Abstract
The increasing availability of biobanks is changing the way individuals are identified for genomic research. This study assesses the validity of a self-reported clinical diagnosis of schizophrenia. The study included 1744 clinically-ascertained participants with schizophrenia or schizoaffective disorder depressed-type (SA-D) diagnosed by self-report and/or research interview and 1453 UK Biobank participants with self-reported and/or medical record diagnosis of schizophrenia or SA-D. Unaffected controls included a total of 501,837 participants. We assessed the positive predictive values (PPV) of self-reported clinical diagnoses against research interview and medical record diagnoses. Polygenic risk scores (PRS) and phenotypes relating to demographics, education and employment were compared across diagnostic groups. The variance explained (r2) in schizophrenia PRS for each diagnostic group was compared to samples in the Psychiatric Genomics Consortium (PGC). In the clinically-ascertained participants, the PPV of self-reported schizophrenia for a research diagnosis of schizophrenia was 0.70, which increased to 0.81 after expanding the research diagnosis to schizophrenia or SA-D. In UK Biobank, the PPV of self-reported schizophrenia for a medical record diagnosis was 0.74. Compared to participants who self-reported, participants with a clinically-ascertained research diagnosis were younger and more likely to have a high school qualification. Participants with a medical record diagnosis in UK Biobank were less likely to be employed or have a high school qualification than those who self-reported. Schizophrenia PRS did not differ between participants that had a diagnosis from self-report, research diagnosis or medical records. Polygenic liability r2, for all diagnosis definitions, fell within the distribution of PGC schizophrenia cohorts. Self-reported measures of schizophrenia are justified in genomic research to maximise sample size and reduce the burden of in-depth interviews on participants, although within sample validation of diagnoses is recommended.
Similar content being viewed by others
Introduction
Schizophrenia is a severe mental health condition characterised by positive, negative, and disorganised symptoms as well as cognitive deficits1 and has a lifetime prevalence of 0.32% worldwide2. In research studies, schizophrenia diagnoses are now determined from various sources; for example, research interview and/or clinical note review, electronic heath records, or diagnoses can be based on a self-report of a clinical diagnosis made by a health professional. Methods combining diagnostic interviews and note reviews are considered the gold-standard for defining cases in research3, but are resource intensive and often associated with ascertainment biases; excluding those severely affected due to the lengthy interview but also milder cases that are not in contact in secondary health services leading to unrepresentative samples4,5.
Traditionally, genomics research has been founded on clinically-ascertained samples that provide a clinical or research interview diagnosis. The availability of large-scale biobanks only sets to increase in the coming years6 and holds great potential for psychiatric genomics, but the appropriateness and validity of these different diagnostic sources is unclear.
Diagnoses generated from medical records have been shown to have good concordance with research interview diagnoses7,8, with particularly high convergence seen in schizophrenia9,10,11. Ascertainment through medical records overcomes some of the practical limitations for participation, but still hinders representation by relying on records typically from secondary care3 and thus under-represents patients less severely affected who are less likely to be admitted to hospital. Alternatively, a self-reported diagnosis from a health professional could be one approach to improving representativeness and increasing sample size in genomic research, circumventing the need for a labour-intensive research interview. However, the validity of self-reported diagnoses is likely to differ between psychiatric disorders, contexts, and cultures. Research using self-reported medical diagnoses from large-scale genomic datasets such as 23andMe12,13,14,15,16,17, UK Biobank13,16,18,19,20,21 and the Million Veterans Programme16,22 are now common, but the reliability and validity of self-reported diagnoses is unclear23.
It is also unknown what impact different diagnostic methodologies such as self-report and medical records have on the outcome of genetic studies24. In order to enhance power in GWAS individuals with a self-reported diagnosis have been included13,17, and are likely to be increasingly so, despite some studies suggesting that individuals defined using minimal phenotyping approaches show genetic differences to participants who are strictly defined23,25,26. In one study, the effect sizes for schizophrenia polygenic risk scores (PRS) were reported to be smaller in samples where diagnoses are derived from electronic health records compared to clinically-ascertained case-control research cohorts in the Psychiatric Genomics Consortium (PGC)27. However, analyses comparing samples from the Schizophrenia Working Group of the PGC found no differences in PRS across consensus DSM/ICD diagnosis (by psychiatrists), diagnostic interview, medical records, and mixed methods28. To our knowledge, there is no published research comparing a self-reported clinical diagnosis of schizophrenia from a health professional against a gold-standard research interview diagnosis. In this study, we address this knowledge gap and assess whether a self-reported clinical diagnosis of schizophrenia is a valid approach to identify relevant individuals for genomic research.
Methods
Participants
Study participants came from two clinically-ascertained Cardiff University cohorts, the National Centre for Mental Health (NCMH) and CardiffCOGS, and from the UK Biobank. All participants provided written informed consent. Table 1 provides information on the assessments used to determine diagnosis in each sample. A flowchart of the samples, methods and number of participants recruited is shown in Fig. 1.
Flowchart showing the methods used to assess participants in each sample (NCMH, CardiffCOGS and UK Biobank), along with information on the number of participants in each sample, broken down by method of diagnosis. Numbers are provided for schizophrenia. In NCMH, of the 1943 participants who self-reported psychosis: 493 reported schizoaffective disorder, 1001 reported psychosis and 634 reported bipolar disorder. Note that participants could select all diagnoses that they had ever been given so some participants reported more than one psychotic diagnosis.
