Abstract
RNAi technology, which can induce mortality by disrupting the transcription of essential growth and development-related genes in insects, has emerged as a groundbreaking pest control method. However, insects have developed defense mechanisms to counteract the efficiency of RNAi. The specific role of symbiotic microorganisms in this process remains poorly understood and requires further exploration. This study examines the reduced RNAi efficiency in Lepidopteran pest Helicoverpa armigera. Through screening, six Bacillus strains exhibiting dsRNA-degrading activity were identified through in vitro assays. Further investigation into one representative strain Ba 6 revealed that it significantly decreased RNAi efficiency by secreting ribonuclease into the insect gut fluid, directly degrading dsRNA, thus reducing its accumulation and blocking RNAi effects. These findings clarify the mechanism by which symbiotic bacteria influence the host’s RNAi efficiency and provides a valuable reference for the development and large-scale implementation of RNA biopesticides targeting H. armigera and other lepidopteran pests.
Similar content being viewed by others
Introduction
RNAi is widely recognized as a potent mechanism for silencing transposon activity, counteracting viral infections, and regulating gene expression. This pathway, conserved throughout biological evolution, has been extensively leveraged in gene function studies, biomedicine, and pest control1,2. As an innovative generation of pesticides, RNAi-based pest control offers significant potential for widespread application. The first RNAi-based transgenic maize SmartStax® PRO and the first spray-based RNA biopesticide CalanthaTM were approved for registration by the US EPA in 2017 and 2023, respectively. Several other products in this category are currently pending approval. The core principle of this approach involves the introduction of dsRNA into the insect, where it is transported to tissue cells to inhibit the expression of target genes, thereby disrupting the insect’s growth and development. However, the varying efficacy of RNAi across different insect taxa limits its broader utility in gene function research and pesticide development3,4. Most studies have shown that RNAi efficiency is relatively low in Lepidoptera insects5,6,7. Since Lepidoptera are among the most widespread and economically damaging agricultural pests, their reduced sensitivity to RNAi poses a significant challenge to the broader application of this technology8,9. Recent studies suggest that factors such as the insect’s physiological state, cellular mechanisms for dsRNA uptake and transport, and the activity of dsRNA-degrading nucleases can impact RNAi efficiency10,11.
Upon ingestion by insects, dsRNA is absorbed by gut cells and transported within the body, making its stability in insect body fluids critical to its effectiveness. Insect body fluids host a substantial number of symbiotic microorganisms, and through long-term co-evolution, these microbes and their insect hosts have developed a mutually beneficial relationship. Insects provide a stable habitat and nutritional resources, while symbiotic bacteria contribute to essential physiological processes, including growth, development, reproduction, and immunity12,13. Given the ubiquity of symbiotic bacteria in insect body fluids, dsRNA inevitably interacts with these microorganisms upon entering the insect, although the exact nature of these interactions remains unclear. Xie et al. demonstrated that dsRNA-mediated gene knockdown can disrupt the intestinal commensal bacteria of locusts14. Once introduced into the insect gut, dsRNA is degraded into ribonucleotides, which in turn stimulate the proliferation of the pathogenic bacterium Pseudomonas putida, accelerating insect mortality15. Additionally, feeding P. putida has been shown to enhance host RNAi efficiency16. While most research has focused on the effects of dsRNA on commensal bacteria, the potential impact of these bacteria on dsRNA stability remains unexplored.
In this study, the relationship between symbiotic bacteria in the cotton bollworm and RNAi efficiency was investigated. Through cultivation and screening, six symbiotic bacterial strains capable of degrading dsRNA were identified. Further analysis of strain Ba 6 revealed that it decreases RNAi efficiency by secreting the nuclease Ribonuclease, which directly degrades dsRNA. This discovery offers new insights into factors influencing RNAi efficiency in insects, providing a valuable reference for enhancing the application of RNAi technology in pest management.
Results
dsRNA is extremely unstable in body fluids of cotton bollworms
The cotton bollworm’s midgut hosts a rich community of symbiotic bacteria. To explore their influence on dsRNA stability, metagenomic sequencing was performed on the guts of fourth and fifth instar larvae (Tables S1, S2). The microbiota profiles across both instars were notably consistent. At the genus level, Enterococcus was the dominant taxon, with other prominent symbionts including Saccharomyces, Betabaculovirus, Fusarium, and Bacillus (Fig. 1a). At the species level, Enterococcus mundtii, Saccharomyces cerevisiae, Fusarium venenatum, and Bacillus cereus were the most abundant (Fig. 1b). KEGG analysis of the functional genes revealed that the symbiotic bacteria primarily participated in metabolism-related pathways, with enrichment in pathways such as carbohydrate metabolism, amino acid metabolism, nucleotide metabolism, and energy metabolism (Fig. 1c).
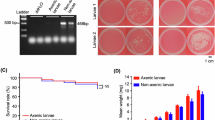
a Relative abundance of gut microbiota at the genus level in 4th and 5th instar cotton bollworm larvae. b Relative abundance of gut microbiota at the species level. c KEGG pathway analysis showing major metabolic pathways of macrogenomic genes. d Stability of dsRNA in undiluted hemolymph and midgut fluid. e Stability of dsRNA in diluted hemolymph and midgut fluid. M: DNA marker 2000; CK: dsRNA incubated in PBS; 1: hemolymph; 2: midgut fluid.
The midgut fluid and hemolymph of H.armigera exhibit significant enzymatic activity, with dsRNA (dsEGFP) being rapidly degraded even at diluted concentrations (Fig. 1d, e, Fig. S1). This highlights the critical role of the insect’s internal environment in maintaining dsRNA stability.
