Abstract
PROTEOLYSIS1 (PRT1), an N-recognin of Arabidopsis thaliana, recognizes the N-terminal aromatic hydrophobic residue (Tyr/Phe/Trp) of its substrates and ubiquitylates them for degradation by the ubiquitin-proteasome system. Herein, we report the structures of the ZZ domain of PRT1 (PRT1ZZ) in complex with bulky hydrophobic N-degron peptides. Unlike other ZZ domains, PRT1ZZ has an unusual binding site with two hydrophobic regions. The N-terminal aromatic residues of N-degrons interact with Ile333 and Phe352 in the flexible loops, which undergo a conformational change. Notably, we identify a third residue from the N-terminus of the substrate that participates in the hydrophobic network with PRT1ZZ. Moreover, AlphaFold prediction and biochemical assays revealed that the tandem RING1 and RING2 domains of PRT1 interact intramolecularly. The dimeric RING domains in a single protein represent a unique feature among the RING-type E3 ligases. The biochemical assays using the N-terminal tyrosine-exposed substrate, BIG BROTHER, show that the intramolecular RING dimer is essential for PRT1’s robust activity. Therefore, this study expands our knowledge of the structural repertoire in the N-degron pathway and provides insights into the regulation of E3 ligases containing tandem RING domains.
Similar content being viewed by others
Introduction
The ubiquitin-proteasome system regulates protein homeostasis in all species, including plants. The polyubiquitin (poly-Ub) chain acts as a degradation marker by delivering ubiquitylated substrates to the proteasome1,2. E3 Ub ligases generate a poly-Ub chain coupled with E1 Ub activating and E2 Ub conjugating enzymes1,3,4. Through this mechanism, the half-life of various substrates is determined by the characteristics of their N-terminal (Nt) residues, which are specifically recognized by a class of E3 Ub ligases known as “N-recognins”5,6,7. In this case, the Nt residues function as a degradational signal known as “N-degrons”7. Since the first N-recognin Ubr1 was characterized8, many N-degron pathways such as those of Arg/N-degron, Ac/N-degron, Pro/N-degron, Gly/N-degron, and fMet/N-degron have been extensively studied6,9,10,11,12,13,14,15,16. The Arg/N-degron pathway was the first to be characterized, targeting at least type-1 (positively charged Nt residues, Arg/Lys/His) and type-2 (hydrophobic Nt residues, Leu/Ile/Phe/Tyr/Trp) substrates7,17. The first and best-characterized N-recognin is yeast Ubr1, which consists of a UBR box and a ClpS-homology domain for recognizing type-1 and type-2 substrates, respectively8,18,19,20,21,22.
Yeast Ubr1 N-recognin shares sequence and functional homology with mammalian UBR1/2 (Ubiquitin Protein Ligase E3 Component N-Recognin 1/2), which also contain the UBR box and ClpS-homology domain18,22. However, the closest homolog in plants is the PROTEOLYSIS6 (PRT6), which only has a UBR box23. No other defined N-recognin ortholog containing the ClpS-homology domain has been identified in plants. However, an E3 ligase known as “PROTEOLYSIS1” (PRT1), which had been isolated from Arabidopsis thaliana, has been defined as an N-recognin recognizing type-2 hydrophobic aromatic Arg/N-degrons24,25. BIG BROTHER (BB), a RING-type E3 Ub ligase that controls organ size in A. thaliana, has been identified as a substrate of PRT126. Although BB controls plant cell proliferation, the downstream regulator remains unknown. BB is negatively regulated by the endoprotease DA1 from A. thaliana (DA1), which cleaves the peptide bond between the 60th Ala residue and the 61st Tyr residue of BB27. As a consequence, the Tyr residue, a neo-Nt residue in BB, exposes the type-2 N-degron. Notably, PRT1 from A. thaliana (AtPRT1) interacts with the cleaved BB (hereafter Tyr61-BB) through the bulky hydrophobic N-degrons and ubiquitylates it24,25,28.
The recognition mechanism of bulky hydrophobic residues by PRT1 is not clearly understood because PRT1 consists of tandem RING domains, a pair of canonical (RING1) and non-canonical (RING2) RING domains, and a C-terminal ZZ domain, instead of the ClpS-homology domain typically associated with type-2 N-degron recognition (Supplementary Fig. 1a)25,28. As noted above, PRT1 lacks conventional N-degron recognition domains. The ZZ domain was presumably predicted to be the recognition domain of bulky hydrophobic N-degrons, particularly after the ZZ domain of p62/SQSTM1 interacts with type-1 and -2 N-degrons29,30. Here, we specify the ZZ domain of PRT1 (PRT1ZZ) as an N-degron-binding region and describe the high-resolution structures of PRT1ZZ in complex with type-2 bulky hydrophobic N-degrons. Biochemical and mutational analyses confirm the key interaction determinants between N-recognin PRT1 and the N-degron substrate Tyr61-BB. We also compare the structure of PRT1ZZ with those of other ZZ domain proteins29,31,32,33,34,35. Furthermore, we find that the tandem RING organization in PRT1 is critical for E3 ubiquitylation activity, as verified using the N-degron substrate Tyr61-BB. These results reveal how the plant ZZ domain recognizes bulky hydrophobic N-degron substrates and regulates the ubiquitylation activity of the tandem RING structures in PRT1. Hence, this study expands our knowledge of the structural repertoire of the N-degron pathway, particularly within plant systems, and provides insights into the regulation of E3 Ub ligases with tandem RING domains.
Results
N-degron recognition by the ZZ domain of AtPRT1
AtPRT1ZZ is responsible for recognizing type-2 bulky hydrophobic N-degrons24,25,28; however, the binding affinity between PRT1 and N-degrons has not been quantitatively analyzed. First, we measured the dissociation constant (KD) between PRT1ZZ and a Tyr N-degron peptide (Tyr-Lys-Phe) using isothermal titration calorimetry (ITC), yielding a KD value of 10.1 μM (Supplementary Fig. 1b). However, no binding affinity was detected between ZZ domain-deleted RING domains (RING1 + RING2) and the same N-degron peptide (Supplementary Fig. 1c). Thus, these results show that type-2 N-degrons are solely recognized by PRT1ZZ, with dissociation constants in the micromolecular range, depending on the Nt residue.
Crystal structure of PRT1ZZ
The structure of PRT1ZZ (residues 303–366) was determined using the single-wavelength anomalous dispersion (SAD) method at the Zn absorption edge (Supplementary Table 1). PRT1ZZ has a canonical “Zig-Zag fold”36, comprising three β-strands and an α-helix (Fig. 1a–c). The ZZ domain has two conserved Asp residues (Asp312 and Asp336) that belong to the negatively charged patch (Fig. 1b), which recognizes the α-NH3+ group of N-degrons. The overall structure of PRT1ZZ has a highly conserved folding pattern that coordinates two Zn2+. The first Zn2+ (Zn1) is coordinated by four Cys residues, and the second Zn+2 (Zn2) by two Cys and two His residues (Fig. 1d). In this arrangement, the Zn2+ coordinating Cys residues form the CxxC motif (“x” represents any amino acid residue). However, the number of amino acid residues between Zn-coordinating His residues differs from that of other ZZ domains29,31,32,33,34,35. Notably, PRT1ZZ features two long loops, Loop 2 (L2) connecting β1 and β2 strands and Loop 3 (L3) connecting the α1 helix and β3 strand (Fig. 1a, c), which are characteristic features compared with other ZZ domains. Because of the flexibility of these loops, building models at these positions is challenging. The three protruding residues (Tyr317, Ile333, and Phe352) around the negatively charged patch potentially participate in N-degron recognition. However, these hydrophobic residues are far from each other in the apo structure of the ZZ domain (Fig. 1a).
a Structure of the apo-form of PRT1ZZ in a ribbon diagram. Three loops (1: red, 2: purple, and 3: green) are indicated and labeled. Key residues for N-degron recognition are drawn using a stick model with labels. N* and C* denote the first and last residues of the construct. Bound zinc atoms (Zn1 and Zn2) are shown as gray balls. The estimated position of F352 is represented by the transparent stick model (invisible phenyl side chain in apo structure). b Transparent molecular surface of apo-form ZZ domain representing electrostatic potential. Red and blue colors represent negatively and positively charged areas, respectively. The N-degron binding pocket is highlighted as a red circle. c Topology diagram of PRT1ZZ. The ZZ domain of PRT1 has a conserved ZZ domain topology. The construction comprises three β-sheets and one α-helix. The loops (L1–L3) containing the key residues are represented with the same colors as those in (a). d Schematic diagram showing Zn ion coordination of PRT1ZZ. Zn1 and Zn2 ions coordinate with C4 and C2H2 Zn-finger motifs, respectively.