NCMH
NCMH participants were recruited via health care services, voluntary organisations or via public advertisement29. Trained researchers administered a brief standardized assessment to gather demographic and clinical information and participants were asked to provide a sample for DNA extraction and genetic analyses. Participants self-reporting a schizophrenia, psychosis or affective diagnosis were invited to take part in a research interview based on the Schedules for Clinical Assessment in Neuropsychiatry (SCAN)30. NCMH received approval from Health Research Authority and Wales Research Ethics Committee (REC) 2 (16/WA/0323).
CardiffCOGS
CardiffCOGS participants were recruited from community, in-patient and voluntary sector mental health services across the UK31. All participants completed a SCAN-based research interview30, underwent a case-note review and were asked to provide a sample for DNA extraction and genetic analyses. CardiffCOGS received approval from Southeast Wales REC (07/WSE03/110). CardiffCOGS participants were included to increase the sample size for the genetic analysis. These participants were not included in comparisons of self-report and research diagnoses, as self-report diagnoses are not available in this sample.
UK Biobank
UK Biobank is a population-based UK cohort of around 500,000 participants, aged between 40–69 at recruitment32. Participants completed a range of assessments and provided a sample for genetic analysis. Ethical approval was granted by the Northwest Multi-Centre Ethics Committee. This study was conducted under UK Biobank project number 13310.
Diagnosis definitions
Table 1 provides an overview of the self-reported, research interview and medical record diagnosis definitions used in this study.
Self-reported diagnosis
In NCMH, participants were asked whether a doctor or health professional had ever told the participant that they had a mental health diagnosis and prompted with a list of psychiatric diagnoses to choose from (Supplementary Fig. 1). In UK Biobank, participants were asked to report if a doctor had told them they had any serious medical condition in the initial assessment. A subset of participants in the UK Biobank (31%) completed the Mental Health Questionnaire (MHQ), where they were prompted with a list of psychiatric diagnoses to choose from (Supplementary Fig. 2). For both NCMH and UK Biobank, if the participant chose schizophrenia from the list or they verbally self-reported a schizophrenia diagnosis, they were assigned a schizophrenia self-reported diagnosis in this study. Table 1 describes the subtypes of self-reported diagnoses available in the clinically-ascertained sample. A self-reported schizoaffective disorder diagnosis was excluded from analyses, as it was not possible to differentiate between the depressed and manic subtypes.
Research interview diagnosis
In the clinically-ascertained samples (NCMH and CardiffCOGS), DSM-IV, DSM-5, and ICD-10 research diagnoses were derived from a SCAN-based clinical interview and note review where available. A research interview diagnosis of schizophrenia was given in this study if either a DSM or ICD schizophrenia criteria were met. If participants met criteria for schizoaffective disorder depressed-type (SA-D), they were also included alongside participants with schizophrenia given evidence that these participants do not differ on a range of phenotypic and genotype measures, including symptoms, cognition and polygenic risk33. ‘Other psychotic disorders’ in this study refer to the following diagnoses: psychosis not otherwise specified, schizophreniform disorder, delusional disorder, brief psychotic disorder, acute polymorphic disorder, and other psychotic illness.
Medical record diagnosis
In UK Biobank, a medical record diagnosis of schizophrenia and SA-D were defined as a F20/F25.1 ICD-10 code from national hospital admission records or death records, or an equivalent read code from primary care (Supplementary Table 1). Hospital records date back to 1997 for England, 1998 for Wales and 1981 for Scotland and contain coded data on admissions, operations, and procedures. Primary care data was obtained for approximately 45% of the UK Biobank cohort. In secondary analyses, hospital admissions for schizophrenia were further subdivided into primary and secondary admissions. Primary ICD-10 codes represent conditions that caused the admission and secondary ICD-10 codes represent conditions that coexist at the time of admission, affect the treatment received, or develop after admission.
Unaffected controls
Unaffected controls for the clinically-ascertained samples were NCMH participants with no history of a mental health diagnosis and who were recruited through participants with a psychiatric diagnosis (e.g., a family member/partner) or via advertisements. Unaffected controls for the UK Biobank analyses consisted of participants in UK Biobank who did not have a psychotic disorder diagnosis (F21-F29 inclusive) from admission records, death records, primary care records, or from self-reported sources.
Phenotypic data
The phenotypes compared across diagnostic groups included sex, age at interview (in years), educational attainment, and employment status. Educational attainment was dichotomised to GCSE (General Certificate of Secondary Education) and above, usually achieved at 16 years upon completing high school, or below GCSE/no qualification, consistent with previous research34, in addition to degree/no degree. Employment status was dichotomised to in current paid employment or not and restricted to participants under the age of 65 who did not report being retired.
