Abstract
Computational research tools have reached a level of maturity that enables efficient simulation of neural activity across diverse scales. Concurrently, experimental neuroscience is experiencing an unprecedented scale of data generation. Despite these advancements, our understanding of the precise mechanistic relationship between neural recordings and key aspects of neural activity remains insufficient, including which specific features of electrophysiological population dynamics (i.e., putative biomarkers) best reflect properties of the underlying microcircuit configuration. We present ncpi, an open-source Python toolbox that serves as an all-in-one solution, effectively integrating well-established methods for both forward and inverse modeling of extracellular signals based on single-neuron network model simulations. Our tool serves as a benchmarking resource for model-driven interpretation of electrophysiological data and the evaluation of candidate biomarkers that plausibly index changes in neural circuit parameters. Using mouse LFP data and human EEG recordings, we demonstrate the potential of ncpi to uncover imbalances in neural circuit parameters during brain development and in Alzheimer’s Disease.
Similar content being viewed by others
Introduction
In the last few decades, numerous research initiatives have focused on developing the research infrastructures and computational engines required for the effective simulation of brain activity at multiple levels, ranging from individual neuron activity to large-scale brain signals1,2,3. Network models composed of single neurons, based on leaky integrate-and-fire (LIF) spiking neurons and multicompartment neurons, have been employed to address fundamental questions about neural dynamics, cortical oscillations, sensory processing, working memory, behavior, cortical connectivity, and neurological diseases4,5,6,7,8,9,10,11,12,13,14. Moreover, the recent surge in both the quantity and quality of available data, coupled with rapid advancements in computational power, has enabled the development of spatially extended data-driven cortical networks composed of LIF and multicompartment neuron models15,16,17,18,19,20,21. Compared to more abstract, idealized models, the benefits of using single-neuron models are: (i) a more realistic accounting of the biophysics of neurons and their interrelationships in the cortical circuit, including, for example, their spiking activity and non-linear synaptic interactions; and (ii) a more precise comparison between model parameters and experimentally accessible in vivo features, using tools such as electrophysiology and fluorescent calcium imaging. Single-cell network models have limitations, including higher computational demands and a potentially large number of parameters that need to be tuned. Additionally, a key drawback of LIF neuron models is their representation as a single compartment, which prevents them from capturing the spatial distribution of transmembrane currents necessary for generating extracellular potentials22. However, progress made in recent years has enabled the accurate approximation of extracellular signals by leveraging variables directly measured from LIF network simulations23,24,25,26,27. By using an appropriate forward head model, such as the New York head model28, these network models can generate realistic representations of large-scale field potentials, such as electroencephalogram (EEG) and magnetoencephalography (MEG) signals, paving the way for applying single-cell network models in clinical settings.
Multicompartment and spiking neural network models have been applied to explore the links between electrophysiological population dynamics and the underlying microscopic biophysics-based understanding of neuron dynamics10,13,17,26,29,30. Recently, single-neuron network models have been utilized to elucidate potential disruptions in cortical circuit mechanisms that lead to atypical patterns in neuroimaging recordings measured in patients with neurological disorders5,31,32. Specifically, these studies have investigated the causal relationship between specific biomarkers, such as the aperiodic component of neural power spectrum, often referred to in the literature as the 1/f slope33, and imbalances of the excitation/inhibition (E/I) ratio in clinical populations. There is a growing body of evidence indicating that E/I imbalances in a variety of medical conditions are mirrored by changes in certain neuroimaging biomarkers5,31,32,34,35,36,37, suggesting that these readouts could serve as indirect indicators of the underlying neural circuit dysregulation. These markers have also been correlated with changes in cortical circuit activity associated with brain development and healthy ageing38,39,40. Unfortunately, we currently lack validated noninvasive methods to evaluate the effectiveness of each marker in measuring the E/I ratio and other circuit parameters34,41. Before these markers can be reliably used in clinical practice, further efforts are needed to refine them and thoroughly investigate their potential as indicators of circuit parameter imbalances.
A critical challenge to achieve this is the absence of standardized methods and tools for systematically assessing the markers’ effectiveness in tracking specific attributes of the neural circuit configuration. For example, previous modeling approaches have primarily focused on the forward modeling of the brain signal stemming from simulated activity of cortical circuit models, manually exploring a limited set of parameter combinations and evaluating their effects on electrophysiological patterns17,26,31,32. They do not exploit the enormous potential of machine learning to improve automatic discovery of relationships between model parameters and neural data, effectively addressing the inverse modeling problem. A successful approach for integrating mechanistic modeling with machine learning methods is the use of a machine-learning-assisted simulation framework, also known as surrogate modeling42,43,44. This methodology has been widely used across diverse fields, from engineering and environmental sciences to finance and medicine, to approximate complex, computationally expensive simulations42,43,44,45,46. The learning model can be trained on simulation data as an inverse surrogate to infer model parameters from empirical data47,48. Inverse surrogate models are often built using well-established regression algorithms, e.g., a multilayer perceptron (MLP) or Ridge regression models, which provide individual estimates of model parameters. On the other hand, simulation-based inference (SBI) offers a powerful alternative to traditional model inversion by leveraging deep neural density estimators to learn a probabilistic Bayesian mapping between observed data and model parameters. Unlike conventional methods, SBI techniques are capable of identifying the full landscape of parameter distributions that are consistent with the observed phenomena49,50,51,52. This enables a thorough exploration of the parameter space, capturing intricate and nonlinear relationships between model parameters. SBI methods are particularly advantageous in inference scenarios where the likelihood function is analytically intractable or computationally prohibitive, such as in the case of neural mass modeling at whole-brain level53,54,55. Yet, the use of deep learning density estimators in biophysically detailed single-cell network models is challenging, and techniques for doing so have only recently started to be developed56.