ZZ domain structure in complex with N-degron of Tyr61-BB
To determine the structure of the ZZ domain in complex, we fused the N-degron of Tyr61-BB (Y61-K62-F63-G64) to the N-terminus of the ZZ domain and then exposed the Tyr residue using the LC3B-fusion technique (Fig. 2a)37,38. Utilizing the Tyr-exposed PRT1ZZ protein, we determined the structure of PRT1ZZ in complex with Tyr61-BB N-degron at a resolution of 1.74 Å (Supplementary Table 1). Each N-degron attached to the ZZ domain mutually interacted with a neighboring ZZ domain in the crystal, forming a dimer in the crystalline lattice (Supplementary Fig. 2). The binding site of the ZZ domain revealed interactions between the positively charged Nt NH3+ group of Tyr61 and the side chains of Asp312 and Asp336. The main chains of the N-degron peptide formed a hydrogen-bond network with Gly308, Gly310, and Ile333 (Supplementary Figs. 3a-c). The aromatic hydrophobic side chain of the Tyr61 residue nests in the hydrophobic pocket and is formed by three key residues: Ile333, Tyr317, and Phe352 (Fig. 2b, c). In addition, it comprises two hydrophobic regions (sites 1 and 2). The Tyr317 residue is at the site 1 formed by L1, whereas the Ile333 and Phe352 residues are at the site 2 formed by L2 and L3. In the apo structure, the putative N-degron-binding region was wide open (Fig. 1b) because these loops (L2 and L3) were dynamic and flexible, corresponding to a weak electron density. However, when PRT1ZZ recognized the N-degron substrate, the electron density required for recognizing the residues and loops became clear (Supplementary Figs. 3d–f). Nt Tyr61 and the third Phe63 of the N-degron residues are snugged into the narrow pocket, and the second Lys62 stretches outside (Fig. 2c). A dramatic conformational change in which the two loops, L2 and L3, moved toward the incoming substrate as shown in the induced-fit model (Fig. 2d). To evaluate a possible conformational change affected by the crystal contact, the individual asymmetric units within the crystalline lattice were analyzed (Supplementary Fig. 4a). The loops were not involved in the crystalline packing, and regardless of the packings, all different N-degron-bound PRT1ZZ structures were almost identical, strongly supporting the conformational change derived from the complex formation. When comparing the B-factor putty representation of the apo and the ligand-bound structures, the B-factors of key residues in PRT1ZZ were notably reduced in the ligand-bound form (Supplementary Fig. 4b). Unlike the other flexible loops (L2 and L3), L1, containing Tyr317, maintained the same position with or without the N-degron (Fig. 2e). While Ile333 exhibited a relatively modest conformational change of 2.9 Å, Phe352 showed the largest conformational change of up to 9.5 Å (distance between the equivalent Cα atoms) (Fig. 2e). Following this conformational change, hydrophobic site 2 established favorable interactions with Tyr61 of Tyr61-BB.
a A. thaliana BB has a proteolytic site cleaved by DA1 protease. As a result, cleaved BB (Tyr61-BB) has an exposed tyrosine residue at the N-terminus, which is an N-degron. To mimic this N-degron, the YKFG amino acid sequence was added between LC3B and PRT1ZZ by mutagenesis. The N-degron residues (Tyr, Phe, and Trp) are exposed by cleaving LC3B using ATG4B protease. b Complex structure of PRT1ZZ with the N-degron of Tyr61-BB. Tyr317 at site 1 (red dotted box) and Ile333 and Phe352 at site 2 (blue dotted box) are key residues interacting with N-degron. c Close-up view of N-degron-binding region with a transparent electrostatic surface. d Structural superposition of N-degron-bound PRT1ZZ (cyan) and apo-form (orange). L2 and L3 loops are very flexible without bound N-degron. e Close-up view of the superposition of the N-degron-binding region. Compared with the apo-form ZZ domain, the flexible loops underwent a large conformational change upon complex formation. The movements of the residues upon complex formation are as follows: Phe352 − 9.5 Å and Ile333 − 2.9 Å.
Unique flexible loops of PRT1ZZ
L3, containing Phe352, is a unique structure exclusively present in the ZZ domain of PRT1. This long loop is conserved among PRT1 proteins from various species (Supplementary Fig. 5) but shares no similarity among the structurally determined ZZ domains (Fig. 3a). According to our sequence alignment, ZZ domains can be classified depending on the number of residues between the two His residues that coordinate the second Zn2+ (Fig. 3a). The His-x-x-x-His motif (HxxxH) has a more curved turn for coordinating Zn2+ compared with the His-x-x-His (HxxH) or His-x-His motif (HxH). When more residues exist between the two His residues coordinating Zn2+, a longer loop (L3) between the α1-helix and the last Zn-coordinating His residue (before the β3-strand) is required. This tendency is also observed in the reported ZZ domains from the PROSITE database39 (Supplementary Fig. 6). This clearly shows the correlation between the length of the loops and the type of Zn coordination (Fig. 3a). PRT1 has long loops with three more residues in L2 and an exceptionally long L3, which is five more residues among the HxxxH-type ZZ domains (Fig. 3b). Considering that the key residues responsible for establishing hydrophobic networks are positioned in these loops, these are distinctive features of PRT1. Consequently, the Phe352 side chain can reach the binding pocket and interact with the Nt hydrophobic bulky residue of the substrate. We compared the structure of PRT1ZZ with that of the p62/SQSTM1 ZZ domain, which contains an HxxH motif and a short L3 (Fig. 4a). p62/SQSTM1 has only a negatively charged patch on its shallow binding site and thus utilizes only limited interacting residues with type-1 Arg/N-degrons (R-BiP), such as Asn132 and Asp147 (Fig. 4b). Unlike p62/SQSTM1, PRT1ZZ forms a narrow and deep hydrophobic binding pocket consisting of more residues, including Tyr317, Ile333, and Phe352, to recognize its substrates (Fig. 4). PRT1 displayed a stronger affinity for its substrates than that of p62/SQSTM129,30 (Supplementary Figs. 1b and 8a).
a Sequence alignment of structurally available ZZ domains. PDB IDs and types of Zn ion coordination are indicated on the left. Conserved zinc-coordinating cysteine and histidine residues are marked with red and green boxes, respectively. The key determinants of hydrophobic type-2 N-degron in the loops are circled and labeled in red. b, c Structure superposition of PRT1ZZ with other known ZZ domain structures. Superpositions with ZZ domains possessing HxxxH-type ZZ domains are shown in (b), and HxxH and HxH-type ZZ domains are presented in c. PDB IDs: 6e86 (ZZZ3; white), 2e5r (Dystrobrevin alpha; blue), 4xi6 (Mindbomb1; orange), 6ww3 (HERC2; green), 5yp7 (p62/SQSTM1; magenta), 2dip (SWIM; gray), 1tot (CBP; salmon), and 6ds6 (p300; yellow).
a Electrostatic surface of PRT1 and p62/SQSTM1 ZZ domains (PDB IDs: PRT1 [8zg9], p62/SQSTM1 [5yp7]). A negatively charged region of PRT1ZZ (left) is enclosed by L3, while that of the p62/SQSTM1 ZZ domain (right) is exposed. b Close-up view of structural comparison. The α-NH3 group of both N-degrons (YKFG and R-BiP peptides) is recognized by two conserved Asp residues. The non-conserved key residues of the PRT1ZZ participate in the interaction with the hydrophobic bulky N-degron.