Genetic data
Clinically-ascertained sample
The clinically-ascertained participants were genotyped on the Illumina OmniExpress (Infinium OmniExpress-24 Kit), Illumina PsychArray (Infinium PsychArray-24 Kit) or Illumina GSA (Infinium Global Screening Array-24 Kit) genotyping platforms. Quality control and imputation using the Haplotype Reference Consortium (HRC)35 was performed as part of the DRAGON-Data protocol36. Datasets containing participants from the clinically-ascertained samples were restricted to those with the diagnoses described above and who did not carry a neurodevelopmental CNV36. These samples were combined with samples from 1000 Genomes European phase 337 using PLINK v1.938 after restricting to overlapping SNPs. The 1000 Genomes sample was included to provide a population reference to allow studies using different arrays to be directly compared39. The following quality control exclusion criteria were subsequently applied to SNPs: minor allele frequency (MAF) < 0.05, genotyping rate < 0.05, and Hardy-Weinberg equilibrium p ≤ 10−6. Linkage disequilibrium-pruned SNPs (500 variant count window size, 20 variant count to shift the window at the end of each step, a pairwise r2 threshold of 0.2) were used to identify related individuals and to derive principal components (PC). One individual from each pair assumed to be duplicates (kinship coefficient > 0.98) or related (kinship coefficient > 0.1875) was removed at random. The first 5 PCs were used to perform multi-dimensional clustering to identify an ancestrally-homogenous subsample of individuals40. The first 5 PCs explained the majority of the variance in the principal components, adding additional PCs did not change the classifications. Individuals within a 90% threshold from the most central point were included for analyses. There were insufficient numbers of participants of non-European ancestries in NCMH and CardiffCOGS to allow us to analyse PRS in different ancestries.
UK Biobank
Imputed genetic data were provided by UK Biobank. Pre-imputation quality control and imputation have been described elsewhere41. Briefly, participants were assayed at the Affymetrix Research Services laboratory using the UK Biobank Axiom or UK BiLEVE Axiom purpose-built arrays. Imputation was completed using the HRC panel35. We applied additional quality control procedures using the same thresholds used in our clinically-ascertained sample and detailed elsewhere39,42. Genetic analyses were restricted to participants with European ancestry, to mirror the clinically-ascertained sample, using the method described above, see also Legge et al42.
Polygenic risk scores
In the clinically-ascertained sample and UK Biobank, PRSicev243 was used to calculate PRS for schizophrenia using GWAS de-duplicated summary statistics that were derived separately from our clinical sample and UK Biobank28. PRS were also calculated for bipolar disorder13 and major depressive disorder44. Summary statistics underwent quality control36 and SNPs with MAF > 0.01 outside of the major histocompatibility complex region were used in the PRS analysis. PRS were calculated, using relatively independent SNPs (r2 < 0.1, within 500 kb window), at a p-value threshold of 0.0528. Polygenic risk scores were standardised within samples prior to analysis.
Analysis
In NCMH, positive predictive values (PPV) were used to assess the ratio of participants with a self-reported schizophrenia diagnosis from a health professional who had a concordant DSM/ICD research interview diagnosis. We also considered a research interview diagnosis of schizophrenia and schizoaffective disorder depressive-type (SA-D) together as there is evidence these two groups do not substantially differ with respect to genetic liability to schizophrenia28,33. It was not possible to assess negative predictive values (NPV), sensitivity and specificity in the clinically-ascertained sample due to the recruitment methods; participants were only approached to complete a SCAN-based research interview if they self-reported a mood or psychotic disorder diagnosis.
In the UK Biobank, PPV, NPV, sensitivity and specificity were used to assess how predictive a self-reported clinical diagnosis from a health professional was of a medical record diagnosis. We scaled the PPV and NPV to the population point prevalence of schizophrenia (0.6%) (Supplementary Note 1). We could not calculate PPV related to a medical record diagnosis of schizophrenia and SA-D together due to a very low prevalence of SA-D in the UK Biobank.
In both NCMH and the UK Biobank, logistic regressions were used to test for phenotypic differences between individuals that only self-reported a diagnosis and those who had a research interview diagnosis/medical record diagnosis (some of whom also self-reported). Year of birth and sex were included as covariates.
Due to the limited number of genotyped participants in NCMH, the genetic analyses included participants from both NCMH and CardiffCOGS. In both the clinically-ascertained sample and the UK Biobank logistic regressions were used to test for genetic differences in schizophrenia between self-report-only and the research interview diagnosis/medical record diagnosis groups.
We compared the variance explained by schizophrenia PRS on the liability-scale (r2, assuming 1% lifetime risk) in schizophrenia case/control status in the clinically-ascertained sample and UK Biobank, separated by diagnosis definitions, against the variances reported by other samples of European genetic ancestry in the PGC3 schizophrenia GWAS. The r2 values refer to the variance explained by the schizophrenia PRS in comparison to a covariates-only baseline model. In addition, we calculated the variance explained in schizophrenia case/control status in UK Biobank for bipolar disorder13 and major depressive disorder44 PRS.
In the UK Biobank sample, further logistic regressions were used to assess if schizophrenia PRS was associated with the number of times a diagnosis was reported, the number of admissions and type of admission (primary and secondary). These PRS analyses were covaried for the first 5 PCs, array, age at assessment, and sex.
All statistical tests were two-sided. Unless otherwise specified, data analysis was conducted in R.
Results
We identified participants with a self-reported, research or medical record diagnosis of either schizophrenia or schizoaffective disorder (SA-D) from across the three samples. This included 1112 participants from NCMH, 632 participants from CardiffCOGS and 1453 participants from the UK Biobank. Unaffected controls included 749 participants from NCMH and 501,088 participants from UK Biobank. Demographic information for each sample is provided in Table 2. The number of participants included in each analysis varies according to the availability of data (e.g. number with both self-report and research/medical record diagnoses for PPI analyses and number of participants with genotype data) and are detailed in the relevant sections.
