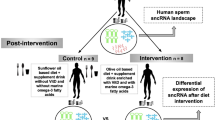
Abstract
The periconception period is critical for embryo development, pregnancy outcomes, and offspring health. During this stage, oviductal and uterine fluids facilitate embryo-maternal interactions and support early embryonic development. Using PANDORA-seq, we identify a diverse repertoire of small non-coding RNAs in female mouse oviduct fluid and uterine fluid during preimplantation, with tRNA-derived small RNAs and rRNA-derived small RNAs being predominant. Maternal high-fat diet during preimplantation period significantly alters tsRNA and rsRNA expression in oviduct fluid and uterine fluid compared to normal diet, disrupting blastocyst metabolic gene expression. While implantation remained unaffected, these alterations impair mid-gestation embryonic and placental growth, resulting in reduced birth weight and length, as well as metabolic disorders in offspring. Furthermore, transfecting embryos with uterine fluid-derived sncRNAs altered by maternal high-fat diet mimics the in vivo effects. These findings suggest that tsRNAs and rsRNAs in reproductive fluids may reflect maternal metabolic status and transmit dietary information to the early embryo, which might influence pregnancy outcomes and offspring health.
Similar content being viewed by others
Introduction
Increasing evidence demonstrates that maternal lifestyle and environmental exposure during pregnancy have profound effects on the long-term health of offspring and are related to disease risk in adulthood, summarized under the concept of Developmental Origins of Health and Disease (DOHaD)1,2. Although the origins of adult diseases can be traced to germ cell generation, gestation, and lactation, the periconception period, especially the preimplantation stage, is particularly critical3,4. During this time, the early embryo undergoes significant genetic and epigenetic changes, including zygote genome activation (ZGA)5, DNA methylation6, histone modification reprogramming7, and the first cell fate decision8. Meanwhile, the maternal reproductive system adapts to support early embryo implantation. Epidemiological studies reported that early pregnancy is the most vulnerable stage of pregnancy and food shortages during this period are associated with increased diabetes risks9. A series of works demonstrated that maternal low-protein diet (LPD) during this stage adversely affects pregnancy10,11 and negatively influences neurogenesis12, behavior13,14, cardiovascular development13,14,15 and metabolism16 in offspring. Additionally, in assisted reproductive processes, techniques like intracytoplasmic sperm injection (ICSI) and in vitro early embryo culture can also induce abnormalities in offspring9,17. Thus, this period is sensitive to environmental changes and plays a crucial role in long-term effects18.
Uterine fluid (UF) has long been recognized for providing a suitable intrauterine environment for early embryo transport and development in early pregnancy. It serves as the medium facilitating crosstalk between the embryo and maternal uterus, influencing embryo implantation. Disruption of uterine glands, which affects the uterine fluid environment, impacts embryo implantation19, endometrial decidualization20,21, and placental development21,22,23, leading to infertility and recurrent pregnancy loss in rodent and sheep models. These findings emphasized the importance of uterine fluid in early pregnancy. It is well known that the contents of uterine fluid are regulated after fertilization, and pioneering analyses of oviduct and uterine fluid compositions found ions, proteins, amino acids, and nucleic acids24,25, which are differentially expressed in cases of recurrent pregnancy loss26. Among these, nucleic acids have been shown to mediate embryo-uterine crosstalk. For instance, miR-30d, secreted from the human endometrium into uterine fluid, can be absorbed by the trophectoderm of blastocysts, aiding in blastocyst adhesion27, and miRNAs have also been suggested to be involved in the regulation of endometrial receptivity28. Given the importance of uterine fluid in connecting the early embryo and maternal uterus, we propose that its contents may mediate the effects of maternal environmental exposure on early embryonic and subsequent development.
In our previous work, we developed a panoramic small noncoding RNA sequencing method, PANDORA-seq, which expanded our understanding of sncRNA diversity across various tissues and cell lines29,30. Beside microRNAs (miRNAs), various small non-coding RNAs (sncRNAs), such as tRNA-derived small RNAs (tsRNAs) and rRNA-derived small RNAs (rsRNAs), have been identified and are suggested to play roles in diverse physiological and pathological processes29. Importantly, our previous study demonstrated that sperm tsRNAs serve as mediators transferring paternal high-fat diet (HFD) information to next generation31. In this study, we discovered the enrichment of tsRNAs and rsRNAs in oviduct and uterine fluid, which dynamically changed under maternal HFD exposure. These changes might lead to disrupted embryonic metabolic gene expression, intrauterine growth restriction (IUGR) and low birth weight. Transfection of altered UF sncRNAs into early embryos impaired blastocyst metabolic gene expression, suggesting UF-derived sncRNAs might transfer maternal HFD information to the early embryo.
Results
PANDORA-seq reveals a dynamic tsRNA/rsRNA expression in OF and UF
After fertilization, early embryos develop in the oviduct for the first 3 days and move to the uterus on the 4th day. During this process, reproductive fluids support early embryo development and transport. To investigate changes in reproductive fluids during this period, we collected oviduct fluid (OF) from days 1–3 and UF on day 4 of pregnant mice using a method adapted from previous study32. Following embryo removal, RNA was extracted (Fig. 1a), yielding ~100 ng of total RNA per uterus/oviduct. Urea-PAGE gel analysis revealed enriched small RNAs in both UF and OF during early pregnancy (Fig. 1b). We then applied PANDORA-seq to profile 15–50 nt sncRNAs across day 1–3 OF and day 4 UF, and found that tsRNAs and rsRNAs were dominantly expressed, comprising over 80% of total sncRNAs in OF and 79.6% in UF (Fig. 1c, d). In contrast, miRNAs, PIWI-interacting RNAs (piRNAs), and YRNA-derived small RNAs (ysRNAs) were also present in the fluid, but each accounted for less than 5% of total sncRNAs (miRNAs <0.5%, piRNAs <5%, ysRNAs <1%).
a Schematic of detection process. b Views of small RNA expression in OF and UF by urea-PAGE gel, n = 3 biologically independent experiments. c Comparison of reads per million (RPM) and length distribution of different small non-coding RNA (sncRNA) categories in OF and UF. d Relative ratios of sncRNAs. e Changes of sncRNA expression throughout day 1 OF to day 4 UF. f Relative ratios of tRNA-derived small RNAs (tsRNAs) to rRNA-derived small RNAs (rsRNAs) throughout day 1 OF to day 4 UF. n = 3 biologically independent experiments, each using pooled samples from 4 mice. Statistical source data are provided in the Source Data file.
Given comparable read depths across samples, we compared sncRNA proportions between day 1–3 OF and day 4 UF (Fig. 1d, Supplementary Fig. 1a, b). During the first 3 days of pregnancy, miRNAs, piRNAs, and ysRNAs exhibited relatively minor variations in expression levels (Supplementary Fig. 1c–e), whereas tsRNAs and rsRNAs exhibited more dynamic shifts. Notably, day 4 UF exhibited a distinct sncRNA expression pattern, with higher rsRNA and lower tsRNA levels than OF. Specifically, tsRNAs increased from 30.8% (day 1) to 40.1% (day 3) but dropped to 21.3% in day 4 UF. Conversely, rsRNAs fluctuated from 53.6% (day 1) to 39.7% (day 2), and increased to 45% on day 3, then reached 58.3% in day 4 UF (Fig. 1e). Interestingly, the tsRNA-to-rsRNA ratios remained stable between day 2 and day 3 OF, but significantly decreased in day 1 OF and day 4 UF (Fig. 1f), suggesting critical environmental changes on these two days. Further analysis revealed that nearly 50% of rsRNA and tsRNA sequences were shared across days 1–3 OF, with many also present in day 4 UF. Genomic tRNA 5ʹ-end-derived tsRNAs constituted a significant proportion of tsRNAs in both OF and UF (Supplementary Fig. 2). However, UF contained a greater diversity of unique tsRNAs and rsRNAs than OF (Supplementary Fig. 1a, b), suggesting distinct fluid environments between the oviduct and uterus, possibly reflecting their different origins. These findings indicated that sncRNA regulation in OF and UF may be independent, which could be critical for early embryo development.