Type-2 Arg/N-degron selectivity for PRT1
To further characterize the binding mode to type-2 Arg/N-degrons, we crystallized the ZZ domain fused to FKFG and WAAG. The binding mode of the Phe N-degron was similar to that of the Tyr N-degron, whereas that of the Trp N-degron partially differed (Supplementary Fig. 2b). Specifically, the Tyr317 residue shifts approximately 2.1 Å backward to accommodate the bulky side chain of Trp into the narrow binding pocket (Fig. 5a). These bulky hydrophobic Nt residues reached the key PRT1ZZ residues—Tyr317, Ile333, and Phe352—within a distance of 4 Å (Fig. 5b). Type-1 Arg/N-degrons (e.g., Arg) cannot interact with key residues because of their positive charge. Similarly, non-bulky hydrophobic residues (such as Leu and Ile) were too short to be in contact with these key residues (Supplementary Fig. 7a). Therefore, our inference from the structure was that only the type-2 bulky Arg/N-degrons fit PRT1ZZ. This coincides with the affinity measurements obtained by ITC experiments with N-degron peptides containing x1AA sequences (x1; Tyr, Phe, Trp, Leu, Ile, and Arg). The ZZ domain showed a similar binding affinity range to the exposed Nt hydrophobic bulky residues, regardless of the type (Fig. 5c). Conversely, non-bulky hydrophobic residues and type-1 Arg/N-degrons showed no affinity for the ZZ domain (Supplementary Fig. 7). These non-bulky and polar residues may fail to induce conformational changes in Phe352 and L3 after docking the N-degron into the ZZ domain.
a Superposition of ZZ domain complexes with peptides, YKFG (colored cyan for protein and white for peptide), FKFG (magenta for protein and slate for peptide), and WAAG (green for protein and salmon for peptide). b Close-up view of the binding mode of the ZZ domain with Nt Tyr, Phe, and Trp residues. Type-2 bulky hydrophobic N-degrons are positioned near the key residues of the ZZ domain. The hydrophobic network is highlighted as a transparent brownish oval. The colors are consistent with those in (a). c Isothermal calorimetry results of the FL PRT1WT with type-2 bulky hydrophobic Arg/N-degrons. KD values: YAA = 23.3 ± 0.958 μM; FAA = 5.45 ± 0.212 μM; WAA = 2.85 ± 0.164 μM. The detailed parameters are listed in Supplementary Table 3.
Hydrophobic network between the PRT1ZZ and N-degron
Tyr317 and Ile333 also established hydrophobic interactions with the third residue of Tyr61-BB, Phe63. Because of these interactions, the aromatic ring of the Phe63 residue covered the outside of the binding site, similar to a roof (Fig. 5a). To investigate the contribution of each residue to the N-degron sequence, we measured the binding affinity between full-length (FL) PRT1 wild-type (WT) and various Tyr-degron peptides (YKF, YKA, YAF, and YAA) using the ITC approach (Supplementary Figs. 8a–c and Fig. 5c). The hydrophobic interaction between the third N-degron residue and PRT1WT was not critical; however, the presence of a bulky aromatic ring was partially preferred. This preference agreed with the ITC data obtained using F-peptides. When comparing the binding affinities of FKF and FKA peptides, FKF showed a stronger affinity with FL PRT1 than FKA (Supplementary Figs. 8d, e). Unfortunately, due to its poor solubility in the buffer, the binding affinity of FAF peptide could not be measured.
However, the third Phe of N-degron peptide significantly influenced the binding affinity with the FL PRT1 mutants (Y317A, I333A, and Y317A/I333A). According to the binding affinity measurement, the YKA peptide was not bound to the PRT1Y317A and PRT1I333A mutants (Fig. 6a). In contrast, the YKF peptide maintained its binding affinity with the PRT1Y317A and PRT1I333A mutants at levels similar to those of PRT1WT (Fig. 6b). Moreover, the PRT1Y317A/I333A double mutant of PRT1 showed a considerably low affinity for the YKF peptide (Fig. 6b). These results show that the Tyr317 and Ile333 residues of PRT1 and Phe63 of Tyr61-BB form a hydrophobic network to stabilize the N-degron in the binding pocket of PRT1ZZ. As a result, the Tyr317 and Ile333 residues support positioning hydrophobic N-degrons and work in concert with Phe352, the primary residue for recognizing the type-2 bulky hydrophobic Nt residue. Next, we investigated whether the presence of a third Phe residue forms a hydrophobic network with non-bulky hydrophobic N-degrons, such as Leu or Ile. We measured the binding affinity of the LKF peptide and found no detectable affinity (Supplementary Fig. 7b). Non-bulky hydrophobic residues were insufficient to establish a hydrophobic network. These results indicate that Phe352 of PRT1 is the primary determinant of the interaction with the type-2 bulky hydrophobic Nt residue of N-degrons. Moreover, additional Tyr317 and Ile333 residues are essential for establishing the hydrophobic network, which prefers a bulky hydrophobic residue at the third position of the N-degron (Figs. 5 and 6).
a Isothermal titration calorimetry (ITC) results of PRT1 mutants (PRT1Y317A and PRT1I333A) with the YKA peptide. No affinity or very weak affinity were determined. b ITC results of PRT1 mutants (PRT1Y317A, PRT1I333A, PRT1Y317A/I333A, and PRT1F352A) with the YKF peptide. KD values: PRT1Y317A = 41.3 ± 0.799 μM; PRT1I333A = 13.7 ± 1.68 μM; PRT1Y317A/I333A = N.D. (not determined); PRT1F352A = N.D. The detailed parameters are listed in Supplementary Table 3. c Ubiquitylation activity of FL PRT1 depending on reaction time (each lane at 0, 0.5, 1, 3, 5, and 7 h). The poly-Ub pattern of Tyr61-BBV198E was detected in the presence (left) and absence (right) of PRT1. See Supplementary Fig. 9 for the details of the diminished auto-ubiquitylation activity of the Tyr61-BBV198E. d Ubiquitylation activity of the PRT1 mutants with an authentic Nt sequence-containing substrate, YKF-BBV198E. Ctrl: no PRT1 protein. e Ubiquitylation activity of the PRT1 mutants with an Nt permutated substrate, YAA-BBV198E (alanine at the second and third position). The key residue mutants of PRT1 showed reduced or no ubiquitylation activity compared with that of the WT. These loss-of-function effects were more reinforced when the N-terminus of Tyr61-BBV198E was permutated to YAA. Ctrl: no PRT1 protein. The ubiquitylation reactions are quenched at 3 h. All images probed with anti-His Ab were detected under intensive conditions (2.5× loading volume and longer exposure time) compared to those probed with anti-FLAG or anti-Ub. The original western (same loading volume and exposure intensity) images with anti-His Ab are shown in Supplementary Fig. 10b. These ubiquitylation assays were performed at least three times.
A ubiquitylation-defective mutant of Tyr61-BB substrate
To assess the relationship between the binding and ubiquitylation of N-degron substrates by PRT1, we performed in vitro ubiquitylation assays using the Tyr61-BB protein. However, BB is also an E3 Ub ligase with a RING domain at its C-terminus and in vitro auto-ubiquitylation activity, resulting in robust poly-Ub chain formation (Supplementary Figs. 9a, b). Therefore, we designed a ubiquitylation-defective Tyr61-BB mutant by changing the two Val residues of the E2~Ub binding motif to Glu residues, where “~” denotes the covalent binding (V198E and V236E)40. These Tyr61-BB mutants showed diminished auto-ubiquitylation activity originated from BB compared with that of the BBWT, and the BBV198E mutant was more effective in reducing this activity than BBV236E (Supplementary Fig. 9a). Although the V198E mutant of Tyr61-BB (Tyr61-BBV198E) showed low auto-ubiquitylation activity, it showed a poly-ubiquitylation pattern in the presence of PRT1WT (Fig. 6c). Therefore, it is a useful substrate for evaluating the ubiquitylation activity of PRT1WT and its mutants.