Positive predictive values
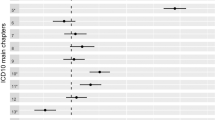
In NCMH, the proportion of participants with a self-reported schizophrenia diagnosis from a health professional who had a research interview diagnosis was used to estimate PPV (n = 273, Table 3). A self-reported current clinical diagnosis of schizophrenia had a PPV of 0.74 for receiving a research interview diagnosis of schizophrenia, 0.85 of receiving a diagnosis of schizophrenia or SA-D and 0.9 of receiving a diagnosis of schizophrenia, SA-D or any other psychotic disorder. A self-reported current clinical diagnosis produced slightly higher PPVs than self-reported lifetime clinical diagnoses or participant opinion diagnoses (Table 3). Secondary PPV analyses indicated that a self-reported psychotic disorder was less predictive of receiving a research diagnosis of schizophrenia, SA-D or other psychotic disorder (PPVs range: 0.27–0.65, Supplementary Table 2). All participants who self-reported schizophrenia but did not proceed to get a research interview diagnosis of schizophrenia received other mood or psychotic research diagnoses, except one participant where there was insufficient data to make a research interview diagnosis (Supplementary Table 3).
In UK Biobank, predictive values were calculated in participants who had both a self-reported diagnosis and a medical record diagnosis. After correction for the point prevalence45 of schizophrenia (0.6%), the PPV of having a medical record diagnosis of schizophrenia for those who self-reported a schizophrenia diagnosis was 0.83, the NPV 0.996, the specificity 0.9995 and the sensitivity 0.383 (Table 4). When including a medical record diagnosis of schizophrenia or any other psychotic-related disorder the specificity decreased to 0.77 and sensitivity to 0.21 (Supplementary Table 4).
Phenotypic and genetic differences across diagnosis source
Clinically-ascertained sample
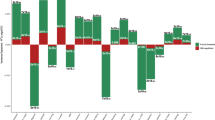
We compared 458 participants in NCMH with a research interview diagnosis of schizophrenia or SA-D with 654 participants whose only source of diagnosis was a self-reported diagnosis of schizophrenia. Participants who had a research interview diagnosis were younger (mean age 43 vs. 47; OR = 0.77; 95% CI = 0.67, 0.88; p = 9.11 × 10-5) and more likely to have a high school qualification (GCSE) or above (OR = 1.61; 95% CI = 1.13, 2.29; p = 0.008) than self-reporting only participants. Having a degree did not significantly differ across self-report only and research interview groups (OR = 1.15, 95%CI = 0.79, 1.67, p = 0.47). No significant differences were detected in employment (OR = 1.35; 95% CI = 0.87, 2.08; p = 0.18) or sex (OR = 1.07; 95% CI = 0.83, 1.37; p = 0.61) (Fig. 2).
For the genetic analyses, we added participants from the CardiffCOGS cohort (who all had a research diagnosis) to the NCMH sample to increase the sample size. Therefore, the genetic analyses included 803 participants with a research diagnosis, 449 participants who exclusively had a self-report diagnosis and 710 controls. We found no significant difference in schizophrenia PRS between participants who had a research interview diagnosis and those who only self-reported a diagnosis (OR = 0.97; 95% CI = 0.86, 1.09; p = 0.59) (Fig. 3).
CardiffCOGS samples were genotyped on a different array platform (OmniExpress) to NCMH cases and controls, which were split across GSA and PsychChip (Supplementary Table 5). To test whether there were any batch effects, we removed the CardiffCOGS samples from the case/control analysis. We found a consistent effect for the association between PRS and schizophrenia case/control status (OR = 1.70; 95%CI = 1.44-2.02; P = 1.91 × 10−10, r2 = 0.036; se = 0.011; AUC = 0.64). This finding, alongside Supplementary Fig. 3 suggests that there were no batch effects in the genetic data.
UK Biobank sample
Compared to participants whose basis for a diagnosis was solely self-report (n = 252), participants who had a medical record diagnosis of schizophrenia or SA-D (n = 1201) were less likely to be in paid employment (OR = 0.55; 95% CI = 0.39, 0.79; p = 0.001), and less likely to have a GCSE (high school) or higher qualification (OR = 0.70; 95% CI = 0.51, 0.95; p = 0.02). Furthermore, participants with a medical record diagnosis were less likely to have a degree (OR = 0.59, 95%CI = 0.44, 0.79, p = 0.0005). There were no differences in sex (OR = 0.95; 95% CI = 0.72, 1.26; p = 0.75) or age across the groups (OR = 0.91; 95% CI = 0.80, 1.04; p = 0.18) (Fig. 2).
The genetic analysis included 181 participants with only a self-report and 809 participants with a medical record diagnosis. No significant difference in schizophrenia PRS was found between participants who had a medical record diagnosis and a self-report diagnosis (OR = 1.01; 95%CI = 0.87,1.19; p = 0.85; Fig. 3).
