Abstract
The efficacy of biologics, such as peptide and protein drugs, is often limited by their short half-lives in vivo, necessitating repeated infusions to maintain therapeutic effects. Here, we demonstrate that long-lived CAR T cells can be leveraged as a versatile platform for long-term delivery of biologics. Our recent findings show that the depletion of BCOR and ZC3H12A induces GD2 CAR T cells into an immortal-like and functional state, which we have termed GD2TIF cells. These GD2TIF cells expanded in immunocompetent mice without the need for chemotherapeutic conditioning and persisted as a polyclonal population, safely and almost indefinitely. In leptin-deficient ob/ob mice, a single infusion of GD2TIF cells producing leptin effectively and durably corrected leptin deficiency. Furthermore, a single infusion of GD2TIF cells secreting GLP-1 prevented obesity and diabetes in mice fed on a high-fat diet. The longevity, safety, and adaptability of GD2TIF cells suggest their potential as a general platform for long-term delivery of biologics, offering durable therapeutic efficacy with a single infusion.
Similar content being viewed by others
Introduction
Biologics, particularly recombinant proteins, are a cornerstone of modern biomedicine. While recombinant proteins are effective in treating various diseases, a major limitation is their short half-life in vivo, and engineering cells as a platform for biologics delivery may overcome the related pitfalls1,2,3. Due to the progressive and often irreversible nature of chronic diseases, most, if not all, protein-based drugs require repeated infusions over many years or even lifelong, to maintain therapeutic efficacy.
For instance, biologics modulating glucagon-like peptide-1 (GLP-1) or GLP-1 receptor (GLP-1R) activities are effective in treating obesity, but these drugs require administration on a weekly or even daily basis4. Upon discontinuation, obesity relapses quickly5, indicating that long-term medication is necessary to maintain efficacy. This requirement negatively impacts patient adherence. Studies have shown that patients on biologic therapies have lower adherence compared to those on non-biologic treatments6,7. A comparison between two well-known GLP-1R agonists, the once-daily liraglutide and the once-weekly semaglutide, demonstrates that the latter, with its longer half-life, is associated with higher patient adherence8.
Repeated infusion of protein drugs also leads to the development of anti-drug immunity. For example, mouse monoclonal antibodies have a high incidence of inducing anti-antibody responses in humans, and even humanized or fully human antibodies can trigger similar reactions9. Once anti-drug immunity forms, further infusions of biologics may result in reduced or no efficacy and can cause immunopathology10. Therefore, a one-time and long-lasting treatment is highly desirable for biologics treating chronic diseases.
To achieve this goal, several strategies have been proposed and tested in animal models and clinic trials. One approach involves using AAV-based vectors to deliver DNA encoding therapeutic proteins, such as FIX in hemophilia11 and antibodies neutralizing HIV in AIDS12,13. However, due to the non-integrating nature of AAV vectors, the injected vectors gradually decay over time, typically lasting less than two years14. Consequently, AAV-based delivery of biologics cannot provide long-term efficacy. More recently, immune cells, including CAR T cells, have been engineered to secrete therapeutic proteins15,16,17. However, the limited number and persistence of these immune cells in previous studies make them unsuitable for long-term delivery of biologics15,16,17.
There are three prerequisites for CAR T cells to serve as a practical platform for delivering biologics in chronic diseases. First, the chemotherapy-based preconditioning regimen currently used in CAR T cell therapy must be eliminated, as chemotherapy is not suitable for patients with noncancerous diseases. Second, CAR T cells must persist in vivo at sufficient numbers and for a sufficient duration of time, ideally lifelong, since most chronic diseases are lifelong persistent. Third, the target of CAR T cells must be carefully selected to avoid damaging normal cells or tissues.
The disialoganglioside GD2 is a tumor-associated antigen that is overexpressed in tumors like neuroblastoma but is rarely expressed in normal tissues18,19. CAR T cells targeting GD2 have demonstrated promising efficacy in clinical settings without significant off-tumor effects20. A unique characteristic of the anti-GD2 CAR derived from the 14g2a monoclonal antibody is its tonic signaling in the absence of target cells21. Although tonic signaling is typically considered detrimental for CAR T cells, we have previously shown that GD2 CAR-induced tonic signaling was able to induce CAR T cells into an immortal-like and functional state (TIF) in the absence of GD2+ tumor cell22. These GD2TIF cells were able to expand in immunocompetent mice without conditioning, persisted in primary hosts for three months, and could be serially transferred to immunocompetent hosts22.
The induction and maintenance of GD2TIF cells require the expression of the GD2 CAR and the simultaneous depletion of two factors: ZC3H12A and BCOR22. ZC3H12A is an RNase that regulates mRNA decay, negatively controlling T cell activation and expansion23, while BCOR is a transcriptional co-repressor that modulates gene expression24. The depletion of both ZC3H12A and BCOR, but not either alone, produces a synergistic effect that imparts TIF features to various CAR T cells22,25, including robust expansion in immunocompetent mice without conditioning, minimal contraction after the peak response, and long-term in vivo persistence with a favorable safety profile.
In this study, we repurpose GD2TIF cells as a general platform for the long-term delivery of biologics in vivo, with the goal of achieving long-term therapeutic efficacy by a single dose of CAR T cells.
Results
Long-term persistence and stability of GD2TIF cells in vivo
To evaluate the potential of GD2TIF cells as a platform for biologic delivery, we conducted a comprehensive characterization of these cells. Previously, we showed that wild-type, BCOR-knockout, and ZC3H12A-knockout GD2 CAR T cells were undetectable in the spleens of C57BL/6 mice one month after infusion22. To extend these findings, we generated wild-type, BCOR-knockout, ZC3H12A-knockout, and BCOR/ZC3H12A double-knockout GD2 CAR T cells using the CRISPR/Cas9 system (Supplementary Fig. 1a) and transferred them into C57BL/6 mice without any conditioning regimen (Supplementary Fig. 1b). Two months post-transfer, only the BCOR/ZC3H12A double-knockout GD2 CAR T cells (GD2TIF) were detectable in the spleen of recipient mice, whereas wild-type, BCOR-knockout, and ZC3H12A-knockout cells were undetectable (Supplementary Fig. 1c, d). These results corroborate our previous conclusion that depletion of both BCOR and ZC3H12A, but not either one alone, is required to induce GD2TIF cells.
Serial blood sampling revealed that GD2TIF cells persisted in recipient mice for at least 12 months (the endpoint of the experiment), consistently comprising approximately 5% of peripheral white blood cells (Fig. 1a). By six months post-transfer, most GD2TIF cells in the spleen exhibited a CD44hiCD62Lhi phenotype (Fig. 1b, c), and expressed TCF1 at levels comparable to endogenous CD8+ T cells (Supplementary Fig. 1i, j). GD2TIF cells also expressed PD-1, CXCR5, TOX, and EOMES (Supplementary Fig. 1i, j). Additionally, splenic GD2TIF cells produced IFNγ upon ex vivo stimulation with PMA and ionomycin (Supplementary Fig. 1g, h). These phenotypes of GD2TIF cells at six months post-transfer were similar to those observed at one month22, suggesting that GD2TIF cells maintain a stable phenotype in vivo. Furthermore, GD2TIF cells were detected in non-lymphoid tissues, including liver, lung, and brain (Supplementary Fig. 1i, j), indicating their broad distribution in vivo.
a One million GD2TIF cells were intravenously transferred into male C57BL/6 mice, and their persistence in peripheral blood was monitored over time through serial bleeding (n = 5). b, c FACS analysis of CD62L expression on splenic GD2TIF cells at 2 months post-transfer. Representative plot (b) and quantification (c) are shown (n = 6). d Uniform Manifold Approximation and Projection (UMAP) of 10,290 single GD2TIF cells pooled from 4 mice. e Normalized expression levels of key markers across identified clusters. f Clustered heatmap showing representative marker genes in annotated CD62Lhi and CD62Llo populations. g–i Gene Set Enrichment Analysis (GSEA) comparing CD62Lhi and CD62Llo populations. Exact P values: g 3.64 × 10-7; h 2.10 × 10-5; i 6.45 × 10-11. j Clonotype distribution of GD2TIF cells based on scTCR-seq. Individual clones were identified by unique CDR3 sequences, and the percentages of cells with identical CDR3 sequences among all sequenced cells were calculated. k UMAP visualization of GD2TIF cells, with individual TCR clonotypes distinguished by color. l Distribution of the top three clonotypes across clusters in the UMAP plot, with all other clonotypes grouped as “common clone”. Data represent mean ± s.e.m. from one of three independent experiments. n indicates the number of mice. Exact P values were determined by two tailed paired Student’s t test in (c), and one-sided permutation tests in (g–i), as implemented in the clusterProfiler package (v4.4.4). Source data are provided as a Source Data file.