Various approaches from brain research and artificial intelligence have studied the relationship between features from neural data and cortical circuit properties, yet key aspects remain unclear. We introduce the neural circuit parameter inference (ncpi) toolbox, an all-in-one platform integrating state-of-the-art methods from both disciplines to advance research in inverse modeling. Our toolbox serves as a model-driven testbench for evaluating candidate biomarkers in population-level electrophysiological data, enabling rapid, interpretable, and systematic comparisons of circuit parameter predictions for individual biomarkers (or a combination of biomarkers) on a given dataset. To demonstrate the applicability of the ncpi toolbox, we created a dataset of 2 million simulated samples generated from a LIF network model and computed local field potentials (LFPs) and EEG signals from this data. We used the simulation data to analyze the relationships between key cortical circuit parameters (e.g., E/I ratio) and field potential features (i.e., candidate biomarkers). We then trained a series of regression-based inverse surrogate models (i.e., MLP and Ridge), and SBI models, and assessed the predictive power of these approaches trained on individual features and their combinations. Finally, we explored ncpi’s capabilities in revealing mechanistic imbalances in circuit parameters using real data, employing a bottom-up approach based on LFP and EEG recordings, starting with mice and extending to humans. We show that our approach has the potential to: (i) reveal the relationship between local circuit properties and population-level brain dynamics using field potential recordings, (ii) be applied across different spatial scales, and (iii) track the pathological progression and variability of circuit parameters in clinical populations.
Results
The results are organized as follows: first, we introduce the ncpi software platform and its features; next, we analyze the characteristics of the simulation data generated for this study and the predictive power of various learning algorithms and biomarker configurations; finally, we apply our approach to two distinct experimental datasets and present the resulting inferences.
Overview of the ncpi toolbox
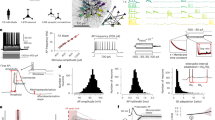
We begin by describing the developed toolkit and key techniques integrated in the software platform. Figure 1 shows the structure of ncpi, illustrating the toolbox’s approach for providing an all-in-one computational framework that unifies methods for both forward and inverse modeling of population neural signals (referred to here as field potentials). The Python classes included in ncpi are designed to encompass the entire workflow for simulating brain models (Simulation), computing extracellular signals (FieldPotential), extracting features (Features), training inverse surrogate models, and utilizing them to compute predictions on real data (Inference), and analyzing results (Analysis). These classes integrate a broad spectrum of state-of-the-art methods based on established software toolkits, e.g., NEST57, NEURON58, LFPy59, scikit-learn (sklearn)60 and SBI61, enabling users to easily derive model-based inferences of circuit parameters with minimal coding. Additionally, all classes are designed to support parallel computing, allowing for faster execution by leveraging all available computational resources, whether natively provided by the software tool (such as NEST) or integrated into the ncpi toolbox using the pathos framework62.
The diagram shows the Python classes, their primary methods, and the included software libraries, and provides a representation of the processing flow. From left to right: simulation of neural activity, computation of field potentials, feature extraction, training of regression models, predictions computed on real data, and analysis and visualization of results. For further information, please refer to Methods.
Briefly, the Simulation class provides methods for constructing a network model of individual neurons, simulating its behavior, and collecting simulation outputs such as spike times, synaptic currents, or membrane potentials. As an example, we provide the code to simulate, using NEST, a two-population network (excitatory, E, and inhibitory, I) of LIF neuron models (iaf_psc_exp neuron model) recurrently connected with current-based synapses. The network parameters (e.g., \({J}_{{IE}}\), the synaptic weight between presynaptic population E and postsynaptic population I, or \({N}_{E}\) and \({N}_{I}\), the population sizes) correspond to the best fit provided in a recent modeling study23. Outputs recorded from the cortical circuit model simulation are utilized by methods of the FieldPotential class. Extracellular signals are derived for LIF network models by convolving population spike rates with spatiotemporal filter kernels. These kernels are previously obtained using a hybrid modeling framework23,27 built on NEURON multicompartment neuron simulations (Fig. 1 and Methods). Alternatively, we can approximate the extracellular signals by combining variables directly measured from network simulations through proxies (i.e., estimates of the signal), such as the average firing rate or the average sum of AMPA and/or GABA currents. The kernel method can be employed to calculate LFP signals across different recording sites or to estimate the current dipole moment (CDM). The CDM defines EEG and MEG signals, both of which can be calculated using a suitable forward head model, such as the New York head model28. Although the FieldPotential methods were initially designed for computing model-based inferences with point-neuron models, they can also be used with multicompartment models, leveraging the packages available from the LFPy workspace59,63 for computing extracellular potentials from multicompartment neuron networks.
Next, features from field potentials (i.e., putative biomarkers) are computed using well-established scientific time-series analysis techniques included in the Features class. Two time-series feature libraries are included: hctsa64 and catch2265, which provide a comprehensive set of data-driven feature-based representations. These feature sets have demonstrated success in solving a broad spectrum of time-series problems across diverse applications in the life sciences, particularly in neuroscience, e.g., for investigating neurophysiological signatures related to cortical micro-architecture66 or exploring multiple representations of cortical connectivity67. We have also integrated two additional well-known feature extraction methods shown to effectively assess changes in the E/I balance: the specparam library for power spectrum parametrization68, which separates power spectra into aperiodic and periodic components, and the functional E/I ratio (fE/I), a fluctuation-based metric that was developed to infer the state of dynamics concerning the critical point36. Features extracted from simulated data are incorporated into the Inference class and used for training inverse surrogate models, accessible through two prominent machine learning libraries integrated into the toolbox: sklearn and SBI. By employing the RepeatedKFold method, available in Inference, the user can evaluate the model’s stability and generalization across multiple training and testing splits. Once the inverse surrogate model is trained on simulation data, it can be applied to real-world data to predict the neural parameters associated with each dataset. Finally, the Analysis class provides methods for automatically performing statistical analyses on features and predictions, e.g., using linear mixed-effects (LME) models, as well as for creating appealing and informative statistical graphics.
Figure 2 presents a Python code example leveraging all ncpi’s classes, demonstrating the toolbox’s full potential to create in-silico brain models, simulate neural activity, extract features, train an inverse model, and make predictions on real data, all with just a few lines of code. This example showcases how ncpi can streamline complex workflows, providing an easy-to-use and intuitive platform for testing hypotheses about the neural origins of electrophysiological data.