Screening and characterization of intestinal symbiotic bacteria with dsRNA degrading activity
The intestinal symbiotic bacteria of cotton bollworms were isolated and cultured in vitro. Single colonies were co-incubated with dsRNA, and six strains (Ba 1, Ba 2, Ba 3, Ba 4, Ba 5, Ba 6) capable of degrading dsRNA were identified through gel electrophoresis, which assessed dsRNA stability and integrity (Fig. 2a, Figs. S2, S3a, S4a). Nuclease activity of these strains was further analyzed using an RNase activity testing plate designed to screen RNase-producing bacteria. Ba 3 and Ba 6 exhibited the highest nuclease activity, as indicated by the larger aperture diameters (Fig. 2b, c). Supernatants from all six strains were subsequently collected via low-speed centrifugation at low temperatures and co-incubated with dsRNA. Gel electrophoresis results demonstrated that these supernatants also degraded dsRNA (Fig. 2d, Figs. S3b, S4b), suggesting the secretion of extracellular nucleases by these bacteria.
a Gel electrophoresis of dsRNA (2000 ng/μL) stability after incubation with six symbiotic bacteria; M: DNA marker 2000; CK: dsRNA incubated in LB. b, c RNase activity assay results for the six symbiotic bacteria. d Gel electrophoresis showing dsRNA (2000 ng/μL) degradation in the supernatants of the six symbiotic bacteria. e Phylogenetic tree construction of the six symbiotic bacteria. f Nuclease gene distribution among the six bacterial genomes. g Extracellular nuclease gene distribution. h Phylogenetic tree of extracellular nucleases identified in the six bacterial strains.
Characterization of symbiotic bacteria
To further elucidate the functions of these strains, whole genome sequencing was conducted (Figs. S5–10, Table S3). Strains Ba 1 and Ba 5 were identified as Bacillus altitudinis, while Ba 2, Ba 3, Ba 4, and Ba 6 were classified as Bacillus cereus (Table S4). All six strains belong to the Bacillus genus. Phylogenetic analysis (Fig. 2e) and synteny analysis (Fig. S11) studies showed that Ba 1 and Ba 5 clustered into one branch, while Ba 2, Ba 3, Ba 4, and Ba 6 clustered into another.
Building on these findings, it was hypothesized that symbiotic bacteria degrade dsRNA by secreting nucleases. Genome analysis of the six strains revealed a significant number of nuclease genes (Fig. 2f). Examination of signal peptide sequences identified extracellular nucleases in all strains. Specifically, Ba 1 and Ba 5 were found to harbor three types of extracellular nucleases, Ba 2, Ba 3, and Ba 4 contained two, and Ba 6 contained one (Fig. 2g). These nucleases were classified into the DNase_NucA_NucB, EndA, microbial_RNases, and COG2374 superfamilies (Table S5). A phylogenetic tree was constructed, grouping nucleases from the same subfamily together (Fig. 2h).
Ba 6 has no effect on the growth and development of cotton bollworm
The supernatant of Ba 6 exhibited strong dsRNA degradation activity but showed only weak DNA degradation (Fig. 3a, Fig. S12). As a result, Ba 6 was selected for further investigation of its effects on the host. SEM revealed that Ba 6 cells are rod-shaped with blunt, rounded ends, reproducing via cell division (Fig. 3b). Multilocus sequence typing (MLST) confirmed that Ba 6 clustered within the same evolutionary branch as Bacillus cereus (Tables S6–S7, Fig. S13).
a Effect of Ba 6 suspension and supernatant on dsRNA and dsDNA stability. b Scanning electron microscopy image of Ba 6 (scale bar: 5 µm). c qRT-PCR analysis showing Ba 6 colonization in the cotton bollworm gut. d Relative abundance of intestinal bacteria at the phylum level in cotton bollworms. e Relative abundance at the genus level. f Simpson analysis of α diversity index values. g Principal component analysis (PCoA) of Bray-Curtis distances between intestinal bacterial communities. CK midgut of cotton bollworms fed a normal diet, LB LB-supplemented diet, B6 Ba 6-supplemented diet; P-values calculated by the Kruskal-Wallis H test: ***P < 0.001; **P < 0.01; *P < 0.05. The length (h), weight (i), and phenotype (j) of the cotton bollworms colonized with Ba 6; Differences between data were analyzed by Student’s t-test: ***P < 0.001; **P < 0.01; *P < 0.05.
Feeding cotton bollworms with a Ba 6 suspension applied to the food surface led to a significant increase in Ba 6 abundance in the gut after three days, as confirmed by qRT-PCR analysis (Fig. 3c). Subsequent 16S rRNA sequencing of the gut microbiota revealed marked shifts in bacterial composition. At the phylum level, the dominant bacteria shifted from Proteobacteria to Firmicutes (Fig. 3d), while at the genus level, Pseudomonas was replaced by Bacillus as the dominant genus (Fig. 3e). α-diversity analysis indicated that bacterial richness and diversity were significantly reduced in the treatment group compared to the two control groups, although no significant difference in diversity was observed between the control groups themselves (Fig. 3f, Fig. S14a, b). PCoA analysis further highlighted distinct shifts in community composition between the treatment group and the controls (Fig. 3g). Despite these microbiota changes, no significant differences were observed in survival rate (Fig. S14c), body length (Fig. 3h), body weight (Fig. 3i), or other developmental parameters between Ba 6-colonized cotton bollworms and the control group (Fig. 3j).
Ba 6 can reduce the RNAi efficiency of the host cotton bollworm
To assess the impact of the Ba 6 strain on dsRNA stability in vivo, fluorescently labeled dsRNA was injected into cotton bollworms, and fluorescence intensity was monitored after 1 and 6 hours. A significant reduction in fluorescence intensity was observed in Ba 6-colonized cotton bollworms (Ba 6 + Cy3-dsEGFP) compared to controls (LB+Cy3-dsEGFP) (Fig. 4a), indicating that Ba 6 can degrade dsRNA within the host.
a dsRNA degradation capability in Ba 6-colonized cotton bollworms. LB + dsEGFP, LB + dsEGFP-Cy3, Ba 6 + dsEGFP-Cy3: Cotton bollworms treated with LB and dsEGFP, LB and dsEGFP-Cy3, and Ba 6 and dsEGFP-Cy3, respectively. BF Bright field image; 540 nm: Excitation wavelength; scale bar: 0.5 cm. b, c Relative expression of target genes CarE and BtR after dsCarE and dsBtR injection in Ba 6-colonized cotton bollworms. d Expression levels of Ago-2 and Dicer in different treatment groups (CK, LB, Ba 6). e Changes in Ago-2 and Dicer expression in Ba 6-colonized cotton bollworms and LB medium-treated controls after dsRNA injection. Effects on target gene inhibition efficiency (f, h, j, l) and cotton bollworm survival rate (g, i, k, m) following feeding with Ba 6 suspension or supernatant, and subsequent feeding with dsCarE or dsTm-1-containing diet. Data analysis in (b–e, f, h, j, l) was performed using Student’s t-test: ***P < 0.001; **P < 0.01; *P < 0.05. Survival data in (g, i, k, m) were analyzed by Log-rank (Mantel-Cox): ***P < 0.001; **P < 0.01; *P < 0.05.