Liability explained in case/control status
The proportion of variance on the liability scale attributable to schizophrenia PRS in both diagnostic groups in the clinically-ascertained sample and UK Biobank studies fell within the distribution of studies in the latest PGC analysis (Fig. 4, Table 5). In the clinically-ascertained sample, the schizophrenia PRS explained 5.0% of the variability in the self-reported-only group, and 4.7% in the research interview diagnosis group. In the UK Biobank sample, the schizophrenia PRS explained 6.5% of the variability in the self-reported only-group and 6.1% in the medical record diagnosis group (Table 5).
The variance explained by schizophrenia PRS in schizophrenia case/control status on the liability scale assuming a 1% population prevalence of European genetic ancestry PGC cohorts. The lines plotted on the graph represent the r2 of each diagnostic group. SZ, MDD and BP PRS are plotted as a reference. Clinical clinically-ascertained sample, UKBB UK Biobank, MDD major depressive disorder, BP bipolar disorder, SZ schizophrenia, PGC Psychiatric Genomics Consortium.
Further examination of diagnosis source in the UK Biobank
Schizophrenia PRS increased with the number of times a schizophrenia diagnosis was reported; OR = 1.82 (95%CI = 1.67, 1.99) for 1 endorsement compared to controls and OR = 2.11 (95%CI = 1.92, 2.32) for 2 or more endorsements (Supplementary Fig. 4). Participants who had two or more diagnosis endorsements had a significantly higher schizophrenia PRS than participants who only had one diagnosis endorsement (OR = 1.15; 95%CI = 1.01, 1.31; P = 0.03). The schizophrenia PRS also increased as the number of schizophrenia hospital admissions increased from OR = 1.85 (95%CI = 1.72, 2.00) for 0 admissions (participants had an alternative source of schizophrenia diagnosis), to OR = 1.92 (95%CI = 1.77, 2.08) for 1 admission, and OR = 2.28 (95%CI = 2.01, 2.58) for 2 or more admissions (Supplementary Fig. 5).
Schizophrenia cases with a primary ICD-10 admission code had a higher schizophrenia PRS than those who had schizophrenia as a secondary ICD-10 admission code (OR = 1.28; 95%CI = 1.10, 1.49; P = 0.002). Participants identified with a schizophrenia diagnosis from a secondary code only, on average, had lower schizophrenia PRS than those identified from self-reported or a primary hospital admission code (Supplementary Fig. 6). These findings did not appear to be related to the secondary code only participants having different associated diagnoses (Supplementary Table 6 and 7).
Discussion
In this study, we demonstrated that participants who self-reported a clinical diagnosis of schizophrenia were likely to be given a subsequent research interview diagnosis of schizophrenia, SA-D or other psychotic disorder (PPVs between 0.70 and 0.90). Furthermore, we found that participants in UK Biobank who self-reported a clinical schizophrenia diagnosis were likely to have a medical record diagnosis of schizophrenia or another psychotic disorder (PPV = 0.80). Although we found some phenotypic differences, genetic liability to schizophrenia did not significantly differ between participants with a self-reported diagnosis compared to those diagnosed via research interview or medical records. The variance explained by the schizophrenia PRS for all diagnostic methods fell within the distribution of PGC studies. These findings suggest that using a self-reported clinical diagnosis of schizophrenia is a valid approach for identifying participants for large-scale genomic research.
In the clinically-ascertained sample, participants who self-reported schizophrenia were likely to receive a research diagnosis of schizophrenia, SA-D or other psychotic disorder, however, participants who self-reported a lifetime clinical diagnosis of psychosis (without schizophrenia and bipolar) were much less likely to obtain a research interview diagnosis of schizophrenia, SA-D, or other psychotic disorder (PPVs 0.27–0.65). Previous research has shown that a schizophrenia diagnosis has much better agreement between diagnostic methods (PPVs 0.69-1.00) than other diagnoses such as bipolar, depression and other psychotic disorders9. Although self-reported diagnoses have generally been shown to have poor predictive accuracy when it comes to obtaining a gold-standard research interview diagnosis46,47, our results suggest that for schizophrenia specifically, self-reported diagnoses could be used in place of a research interview diagnosis to identify participants in genomic research. No research diagnostic method, including clinical interviews, is totally free of bias and recruiting participants via a range of sources is likely to lead to more representative studies as a whole.
In UK Biobank, participants who self-reported schizophrenia were likely to have a medical record diagnosis of schizophrenia. However, the low sensitivity values indicate that a self-report in the UK Biobank did not capture everyone who had a medical record. This could be for many different reasons including later onset of illness, the stigma associated with reporting a schizophrenia diagnosis, or the non-specific nature of the question in the initial assessment (“Has a doctor ever told you that you have any other serious medical conditions?”). Participants were not specifically asked about mental health and prompts, if any, were only given for physical health conditions. This may have led to under-reporting of schizophrenia and underscores the importance of ensuring self-report questions specifically reference the diagnosis of interest. The high negative classifications in UK Biobank illustrated that participants who did not self-report schizophrenia also did not have a medical record of schizophrenia and vice versa, demonstrating that, although certainly enhanced by the low prevalence, self-reported diagnoses are effective at ruling out non-cases.