To investigate the role of GD2 CAR in shaping the phenotype of GD2TIF cells, we analyzed CD8+ T cells expressing sgRNAs targeting Bcor and Zc3h12a, either in the presence (GD2TIF) or absence (DKO) of GD2 CAR (Supplementary Fig. 2a). Cells were labeled with CTV before transfer to track cell division and analyzed on day 4 (blood) and day 7 (spleen) post-transfer (Supplementary Fig. 2b, c). Both GD2TIF and DKO CD8+ T cells exhibited extensive CTV dye dilution (Supplementary Fig. 2c–e), indicating robust in vivo proliferation. However, by day 7 post-transfer, GD2TIF cells were more abundant in the spleen than DKO cells (Supplementary Fig. 2f, g), underscoring the critical role of GD2 CAR signaling in GD2TIF cell survival and maintenance. Notably, GD2TIF cells, but not DKO CD8+ T cells, exhibited increased expression of PD-1, CXCR5, TOX, TCF1, and EOMES compared to endogenous CD8+ T cells (Supplementary Fig. 2h, i). These findings demonstrate that deleting Bcor and Zc3h12a alone, without GD2 CAR expression, is insufficient to induce the expression of GD2TIF-associated markers in CD8+ T cells.
Transcriptomic and clonality analysis of GD2TIF cells
To further characterize GD2TIF cells, we performed single-cell RNA sequencing (scRNA-seq) and single-cell T cell receptor sequencing (scTCR-seq) analysis 30 days post-transfer. A total of 10,378 GD2TIF cells from the spleens pooled from 4 mice were sequenced and grouped into 10 clusters using unsupervised clustering (Supplementary Fig. 3a–c). These clusters were primarily classified into CD62Lhi stem-like and CD62Llo effector/exhaustion-like populations (Fig. 1d and Supplementary Fig. 3d). Cells in CD62Lhi population expressed high levels of Sell and Tcf 7 (Fig. 1e, f), indicating stem-like characteristics. In contrast, cells in CD62Llo population expressed low levels of Sell and Tcf7 but were positive for Pdcd1, Havcr2, Gzmb, and Gzmk (Fig. 1e, f), signifying an effector/exhaustion phenotype. Gene set enrichment analysis (GSEA) suggested that GD2TIF cells exhibited naïve-like features compared to exhausted, memory, and effector CD8+ T cells (Fig. 1g–i, Supplementary Fig. 3e), consistent with the stem-like nature of GD2TIF cells22.
We also compared GD2TIF cells with recently reported stem-like exhausted T (TSLEX) cells26. After harmonization, unsupervised clustering revealed 11 clusters in the merged sample (Supplementary Fig. 3f). GD2TIF cells distinctly segregated from TSLEX cells, supporting the idea that CAR T cells exist in a synthetic state distinct from TCF1+ TSLEX cells27. While both GD2TIF and TSLEX cells expressed high levels of Tcf7 and Tox, GD2TIF cells expressed much higher levels of Sell compared to TSLEX cells (Supplementary Fig. 3g, h), aligning with the stem-like feature of GD2TIF cells observed in serial transfer assays22.
scTCR-seq analysis identified 1,250 clonotypes among the 10,378 sequenced GD2TIF cells (Fig. 1j). The top three clones (clones 1, 2, and 3) constituted 9.6%, 5.5%, and 3.0% of the total cells, respectively (Fig. 1j). This level of clonality in GD2TIF cells is more diverse than what is observed in CAR T cells without gene knockout28. Mapping TCR clonotypes to the scRNA-seq data (Fig. 1k), we found that approximately 70% of the cells in the top 1 clone were located in CD62Llo population (Fig. 1l). Since cells in CD62Llo population exhibited an effector-like phenotype (Fig. 1e), it is possible that this clone originated from a common progenitor. Apart from the dominant clones, the majority of common GD2TIF cell clones were distributed across all clusters (Fig. 1l). These findings indicate that the induction of GD2TIF cells is not the result of selecting and expanding a few aberrant CAR T cell clones; rather, GD2TIF cells are highly diverse, resembling memory T cells29,30.
In summary, these data demonstrate that GD2TIF cells persist in vivo in a stable, polyclonal state with stem-like features.
Tonic signaling plays an important role in GD2TIF cell induction
The GD2 CAR, derived from the 14g2a monoclonal antibody, is known to induce tonic signaling in the absence of antigen stimulation, often leading to T cell exhaustion21. To determine whether tonic signaling alone is sufficient for GD2TIF cell induction, we mutated all complementary determining regions (CDRs) of the GD2 CAR (GD2mut) to abolish antigen binding, as previously reported21 (Supplementary Fig. 4a, b). Despite lacking antigen binding21, the depletion of BCOR and ZC3H12A still promoted the expansion and persistence of GD2mut CAR T (GD2mutTIF) cells, albeit at slightly lower levels than GD2TIF cells (Supplementary Fig. 4c, d). This suggests that tonic signaling alone is sufficient to drive GD2TIF cell induction. Notably, the expression of EOMES, PD-1, and TOX, markers associated with CAR tonic signaling and T cell exhaustion, was higher in GD2TIF cells compared to GD2mutTIF cells (Supplementary Fig. 4e, f), indicating that mutating the CDRs of GD2 CAR may modulate the strength of tonic signaling.
The costimulatory domain of our GD2 CAR is CD28, which is known to drive tonic signaling in this CAR21. It has been shown that using a 4-1BB costimulatory domain can diminish the tonic signaling of GD2 CAR21. To investigate whether tonic signaling is indeed responsible for the induction of GD2TIF cells, we replaced the CD28 costimulatory domain with 4-1BB in the GD2 CAR (GD2-BBζ). The depletion of BCOR and ZC3H12A did not promote the expansion of GD2 CAR T cells with the 4-1BB costimulatory domain (Supplementary Fig. 4g-i), suggesting that the tonic signaling mediated by the CD28 costimulatory domain is important for the induction of GD2TIF cells.
Safety of GD2TIF cells in vivo
We thoroughly evaluated the safety of GD2TIF cells in vivo. The presence of GD2TIF cells did not affect the survival of recipient mice (Supplementary Fig. 5a). Six months post-infusion, the levels of endogenous immune cells in the spleen and bone marrow were comparable between mice infused with GD2TIF cells and those receiving PBS (Supplementary Fig. 5b-g). Furthermore, endogenous T cells in mice harboring GD2TIF cells did not exhibit any signs of aberrant activation (Supplementary Fig. 5h, i). Hematoxylin and eosin (H&E) staining of sections from key organs, including the heart, lung, liver, and kidney, revealed no abnormalities in mice with GD2TIF cells (Supplementary Fig. 5j). These data demonstrate that the presence of GD2TIF cells does not disrupt the homeostasis of the endogenous immune system under steady-state.
When mice with GD2TIF cells were infected with the lymphocytic choriomeningitis virus (LCMV) Armstrong strain, there were no alterations in the endogenous CD8+ T cell response to the infection (Supplementary Fig. 6a-d). In the classic experimental autoimmune encephalomyelitis (EAE) model, mice bearing GD2TIF cells showed no differences in disease induction or progression compared to control mice (Supplementary Fig. 6e, f).
Finally, the administration of an anti-Thy1.1 antibody rapidly cleared GD2TIF cells from recipient mice (Supplementary Fig. 6g-i), indicating that these cells can be eliminated as needed.
In summary, these data demonstrate that the presence of GD2TIF cells does not compromise the health of recipient mice under both steady-state and immune-stressed conditions. Furthermore, GD2TIF cells can be effectively removed, establishing a foundation for repurposing these cells as a safe platform for drug delivery.