Maternal preimplantation HFD alters tsRNA/rsRNA expression
Since sperm tsRNAs and rsRNAs have been shown to mediate the transmission of paternal HFD/LFD exposure into offspring through pregnancy31,33, we investigated whether sncRNAs in OF and UF similarly reflect maternal metabolic changes during early pregnancy. We established a pre-implantation maternal HFD model by exposing female mice to HFD throughout the first 4 days of pregnancy (Fig. 2a). Despite no significant differences were observed in body weight or glucose tolerance between control (ND) and HFD mice (Supplementary Fig. 3), PANDORA-seq revealed variations in sncRNA expression levels in UF and OF under HFD (Supplementary Data 1). While rsRNAs and tsRNAs remained the most abundant sncRNAs, they exhibited dynamic changes in response to HFD. Specifically, tsRNAs in HFD OF showed consistency from day 2 to day 3 but a marked decrease in day 4 UF, dropping from 21.3% to 9.6% (P = 0.026) (Fig. 2b). Similarly, rsRNAs in HFD OF were comparable to the control on days 2 (43.4%, P = 0.084) and 3 (41.6%, P = 0.156) but significantly decreased on day 1 (47.6 %, P = 0.008) and increased in day 4 UF (72.3%, P = 0.036) (Fig. 2c). These shifts altered tsRNA/rsRNA ratio, increasing on day 1 in HFD (Fig. 2d). Additionally, a heatmap analysis of tsRNAs and rsRNAs from day 2 to day 3 OF indicated that although the tsRNA/rsRNA ratio was consistent, HFD induced dynamic sequence changes (Supplementary Fig. 4).
a Schematic overview of the experimental workflow. b–c Comparison of tsRNAs (b) and rsRNAs (c) expression in OF and UF under normal diet (ND) and HFD. Box plots display the median (center line), 25th and 75th percentiles (box bounds), and whiskers extending to the minimum and maximum values within 1.5× the interquartile range (IQR). Individual data points represent each sample and are overlaid as dots on the box plots, two-tailed Student’s t-tests, n = 3 biologically independent experiments, each using pooled samples from 4 mice. d Relative ratio of tsRNAs to rsRNAs in different OF and UF, two-tailed Student’s t-tests, n = 3 biologically independent experiments, each using pooled samples from 4 mice. e–f Changes of mitochondrial and genomic tsRNAs (e) and rsRNAs (f), mt-tsRNA: mitochondrial tsRNA, cyto-tsRNA: genomic tsRNA, mt-rsRNA: mitochondrial rsRNA, cyto-rsRNA: genomic rsRNAs, two-tailed Student’s t-tests, n = 3 biologically independent experiments, each using pooled samples from 4 mice. g Changes of amino acid corresponds tRNA origins. h Changes of rsRNAs origin loci, RPM: Reads Per Million mapped reads. i Changes of UF RNA modifications in response to HFD exposure. The red font indicates upregulated RNA modifications, blue font indicates downregulated RNA modifications. n = 3 biologically independent experiments, each using pooled samples from 4 mice. The data represent means ± s.e.m. Two-tailed multiple t-tests, *P ≤ 0.05, **P ≤ 0.01. Statistical source data, precise P value and unprocessed blots are provided as in the Source Data file.
Interestingly, HFD exposure significantly reduced the tsRNA/rsRNA ratio in day 4 UF from 37.7% to 13.3% (Fig. 2d), highlighting the sensitivity of UF sncRNAs to maternal metabolic changes. Further analysis revealed that over 99% of UF tsRNAs and rsRNAs originate from the cytoplasm, while only a small portion is derived from mitochondria. The altered tsRNAs and rsRNAs were predominantly genomic rather than mitochondrial in origin (Fig. 2e, f). Among these, tRNA-Aspartate (Asp), Valine (Val), and Glutamate (Glu) derived tsRNAs were the most expressed and decreased in HFD UF (Fig. 2g). Meanwhile, rsRNAs derived from genomic 5S, 5.8S, 18S, and 28S rRNAs showed global changes in origin sites, with slight changes in mitochondrial 12S and 16S rRNA-derived rsRNAs (Fig. 2h). Previous studies have shown that sncRNAs, such as tsRNAs, retain RNA modifications from their precursor tRNAs, which regulated tsRNA generation34. Additionally, paternal HFD has been shown to alter RNA modifications in sperm sncRNAs35. To assess RNA modifications in UF sncRNAs, we used liquid chromatography–mass spectrometry (LC-MS/MS) to quantify 19 RNA modifications in UF from HFD and control mice. Pseudouridine (Ψ) was the most abundant, followed by m1A, while m6A was least abundant (Supplementary Fig. 5). HFD exposure altered the expression of most RNA modifications, notably decreasing m1A and m2,2G while increasing ac4C, Um, m5U, and m3U (Fig. 2i). These changes likely reflect the metabolic response of the uterus, which plays a crucial role not only in extracellular sncRNA formation, but also in early embryo development. These findings suggest that the uterine environment is finely tuned to respond to maternal dietary conditions.
sncRNA links to aberrant embryonic gene expression and IUGR
As the early embryo develops within the reproductive fluid, we investigated whether changes of sncRNAs in OF and UF are associated with early embryo development. Blastocysts were collected from both HFD and normal diet (ND) groups. Morphologically, no significant differences were observed in blastocyst number or appearance between the two groups (Fig. 3a). However, gene expression analysis revealed distinct profiles between HFD and ND blastocysts. Principal component analysis (PCA) showed clear separation between the two groups (Fig. 3b), and volcano plot data highlighted significant differential gene expression in HFD blastocysts compared to ND (Fig. 3c). Gene Ontology (GO) analysis of Differentially Expressed Genes (DEGs) revealed that the most altered transcriptomic clusters were related to metabolic pathways. For example, amide metabolic process (adjusted P = 0.00079), sulfur compound metabolic process (adjusted P = 0.0048), nucleoside phosphate biosynthetic process (adjusted P = 0.0078), glycerolipid metabolic process (adjusted P = 0.021), purine nucleotide metabolic process (adjusted P = 0.021), and pigment metabolic process (adjusted P = 0.021) (Fig. 3d). These findings suggested that preimplantation HFD exposure disrupts metabolic gene expression in blastocysts.
a Representative morphology of blastocysts in ND and HFD groups, scale bar = 20 μm, n = 3 biologically independent experiments. b Principal component analysis (PCA) of transcriptome profiles of blastocysts in the ND and HFD groups. c Differential gene expression in blastocysts of ND and HFD groups. NS not significant. d Top 10 enriched Gene Ontology (GO) terms among significantly altered genes. The red font signifies the metabolic pathways. n = 3, BP Biological Process, CC Cellular Component, MF Molecular Function. Statistical source data and precise P value are provided as in the Source Data file.