Ubiquitylation activity of PRT1 and its mutants
Although PRT1 also exhibited minimal in vitro auto-ubiquitylation activity, regardless of the presence or absence of the substrate (Fig. 6c and Supplementary Fig. 10a), the Tyr61-BBV198E mutant substrate was used to investigate the poly-ubiquitylation activity of the PRT1 E3 Ub ligase. The input amounts of the PRT1WT and its variants were relatively lower than those of other components, making the detection of their bands difficult under standard conditions (Supplementary Fig. 10b). Therefore, each corresponding sample was loaded at a 2.5-fold greater volume and detected under intensive exposure conditions to improve visibility (His-tag Ab detection). To confirm the relationship between in vitro binding affinity and the enzymatic activity of PRT1, we performed a ubiquitylation assay using PRT1WT and various mutants (Fig. 6d). A similar tendency in the ubiquitylation pattern was observed, consistent with the binding affinity. The PRT1WT showed the most robust poly-Ub band, and the PRT1Y317A and PRT1I333A mutants and PRT1Y317A/I333A double mutant showed decreased ubiquitylation activity compared with that of the WT (Fig. 6d). The Tyr317 residue of PRT1 is more crucial than Ile333 for binding to the substrate, which can be expected in the complex structure (Supplementary Fig. 3). This was further confirmed by the ubiquitylation activity of the mutants (Fig. 6d). Ubiquitylation activity was completely abolished in the PRT1F352A mutant, confirming that Phe352 is the primary determinant for recognizing the hydrophobic Nt residue (Fig. 6d). A similar pattern was also observed in ubiquitylation assay using the Tyr-CP8GFP model substrate41 (Supplementary Fig. 11). Since PRT1WT and its variants were auto-ubiquitylated in vitro (Fig. 6c), the western blotting was performed at initial time point to compare input amount of PRT1WT and its variants (Fig. 6d, e and Supplementary Fig. 11).
To verify the effect of the third N-degron residue, Phe63, we performed an in vitro ubiquitylation assay with Tyr61-BBV198E, in which the residue in the N-degron was changed to an Ala residue (YAA-BBV198E) using the same batch of PRT1WT and its mutants (Fig. 6e). In all cases, the YAA-BBV198E substrate showed diminished ubiquitylation activity compared with that of Tyr61-BBV198E (YKF-BBV198E), which has the original N-degron (Fig. 6d, e). Again, the PRT1Y317A mutant showed lower ubiquitylation activity than the PRT1I333A mutant. The disruption of the hydrophobic network by mutating the hydrophobic residues, Tyr317 and/or Ile333, dramatically decreased the ubiquitylation of YAA-BBV198E more than YKF-BBV198E (Fig. 6d, e). These results indicate that the key hydrophobic residues in the binding pocket of PRT1 exert differential effects in constructing the hydrophobic network and fine-tuning the substrates, depending on the third position of the N-degron sequence.
Overall shape of PRT1 and characteristics of tandem RING domains
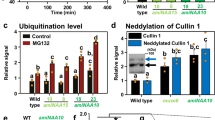
Although PRT1 has tandem RING domains (RING1 and RING2), RING1 is essential for the ubiquitylation of the substrate (Supplementary Fig. 12). To gain insights into the role of the RING2 domain, the FL PRT1 structure was predicted using AlphaFold (AF)42,43. The AF model exhibited a compact structure with two RINGs and one ZZ domain (Supplementary Figs. 13a, b). However, the AF model contradicts the ‘L’ shaped molecular envelope of FL PRT1 of the small-angle X-ray scattering (SAXS) method (Fig. 7a and Supplementary Fig. 13c). As expected, FL PRT1 exhibited a monomeric state confirmed by both SEC-MALS (Supplementary Fig. 14a) and SEC-SAXS analyses (Supplementary Table 2). Therefore, we dissected the monomeric FL PRT1 into RING and ZZ domains, and investigated these interactions using separate domains. The C-terminal ZZ domain behaved as an independent domain in solution, based on individual migration in size-exclusion chromatography (Supplementary Fig. 14b), as expected from the SAXS envelope model (Fig. 7a). Meanwhile, the AF model of RING1 + 2 showed an intramolecular heterodimer due to the hydrophobic network of their four Leu residues (Leu79 and Leu82 in RING1 and Leu246 and Leu249 in RING2). Similar dimer formation was observed in other intermolecular homo- and heterodimeric RING–RING structures (Fig. 7b and Supplementary Fig. 15)44,45,46,47,48,49,50,51,52. As expected, the two RING domains co-migrated in size-exclusion chromatography, suggesting intramolecular heterodimeric formation between RING1 and RING2 (Fig. 7c, d and Supplementary Fig. 14c). RING dimerization reportedly is crucial in ubiquitylation activity44,49,53,54. Therefore, we verified whether the PRT1 mutant that disrupted the RING intramolecular heterodimer showed significantly reduced ubiquitylation activity (Fig. 7e). The core hydrophobic leucine residues were replaced with hydrophilic serine residues (2 L/S—one of the RINGs: L79S/L82S in RING1 and L246S/L249S in RING2; 4 L/S—both of the RINGs: L79S/L82S/L246S/L249S) at the dimeric interface. Unlike the RING2WT, the RING22L/S mutant did not co-migrate with RING1WT (Fig. 7c, d). Moreover, RING2WT acted as a dimer in solution, whereas RING22L/S behaved as a monomer (Fig. 7c and Supplementary Fig. 14c). Therefore, it is clear that the RING1 and RING2 of PRT1 form an intramolecular dimer. As expected, the RING dimer-disrupted mutant, PRT14L/S exhibited little ubiquitylation activity (Fig. 7e). In addition, to assess the loss of function in the PRT14L/S mutant, we performed FRET-based Ub discharge assay. As a result, the PRT14L/S mutant showed 10-fold lower activity compared to PRT1WT (Fig. 7f). These results indicate that while disruption of the intramolecular RING1+2 dimer organization substantially reduces Ub discharge activity, it does not completely abolish it. These findings support that the correct orientation of the dimeric RING organization is critical for full E3 ligase activity.
a SEC-SAXS envelope model of PRT1. PRT1 has an L-shaped structure. The upper part of the model corresponds to the C-terminus, where the ZZ domain is located. The dimeric RING domain is positioned at the curved body. The ZZ domain is colored cyan, and the AlphaFold2 model of the dimeric RING domain is colored blue (RING1) and salmon (RING2) in the atomic models fitted into the SAXS molecular envelope. b RING1 and RING2 heterodimer formation. This model was obtained using AlphaFold, and the mutated residues (4 L/S: L79S/L82S in RING1 and L246S/L249S in RING2) for disrupting dimers are shown as yellow sticks. c Peak migration profile for characterizing tandem RING domains. RING2WT co-migrates with RING1WT, whereas RING22L/S (L246S/L249S double mutant) behaves independently. Both species in peak 1 are too low a concentration to calculate experimental molecular weights. The detailed SEC-MALS analysis for peak 2 is shown in Supplementary Fig. 14c. UV absorption was measured at a 280-nm wavelength. The results for the RING1WT + RING2WT mixture are represented by a yellow line, and those for RING1WT + RING22L/S by a blue line. Schematic oligomeric states of RING domains are shown below the UV profile. d Corresponding gel images for the fractions in (c). RING1WT and RING2WT are shown at 20 and 10 kDa, respectively. We observed consistent results at least three times. e Ubiquitylation assay of PRT1WT and 4 L/S mutant. When the dimeric interface of RING domains was disrupted, the ubiquitylation activity of PRT1 was lost. Each of the samples was incubated at 30 °C for 3 h. Ctrl: No PRT1. The ubiquitylation assays were performed at least three times. f FRET-based Ub discharge assay. The E2~Ub discharge activity of the 4 L/S mutant was decreased by 10-fold compared to the WT. The normalized FRET data was analyzed by one-phase decay analysis with GraphPad Prism 5. The data are represented by mean ± SEM from three independent experiments.
