GSE182229 and GSE182231 for experiments performed in adipocytes (sh-OLFM2 and si-OLFM2, respectively), and GSE214900 for mice adipose tissues. Original data was also downloaded from GEO to retrieve public available transcriptomes of human adipose tissue upon weight loss (GSE53378 and GSE199063)12,13, treatments of LPS (GSE39117)14, ex vivo isolated fat cells (GSE135776)20, and adipocyte cultures (GSE77532 and GSE14312)21,22 were reanalyzed to evaluate the expression of transcripts coding for Olfactomedins 1–4. The array data of GSE53378 was comprised of 32 human SC adipose tissue samples from women with obesity before and ~2 years (1:1) after bariatric surgery12. GSE199063 included SC adipose tissue biopsies obtained before (n = 50) Roux-en-Y gastric bypass (RYGB), and 2 (n = 49) and 5 (n = 38) years thereafter, and also samples provided by 28 age-matched nonoperated women13. In GSE39117, 3 ng/kg of LPS were administrated to evoke endotoxemia in 14 healthy volunteers monitored during 24 h14. In these studies, samples of SC abdominal adipose tissue were obtained from the same region each time (lateral to the umbilicus), under local anesthesia (5% lidocaine), by needle aspiration. By default, the adjusted P values were applied to correct the occurrence of false positive results using the Benjamini and Hochberg FDR method. With regard to proteomics, the full list of mass spectrometry settings can be found in the “report.log.txt” uploaded along with the mass spectrometric raw data to the ProteomeXchange Consortium (https://www.proteomexchange.org) with dataset identifier: PXD065388, via the MassIVE partner repository (https://massive.ucsd.edu/, MassIVE ID: MSV000098300; https://doi.org/10.25345/C5Z31P19P). All data supporting the findings described in this manuscript are available in the article and in the Supplementary Information and from the corresponding author upon request. A Source data file is provided with this paper and available online.
References
Anholt, R. R. H. Olfactomedin proteins: central players in development and disease. Front. Cell Dev. Biol. 2, 6 (2014).
Stankiewicz, T. R., Gray, J. J., Winter, A. N. & Linseman, D. A. C-terminal binding proteins: central players in development and disease. Biomol. Concepts 5, 489–511 (2014).
Li, Q., Liu, A., Gu, X. & Su, Z. Olfactomedin domain-containing proteins: evolution, functional divergence, expression patterns and damaging SNPs. Mol. Genet. Genom. 294, 875–885 (2019).
Zeng, L. C., Han, Z. G. & Ma, W. J. Elucidation of subfamily segregation and intramolecular coevolution of the olfactomedin-like proteins by comprehensive phylogenetic analysis and gene expression pattern assessment. FEBS Lett. 579, 5443–5453 (2005).
Tomarev, S. I. & Nakaya, N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol. Neurobiol. 40, 122–138 (2009).
Lee, J.-A., Anholt, R. R. H. & Cole, G. J. Olfactomedin-2 mediates development of the anterior central nervous system and head structures in zebrafish. Mech. Dev. 125, 167–181 (2008).
Funayama, T. et al. SNPs and interaction analyses of noelin 2, myocilin, and optineurin genes in Japanese patients with open-angle glaucoma. Investig. Ophthalmol. Vis. Sci. 47, 5368–5375 (2006).
Shi, N., Guo, X. & Chen, S.-Y. Olfactomedin 2, a novel regulator for transforming growth factor-β-induced smooth muscle differentiation of human embryonic stem cell-derived mesenchymal cells. Mol. Biol. Cell 25, 4106–4114 (2014).
Shi, N., Li, C.-X., Cui, X.-B., Tomarev, S. I. & Chen, S.-Y. Olfactomedin 2 regulates smooth muscle phenotypic modulation and vascular remodeling through mediating runt-related transcription factor 2 binding to serum response factor. Arterioscler. Thromb. Vasc. Biol. 37, 446–454 (2017).
González-García, I. et al. Olfactomedin 2 deficiency protects against diet-induced obesity. Metabolism 129, 155122 (2022).
Sultana, A. et al. Deletion of olfactomedin 2 induces changes in the AMPA receptor complex and impairs visual, olfactory, and motor functions in mice. Exp. Neurol. 261, 802–811 (2014).
Ortega, F. J. et al. Surgery-induced weight loss is associated with the downregulation of genes targeted by MicroRNAs in adipose tissue. J. Clin. Endocrinol. Metab. 100, E1467–E1476 (2015).
Kerr, A. G., Andersson, D. P., Rydén, M., Arner, P. & Dahlman, I. Long-term changes in adipose tissue gene expression following bariatric surgery. J. Intern. Med. 288, 219–233 (2020).