Next, we assessed the impact of these metabolic gene changes on later pregnancy. The implantation rates, embryo spacing, and implantation numbers were comparable between the HFD and ND groups (Fig. 4a). Similarly, at mid-gestation (day 12), implantation numbers and survival rates were comparable between the two groups (Fig. 4c, d), suggesting that preimplantation HFD exposure may not affect embryo implantation in mice. However, the weight of implantation sites on day 12 was significantly reduced in the HFD group compared to ND (Fig. 4e), with lower weights observed in both embryos and placentas (Fig. 4f, g). Although HFD embryos appeared morphologically normal at mid-gestation, their sizes were noticeably smaller than those in the ND group (Fig. 4b). By the end of pregnancy, offspring from the HFD group exhibited lower birth weights and shorter lengths compared to the ND group, despite having similar overall sizes (Fig. 4h–k). These findings suggest that pre-implantation maternal HFD exposure affects early embryo development, leading to intrauterine growth restriction. We then evaluated the metabolic conditions of the offspring. Although no differences in body weight and metabolic rates were observed at 2 and 4 months, 6-month-old offspring from HFD group exhibited a significant increase in body weight (Supplementary Fig. 6), and compromised glucose tolerance compared to ND offspring (Fig. 4l, m), which was observed in both males and females. These results suggest that maternal HFD exposure during pre-implantation induced intrauterine growth restriction and offspring metabolic disorder in their mid-age stage.
a Representative images of implantation sites from dams fed either ND or HFD, n = 3 biologically independent experiments. b Representative images of embryos at embryonic day 12 from ND and HFD groups. c Quantification of the number of implantation sites at day 12 in both ND and HFD groups (n = 3 mice). d Comparison of embryo survival rates at day 12 between ND and HFD groups (n = 3 mice), IS: implantation sites. e Average weight of implantation sites at day 12 (ND: n = 46 implantation sites, HFD: n = 51 implantation sites), pooled from 3 dams per group. f Average weight of surviving embryos at day 12 (ND: n = 44 implantation sites, HFD: n = 48 implantation sites), pooled from 3 dams per group. g Average placental weight at day 12 (ND: n = 44 implantation sites, HFD: n = 48 implantation sites), pooled from 3 dams per group. h Representative images of newborn pups from the ND and HFD groups. i Average litter size at birth (ND: n = 8 dams, HFD: n = 9 dams). j Average body weight of newborn pups (ND: 67 pups from 5 dams, HFD: 59 pups from 5 dams). k Average length of newborn pups (ND: 42 pups from 3 dams, HFD: 67 pups from 4 dams). l Glucose Tolerance Test (GTT) results for 6-month-old male (left) and female (right) F1 offspring, M month. m GTT results represented as the area under the curve (AUC) for 2-, 4-, and 6-month-old male (left) and female (right) F1 offspring. ND-F1-male (n = 14 mice), HFD-F1-male (n = 11 mice), ND-F1-female (n = 8 mice), HFD-F1-female (n = 15 mice). Blue represents the ND group, and orange represents the HFD group. Two-tailed Student’s t-tests was used and data represent as mean ± s.e.m. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Statistical source data and precise P value are provided as in the Source Data file.
tsRNAs /rsRNAs regulate embryonic metabolic gene expression
Were the altered tsRNAs and rsRNAs under HFD involved in metabolic changes in blastocysts? Due to the insufficient in vivo quantities of specific tsRNAs and rsRNAs for isolation, we synthesized eleven FAM-labeled sncRNAs that were altered by HFD, including tsRNA-Glu-CTC, tsRNA-Ser-GCT, tsRNA-His-GTG, tsRNA-Trp-CCA, tsRNA-Leu-AAG, mt-tsRNA-Val-TAC, mt-tsRNA-Trp-TCA (the standardized nomenclature of these tsRNAs showed in Supplementary Table 1), rsRNA-28S, rsRNA-18S, miR-24 and miR-127, that exhibited significant changes following HFD exposure. To investigate their function, we transfected these synthesized sncRNAs into NIH/3T3 cells, both individually and in combination, and observed a marked decrease in cell proliferation post-transfection (Supplementary Fig. 7), suggesting these sncRNAs might play a role in cellular functions.
Next, we transfected these tsRNAs, rsRNAs, miRNAs, and a combined pool of these RNAs into zona-free morulae from ND mice and cultured them to the blastocyst stage. The transfected RNAs localized primarily in trophoblast cells, with minor uptake in the inner cell mass (Fig. 5a; Supplementary Fig. 8a). No overt morphological abnormalities were observed (Fig. 5a). Subsequently, transcriptomic RNA sequencing results revealed that sncRNA pool transfection shifted the gene expression profiles of blastocysts (Fig. 5b, c). Gene set enrichment using a rank-weighted gene expression algorithm29 identified significant changes in metabolic pathway from Gene Ontology Biological Process (GOBP) terms, including glucan biosynthetic processes (P = 0.00059), regulation of glycogen metabolic processes (P = 0.00353), glutamate catabolic processes (P = 0.01225), aminoglycan metabolic processes (P = 0.00628) and propionate metabolic processes (P = 0.04206), which showed significant differences between control and sncRNA pool transfected blastocysts (Fig. 5d). The specific genes impacted within these pathways are presented in Fig. 5e. These altered genes also showed changes in blastocysts collected from HFD and ND groups (Supplementary Fig. 8b). Interestingly, when separately transfected early embryos with either tsRNA or rsRNA pools, we found that they may regulate overlapping biological processes via distinct molecular pathways (Supplementary Fig. 9). Notably, small RNA target prediction indicated that the sequences of selected tsRNAs and rsRNAs were targeted to these altered metabolic genes (Fig. 5f, g). Similar regulatory relationships between sncRNAs and genes were observed in HFD-exposed blastocysts. By analyzing the correlation patterns between UF sncRNA expression changes and selected altered genes in blastocysts, we identified significant associations between tsRNA-Leu-AAG and Hs6st1, mt-tsRNA-Trp-TCA and Ctdspl, and rsRNA-18S and B3gnt7 (Fig. 5h–j). Validation experiments in NIH/3T3 cells showed that transfected selected individual sncRNAs (tsRNA-Leu-AAG, mt-tsRNA-Trp-TCA, and rsRNA-18S) modulated their respective target gene expression (Fig. 5h–j), supporting their functional involvement in metabolic regulation during early development.
a Representative images of blastocysts after transferring synthesized FAM-labeled sncRNAs (green). Scale bar = 10 μm, n = 3 biologically independent experiments. b PCA plot of blastocysts transfected with empty liposome (control) or sncRNA pool. c Expression heatmaps of genes in control and sncRNA pool transfected blastocysts. d Pathway changes after transfection, two-tailed Student’s t-tests, n = 3 biologically independent experiments. e Specifically changed genes. f–g Predicted interactions between transfected tsRNAs (f) and rsRNAs (g) with changed genes. h–j Significant correlations between sncRNAs in UF and altered genes within blastocysts, tsRNA-Leu-AAG and Hs6st1 (h), mt-tsRNA-Trp-TCA and Ctdspl (i), and rsRNA-18S and B3gnt7 (j), were detected by both correlation patterns analysis and individual sncRNAs transfection in NIH/3T3 cells, n = 3 biologically independent experiments. Two-tailed Student’s t-tests, and the data represent means ± s.e.m. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Statistical source data and precise P value are provided as in the Source Data file.