Currently, using a research interview and note review to obtain a diagnosis is considered gold standard, although we find the requirement to attend and undergo a detailed and time-consuming interview may induce recruitment biases, with those participating in such an interview (after the majority having a brief interview first) being younger and more likely to have a high school qualification (GCSEs) than those who only have self-reported. No difference in degree qualification was observed across groups, however, this is likely due to the small proportion of individuals with a degree in the clinical sample (research interview = 18% vs self-report = 16%). Research interviews may exclude participants who are more acutely unwell or cognitively impaired and unable to complete a long assessment. In the UK Biobank, participants with a medical record diagnosis were less likely to have GCSEs and to be employed. Furthermore, participants with a medical record diagnosis were less likely to have a degree (medical record = 25% vs self-report = 36%). This suggests that these participants may have more impaired functioning than those with a self-report only, and by using medical records only as researchers we may be missing participants who are functioning well and/or have not been admitted to hospital. An alternative explanation is that those who did not self-report schizophrenia in UK Biobank may not have sufficient insight to verbally report their diagnosis to the research nurse or may not have been able to complete the online Mental Health Questionnaire due to poorer functioning or education. Thus, the self-report sample in UK Biobank may be missing participants with more impaired functioning. Taken together, our results highlight some phenotypic differences between different methods of identifying a diagnosis of schizophrenia, particularly within UK Biobank where sample representativeness is a known issue. However, for clinically ascertained samples, our results suggest that self-report of a diagnosis made by a health professional may be sufficient given the limited phenotypic differences between self-report and research interview diagnosis, and the potential to include more participants with lower education using self-report.
In contrast to a depression study in UK Biobank which found participants defined by minimal phenotyping (self-report, help-seeking, and symptom-based) had lower SNP-derived heritability than the strictly defined participants (Composite International Diagnostic Interview)23, we did not find a difference in schizophrenia PRS between the self-report and research interview/medical record diagnosis groups. This may reflect differences in obtaining clinical diagnoses of schizophrenia and depression. Depression can be diagnosed in many settings including primary care, whilst schizophrenia is almost universally diagnosed in secondary care following comprehensive assessment. There may be hesitancy to disclose a diagnosis of schizophrenia to patients due to the associated stigma, as there is evidence that health professionals are more likely to communicate diagnoses of depression than schizophrenia to patients, are more likely to diagnose schizophrenia after recurrent episodes than during the first episode and show a preference for using alternative terms to schizophrenia such as psychosis48,49,50. As such, a self-reported diagnosis of schizophrenia may have higher validity.
Our results highlight the potential, especially for genomic studies, of using this self-report method to identify participants for schizophrenia research. However, we did find differences in schizophrenia PRS within hospital admission diagnoses (in primary and secondary admissions). We also found the number of diagnosis reports and admissions were associated with higher PRSs in UK Biobank, as has been reported in previous literature51,52. This is consistent with findings from the PGC, who reported schizophrenia PRS to be higher in patients who were recruited from inpatient settings28. These findings could indicate greater severity or improved accuracy of diagnosis, or both. We also found participants whose primary reason for admission was schizophrenia, and those who only self-reported schizophrenia, had a higher schizophrenia PRS than those with a secondary admission diagnosis. One explanation of the difference in schizophrenia PRS could be that the secondary admission group were participants who were not admitted primarily for psychosis because their symptoms were milder or were well treated. Alternatively, the accuracy of a secondary diagnosis may be more prone to error than a diagnosis given for a primary admission to hospital (e.g., if admitted for a heart attack), although this did not appear to be the case when looking specifically at psychiatric comorbidities.
Limitations
It is important to note limitations of the current study. Participants were invited to complete a SCAN-based research interview if they self-reported psychosis or a schizophrenia diagnosis. This study design prevented us from assessing other metrics (negative predictive value, sensitivity, and specificity) in the clinically-ascertained sample. This also meant we were unable to adjust the PPV to the population point prevalence of schizophrenia. As a result, the PPV could have been inflated by the high proportion of schizophrenia participants in our clinically-ascertained sample. Additionally, some participants had their diagnosis confirmed by a clinician if systematically recruited, which could have increased the positive predictive values, and only a subset of the sample were asked their own opinion of their diagnosis (n = 99). Despite these limitations, our clinically-ascertained sample is one of the only psychosis-based samples with both self-report diagnosis data and a gold-standard research diagnosis. In the UK Biobank, 93% of the participants with a medical record diagnosis of schizophrenia have a hospital admission, therefore the predictive values primarily reflect how predictive a self-report was of a hospital admission.
Both the clinically-ascertained sample and UK Biobank were sampled from the UK; therefore, the findings may not apply to other countries. The generalisability of the UK Biobank findings are also hindered because this sample is not wholly representative of the UK population53. Furthermore, the primary and secondary admission diagnosis in the UK Biobank may have differed depending on the location of the admission (e.g., psychiatric hospital vs general) or by the clinician’s expertise. Polygenic risk scores were restricted to participants from a European genetic ancestry. We were unable to investigate whether the polygenic risk scores differ across diagnostic groups in non-European genetic ancestries due to a limited number of participants in our samples from non-European genetic ancestries. Lastly, the recruitment methods in the NCMH study could have enriched for relatives of those with mood and psychotic disorders, although only 5% (n = 33) of our controls reported having a family history of bipolar disorder or schizophrenia. Nonetheless, and although the effect of this would be conservative, this could weaken the variance explained in our schizophrenia PRS analyses. Our findings apply to large-scale genomic studies and caution should be applied in applying self-report methods of diagnosis to studies such as clinical trials where diagnostic accuracy for individual participants is paramount and where participants may receive financial reimbursement for their time.
