Abstract
The folding nucleus (FN) initiates and enables an efficient protein folding pathway. Despite its essential role, the FN has long remained cryptic. Here we directly visualize the tubulin FN consisting of a nonnative, partially assembled Rossmann fold, in the closed chamber of human chaperonin TRiC. Chaperonin TRiC interacts with nonnatively folded secondary structure elements of tubulin, stabilizing the nucleus poised for transition into its first native domain tertiary structure. Through progressive folding into the native state, we observe that the unfolded sequence of tubulin undergoes drastic spatial rearrangement in the TRiC chamber to sample the conformational space, mediated by the highly dynamic CCT tails. The observed presence of individual nonnative secondary structure elements first in the nonnative FN and then around the incrementally folded native domains supports the hypothesis that tubulin folding in TRiC is a hierarchical process of nucleation, condensation and propagation in cooperation with TRiC subunits.
Similar content being viewed by others
Introduction
A newly synthesized polypeptide needs to fold into its native conformation to function properly. Despite the seemingly astronomical number of possible conformations a protein can theoretically explore, the Levinthal Paradox and Kinetic Partitioning suggested that proteins sample a subset of fast, directed folding pathways1,2. The Energy Landscape Theory further proposed that proteins explore various conformations while descending a funnel-shaped energy landscape, eventually reaching a thermodynamically stable, low-energy native state3,4,5. Both theories accommodate the diversity of folding pathways but neither clearly define them.
Several models have been proposed to explain the detailed mechanisms of protein folding. The Framework Model emphasizes the early formation of local native secondary structures, followed by their diffusion and collision to produce the correct tertiary structure6,7. In contrast, the Hydrophobic Collapse Model proposes a rapid, nonspecific collapse of hydrophobic residues into a disordered core, forming a molten globule, followed by structural rearrangement into the native state8,9,10,11. The Nucleation-Condensation Model posits that an extended folding nucleus with weak secondary and tertiary interactions forms first. Around this nucleus, a significant portion of the protein structure condenses into an approximately correct conformation, reaching a transition state before quickly forming the final structure12. Alternatively, the Nucleation-Propagation Model suggests that folding starts with the formation of a nucleus consisting of a small number of nascent secondary structures. This nucleus then propagates, extending to adjacent sequences until the entire protein adopts its native tertiary structure13. Each of these models offers a perspective on the folding process, providing insights into how specific proteins achieve their functional conformations.
While some proteins fold spontaneously, many require the assistance of chaperones to reach their native state. The human chaperonin TRiC (also known as CCT), a two-ring stacked complex with eight paralogous subunits in each ring, uses ATP to fold approximately 10% of the proteome14, including the cytoskeletal protein tubulin15. As tubulin emerges from the ribosome in an unfolded state, it is initially captured by the chaperone prefoldin16 and subsequently transferred to TRiC for proper folding17. Upon ATP binding and hydrolysis, TRiC transitions from an open to a closed conformation, encapsulating tubulin within its chamber for folding. In our previous study, we identified three progressive folding intermediates and one final native state of tubulin within the closed TRiC chamber. Electrostatic interactions between the TRiC interior wall and the folded tubulin domains demonstrate that TRiC actively guides tubulin folding as a scaffold, rather than serving merely as a passive Anfinsen cage17. In that study, we defined four non-linear progressively folded domains in tubulin: the N domain (comprising 84% of tubulin’s Rossman fold), the C domain, the core helix domain, and the middle domain (Fig. S1)17. Despite this, the tubulin folding nucleus that emerges early and seeds tubulin’s correct folding into its functional state has remained cryptic. In addition, throughout the tubulin folding pathway, the mechanism whereby unfolded tubulin sequence transitions into the nascent structure elements remains unknown.
Here we combine methods of conventional reconstruction with reference-free deep learning to reveal tubulin folding states from cryo-EM images of the TRiC/tubulin complex in its closed state, including a subset of particle images previously dismissed as “junk.” Using deep learning to analyze cryo-EM images in high-dimensional space, we visualize the particle-to-particle heterogeneity in tubulin folding by TRiC. This analysis led to the identification of nonnative tubulin folding nucleus in TRiC and allowed us to visualize the dynamic transition in folding individual secondary structure elements within the folding nucleus and on the natively folded domains of folding intermediates. Our findings suggest that tubulin folding within TRiC follows a hierarchical process of nucleation, condensation, and propagation, combining elements from several protein folding models. This work provides a dynamic view of tubulin folding, with potential implications for drug discovery targeting tubulin folding, particularly in cancer therapy.
Results
The compositional heterogeneity in tubulin encapsulation by closed TRiC chamber
The human chaperonin TRiC closes its rings upon ATP binding and hydrolysis to assist with substrate protein folding. However, exactly how the two rings function in a coordinated manner is not well understood. Whether the two rings function independently and/or are simultaneously active in folding remains unknown. To understand how TRiC coordinates its two rings for tubulin encapsulation and folding, we investigate the TRiC binding valency of tubulin.
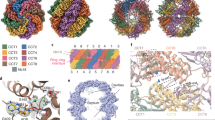
In our previous study, we induced the ring closure of purified TRiC/tubulin binary complex under ATP/AlFx condition. Focused analysis of the cryoEM closed state particle images revealed TRiC rings in the apo state (i.e. no tubulin) and rings occupied by tubulin in various folding states, including three intermediate states (I, II, III) and one native state (IV) (Figs. 1A and S2)17. Building on that analysis, we trace each ring back to the original 529,181 TRiC particle images to determine the compositional heterogeneity within each double-ring TRiC complex (Fig. 1B, C). We find many TRiC molecules encapsulate either fully or partially folded tubulin in only one ring, while the opposite ring remains in the apo state (Fig. 1C, diamond symbols). However, a subset of TRiC molecules encapsulates tubulin in both rings, suggesting that both rings are folding-active and can fold tubulin simultaneously (Fig. 1C). The presence of identical or different tubulin folding states (I through IV) across the two rings illustrates both synchronized and non-synchronized folding in TRiC’s rings (examples shown in Fig. 1D). This observation suggests the possibility of simultaneous encapsulation, but non-synchronized folding across the rings, or chamber reopening during the folding of the first tubulin to accommodate a second tubulin.