Discussion
In this study, we identified abundant tsRNAs and rsRNAs in uterine and oviductal fluids, which displayed dynamic expression patterns during early pregnancy in mice. Maternal exposure to HFD before implantation altered the expression and RNA modifications of these sncRNAs, particularly in the UF. This disruption in sncRNA expression adversely affected blastocyst metabolic signaling, which might lead to intrauterine growth restriction and subsequent metabolic disorders in the offspring.
Historically, research on the composition of uterine and oviductal fluids played a critical role in developing embryo culture media and advancing in vitro fertilization (IVF) techniques. The understanding of uterine gland development21, which can be disrupted by progesterone treatment36 or FOXA2 knockout20, has expanded the role of uterine secretions beyond merely supporting early embryo survival. It is now recognized that these secretions are essential for regulating uterine receptivity22, decidualization, and placental development23. While ions, amino acids, proteins, and hormones have long been known to constitute these fluids24,25,37, more recently, components discovered including extracellular vesicles (EVs)38,39,40, miRNAs26,41, and metabolites42,43. Our study adds to this expanding field by identifying a significant presence of extracellular sncRNAs, particularly tsRNAs and rsRNAs, in reproductive fluids. A critical question is: what roles do these tsRNAs and rsRNAs play?
It is well established that in vitro embryo development differs significantly from in vivo conditions. In vitro cultured embryos often exhibit delayed development, reduced blastocyst formation rates, altered cell numbers, and differences in metabolism and gene expression compared to in vivo embryos44,45. Moreover, offspring derived from in vitro cultured embryos may experience long-term effects, including abnormal developmental rates, specific behavioral changes46, and even potential transgenerational risks47. Our findings of dynamic changes in sncRNA expression throughout early pregnancy indicate that embryos might have varying requirements at different developmental stages. Therefore, the identification of these sncRNAs in reproductive fluids may present a novel opportunity to refine in vitro culture conditions, potentially reducing the developmental discrepancies between in vitro and in vivo embryo growth.
In both OF and UF, tsRNAs and rsRNAs are the predominant sncRNAs. tsRNAs were first identified as being generated in cells under stress48,49 and have been shown to participate in various biological processes50, including hematopoietic stem cell differentiation51, immunity52, cancer progression53, and mediating epigenetic inheritance31. However, the role of extracellular tsRNAs remains largely unexplored. In this study, we proposed following potential roles for extracellular tsRNAs. Firstly, our previous research demonstrated that serum tsRNAs are highly responsive, with significant elevation following lipopolysaccharide (LPS) treatment54. We also observed a marked increase of tsRNAs in day 1 OF after dietary changes in present study, suggesting that extracellular tsRNAs may serve as sensitive indicators of physiological changes. Besides, components of uterine fluid, such as metabolites and proteins, have been proposed as potential biomarkers for disease detection. For example, the metabolome of uterine fluid may aid in the early detection of ovarian cancer55, and proteomic profiles could be useful in diagnosing endometriosis56. Similarly, extracellular tsRNAs may also serve as potential biomarkers during early pregnancy. Moreover, miRNAs, piRNAs and ysRNAs were also found in reproductive fluid, although their ratios are much lower than tsRNAs and rsRNAs, they might also play critical function in regulating early embryo development and mediate maternal environmental exposure information. Consequently, these sncRNAs could serve as novel biomarkers for assessing metabolic status before IVF, potentially improving pregnancy outcomes.
Additionally, both tsRNAs and rsRNAs are also highly expressed in sperm29, where their expression patterns and RNA modifications form a signature that mediates the intergenerational transmission of paternal metabolic disorders35. This phenomenon is observed in UF, where we detected changes in sncRNA expression patterns and RNA modifications following HFD exposure, suggesting their potential role in conveying environmental information. Previous studies have shown that sperm tsRNAs mediate the transmission of HFD31,35 and low-protein diet (LPD)33 information to offspring in both mouse models and human subjects57, with HFD-altered tsRNAs targeting genes involved in early embryonic development31. Our findings indicate that maternal HFD might also induce changes in reproductive fluid tsRNAs, which targeted genes related to metabolic pathways, suggesting that these sncRNAs in reproductive fluid might serve as vehicles for conveying maternal dietary information to the early embryo. Interestingly, earlier research has shown that in addition to sperm, seminal plasma can transmit paternal LPD information to offspring16,58, and tsRNAs have also been identified in seminal plasma59, suggesting that seminal tsRNAs may play a role in transmitting paternal diet-induced traits to the next generation. Thus, in the reproductive fluid, including UF, OF and seminal plasma, sncRNAs may serve as epigenetic information carriers, mediating the transfer of environmental and dietary information across generations.
The mechanism of sncRNAs is also critical. While the functional roles of cellular rsRNAs are still being elucidated, tsRNAs have been more extensively studied and are known to participate in various cellular processes, including ribosome biogenesis, translation regulation, and retrotransposon suppression50,60,61. In this study, we found that sncRNA transfection might affect both trophectoderm and ICM. Through transfection experiments, we demonstrated that individual tsRNA and rsRNA may directly target metabolic genes, suggesting a regulatory mechanism similar to RNAi, as reported in previous studies62,63. Thus, tsRNAs and rsRNAs together may regulate the expression of important developmental or metabolic genes, thereby influencing embryonic and placental development. However, it is noticeable that the synthesized tsRNAs lacked modifications. This limitation implies that the biological function of in vivo tsRNAs might not be fully replicated by their synthetic counterparts, as their function may depend not only on their sequences but also on their specific RNA modifications. RNA modifications in tsRNAs have been shown to influence their structures and, consequently, their functions35. Our findings of altered RNA modifications in tsRNAs from HFD-exposed UF, alongside sequence changes, support the hypothesis that these extracellular tsRNAs carry modifications that allow them to assume different structural forms. Thus, tsRNAs might regulate biological events through both sequence-dependent and aptamer-like mechanisms64. Further research is needed to confirm these functional roles and to understand the full extent of their regulatory capacities.
Interestingly, although maternal HFD did not alter blastocyst morphology or numbers, it caused significant transcriptomic and developmental changes, leading to IUGR. The adverse effects of parental environmental exposures during the periconception period have garnered increasing attention in recent years3,65,66, underscoring the critical importance of this developmental window. Unlike certain stimuli that can severely impair early embryo development, maternal dietary exposure typically exerts more subtle effects on the embryo initially, with significant consequences manifesting later in pregnancy. For instance, maternal LPD during this period has been shown to alter the blastocyst’s growth trajectory, increasing trophectoderm proliferation12,13 and activating compensatory responses in the primitive endoderm, ultimately leading to abnormalities in adult offspring, which were associated with alterations in circulating insulin and branched-chain amino acids in uterine fluid67. Our study not only demonstrated the sensitivity response of uterine environment during periconception, but also further highlights the serious impact of periconceptional exposure, which can cause IUGR without observable effects on blastocyst morphology or embryo implantation. Notably, IUGR is typically characterized by low birth weight, a condition linked to an increased risk of cardiovascular disease68, metabolic disorders69, and neurodevelopmental problems70 in offspring. Thus, maternal diet during the periconception period might not only affect pregnancy outcomes but also have long-term implications for offspring health. The preimplantation period is a particularly sensitive stage that often goes undetected in clinical settings, emphasizing the importance of maintaining a healthy lifestyle for women preparing to conceive. It is important to note that maternal-fetal communication during early pregnancy is a highly complex process involving multiple layers of regulation. Consequently, various mediators are likely to participate in this process, and the observed pregnancy outcomes and effects on offspring health may result from the interplay of these regulatory mechanisms. This study also has limitations. For instance, the extent to which extracellular tsRNAs and rsRNAs are absorbed by early embryos remains unclear. Additionally, by which tsRNAs and rsRNAs regulate gene expression in early embryos, as discussed above, require further investigation. Future studies are needed to elucidate these aspects and provide a more comprehensive understanding of the roles of sncRNAs in early embryonic development and maternal-fetal communication.