Conclusion
Self-reporting a clinical schizophrenia diagnosis may be a valid method for identifying cases in schizophrenia genomic research, providing systematic differences of methodologies are transparently noted. Participants who only self-reported a schizophrenia diagnosis showed differences in age, education, and employment but crucially, they did not differ in relation to schizophrenia genetic liability. These findings provide preliminary evidence for using less stringent methods of ascertaining diagnoses in schizophrenia genomic research, which could reduce the burden on participants and researchers to complete extensive interviews, and thereby potentially improve the representativeness of future samples and increase sample sizes.
Data availability
UK Biobank data can be obtained upon application from https://www.ukbiobank.ac.uk/enable-your-research. The de-duplicated PGC summary stats for schizophrenia are available on the PGC website https://figshare.com/articles/dataset/scz2022/19426775. All code will be made available upon request. Data from the clinically-ascertained cohorts cannot be made publicly available due to restrictions in ethical approvals.
References
American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5™, 5th ed. Arlington, VA, US: American Psychiatric Publishing, Inc.; 2013.
World Health Organisation. Schizophrenia. https://www.who.int/news-room/fact-sheets/detail/schizophrenia. Published 2022. Accessed 20th December 2022.
Harvey, P. D. et al. Diagnosis of schizophrenia: consistency across information sources and stability of the condition. Schizophr. Res. 140, 9–14 (2012).
Haapea, M. et al. Non-participation may bias the results of a psychiatric survey. Soc. Psychiatry Psychiatr. Epidemiol. 42, 403–409 (2007).
Reinikainen, J. et al. Participation rates by educational levels have diverged during 25 years in Finnish health examination surveys. Eur. J. Public Health 28, 237–243 (2017).
O’Connell, K. S. et al. Genomics yields biological and phenotypic insights into bipolar disorder. Preprint at https://www.medrxiv.org/content/10.1101/2023.10.07.23296687v3 (2024).
Weiser, M. et al. Sensitivity of ICD-10 diagnosis of psychotic disorders in the Israeli National Hospitalization Registry compared with RDC diagnoses based on SADS-L. Compr. Psychiatry. 46, 38–42 (2005).
Williams, J., Farmer, A., Ackenheil, M., Kaufmann, C. & McGuffin, P. Group ORR. A multicentre inter-rater reliability study using the OPCRIT computerized diagnostic system. Psychol. Med. 26, 775–783 (1996).
Vares, M., Ekholm, A., Sedvall, G. C., Hall, H. & Jönsson, E. G. Characterization of patients with schizophrenia and related psychoses: evaluation of different diagnostic procedures. Psychopathology 39, 286–295 (2006).
Ekholm, B. et al. Evaluation of diagnostic procedures in Swedish patients with schizophrenia and related psychoses. Nord. J. psychiatry 59, 457–464 (2005).
Davis, K. A., Sudlow, C. L. & Hotopf, M. Can mental health diagnoses in administrative data be used for research? A systematic review of the accuracy of routinely collected diagnoses. BMC Psychiatry 16, 1–11 (2016).
Pasman, J. A. et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal effect of schizophrenia liability. Nat. Neurosci. 21, 1161–1170 (2018).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Demontis D., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51:63 (2019).
Dalby, M. et al. Characterizing mood disorders in the AFFECT study: a large, longitudinal, and phenotypically rich genetic cohort in the US. Transl. Psychiatry 12, 121 (2022).
Levey, D. F. et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat. Neurosci. 24, 954–963 (2021).
Hyde, C. L. et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 48, 1031–1036 (2016).
Davis, K. A. et al. Indicators of mental disorders in UK Biobank—A comparison of approaches. Int. J. Methods Psychiatr. Res. 28, e1796 (2019).
Mallard, T. T. et al. Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities. Cell Genom. 2, 100140 (2022).
Wainberg, M., Jacobs, G. R., di Forti, M. & Tripathy, S. J. Cannabis, schizophrenia genetic risk, and psychotic experiences: a cross-sectional study of 109,308 participants from the UK Biobank. Transl. Psychiatry 11, 211 (2021).
Jermy, B. S., Hagenaars, S., Coleman, J. R., Vassos, E. & Lewis, C. M. Risk factor profiles for depression following childbirth or a chronic disease diagnosis: case–control study. BJPsych Open. 8, e182 (2022).
Levey, D. F. et al. Reproducible Genetic Risk Loci for Anxiety: Results From similar to 200,000 Participants in the Million Veteran Program. Am. J. Psychiatry 177, 223–232 (2020).
Cai, N. et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat. Genet. 52:437 (2020).
Waszczuk, M. A. et al. Dimensional and transdiagnostic phenotypes in psychiatric genome-wide association studies. Mol. Psychiatry 28, 4943–4953 (2023).
Liu, H. et al. Mendelian randomization highlights significant difference and genetic heterogeneity in clinically diagnosed Alzheimer’s disease GWAS and self-report proxy phenotype GWAX. Alzheimer’s. Res. Ther. 14, 17 (2022).
Schoeler, T., Pingault, J.-B. & Kutalik, Z. Self-report inaccuracy in the UK Biobank: Impact on inference and interplay with selective participation. Preprint at https://www.medrxiv.org/content/10.1101/2023.10.06.23296652v1 (2023).