Discussion
Well-known N-recognins, such as those of the UBR family, p62/SQSTM1, and ClpS, can recognize corresponding N-degron substrates using their unique domains, such as the UBR box and ZZ and ClpS domains, to bind with N-degrons. In contrast to UBR proteins in yeast and mammals, no defined N-recognin ortholog containing a ClpS-homology domain for recognizing type-2 N-degron has been identified in plants23. However, PRT1 is a well-known N-recognin that has been reported to ubiquitylate the F-DHFR model5 and Tyr61-BB physiological substrates27. PRT1 has a ZZ domain that explicitly recognizes only type-2 Arg/N-degrons, particularly the bulky hydrophobic residues, whereas the ZZ domain of p62/SQSTM1 prefers positively charged type-1 Arg/N-degrons29,55. Therefore, the mechanism of type-2 N-degron recognition by PRT1ZZ is a long-standing mystery in plant Ub biology. Furthermore, the N-degron substrate specificity of the ZZ domain is notable. Our structural and biochemical studies characterized the unique features of PRT1ZZ in comparison to other ZZ domains and identified the key residues that participate in recognizing type-2 bulky hydrophobic Arg/N-degrons. In contrast to the N-degron recognition domains in the N-recognin E3 ligases, PRT1ZZ underwent dramatic conformational changes upon N-degron complex formation (Fig. 8a). The key residues, Tyr317, Ile333, and Phe352, form hydrophobic networks not only with the first residue of the N-degrons but also with the third Phe. These hydrophobic residues in PRT1 are at the three loops (L1, L2, and L3). These dynamic loops, especially L2 and L3, are long enough to participate in the hydrophobic network, which is a unique feature of PRT1ZZ relative to the p62/SQSTM1 ZZ domain for recognizing basic type-1 N-degrons as well as other ZZ domains, as well as for binding to proteins involved in various cellular processes such as transcriptional regulation, chromatin remodeling, and ubiquitylation29,31,32,33,34,35.
a Conformational change of two PRT1ZZ loops, L2 and L3, upon complex formation with the N-degron substrate, while L1 maintains position as a rigid frame. b Active RING1-RING2 heterodimer interacts with the UBC8 E2 Ub-conjugating enzyme and Ub, which forms a competent ternary complex for ubiquitylation. The ZZ domain at the C-terminal region of PRT1 grabs the Tyr61-BB substrate through its bulky hydrophobic N-degron (Nt tyrosine). The flexibility between ZZ and RING domains enables the efficient delivery of the substrate to RING1, which interacts with the E2 enzyme and Ub. The red star represents the ubiquitylation reaction, and several rounds of reaction generate the poly-ubiquitylated Tyr61-BB.
Bulky hydrophobic N-degrons are sufficient for maintaining the hydrophobic network at the binding site; however, small hydrophobic and charged N-degrons are unlikely to do so. The hydrophobic networks between PRT1ZZ and type-2 N-degrons significantly affected ubiquitylation activity. Next, we investigated how PRT1 ubiquitylates recognized type-2 N-degron substrates. Based on our SAXS data of PRT1, an overall ‘L’ shape was observed, where the heterodimeric RING domains are at the bottom and the ZZ domain is at the top (Fig. 7a). RING1 + 2 in PRT1 appears to have an active RING dimer conformation, as seen in other RING-type E3 Ub ligases44,45,46,47,48,49,50,51,52. Various RING homodimerizations activate E3 ubiquitylation, and provide several examples of intermolecular RING heterodimerization (Supplementary Fig. 15). In the case of PRT1, intramolecular RING heterodimerization may occur based on AF2 prediction; more importantly, the disruption mutant for dimerization abolishes ubiquitylation activity. Therefore, we speculate that monomeric PRT1 becomes an active E3 ligase via its unique tandem RING domain structure. Notably, the functional RING domains and N-degron recognizing the ZZ domain were far from each other (Fig. 8b). Although a dynamic structural change in PRT1 must precede the ubiquitylation reaction, the PRT1 substrate, Tyr61-BB, must have a stretched shape that allows it to interact with the ZZ domain and simultaneously reach the RING1 domain for ubiquitylation (Fig. 8b). Indeed, Tyr61-BB has an unstructured region consisting of approximately 130 residues in front of the C-terminal RING domain, which contains many lysine residues for ubiquitylation.
We propose the following two possibilities for the relevance of the intramolecular RING dimerization for the ubiquitylation activity of PRT1. First, the dimeric interface is critical for maintaining protein stability (Supplementary Fig. 16). The mixture of RING1WT and RING2WT formed a stable complex, whereas RING1WT in the presence of RING22L/S exhibited a degradation pattern without complex formation (Fig. 7d). These results suggest that the RING1 and RING2 dimeric organization is essential for the stability of PRT1. Second, the disruption of the intramolecular dimerization reduces the probability of forming a functional ubiquitylation complex involving RING1-RING2-E2~Ub-substrate (Supplementary Fig. 16). As mentioned above, the Ub discharge activity of 4 L/S mutant 10-fold weaker than that of the WT (Fig. 7f). Considering that PRT1 contains an intrinsically disordered region consisting of approximately 80 amino acid residues between the two RING domains, PRT14L/S mutant must be a spatially distributed contact site on both RING domains to interact with E2~Ub conjugate. Especially once the dimeric interface of PRT1 RING domains is disrupted, the functional RING1 domain exhibits increased conformational flexibility. In conclusion, even when the E2~Ub conjugate contacts the RING domain, the increased mobility of mutant RING1 impairs the precise alignment required for the Ub transfer to the substrates (Supplementary Fig. 16). Together, these results highlight the importance of intramolecular RING organization for maintaining the ubiquitylation activity of PRT1 (Fig. 8b). Several tandemly duplicated RING proteins from various species, including Arabidopsis, have been identified56, and several tandemly repeated RING constructs have demonstrated enhanced ubiquitylation activity50,57. PRT1 represents a single-chain tandem RING domain, an active and robust E3 ligase without intermolecular dimerization. Thus, our results represent a prototype case of tandem RING domain architecture, which might help interpret the activation mechanism. In the current study, essential structural inquiries concerning the Arg/N-degron pathway of plants were addressed, in addition to AtClpS1 and AtPRT658,59,60, expanding our understanding of the structural repertoire of N-degron recognition.
Methods
Cloning
The PRT1ZZ domain (residues 302–366) coding gene was amplified by PCR using codon-optimized gBlock as a synthesized gene template (Integrated DNA Technologies, Coralville, IA, USA). The amplified ZZ domain gene was inserted into a pET-His-LC3B vector, a modified pET-His vector containing the Kpn1 and Xho1 restriction enzyme sites37. The resulting plasmids were transformed into Escherichia coli BL21(DE3) cells. The cloning process for FL PRT1 was the same, except that the coding gene was inserted into FP-His, a modified pET-His6 vector. The N-degron sequence of BB (YKFG) and variations in its N-terminal sequence were introduced between the LC3B and ZZ domain to generate various N-degron-fused ZZ domains. Key mutations (Y317A, I333A, and F352A) were introduced into the FL PRT1 plasmid. These mutations were induced by PCR-based site-directed mutagenesis. PRT1 constructs (FL, 1–273, and 308–365) were cloned into the pET-bRIP (barley ribosome-activating protein) vector, a bRIP-tag-inserted version of the pET-His vector61.