GD2TIF cells secreting inflammatory cytokine rapidly establish disease models in mice
After thoroughly characterizing and evaluating the safety of GD2TIF cells, we investigated their potential as a platform for in vivo production of biologics. Inflammatory cytokines play crucial roles in various diseases, and the traditional approach to establishing mouse models for these diseases involves creating cytokine transgenic mice, such as the TNF transgenic mice widely used in mechanistic studies and drug evaluations31. However, constructing and maintaining transgenic mice is labor-intensive and resource-demanding. Therefore, we engineered GD2TIF cells to secrete TNF (Fig. 2a). Infusion of TNF-secreting GD2TIF (GD2TIF-TNF) cells into C57BL/6 mice induced rapid and dramatic expansion of CD11b+ myeloid cells (Fig. 2b-f), accompanied by a reduction in body weight (Fig. 2g). These features in GD2TIF-TNF-bearing mice resemble those observed in TNF transgenic mice31.
a Experimental design for generating GD2TIF-TNF cells. One million GD2TIF cells and GD2TIF-TNF cells were transferred into six-week-old female C57BL/6 mice, which were subsequently monitored for GD2TIF-TNF cell persistence and monocyte expansion. GFP: green fluorescent protein; 2 A: P2A autoclevage peptide sequence; TNF: tumor necrosis factor alpha. b, c FACS analysis of GD2TIF-TNF cells in peripheral blood at 4 weeks post-transfer. Representative plot (b) and quantification (c) are shown (n = 3). d–f FACS analysis of CD11b+ myeloid cells and Thy1.1+ GD2TIF-TNF cells in peripheral blood. Representative plot (d) and quantification (e, f) are shown (n = 3). g, Body weight changes in mice transferred with GD2TIF-TNF cells or PBS over 5 weeks post-transfer (n = 3). h, Experimental design for generating GD2TIF-IL-5 cells. PBS or GD2TIF-IL-5 cells were transferred into six-week-old female C57BL/6 mice, which were monitored for GD2TIF-IL-5 cell persistence and eosinophil expansion. IL-5: interleukin-5. i, j, FACS analysis of GD2TIF-IL-5 cells in peripheral blood at 4 weeks post-transfer. Representative plot (i) and quantification (j) are shown (n = 4). k–m FACS analysis of SiglecF+ eosinophils and Thy1.1+ GD2TIF-IL-5 cells in peripheral blood. Representative plot (k) and quantification (l, m) are shown (n = 4). Data represent mean ± s.e.m. from one of two independent experiments. n indicates the number of mice. Exact P values were determined by two tailed unpaired Student’s t test in (c, j) and two-way ANOVA with Bonferroni’s post hoc test in (e–g, l, m). Source data and exact P values are provided as a Source Data file.
Hypereosinophilic syndrome (HES) is a disease characterized by the aberrant expansion of eosinophils32, which can be modeled by the transgenic overexpression of IL-533, an inflammatory cytokine crucial for eosinophil expansion34. Infusion of GD2TIF cells secreting mouse IL-5 (GD2TIF-IL-5) into C57BL/6 mice induced a rapid and sustained expansion of SiglecF+SSChi eosinophils in peripheral blood (Fig. 2h-m), effectively establishing a mouse model of HES within two weeks.
The dramatic biological effects observed in mice infused with GD2TIF-TNF and GD2TIF-IL-5 cells demonstrate that GD2TIF cells are capable of stably secreting cytokines in vivo.
Rescue of obesity in ob/ob mice by a single dose of GD2TIF cells secreting leptin
Leptin, a protein that regulates food intake, is deficient in ob/ob mice due to a genetic mutation, resulting in severe obesity from uncontrolled eating35. To address this, we engineered GD2TIF cells to secrete leptin (GD2TIF-Leptin) and tested their ability to correct the leptin deficiency in ob/ob mice (Fig. 3a-c).
a, Schematic representation of constructs used to generate DKO, GD2TIF, and GD2TIF-Leptin cells, DKO: Bcor/Zh3c12a genes double knock out without GD2 CAR expression CD8⁺ T cells; GFP: green fluorescent protein; 2A: P2A autoclevage peptide sequence. b, Illustration depicting GD2TIF-Leptin cells secreting leptin. c, Experimental design for rescuing leptin deficiency in male ob/ob mice using GD2TIF-Leptin cells. One million DKO, GD2TIF, or GD2TIF-Leptin cells were intravenously transferred into four-week-old ob/ob mice. Body weight, cell kinetics, metabolic phenotyping, Insulin Tolerance Test (ITT), and Glucose Tolerance Test (GTT) were assessed at specified time points. GD2TIF-Leptin cell distribution, serum leptin levels, and ex vivo restimulation assays were performed at the study endpoint. Age-matched C57BL/6 mice fed a chow diet served as the reference group. d, Simple linear regression analysis of serum leptin levels (log2) and the percentage of GD2TIF-Leptin cells in the spleen (n = 8), R2 = 0.9063. e, Body weight curves for mice receiving the indicated treatments, n indicates mice numbers. f, Body fat ratio before (week 0) and after (week 8) treatment in the indicated groups (n = 6). g, h, ITT results, including glucose levels (g) and relative area under the curve (AUC) (h) for the indicated groups (n = 6). i, j, GTT results, including glucose levels (i) and relative AUC (j) for the indicated groups (n = 6). k-n, Metabolic phenotyping of mice following treatment, with 48-hour kinetic measurements post-acclimation (n = 6). k, Respiratory exchange ratio (RER). l, Total energy expenditure. m, Accumulated food intake. n, Physical activity. Data represent the mean ± s.e.m. n indicates the number of mice. Exact P values were determined by simple linear regression (d), two tailed paired Student’s t test (f), one-way ANOVA with Tukey’s multiple-comparisons test (h, j), and two-way ANOVA with Bonferroni’s post hoc test (e, g, i, k-n). Source data and exact P values are provided as a Source Data file.
After infusion into ob/ob mice, GD2TIF and GD2TIF-Leptin cells reached a plateau around four weeks post-transfer, stabilizing thereafter with a gradual upregulation of CD62L (Supplementary Fig. 7a-d). Both GD2TIF and GD2TIF-Leptin cells were detected in the liver, lung, and brain tissues of ob/ob mice (Supplementary Fig. 7e, f). GD2TIF-Leptin cells constitutively produced leptin ex vivo, with increased secretion upon TCR stimulation (Supplementary Fig. 7g). In vivo, serum leptin levels in ob/ob mice correlated with the GD2TIF-Leptin cell number in the spleen (Fig. 3d).
Notably, a single infusion of GD2TIF-Leptin cells completely reversed obesity in ob/ob mice (Fig. 3e), whereas GD2TIF or DKO cells had no therapeutic effect (Fig. 3e). EchoMRI measurements confirmed a significant reduction in body fat in ob/ob mice treated with GD2TIF-Leptin cells (Fig. 3f). Furthermore, insulin tolerance tests (ITT) and glucose tolerance tests (GTT) demonstrated that GD2TIF-Leptin cells, but not GD2TIF or DKO cells, effectively restored insulin sensitivity and glucose homeostasis in ob/ob mice (Fig. 3g-j). In addition, GD2TIF-Leptin cells reduced the respiratory exchange ratio, energy expenditure, and food intake in ob/ob mice (Fig. 3k-m). Finally, GD2TIF-Leptin-treated mice exhibited a significant increase in movement (Fig. 3n).
Collectively, these findings demonstrate that a single infusion of GD2TIF-Leptin cells effectively corrects leptin deficiency in ob/ob mice.
A single dose of GD2TIF cells secreting GLP-1 provides durable anti-obesity effects in mice fed a high-fat diet (HFD)
Obesity is a global health issue associated with various pathological conditions, including diabetes, hyperlipidemia, and cancers36,37. Biologics targeting the GLP-1/GLP-1R axis are effective in treating obesity38,39. However, due to the short half-life of GLP-1, existing GLP-1 analogs and GLP-1R agonists require daily or weekly administration to maintain efficacy4, with rapid relapse upon discontinuation5.
To address this, we engineered GD2TIF cells to continuously deliver GLP-1 (GD2TIF-GLP-1) in vivo (Fig. 4a, b). C57BL/6 mice were fed a high-fat diet (HFD) for one week before receiving an infusion of either DKO or GD2TIF-GLP-1 cells (Fig. 4c). GD2TIF-GLP-1 cells expanded in HFD-fed mice, reaching a plateau four weeks post-transfer, accompanied by gradual upregulation of CD62L (Supplementary Fig. 8a-d). These cells were detected in the liver, lung, and brain tissues of HFD-fed mice (Supplementary Fig. 8e, f).
a Schematic representation of constructs used to generate DKO and GD2TIF-GLP-1 cells, DKO: Bcor/Zh3c12a genes double knock out without GD2 CAR expression CD8⁺ T cells; GFP: green fluorescent protein. b Illustration depicting GD2TIF-GLP-1 cells secreting GLP-1. To increase the half-life of GLP-1, the GLP-1 construct used in this study included an IL-2 signal sequence, GLP-1(7-38) with three mutations (A8G, G22E, and R36G), followed by a 3×GGGGS linker and a mouse IgG2a Fc sequence. c Experimental design for the treatment of HFD-induced obesity and diabetes using GD2TIF-GLP-1 cells. Male C57BL/6 mice were fed a HFD for one week before the intravenous transfer of one million DKO or GD2TIF-GLP-1 cells. Body weight, cell kinetics, metabolic phenotyping, Insulin Tolerance Test (ITT), and Glucose Tolerance Test (GTT) were assessed at specified time points. GD2TIF-GLP-1 cell distribution was analyzed at the study endpoint. Age-matched chow-fed mice served as a reference group. HFD: High Fat Diet. d Body weight curves for mice receiving the indicated treatments (n = 6). e Body fat ratio before (week 0) and after (week 8) treatment in the indicated groups (n = 6). f, g ITT results, including glucose levels (f) and relative area under the curve (AUC) (g) for the indicated groups (n = 6). h, i GTT results, including glucose levels (h) and relative AUC (i) for the indicated groups (n = 6). j-m, Metabolic phenotyping of mice following treatment, with 48-hour kinetic measurements post-acclimation (n = 6). j, Respiratory exchange ratio (RER). k, Total energy expenditure. l, Accumulated food intake. m, Physical activity. Data represent the mean ± s.e.m. n indicates the number of mice. Exact P values were determined by two tailed paired Student’s t test (e), one-way ANOVA with Tukey’s multiple comparisons test (g, i), and two-way ANOVA with Bonferroni’s post hoc test (d, f, h, j-m). Source data and exact P values are provided as a Source Data file.