This code snippet makes a few assumptions, such as the presence of ‘network.py’ and ‘simulation.py’ as files for creating and simulating the LIF network model, which are included in the ncpi library as examples, the availability of simulated firing rates as LIF_spike_rates, or the preprocessing of empirical data.
Simulation of neural activity and extracellular potentials using a LIF network model
We next describe the characteristics of the neural dynamics produced by the network model of spiking neurons used in our study, along with the generation of extracellular signals and the computation of relevant features. In brief, the model is a generic current-based cortical network model consisting of recurrently connected populations of excitatory and inhibitory LIF point neurons (8192 and 1024 neurons, respectively), driven by external Poisson processes with fixed rates, as described previously23 (see Methods for further details). In addition to simulating the spiking output, we computed the CDM by convolving population firing rates (\({\upsilon }_{X}\)) with spatiotemporal kernels obtained using the methods of the FieldPotential class. Here, for the sake of simplicity, we present changes in the behavior of the network in response to a varying external input current (Fig. 3). Effects of changes in the external input are well-documented in the literature and illustrate how the model’s parameters modulate the output of a recurrent spiking network model8,10,11,14,30,69,70. A key property of excitatory-inhibitory recurrent networks is that increasing the strength of the external input leads to an increase in spiking output and enhancement of synchrony (Fig. 3A). This phenomenon is also observed in the CDM: at a low external input level (\({J}_{{syn}}^{{ext}}=28{nA}\)), the network remains in an asynchronous irregular state, characterized by weak and strongly damped oscillations. In contrast, increasing the external input leads to more synchronized oscillatory activity. This increase in synchronized oscillatory activity is clearly reflected in the power spectrum (Fig. 3B), which shows how increasing external input values from a low level (\({J}_{{syn}}^{{ext}}=28{nA}\)), a state predominantly characterized by 1/f dynamics, lead to an entrainment of gamma oscillations. With an increase in external input, gamma-band oscillation frequencies are also elevated.
A Spike raster plots of the excitatory (E) and inhibitory (I) populations spanning 100 ms of spontaneous activity (top), spike counts in bins of width Δt (middle) and current dipole moment along the z-axis (Pz, bottom) for one of the six separate trials that were computed for this example. B Trial-averaged normalized power spectra of CDMs for the different values of external synaptic currents. C Features extracted from the CDMs across trials using the specparam library (1/f slope) and catch22 (dfa, rs range, and high fluctuation). For illustration, we focus on these three features from the catch22 subset, although we noted that most of the catch22 features exhibited marked trends in response to variations of the external input. A comprehensive description of the various catch22 features and their properties can be found in the associated publication65.
Next, using the tools provided in the Features class, we extracted features from the simulated CDMs by applying the aperiodic 1/f slope and the catch22 library. We chose these feature sets because we believe that comparing these two methods offers a valuable opportunity to contrast a general-purpose time-series feature extraction library with a specific marker, the 1/f slope, that has been shown to reliably track circuit parameter imbalances. Clearly, both the 1/f slope and the three representative features selected from the catch22 set show marked changes in response to alterations in the external input (Fig. 3C). These initial findings indicate that alterations in specific network model parameters are strongly mirrored in the extracellular signal and could potentially be extracted from its readout. However, extrapolating these findings to more complex contexts, such as the joint effects of various parameters in the signal, may not be straightforward, as demonstrated below.
Characterization of features in the simulation dataset
The spiking network model introduced above was used to develop a comprehensive training dataset containing two million individual simulation samples. To create this dataset, we systematically explored key parameters that define recurrent and external synaptic connectivity (\({J}_{{synEE}}\), \({J}_{{synIE}}\), \({J}_{{synEI}}\), \({J}_{{synII}}\), \({\tau }_{{syn}}^{{exc}}\), \({\tau }_{{syn}}^{{inh}}\), and \({J}_{{syn}}^{{ext}}\)), generating simulation data that fully characterize how the model’s synaptic mechanisms contribute to cortical circuit activity and, ultimately, the resulting extracellular signals (for more details, refer to the Methods section). We chose to focus on the synaptic parameters of the model, maintaining the others unchanged, because of their relevance to brain development and ageing38,39,40, and their vital role in neurodegenerative and neurodevelopmental diseases71,72,73,74. Once we generated the simulated spiking activity for the dataset, we calculated the corresponding extracellular signals and computed features using the 1/f slope method and the catch22 library (Fig. 4).
Distributions of features computed on simulation data are shown as a function of synaptic parameters of the LIF network model (E/I, \({\tau }_{{syn}}^{{exc}}\), \({\tau }_{{syn}}^{{inh}}\) and \({J}_{{syn}}^{{ext}}\)). Values of the parameters were binned into equal size intervals. In each bin, for a specific parameter, parameter values were within the limits of the bin, while the other synaptic parameters were free to vary without restriction. Each panel includes η2 as a measure of effect size (ANOVA model).
Distributions of features extracted from the simulation dataset are presented as a function of the parameters of the spiking network model used to generate them (Fig. 4). It is important to note that in each plot for a specific parameter, the other synaptic parameters were permitted to vary without constraints. This data visualization was intentionally selected to illustrate the challenge of distinguishing changes in parameters from feature variations when all parameters simultaneously impact the model’s output and may lead to similar and/or conflicting shifts in the features. Indeed, the analysis of features revealed small effect sizes across most features (η2, ANOVA test). This challenge is exemplified by the pair dfa and \({\tau }_{{syn}}^{{exc}}\), which displays diverse trends based on the interval of \({\tau }_{{syn}}^{{exc}}\) considered, complicating the interpretation of the feature-parameter relationship. In this case, if we consider dfa as the only measured feature, it would be very difficult to accurately deduce the value of \({\tau }_{{syn}}^{{exc}}\). However, other features like high fluctuation, which provides a clearer and more distinguishable trend, could help resolve the ambiguities associated with dfa and offer more accurate estimates of \({\tau }_{{syn}}^{{exc}}\). Nevertheless, although some feature-parameter pairs appear to exhibit a clearer (even quasi-linear) trend, it is evident that, in general, estimating the different intervals of the parameter distribution based on a single feature in isolation is challenging when multiple circuit parameters vary together.