Injections of dsCarboxylesterase (dsCarE) and dscadherin (dsBtR) into Ba 6-colonized cotton bollworms failed to effectively suppress the expression of target genes, resulting in reduced RNAi efficiency compared to control insects (Fig. 4b, c). Notably, feeding Ba 6 did not alter the baseline expression of key RNAi pathway genes, Argonaute-2 (Ago-2) and Endoribonuclease Dicer (Dicer) (Gene ID:110381609) (Fig. 4d). However, upon dsRNA treatment, the expression of these genes in Ba 6-colonized cotton bollworms was significantly lower than in the control group (Fig. 4e), potentially compromising RNAi efficiency.
The effect of Ba 6 colonization on RNAi efficiency was further confirmed by dsRNA feeding experiments. After Ba 6 colonization, cotton bollworms fed with dsCarE and dsTropomyosin (dsTm-1) exhibited poor suppression of the target genes (Fig. 4f, h). Moreover, mortality rates due to gene interference were significantly lower in Ba 6-colonized cotton bollworms compared to controls (Fig. 4g, i).
Further experiments where Ba 6 supernatant was fed to cotton bollworms demonstrated that the impact of the bacterial supernatant on RNAi efficiency was comparable to that of the Ba 6 bacteria (Fig. 4j–m). These results suggest that both Ba 6 bacteria and their secreted products play a role in modulating RNAi efficiency in cotton bollworms.
Screening and identification of Ba 6 effectors and their effects on RNAi efficiency
Based on prior experimental findings, Ba 6 likely degrades dsRNA through the secretion of specific nucleases. Subsequent tests confirmed that dsRNA degradation activity was only observed in the supernatant and suspension of Ba 6, while the bacterial cells themselves did not exhibit such activity (Fig. 5a, Fig. S15a). When the Ba 6 supernatant was inactivated by high temperature, its dsRNA-degrading capability was lost (Fig. 5b, Fig. S15b), suggesting that the active component is likely an extracellular nuclease.
a Gel electrophoresis analysis of dsRNA incubated with Ba 6 cells, suspension, and supernatant for 4 hours. b dsRNA degradation by Ba 6 supernatant following heat treatment. c Western blotting analysis detecting Ribonuclease in Ba 6 supernatant. d Ribonuclease protein degradation of dsRNA. e, f Relative expression of target genes CarE and BtR after dsCarE and dsBtR injection in cotton bollworms treated with Ribonuclease. g Fluorescence observation of dsRNA degradation by Ribonuclease in cotton bollworms. CK + dsEGFP, CK + dsEGFP-Cy3, Ribonuclease + dsEGFP-Cy3: Cotton bollworms treated with dsEGFP, dsEGFP-Cy3, and Ribonuclease + dsEGFP-Cy3, respectively. BF: Bright field image; 540 nm: Excitation wavelength; scale bar: 0.5 cm. h, j Effect of Ribonuclease feeding followed by dsCarE or dsTm-1 on target gene expression. i, k Effect on insect survival rate post-Ribonuclease feeding followed by dsCarE or dsTm-1. Data analysis in (e, f, h, j) was performed using Student’s t-test: ***P < 0.001; **P < 0.01; *P < 0.05. Survival data in (i, k) were analyzed by Log-rank (Mantel-Cox): ***P < 0.001; **P < 0.01; *P < 0.05.
Genome analysis of Ba 6 identified a single extracellular nuclease, Ribonuclease, which may be critical in destabilizing host dsRNA (Table S5). A polyclonal antibody against Ribonuclease was generated, and the presence of the protein in the Ba 6 supernatant was confirmed via Western blotting (Fig. 5c, Figs. S15c, S16). Additionally, recombinant Ribonuclease produced using a prokaryotic expression system demonstrated rapid dsRNA degradation in vitro (Fig. 5d, Fig. S15d), further establishing Ribonuclease as the primary effector responsible for the dsRNA degradation activity in Ba 6.
Feeding cotton bollworms with Ribonuclease protein accelerated the degradation of Cy3-labeled dsRNA, with fluorescence disappearing entirely after 6 hours (Fig. 5g). Injections of dsBtR and dsCarE similarly failed to suppress the expression of target genes effectively (Fig. 5e, f). To further assess RNAi efficiency, dsRNA feeding experiments were conducted. Cotton bollworms that consumed Ribonuclease protein exhibited significantly reduced RNAi efficiency, resulting in insufficient suppression of target genes (Fig. 5h, j). This was accompanied by a marked reduction in mortality rates due to ineffective gene suppression post-treatment (Fig. 5i, k). These results clearly demonstrate that Ribonuclease impairs host RNAi by degrading dsRNA in vivo.
Based on our hypothesis, the Ba 1, Ba 2, Ba 3, Ba 4, and Ba 5 strains isolated from the cotton bollworm gut may function similarly to Ba 6. Assuming a potential functional similarity, these strains likely colonize the host’s intestines and secrete nucleases into the intestinal cavity. Upon the introduction of exogenous dsRNA, these bacterial-secreted nucleases degrade it, preventing dsRNA from effectively penetrating the intestinal epithelial cells and thereby inhibiting the host’s RNAi efficiency (Fig. 6).