Due to the attached Fc fragment from mouse IgG2a in the GLP-1 molecule (Fig. 4b), the accurate circulating GLP-1-Fc level remains to be determined. However, GD2TIF-GLP-1 cell infusion, but not DKO cell infusion, significantly reversed HFD-induced obesity (Fig. 4d). EchoMRI analysis confirmed a reduction in body fat in HFD-fed mice treated with GD2TIF-GLP-1 cells (Fig. 4e). In ITT and GTT, GD2TIF-GLP-1 cells effectively restored insulin sensitivity and glucose homeostasis, whereas DKO cells had no effect (Fig. 4f-i). Additionally, GD2TIF-GLP-1 cells reduced the respiratory exchange ratio, energy expenditure, and food intake in HFD-fed mice (Fig. 4j-l). Notably, GD2TIF-GLP-1 cell-treated mice exhibited increased movement compared to those receiving DKO cells (Fig. 4m).
We also evaluated a preventive model in which GD2TIF-GLP-1 cells were infused before initiating the HFD (Supplementary Fig. 8g). A single infusion completely prevented HFD-induced obesity (Supplementary Fig. 8h). Fasting blood glucose levels were significantly lower in GD2TIF-GLP-1-treated mice compared to PBS-treated controls (Supplementary Fig. 8i). At the final analysis, GD2TIF-GLP-1 cells were still detectable in the spleen (Supplementary Fig. 8j, k).
In clinical settings, GLP-1R agonists have been reported to cause side effects, including delayed gastric emptying4. To determine whether GD2TIF-GLP-1 cells induce similar effects, we performed in vivo gastric emptying assay. The results showed that gastric emptying was comparable between control mice and GD2TIF-GLP-1-treated mice on a HFD (Supplementary Fig. 9a, b).
We further employed the conditioned taste avoidance test to evaluate potential gastrointestinal side effects of GD2TIF-GLP-1 cells in mice. After 12 weeks of HFD feeding, mice were trained, conditioned, and tested for their avoidance of hedonic saccharin water. Unlike Exendin-4 treated mice (positive control), mice bearing GD2TIF-GLP-1 cells did not exhibit avoidance of saccharin water after conditioning (Supplementary Fig. 9c, d), suggesting that GD2TIF-GLP-1 cells do not induce overt gastrointestinal side effects.
To further rule out the possibility that the observed weight loss was due to an unhealthy physiological state, we assessed the effects of GD2TIF-GLP-1 cells on body weight in mice fed a normal chow diet (Supplementary Fig. 9e-h). GD2TIF-GLP-1 treatment did not reduce the body weight of these chow-fed mice (Supplementary Fig. 9h). This finding supports the notion that GD2TIF-GLP-1-treated mice experience a targeted reduction in body weight under HFD condition, with final weights comparable to those of chow-fed controls and without excessive weight loss (Fig. 4d and Supplementary Fig. 8h).
Collectively, these findings demonstrate that a single dose of GD2TIF-GLP-1 cells provides long-term protection against diet-induced obesity and diabetes in mice.
Discussion
Chronic diseases are typically considered incurable. While current treatments can alleviate symptoms and slow disease progression, they often require repeated administration of small molecule or protein-based drugs, ranging from daily to monthly, for the duration of the patient’s life. This endless treatment is necessary due to the limited half-lives of these drugs, ranging from hours to days.
For protein-based therapies, one potential approach to achieving a single-dose “cure” is to enable the patient’s body to produce the therapeutic proteins. In this study, we demonstrate that obesity, whether caused by genetic leptin deficiency or, more commonly, by overnutrition, can be effectively and durably treated by a single dose of long-lived CAR T cells that stably secrete the therapeutic biologics leptin and GLP-1. These findings suggest that single-dose therapies for chronic diseases are feasible. Prior to our study, various cells, including CAR T cells, have been engineered to produce therapeutic peptides or proteins in vivo. However, the limited persistence and numbers of these engineered cells have prevented such approaches from offering a “cure” for chronic diseases15.
GD2TIF cells possess three key features that make them well-suited for in vivo delivery of biologics. First, unlike traditional CAR T cell therapies that require preconditioning with chemotherapeutic regimens such as cyclophosphamide and/or fludarabine, GD2TIF cells can expand and persist in vivo without any such preconditioning. This is a significant advantage because these chemotherapeutic agents are associated with various toxicities, including oncogenic risks40,41,42, which are acceptable for cancer patients but not for those with non-cancerous diseases like genetic disorders and obesity. GD2TIF cells are unique in their ability to expand and persist in fully immunocompetent mice without conditioning, making them a promising option for treating common non-cancerous diseases.
Secondly, GD2TIF cells exhibit long-term persistence in vivo at relatively high numbers. While most CAR T cells become undetectable in peripheral blood by flow cytometry within two months post-infusion43, GD2TIF cells maintain approximately 5% of peripheral blood cells in recipient mice for at least one year in mice. This durability is crucial for their function as a continuous source of recombinant proteins. The persistence of GD2TIF cells correlates well with the high expression levels of CD62L and TCF1, two key markers of T cell stemness26,44. Although the lack of expansion of control GD2 CAR T cells, including wild-type, BCOR-deficient, and ZC3H12A-deficient cells has limited our ability to fully dissect the mechanisms underlying GD2TIF cell induction and maintenance, GD2TIF cells share many features with other CAR TIF cells22. This suggests that GD2TIF cells operate through mechanisms similar to those of other CAR TIF cells.
Another distinguishing feature of GD2TIF cells is their unique CAR signaling. GD2TIF cells cannot be induced when 4-1BB is used as the costimulatory domain, indicating that tonic signaling is crucial for GD2TIF cell induction. Indeed, GD2TIF cells can be induced even with an antigen-binding deficient GD2 CAR, though at slightly lower percentages compared to the wild-type GD2 CAR. These findings suggest that tonic signaling is the primary driver for GD2TIF cell induction and maintenance, with interaction with GD2+ cells contributing to a modest increase in cell numbers. GD2, a disialoganglioside antigen, is overexpressed in tumors such as neuroblastoma but is also present at low levels in normal cells45. While targeting GD2 with monoclonal antibodies has been associated with on-target off-tumor toxicities46, GD2 CAR T cells derived from the 14g2a antibody have demonstrated high safety in multiple clinical trials20,47,48. In our study, mice bearing GD2TIF cells did not exhibit abnormalities for at least a year post-infusion, suggesting that GD2TIF cells do not harm normal cells in vital organs or tissues.
Furthermore, the presence of GD2TIF cells does not disrupt the homeostasis of the endogenous immune system or affect susceptibility to viral infections or autoimmune diseases. GD2TIF cells are highly polyclonal, with clonality comparable to that of memory T cells, indicating that their induction does not involve the selection of aberrant or transformed T cell clones. Flow cytometry and scRNA-seq analysis revealed that while the majority of GD2TIF cells exhibit a CD62Lhi stem-like phenotype, a portion are CD62Llo effector-like cells. It remains unclear whether CD62Llo cells originate from CD62Lhi cells or if they maintain themselves independently. Further investigations are needed to determine whether these two populations have equal capabilities for producing biologics.
Using GD2TIF cells as a platform, we achieved long-term in vivo delivery of four different biologics. A single infusion of GD2TIF cells secreting TNF or IL-5 rapidly established mouse models of TNF-induced monocytic inflammation and HES, respectively. Traditionally, these disease models are created through transgenic overexpression of the relevant cytokines, a process that can take months to years and is resource-intensive. Our approach offers a simpler, faster, more cost-effective, and adaptable method for establishing these classic disease models. Furthermore, in leptin-deficient ob/ob mice, a single dose of GD2TIF cells produced leptin and maintained leptin levels at ng/mL concentrations, effectively correcting the obese phenotype of these mice.
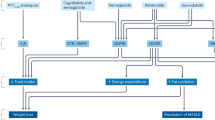
Obesity, which is highly prevalent in modern society and associated with numerous health conditions, is often treated with recombinant GLP-1 or GLP-1R agonists. However, due to their short half-lives, these biologics require daily or weekly administration4, with rapid relapse upon discontinuation5. In a mouse model of diet-induced obesity, a single dose of GD2TIF cells secreting GLP-1 reversed both obesity and diabetes in recipient mice, suggesting a potentially “curative” effect of these cells.