Color coding for A–E: TRiC in dark gray; undefined density in orange; N domain in cyan; C domain in green; Core helix domain in blue; Middle domain in red. A 529,181 TRiC particles display six types of compositional heterogeneity within their rings, including the apo state, occupied by undefined density, tubulin folding states I, II, III, IV. B The diagram shows that each of the two rings in a TRiC can possibly be one of six scenarios in (A). C The histogram shows the statistical distribution of TRiC particles with compositional heterogeneity from both rings. * The particle set for Fig. 2. Δ The particle set for Fig. 3. ◊ The particle set for Fig. 4. Color coding: Apo TRiC ring in gray; rings with undefined density in orange, state I in cyan, state II in green, state III in blue and state IV in red, respectively. Source data are provided as a Source Data file. D Representative reconstruction of TRiC with synchronous folding between two rings (left) and representative reconstruction of TRiC with the nonsynchronous folding between two rings (right). E The reconstructions of TRiC when one ring is occupied by undefined density and the opposite ring is either in the apo state or occupied by a tubulin folding intermediate I.
In addition to TRiC rings in the apo state or encapsulating one of the four tubulin folding states, a significant portion—45%—of TRiC rings display an undefined density, lacking resolvable secondary structures in their average (Fig. 1A). The opposite ring in this population of TRiC can either be in the apo state or be occupied by one of the four tubulin folding states (Fig. 1C, E). We will characterize this undefined density further in Fig. 3.
The spatial occupancy and dynamics of CCT tails in apo TRiC
The CCT tails at the inter-ring interface interact with tubulin during its loading and folding process. Here we reconstruct the apo state TRiC to 3.7 Å resolution from 40,464 both-rings-apo (Apo/Apo) TRiC particle images (Figs. 2A and S3A). Due to their structural flexibility, many CCT tails are only partially resolvable in the average reconstruction (Fig. S3B, S3D, S3E). To gain insight into the locations of CCT tails in TRiC, we build the full sequence models of the CCT tails based on the average reconstruction, using two constraints (see Methods): 1) extend each CCT tail in the direction of its resolved sequence; 2) avoid overlapping with the resolved TRiC density. The density is resolved for the majority of the CCT N tails but less so for C tails (Tables S1 and S2), thus the structural models for N tails are built with more confidence than those for C tails. From these models, we find that CCT N tails either extend towards the opposite ring (CCTs 1,2,6,7,8) or project out of TRiC through the inter-ring interface (CCTs 4,5) (Figs. 2B and S3C). In contrast, CCT3’s N tail and all CCT C tails (i.e., CCTs 1,2,3,5,6,7,8; CCT 4 does not have a C tail) likely extend toward the center of the chamber, possibly engaging in intra-ring interactions among these tails (Fig. 2B).
A The sliced side view of apo TRiC reconstruction in dark gray color without B factor sharpening. B The complete modeling of CCT tails. The left figure shows the CCT tails in a single TRiC ring of the bottom view, with the location of the first N tail residue (orange circle) and the last C tail residue (purple diamond). The resolved CCT tails are highlighted in thicker lines. The right figure shows the transparent side view of TRiC with CCT C tails in orange and N tails in green. C The workflow of CryoDRGN training and analysis of heterogeneity in apo TRiC particles. D The sliced side view of TRiC volume reconstructed from latent space embedding shows the CCT tails density in orange color indicating intra-ring termini interaction. E The sliced side view of TRiC volume reconstructed from latent space embedding shows the CCT tails density at the inter-ring interface, indicating inter-ring tails interaction. F The single TRiC ring model from the bottom view displaying the electrostatic distribution on the TRiC chamber wall. Blue color indicates positively charged distribution while red color indicates negatively charged distribution. G The single TRiC ring model from the bottom view displaying the electrostatic distribution on the CCT tails. Blue color indicates positively charged distribution while red color indicates negatively charged distribution. H The segregation of CCT tails into a positively charged cluster and a negatively charged cluster across the two rings of TRiC. I The schematic shows positively charged C tails (CCTs 3,6,8,7) adjacent to positively charged wall (CCTs 1,3,6,8), and negatively charged C tails (CCTs 5,2,4,1) adjacent to negatively charged wall (CCTs 7,5,2,4).
To validate the CCT tail modeling, we extracted the particle stack of apo TRiC and investigated the spatial occupancy of CCT tails using cryoDRGN, a deep-learning method for heterogeneity analysis18 (Fig. 2C). K-means clustering and volume reconstruction from the latent space embedding revealed additional density at the inter-ring interface of apo TRiC (Fig. 2D, E and Supplementary Movie 1), in contrast to the conventional reconstruction (Fig. 2A). The extra density observed at the positions of the modeled CCT C tails in many volume reconstructions supports the overall modeling of CCT C tails and their intra-ring contacts (Supplementary Movie 1 and Fig. 2B, D). Additionally, density observed across rings at the inter-ring interface in many other TRiC volume reconstructions indicates that inter-ring tails interactions occur in these apo TRiC molecules (Fig. 2E, Supplementary Movie 1). The long, unresolvable C tails of CCTs 1, 3, 7, 8 may cross the ring to participate in these inter-ring tails interactions (Fig. 2I and Table S2).