Inverse model predictions using single vs. combined features
Using the dataset of simulated features, we developed inverse surrogate models to recover circuit parameters. As an initial analysis, our goal was to assess the performance of inverse models trained on individual features versus their combinations to determine whether combining features enhances inference accuracy or if certain features are redundant. To do so, we trained a range of neural network regression models varying the feature sets used for training. For each combination, we then compared the predicted and actual parameter values on a held-out subset (Fig. 5A). The predicted parameters from multi-feature models, such as catch22 or catch22 + 1/f slope, showed a linear or quasi-linear trend when compared to the actual values, with the distributions of predicted values closely matching the actual ones. Single-feature model predictions, on the other hand, struggled more to accurately capture actual parameters. However, in most cases, the predicted vs. actual value distributions for single-feature models showed a monotonically increasing trend. Inverse models can be used to compare clinical groups in empirical data or across different experimental conditions. In such cases, this suggests that single-feature models may have the potential to correctly track increases or decreases in a parameter, even if the actual parameter value is not accurately decoded.
A Predicted versus actual values. B Probability functions of absolute errors. DH,1 represents the Hellinger distance between the probability distributions of models trained with catch22 and catch22 + 1/f slope. DH,2 represents the Hellinger distance between all the models trained on a single feature. DH,3 represents the distance between catch22 and the models trained on a single feature. All prediction results shown here were computed on a held-out test dataset representing 10% of the whole simulation dataset.
The differences between single- and multi-feature models in capturing the real circuit parameters are reflected in the distributions of absolute errors (Fig. 5B). Error distributions from multi-feature models tend to cluster more around lower values than those from single-feature models. To further investigate the differences in errors across various feature combinations, we computed the Hellinger distance between the corresponding distributions (where 0 indicates that the two distributions are identical, and 1 indicates that they are completely dissimilar). Our first observation is that the distance between models trained with catch22 and those with catch22 + slope (DH,1) is minimal, indicating that including the slope in the catch22 feature set does not significantly improve predictive accuracy. The distance between all models trained on a single feature (DH,2) is slightly larger, though still small, which may suggest that no single-feature method clearly outperforms the others. Finally, the distance between catch22 and the models trained on a single feature (DH,3) is the largest, indicating that multi-feature models and single-feature models produce the most distinct error distributions. We also trained linear regression models and repeated the same analysis, drawing similar conclusions (Supplementary Fig. 1). However, in this case, the distances between single- and multi-feature models were smaller, suggesting that linear models may not capture all the non-linear relationships among multiple features. Finally, we trained an SBI model, i.e., a Neural Posterior Estimation (NPE) model, on the different feature configurations and evaluated the posterior fit of each method by comparing the posterior z-score and posterior shrinkage (Supplementary Fig. 2). Posterior estimates for all approaches were concentrated toward large shrinkages, indicating that the posteriors in the inversion are quite informative. Additionally, the small z-scores suggest that the true values are accurately captured within the posteriors. However, the multi-feature models provided the best Bayesian inversions, as they exhibited lower z-scores and larger posterior shrinkages across all parameter components.
To apply inverse models to empirical data, we opted to compare a multi-feature model with a single-feature model. Based on previous results, catch22 has been shown to perform well without the need for additional features, so we chose it as the multi-feature model. For the single-feature model, we selected the 1/f slope, a well-established feature that has been widely studied in recent years for analyzing E/I34. We believe these two configurations illustrate the contrast between single- and multi-feature models, enabling a clear comparison of their differences.
Estimated shifts in cortical circuit parameters during early postnatal development
The applicability of the toolbox to real-world scenarios was first assessed using a dataset of resting-state LFP recordings obtained in vivo from the prefrontal cortex (PFC) of unanesthetized mice during their early postnatal days38. Using the same feature extraction methods applied to the simulation data, we began by computing features from the empirical data. Our analysis of LFP data features revealed consistent trends across features (except rs range) within the first two weeks after birth (Supplementary Fig. 3). Essentially, we found that the 1/f slope and high fluctuation exhibited a clear upward trend during the first postnatal days, while dfa showed a decrease. Indeed, the increasing trend of 1/f slope with respect to postnatal days aligns with findings from earlier studies38. Upon completing feature extraction, we trained two neural network regression models using repeated 10-fold cross-validation: one incorporating the full set of catch22 features and the other relying exclusively on the 1/f slope. We then used the trained models, serving as inverse models, on features derived from the LFP data to obtain parameter predictions (Fig. 6).
Fits of parameters of the LIF network model (E/I, \({\tau }_{{syn}}^{{exc}}\), \({\tau }_{{syn}}^{{inh}}\) and \({J}_{{syn}}^{{ext}}\)) were estimated using a MLP regressor based on the complete set of catch22 features (A) or the 1/f slope alone (B). The last column shows estimated normalized firing rates. Asterisks indicate significant differences between ages based on p-values obtained using the Holm-Bonferroni method with the LME and linear models defined in Eqs. 2 and 3 (see Methods). The significance levels are categorized as follows: a p-value between 0.01 and 0.05 is denoted by one asterisk (*), between 0.001 and 0.01 by two asterisks (**), between 0.0001 and 0.001 by three asterisks (***), less than 0.0001 by four asterisks (****), and p-values of 0.05 or greater are considered not significant (n.s.). To compute firing rates, 50 samples were drawn from each parameter prediction within the first and third quartiles of each age, and the firing rates were then normalized by the maximum firing rate of all samples.