In the clinic, some patients receiving GLP-1R agonists experience gastrointestinal side effects49, likely due to drug overdosing. In our study, mice treated with GD2TIF-GLP-1 cells did not exhibit delayed gastric emptying. The weight loss observed in GD2TIF-GLP-1 treated mice fed on a HFD is unlikely to reflect an unhealthy state, as their body weight decreased only to levels comparable to those of chow-fed mice, without further reduction. Importantly, GD2TIF-GLP-1 cells did not reduce the body weight of chow-fed mice. These findings suggest that GLP-1 was not overdosed in our study. Although we validated that GD2TIF-GLP-1 cells inhibited food intake (Fig. 4l) without causing delayed gastric emptying or an aversive effect toward hedonic saccharin water consumption in mice (Supplementary Fig. 9a-d), fully confirming the central effect of GLP-1-Fc secreted by GD2TIF cells would require the use of a central nervous system-specific GLP-1R conditional knockout mouse model.
Nevertheless, careful titration of GLP-1 secretion may be necessary to achieve an optimal therapeutic response in clinical settings. Several strategies could be employed to control the dose of GLP-1 secreted by GD2TIF-GLP-1 cells in vivo. First, the number of infused GD2TIF-GLP-1 cells can be adjusted. Although these cells proliferate in vivo, a higher cell dose is expected to result in greater GLP-1 production. Second, transcription of GLP-1 in our study is driven by the EFS promoter, which has moderate transcriptional activity. Replacing EFS with a stronger promoter (e.g., EF1α) or a weaker one (e.g., PGK) would correspondingly increase or decrease GLP-1 expression. Third, the GLP-1 peptide used in our study contains three mutations to reduce degradation and is fused to an Fc fragment to extend its half-life. If GD2TIF-GLP-1 activity remains excessive, using wild-type GLP-1 peptide or removing the Fc portion could lower its in vivo activity. Finally, in cases of severe side effects, GD2TIF-GLP-1 cells can be eliminated using antibody-mediated CAR T cell depletion. Together, these strategies provide flexibility for fine-tuning GLP-1 production by GD2TIF-GLP-1 cells in vivo, warranting further investigation.
Building on the success of CAR T cells in cancer treatment50, CAR T cell therapy has recently expanded to treat noncancerous diseases such as autoimmune conditions51,52,53. In this study, we demonstrate that long-lived CAR T cells can serve as a versatile platform for the delivery of biologics. Theoretically, any biologic encoded by DNA can be delivered long-termly by GD2TIF cells. B cells (plasma cells), known for their ability to produce large quantities of antibodies, have been engineered to produce therapeutic antibodies in vivo, such as broad neutralizing antibodies (bnABs) against HIV54,55. These engineered B cells require antigen vaccinations monthly or fortnightly to maintain their persistence in vivo54,55. This requirement undermines the initial goal of cell therapy, as monthly injections of bnABs is likely more effective. Therefore, to overcome the challenge of in vivo persistence of engineered B cells is crucial for their use as a platform for antibody production.
In our study, GD2TIF cells were used to produce four biologics: TNF (235 amino acids), IL-5 (133 amino acids), leptin (167 amino acids), and GLP-1-Fc (285 amino acids with a Fc domain), all of which are relatively small proteins. This may account for their efficient production in vivo. However, the efficiency of GD2TIF cells in delivering larger proteins, such as full-length antibodies, remains to be investigated.
In summary, we have successfully repurposed GD2TIF cells as a robust platform for the stable production of biologics in vivo. This approach offers a single-dose, “curative” cell therapy for chronic diseases that typically require long-term, repeated biologic injections. Given the wide range of diseases currently treated with biologics, our method could provide a one-time solution for multiple conditions. While the current costs of CAR T cell therapy are high, we anticipate that as the price of cell therapy decreases, long-term biologic delivery via CAR T cells will become more cost-effective compared to repeated biologic infusions. Most importantly, this one-time solution could enable patients to live normal lives without the need for ongoing drug administration and frequent hospital visits.
Methods
Mice and cell lines
C57BL/6 mice (000664, Jackson Laboratory), Cas9 transgenic mice (026430, Jackson Laboratory) and ob/ob mice (T001461, GemPharmatech) were maintained under specific pathogen-free conditions at the Laboratory Animal Research Center of Tsinghua University (Beijing, China). Mice were maintained under standard housing conditions with ad libitum access to food and water, except for the fasting blood glucose testing or water restriction. In some experiments (Fig. 4, Supplementary Fig. 8), mice were fed with high-fat diet (60% kCal fat, D12492, Wohua Biologicals Technology). The sex of mice was indicated in the figure legends. Sex was not considered as an independent variable in the experimental designs.
Phoenix-Eco cells (CRL-3214, American Type Culture Collection) were cultured in DMEM (Gibco) containing 5% FBS, 2 mM glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin in a humidified incubator at 37 °C. All cell lines tested negative for mycoplasma.
Plasmids
The TIF induction vector was constructed as described previously (Supplementary Fig. 1)12. The GD2mut CAR was generated by mutating the CDRs of the original GD2 CAR (Supplementary Fig. 4a). The GD2-BBζ CAR was created by replacing the CD28 costimulatory domain in the original GD2 CAR with the mouse 4-1BB transmembrane and signaling domains (Supplementary Fig. 4e).
For the overexpression vectors, a GFP-P2A-ORF cassette was cloned into a pMSCV backbone (74056, Addgene). The cDNAs of mouse TNF, IL-5, leptin, or GLP-1-Fc were cloned into the pMSCV-GFP-P2A-ORF vector by Gibson Assembly. Primers: HpaI-TNF-F:CTGGACCGACCGAGTACGTTAACATGTACAGGATGCAACTCCTG; SalI-TNF-R: CGATAAGCTTGGCTGCAGGTCGACTCACAGGGCAATGATCCCAAAG; HpaI-IL-5-F:CTGGACCGACCGAGTACGTTAACATGAGAAGGATGCTTCTGC; SalI-IL-5-R: CGATAAGCTTGGCTGCAGGTCGACTCAGCCTTCCATTGCCCACTCTG; HpaI-GLP-1-F: CTGGACCGACCGAGTACGTTAACATGTACAGGATGCAACTCC; SalI-GLP-1-R: CGATAAGCTTGGCTGCAGGTCGACTCATTTACCCGGAGTCCGGG; HpaI-Leptin-F: CTGGACCGACCGAGTACGTTAACATGTGCTGGAGACCCCTGTG; SalI-Leptin-R: CGATAAGCTTGGCTGCAGGTCGACTCAGCATTCAGGGCTAACATCC. The GLP-1 cDNA was constructed with an IL2 signal sequence, GLP-1(7-38) with three mutations (A8G, G22E, and R36G), similar to the approved GLP-1R agonist Dulaglutide56, followed by a (GGGGS) × 3 linker and a mouse IgG2a Fc sequence. All sequences were validated by Sanger sequencing.
Production of retrovirus
Retrovirus was produced as described previously22. Briefly, Phoenix-Eco cells were co-transfected with expression plasmids and pCL-Eco helper plasmids using ChemifectTM (#51985, Fengrui Bio). Viral supernatants were collected 48 hours post-transfection, filtered through a 0.45 μm filter, aliquoted, and stored at -80 °C.
T cell activation, transduction and adoptive cell transfer
The isolation, activation, spin-infection, and culture of mouse primary T cells were conducted as previously described12. For co-infection with the TIF induction virus and the overexpression virus, the two viruses were mixed in equal volumes before spin-infection. The T cell culture medium (TCM) consisted of RPMI1640 (Gibco) supplemented with 5% fetal bovine serum (FBS), 2 mM glutamine, 55 μM β-mercaptoethanol, 1 mM sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Twenty-four hours after spin-infection, CD8+Thy1.1+ cells (DKO cells: Fig. 3, Fig. 4, Supplementary Fig. 2, Supplementary Fig. 7; GD2TIF cells: Fig. 1, Supplementary Fig. 1, Supplementary Fig. 3, Supplementary Fig. 4, and Supplementary Fig. 5) or CD8+Thy1.1+GFP+ cells (GD2TIF-TNF: Fig. 2, GD2TIF-IL-5: Fig. 2; GD2TIF-Leptin: Fig. 3, Supplementary Fig. 7; GD2TIF-GLP-1: Fig. 4, Supplementary Fig. 8) were sorted by FACS and transferred into mice via the tail vein injection (1 × 106 cells per mouse). All adoptive cell transfers were performed without any conditioning regimen.
Tissue distribution analysis
Mice were euthanized and perfused with 30 mL of 1× PBS containing 2 mM EDTA, then dissected to isolate fresh liver, lung, and brain tissues. Liver and lung tissues were digested with 1 mg/mL Collagenase IV (#17104-019, Thermo Scientific), passed through a 70-μm strainer (#352350, BD), and resuspended in 45% Percoll (#17089101, Cytiva) for gradient centrifugation at 1000 g, 4 °C, for 15 min. Brain tissues were passed directly through a 70-μm strainer and subjected to the same gradient centrifugation. The sedimented cells were resuspended for FACS staining (Supplementary Fig. 1g,h; Supplementary Fig. 7e,f; Supplementary Fig. 8e, f).