The closed TRiC chamber comprises a positively charged half-hemisphere formed by CCTs 1, 3, 6, and 8, and a negatively charged half-hemisphere formed by CCTs 7, 5, 2, and 4 (Fig. 2F). We observe a similar charge distribution pattern in the CCT C tails as seen on the chamber wall (Fig. 2G). The C tails from neighboring CCTs 3, 6, 8, and 7 form a positively charged cluster near the positive wall hemisphere, while C tails from neighboring CCTs 5, 2, 4, and 1 form a negatively charged cluster near the negative wall hemisphere (Fig. 2F–I). This electrostatic distribution of CCT tails may play a crucial role in orienting unfolded tubulin by attracting the negatively charged sequence in its N and C domains to the positively charged hemisphere of CCTs 1, 3, 6, and 8 during tubulin recruitment and relocation to the TRiC chamber. In summary, both the modeling and cryoDRGN analysis suggest interactions of C tails both within and between rings in TRiC.
Tubulin folding nucleus in closed TRiC chamber
The three progressive folding intermediates of tubulin (States I, II, III), along with the fully folded state (IV) (Fig. S1), are situated near or within the native state on the energy landscape17. The polypeptide chain of tubulin, synthesized by the ribosome, collapses into a compact molten globule driven by hydrophobic burial. However, the mechanism by which tubulin folding initiates from the molten globule remains unknown.
To understand the molten globule state of tubulin before amino acid sequence 1-170 (84% of tubulin’s Rossman fold) is folded in State I, we examined TRiC particles where one ring was occupied by undefined density while the opposite ring was in the apo state, using cryoDRGN. Compositional heterogeneity analysis and K-means clustering (Fig. S4A) distinguishes TRiC particles with undefined high density from those with undefined low density (Fig. S4B). We then reconstruct a 3D map by RELION19 from the pooled particle images with undefined high density in the TRiC chamber (Figs. 3A and S4C, S4D). After fitting the native state tubulin model (State IV, PDB: 7TUB) to this map (Fig. 3B), we could attribute the density at the CCTs 1,4 pocket as the tubulin E-hook tail, and the undefined density to the full-length tubulin (Fig. 3B, C).
A The sliced side view of TRiC (in gray color) with one ring encapsulating tubulin density (in orange color) and the opposite ring in the apo state. B The tubulin density segmented from the map in A is displayed (in orange color) in the TRiC chamber of CCTs 1, 3, 6, 8, 7. C The native tubulin model (7TUB) is fit into the tubulin density in orange color, highlighting the tubulin E hook density at positively charged CCTs 1/4 pocket. Surface representations of TRiC subunits are colored by their electrostatic potential. D Top right: Three layers (segregated by two diagonal dotted lines) of bulky density in State 0.5 (in orange color) is the typical Rossman fold with a β sheet (lower left panel) sandwiched by two layers of alpha helices (upper left and lower right panel). Note: The native tubulin model is used only to illustrate the three layers of Rossman fold; registration is uncertain for the individual loosely formed β-strands. The view angle for each of three layers is as illustrated. E The topology diagram of tubulin highlighting the loosely folded secondary structure elements (highlighted in dark cyan) of State 0.5. The CCT residues participating in the salt bridge and hydrogen bond interactions with native tubulin are colored as the CCTs. The individual domains of tubulin are colored as in Fig. 1A. Background adapted from Cell 185, 4770–4787.e20 (2022), © Elsevier, used with permission. F i, ii, iii show the density in orange color of individual nonnative structure elements (in cyan color of N domain) that engage with CCT subunits and iv shows the absence of density for T3 loop in State 0.5, compared to that in State I (model in cornflower blue). The view angle for each of the four panels are illustrated in (G ii). G i shows the loosely assembled sequence of the folding nucleus (in orange) as part of the N domain (in cyan). ii shows the sites of the folding nucleus amino acid sequence (diamond) that engage with TRiC. iii shows the sequence of tubulin within the folding nucleus (in orange).
Using the native state tubulin model as a reference, we determined that the N domain of tubulin (i.e. residues 1–170) is only partially folded in this reconstruction, hence we designate this tubulin state as State 0.5. In State 0.5, the sequence corresponding to β strands S1/S4/S5 and helices H1/H3(C half)/H4, are loosely folded, representing about half of the six-stranded Rossmann fold (321456) in tubulin (Fig. 3D, E, G). Their diffuse density in contrast to that of the natively folded protein suggests that these secondary structure elements are in a nonnative state with irregular folding.
Although in a nonnative state, helix H4, with full density coverage, can be seen to engage with subunit CCT3, while helix H3, partially included in density at its C end, contacts subunit CCT6 (Fig. 3F). While it is unclear whether the assembly of the continuous sequence H3(C half)-S4-H4-S5 occurs before these interactions with TRiC, such interactions could help stabilize the engaged sequences and facilitate the transition from the nonnative state to fully native structures. The density ascribed to helix H1 is visibly displaced compared to its native state (Fig. 3D). However, the short β-strand S11 of a β hairpin insertion immediately following helix H1, is included in density at its native position while it interacts with subunit CCT8 as in State I (Fig. 3F). The stabilization oef β strand S11 on the TRiC wall could facilitate the formation of the hairpin and likely brings the β strand S2 sequence into proximity with the β strand S1, enabling its annexation. Compared to State I, where the T3 loop is fixed on the TRiC wall17, the absence of the T3 loop density in State 0.5, along with its end motifs–the N half of helix H3 and β strand S3–suggests that the attachment of the T3 loop to the TRiC wall may facilitate the folding of β strand S3. This attachment could help bring the sequence of β strand S3 close to β strand S2, promoting their interaction and annexation (Fig. 3E).