The analysis of the prediction results obtained using the catch22 subset (Fig. 6A) revealed a moderate reduction in the E/I ratio during the first postnatal days. Notably, the greatest number of significant pairwise differences was observed for \({\tau }_{{syn}}^{{inh}}\), \({J}_{{syn}}^{{ext}}\) and firing rates (LME and linear models, Holm-Bonferroni test). Changes in these parameters during early development were primarily characterized by a shortening of \({\tau }_{{syn}}^{{inh}}\) accompanied by higher \({J}_{{syn}}^{{ext}}\) values and increased firing rates. Following this, we examined the prediction results using the 1/f slope (Fig. 6B). Nearly all pairwise differences for all parameters were statistically significant, excluding \({J}_{{syn}}^{{ext}}\). Compared to the results observed using the catch22 feature set, the estimated parameters (\({\tau }_{{syn}}^{{exc}}\), \({\tau }_{{syn}}^{{inh}}\) and firing rates) exhibited opposite trends except for the E/I ratio. We also trained Ridge and NPE models, applying them to the LFP dataset (Supplementary Figs. 4 and 5). The prediction results were largely consistent with those shown in Fig. 6, particularly for the NPE model. However, some results from the Ridge regression model exhibited different or less significant trends, such as those observed for the E/I ratio and firing rates using the catch22 feature set.
Predictions of the spatiotemporal progression of circuit parameter alterations in Alzheimer’s Disease
Finally, we tested the applicability of our toolbox on a larger-scale type of field potential recording, i.e., EEG data. To this end, we selected a comprehensive EEG dataset that includes both healthy controls (HCs) and patients clinically diagnosed with dementia due to Alzheimer’s Disease (AD) at various stages of the disease (i.e., mild AD patients, ADMIL; moderate AD patients, ADMOD; and severe AD patients, ADSEV)75. Eyes-closed resting-state EEG recordings were acquired from 19 electrodes according to the International 10-20 system75. We employed a morphologically realistic forward model, the New York head model28, to simulate EEG signals for comparison with empirical measurements. We assumed that the signal from each EEG electrode originated from an independent, single local cortical circuit model (i.e., the LIF network model). Simulated EEG data were used to train the inverse surrogate models through repeated 10-fold cross-validation.
As shown in Fig. 7, the prediction results were calculated using the same feature sets as those applied to the LFP data presented above. Initially, focusing on the predictions of E/I imbalances, we observed that the 1/f slope-based predictions revealed a significant shift towards inhibition in many electrode positions of AD patients, while catch22 showed only minimal changes (LME and linear models, Holm-Bonferroni test). Conversely, predictions related to changes in the external input \({J}_{{syn}}^{{ext}}\) were more evident for catch22, indicating a decline in \({J}_{{syn}}^{{ext}}\) starting with ADMOD patients and spreading into the later stages of the disease. Finally, the 1/f slope and catch22 generated predictions of synaptic time constants that conflicted with each other, much like the results from the LFP data. Interestingly, catch22-based predictions revealed an increase in \({\tau }_{{syn}}^{{exc}}\), which was more prominent during the later stages of the disease (i.e., ADSEV group), while \({\tau }_{{syn}}^{{inh}}\) showed a decrease. Opposing directional trends were observed for the 1/f slope predictions of synaptic time constants, though the affected areas did not align with predictions of catch22.
Topographic representation of group differences in parameter fits based on the complete set of catch22 features (A) or the 1/f slope alone (B). The groups are classified as healthy controls (HCs), mild AD patients (ADMIL), moderate AD patients (ADMOD), and severe AD patients (ADSEV). Significant differences between groups were assessed using the Holm-Bonferroni method, applying the LME and linear models as defined in Eqs. 4–7. Only z-ratio values with an associated p-value ≤ 0.01 are plotted. The color range for the z-ratio values was selected such that positive differences relative to the control group were represented by shades of red, while negative differences were indicated by shades of blue.
Discussion
Electrophysiology approaches for recording neural population activity, extending from LFP to MEG and EEG, are tremendously valuable for examining neuronal activity and offer enormous opportunities for treating the human brain76,77,78. A key challenge in neuroscience is identifying the neural circuit configurations that generate the spatial, spectral, and temporal features of brain signals associated with relevant neural processes in both the healthy and diseased brain. Various influential strategies have been applied to interpret electrophysiological signals in terms of their biophysical substrate, yet substantial challenges continue to impede further progress. The computational framework presented here has been created to effectively integrate advancements from the different disciplines, tackling these challenges and advancing existing techniques for exploring the inverse modeling of electrical and magnetic brain signals. In this work, we present the results of a first study using ncpi, highlighting its ability to simulate realistic in-silico field potential data from a LIF network model, extract key neurophysiological features, train different inverse model approaches and, ultimately, compare how different features (or set of features) influence parameter predictions in both simulated and real data. Below, we discuss our findings and their significance.
Our initial simulation results using the ncpi toolbox (Fig. 3) reveal that variations in individual network parameters have a significant impact on the extracellular signal, which supports the hypothesis that changes in neural parameters could potentially be estimated from the signal’s readout. Following this, we assessed a more realistic case where all synaptic parameters (\({J}_{{synEE}}\), \({J}_{{synIE}}\), \({J}_{{synEI}}\), \({J}_{{synII}}\), \({\tau }_{{syn}}^{{exc}}\), \({\tau }_{{syn}}^{{inh}}\), and \({J}_{{syn}}^{{ext}}\)) were allowed to vary together (Fig. 4). A key insight was that accurately inferring parameter changes from a single feature is challenging when multiple parameters simultaneously influence the model’s output, as their effects can overlap or counteract, leading to confounding shifts in the features. Since different features capture distinct aspects of the signal, these findings suggest that a combination of features could provide a more comprehensive representation of the underlying neural processes. Moreover, many features are known to be intercorrelated, as demonstrated in our study, suggesting they may reflect redundant or overlapping aspects of the same phenomenon34,66. Thus, examining multiple biomarkers simultaneously may provide a richer and more detailed understanding of brain dynamics. Our results show that ncpi is perfectly tailored for these objectives, facilitating quick and efficient investigation of all possible links between circuit parameters and individual biomarkers or combined configurations of biomarkers.