Discussion
Insects host a substantial number of commensal microorganisms within their internal environments, accounting for 1–10%17 of the insect’s dry weight. These microorganisms are vital for regulating various physiological processes, including host growth, development, reproduction, and immunity, significantly contribute to the survival and evolution of insects12,13. Insects manage external invaders and initiate innate immune responses by modulating the structure of their microbial communities and controlling the metabolites produced by these microbes18. The secretions from symbiotic microorganisms can directly degrade exogenous substances19. For example, Pseudomonas in Rhagoletis pomonella20, Burkholderia in Riptortus pedestris21, and Bacillus cereus in Plutella xylostella22 have demonstrated the ability to break down toxic compounds, such as pesticides.
The RNAi mechanism, triggered by dsRNA, is recognized as an exogenous toxin to insects and plays an essential role in the antiviral response. In lepidopteran insects, which are highly susceptible to viral infections, symbiotic bacteria may modulate responses to viral threats through their initial interactions with the host.
This study focused on intestinal symbiotic bacteria that influence the host’s RNAi pathway. Six Bacillus strains isolated from the cotton bollworm were found to secrete nucleases into the extracellular environment, which directly degrade dsRNA in the insect gut, thereby affecting RNAi efficiency. Bacillus species are known for their robust capacity to secrete nucleases, with over 20 species possessing this ability. The predominant nucleases secreted by Bacillus belong to the N1/T1 ribonuclease family (EC 3.1.27.3), characterized by small extracellular basic proteins of approximately 12.5 kDa. This family includes balifase from Bacillus licheniformis, barnase from Bacillus amyloliquefaciens and Bacillus circulans, and binase produced by Bacillus pumilus, Bacillus coagulans, and Bacillus thuringiensis23,24,25,26,27,28. Nucleases secreted by Ba 1 and Ba 5 belong to the DNase_NucA_NucB superfamily, containing the conserved barnase domain. Another extracellular nuclease secreted by Bacillus, with a high molecular mass of approximately 30 kDa, includes members such as Bsn from Bacillus subtilis and binase II from Bacillus pumilus29,30. The nucleases secreted by Ba 1, Ba 2, Ba 3, Ba 4, Ba 5, and Ba 6 contain the conserved EndA domain, which is characteristic of this class. These nucleic acid-degrading bacteria, found in the body fluids of insects, can directly degrade dsRNA.
Previous studies have identified numerous nucleases present in the gut, hemolymph, and salivary glands of insects that can degrade dsRNA. These nucleases are often upregulated in response to dsRNA stimulation, thereby influencing the efficiency of RNAi31,32,33,34,35,36. Analysis of insect-expressed and bacterially-secreted nucleases reveals that the majority of insect nucleases belong to the Endonuclease_NS domain (DsNucleases, PF01223)35,37,38. Interestingly, the EndA superfamily, microbial_RNases superfamily and COG2374 superfamily nucleases secreted by Bacillus are also classified as endonucleases (Table S5). This indicates potential parallels in their biochemical activities and biological roles between insect endogenous nucleases and bacterial secreted nucleases, underscoring their pivotal role in modulating the host RNAi pathway. These results enhance our comprehension of the factors influencing RNAi efficiency.
The structure of an insect’s gut microbiota is highly variable. At the phyla level, the symbiotic bacteria in insects mainly belong to Firmicutes and Proteobacteria, but at the genus level, the bacterial community composition varies significantly among insect species39. Genera such as Enterococcus, Staphylococcus, Lactococcus, Lactobacillus, Streptococcus, Bacillus, Methylobacterium, Pseudomonas, Acinetobacter, Sphingomonas, Klebsiella, and Enterobacter are widely distributed in Lepidoptera40. In the Coleoptera species Phalacrognathus muelleri, the dominant symbiotic genera include Dysgonomonas, Enterococcus, Lactococcus, Prevotella, Turicibacter, Sporomusa, Candidatus, and Coprococcus41. For Hemiptera, the predominant symbiotic bacteria are Chloroplast_unclassified, Serratia, Lactococcus, Lactobacillus, and Klebsiella42. These bacterial variables can be shaped by feeding habits, environmental interactions, and contact with other organisms43,44. Furthermore, the gut pH is also a significant factor. The gut fluid of Coleoptera and Hemiptera tends to be weakly acidic, while in Orthoptera, Diptera, and Hymenoptera, it is mildly alkaline. In contrast, the midgut of Lepidopteran larvae is highly alkaline, with pH levels reaching 11–1245. This alkaline environment is conducive for the proliferation of Bacillus, which is an alkaliphilic bacterium46, contributing to the higher prevalence of Bacillus in Lepidoptera. Furthermore, researches have shown that dsRNA stability is pH-dependent, being most stable between pH 4.0–5.0, but hydrolyzing when the pH rises above 6.0 or falls below 2.0. Depurination occurs at pH levels below 3.011,47,48. The alkaline environment of the Lepidoptera gut leads to dsRNA instability, impairing its absorption by the epithelium49. Thus, both the midgut environment and symbiotic bacteria of Lepidoptera contribute to the instability of dsRNA, as well as low RNAi efficiency.
In this study, we utilized both feeding and injection methods to assess the effectiveness of Ba 6. Both these techniques constitute fundamental dsRNA delivery strategies, but the RNAi efficacy of these methods varies greatly in different insect orders8. In the dsRNA feeding experiments, it was observed that the dsRNA was swiftly degraded by gut microbiota upon its entry into the intestinal tract, leading to low stability levels. However, we also observed that Bacillus in the host gut had an impact on RNAi efficiency in the dsRNA injection experiments. Research indicates that infection, stress, or external stimuli can trigger gut bacterial translocation into hemolymph15,16. We speculate that Ba6 may translocate from the gut into the hemolymph, thereby potentially influencing the stability of dsRNA. In addition, metabolites from the gut microbiota may also infiltrate the hemolymph50. Furthermore, introduced dsRNA is likely transferred between various cells and tissues, a phenomenon known as systemic RNAi7. Shukla et al. confirmed that the injected dsRNA successfully reached both the gut and fat body tissues49. This principle entails uptake of dsRNA from the environment and subsequent transport of the RNAi signal between cells and tissues in the body. To date, a reasonable understanding toward this process in insects remains elusive3,7,8,48.