In State 0.5, β strands S1/S4/S5 and helices H1/H3(C half)/H4, although in their nonnative states, assemble into a loose tertiary structure that resembles the native state of tubulin (Fig. 3D). This intermediate state supports the nucleation and condensation model of protein folding, with concurrent buildup of nonnative secondary and tertiary structural contacts. Consolidation into their native structures, like state I, may require the condensation of additional secondary structure elements in the N domain (Fig. 3G). The nucleation from several secondary structure elements (H3/H4/S11) contacting the TRiC wall suggests that TRiC actively guides tubulin folding from State 0.5 toward State I (Fig. 3F).
The propagation dynamics in tubulin folding within TRiC examined in latent space
Understanding the spatial arrangement of the unfolded sequence is crucial for comprehending how a polypeptide transitions from a disordered to a progressively ordered structure and for characterizing the kinetic barriers in the protein folding process. The three tubulin intermediate states (States I, II, III) indicate a domain-wise folding pathway, where discontinuous sequence elements of the tubulin polypeptide fold progressively (Fig. S1). However, the precise mechanism by which sequence of each unfolded domain transitions into its native state during the folding process remains unclear.
To gain insight into the dynamics of the unfolded sequence, we investigate their spatial arrangement in each intermediate state. We pool particle images of TRiC with one ring occupied by each intermediate state while the opposite ring remains in the apo state (State x/Apo). We then reconstruct a 3D map of TRiC encapsulating each tubulin intermediate from these pooled particle images (Figs. 4A and S5). Additionally, we embed the pooled images of TRiC encapsulating each state into a separate latent space and reconstruct volumes from the sampled embeddings to examine the conformational heterogeneity in the unfolded sequence for each intermediate state (Supplementary Movies 2, 3, 4, 5, “Methods”).
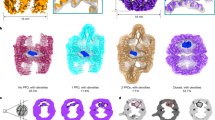
Color coding for Fig. 4: TRiC in dark gray; CCT tails and unfolded tubulin sequence in orange; N domain in cyan; C domain in green; Core helix domain in blue; Middle domain in red. A The reconstructions of TRiC with one ring encapsulating tubulin intermediate State I, II, III or native state IV, while the opposite ring is in the apo state. B Zoom in view of tubulin density in (A). i shows the native position of C domain exhibit only little density in State I; ii shows the helix H7 in Core helix domain display visibly displaced density in State II; iii shows the Middle domain density in state III condenses around native position; iv shows that the fully folded tubulin density in State IV covers all the secondary structures of native state tubulin. The view angle for i and ii are demonstrated in A i and ii, respectively. The view angle for iii and iv are the same as A iii and iv, respectively. C The examples show the density in orange color for tubulin unfolded sequences and the CCT tails in the TRiC chamber from state I to State IV by cryoDRGN analysis. D An example of cryoDRGN volumes indicates the density is displaced from the C domain helices H11 and H12 in State I; E An example of cryoDRGN volumes shows the helix H7 from core helix domain is partially covered by density in State II; E is shown from the same view angle as (C ii). F An example of cryoDRGN volumes indicates the density of unfolded middle domain is displaced from the native position in State III. G The illustration of the view angles for (D) and (F). D is the view from the back of tubulin, and F is the view from front left of tubulin.
In the average reconstruction of State I/Apo, the folded N domain, accounting for 38% of the tubulin sequence, is well-resolved (Figs. 4Ai and S1). However, the average density for the remaining 62% unfolded sequence appears weak due to conformational heterogeneity (Fig. 4Bi). Superimposing the native state tubulin model shows that the C domain, which appears folded in State II, is barely covered by density (Fig. 4Bi), indicating that sequence of this domain infrequently occupies the native state position among the analyzed particles in State I/Apo. Latent space sampling reveals that the unfolded sequence undergoes extensive spatial rearrangement, despite being constrained by the TRiC chamber (Supplementary Movie 2, and Fig. 4Ci). This diverse spatial arrangement enhances the conformational space sampling of the unfolded sequence. The flexible CCT tails from the same or across the chamber perturb the unfolded sequence through direct contacts (Supplementary Movie 2 and Fig. 4Ci), likely driving it out of local energy minima. Examination of the latent space volumes for State I/Apo reveals the absence of resident density for the C domain helices H11 and H12 at their native positions, although the tubulin C-terminal E-hook tail immediately following these helices is observed still attached to the CCT1/4 pocket of TRiC (Supplementary Movie 6 and Fig. 4D). Condensation of the C domain to the approximate native position seems challenging and given the relatively small percentage of State II among the folding intermediates (Fig. 1A), the final folding of the C domain to State II is likely a rate-limiting step in the folding process.
In the average reconstruction of State II/Apo, further folding of the C domain leaves 37% of the tubulin sequence still unfolded (Figs. 4Aii and S1). However, diffuse density is observed at the position of native helix H7 in the tubulin Core helix domain, which appears folded in State III, although it is visibly displaced toward its C end (Fig. 4Bii). Latent space volumes of State II/Apo reveal that the unfolded sequence remains highly dynamic within the TRiC chamber (Supplementary Movie 3 and Fig. 4Cii). Despite the overall extensive movement of the unfolded sequence, persistent density coverage is observed at the N end of helix H7, indicating a nonnative fold in this region (Supplementary Movie 7 and Fig. 4E). In contrast, the C end of helix H7 shows variable density coverage among the volumes (Supplementary Movie 7), indicating the C end is actively condensing on the surface of the folded N/C domain. The folding of helix H7 propagates from the N to the C end, arguing against the framework model which posits pre-established secondary structures. Instead, the folding of the Core helix domain is consistent with the canonical nucleation and propagation model of protein folding, involving the gradual formation and growth of secondary structures around a native nucleus. Compared to the C domain in State I/Apo, helix H7—the major secondary structure element of the Core helix domain—restricts the spatial search to the vicinity of its native state position, suggesting an easier condensation of Core helix domain, likely leading to a fast progression from State II to State III, supported by the relatively high percentage of State III among the folding intermediates (Fig. 1A).