Shah, R. et al. Gene profiling of human adipose tissue during evoked inflammation in vivo. Diabetes 58, 2211–2219 (2009).
Morrison, D. C. & Ulevitch, R. J. The effects of bacterial endotoxins on host mediation systems. A review. Am. J. Pathol. 93, 526–618 (1978).
Hersoug, L.-G., Møller, P. & Loft, S. Gut microbiota-derived lipopolysaccharide uptake and trafficking to adipose tissue: implications for inflammation and obesity. Obes. Rev.17, 297–312 (2016).
Cani, P. D. et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772 (2007).
Ferguson, J. F. et al. Integrative genomics identifies 7p11.2 as a novel locus for fever and clinical stress response in humans. Hum. Mol. Genet. 24, 1801–1812 (2015).
Moreno-Navarrete, J. M. et al. A role for adipocyte-derived lipopolysaccharide-binding protein in inflammation- and obesity-associated adipose tissue dysfunction. Diabetologia 56, 2524–2537 (2013).
Vijay, J. et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab. 2, 97–109 (2020).
Ambele, M. A., Dessels, C., Durandt, C. & Pepper, M. S. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 16, 725–734 (2016).
O’Hara, A., Lim, F.-L., Mazzatti, D. J. & Trayhurn, P. Microarray analysis identifies matrix metalloproteinases (MMPs) as key genes whose expression is up-regulated in human adipocytes by macrophage-conditioned medium. Pflug. Arch. 458, 1103–1114 (2009).
Reggio, S. et al. Increased basement membrane components in adipose tissue during obesity: links with TGFβ and metabolic phenotypes. J. Clin. Endocrinol. Metab. 101, 2578–2587 (2016).
Zhang, Q. et al. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genom. Proteom. Bioinform. 18, 120–128 (2020).
Norreen-Thorsen, M. et al. A human adipose tissue cell-type transcriptome atlas. Cell Rep. 40, 111046 (2022).
Saltiel, A. R. & Olefsky, J. M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 127, 1–4 (2017).
Fryklund, C., Morén, B., Neuhaus, M., Periwal, V. & Stenkula, K. G. Rosiglitazone treatment enhances intracellular actin dynamics and glucose transport in hypertrophic adipocytes. Life Sci. 299, 120537 (2022).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Sonnenberg, G. E., Krakower, G. R. & Kissebah, A. H. A novel pathway to the manifestations of metabolic syndrome. Obes. Res. 12, 180–186 (2004).
Tabata, M. et al. Angiopoietin-like protein 2 promotes chronic adipose tissue inflammation and obesity-related systemic insulin resistance. Cell Metab. 10, 178–188 (2009).
Pfeiffer, J. R., McAvoy, B. L., Fecteau, R. E., Deleault, K. M. & Brooks, S. A. CARHSP1 is required for effective tumor necrosis factor alpha mRNA stabilization and localizes to processing bodies and exosomes. Mol. Cell. Biol. 31, 277–286 (2011).
Markovics, J. A. et al. Interleukin-1beta induces increased transcriptional activation of the transforming growth factor-beta-activating integrin subunit beta8 through altering chromatin architecture. J. Biol. Chem. 286, 36864–36874 (2011).
Kitamura, H. et al. Mouse and human lung fibroblasts regulate dendritic cell trafficking, airway inflammation, and fibrosis through integrin αvβ8-mediated activation of TGF-β. J. Clin. Investig. 121, 2863–2875 (2011).
Li, Q. et al. Obesity and hyperinsulinemia drive adipocytes to activate a cell cycle program and senesce. Nat. Med. 27, 1941–1953 (2021).
Garrison, J. L. & Knight, Z. A. Linking smell to metabolism and aging. Science 358, 718–719 (2017).
Jovanovic, P. & Riera, C. E. Olfactory system and energy metabolism: a two-way street. Trends Endocrinol. Metab. 33, 281–291 (2022).
López, M., Fernández-Real, J. M. & Tomarev, S. I. Obesity wars: may the smell be with you. Am. J. Physiol. Endocrinol. Metab. 324, E569–E576 (2023).
Wang, Y. et al. The role of somatosensory innervation of adipose tissues. Nature 609, 569–574 (2022).
Guilherme, A., Henriques, F., Bedard, A. H. & Czech, M. P. Molecular pathways linking adipose innervation to insulin action in obesity and diabetes mellitus. Nat. Rev. Endocrinol. 15, 207–225 (2019).