To further investigate the relationship between features and model parameters in the simulation dataset, we trained various inverse models on individual features and their combinations (Fig. 5). This approach helped us assess whether combining features improves inference accuracy or if multi-feature models offer no added value. A few important findings were revealed through this analysis. First, we observed that multi-feature models outperform single-feature models in capturing the real values of all parameters. However, single-feature models exhibited a monotonically increasing trend in the predicted vs. actual value distributions. Inverse models may be designed to compare clinical groups or experimental conditions. In such cases, this result suggests that single-feature models may still be able to identify whether a parameter is increasing or decreasing, even if they fail to accurately decode the actual value. Furthermore, we discovered that adding the slope to the catch22 feature set does not enhance predictive accuracy, implying that the catch22 subset is already sufficiently informative to accurately predict all cortical circuit parameters. Finally, our evaluation revealed that models trained on individual features exhibited comparable predictive performance, suggesting that no single-feature approach consistently outperformed the others.
The toolbox has been validated using two distinct experimental datasets containing electrophysiological recordings captured at different spatial scales and in different species. We began by calculating predictions of neural parameters from a dataset of in vivo LFP recordings taken from the PFC of unanesthetized mice during the first days after birth38. As the brain develops, the parameters that define neural activity change progressively, with alterations in inhibitory neurons primarily driving changes in the excitatory and inhibitory balance during prefrontal cortex development38,79,80. Moreover, E/I dysregulation during brain development has been linked to the pathophysiology of neurodevelopmental disorders74. Given the well-established role of excitatory and inhibitory processes and their significance in neurodevelopment, this dataset is essential for evaluating whether our methods can reveal how these neural parameters evolve during development.
Using all catch22 features, we found a reduction in the E/I ratio during the first postnatal days (Fig. 6A), a result which is consistent with previous findings suggesting a concomitant shift in the synaptic and functional excitation-inhibition balance favoring inhibition38,80. In contrast, \({\tau }_{{syn}}^{{exc}}\) exhibited a significant upward trend after the tenth postnatal day, which might align with electrophysiological reports of increasing spontaneous excitatory postsynaptic current (sEPSC) time constants as measured in pyramidal neurons during the first postnatal weeks of age81. Remarkably, the empirical literature consistently aligns with our tool’s predictions regarding the directional changes in \({\tau }_{{syn}}^{{inh}}\), \({J}_{{syn}}^{{ext}}\) and firing rates throughout postnatal development, i.e., a shortening in \({\tau }_{{syn}}^{{inh}}\)81,82 coupled with higher firing rates38. Though empirical measures of \({J}_{{syn}}^{{ext}}\) are more difficult to obtain, we expect this parameter to correlate with predicted firing rates and reflect the increase in firing inputs from other neurons in the network.
We next computed results using the 1/f slope alone (Fig. 6B). While the E/I predictions aligned well with the hypothesis of reduced E/I during early development, the predictions for other parameters conflicted with prior empirical evidence. Notably, the slowing in GABA kinetics (i.e., higher \({\tau }_{{syn}}^{{inh}}\)) and the decrease in firing rates are largely inconsistent with prior assumptions. Our predictions based solely on the 1/f slope indicate that, although this marker is effective in reflecting changes in E/I imbalances, as previously documented5,31,32,33,34,35,37,38, it may struggle to predict other parameters accurately when those predictions might contradict the neuronal effects they generate (e.g., a shift in E/I favoring inhibition combined with a rise in firing rates). In these contexts, predictions developed using a combination of features (such as catch22) may incorporate complementary insights from diverse features, producing more accurate estimates for the whole set of parameters.
The second dataset that we analyzed consists of EEG resting-state recordings from healthy controls and clinical AD patients at various severity stages83. This diversity in disease stages presents a unique opportunity to evaluate potential predictions regarding the progression and variability of circuit parameters based on diverse biomarkers across the course of the pathology. A key finding in this study was that predictions based on catch22 features indicated a significant reduction in the external input of AD patients progressing at later stages of the disease (Fig. 7A), while the 1/f slope results (Fig. 7B) exhibited a significant shift of E/I towards inhibition. Predictions derived from these two distinct feature metrics may index neuronal and neurodegenerative mechanisms mediated by the tau protein, albeit from different perspectives: one indicating a decrease in E/I71,72,73,84 and the other reflecting the accelerated breakdown of synaptic connectivity associated with the widespread of this protein in advanced stages of the disease71,73,85,86. Although both tau and amyloid-β (Aβ) exhibit synaptotoxic effects85,87, recent studies suggest that tau burden is more strongly associated with synaptic loss, independent of Aβ plaques88,89. As tau spreads throughout the cortex, particularly in later disease stages, it may functionally manifest as a reduction in net E/I balance at the network level, overcoming the excitatory effects of amyloid-β, and as a significant deterioration in external input at the synaptic connectivity level. Another important result here, derived from the analysis of predicted time constants, is the progressive increase of \({\tau }_{{syn}}^{{exc}}\) revealed by catch22, suggesting a loss of efficacy in the excitatory synapses. This result aligns with earlier modeling work that fitted mass model parameters to MEG data and suggested that this increase in \({\tau }_{{syn}}^{{exc}}\) may be tied to tau-mediated neuronal effects90. In fact, this change in \({\tau }_{{syn}}^{{exc}}\) was more prominent in the later stages of the disease, matching well with the timeline of tau protein dissemination73. This previous work also revealed a mild decrease in \({\tau }_{{syn}}^{{inh}}\) in some frontal and parietal areas, consistent with our predictions using catch22; however, no significant correlation with the PET images of amyloid beta or tau was identified90.