In the average reconstruction of State III/Apo, where the Core helix domain is further folded, density corresponding to the remaining 14% unfolded sequence is observed around the position of the Middle domain of native state tubulin (Figs. 4Aiii, Biii and S1). Many latent space volumes of State III/Apo also reveal similar spatial arrangement in the unfolded density (Fig. 4Ciii and Supplementary Movie 4). However, many other volumes have the unfolded density located away from the native state position (Fig. 4Ciii, F and Supplementary Movie 4). The flexible CCT tails may help propel the unfolded sequence toward the native state position for folding. Overall, the unfolded sequence has a more restricted search space due to its shorter length and stabilization at both ends by the already folded regions (Fig. S1).
In the average reconstruction of State IV/Apo, the final folding of the Middle domain clears the TRiC chamber (Fig. 4Aiv, Biv). The latent space volumes for State IV/Apo align with the average reconstruction, though occasional residual density likely corresponding to the flexible CCT tails is observed elsewhere in the chamber (Figure 4Civ and Supplementary Movie 5). In these latent space volumes, the CCT tails are observed below the tubulin at the inter-ring interface, engaging the intra-ring or inter-ring tails interaction (Fig. 4Civ). Throughout the progressive folding of tubulin from State I until its native State IV, the CCT tails consistently interact with the tubulin sequence, facilitating the conformational sampling by spatial rearrangement (Fig. 4C).
Discussion
Tubulin folding mechanism
The tubulin polypeptide collapses into a molten globule state during ribosome translation. However, the mechanisms of folding initiation and propagation remain unclear. In this study, we use cryo-EM in combination with deep learning image processing technique to dissect in detail the complex folding mechanism of tubulin within the closed chamber of chaperonin TRiC. We move beyond static snapshots of tubulin folding intermediates to describe the condensation and propagation of individual structural elements within the folding nucleus (State 0.5) and on the natively folded domains (States I, II, III). We demonstrate the tubulin folding nucleus as a nonnative, partially assembled Rossmann fold consisting of irregularly or partially folded secondary structure elements, and/or nonnative tertiary structure interactions between them. TRiC interacts with the loosely folded secondary structure elements of this nucleus, stabilizing it to facilitate its transition into the first native domain (the N domain). Furthermore, we examine the range of spatial rearrangements of the unfolded tubulin sequence in each intermediate state within the TRiC chamber, revealing that CCT tails of the chaperonin TRiC actively promote the conformational space sampling during tubulin folding. These observations provide detailed insights into the tubulin folding pathway (Fig. 5) and highlight the potential kinetic barriers in the folding process.
A The energy landscape of tubulin folding, showing the folding nucleus, intermediate states, and the fully folded tubulin positioned at distinct energy minima. B The hierarchical mechanism of tubulin folding, illustrating the sequential progression through a nonnative folding nucleus, three intermediate states, and the fully folded tubulin.
Challenges in studying folding nucleus
Folding nuclei (FN) refers to a minimal compact region of protein at the early stage of the folding process that establishes a framework for subsequent rapid assembly of the native state. The existence and nature of FN are central to understanding how proteins fold. Over the past few decades, experimental identification of FN has been attempted using several techniques, including Φ-value analysis12,20,21, Nuclear Magnetic Resonance (NMR) spectroscopy22, Single-Molecule Fluorescence Resonance Energy Transfer (smFRET)23 and Single-molecule force spectroscopy24. While these methods provide valuable insights, they are indirect and subject to limitations such as the unknown impact of mutations, resolution, size limitations, complexity of multi-domain proteins, and signal overlap. In contrast, theoretical experiments allow simulation of the protein folding process to directly visualize the FN by molecular dynamics (MD) Simulations, Coarse-Grained Simulations and Energy Landscape Modeling. However, issues such as time scale limitations, force field accuracy, conformational sampling, solvent effects, and computational cost make the accurate prediction of the location of the FN challenging for even small proteins.
Visualizing tubulin folding nucleus: an alternative perspective
In our study, we directly visualize the tubulin FN as a loosely and partially assembled Rossman fold in the closed state of the chaperonin TRiC by cryoEM (Fig. 5). The assembly of nonnative secondary structure elements into a tertiary structure resembling part of the native state Rossman fold suggests the folding nucleus as a dry molten globule state25,26. The canonical nucleation-condensation model posits that a metastable and short-lived nucleus attracts rapid condensation of a large portion of the sequence around it, leading to the native state. However, our observation of the tubulin folding nucleus within a large population of TRiC molecules contrasts this assumption. TRiC contributes to stabilizing the folding nucleus by interacting with its nonnative structure elements. The condensation of the remaining N domain sequence into an approximately correct conformation is expected to transition the full N domain into its native state, as observed in State I.