In conclusion, we identified a significant enrichment of tsRNAs and rsRNAs in uterine and oviductal fluid, with their expression patterns and RNA modifications being highly responsive to maternal HFD exposure. These altered sncRNAs might disrupt early embryo development by affecting metabolic gene expression, leading to intrauterine growth restriction. Therefore, sncRNAs in uterine and oviductal fluid might serve as critical carriers of maternal environmental information to the early embryo, and might influence pregnancy outcomes.
Methods
Animals
All experimental procedures involving Crl:CD1 (ICR) mice (Charles River Laboratories China) were conducted in strict compliance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Beijing Normal University (CLS-EAW-2021-005). The study adhered to established guidelines aimed at minimizing animal suffering. 8-week-old mice were housed under standard laboratory conditions with unrestricted access to food and water, maintained on a 12:12-h light/dark cycle at 22–25 °C and 40–60% relative humidity. Eight-week-old female ICR mice were co-housed with male ICR mice, and pregnancy was confirmed by the presence of a vaginal plug, designated as embryonic day 1 (day 1). Pregnant females were then housed individually. In the HFD group, pregnant females were fed a high-fat diet (HFD, 60% fat, 20% protein and 20% Carbohydrate, D12492, Research Diets Inc.), or normal diet (ND, 5% fat, 17% protein, Beijing HFK Bioscience Co) from day 1 to day 4 of pregnancy, after which they were switched to ND starting on day 5 and maintained on ND until parturition. The control group received a normal diet throughout the entire pregnancy. Blastocysts of both groups were collected on D4 from the uteri for transcriptomic sequencing analysis. On day 5, implantation sites were identified by visualizing blue-stained regions in the uterus following intravenous injection of 0.1 mL of 0.1% trypan blue. On day 12, pregnant mice were humanely euthanized, and their uteri were harvested, fixed overnight in Bouin’s solution, and weighed. Litter sizes and neonate weights were recorded at birth, and fetal lengths were measured.
Glucose tolerance test (GTT)
GTT was performed on dams on day 4 of pregnancy, which were fed either ND or HFD, as well as on their offspring at 2, 4, and 6 months of age. Both male and female progeny were included in the analysis. For the GTT, mice were fasted for 12 h prior to testing. Baseline blood glucose levels were measured from tail vein samples using a glucometer. Mice were then administered glucose intraperitoneally at a dose of 1.5 g/kg body weight. Blood glucose levels were subsequently measured at 0, 30, 60, 90, and 120 min following glucose administration. Data were presented as line graphs, and the area under the curve (AUC) was calculated to assess total glucose exposure over time.
Oviductal fluid and uterine fluid collection
Oviductal fluid (OF) was collected at 18:00 on days 1–3, and UF was collected at 18:00 on day 4, using methods adapted from previous studies32. Briefly, after anesthetizing the mice, uteri or oviducts were excised, and carefully trimmed of excess mesometrium and adipose tissue, rinsed twice in pre-cooled Dulbecco’s Phosphate-Buffered Saline (DPBS. RNase-free), and blotted dry with filter paper. For UF collection, the uterine horns were incised, and cold DPBS was gently injected through the cervical end using a 1 mL syringe. OF was collected by flushing cold DPBS through the oviductal opening. Collected fluids were transferred to sterile glass dishes, where embryos were carefully removed under a stereomicroscope using a mouth pipette. The fluids were then transferred to 1.5 mL centrifuge tubes and centrifuged at 300 × g for 20 min at 4 °C to remove tissue debris, followed by a second centrifugation at 1000 × g for 20 min at 4 °C to eliminate blood cells and other impurities. The supernatant was mixed with three volumes of TRIzol LS reagent, vortexed vigorously, incubated at room temperature for 5 min, and stored at −80 °C for subsequent analysis.
Isolation of 15–50 nt small RNAs
Total RNA from oviductal and uterine fluids was extracted using the TRIzol LS protocol. The mixture was thoroughly vortexed, incubated at room temperature for 15 min, and phase-separated by adding 200 μL of chloroform per mL of solution, followed by vigorous shaking for 10 s and incubation at room temperature for 10 min. Samples were centrifuged at 12,000 × g for 15 min at 4 °C, and the upper aqueous phase (~500 μL) was carefully transferred to a new tube. RNA was precipitated by adding an equal volume of isopropanol and 1 μL of glycogen, followed by gentle inversion and overnight incubation at −20 °C. The RNA pellet was obtained by centrifugation at 12,000 × g for 15 min at 4 °C, washed twice with 1 mL of 75% ethanol, air-dried, and resuspended in 10 μL of RNase-free water. RNA concentration and purity were measured using a NanoDrop 2000C spectrophotometer. RNA fragments ranging from 15 to 50 nucleotides (nt) were isolated using a 10% denaturing polyacrylamide gel, following a protocol adapted from our previous work29. Total RNA was electrophoresed on a 10% urea-polyacrylamide gel at 200 V for 40 min in 1× Tris-Borate-EDTA (TBE) buffer (Thermo Fisher Scientific). The gel was stained with SYBR Gold nucleic acid gel stain (Thermo Fisher Scientific), visualized using a UV transilluminator, and RNA fragments within the 15–50 nt range were excised using RNA molecular weight markers (New England Biolabs) as guides.
Quantitative analysis of RNA modifications in mouse uterine fluid using LC-MS/MS
Uterine fluid samples were collected from pregnant mice at gestation day 4, with three biological replicates analyzed per dietary group (ND and HFD). Each biological replicate comprised pooled samples from four mice to ensure sufficient samples. 15–50 nt RNA isolated from mouse uterine fluid was enzymatically digested into mononucleotides in a 50 μL reaction mixture containing 5 μL of 10× reaction buffer (2.5 M Tris-HCl, pH 8.0, 50 mM MgCl2, 5 mg/mL bovine serum albumin [BSA]), 1 IU Benzonase (Sigma-Aldrich), 0.2 IU alkaline phosphatase (Sigma-Aldrich), 0.01 IU phosphodiesterase I (USB), and nuclease-free water, and incubated at 37 °C for 3 h. The digest was filtered using a Nanosep 3 K device with Omega membrane (Sigma-Aldrich) at 14,000 rpm for 20 min at 4 °C, then the flow-through was transferred to a chromatographic vial for LC-MS/MS analysis as described previously71,72. Nucleoside standards (cytidine, adenosine, guanosine, uridine, m¹A, m6A, Am, I, Im, hm5C, m5C, ac4C, Cm, m¹G, m2G, m7G, m²,²G, Gm, Um, m3U, m5U, m5Um, and Ψ) served as quantification controls, with calibration curves generated from a gradient dilution series. Replications were individually processed and subjected to single-injection analysis. Chromatography was performed on an Agilent 1200 series LC, using a 2.1 mm × 150 mm Hypersil GOLD aQ C18 column, 3 µm particles (Thermo Fisher) held at 40 °C. MS analysis was performed by a triple quadrupole mass spectrometer (ThermoFisher TSQ Vantage) with an electrospray ionization source in a positive ion mode using multiple reaction monitoring. Raw data from LC-MS/MS were acquired and subsequently processed with Thermo Xcalibur 3.1 data system. Absolute amounts of each ribonucleoside were back-calculated from the contemporaneous calibration curve. To control for variation in sample loading, the molar concentration of every modified ribonucleoside was normalized to the total amount of nucleosides sharing the same nucleobase (e.g., m5C% = m5C/[m5C + Cm + hm5C + ac4C + C]).