Ex vivo restimulation of GD2TIF and GD2TIF-Leptin cells
Mice were euthanized, and spleens were isolated. Single-cell suspensions were prepared by grinding the tissue through a 70-μm strainer (#352350, BD). For restimulation, 1 × 106 GD2TIF cells were stimulated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA, #HY-18739, MCE) and 1 μM ionomycin (#HY-13434, MCE) for 4 hours in the presence of GolgiStop (#554724, BD) in a 96-well plate (Corning). IFNγ production was analyzed by intracellular staining (Supplementary Fig. 1e, f).
GD2TIF and GD2TIF-Leptin cells were restimulated with 1 μg/mL soluble anti-CD3 and 1 μg/mL anti-CD28 antibodies for 24 hours in a 96-well plate. The supernatant was collected for Leptin quantification by ELISA (Sangon, #D721199), and Leptin concentration was normalized to the number of input cells (Supplementary Fig. 7g).
Cell proliferation assay
Single-cell suspensions (1 × 107 cells/mL in 1× PBS) were stained with CellTraceTM Violet (#C34571, Invitrogen) at a final concentration of 5 μM. Cells were incubated in the dark at 37 °C for 10 minutes in a water bath. Samples were analyzed by flow cytometry (BV421 channel, 405 nm) before and after labeling (Supplementary Fig. 2c). Blood and splenic Thy1.1+ cells were examined on day 4 or day 7 to assess cell proliferation (Supplementary Fig. 2d).
Flow cytometry
Single-cell suspensions were prepared from blood, spleen, bone marrow, liver, lung and brain. Surface protein staining was performed using the indicated antibodies mixed with Fc receptor block (#BE0307, InVivoMab) in FACS buffer (PBS containing 1% calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin) at 4 °C for 15 minutes. Except for listed antibodies below, H-2Db LCMV gp33 Tetramer-KAVYNFATC-PE (#TB-5002-1) was purchased from MBL company. DAPI (#62248, Thermo) was added immediately before flow cytometric analysis to exclude dead cells.
For transcription factor staining, cells were processed using the Transcription Factor Fix/Perm Kit (BD, #562574) following the manufacturer’s instructions (Supplementary Fig. 1e, Supplementary Fig. 2h, Supplementary Fig. 4e). For cytokine staining, T cells were stimulated with 0.2 μg/mL gp33 peptide (QYAOBIO) in the presence of GolgiStop (#554714, BD Pharmingen) in T cell medium at 37 °C for 4.5 hours. After surface staining and live/dead exclusion using fixable Near-IR dye (L34976, Invitrogen), cells were fixed and permeabilized using the BD CytoFix/Perm Kit (#554714, BD Pharmingen). The indicated cytokines were then stained according to the manufacturer’s instructions (Supplementary Fig. 5d). Samples were acquired using an LSRFortessa cytometer (BD). Cell sorting was performed with an S3e sorter (Bio-Rad). Flow cytometry data were analyzed with FlowJo (v10.8.1).
For t-SNE reduction of immune cell profiling, 10,000 single live cells from individual samples were extracted using the DownSample plugin, and multiple sample files were merged by concatenating. Gated cell populations were identified based on surface protein expression and visualized using the built-in KNN algorithm with t-SNE reduction in FlowJo (v10.8.1) (Supplementary Fig. 5d, f).
Antibodies
Anti-mouse CD3ε, Bio X Cell, BE0001-1, RRID: AB_1107634; anti-mouse CD28, Bio X Cell, BE0015-1, RRID: AB_1107624; anti-mouse Thy1.1(CD90.1), Bio X Cell, BE0214, RRID: AB_2687700; anti-mouse Thy1.1 (OX-7), biotin, BioLegend (1:500 dilution), 202510, RRID: AB_2201417; anti-mouse Thy1.1 (OX-7), PE, BioLegend (1:500 dilution), 202524, RRID: AB_1595524; anti-mouse Thy1.1 (OX-7), APC, BioLegend (1:300 dilution), 202526, RRID: AB_1595470; anti-mouse CD8α (53-6.7), Brilliant Violet 421, Thermo Fisher Scientific (1:300 dilution), 100725, RRID: AB_493425; anti-mouse CD4 (RM4-5), APC, BioLegend (1:400 dilution), 100526, RRID: AB_312727; anti-mouse CD11b(M1/70), APC/Cyanine7 (1:600 dilution), BD Biosciences, 561039, RRID: AB_2033993 anti-mouse CD44 (1M7), PerCP/Cyanine5.5, BioLegend (1:300 dilution), 103032, RRID: AB_2076204; anti-mouse CD62L (MEL-14), PE/Cyanine7, BioLegend (1:100 dilution), 104418, RRID: AB_313103; anti-mouse TCF1 (C63D9), PE, Cell Signaling Technology (1:800 dilution), 14456, RRID: AB_2798483; anti-mouse TOX (TXRX10), PE, Thermo Fisher Scientific (1:200 dilution), 12-6502-82, RRID: AB_10855034; anti-mouse EOMES, APC, Thermo Fisher Scientific (1:200 dilution), 17-4875-82, RRID: AB_2866428; anti-mouse Ly108 (13G3-19D), PE, Thermo Fisher Scientific (1:300 dilution), 17-1508-82, RRID: AB_10717668; anti-mouse CXCR5, biotin (L138D7), BioLegend (1:500 dilution), 145510, RRID: AB_2562126; anti-mouse IFN-γ, PE (XMG1.2), BioLegend (1:200 dilution), 505808, RRID: AB_315402; anti-mouse B220, Brilliant Violet 421(RA3-6B2), BioLegend (1:300 dilution), 48-0452-82, RRID: AB_1548761; anti-mouse Siglec-F, PE (1RNM44N), Thermo Fisher Scientific (1:500 dilution), 12-1702-82, RRID: AB_2637129; Streptavidin, PE, BioLegend (1:100 dilution), 405204; Streptavidin, APC, BioLegend (1:100 dilution), 405243.
Single cell RNA-TCR sequencing and data analysis
CD8α+Thy1.1+ GD2TIF cells were isolated from the pooled spleens of 4 mice at 2 months post-transfer for scRNA-seq. scRNA-seq libraries were constructed using the Singleron Matrix® Single Cell Processing System. TCRα/β sequences were amplified using 3’ primers and sequenced. All sequencing was performed on an Illumina NovaSeq 6000 platform with paired-end reads.
Raw data were processed using the CeleScope pipeline (v1.9.0; https://github.com/singleron-RD/CeleScope). Reads were mapped to the GRCm38 reference genome using STAR (v2.6.1a)57. UMI (Unique Molecular Identifier) counts and gene counts for each cell were generated with featureCounts (v2.0.1)58, and expression matrix files were extracted. Downstream analysis was performed using Seurat (v3.1.2)59. Cells were excluded if they had fewer than 200 or more than 4000 genes, or if mitochondrial gene content exceeded 20%. Principal components (PCs) 1-10 were selected for t-SNE, and clustering was determined using the Jackstraw elbow plot with a resolution of 0.5. Unsupervised UMAP reduction was performed, and cells were annotated as CD62Lhi or CD62Llo sub-populations based on differential feature gene expression (Fig. 1d, Supplementary Fig. 3a, d).
Markers for all unsupervised clusters were identified by the FindAllMarker function (Supplementary Fig 3 b). The DoHeatmap function was used to plot top 20 differentially expressed genes between the CD62Lhi and CD62Llo sub-populations (Fig. 1f). GSEA analysis for differentially expressed genes between the CD62Lhi and CD62Llo sub-populations was conducted using clusterProfiler (v4.4.4)60, employing the immunologic signature gene sets ‘C7’ from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb). For scRNA-TCR analysis, uniquely barcoded CDR3_nt sequences were annotated as clonotypes in the Seurat object using the AddMetaData function, and UMAP plots were generated grouped by CDR3_nt. Shannon and Simpson diversity indices were calculated using the vegan package61, with inputs from the ‘clonotypes.csv’ file generated by Cell Ranger.
The expression matrix of splenic TSLEX cells was downloaded from NCBI Gene Expression Omnibus (GEO) library (GSM5988328)26. GD2TIF and TSLEX cells were merged for analysis and harmonization was performed after PCA. Feature gene expression levels were visualized by DotPlot and VlnPlot functions (Supplementary Fig. 3f-h).