Evolutionary perspective on the Rossmann fold in tubulin
The Rossmann fold is one of the most primitive protein folds, emerging early in evolution and found across all domains of life, including bacteria, archaea, and eukaryotes. Its simplicity and stability likely facilitate spontaneous and efficient folding, as exemplified by proteins like the tubulin paralog FtsZ in E. coli. However, in tubulin, the Rossmann fold fails to fold spontaneously during ribosomal translation, as indicated by intermediate State 0.5 (Fig. 3). An additional sequence insertion in tubulin compared to the Rossman fold in FtsZ consisting of a β hairpin between helix H1 and β strand S2 may hinder the rapid condensation of β strand S2 onto the folding nucleus, thereby possibly impeding the proper folding of the N domain. Notably, this insertion is partially stabilized in intermediate State 0.5 and further stabilized in State I by the TRiC chaperonin complex. This suggests that TRiC assists tubulin in achieving the native folding of the N domain by sequestering the insertion sequence away from the core structure elements of the Rossmann fold. Previous study of tubulin folding reported that 80–90% of tubulin intermediate could be kinetically trapped and rebound to TRiC for cycling, with only 10% tubulin reaching quasi-native configuration per cycle27. Our observation of the nonnative folding nucleus could represent such a kinetically trapped state that requires the chamber reopening and closure for efficient transition into the State I.
TRiC’s multifaceted role in tubulin folding
TRiC’s role extends beyond merely providing a closed confinement, i.e. Anfinsen cage, for folding. In the previous study17, we demonstrated that TRiC actively guides tubulin folding through electrostatic interactions between its interior wall and the natively folded tubulin domains. Our current findings reveal that TRiC stabilizes the nonnative tubulin FN and assists in its transition to the first natively folded domain. The observation that TRiC’s CCT tails actively mediate the drastic spatial rearrangement in the unfolded sequence adds a layer to our understanding of TRiC’s function, where the flexible CCT tails likely drive the folding of the unfolded sequence out of local energy minima. A recent study28 reported the mutation in the N tail or the truncation of the C termini at R518 of CCT3 in human TRiC are correlated with brain diseases and the latter truncation is deleterious in yeast. These findings echo our observation of the active interaction between the flexible CCT tails and the unfolded tubulin sequences.
Both the average reconstruction and volume reconstructions generated from latent space embedding reveal the complete relocation of the tubulin sequence from the inter-ring space to the TRiC chamber upon TRiC ring closure. In State 0.5, the TRiC chamber is almost entirely occupied by unfolded tubulin, which would complicate a large reorientation if the N and C domain sequence were initially positioned incorrectly, e.g. facing hemisphere of CCTs 7/5/2/4 instead. This observation supports our hypothesis that the electrostatic distribution in the CCT tails plays a crucial role in orienting the unfolded tubulin by attracting the negatively charged sequences of the N and C domains to the CCT1/3/6/8 hemisphere during the tubulin recruitment and relocation stage.
In this study, we observe double occupancy of tubulin within the closed TRiC, indicating simultaneous folding in both rings. This contrasts with previous studies in which plp2, a co-chaperone, occupies one ring to assist tubulin folding in the opposite ring29. Three hypotheses that could explain this observation warrant further investigation: (1) plp2 may be dispensable but significantly accelerates tubulin folding; (2) plp2, as observed in previous studies, may be on standby to assist in the folding of actin rather than tubulin; and 3) plp2 may participate in tubulin folding specifically when TRiC is in its open state.
Future direction
As a building block of microtubules, tubulin is integral to processes such as mitosis, intracellular transport, and cell signaling. The proper folding of tubulin is essential for cell proliferation and survival. Our study provides a comprehensive framework for understanding the TRiC-mediated tubulin folding mechanism, laying the groundwork for future studies exploring tubulin folding in greater detail. Furthermore, these insights create promising opportunities for drug development targeting TRiC-assisted tubulin folding, presenting a strategy for cancer therapy.
Protein folding, evolution, and de novo design are interconnected fields that rely on a thorough understanding of how a polypeptide’s primary structure dictates an efficient folding pathway, ultimately leading to a well-defined tertiary structure. Protein folding—whether spontaneous or chaperone-mediated—remains a frontier in molecular biology. Our findings advance this understanding by revealing a hierarchical folding mechanism that integrates nucleation, condensation, and propagation, combining aspects of multiple protein folding models. Extending cryo-EM and deep learning methodologies to study the folding of other substrates by TRiC or similar chaperonin systems holds the potential to uncover insights, enriching our knowledge of the fundamental principles governing protein folding.
Methods
Data description
The dataset analyzed in the current study comes from our previous study30. TRiC was co-expressed with tubulin from insect cells, and the binary complex was purified and incubated under ATP/AlFx condition to induce TRiC ring closure. The reaction mixture was vitrified on EM grids, and data was collected on Titan Krios (Thermo Fisher Scientific) equipped with the K2 Summit (Gatan) detector at 1.1 Å/pix. 21,486 movies were aligned using MotionCor231, and CTF parameters were estimated with CTFFIND31,32 (v 4.1). Particles picked from template matching were filtered by 2D classification and 3D classification. 529,181 particles of closed state TRiC were used to reconstruct a map with D1 symmetry to 2.5 Å, followed by symmetry expansion to classify the tubulin in each TRiC chamber with a tubulin mask using RELION 333.
The compositional heterogeneity in closed TRiC chamber
Rings in the apo state, occupied by undefined density, or tubulin intermediate state I, II, III, IV are traced back to the original double-ring TRiC particles using the particle information registered in the intermediate star file. All maps described in Fig. 1 are reconstructed using RELION 419. A map at 7.5 Å resolution (without B-factor sharpening) was reconstructed using 5342 TRiC particles encapsulating tubulin intermediate State I within both rings. A map at 7.2 Å resolution (without B-factor sharpening) was reconstructed using 7515 TRiC particles encapsulating tubulin intermediate State I in one ring and State III in the opposite ring. A map at 4.2 Å resolution (without B-factor sharpening) was reconstructed using 130,232 TRiC particles with one ring encapsulating undefined density and the opposite ring in the apo state. TRiC particles with undefined density in one ring and tubulin intermediate State I in the opposite ring are filtered using cryoDRGN (v2)18 heterogeneity analysis and a map at 7.7 Å resolution (without B-factor sharpening) was reconstructed using 6,784 TRiC particles encapsulating undefined high density in one ring and tubulin intermediate State I in the opposite ring.