Additionally, the composition of gut microbiota in insects fluctuates throughout their developmental stages. During the larval stage, insects primarily accumulate energy through feeding, leading to a highly developed intestinal tract rich in symbiotic microorganisms. However, as the larval gut degrades before pupation, the abundance of symbiotic microbe declines, thereby reducing their influence on the host’s physiological functions40. Available evidence suggests that in in some lepidopteran species, dsRNA-mediated RNAi effects appear to be more pronounced during later developmental stages (such as terminal larval, pupal or adult phases), though the efficiency remains highly variable across species and experimental systems33, and the reduction of symbiotic bacteria during these stages may enhance the insect’s susceptibility to RNAi.
The surrounding environment plays a pivotal role in shaping the microbial community of insects, which in turn can influence the insect’s physiological processes51. For example, under semi-field conditions, Rosenbergiella_YN46 was successfully introduced into mosquitoes, effectively blocking the transmission of DENV2 in newly emerged adults52. This demonstrates that specific microorganisms can be introduced into the environment to modulate host physiology. Therefore, when insects ingest bacteria capable of secreting nucleases from the environment, it can directly affect the RNAi machinery of the host. This suggests that large-scale application of RNA biopesticides in the field must account for the complex microbial ecosystem, in addition to environmental factors like temperature, UV exposure, and rainfall. This is especially important as Bacillus spp., commonly released in large quantities for agricultural purposes, may interfere with RNAi efficacy due to their nuclease-secreting capabilities.
This study employed Ba 6 as a model organism and demonstrated that Ba 6 suspension, their supernatants, and the secreted Ribonuclease efficiently degraded dsRNA both in vivo and in vitro, leading to reduced RNAi efficiency in cotton bollworms. Symbiotic bacteria capable of secreting nucleases degrade dsRNA in the insect gut, thus impairing RNAi efficiency. Future improvements in dsRNA stability could involve modifying the insect’s microbiota by reducing the abundance of symbiotic bacteria with nucleic acid degradation capabilities. Whether other symbiotic factors influence the RNAi pathway or whether additional microorganisms with similar effects exist in insects remains an area for further exploration.
In conclusion, intestinal commensal bacteria significantly modulate host RNAi efficiency and contribute to the low RNAi efficacy observed in insects. This research sheds new light on the physiological barriers impacting the RNAi mechanism in insects. It also highlights the role of symbiotic bacteria in degrading exogenous dsRNA via their metabolic products, thus offering protection to the host. Furthermore, this study deepens our understanding of the mutualistic relationships between insects and their symbiotic bacteria, alongside the physiological roles these microorganisms play in their hosts.
Methods
The capability of cotton bollworm body fluids to degrade dsRNA
Cotton bollworm eggs were sourced from Jiyuan Baiyun Industry Co., Ltd. and reared in an artificial climate chamber under controlled conditions: a 14 h:10 h photoperiod, a temperature of 25 ± 1 °C, and 75% relative humidity. Larvae were provided with an artificial diet (soybean flour (160 g), wheat germ flour (300 g), yeast powder (60 g), sucrose (40 g), sorbic acid (6 g), nipagin methyl ester (6 g), agar powder (40 g), casein (80 g), vitamin C (6 g), multivitamin (25 mL), formaldehyde (2 mL), acetic acid (2 mL), and distilled water (2.7 L)), while adults were fed 10% honey water53.
Hemolymph and midgut fluid were collected from third instar cotton bollworm larvae under sterile conditions. Hemolymph was extracted by puncturing the abdominal proleg of five larvae with a sterile needle. The midguts of five larvae were dissected and homogenized in 200 μL of sterile PBS. Both hemolymph (38 mg/mL) and midgut fluid stock (54 mg/mL) solutions were diluted 50 times with 1x PBS for subsequent experiments. A 10 μL aliquot of the original hemolymph, midgut fluid, and their respective dilutions was incubated with 1 μL of 2000 ng/μL dsRNA. The dsRNAs were synthesized by Shanghai Plant Science Biotechnology Co., Ltd. (https://www.zsygbio.com). Following incubation for a specified duration, dsRNA degradation was analyzed via agarose gel electrophoresis.
Isolation, cultivation, and identification of symbiotic bacteria with dsRNA degrading activity
To isolate symbiotic bacteria, the midgut fluid of third instar larvae was diluted and plated on an LB solid medium. Monoclonal colonies with distinct morphologies were selected, purified, and identified through 16S rRNA sequencing using 27 F/1492 R primers (Table S8). The selected strains were then grown in an LB liquid medium at 37 °C until reaching the logarithmic growth phase. Bacterial supernatants were obtained by low-speed centrifugation and co-cultured with dsRNA for a specified time. The integrity of dsRNA was then examined using gel electrophoresis to identify bacteria capable of degrading dsRNA.
For further analysis, bacterial suspensions of strains Ba 1 through Ba 6 (OD600 = 1.0) were centrifuged at 4 °C at 4500 rpm for 10 minutes to isolate the supernatants. Both bacterial suspensions and supernatants (10 μL each) were incubated with 1 μL (2000 ng/μL) of dsRNA. Samples were collected at intervals of 5 min, 10 min, 30 min, 1 h, 2 h, 3 h, 4 h, 7 h, and 10 h, with dsRNA degradation assessed through agarose gel electrophoresis.
For Ba 6 cell preparation, the bacterial suspension was centrifuged and the supernatant was removed. The resulting pellet was then washed with PBS, resuspended, centrifuged, and washed three times consecutively. Finally, the cells were resuspended in fresh PBS to obtain Ba 6 cell.
To confirm the RNase activity of strains Ba 1 - Ba 6, an RNase activity assay was performed on an RNase activity testing plate. A 40 μL bacterial suspension was inoculated onto RNase medium using a sterilized Oxford cup and incubated at 37 °C. Equal volumes of LB medium and RNase A (Yeasen, Cat No: 10406ES03) served as negative and positive controls, respectively. After 3 days of incubation, plates were treated with 70% perchloric acid for 20 seconds, and the diameter of the halo on the RNase medium was measured54,55. This experiment was repeated three times to ensure accuracy.