At present, there is no systematic methodology to assess the accuracy, reproducibility, reliability, and utility of electrophysiological data biomarkers, particularly in terms of the mechanistic link between each marker and the cortical circuit attribute under investigation. Importantly, there is a need for noninvasive and robust metrics that can detect circuit imbalances in a variety of neurodevelopmental, neurodegenerative, and psychiatric conditions. Our computational framework makes a significant contribution to the development of software tools that integrate all stages of the model-based inference pipeline, from the forward modeling of field potentials to parameter estimation, serving as a benchmarking resource for evaluating candidate biomarkers of neural circuit parameters. Our model-driven approach has proven effective in capturing local changes in cortical dynamics during brain development and identifying pathological alterations in circuit parameters within clinical populations in AD. The ncpi toolbox has been tested on a spiking microcircuit model of recurrent excitatory and inhibitory neuronal populations, which approximates key circuit phenomena at the local scale. However, this approach does not account for macroscopic network dynamics, such as long-range cortico-cortical interactions, which are known to play a critical role in modulating low-frequency cortical oscillations91. These dynamics are explicitly incorporated in other modeling frameworks, such as mean-field or neural mass models53,54,92,93,94. To partially address this, we included an external input representing inputs from cortico-cortical connections, allowing us to approximate macroscopic influences on local predictions, while also managing the complexity of the learning model to avoid overfitting. Future investigations may focus on larger-scale single-cell network models, for instance, integrating connectivity among different brain areas19,95, to offer a more comprehensive understanding of global processes that influence predictions of local dynamics. Moreover, time-series feature libraries such as catch22 and hctsa, and spectral measures like the 1/f slope, provide interpretable features of neural activity that have consistently been linked to underlying cortical mechanisms66,67,96. However, achieving a more comprehensive understanding of circuit parameter imbalances will likely require assessing biomarker interactions and further exploring how these measures relate to one another and reflect different properties of the data. The ncpi toolbox provides a powerful platform for exploring different hypotheses about neural circuit biomarkers, offering opportunities to reassess past findings and identify new avenues for future research.
Methods
Modeling and simulation of single-neuron network models
Given the variety of simulation tools available for creating neural network models and the vast range of possible network configurations and modeling parameters, it is challenging to define a class with fixed, parameterized methods for setting up network simulations. Instead, the Simulation class includes methods to configure and simulate the network model by loading custom external scripts and parameter files, adhering to the workflow typically used in brain modeling projects, which includes distinct code segments for network setup, simulation, and analysis19,95. We provide example scripts within the library that are specifically designed to simulate a current-based cortical network model composed of recurrently connected populations of excitatory and inhibitory LIF point neurons (8192 and 1024 neurons, respectively) driven by external fixed-rate Poisson processes. Network parameters correspond to the best-fit parameters as described in the associated publication23 (Table 1). We defined a 12,000 ms simulation with a 0.0625 ms time step, discarding the first 2000 ms as transient time in the analysis. Model simulation is configured for execution in NEST, utilizing its multithreading capabilities97. Throughout the simulation, variables like spike times, synaptic currents, and membrane potentials are recorded by multimeters and stored for later computation of field potentials.
Additionally, we conducted extensive simulations of this model, systematically varying key parameters that define recurrent and external synaptic connectivity (\({J}_{{synEE}}\), \({J}_{{synIE}}\), \({J}_{{synEI}}\), \({J}_{{synII}}\), \({\tau }_{{syn}}^{{exc}}\), \({\tau }_{{syn}}^{{inh}}\) and \({J}_{{syn}}^{{ext}}\)), generating a dataset of about 2 million distinct simulations. These parameters fully characterize how synaptic mechanisms of the model contribute to cortical circuit activity and, ultimately, the resulting macroscopic brain signals. Furthermore, since synaptic alterations play a crucial role in neurodegenerative and neurodevelopmental diseases71,72,73,74, key clinical targets of our toolbox, we chose to focus on studying these parameters while keeping the others unchanged. Table 2 details the parameter ranges chosen to ensure that the network does not enter either an inactive or an excessively high activity state, and resulting simulation output values are biologically plausible. This extensive dataset of simulation data has been made publicly available (see Data and code availability), allowing researchers to explore the intricate relationships between synaptic parameters of a cortical model and field potential features without the lengthy wait associated with months of simulations. Importantly, the parameter search encompasses all possible combinations within the specified ranges for each parameter, providing rich insights into their interrelations. For example, one could compare the combined effects of modifying recurrent and external synaptic connection parameters on network activity to determine whether they are distinguishable or whether specific features can reveal the independent contribution of each parameter.
Computation of field potentials
Extracellular signal predictions can be obtained by convolving population spike rates with appropriate spatiotemporal causal filters or by using predefined proxies. In the case of kernel-based methods, ncpi integrates the LFPykernels package23, which incorporates efficient forward-model calculations of causal spike-signal impulse response functions for finite-sized neuronal network models. Let \({\upsilon }_{X}(t)\) and \({H}_{{YX}}({\boldsymbol{R}},\tau )\) describe the presynaptic population spike rates in units of spikes/dt and the predicted spike-signal kernels for the connections between presynaptic populations \(X\) and postsynaptic populations \(Y\). The full extracellular signal, \(\psi \left({\boldsymbol{R}},t\right)\), may then be computed via the sum over linear convolutions:
To compute the kernels \({H}_{{YX}}({\boldsymbol{R}},\tau )\), a hybrid scheme can be used, which simulates ‘ground-truth’ multicompartment neuron populations and computes the responses to synchronous activation of all neurons in each population27. The recent kernel computation method introduced in LFPykernels replaces the simulation of multicompartment neuron populations with the simulation of a single linearized multicompartment neuron representing each population, while accounting for the known spatial distributions of cells and synapses in space, recurrent connectivity parameters, and biophysical membrane properties23. The example multicompartment network description provided in ncpi for computing the kernels is based on the reference network of simplified ball-and-stick neurons provided in Hagen et al23. Tables 3 and 4 include information on the parameters of individual neurons, network configuration, and synaptic connectivity of the multicompartment network model. Additional details on the multicompartment neuron model specifications, including specific ionic current dynamics, are available in the corresponding papers23,98.
A kernel can be used to generate two types of signals, namely the LFP signals across recording sites, or the CDM, using the GaussCylinderPotential and KernelApproxCurrentDipoleMoment methods from LFPykernels23, which assume radial symmetry of the cell population around the z-axis. We have used the CDM to approximate the summed signal of extracellular potentials across depths (i.e., the LFP signal). Based on the kernel method, we computed a CDM for each of the two million simulation samples in the dataset and stored them along with the simulation parameters. The CDM can then be used to derive the EEG and MEG signals using an appropriate forward head model, such as the New York head model28. This head model enabled us to generate simulated EEG signals configured according to the 10-20 system using the NYHeadModel class from the LFPy suite59,63. Specifically, we positioned a CDM beneath each electrode in the 10–20 system and, at each instance, calculated the resulting EEG signal at each electrode site, assuming independent contributions from the simulated activity of the local cortical model. EEG signals for each simulation were stored alongside the corresponding CDMs in the dataset.