The heterogeneity analysis of CCT tails in apo TRiC
40,464 particles were used to reconstruct an apo TRiC map at 4.5 Å resolution, which improved to 3.7 Å after B-factor sharpening with RELION 4. The same set of particles are subject to heterogeneity analysis using cryoDRGN (v2). Particles were downsampled to 160 × 160 (2.2 Å/pixel) and fed to a neural network with an 8-dimensional latent variable and 1024 × 3 encoder and 1024 × 3 decoder architectures. After 40 epochs of training, 500 volumes were systematically sampled at k-means cluster centers of the latent embeddings. These volumes were inspected in ChimeraX carefully to confirm the set of particles analyzed were indeed in apo state.
The tubulin State 0.5 in closed TRiC
130,232 particles with TRiC encapsulating undefined density were downsampled to 160 × 160 (2.2 Å/pixel) and fed to a neural network with an 8-dimensional latent variable and 1024 × 3 encoder and 1024 × 3 decoder architectures for heterogeneity analysis. After 40 epochs of training, 500 volumes were systematically sampled at k-means cluster centers of the latent embeddings and examined in ChimeraX to filter the particles into two classes: TRiC with undefined low density, and TRiC with undefined high density. Each subset of particles was individually analyzed in a second run of cryoDRGN training with the same neural network architecture to confirm the successful separation of two classes. In the end, 71,954 particles with undefined high density were used to reconstruct a map to 4.3 Å and 3.8 Å resolution after B factor sharpening by RELION.
The conformational heterogeneity analysis of TRiC encapsulating tubulin folding states I, II, III and IV
40,844 particles of TRiC with one ring encapsulating tubulin intermediate State I and the opposite ring in the apo state were used to reconstruct a map at 4.1 Å resolution without B factor sharpening by RELION. The same set of particles was downsampled to 160 × 160 (2.2 Å/pixel) and fed to a neural network with an 8-dimensional latent variable and 1024 × 3 encoder and 1024 × 3 decoder architectures for heterogeneity analysis. After 50 epochs of training, 300 volumes were systematically sampled at k-means cluster centers of the latent embeddings. These volumes were inspected in ChimeraX carefully to confirm the set of particles analyzed were indeed with tubulin intermediate of State I in one ring and the opposite ring is in the apo state. The location of the unfolded tubulin sequence together with CCT tails was further examined in these volumes.
18,060 particles of TRiC with one ring encapsulating tubulin intermediate state II and the opposite ring in the apo state were used to reconstruct a map at 4.5 Å resolution without B factor sharpening by RELION. The same set of particles were downsampled to 160 × 160 (2.2 Å/pixel) and fed to a neural network with an 8-dimensional latent variable and 1024 × 3 encoder and 1024 × 3 decoder architectures for heterogeneity analysis. After 40 epochs of training, 200 volumes were systematically sampled at k-means cluster centers of the latent embeddings. These volumes were inspected in ChimeraX carefully to confirm the set of particles analyzed were indeed with tubulin intermediate of State II in one ring and the opposite ring is in the apo state. The location of the unfolded tubulin sequence together with CCT tails were further examined in these volumes.
42,695 particles of TRiC with one ring encapsulating tubulin intermediate state III and the opposite ring in the apo state were used to reconstruct a map at 4 Å resolution without B factor sharpening by RELION. The same set of particles were downsampled to 160 × 160 (2.2 Å/pixel) and fed to a neural network with an 8-dimensional latent variable and 1024 × 3 encoder and 1024 × 3 decoder architectures for heterogeneity analysis. After 50 epochs of training, 300 volumes were systematically sampled at k-means cluster centers of the latent embeddings. These volumes were inspected in ChimeraX carefully to confirm the set of particles analyzed were indeed with tubulin intermediate of State III in one ring and the opposite ring is in the apo state. The location of the unfolded tubulin sequence together with CCT tails were further examined in these volumes.
10,202 particles of TRiC with one ring encapsulating tubulin intermediate state IV and the opposite ring in the apo state were used to reconstruct a map at 6.5 Å resolution without B factor sharpening by RELION. The same set of particles was downsampled to 160 × 160 (2.2 Å/pixel) and fed to a neural network with an 8-dimensional latent variable and 1024 × 3 encoder and 1,024 ×3 decoder architectures for heterogeneity analysis. After 40 epochs of training, 200 volumes were systematically sampled at k-means cluster centers of the latent embeddings. These volumes were inspected in ChimeraX carefully to confirm the set of particles analyzed were indeed with tubulin intermediate of State IV in one ring and the opposite ring is in the apo state. The location of the CCT tails was further examined in these volumes.
Model building of Apo TRiC
Model of TRiC (PDB: 7TUB) was fitted into the apo state TRiC map (Table S2), and the unresolved CCT tails are modeled with ChimeraX34 using two constraints: 1) extend each CCT tail in the direction of its resolved sequence; 2) avoid overlapping with the resolved TRiC density. TRiC in PDB model of TRiC/tubulin complex (PDB: 7TUB) was rigidly fitted into the density of TRiC in State 0.5 map to illustrate the tubulin folding nucleus position. Local resolution of tubulin in State 0.5 was characterized by Resmap35, and the resolvability of CCT tails are described with Q score36.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryoEM data in this study are available in the EMPIAR under accession code EMPIAR-12849. The model and maps are accessible in EMDB and PDB (accession numbers: EMD-47463, Apo TRiC; EMD-47464, State 0.5/Apo; EMD-47465, State I/Apo; EMD-47466, State II/Apo; EMD-47467 [https://www.ebi.ac.uk/emdb/EMD-47466], State III/Apo; EMD-47468, State IV/Apo; PDB: 9E2S, Apo TRiC). The PDB structure used for modeling is 7TUB. The source data underlying Fig. 1C are provided as a Source Data file.