Protein expression and purification
LC3B-N-degron-fused PRT1ZZ and FL His6-PRT1 plasmid transformed E. coli BL21(DE3) cells were cultured at 37 °C in LB medium in a shaking incubator until OD600 = 1.0. Thereafter, protein expression was induced by adding 0.5 mM isopropyl β-D-1-thiogalactopyranoside, and 400 μM of ZnSO4 was also added to the culture medium simultaneously. The induced cells were further incubated at 18 °C for 20 h. The cultured cells were harvested by centrifugation (10,000 × g for 20 min) and resuspended in a lysis buffer containing 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl), and 10 mM β-mercaptoethanol. After cell lysis by ultrasonication, the soluble fraction was separated from the pellet by centrifugation (35,000 × g for 1 h). The LC3B-N-degron-fused PRT1ZZ was purified using Ni-NTA affinity chromatography by loading the supernatants onto a HisTrapTM HP column (17524802; Cytiva, Marlborough, MA, USA) and eluted by increasing the concentration of His elution buffer containing 50 mM Tris-HCl pH 8.0, 100 mM NaCl, and 0.5 mM TCEP. The His6-LC3B tag was cleaved by incubating with His6-tagged ATG4B protease (20 °C, overnight)62, after which the His6-ATG4B protease and His6-LC3B tag were removed using HisTrapTM HP and HiTrapTM Q HP (17115301; Cytiva) columns. As a final step, the N-degron-fused PRT1ZZ domain was loaded onto a HiLoadTM SuperdexTM 75 16/600 pg column (28989333; Cytiva) pre-equilibrated with crystallization buffer comprising 25 mM Tris-HCl pH 8.0, 150 mM NaCl, and 0.5 mM TCEP. FL PRT1 and its mutants were purified following Ni-NTA affinity chromatography, His6-TEV protease incubation, a second Ni-NTA affinity chromatography, and size-exclusion chromatography with HiLoadTM SuperdexTM 75 16/600 pg pre-equilibrated with the ITC buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 1 mM TCEP) or Ub assay buffer (50 mM Tris-HCl pH 7.5 and 150 mM NaCl). The detailed processes for each purification step were the same as those described above. The bRIP-tagged PRT1 construct was expressed as described previously and purified through Ni-NTA and HiTrapTM Blue HP (Cytiva, 17041301) affinity chromatography and size-exclusion chromatography with HiLoadTM SuperdexTM 75 16/600 pg pre-equilibrated with the ITC buffer.
Crystallization
The purified N-degron-fused PRT1ZZ was concentrated to approximately 20 mg/mL and crystallized at 20 °C using the sitting-drop vapor diffusion method with a Gryphon machine (Art Robbins Instruments, Sunnyvale, CA, USA). The N-degron-fused PRT1ZZ was mixed at a ratio of 1:1 with crystallization solutions (apo—4 M sodium formate [Crystal Screen, C9 condition: HR2-130; Hampton Research, Aliso Viejo, CA, USA]; Y(F)KFG-AtPRT1ZZ—32.5–37.5% (v/v) precipitant mix 4, 0.1 M buffer system 2 pH 7.5, and 0.06 M divalent [Morpheus, A8 condition: MD1-46; Molecular Dimensions, Sheffield, UK]; WAAG-AtPRT1ZZ—1.4 M lithium sulfate, 100 mM Tris-HCl pH 8.5, and 2% (w/v) PEG 400 [Wizard Classic 3, E8 condition: 1009533; Rigaku, Tokyo, Japan]). Next, 20% (v/v) glycerol and 20% (v/v) ethylene glycol were mixed with mother liquors as Y(F)KFG-AtPRT1ZZ and WAAG-AtPRT1ZZ crystal cryoprotectants, respectively. The crystals were then flash-frozen in liquid nitrogen.
Data collection and structure determination
The X-ray data of the N-degron-fused PRT1ZZ domains (apo and YKFG, 1.7-Å; FKFG, 2.1-Å; WAAG, 2.8-Å resolution) were obtained at Pohang Accelerator Laboratory (PAL) in South Korea and Spring-8 in Japan (Supplementary Table 1). A single-anomalous dispersion dataset of apo-crystal was obtained at the absorption edge of the zinc atom (λ = 1.28 Å) at beamlines 11 C (PAL)63 and BL44XU (Spring-8)64. Zinc site determination, phasing, and model auto-building (Autosol and AutoBuild modules) were performed using Phenix software65 with the apo PRT1ZZ dataset. After the initial model was generated using AutoBuild, the remaining parts of the ZZ domain model were manually built using Coot66. After completing the apo PRT1ZZ model, the phases of the other crystal data were determined by molecular replacement using the apo-model. The models were rebuilt manually and refined in iterative cycles using Coot66. The final structural models were validated using MolProbity67.
Isothermal titration calorimetry (ITC)
The purified FL PRT1 and its mutations (Y317A, I333A, F352A, ΔRING1 + RING2, and ΔZZ domain) were concentrated to 45 ~ 55 μM in an ITC buffer. The N-degron peptides (synthesized by PeptronTM, Daejeon, South Korea) were dissolved in the same buffer and diluted to a concentration of ~550 μM. The ITC experiment was executed at 25 °C using a Microcal PEAQ-ITC (Malvern Panalytical, Malvern, UK). Each N-degron peptide was injected 19 times (2 μL each) into a 280-μL sample of each protein. ITC data were processed using a one-site binding model with the MicroCal PEAK-ITC analysis software. All ITC experiments were conducted at least thrice.
In vitro ubiquitylation assay
To assess the auto-ubiquitylation activity of Tyr61-BB and its mutants, we used purified and concentrated components as follows: 0.1 μM HsUBA1 (E1), 0.5 μM AtUBC8 (E2), 60 μM Ub, 3 μM Tyr61-BB WT and its mutants, 10 μM ATP, and 10 μM MgCl2 as the final concentrations in a 50-μL mixture. Each sample was incubated at 30 °C overnight and collected for activity monitoring every 1 h. To perform the Tyr61-BB ubiquitylation assay with PRT1 over different time courses, all components were mixed as follows: 0.1 μM HsUBA1, 0.5 μM AtUBC8, 1 μM PRT1 FL, 60 μM Ub, 3 μM Tyr61-BB V198E mutant, 10 μM ATP, and 10 μM MgCl2 as the final concentrations in a 50-μL mixture. These samples were incubated at 30 °C. To compare the ubiquitylation activity of PRT1WT and its mutants, the same concentrations of all components except 0.5 μM PRT1WT and mutants in a 100-μL mixture were incubated for 3 h (or over 12 h) at 30 °C. Ubiquitylated Tyr61-BB was detected using an HRP-conjugated anti-FLAG tag (sc-166355; Santa Cruz Biotechnology, Dallas, TX, USA, 1:2000 ratio) and anti-Ub antibodies (sc-8017; Santa Cruz Biotechnology, 1:2000 ratio), and PRT1 was detected using an HRP-conjugated anti-His antibody (sc-8036; Santa Cruz Biotechnology, 1:1000 ratio). Chemiluminescent signals were captured by brief incubation with 2 mL ECLTM Prime Western Blotting Detection Reagent (RPN2232; Cytiva). The 3 μM YKF-CP8GFP was used as a model substrate, which contained no RING domains41. The GFP was probed by anti-GFP antibodies (sc-9996; Santa Cruz Biotechnology 1:2000 ratio) and HRP-conjugated Goat anti-Mouse IgG antibodies (31430, Invitrogen, 1:5000 ratio). The anti-His chemiluminescence signal for input PRT1 was detected under stronger exposure conditions (high exposure mode, 50 s), while the others were acquired under standard conditions (high-resolution mode, 100 s). The other conditions are the same as mentioned above.
Size-exclusion chromatography coupled with small-angle X-ray scattering (SEC-SAXS)
For the SEC-SAXS experiment, FL PRT1 (10 mg/mL, 250 μL) was loaded onto an S200 increase 10/300 GL column (28990944; Cytiva) pre-equilibrated with a buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM TCEP, and 2% (w/v) glycerol. The scattering data from solutions of PRT1 in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM TCEP, and 2% (w/v) glycerol were collected at a wavelength of 1.5 Å and a distance of 3.0 m between the sample and detector at BL10C, Photon Factory, Japan (Supplementary Table 2). A PILATUS3 2 M (DECTRIS, Täfernhof II, Switzerland) was used as the X-ray detector at a sample-detector distance of 3.0 m and at a wavelength of λ = 0.15 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). The SEC parameters were as follows: A 250.00 μL sample at 10 mg/mL was injected at different flow rates in the protein peak (20 sec/image with flow at 0.05 mL/min) and the baseline (20 sec/image with flow at 0.5 mL/min) using a GE Superdex 200 Increase 10/300 column at 25 °C, and 332 successive 20-second frames were collected. Raw data were initially subtracted from the background signals using SAngler68. Protein signal peaks and baselines were selected using Chromixs69. Next, the data were analyzed using PRIMUS to obtain structural parameters and molecular weight deviations70. GNOM71 was used to calculate the distance distribution function [P(r)] and maximum particle dimension (Dmax). Ab initio modeling was performed using DAMMIF72 with 10 calculations with q range (0.0157–0.1657) and DAMMIN73 in PRIMUS program suit. The DAMMIN model was represented using ChimeraX74. The AlphaFold model fit was performed by CRYSOL75. Detailed parameters and results are described in Supplementary Table 276,77,78.