Detection of small RNA expression patterns using PANDORA-seq
Small RNAs were treated following the PANDORA-seq protocol, as previously described29,30. Small RNAs were treated with T4 polynucleotide kinase (T4PNK) reaction mixture (5 μL 10 × PNK buffer, 1 mM ATP, 10 U T4PNK) followed by RNA isolation. The collected RNAs were then treated with α-ketoglutarate-dependent hydroxylase (AlkB) mixture (50 mM HEPES, 75 μM ferrous ammonium sulfate, 1 mM α-ketoglutaric acid, 2 mM sodium ascorbate, 50 mg/L BSA, 4 μg/mL AlkB, 2,000 U/mL RNase inhibitor) followed by RNA isolation with TRIzol LS. Then the small RNA libraries were constructed and sequenced using DNBSEQ platform (BGI Genomics Co., Ltd., Shenzhen, China) with 50-bp single-end reads.
Then small RNA sequences were annotated using the software SPORTS1.173 with a one-mismatch tolerance. Reads were then mapped to several non-coding RNA databases, including: (1) miRNA database miRBase 22.1; (2) genomic tRNA database GtRNAdb (mm39 for mouse); (3) mitochondrial tRNA database mitotRNAdb; (4) rRNA and YRNA databases assembled from the National Center for Biotechnology Information nucleotide and gene database; (5) piRNA database piRBase v3.0; and (6) non-coding RNAs defined by Ensembl and Rfam 14.7.
Differential expression analysis was conducted using the R package DESeq2, identifying significantly differentially expressed small RNAs (defined based on an average RPM > 1) with a threshold of adjusted P ≤ 0.05 and an absolute |log2 fold change| ≥ 1.
Selection of sncRNAs for transfection
Differential expression analysis was conducted to identify sncRNAs significantly upregulated in the HFD group relative to the ND group, focusing on four major classes defined by molecular origin and annotation: genomic tRNA-derived small RNA (cyto-tsRNA), rRNA-derived small RNAs (rsRNAs), mitochondrial tRNA-derived small RNAs (mt-tsRNAs), and microRNAs (miRNAs). Candidate miRNAs were selected based on prior reports of functional relevance in metabolic regulation. To refine tsRNA and rsRNA selection, class-specific abundance thresholds were applied, retaining sequences with log2FC > 1, baseMean > 500 and adjusted P ≤ 0.01. Only two mt-tsRNAs meet the criterion, which were chosen. As cyto-tsRNAs predominantly originated from Leu, Trp, His, Glu, and Ser tsRNAs, while rsRNAs were mainly derived from 18S and 28S rRNAs, we selected the top sequences from tsRNA-Leu, tsRNA-Trp, tsRNA-His, tsRNA-Glu, tsRNA-Ser, mt-tsRNA, 18S-rsRNA, 28S-rsRNA separately, by considering the lower adjusted P, higher log2FC, higher baseMean, smaller lfcSE (the standard error estimate for the log2 fold change estimate) and lower sem (standard error of the mean).
Transfection of murine morulae with sncRNAs
On the morning of day 3, morulae were isolated from mouse oviducts and subjected to four washes in M2 medium. The embryos were then transferred into acidic Tyrode’s solution in a covered dish, ensuring that they remained suspended near the solution’s surface throughout the process. The zona pellucida was dissolved within 15–30 s under continuous microscopic observation, taking care to prevent embryos from adhering to the dish. Following complete dissolution, embryos were transferred to fresh M2 medium, washed 1–2 times to remove any residual acidic solution, and cultured in KSOM medium. Transfection procedures followed established protocols, with four experimental groups corresponding to different RNA types and an empty liposome control group that underwent the same treatment process but without the addition of sncRNAs. The transfected sncRNA groups included: sncRNA pool, tsRNA pool, rsRNA pool, miRNA pool. Sequences of the sncRNAs used were listed in Supplementary Table 2. Transfections were performed using Lipofectamine™ (STEM00001). Solution A was prepared by mixing 0.5 μL of Lipofectamine™ with 25 μL of KSOM medium, while a 5 nM concentration of 5′ FAM-labeled RNA pool was mixed with 25 μL of KSOM medium to prepare Solution B. Solutions A and B were combined, incubated for 10–15 min to allow complex formation, and then applied in 50 μL droplets covered with mineral oil to prevent evaporation. The droplets were equilibrated in an incubator at 37 °C for at least 30 min before 10 morulae were cultured in one droplet of transfection mixture for 24 h. Transfection efficiency was assessed by fluorescence microscopy, evaluating the localization of the fluorescent signal within the morulae. Following transfection, blastocysts were collected and stored in RNA preservation solution for subsequent sequencing analysis. Each transfection experiment was independently repeated three times.
NIH/3T3 cell transfection and QPCR detection
In the NIH/3T3 cell transfection experiments, NIH/3T3 cells with a passage number below 20 were sourced from the National Cell Resource Center (1101MOU-PUMC000018, Beijing, China). Transfection procedures were carried out according to established protocols. Specifically, the following were transfected into NIH/3T3 cells separately: sncRNA pool, tsRNA pool, rsRNA pool, miRNA pool, individual sncRNAs (tsRNA-Leu-AAG: GGUAGCGUGGCCGAGC, mt-tsRNA-Trp-TCA: AGUCCGCGAGCCUUC, and rsRNA-18S: GAGCGGUCGGCGUCCC), and controls (empty liposome transfection without sncRNAs). Transfection efficiency was evaluated 48 h post-transfection using fluorescence microscopy, and cell proliferation was assessed using the MTT assay. Cells transfected with individual sncRNAs and controls were harvested 48 h after transfection. Total RNA was extracted and reverse-transcribed into cDNA. Quantitative PCR was then performed using the primers, which listed in Supplementary Table 3.
Transcriptome sequencing of blastocysts and data analysis
Total RNA from blastocysts was isolated and enriched for polyadenylated mRNA using Oligo (dT) magnetic beads, followed by reverse transcription and amplification according to the Smart-seq2 protocol. The resulting double-stranded complementary DNA (cDNA) was treated with Tn5 transposase for fragmentation and adapter ligation, constructing libraries suitable for high-throughput sequencing. Library quality was assessed using an Agilent 2100 Bioanalyzer and quantitative PCR (qPCR) before sequencing on the DNBSEQ platform, generating paired-end 100 base pair (PE100) reads, providing a minimum of 6 gigabytes (GB) of high-quality data per sample. Raw sequencing data underwent rigorous quality control using SOAPnuke (v1.5.6), which included removing reads containing adapters, reads with more than 5% unknown bases (N), and low-quality reads where more than 20% of bases had a quality score below 15. Clean data were aligned to the mouse reference genome using Bowtie2 (v2.3.4.3), and gene expression was quantified using RNA-Seq by Expectation-Maximization (RSEM) software (v1.3.1). Principal component analysis (PCA) was performed using the FactoMineR package, with visualizations generated by factoextra. Differentially expressed genes (DEGs) were identified using the DESeq2 package, with significance thresholds set at |log2 fold change| ≥ 1 and adjusted P ≤ 0.05. Volcano plots of differential gene expression were created using ggplot2, and gene expression clustering across samples was visualized using heatmap. Functional annotation and pathway enrichment analyses of DEGs were performed using the clusterProfiler package. The Benjamini-Hochberg method was used to correct for multiple testing. Gene Ontology (GO) term enrichment analyses were conducted, with significance thresholds set at an adjusted P ≤ 0.05.