Morphological analysis of Ba 6 bacteria and examines its impact on the growth and development of cotton bollworm
The morphology of Ba 6 bacteria was examined using a scanning electron microscope (SEM) (Zeiss Merlin Compact). A phylogenetic tree for Ba 6 and closely related strains was constructed using the MLST method, following the protocol described by Helgason et al.56 Ba 6 bacteria were inoculated into an LB liquid medium at a 1:100 ratio and cultured at 37 °C with shaking at 200 rpm. The bacterial growth curve was plotted by measuring OD600 at various time intervals using a spectrophotometer (NanoDropTM One, Thermo Scientific, USA).
To investigate the effects of Ba 6 on the growth and development of cotton bollworms, 200 μL of Ba 6 bacterial suspension (OD600 = 1.0) was evenly spread onto 1 cm × 1 cm pieces of artificial diet. Newly hatched larvae of similar size and condition were fed with this diet, which was replaced daily along with fresh bacterial suspension. Body weight and length measurements were obtained from 5 biological replicates, each containing 5 larvae. Survival rates were assessed from 5 biological replicates, each with 10 larvae.
Effect of feeding Ba 6 bacterial suspension/supernatant/effector protein Ribonuclease on dsRNA stability and RNAi efficiency
A 200 μL aliquot of Ba 6 or Ribonuclease protein was evenly applied to the surface of the artificial diet. Newly hatched cotton bollworm larvae of uniform growth status were placed on this diet for a single feeding session. The diet was refreshed daily until the larvae reached the third instar, at which point they were used for subsequent experiments.
For subsequent experiments, larvae were starved for 1 hour, then injected with 4.5 μL of fluorescently labeled dsRNA (Silencer® siRNA Labeling Kit, AM1632, Thermo Scientific) at a concentration of 900 ng/μL. Fluorescence intensity was observed after 1 hour and 6 hours using a confocal microscope (Nikon, AZ100).
In the dsRNA injection experiment, 5 μL of dsRNA (2000 ng/μL) was injected into each treated larva, with dsEGFP injected as a negative control. Samples were collected 6 hours post-injection, with three biological replicates of three insects per treatment group.
For dsRNA feeding experiments, 200 μL of Ba 6 bacterial suspension (OD600 = 1.0), its supernatant, or Ribonuclease protein (1 mg/mL) was spread on 1 cm×1 cm pieces of artificial diet, which was then fed to newly hatched cotton bollworms until they reached the third instar. The larvae were subsequently fed with 20 μL of dsRNA (1000 ng/μL) incorporated into the diet. Samples were collected 12 and 24 hours post-feeding to evaluate RNAi efficiency. Three treatments were combined for each biological replicate, and three biological replicates were conducted per treatment.
Ribonuclease protein expression, purification, and in vitro functional validation
The Ribonuclease gene sequence was cloned into a pET expression vector and transformed into an Escherichia coli BL21 (DE3) strain. Protein expression was induced at 15 °C with 0.4 mM IPTG, and the Ribonuclease protein was purified using Ni-NTA affinity chromatography. A 10 μL aliquot of the purified Ribonuclease protein (1 mg/mL) was co-incubated with 1 μL of dsRNA (2000 ng/μL), and dsRNA degradation was evaluated at specific time intervals through agarose gel electrophoresis.
The purified Ribonuclease protein was then sent to Hangzhou HuaAn Biotechnology Co., Ltd. for the generation of polyclonal antibody. The antibody was employed in Western blotting to detect Ribonuclease levels in the Ba 6 supernatant.
High-throughput sequencing and data analysis
Metagenomics sequencing
Midgut samples from five L4 and L5 instar cotton bollworms, reared under identical conditions, were pooled into a single sample and each treatment included two biological replicates. The samples were sent to BGI for metagenomic sequencing. Standardized analysis was performed using the MGISEQ-2000 platform. Kraken2 software was employed to classify the sequences corresponding to each species in the sample, while Bracken2 was utilized to estimate the actual abundance of these species, providing comprehensive species annotation.
16S rRNA sequencing
Newly hatched cotton bollworms were fed with a Ba 6 bacterial suspension until reaching the third instar, after which midgut samples were collected for 16S rRNA sequencing. The sequencing was performed on the Illumina MiSeq platform at Shanghai Majorbio Bio-pharm Technology Co., Ltd. Cotton bollworms fed with LB-containing artificial diets and regular artificial diets served as control groups. Three midguts were pooled for each biological replicate, and each treatment group consisted of 3 biological replicates. The data were analyzed using tools available on the Majorbio Cloud Platform (https://cloud.majorbio.com/page/tools/).
Bacterial whole genome sequencing
The Ba 1 to Ba 6 bacterial strains were sent to BGI Genomics Co., Ltd. for whole genome sequencing using the Illumina HiSeq 4000 platform. Read assembly, gene prediction, and gene function annotation were conducted using Unicycler, Glimmer, and Diamond software, respectively. Multicollinearity among the genomes of the six symbiotic bacteria was analyzed using MCscanX, which also identified single nucleotide polymorphisms (SNPs), insertions and deletions (Indels), and structural variations (SVs).
Potential nuclease prediction
The genome sequences were annotated using the eggNOG database to identify potential nuclease genes. Predicted nuclease genes were further analyzed using SignalP 6.0 software to detect signal peptides. The mature protein sequences, excluding the signal peptide regions, were then evaluated using TMHMM-2.0 and PredGPI software to exclude proteins containing transmembrane structures or GPI anchor sites. This process ultimately led to the identification and screening of extracellular nucleases from the six symbiotic bacteria. The phylogenetic tree of these extracellular nucleases were constructed using the maximum likelihood (ML) method in RAxML.