Insulin Tolerance Test (ITT) and Glucose Tolerance Test (GTT)
For ITT, mice were fasted for 6 hours (9:00-15:00). Blood glucose levels were measured at 0, 15, 30, 60, 90 and 120 min after insulin (Novolin® R, Novo Nordisk) injection by a glucometer (Yuwell). Insulin dosage was optimized as 0.8 U/kg for HFD mice (Fig. 3) and 0.6 U/kg ob/ob mice (Fig. 4). For GTT, mice were fasted overnight for 16 hours (18:00-10:00. +1 day), and blood glucose levels were measured at 0, 15, 30, 60, 90 and 120 min after D-glucose injection (1 g/kg) using a glucometer (Yuwell).
Metabolic phenotyping
A 16-channel PhenoMaster system (TSE System) was used for indirect metabolic phenotyping, including calorimetry, food intake and physical activity. After 48 hours of acclimation in separate channels with ad libitum access to food and water, data were collected continuously for 48 hours. The respiratory exchange ratio and total energy expenditure were calculated from real-time VO2 and VCO2, measured every 30 minutes. The longitude and lateral lasers interruptions were recorded to assess physical activity (Fig. 3k-n, Fig. 4j-m). The body fat ratio of mice was determined using an EchoMRI analyzer.
LCMV-Armstrong virus infection
The LCMV-Armstrong virus strain was kindly provided by Dr. Yuncai Liu’s lab at Tsinghua University. Frozen virus stocks were diluted in PBS and administered intraperitoneally to mice (2 × 105 PFU per mouse). Mice were euthanized 7.5 days post-infection, and the samples were analyzed by FACS.
Experimental Autoimmune Encephalomyelitis (EAE)
An emulsion of MOG peptide, Incomplete Freund’s Adjuvant (#263910, Difco), and Mycobacterium (#3114-33, Difco) was prepared using a three-way stopcock (Bio-Rad). On day 0, 50 μL of the emulsion was injected subcutaneously at four spots per mouse. Pertussis toxin (#PHZ1174, Invitrogen) was administered on day 0 and day 2. The EAE clinical score was assessed daily from day 7 to day 30. Scoring criteria were as follows: 0.5 for a partially limp tail, 1 for a fully limp tail, 2 for poor righting ability or single hind limb paralysis, 3 for total hind limb paralysis, 4 for 75% body paralysis, 5 for moribund, and 6 for death.
Genetic and HFD-induced obesity models in mice
For the high-fat diet (HFD)-induced obesity and diabetes model, 6-week-old C57BL/6 mice were transferred with PBS, DKO or GD2TIF-GLP-1 cells. One week later, these mice were placed on an HFD (60% kCal fat, D12492, Wohua Biologicals Technology), while a separate group of mice maintained on a normal chow diet served as a control. Body weight was recorded weekly. After 3 months of HFD feeding, the mice were fasted overnight (12 hours), and blood glucose levels were assessed.
In the genetically obese ob/ob mice model, mice were maintained on a normal chow diet. At 4 weeks of age, they were administered PBS, DKO, GD2TIF or GD2TIF-GLP-1 cells, with body weight measurements taken weekly.
Clearance of GD2TIF cells by anti-Thy1.1 antibody
Following FACS examination of GD2TIF cells in peripheral blood, two doses of anti-Thy1.1 antibodies (#BE0214, Bio X Cell) were administered intraperitoneally at a dose of 250 μg/mouse, with a 3-day interval between injections. Eight days after the initial antibody injection, the presence of GD2TIF cells in peripheral blood was assessed by FACS.
Enzyme-Linked Immunosorbent Assay (ELISA)
The Leptin ELISA kit (#D721199) was obtained from Sangon. Serum and cultured supernatant samples from the indicated assays were prepared and analyzed for leptin levels according to the manufacturer’s instructions.
Histology
Mice were euthanized and perfused with PBS containing 2 mM EDTA. The indicated organs were then fixed immediately by immersion in 4% paraformaldehyde to preserve tissue integrity and morphology. The tissue samples were embedded in paraffin, sectioned at a thickness of 7 μm, and mounted onto glass slides. Hematoxylin and eosin (H&E) staining was performed using a kit (E607318, Sangon Biotech) according to the manufacturer’s instructions.
Gastric emptying assay
Fluorescent bead preparation followed a classic gastric emptying test protocol62 with modified parameters. Tentagel-NH2 polymers (Tentagel MB-NH2, 200-250 μm, 0. 15-0.3 mmol/g, #RAP-MB250002, RAPP Polymer, Germany) were reconstituted in DMF (N,N-Dimethylformamide) and allowed to swell for 30 minutes at room temperature. The beads were then washed three times with DMF and labeled using a Cy7 labeling kit (#E-LK-C005C, ElabScience) at a 0.5% equimolar ratio for 90 minutes at 37 °C with shaking at 150 rpm. After washing, a 5-fold molar excess of acetic anhydride was added to block residual -NH2 groups for 30 minutes at room temperature. The labeled beads were washed and dried using a gentle nitrogen stream for storage. All materials should be kept water-free to prevent hydrolysis of the NHS ester.
Mice were fasted for 6 hours prior to the gastric emptying test. Before gavage, the labeled beads were resuspended in 0.5% methylcellulose in 1× PBS and administered via oral gavage at 200 mg/kg body weight. Anesthesia was induced using 3% isoflurane in oxygen (100-200 mL/min) and maintained using a nose cone during imaging. Fluorescence (excitation: 710 nm; emission: 760 nm) was measured using the IVIS Spectrum in vivo imaging system (PerkinElmer) at 0 and 30 minutes post-gavage. Changes in total radiant efficiency ([p/s]/[µW/cm2]) within the region of interest were quantified using Living Image software (PerkinElmer, version 4.8.0).
Conditioned taste avoidance test
Mice were adapted by water restriction 11 hours and supplementation in an hour with two bottles of water, twice daily, for 5 consecutive days. PBS group mice and GD2TIF-GLP-1 cells bearing mice were conditioned with 0.1% saccharin (HY-Y0272, MCE). A group of age and sex matched mice were adopted as positive control, 10 μg/kg Exendin-4 (HY13443, MCE) was intraperitoneally injected immediately after the conditioning, to induce the aversive reaction to the hedonic saccharin63.
At day 2 post-conditioning, mice were water restricted for 12 hours and supplemented with 0.1% saccharin and non-flavored plain water. After 30 minutes of re-feeding, the position of the the bottles were exchanged. Water consumption (g) within 1 hour of re-feeding was recorded and the hedonic saccharin avoidance was calculated as:
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analysis. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. All numerical data were analyzed using Prism (version 10), with analytical methods specified in the figure legends for each graph. Illustrations and graph layouts were created using Adobe Illustrator (2021). Error bars represent the standard error of the mean (SEM).
Ethical statement
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Tsinghua University (protocol number: 17-PM-1), and were conducted in accordance with institutional guidelines and standard animal welfare regulations.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data from this study have been shown in Figures. Source data are provided with this paper. Raw data of scRNA sequencing and TCR sequencing can be accessed through Gene Expression Omnibus (GEO) by accession number GSE291791. The splenic TCF1+ TSLEX cell scRNA-seq data for merged analysis are available in the GEO database under accession number GSM5988328. Source data are provided with this paper.
References
Vargason, A. M., Anselmo, A. C. & Mitragotri, S. The evolution of commercial drug delivery technologies. Nat. Biomed. Eng. 5, 951–967 (2021).
Cubillos-Ruiz, A. et al. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 20, 941–960 (2021).
Li, Z., Wang, Y., Gu, Z. & Hu, Q. Engineering cells for therapy and diagnosis. Nat. Rev. Bioeng. 2, 770–784 (2024).
Holst, J. J. GLP-1 physiology in obesity and development of incretin-based drugs for chronic weight management. Nat. Metab. 6, 1866–1885 (2024).
Wilding, J. P. H. et al. Weight regain and cardiometabolic effects after withdrawal of semaglutide: The STEP 1 trial extension. Diabetes Obes. Metab. 24, 1553–1564 (2022).
Baryakova, T. H., Pogostin, B. H., Langer, R. & McHugh, K. J. Overcoming barriers to patient adherence: the case for developing innovative drug delivery systems. Nat. Rev. Drug Discov. 22, 387–409 (2023).
Rosenberg, V., Chodick, G., Xue, Z., Faccin, F. & Amital, H. Real-world data of adherence and drug survival of biologics in treatment-naive and treatment-experienced adult patients with rheumatoid arthritis. Adv. Ther. 40, 4504–4522 (2023).
Uzoigwe, C., Liang, Y., Whitmire, S. & Paprocki, Y. Semaglutide once-weekly persistence and adherence versus other GLP-1 RAs in patients with type 2 diabetes in a US real-world setting. Diabetes Ther. 12, 1475–1489 (2021).
Garces, S. & Demengeot, J. The immunogenicity of biologic therapies. Curr. Probl. Dermatol 53, 37–48 (2018).
de Speville, B. D. & Moreno, V. Antidrug antibodies and drug development: challenges in the immunotherapy era. Clin. Cancer Res. 27, 2669–2671 (2021).