Statistics and reproducibility
Mice were randomly sorted into ND and HFD groups. Sample sizes were determined based on previous studies and power calculations. Two-tailed Student’s t-tests were conducted to compare the relative abundance of sncRNAs, embryo/placental weight, survival numbers/rates, litter size, newborn birth weights/lengths, GTT results between the ND and HFD groups, gene set scores for GOBP terms between control and RNA pool transfection, as well as relative gene expression levels between control and individual sncRNA transfections. For the analysis of RNA modification dynamics, a two-tailed multiple t-test was performed using GraphPad Prism. Correlations between sncRNAs and genes were assessed using Spearman’s rank correlation analysis to generate the correlation coefficient (ρ). Statistical significance was set at P ≤ 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequence data generated in this study have been deposited at Genome Sequence Archive (GSA: CRA019244 and GSA: CRA019272) in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences. These data can be accessed by https://ngdc.cncb.ac.cn/gsa/browse/CRA019244 and https://ngdc.cncb.ac.cn/gsa/browse/CRA019272. Source data are provided with this paper.
Code availability
This paper does not report original code. The sncRNA annotation pipeline SPORTS1.1 is available from GitHub (https://doi.org/10.5281/zenodo.16418220). The specific version of the code associated with the publication is archived in Zenodo (https://zenodo.org/records/16418220).
References
Barker, D. J., Forsen, T., Uutela, A., Osmond, C. & Eriksson, J. G. Size at birth and resilience to effects of poor living conditions in adult life: longitudinal study. BMJ. 323, 1273–1276 (2001).
Barker, D., Barker, M., Fleming, T. & Lampl, M. Developmental biology: support mothers to secure future public health. Nature 504, 209–211 (2013).
Fleming, T. P. et al. Origins of lifetime health around the time of conception: causes and consequences. Lancet 391, 1842–1852 (2018).
Velazquez, M. A., Fleming, T. P. & Watkins, A. J. Periconceptional environment and the developmental origins of disease. J. Endocrinol. 242, T33–T49 (2019).
Schulz, K. N. & Harrison, M. M. Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234 (2019).
Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019).
Jambhekar, A., Dhall, A. & Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20, 625–641 (2019).
Chen, Q., Shi, J., Tao, Y. & Zernicka-Goetz, M. Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo. Nat. Commun. 9, 1819 (2018).
Mertens, J. et al. Children born after assisted reproduction more commonly carry a mitochondrial genotype associating with low birthweight. Nat. Commun. 15, 1232 (2024).
Sun, C. et al. Mouse early extra-embryonic lineages activate compensatory endocytosis in response to poor maternal nutrition. Development 141, 1140–1150 (2014).
Watkins, A. J. et al. Maternal nutrition modifies trophoblast giant cell phenotype and fetal growth in mice. Reproduction 149, 563–575 (2015).
Kwong, W. Y., Wild, A. E., Roberts, P., Willis, A. C. & Fleming, T. P. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development 127, 4195–4202 (2000).
Watkins, A. J. et al. Adaptive responses by mouse early embryos to maternal diet protect fetal growth but predispose to adult onset disease. Biol. Reprod. 78, 299–306 (2008).
Watkins, A. J. et al. Low protein diet fed exclusively during mouse oocyte maturation leads to behavioural and cardiovascular abnormalities in offspring. J. Physiol. 586, 2231–2244 (2008).
Watkins, A. J. et al. Maternal low-protein diet during mouse pre-implantation development induces vascular dysfunction and altered renin-angiotensin-system homeostasis in the offspring. Br. J. Nutr. 103, 1762–1770 (2010).
Watkins, A. J., Lucas, E. S., Wilkins, A., Cagampang, F. R. & Fleming, T. P. Maternal periconceptional and gestational low protein diet affects mouse offspring growth, cardiovascular and adipose phenotype at 1 year of age. PLoS ONE. 6, e28745 (2011).
Lo, H., Weng, S. F. & Tsai, E. M. Neurodevelopmental disorders in offspring conceived via in vitro fertilization vs intracytoplasmic sperm injection. JAMA Netw. Open 5, e2248141 (2022).
Zhang, Y., Wang, Q., Wang, H. & Duan, E. Uterine fluid in pregnancy: a biological and clinical outlook. Trends Mol. Med. 23, 604–614 (2017).
Kelleher, A. M., Burns, G. W., Behura, S., Wu, G. & Spencer, T. E. Uterine glands impact uterine receptivity, luminal fluid homeostasis and blastocyst implantation. Sci. Rep. 6, 38078 (2016).
Kelleher, A. M. et al. Forkhead box a2 (FOXA2) is essential for uterine function and fertility. Proc. Natl. Acad. Sci. USA 114, E1018–E1026 (2017).
Kelleher, A. M., DeMayo, F. J. & Spencer, T. E. Uterine glands: developmental biology and functional roles in pregnancy. Endocr. Rev. 40, 1424–1445 (2019).
Moraes, J. G. N. et al. Uterine influences on conceptus development in fertility-classified animals. Proc. Natl. Acad. Sci. USA 115, E1749–E1758 (2018).
Dhakal, P., Kelleher, A. M., Behura, S. K. & Spencer, T. E. Sexually dimorphic effects of forkhead box a2 (FOXA2) and uterine glands on decidualization and fetoplacental development. Proc. Natl. Acad. Sci. USA 117, 23952–23959 (2020).
Leese, H. J. The formation and function of oviduct fluid. J. Reprod. Fertil. 82, 843–856 (1988).
Iritani, A., Nishikawa, Y., Gomes, W. R. & VanDemark, N. L. Secretion rates and chemical composition of oviduct and uterine fluids in rabbits. J. Anim. Sci. 33, 829–835 (1971).
von Grothusen, C. et al. Uterine fluid microRNAs are dysregulated in women with recurrent implantation failure. Hum. Reprod. 37, 734–746 (2022).
Vilella, F. et al. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 142, 3210–3221 (2015).
Ruiz-Alonso, M. et al. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil. Steril. 100, 818–824 (2013).
Shi, J. et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol. 23, 424–436 (2021).
Shi, J. et al. Optimized identification and characterization of small RNAs with PANDORA-seq. Nat. Protoc. https://doi.org/10.1038/s41596-025-01158-4 (2025).
Chen, Q. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 (2016).
Milligan, S. R. & Martin, L. The resistance of the mouse uterine lumen to flushing and possible contamination of samples by plasma and interstitial fluid. J. Reprod. Fertil. 71, 81–87 (1984).
Sharma, U. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 (2016).
Wang, X. Y. et al. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313 (2018).
Zhang, Y. et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 20, 535–540 (2018).