Seoane-Collazo, P. et al. Central nicotine induces browning through hypothalamic κ opioid receptor. Nat. Commun. 10, 4037 (2019).
Lin, Z., Chi, J. & Cohen, P. A clearing method for three-dimensional imaging of adipose tissue. Methods Mol. Biol. 2448, 73–82 (2022).
Tsukiyama-Kohara, K., Katsume, A., Kimura, K., Saito, M. & Kohara, M. 4E-BP1 regulates the differentiation of white adipose tissue. Genes Cells 18, 602–607 (2013).
Tsukiyama-Kohara, K. et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7, 1128–1132 (2001).
Latorre, J. et al. Adipose tissue knockdown of lysozyme reduces local inflammation and improves adipogenesis in high-fat diet-fed mice. Pharmacol. Res. 166, 105486 (2021).
Latorre, J. et al. Specific adipose tissue Lbp gene knockdown prevents diet-induced body weight gain, impacting fat accretion-related gene and protein expression. Mol. Ther. Nucleic Acids 27, 870–879 (2022).
Gupta, R. K. et al. Transcriptional control of preadipocyte determination by Zfp423. Nature 464, 619–623 (2010).
Wu, Z. & Wang, S. Role of kruppel-like transcription factors in adipogenesis. Dev. Biol. 373, 235–243 (2013).
Elks, C. E. et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 42, 1077–1085 (2010).
Perry, J. R. et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 514, 92–97 (2014).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Kichaev, G. et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104, 65–75 (2019).
Zhu, Z. et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 145, 537–549 (2020).
Zhu, X., Zhu, L., Wang, H., Cooper, R. S. & Chakravarti, A. Genome-wide pleiotropy analysis identifies novel blood pressure variants and improves its polygenic risk scores. Genet. Epidemiol. 46, 105–121 (2022).
Pietzner, M. et al. Mapping the proteo-genomic convergence of human diseases. Science 374, eabj1541 (2021).
Emilsson, V. et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 361, 769–773 (2018).
Gudjonsson, A. et al. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat. Commun. 13, 480 (2022).
Surapaneni, A. et al. Identification of 969 protein quantitative trait loci in an African American population with kidney disease attributed to hypertension. Kidney Int. 102, 1167–1177 (2022).
Breeze, C. E. et al. FORGEdb: a tool for identifying candidate functional variants and uncovering target genes and mechanisms for complex diseases. Genome Biol. 25, 3 (2024).
Chen, C. T. L. et al. Replication of loci influencing ages at menarche and menopause in Hispanic women: The Women’s Health initiative SHARe study. Hum. Mol. Genet. 21, 1419–1432 (2012).
Yates, C. M., Filippis, I., Kelley, L. A. & Sternberg, M. J. E. SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J. Mol. Biol. 426, 2692–2701 (2014).
Yokoe, H. & Anholt, R. R. Molecular cloning of olfactomedin, an extracellular matrix protein specific to olfactory neuroepithelium. Proc. Natl. Acad. Sci. USA 90, 4655–4659 (1993).
Snyder, D. A., Rivers, A. M., Yokoe, H., Menco, B. P. & Anholt, R. R. Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry 30, 9143–9153 (1991).
Sakers, A., De Siqueira, M. K., Seale, P. & Villanueva, C. J. Adipose-tissue plasticity in health and disease. Cell 185, 419–446 (2022).
de sá, P. M., Richard, A. J., Hang, H. & Stephens, J. M. Transcriptional regulation of adipogenesis. Compr. Physiol. 7, 635–674 (2017).
Tontonoz, P. & Spiegelman, B. M. Fat and beyond: the diverse biology of PPARgamma. Annu. Rev. Biochem. 77, 289–312 (2008).
Bahrami-Nejad, Z. et al. A transcriptional circuit filters oscillating circadian hormonal inputs to regulate fat cell differentiation. Cell Metab. 27, 854–868.e8 (2018).
Pan, D. Z. et al. Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS. Nat. Commun. 9, 1512 (2018).
Lefterova, M. I. et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 22, 2941–2952 (2008).
Reusch, J. E. B., Colton, L. A. & Klemm, D. J. CREB activation induces adipogenesis in 3T3-L1 cells. Mol. Cell. Biol. 20, 1008–1020 (2000).
Rosenbaum, S. E. & Greenberg, A. S. The short- and long-term effects of tumor necrosis factor-α and BRL 49653 on peroxisome proliferator-activated receptor (PPAR)γ2 gene expression and other adipocyte genes. Mol. Endocrinol. 12, 1150–1160 (1998).