Size-exclusion chromatography coupled with multi-angle laser scattering (SEC-MALS)
For SEC-MALS, RING1WT + RING2WT, and RING1WT + RING22L/S mixtures (1:1 ratio of each component; a final concentration of 3 mg/mL) were loaded after incubation (4 °C, 1 h) onto an S75 Increase 10/300 GL column (29148721; Cytiva) pre-equilibrated with a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM TCEP. FL PRT1 (3 mg/mL) was loaded onto an S200 increase 10/300 GL column (28990944; Cytiva). Light scattering data were collected using a miniDAWN instrument (Wyatt Technology, Santa Barbara, CA, USA). ASTRA software (Wyatt Technology) was used for data analysis with an appropriate extinction coefficient (RING1WT: 0.592 mL/mg·cm; RING2WT and RING22L/S: 0.294 mL/mg·cm; PRT1WT FL: 0.596 mL/mg·cm).
FRET-based ubiquitin discharge assay
The FITC-labeled AtUBC8 was generated by sortase-catalyzed transpeptidation79. A FITC-labeled sortase recognition peptide (FITC-KLPETGG) was synthesized by PeptronTM. The transpeptidation reaction was conducted using 800 μM FITC-labeled peptide, 200 μM GST-sortase A, and 200 μM AtUBC8 (the N-terminal Gly-Ser of AtUBC8 remained after TEV cleavage). The mixture was incubated at 20 °C for 12 h. For AlexaFlorTM 594 labeling of Ub, the N-terminal Cys-Gly-Gly-Gly sequence was inserted by mutagenesis between the N-terminal His6-TEV and Ub sequences (C-UB). The N-terminal His6-tag was cleaved by TEV during purification. C-UB was mixed with AF594 C5 maleimide (A10256; InvitrogenTM) at a 3:1 molar ratio and incubated at 20 °C for 12 h. Excess peptide, GST-sortase, and AF594 dye were removed by size-exclusion chromatography using a S75 Increase 10/300 GL column (29148721; Cytiva). The Fluorescence labeled E2~Ub conjugate was prepared by incubating 0.5 μM hUBA1, 5 μM FITC-AtUBC8, 5.5 μM AF594-UB, 5 μM USP2 catalytic domain, 100 mM MgCl2, and 100 mM ATP in assay buffer (50 mM Tris-HCl pH 7.5 and 150 mM NaCl) for 30 min at 30 °C. The charging reaction was quenched after a 30-minute reaction time by ATP depletion. Discharge reactions were performed by incubating 125 nM E2~Ub mix, 10 μM unlabeled Ub, and 1 μM PRT1 variants (WT or 4 L/S mutant). Fluorescence signals were recorded in a 384-well black flat-bottom plate (3621; CorningTM, NY, US) using a SpectraMax iD5 microplate reader (Molecular Devices, CA, US) at 30 °C for 3 h, with 34-second intervals. The FRET data was obtained by exciting the donor fluorophore (FITC) at 475 nm and measuring emissions at 545 nm (FD; donor intensity) and 600 nm (FA; acceptor intensity). The raw FRET ratio (F°) of acceptor to donor intensity was calculated (1):
The decay in FRET signal was fitted to one-phase exponential decay Eq. (2):
where F°0 is the initial normalized FRET signal, Plateau represents the baseline signal after complete discharge or nonspecific decay, k is the decay rate constant, and x is time. To eliminate background signal reduction that occurs independently of PRT1 activity—such as spontaneous E2~Ub hydrolysis or photobleaching—we performed a normalization step using the no-PRT1 control. Fluorescence curves measured in the absence of PRT1 were used to define the baseline decay behavior. The following transformation was applied to correct for PRT1-independent decay and normalize the experimental curves (3):
Here, F°no PRT1 refers to the FRET signal measured from the control reaction lacking PRT1 under identical conditions. This transformation rescales the data to PRT1-mediated Ub discharge, thereby enabling comparison between WT and mutant variants.
The normalized FRET data (Fn) were refitted using the same exponential decay model (Eq. 2), and the half-life (t1/2) for each reaction condition was calculated using (4):
The equations were modified based on methodologies established in prior experiments80. All reactions were performed in triplicate. Curve fitting and kinetic parameter extraction were conducted using nonlinear regression analysis in GraphPad Prism 5 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and structure factors were deposited in the Protein Data Bank under accession codes 8ZG8 (apo), 8ZG9 (Y-peptide complex), 8ZGA (F-peptide complex), and 8ZGB (W-peptide complex). We deposited our SEC-SAXS data at the Small Angle Scattering Biological Data Bank under accession code SASDWM2 (SAXS data for full-length PRT1). Data Source are provided with this paper. Source data are provided with this paper.
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu Rev. Biochem. 67, 425–479 (1998).
Ciechanover, A. & Schwartz, A. L. The ubiquitin-proteasome pathway: The complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. USA 95, 2727–2730 (1998).
Rechsteiner, M. Ubiquitin-mediated pathways for intracellular proteolysis. Annu. Rev. Cell Biol. 3, 1–30 (1987).
Coux, O., Tanaka, K. & Goldberg, A. L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 (1996).
Bachmair, A., Finley, D. & Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986).
Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 116, 358–366 (2019).
Varshavsky, A. N-degron pathways. Proc. Natl. Acad. Sci. USA 121, e2408697121 (2024).
Bartel, B., Wünning, I. & Varshavsky, A. The recognition component of the N-end rule pathway. EMBO J. 9, 3179–3189 (1990).
Kim, B. H. et al. Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates. Proc. Natl. Acad. Sci. USA 119, e2209597119 (2022).
Kim, M. K., Oh, S. J., Lee, B. G. & Song, H. K. Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway. Proc. Natl. Acad. Sci. USA 113, 12438–12443 (2016).
Oh, J. H., Hyun, J. Y., Chen, S. J. & Varshavsky, A. Five enzymes of the Arg/N-degron pathway form a targeting complex: the concept of superchanneling. Proc. Natl. Acad. Sci. USA 117, 10778–10788 (2020).
Hwang, C. S., Shemorry, A. & Varshavsky, A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010).
Chen S. J., Wu X., Wadas B., Oh J. H. & Varshavsky A. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355, eaal3655 (2017).
Chen S. J., Kim L., Song H. K. & Varshavsky A. Aminopeptidases trim Xaa-Pro proteins, initiating their degradation by the Pro/N-degron pathway. Proc. Natl. Acad. Sci. USA 118, e2115430118 (2021).
Timms R. T., et al. A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science 365, eaaw4912 (2019).
Kim J. M., et al. Formyl-methionine as an N-degron of a eukaryotic N-end rule pathway. Science 362, eaat0174 (2018).
Xia, Z. et al. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J. Biol. Chem. 283, 24011–24028 (2008).
Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011).
Choi, W. S. et al. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat. Struct. Mol. Biol. 17, 1175–1181 (2010).
Matta-Camacho, E., Kozlov, G., Li, F. F. & Gehring, K. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat. Struct. Mol. Biol. 17, 1182–1187 (2010).
Pan, M. et al. Structural insights into Ubr1-mediated N-degron polyubiquitination. Nature 600, 334–338 (2021).
Tasaki, T., Sriram, S. M., Park, K. S. & Kwon, Y. T. The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 (2012).
Garzón, M. et al. PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 581, 3189–3196 (2007).
Potuschak, T. et al. PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc. Natl. Acad. Sci. USA 95, 7904–7908 (1998).
Stary, S. et al. PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol. 133, 1360–1366 (2003).
Disch, S. et al. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279 (2006).
Dong, H. et al. Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes Dev. 31, 197–208 (2017).
Mot, A. C. et al. Real-time detection of N-end rule-mediated ubiquitination via fluorescently labeled substrate probes. N. Phytol. 217, 613–624 (2018).
Kwon, D. H. et al. Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat. Commun. 9, 3291 (2018).
Zhang, Y. et al. ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat. Commun. 9, 4373 (2018).