References
Zwanzig, R., Szabo, A. & Bagchi, B. Levinthal’s paradox. Proc. Natl. Acad. Sci. USA 89, 20–22 (1992).
Levinthal, C. Pathways for protein folding? J. Chem. 65, 44 (1968).
Onuchic, J. N., Wolynes, P. G., Luthey-Schulten, Z. & Socci, N. D. Toward an outline of the topography of a realistic protein-folding funnel. Proc. Natl. Acad. Sci. USA 92, 3626–3630 (1995).
Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997).
Onuchic, J. N., Socci, N. D., Luthey-Schulten, Z. & Wolynes, P. G. Protein folding funnels: the nature of the transition state ensemble. Fold. Des. 1, 441–450 (1996).
Karplus, M. & Weaver, D. L. Protein-folding dynamics. Nature 260, 404–406 (1976).
Anfinsen, C. B. Principles that govern the folding of protein chains. Science 181, 223–230 (1973).
Dill, K. A. Theory for the folding and stability of globular proteins. Biochemistry 24, 1501–1509 (1985).
Gutin, A. M., Abkevich, V. I. & Shakhnovich, E. I. Is burst hydrophobic collapse necessary for protein folding? Biochemistry 34, 3066–3076 (1995).
Agashe, V. R., Shastry, M. C. R. & Udgaonkar, J. B. Initial hydrophobic collapse in the folding of barstar. Nature 377, 754–757 (1995).
Dill, K. A., Fiebig, K. M. & Chan, H. S. Cooperativity in protein-folding kinetics. Proc. Natl. Acad. Sci. USA 90, 1942–1946 (1993).
Itzhaki, L. S., Otzen, D. E. & Fersht, A. R. The structure of the transition state for folding of chymotrypsin inhibitor 2 analysed by protein engineering methods: evidence for a nucleation-condensation mechanism for protein folding. J. Mol. Biol. 254, 260–288 (1995).
Wetlaufer, D. B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc. Natl. Acad. Sci. USA 70, 697–701 (1973).
Yam, A. Y. et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15, 1255–1262 (2008).
Yaffe, M. B. et al. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 358, 245–248 (1992).
Geissler, S., Siegers, K. & Schiebel, E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 17, 952–966 (1998).
Gestaut, D. et al. Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT. Cell 185, 4770–4787.e20 (2022).
Zhong, E. D., Bepler, T., Berger, B. & Davis, J. H. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods 18, 176–185 (2021).
Zivanov, J. et al. A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. https://doi.org/10.7554/eLife.83724 (2022).
Otzen, D. E., Itzhaki, L. S., elMasry, N. F., Jackson, S. E. & Fersht, A. R. Structure of the transition state for the folding/unfolding of the barley chymotrypsin inhibitor 2 and its implications for mechanisms of protein folding. Proc. Natl. Acad. Sci. USA 91, 10422–10425 (1994).
Fersht, A. R. Nucleation mechanisms in protein folding. Curr. Opin. Struct. Biol. 7, 3–9 (1997).
Kazmirski, S. L. et al. Protein folding from a highly disordered denatured state: the folding pathway of chymotrypsin inhibitor 2 at atomic resolution. Proc. Natl. Acad. Sci. USA 98, 4349–4354 (2001).
Borgia, A. et al. Localizing internal friction along the reaction coordinate of protein folding by combining ensemble and single-molecule fluorescence spectroscopy. Nat. Commun. 3, 1195 (2012).
Bauer, D. et al. A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy. Proc. Natl. Acad. Sci. USA 115, 4666–4671 (2018).
Shakhnovich, E. I. & Finkelstein, A. V. Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition. Biopolymers 28, 1667–1680 (1989).
Finkelstein, A. V. & Shakhnovich, E. I. Theory of cooperative transitions in protein molecules. II. Phase diagram for a protein molecule in solution. Biopolymers 28, 1681–1694 (1989).
Lewis, S. A., Tian, G. & Cowan, N. J. The alpha- and beta-tubulin folding pathways. Trends Cell Biol. 7, 479–484 (1997).
Kraft, F. et al. Brain malformations and seizures by impaired chaperonin function of TRiC. Science 386, 516–525 (2024).
Han, W. et al. Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT. Sci. Adv. 9, eade1207 (2023).
Gestaut, D. et al. Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT. Cell 186, 2038 (2023).
Zheng, S. Q., Palovcak, E., Armache, J.-P., Cheng, Y. & Agard, D. A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2013).
Pintilie, G. et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods 17, 328–334 (2020).
Acknowledgements
We acknowledge funding support from the Stanford School of Medicine Dean’s Postdoctoral Fellowship (to Y.Z.) and Silicon Valley Community Foundation CZI Imaging Initiative (2021-234593) to W.C.
Author information
Authors and Affiliations
Contributions
Y.Z. and W.C. conceived the study. Y.Z. developed the method, analyzed the data, and built the models. Y.Z. drafted the original manuscript. Y.Z., M.S., and W.C. edited the manuscript. Y.Z. and W.C. acquired funding for the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Hong-Wei Wang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, Y., Schmid, M.F. & Chiu, W. Visualizing dynamic tubulin folding in chaperonin TRiC from nonnative nucleus to final native state. Nat Commun 16, 7878 (2025). https://doi.org/10.1038/s41467-025-63016-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63016-x