Data availability
The bacterial 16S rRNA sequencing data and metagenomics sequencing data of the cotton bollworm midgut have been deposited in the NCBI Sequence Read Archive (SRA) database, with BioProject numbers PRJNA1203073 and PRJNA1201943, respectively. The bacterial genome sequencing data of Ba 1–Ba 6 were deposited at the NCBI under BioProject PRJNA1203013, PRJNA1203025, PRJNA1203033, PRJNA1203043, PRJNA1203021, and PRJNA1203048, respectively.
Code availability
All data, models, or code generated or used during the study are available from the corresponding author by request.
References
Niu, J., Chen, R. & Wang, J. J. RNA interference in insects: the link between antiviral defense and pest control. Insect Sci. 31, 2–12 (2023).
Tang, Q. & Khvorova, A. RNAi-based drug design: considerations and future directions. Nat. Rev. Drug Discov. 23, 341–364 (2024).
Koo, J. & Palli, S. R. Recent advances in understanding of the mechanisms of RNA interference in insects. Insect Mol. Biol. Online ahead of print. https://doi.org/10.1111/imb.12941 (2024).
De Schutter, K. et al. RNAi-based biocontrol products: market status, regulatory aspects, and risk assessment. Front. Insect Sci. 1, 818037 (2021).
Nitnavare, R. B. et al. Next generation dsRNA-based insect control: success so far and challenges. Front. Plant Sci. 12, 673576 (2021).
Terenius, O. et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 57, 231–245 (2011).
Joga, M. R., Zotti, M. J., Smagghe, G. & Christiaens, O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol. 7, 553 (2016).
Lucena-Leandro, V. S. et al. Current scenario of exogenously induced RNAi for lepidopteran agricultural pest control: from dsRNA design to topical application. Int. J. Mol. Sci. 23, 15836 (2022).
Palli, S. R. RNAi turns 25: contributions and challenges in insect science. Front. Insect Sci. 3, 1209478 (2023).
Zhu, K. Y. & Palli, S. R. Mechanisms, applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 65, 293–311 (2020).
Cooper, A. M., Silver, K., Zhang, J., Park, Y. & Zhu, K. Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 75, 18–28 (2019).
Engel, P. & Moran, N. A. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735 (2013).
Rupawate, P. S. et al. Role of gut symbionts of insect pests: a novel target for insect-pest control. Front. Microbiol. 14, 1146390 (2023).
Xie, J., Li, S., Zhang, W. & Xia, Y. RNAi-knockdown of the Locusta migratoria nuclear export factor protein results in insect mortality and alterations in gut microbiome. Pest Manag. Sci. 75, 1383–1390 (2018).
Xu, L. et al. Synergistic action of the gut microbiota in environmental RNA interference in a leaf beetle. Microbiome 9, 98 (2021).
Zhang, Y. et al. A faster killing effect of plastid-mediated RNA interference on a leaf beetle through induced dysbiosis of the gut bacteria. Plant Commun. 5, 100974 (2024).
Douglas, A. E. Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34 (2015).
Yin, C., Sun, P., Yu, X., Wang, P. & Cheng, G. Roles of symbiotic microorganisms in arboviral infection of arthropod vectors. Trends Parasitol. 36, 607–615 (2020).
Jaffar, S., Ahmad, S. & Lu, Y. Contribution of insect gut microbiota and their associated enzymes in insect physiology and biodegradation of pesticides. Front. Microbiol. 13, 979383 (2022).
Boush, M. G. & Matsumura, F. Insecticidal degradation by pseudomonas melophthora, the bacterial symbiote of the Apple Maggot1. J. Econ. Entomol. 60, 918–920 (1967).
Kikuchi, Y. & Yumoto, I. Efficient colonization of the bean bug Riptortus pedestris by an environmentally transmitted Burkholderia symbiont. Appl. Environ. Microbiol. 79, 2088–2091 (2013).
Ramya, S. L., Venkatesan, T., Murthy, K. S., Jalali, S. K. & Varghese, A. Degradation of acephate by Enterobacter asburiae, Bacillus cereus and Pantoea agglomerans isolated from diamondback moth Plutella xylostella (L), a pest of cruciferous crops. J. Environ. Biol. 37, 611–618 (2016).
Shliapnikov, S. V. & Dement’ev, A. A. Amino acid sequence and catalytic properties of the Bacillus coagulans extracellular ribonuclease. Dokl. Akad. Nauk 332, 382–384 (1993).
Sokurenko, Y., Nadyrova, A., Ulyanova, V. & Ilinskaya, O. Extracellular Ribonuclease from Bacillus licheniformis (Balifase), a New Member of the N1/T1 RNase Superfamily. Biomed. Res. Int. 2016, 4239375 (2016).
Dementiev, A. A., Moiseyev, G. P. & Shlyapnikov, S. V. Primary structure and catalytic properties of extracellular ribonuclease of Bacillus circulans. FEBS Lett. 334, 247–249 (1993).
Hartley, R. W. & Rogerson, D. L. Jr Production and purification of the extracellular ribonuclease of Bacillus amyloliquefaciens (barnase) and its intracellular inhibitor (barstar). I. Barnase. Prep. Biochem. 2, 229–242 (1972).
Golubenko, I. A., Balaban, N. P., Leshchinskaia, I. B., Volkova, T. I. & Kleĭner, G. I. Ribonuclease of Bacillus intermedius 7 P. Purification by chromatography on phosphocellulose and several characteristics of the homogeneous enzyme. Biokhimiia 44, 640–648 (1979).
Dudkina, E. et al. Three-step procedure for preparation of pure Bacillus altitudinis ribonuclease. FEBS Open Bio 6, 24–32 (2016).
Nakamura, A. et al. Gene cloning and characterization of a novel extracellular ribonuclease of Bacillus subtilis. Eur. J. Biochem. 209, 121–127 (1992).
Skvortsova, M. A., Bocharov, A. L., Yakovlev, G. I. & Znamenskaya, L. V. Novel extracellular ribonuclease from Bacillus intermedius-binase II: purification and some properties of the enzyme. Biochemistry 67, 802–806 (2002).