Legge, G. B. et al. ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J. Mol. Biol. 343, 1081–1093 (2004).
Mi, W. et al. The ZZ-type zinc finger of ZZZ3 modulates the ATAC complex-mediated histone acetylation and gene activation. Nat. Commun. 9, 3759 (2018).
McMillan, B. rianJ. et al. A tail of two sites: a bipartite mechanism for recognition of notch ligands by mind bomb E3 ligases. Mol. Cell 57, 912–924 (2015).
Liu, J. et al. Structural insight into binding of the ZZ domain of HERC2 to histone H3 and SUMO1. Structure 28, 1225–1230.e1223 (2020).
Zhang, Y. et al. The ZZ domain of p300 mediates specificity of the adjacent HAT domain for histone H3. Nat. Struct. Mol. Biol. 25, 841–849 (2018).
Ponting, C. P., Blake, D. J., Davies, K. E., Kendrick-Jones, J. & Winder, S. J. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 21, 11–13 (1996).
Kim, L., Kwon, D. H., Heo, J., Park, M. R. & Song, H. K. Use of the LC3B-fusion technique for biochemical and structural studies of proteins involved in the N-degron pathway. J. Biol. Chem. 295, 2590–2600 (2020).
Kim, B. H., Kwon, D. H., Kim, L., Yang, W. S. & Song, H. K. In vitro production of N-degron fused proteins and its application. Methods Enzymol. 686, 99–123 (2023).
Sigrist, C. J. et al. New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 (2013).
Moududee, S. A. et al. Structural and functional characterization of hMEX-3C Ring finger domain as an E3 ubiquitin ligase. Protein Sci. 27, 1661–1669 (2018).
Khmelinskii, A. et al. Incomplete proteasomal degradation of green fluorescent proteins in the context of tandem fluorescent protein timers. Mol. Biol. Cell 27, 360–370 (2016).
Varadi, M. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Sanchez, J. G. et al. Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep. 16, 1315–1325 (2016).
Anandapadamanaban, M. et al. E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2. J. Biol. Chem. 294, 11404–11419 (2019).
Campbell, S. J. et al. Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. J. Biol. Chem. 287, 23900–23910 (2012).
Yudina, Z. et al. RING dimerization links higher-order assembly of TRIM5alpha to synthesis of K63-linked polyubiquitin. Cell Rep. 12, 788–797 (2015).
Keown, J. R., Yang, J., Black, M. M. & Goldstone, D. C. The RING domain of TRIM69 promotes higher-order assembly. Acta Crystallogr. D Struct. Biol. 76, 954–961 (2020).
Koliopoulos, M. G., Esposito, D., Christodoulou, E., Taylor, I. A. & Rittinger, K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 35, 1204–1218 (2016).
Park, S. H. et al. Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane. Nat. Struct. Mol. Biol. 30, 1695–1706 (2023).
Brzovic, P. S., Rajagopal, P., Hoyt, D. W., King, M. C. & Klevit, R. E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8, 833–837 (2001).
Shukla, S. et al. Small-molecule inhibitors targeting polycomb repressive complex 1 RING domain. Nat. Chem. Biol. 17, 784–793 (2021).
Fiorentini, F., Esposito, D. & Rittinger, K. Does it take two to tango? RING domain self-association and activity in TRIM E3 ubiquitin ligases. Biochem. Soc. Trans. 48, 2615–2624 (2020).
Kiss, L., Clift, D., Renner, N., Neuhaus, D. & James, L. C. RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination. Nat. Commun. 12, 1220 (2021).
Cha-Molstad, H. et al. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat. Commun. 8, 102 (2017).
Stone, S. L. et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137, 13–30 (2005).
Fletcher, A. J. et al. Trivalent RING assembly on retroviral capsids activates TRIM5 ubiquitination and innate immune signaling. Cell Host Microbe 24, 761–775 e766 (2018).
Kim, L. et al. Structural basis for the N-degron specificity of ClpS1 from Arabidopsis thaliana. Protein Sci. 30, 700–708 (2021).
Kim L., et al. Structural analyses of the plant PRT6-UBR box in the Cys-Arg/N-degron pathway and insights into the plant submergence response. bioRxiv, 2022.2008.2019.504472 (2022).
Rudi, O. et al. The UBR domain of plant Ubr1 homolog PRT6 accommodates basic and hydrophobic amino termini for substrate recognition. J. Mol. Biol. 437, 168939 (2025).
Lee, B.-G., Kim, M. K., Kim, B.-W., Suh, S. W. & Song, H. K. Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism. Acta Crystallogr. Sect. D 68, 1488–1500 (2012).
Kwon, D. H. et al. The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane. Autophagy 13, 70–81 (2017).
Gu, D.-H. et al. BL-11C micro-MX: a high-flux microfocus macromolecular-crystallography beamline for micrometre-sized protein crystals at Pohang light source II. J. Synchrotron Radiat. 28, 1210–1215 (2021).
Higashiura, A. et al. SPring-8 BL44XU, beamline designed for structure analysis of large biological macromolecular assemblies. AIP Conf. Proc. 1741, 030028 (2016).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D 75, 861–877 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Prisant, M. G., Williams, C. J., Chen, V. B., Richardson, J. S. & Richardson, D. C. New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink “waters,” and NGL viewer to recapture online 3D graphics. Protein Sci. 29, 315–329 (2020).
Shimizu N. et al. Software development for analysis of small-angle x-ray scattering data. AIP Conf. Proc. 1741, 050017 (2016).
Panjkovich, A. & Svergun, D. I. CHROMIXS: automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics 34, 1944–1946 (2017).
Manalastas-Cantos, K. et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355 (2021).
Svergun, D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Franke, D. & Svergun, D. I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009).
Svergun, D. I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Franke, D. et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 50, 1212–1225 (2017).
Kikhney, A. G., Borges, C. R., Molodenskiy, D. S., Jeffries, C. M. & Svergun, D. I. SASBDB: Towards an automatically curated and validated repository for biological scattering data. Protein Sci. 29, 66–75 (2020).
Trewhella, J. et al. 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update. Acta Crystallogr. D. Struct. Biol. 73, 710–728 (2017).
Trewhella, J., Jeffries, C. M. & Whitten, A. E. 2023 update of template tables for reporting biomolecular structural modelling of small-angle scattering data. Acta Crystallogr. D. Struct. Biol. 79, 122–132 (2023).
Theile, C. S. et al. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat. Protoc. 8, 1800–1807 (2013).
Herkules, F. et al. Structural and functional asymmetry of RING trimerization controls priming and extension events in TRIM5alpha autoubiquitylation. Nat. Commun. 13, 7104 (2022).
Acknowledgements
We thank the staff at beamlines 4 C, 5 C, and 11 C at the Pohang Accelerator Laboratory in South Korea, beamline BL44XU at SPring-8, and beamline BL-10C at the Photon Factory in Japan for their assistance with X-ray data collection. We appreciate Dr. Cy Jeffries at EMBL for his advice on SAXS data analysis. This study was partially supported by the International Collaborative Research Program of the Institute for Protein Research at Osaka University (ICR-22-05 and ICR-23-05). Diffraction data were collected at the Osaka University beamline BL44XU at SPring-8 (Harima, Japan) (proposal no. 2023A6873). This study was supported by the National Research Foundation of Korea (NRF) grants from the Korean government (Grant Nos. RS-2020-NR049540, RS-2021-NR056525, RS-2021-NR056577, RS-2022-NR067411, and RS-2025-00521009 to H.K.S.).
Author information
Authors and Affiliations
Contributions
W.S.Y., S.H.K., M.K., H.S., J.L., and A.S. performed biochemical experiments. W.S.Y. prepared the crystals, solved the structures, and conducted SAXS experiments. W. S. Y., O. K. P., N. D., and H. K. S. analyzed the data. W.S.Y. and H.K.S. designed the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, W.S., Kim, S.H., Kim, M. et al. Structural basis for the recognition and ubiquitylation of type-2 N-degron substrate by PRT1 plant N-recognin. Nat Commun 16, 7817 (2025). https://doi.org/10.1038/s41467-025-63282-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63282-9