Zhang, B. et al. Negative regulation of peroxisome proliferator-activated receptor-gamma gene expression contributes to the antiadipogenic effects of tumor necrosis factor-alpha. Mol. Endocrinol. 10, 1457–1466 (1996).
Keuper, M. et al. TRAIL (TNF-related apoptosis-inducing ligand) regulates adipocyte metabolism by caspase-mediated cleavage of PPARgamma. Cell Death Dis. 4, e474 (2013).
Gustafson, B., Hammarstedt, A., Andersson, C. X. & Smith, U. Inflamed adipose tissue: a culprit underlying the metabolic syndrome and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 27, 2276–2283 (2007).
Emont, M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933 (2022).
Barilla, S., Treuter, E. & Venteclef, N. Transcriptional and epigenetic control of adipocyte remodeling during obesity. Obesity 29, 2013–2025 (2021).
Bäckdahl, J. et al. Spatial mapping reveals human adipocyte subpopulations with distinct sensitivities to insulin. Cell Metab. 33, 1869–1882.e6 (2021).
Aguilar, V. & Fajas, L. Cycling through metabolism. EMBO Mol. Med. 2, 338–348 (2010).
Huber, K., Mestres-Arenas, A., Fajas, L. & Leal-Esteban, L. C. The multifaceted role of cell cycle regulators in the coordination of growth and metabolism. FEBS J. 288, 3813–3833 (2021).
Lopez-Mejia, I. C., Castillo-Armengol, J., Lagarrigue, S. & Fajas, L. Role of cell cycle regulators in adipose tissue and whole body energy homeostasis. Cell. Mol. Life Sci. 75, 975–987 (2018).
Liu, R. H. et al. Depletion of OLFM4 gene inhibits cell growth and increases sensitization to hydrogen peroxide and tumor necrosis factor-alpha induced-apoptosis in gastric cancer cells. J. Biomed. Sci. 19, 38 (2012).
Kobayashi, D., Koshida, S., Moriai, R., Tsuji, N. & Watanabe, N. Olfactomedin 4 promotes S-phase transition in proliferation of pancreatic cancer cells. Cancer Sci. 98, 334–340 (2007).
Park, K. S. et al. Olfactomedin 4 suppresses tumor growth and metastasis of mouse melanoma cells through downregulation of integrin and MMP genes. Mol. Cells 34, 555–561 (2012).
Klaas, M. et al. Olfactomedin-4 improves cutaneous wound healing by promoting skin cell proliferation and migration through POU5F1/OCT4 and ESR1 signalling cascades. Cell. Mol. Life Sci. 79, 157 (2022).
Cárdenas-León, C. G. et al. Olfactomedin 4 regulates migration and proliferation of immortalized non-transformed keratinocytes through modulation of the cell cycle machinery and actin cytoskeleton remodelling. Exp. Cell Res. 415, 113111 (2022).
Gao, X. et al. GeneChip expression profiling identified OLFML2A as a potential therapeutic target in TNBC cells. Ann. Transl. Med. 10, 274–274 (2022).
Short, C. A., Suarez-Zayas, E. A. & Gomez, T. M. Cell adhesion and invasion mechanisms that guide developing axons. Curr. Opin. Neurobiol. 39, 77–85 (2016).
Romer, L. H., Birukov, K. G. & Garcia, J. G. N. Focal adhesions: paradigm for a signaling nexus. Circ. Res. 98, 606–616 (2006).
Robles, E. & Gomez, T. M. Focal adhesion kinase signaling at sites of integrin-mediated adhesion controls axon pathfinding. Nat. Neurosci. 9, 1274–1283 (2006).
Lluch, A. et al. Impaired Plakophilin-2 in obesity breaks cell cycle dynamics to breed adipocyte senescence. Nat. Commun. 14, 5106 (2023).
Li, Y. & Burridge, K. Cell-cycle-dependent regulation of cell adhesions: adhering to the Schedule: Three papers reveal unexpected properties of adhesion structures as cells progress through the cell cycle. Bioessays 41, e1800165 (2019).
Jones, M. C., Zha, J. & Humphries, M. J. Connections between the cell cycle, cell adhesion and the cytoskeleton. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 374, 20180227 (2019).
Minokoshi, Y. et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428, 569–574 (2004).
Milbank, E. et al. Small extracellular vesicle-mediated targeting of hypothalamic AMPKα1 corrects obesity through BAT activation. Nat. Metab. 3, 1415–1431 (2021).
Ferreira, V. et al. Modulation of hypothalamic AMPK phosphorylation by olanzapine controls energy balance and body weight. Metabolism 137, 155335 (2022).
Seoane-Collazo, P. et al. SF1-specific AMPKα1 deletion protects against diet-induced obesity. Diabetes 67, 2213–2226 (2018).