Nathwani, A. C. et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 365, 2357–2365 (2011).
Martinez-Navio, J. M. et al. Adeno-associated virus delivery of anti-HIV monoclonal antibodies can drive long-term virologic suppression. Immunity 50, 567–575 e565 (2019).
Casazza, J. P. et al. Safety and tolerability of AAV8 delivery of a broadly neutralizing antibody in adults living with HIV: a phase 1, dose-escalation trial. Nat. Med. 28, 1022–1030 (2022).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
Roybal, K. T. et al. Precision tumor recognition by t cells with combinatorial antigen-sensing circuits. Cell 164, 770–779 (2016).
Kaczanowska, S. et al. Genetically engineered myeloid cells rebalance the core immune suppression program in metastasis. Cell 184, 2033–2052 e2021 (2021).
Adachi, K. et al. IL-7 and CCL19 expression in CAR-T cells improves immune cell infiltration and CAR-T cell survival in the tumor. Nat. Biotechnol. 36, 346–351 (2018).
Nazha, B., Inal, C. & Owonikoko, T. K. Disialoganglioside GD2 expression in solid tumors and role as a target for cancer therapy. Front Oncol. 10, 1000 (2020).
Schulz, G. et al. Detection of ganglioside GD2 in tumor tissues and sera of neuroblastoma patients. Cancer Res 44, 5914–5920 (1984).
Majzner, R. G. et al. GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature 603, 934–941 (2022).
Long, A. H. et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015).
Wang, L. et al. Induction of immortal-like and functional CAR T cells by defined factors. J. Exp. Med. 221, e20232368 (2024).
Matsushita, K. et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 (2009).
Huynh, K. D., Fischle, W., Verdin, E. & Bardwell, V. J. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14, 1810–1823 (2000).
Jin, G. et al. A single infusion of engineered long-lived and multifunctional T cells confers durable remission of asthma in mice. Nat. Immunol. 25, 1059–1072 (2024).
Tsui, C. et al. MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature 609, 354–360 (2022).
Restifo, N. P. & Gattinoni, L. Synthetic soldiers: Turning T cells into immortal warriors. J. Exp. Med. 221, e20240258 (2024).
Sheih, A. et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat. Commun. 11, 219 (2020).
Ahmed, M. et al. Clonal expansions and loss of receptor diversity in the naive CD8 T cell repertoire of aged mice. J. Immunol. 182, 784–792 (2009).
Oakes, T. et al. Quantitative characterization of the T cell receptor repertoire of naive and memory subsets using an integrated experimental and computational pipeline which is robust, economical, and versatile. Front Immunol. 8, 1267 (2017).
Keffer, J. et al. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 10, 4025–4031 (1991).
Requena, G. et al. Clinical profile and treatment in hypereosinophilic syndrome variants: a pragmatic review. J. Allergy Clin. Immunol. Pr. 10, 2125–2134 (2022).
Dent, L. A., Strath, M., Mellor, A. L. & Sanderson, C. J. Eosinophilia in transgenic mice expressing interleukin 5. J. Exp. Med. 172, 1425–1431 (1990).
Kouro, T. & Takatsu, K. IL-5- and eosinophil-mediated inflammation: from discovery to therapy. Int Immunol. 21, 1303–1309 (2009).
Ingalls, A. M., Dickie, M. M. & Snell, G. D. Obese, a new mutation in the house mouse. J. Hered. 41, 317–318 (1950).
Bodirsky, B. L. et al. The ongoing nutrition transition thwarts long-term targets for food security, public health and environmental protection. Sci. Rep. 10, 19778 (2020).
Lee, Y. S., Wollam, J. & Olefsky, J. M. An integrated view of immunometabolism. Cell 172, 22–40 (2018).
Alhabeeb, H. et al. Gut hormones in health and obesity: the upcoming role of short chain fatty acids. Nutrients 13, 481 (2021).
Zhang, T., Perkins, M. H., Chang, H., Han, W. & de Araujo, I. E. An inter-organ neural circuit for appetite suppression. Cell 185, 2478–2494 e2428 (2022).
Marachelian, A., Butturini, A. & Finlay, J. Myeloablative chemotherapy with autologous hematopoietic progenitor cell rescue for childhood central nervous system tumors. Bone Marrow Transpl. 41, 167–172 (2008).
Bhatia, S. Long-term health impacts of hematopoietic stem cell transplantation inform recommendations for follow-up. Expert Rev. Hematol. 4, 437–452 (2011).
Inamoto, Y. et al. Secondary solid cancer screening following hematopoietic cell transplantation. Bone Marrow Transpl. 50, 1013–1023 (2015).
Brentjens, R. J. et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 118, 4817–4828 (2011).
Kaech, S. M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Lammie, G., Cheung, N., Gerald, W., Rosenblum, M. & Cordoncardo, C. Ganglioside gd(2) expression in the human nervous-system and in neuroblastomas - an immunohistochemical study. Int J. Oncol. 3, 909–915 (1993).
Uttenreuther-Fischer, M. M., Huang, C. S., Reisfeld, R. A. & Yu, A. L. Pharmacokinetics of anti-ganglioside GD2 mAb 14G2a in a phase I trial in pediatric cancer patients. Cancer Immunol. Immunother. 41, 29–36 (1995).
Gargett, T. et al. Safety and biological outcomes following a phase 1 trial of GD2-specific CAR-T cells in patients with GD2-positive metastatic melanoma and other solid cancers. J. Immunother. Cancer 12, e008659 (2024).
Del Bufalo, F. et al. GD2-CART01 for Relapsed or Refractory High-Risk Neuroblastoma. N. Engl. J. Med 388, 1284–1295 (2023).
Drucker, D. J. Efficacy and Safety of GLP-1 Medicines for Type 2 Diabetes and Obesity. Diabetes Care 47, 1873–1888 (2024).
June, C. H., O’Connor, R. S., Kawalekar, O. U., Ghassemi, S. & Milone, M. C. CAR T cell immunotherapy for human cancer. Science 359, 1361–1365 (2018).
Wang, X. et al. Allogeneic CD19-targeted CAR-T therapy in patients with severe myositis and systemic sclerosis. Cell 187, 4890–4904.e4899 (2024).
Chung, J. B., Brudno, J. N., Borie, D. & Kochenderfer, J. N. Chimeric antigen receptor T cell therapy for autoimmune disease. Nat. Rev. Immunol. 24, 830–845 (2024).
Baker, D. J., Arany, Z., Baur, J. A., Epstein, J. A. & June, C. H. CAR T therapy beyond cancer: the evolution of a living drug. Nature 619, 707–715 (2023).
Hartweger, H. et al. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J. Exp. Med 216, 1301–1310 (2019).
Nahmad, A. D. et al. In vivo engineered B cells secrete high titers of broadly neutralizing anti-HIV antibodies in mice. Nat. Biotechnol. 40, 1241–1249 (2022).
Glaesner, W. et al. Engineering and characterization of the long-acting glucagon-like peptide-1 analogue LY2189265, an Fc fusion protein. Diabetes Metab. Res Rev. 26, 287–296 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Camb.) 2, 100141 (2021).
vegan: Community Ecology Package. Comprehensive R Archive Network (CRAN).
Gremlich, H. U. et al. Noninvasive assessment of gastric emptying by near-infrared fluorescence reflectance imaging in mice: pharmacological validation with tegaserod, cisapride, and clonidine. Mol. Imaging 3, 303–311 (2004).
Kanoski, S. E., Rupprecht, L. E., Fortin, S. M., De Jonghe, B. C. & Hayes, M. R. The role of nausea in food intake and body weight suppression by peripheral GLP-1 receptor agonists, exendin-4 and liraglutide. Neuropharmacology 62, 1916–1927 (2012).
Acknowledgements
We thank Institute for Immunology at Tsinghua University for providing and maintaining equipment. We thank Dr. Yuncai Liu (Tsinghua University) for providing LCMV Armstrong viruses. This research was supported by the National Natural Science Foundation of China (82350108 and T2495270 to M.P.), Tsinghua University DUSHI Program (52302102323 to M.P.), Tsinghua-Peking Center for Life Sciences (to M.P.), SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine (to M.P.).
Author information
Authors and Affiliations
Contributions
G.Z. performed experiments with ideas from M. P., analyzed the data; N.Y. provided technical help; N.Y. supervised the project; M.P. conceived and supervised the project, analyzed and interpreted data, and wrote the paper with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
A patent application (WO2023082640A1) related to this work has been filed by Tsinghua University, with M.P. and G.Z. listed as inventors. The application pertains to the use of GD2TIF cells for long-term delivery of biologics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Roland Schelker, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, G., Yin, N. & Peng, M. GD2TIF cells as a platform for single-dose and long-term delivery of biologics. Nat Commun 16, 8088 (2025). https://doi.org/10.1038/s41467-025-63427-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63427-w