Filant, J., Zhou, H. & Spencer, T. E. Progesterone inhibits uterine gland development in the neonatal mouse uterus. Biol. Reprod. 86, 146–149 (2012).
Beier, H. M. Oviducal and uterine fluids. J. Reprod. Fertil. 37, 221–237 (1974).
Burns, G. W., Brooks, K. E. & Spencer, T. E. Extracellular vesicles originate from the conceptus and uterus during early pregnancy in sheep. Biol. Reprod. 94, 56 (2016).
Simon, C. et al. Extracellular vesicles in human reproduction in health and disease. Endocr. Rev. 39, 292–332 (2018).
Nguyen, H. P., Simpson, R. J., Salamonsen, L. A. & Greening, D. W. Extracellular vesicles in the intrauterine environment: challenges and potential functions. Biol. Reprod. 95, 109 (2016).
Grasso, A. et al. Endometrial liquid biopsy provides a miRNA roadmap of the secretory phase of the human endometrium. J. Clin. Endocrinol. Metab. 105, 877–889 (2020).
Simintiras, C. A. et al. Capture and metabolomic analysis of the human endometrial epithelial organoid secretome. Proc. Natl. Acad. Sci. USA 118, e2026804118 (2021).
Simintiras, C. A., Drum, J. N., Liu, H., Sofia Ortega, M. & Spencer, T. E. Uterine lumen fluid is metabolically semi-autonomous. Commun. Biol. 5, 191 (2022).
Gualtieri, R. et al. In vitro culture of mammalian embryos: is there room for improvement? Cells 13, 996 (2024).
Sjoblom, C., Roberts, C. T., Wikland, M. & Robertson, S. A. Granulocyte-macrophage colony-stimulating factor alleviates adverse consequences of embryo culture on fetal growth trajectory and placental morphogenesis. Endocrinology 146, 2142–2153 (2005).
Fernandez-Gonzalez, R. et al. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc. Natl. Acad. Sci. USA 101, 5880–5885 (2004).
Calle, A. et al. Long-term and transgenerational effects of in vitro culture on mouse embryos. Theriogenology 77, 785–793 (2012).
Lee, S. R. & Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 280, 42744–42749 (2005).
Thompson, D. M., Lu, C., Green, P. J. & Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103 (2008).
Chen, Q., Zhang, X., Shi, J., Yan, M. & Zhou, T. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem. Sci. 46, 790–804 (2021).
Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216 e1226 (2018).
Yue, T. et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science 372, eaba4220 (2021).
Balatti, V. et al. tsRNA signatures in cancer. Proc. Natl. Acad. Sci. USA 114, 8071–8076 (2017).
Zhang, Y. et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 6, 172–174 (2014).
Wang, P. et al. Profiling the metabolome of uterine fluid for early detection of ovarian cancer. Cell Rep. Med. 4, 101061 (2023).
Ametzazurra, A. et al. Endometrial fluid is a specific and non-invasive biological sample for protein biomarker identification in endometriosis. Hum. Reprod. 24, 954–965 (2009).
Tomar, A. et al. Epigenetic inheritance of diet-induced and sperm-borne mitochondrial RNAs. Nature 630, 720–727 (2024).
Watkins, A. J. et al. Paternal diet programs offspring health through sperm- and seminal plasma-specific pathways in mice. Proc. Natl. Acad. Sci. USA 115, 10064–10069 (2018).
Grosso, J. B. et al. Levels of seminal tRNA-derived fragments from normozoospermic men correlate with the success rate of ART. Mol. Hum. Reprod. 27, gaab017 (2021).
Shi, J. C., Zhang, Y. F., Zhou, T. & Chen, Q. tsRNAs: the Swiss army knife for translational regulation. Trends Biochem. Sci. 44, 185–189 (2019).
Su, Z. L., Wilson, B., Kumar, P. & Dutta, A. Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu Rev. Genet. 54, 47–69 (2020).
Maute, R. L. et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 110, 1404–1409 (2013).
Kuscu, C. et al. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105 (2018).
Chen, Q. & Zhou, T. Emerging functional principles of tRNA-derived small RNAs and other regulatory small RNAs. J. Biol. Chem. 299, 105225 (2023).
Lane, M., Robker, R. L. & Robertson, S. A. Parenting from before conception. Science 345, 756–760 (2014).
Trigg, N. et al. Subchronic elevation in ambient temperature drives alterations to the sperm epigenome and accelerates early embryonic development in mice. Proc. Natl. Acad. Sci. USA 121, e2409790121 (2024).
Kermack, A. J. et al. Amino acid composition of human uterine fluid: association with age, lifestyle and gynaecological pathology. Hum. Reprod. 30, 917–924 (2015).
Skilton, M. R., Evans, N., Griffiths, K. A., Harmer, J. A. & Celermajer, D. S. Aortic wall thickness in newborns with intrauterine growth restriction. Lancet 365, 1484–1486 (2005).
Xiao, X. et al. Low birth weight is associated with components of the metabolic syndrome. Metabolism 59, 1282–1286 (2010).
Hack, M., Klein, N. K. & Taylor, H. G. Long-term developmental outcomes of low birth weight infants. Future Child 5, 176–196 (1995).
Zhang, L. et al. RNA modification signature of peripheral blood as a potential diagnostic marker for pulmonary hypertension. Hypertension 79, e67–e69 (2022).
Zhang, L. et al. Peripheral blood RNA modifications as a novel diagnostic signature for polycystic ovary syndrome. Sci China Life Sci. https://doi.org/10.1007/s11427-024-2913-7 (2025).
Shi, J., Ko, E. A., Sanders, K. M., Chen, Q. & Zhou, T. SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA- and tRNA-derived small RNAs. Genom. Proteom. Bioinform. 16, 144–151 (2018).
Acknowledgements
This work was funded by Natural Science Foundation of China (92357306, 82122027 and 32171110 to Y.Z.), National Key Research and Development Program of China (2019YFA0802600 to Y.Z.), Shandong Province Key Research and Development Program (Major Science and Technology Innovation Projects of Shandong Province) (2023CXGC010505 to L.Y.), the Fundamental Research Funds for the Central Universities (2253200027 to Y.Z.). The authors thank Dr. Chao Xi, Jin Liu and Xi Jin for their technical and timely help. The authors also thank the Experimental Technology Center for Life Sciences of Beijing Normal University and Beijing Normal University Super-Computing Center for supporting this study.
Author information
Authors and Affiliations
Contributions
Y.Z., S.P. and L.Z. designed the project. S.P. and L.Z. performed the experiments under the supervision of Y.Z. S.P. did early embryo transfection experiment with the help from X.Y. S.P. and L.Z. collected UF and OF with the help from L.W. and C.L. S.P. and L.Z. checked the mouse pregnancy phenotypes with the help from X.Y., S.Q. and Y.Q. S.P. detected UF and OF sncRNAs with help from L.T. S.P., X.L. and L.Z. performed the data analyses with the help from J.Z. and R.Z. under the supervision of J.S. Y.Z., S.P., L.Y. and L.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Ling-Ling Zheng who co reviewed with Xiao Feng; Raffaele Teperino, Adam Watkins and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pan, S., Zhang, L., Yang, X. et al. Maternal diet-induced alterations in uterine fluid sncRNAs compromise preimplantation embryo development and offspring metabolic health. Nat Commun 16, 7637 (2025). https://doi.org/10.1038/s41467-025-63054-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63054-5