Hardie, D. G., Ross, F. A. & Hawley, S. A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262 (2012).
Göransson, O., Kopietz, F. & Rider, M. H. Metabolic control by AMPK in white adipose tissue. Trends Endocrinol. Metab. 34, 704–717 (2023).
Bunnell, B. A., Flaat, M., Gagliardi, C., Patel, B. & Ripoll, C. Adipose-derived stem cells: isolation, expansion and differentiation. Methods 45, 115–120 (2008).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, baz046 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 13, 2513–2526 (2014).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Pang, Z. et al. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Yang, J. & Zhang, Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015).
Kim, D. E., Chivian, D. & Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004).
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Sigrist, C. J. A. et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161–D166 (2010).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Li, D. et al. WashU epigenome browser update 2022. Nucleic Acids Res. 50, W774–W781 (2022).
Acknowledgements
This study has been funded by the Instituto de Salud Carlos III (ISCIII), through the projects CP19/00109, PI18/00550, and PI21/00074 (to F.J.O.), the Ministerio de Ciencia e Innovación (PID2022-141115NA-I00 to I.G.-G.; PID2022-142951OB-I00 to S.M.; PID2021-128145NB-I00 and PDC2022-133958-I00, to M.L.), and the Xunta de Galicia (ED431C 2024/10 and ED431F 2024/14, to I.G.-G.), also co-founded by the European Union and the Fondo Europeo de Desarrollo Regional (FEDER). F.J.O. (MS19/00109) is recipient of the Miguel Servet scheme (ISCIII), A.L. (FI19/00045) is recipient of a Contrato Predoctoral de Formación en Investigación en Salud (PFIS, ISCIII), I.G.-G. is supported by a Ramón y Cajal award (RYC2021-031225-I), and N.F.M. is recipient of a Contrato Predoctoral para la Formación de Doctores (PREP2022-000775), Ministerio de Ciencia, Innovación y Universidades, Gobierno de España (ES). N.N. and S.I.T. were supported by the Intramural Research Programs of the National Eye Institute, NIH. The authors thank the CERCA and PERIS programmes (Generalitat de Catalunya) for financial and institutional support, and are indebted to the participants of the FATBANK platform, promoted by the Centro de Investigación Biomédica en Red de la Fisiopatología de la Obesidad y la Nutrición (CIBEROBN, ISCIII) and the Institut d’Investigació Biomèdica de Girona (IDIBGI) Biobank (Biobanc IDIBGI, B.0000872), integrated in the Spanish National Biobanks Network, for their collaboration and coordination. We would like to thank the Animal Facility of the Barcelona Science Park and its staff for the technical assistance provided in carrying out the tasks related to the work with animals. The authors are also indebted to the IMIM Microarray Analysis Service (Hospital del Mar Medical Research Institute, Barcelona, Spain) for technical guidance.
Author information
Authors and Affiliations
Contributions
All authors contributed to data interpretation and revised the manuscript. A.L., J.L., N.O-C., J.M.M.-N., E.C.-I., S.M., E.C., and F.J.O. conducted research (hands on conduct of the experiments and data collection). N.N. and S.I.T. provided the Olfm2-null mouse model. N.F.M., I.G.-G., and M.L. performed animal husbandry maintenance, treatment, characterization, and sample collection. G.S. and A.I.D. performed TH-immunostaining. P.G.-G. and J.L.L.-G. obtained HPLC neurotransmitter measures. I.E. and A.M.-M. participated in high-throughput computing and microarray data assessment. B.Z. and Y.Z. ran cell deconvolution analysis. J.P., M.G.-S., W.S., and J.G. conducted untargeted proteomics, quantitative analysis, and data visualization. J.L.-A. and M.M.M. performed conformational protein analyses. J.M.F-R. and F.J.O. coordinated human samples, clinical information, written consents, and intellectual content collection. F.J.O. designed the research, prepared the manuscript and developed the project conception and administration, visualization, supervision, funding acquisition, and lead contact.
Corresponding author
Ethics declarations
Competing interests
The authors have nothing to disclose. All authors have approved the final version submitted, being listed as authors on the manuscript. The contents of this manuscript have not been copyrighted or published previously. The contents of this manuscript are not under consideration for publication elsewhere.
Peer review
Peer review information
Nature Communications thanks Jose Donato Jr, Minsoo Noh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lluch, A., Latorre, J., Espadas, I. et al. Defective Olfactomedin-2 connects adipocyte dysfunction to obesity. Nat Commun 16, 7154 (2025). https://doi.org/10.1038/s41467-025-62430-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62430-5