Song, H. et al. Contributions of dsRNases to differential RNAi efficiencies between the injection and oral delivery of dsRNA in Locusta migratoria. Pest Manag. Sci. 75, 1707–1717 (2019).
Chen, J.-Z. et al. Double-stranded RNA-degrading enzymes reduce the efficiency of RNA interference in Plutella xylostella. Insects 12, 712 (2021).
Fan, Y. H. et al. A dsRNA-degrading nuclease (dsRNase2) limits RNAi efficiency in the Asian corn borer (Ostrinia furnacalis). Insect Sci. 28, 1677–1689 (2021).
Tayler, A., Heschuk, D., Giesbrecht, D., Park, J. Y. & Whyard, S. Efficiency of RNA interference is improved by knockdown of dsRNA nucleases in tephritid fruit flies. Open Biol. 9, 190198 (2019).
Peng, Y. et al. Identification and characterization of multiple dsRNases from a lepidopteran insect, the tobacco cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 162, 86–95 (2020).
Yoon, J. S., Ahn, S. J., Flinn, C. M. & Choi, M. Y. Identification and functional analysis of dsRNases in spotted-wing drosophila, Drosophila suzukii. Arch. Insect Biochem. Physiol. 107, 21822 (2021).
Abreu Reis, M. et al. Why is oral-induced RNAi inefficient in Diatraea saccharalis? A possible role for DsREase and other nucleases. Pestic. Biochem. Physiol. 186, 105166 (2022).
Peng, Y. et al. Identification of a double-stranded RNA-degrading nuclease influencing both ingestion and injection RNA interference efficiency in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 125, 103440 (2020).
Colman, D. R., Toolson, E. C. & Takacs-Vesbach, C. D. Do diet and taxonomy influence insect gut bacterial communities?. Mol. Ecol. 21, 5124–5137 (2012).
Shao, Y., Mason, C. J. & Felton, G. W. Toward an integrated understanding of the lepidoptera microbiome. Annu. Rev. Entomol. 69, 117–137 (2024).
Wang, M., Xiang, X. & Wan, X. Divergence in gut bacterial community among life stages of the rainbow stag beetle Phalacrognathus muelleri (Coleoptera: Lucanidae). Insects 11, 179 (2020).
Zheng, X. et al. Gut bacterial communities across 12 Ensifera (Orthoptera) at different feeding habits and its prediction for the insect with contrasting feeding habits. PLoS ONE 16, e0250675 (2021).
Juottonen, H., Moghadam, N. N., Murphy, L., Mappes, J. & Galarza, J. A. Host’s genetic background determines the outcome of reciprocal faecal transplantation on life-history traits and microbiome composition. Anim. Microbiome 4, 67 (2022).
Ravenscraft, A., Berry, M., Hammer, T., Peay, K. & Boggs, C. Structure and function of the bacterial and fungal gut microbiota of Neotropical butterflies. Ecol. Monogr. 89, 1346 (2019).
Dow, J. A. pH gradients in lepidopteran midgut. J. Exp. Biol. 172, 355–375 (1992).
Zhu, D. et al. Bacillus ligniniphilus sp. nov., an alkaliphilic and halotolerant bacterium isolated from sediments of the South China Sea. Int. J. Syst. Evol. Microbiol. 64, 1712–1717 (2014).
Choudhary, C., Meghwanshi, K. K., Shukla, N. & Shukla, J. N. Innate and adaptive resistance to RNAi: a major challenge and hurdle to the development of double stranded RNA-based pesticides. 3 Biotech 11, 498 (2021).
Christiaens, O., Whyard, S., Vélez, A. M. & Smagghe, G. Double-stranded RNA technology to control insect pests: current status and challenges. Front. Plant Sci. 11, 451 (2020).
Shukla, J. N. et al. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 13, 656–669 (2016).
Siddiqui, J. A. et al. Role of insect gut microbiota in pesticide degradation: a review. Front. Microbiol. 13, 870462 (2022).
Chandrasegaran, K., Lahondère, C., Escobar, L. E. & Vinauger, C. Linking mosquito ecology, traits, behavior, and disease transmission. Trends Parasitol. 36, 393–403 (2020).
Zhang, L. et al. A naturally isolated symbiotic bacterium suppresses flavivirus transmission by Aedes mosquitoes. Science 384, 293 (2024).
Liu, X. et al. Baculoviruses hijack the visual perception of their caterpillar hosts to induce climbing behaviour thus promoting virus dispersal. Mol. Ecol. 31, 2752–2765 (2022).
Khalaf, E. M. & Raizada, M. N. Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front. Microbiol. 9, 995 (2018).
Hole, R. C., Singhal, R. S., Melo, J. S. & D’Souza, S. F. A rapid plate screening technique for extracellular ribonuclease producing strains. BARC Newsl. 249, 91–97 (2004).
Helgason, E., Tourasse, N. J., Meisal, R., Caugant, D. A. & Kolstø, A.-B. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70, 191–201 (2004).
Acknowledgements
This work was sponsored by Shanghai Agricultural Science and Technology Innovation Program (Grant No. K2023019); Shanghai Rising-Star Program (22QB1405900); Key scientific research projects of colleges and universities in Henan Province (25B210009), the Science and Technology Research Project of Henan Province (252102111110), and State Key Laboratory of Cotton Bio-breeding and Integrated Utilization and Sponsored by State Key Laboratory of Cotton Bio-breeding and Integrated Utilization Open Fund (CB2025A34). The authors declare that no funds, grants, or other support were received during the preparation of this manuscript. We thank Bullet Edits (https://www.bulletedits.cn) for editing the language of a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
H.L. and R.G. conceived the concept and designed the experiments. H.L. and X.H. collected samples. X.H. performed the experiments. X.H. and S.X. analysis the data. R.G., H.L. and X.M. wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Han, X., Li, H., Xu, S. et al. Bacillus secretes nucleases to degrade dsRNA, thereby reducing host’s susceptibility to RNAi. npj Biofilms Microbiomes 11, 127 (2025). https://doi.org/10.1038/s41522-025-00757-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-025-00757-z