Zheutlin, A. B. et al. Penetrance and Pleiotropy of polygenic risk scores for schizophrenia in 106,160 patients across four health care systems. Am. J. Psychiatry 176, 846–855 (2019).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Underwood, J. F. G. et al. Autism spectrum disorder diagnosis in adults: phenotype and genotype findings from a clinically derived cohort. Br. J. Psychiatry 215, 647–653 (2019).
Wing, J. K. et al. SCAN: Schedules for clinical assessment in neuropsychiatry. Arch. Gen. Psychiatry 47, 589–593 (1990).
Legge, S. E. et al. Clinical indicators of treatment-resistant psychosis. Br. J. Psychiatry 216, 259–266 (2020).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Dennison, C. A. et al. Risk factors, clinical features, and polygenic risk scores in schizophrenia and schizoaffective disorder depressive-type. Schizophr. Bull. 47, 1375–1384 (2021).
Escott-Price, V. et al. Genetic liability to schizophrenia is negatively associated with educational attainment in UK Biobank. Mol. Psychiatry 25, 703–705 (2020).
McCarthy, S., Das, S. & Kretzschmar, W. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Lynham, A. J. et al. DRAGON-Data: a platform and protocol for integrating genomic and phenotypic data across large psychiatric cohorts. BJPsych Open. 9, e32 (2023).
The 1000 Genomes Project Consortium, Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Chang C. C., et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:s13742-13015-10047-13748.
Leonenko, G. et al. Identifying individuals with high risk of Alzheimer’s disease using polygenic risk scores. Nat. Commun. 12, 1–10 (2021).
Conomos, M. P. et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 98, 165–184 (2016).
Bycroft, C. et al. Genome-wide genetic data on ~ 500,000 UK Biobank participants. Preprint at https://www.biorxiv.org/content/10.1101/166298v1 (2017).
Legge, S. E. et al. Association of genetic liability to psychotic experiences with neuropsychotic disorders and traits. JAMA Psychiatry 76, 1256–1265 (2019).
Choi, S. W. O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 8, giz082 (2019).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Tenny S., Hoffman M. R. Prevalence. In: StatPearls. Treasure Island (FL) StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC.; 2023.
Davies, M. R. et al. Comparison of symptom-based versus self-reported diagnostic measures of anxiety and depression disorders in the GLAD and COPING cohorts. J. Anxiety Disord. 85, 102491 (2022).
Vieira, L. S. et al. Self-reporting of psychiatric illness in an online patient registry is a good indicator of the existence of psychiatric illness. J. Psychiatr. Res. 151, 34–41 (2022).
Mitchell, A. J. Reluctance to disclose difficult diagnoses: a narrative review comparing communication by psychiatrists and oncologists. Support. Care Cancer 15, 819–828 (2007).
Paccaloni, M., Moretti, F. & Zimmermann, C. Giving information and involving in treatment: what do psychiatrists think? A review. Epidemiol. Psychiatr. Sci. 14, 198–216 (2005).
Clafferty, R. A., McCabe, E. & Brown, K. W. Conspiracy of silence? Telling patients with schizophrenia their diagnosis. Psychiatr. Bull. 25, 336–339 (2001).
Meier, S. M. et al. High loading of polygenic risk in cases with chronic schizophrenia. Mol. Psychiatry 21, 969–974 (2016).
Glanville, K. P. et al. Multiple measures of depression to enhance validity of major depressive disorder in the UK Biobank. BJPsych Open. 7, e44 (2021).
Fry, A. et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Acknowledgements
JTRW, MCO’D and MJO received a research grant to Cardiff University from Takeda Pharmaceuticals that funded this work and GW’s research position. This work was supported by the following grants: Medical Research Council Program (MR/P005748/1), DATAMIND (MR/W014386/1) and a grant from NIH (Award U01MH109514). We thank the participants, clinicians, lab staff and field team for their help with the NCMH, CardiffCOGS and UK Biobank studies.
Author information
Authors and Affiliations
Contributions
G.W. conducted statistical analyses with supervision from S.E.L., A.J.L., I.R.J., and J.T.R.W. S.S. and L.H. processed the clinically-ascertained genomic data with supervision from A.F.P. and J.T.R.W. E.R.D. assisted in the processing of the phenotypic data from the clinically-ascertained sample. J.T.R.W., V.E.P., M.C.O.D. and M.J.O. critically reviewed analyses and results. G.W. drafted the manuscript with input from S.E.L. and A.J.L. All authors read, contributed to revisions, and approved the final draft of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
JTRW, MCO’D and MJO received a research grant to Cardiff University from Takeda Pharmaceuticals that funded this work and GW’s research position. Pharmaceuticals that funded this work and GW’s research position. Takeda Pharmaceuticals have not had any input into the study design, analysis, or interpretation of results. JTRW, MCO’D, MJO, IRJ and AFP have received research funding from Akrivia Health for work unrelated to this study. The remaining authors have nothing to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Woolway, G.E., Legge, S.E., Lynham, A.J. et al. Assessing the validity of a self-reported clinical diagnosis of schizophrenia. Schizophr 10, 99 (2024). https://doi.org/10.1038/s41537-024-00526-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41537-024-00526-5