An alternative approach offered by ncpi for approximating extracellular signals is based on the use of proxies, directly combining variables measured from simulations of point-neuron networks69. We have incorporated most of the ad-hoc and optimized proxies proposed in the literature to estimate field potentials across the different recording scales (Table 5 and Fig. 1). Ad-hoc proxies, such as the sum of membrane potentials or the population spike rate, approximate the field potential by summing (or, optionally, averaging) the corresponding simulation variable across all cells in the population. Proxies such as LRWS, ERWS1, and ERWS2 account for both causal and non-causal relationships between measured extracellular potentials and synaptic currents24,25. Based on a weighted sum of time-shifted synaptic currents, these proxies were optimized using a hybrid modeling scheme27 to capture a broad range of multicompartment network configurations, including variations in cell morphologies, distributions of presynaptic inputs, positions of recording electrodes, and the spatial extent of the network. Unlike kernel methods, these proxies have already been optimized for multiple configurations of a generic recurrent multicompartment network, eliminating the need for new, cumbersome simulations. However, they may yield less accurate approximations of the signal for new network models that differ significantly from the configuration of the network used during optimization.
Feature extraction
We have integrated methods for computing features (i.e., putative biomarkers) from population-level neural signals that are either derived from toolboxes for time-series feature extraction or based on well-known markers of circuit parameters. The two time-series feature libraries that have been incorporated are hctsa64 and its condensed version, catch2265. The hctsa library, developed following a data-driven approach, provides a vast library of over 7000 features, enabling extensive analysis of time-series data across various domains. This includes statistical, temporal, and spectral features that capture different aspects of the data, e.g., distribution of values in the time series, their linear and non-linear autocorrelation, measures derived from power spectral densities, temporal statistics, scaling of fluctuations, and others64. This library has found successful applications across multiple areas of life sciences, particularly in neuroscience, e.g., for investigating neurophysiological signatures related to cortical micro-architecture66 or exploring multiple representations of cortical connectivity67. As we could only find its MATLAB version, we have integrated hctsa into the ncpi toolbox by leveraging the MATLAB engine API for Python. The other time-series feature extraction library implemented is catch2265 (its Python version), which was designed to provide a robust summary of the 22 top-performing features from the hctsa library while offering significantly faster computation.
We have also integrated well-established neuroimaging biomarkers that have been shown to serve as indirect indicators of neural circuit abnormalities in clinical conditions, e.g., using the 1/f slope5,31,32,37, and have also been correlated with changes in cortical circuit activity associated with brain development and healthy ageing38,39,40. The first method we included, the specparam toolbox, is a widely used approach for parameterizing neural power spectra into periodic and aperiodic components68. It conceptualizes the power spectrum as a combination of two distinct functional processes: an aperiodic component, characterized by 1/f-like properties, and a variable number of periodic components, representing putative oscillations as peaks above the aperiodic background. In this model, peaks in the power spectrum are defined by their center frequency, power, and bandwidth, enabling analysis without setting predefined frequency bands, while adjusting for the aperiodic component. The model also outputs a metric for the aperiodic component, often represented as the exponent of the aperiodic fit or 1/f slope. The other marker featured in the library is the functional E/I ratio (fE/I). It integrates two key properties of neuronal network activity, oscillation amplitude and long-range temporal correlations, characterized by the normalized fluctuation function nF(t) (Fig. 1), to provide an estimate of the E/I ratio36. For networks near the critical state, the fE/I metric has been shown to accurately categorize their activity as inhibition-dominated, excitation-dominated, or balanced36.
For this study, we used the catch22 library to extract features from the empirical datasets and to train the machine learning algorithms. Together with the catch22 features, we also focused on analyzing prediction results using the 1/f slope, employing the parameters outlined in Table 6 to configure the specparam algorithm.
Inference of neural circuit parameters
The ncpi library facilitates inverse surrogate model creation by enabling the training of models via two popular libraries: sklearn60 and SBI61. The user can import any regression model from sklearn, e.g., a multi-layer perceptron regressor (MLPRegressor) or a Ridge regression model, define its hyperparameters, and train it using features derived from simulation data. Once trained, the machine learning model can be used to predict neural circuit parameters on real-world data. Additionally, we have included the SBI library61 to provide access to robust simulation-based inference approaches for computing the inverse model. Given that our focus is on training an inverse model applicable to any new dataset, we support neural density estimators that enable amortized inference, such as NPE51, allowing us to evaluate the posterior probability for different observations without the need to re-run the inference process. SBI models can be tailored to user preferences, allowing for the definition of prior distributions (i.e., initial constraints on model parameters), the choice of density estimator (e.g., masked autoregressive flow or neural spline flow), and the configuration of hyperparameters for the neural network, such as the number of hidden units and transformation layers. We assessed the quality of the posterior fits provided by NPE by comparing the posterior z-score and posterior shrinkage, defined as follows99:
where \(\bar{\theta }\) and \({\theta }^{* }\) represent the estimated mean of the posterior and the ground-truth of the estimated parameter, respectively, and \({\sigma }_{{prior}}^{2}\) and \({\sigma }_{{post}}^{2}\) denote the variances (uncertainties) of the prior and posterior, respectively.
Our library offers two options for training the model: (i) the user can either fit the model using the entire dataset, or (ii) conduct a grid search using the RepeatedKFold cross-validation method from sklearn to identify the optimal hyperparameters, assess stability and generalization of machine learning models, and reduce overfitting. The grid search is informed by the mean squared error computed from the regression model’s predictions (in the case of sklearn) or from the mean values derived from the posteriors (in the case of SBI). For repeated cross-validation, once the optimal hyperparameters are identified and a single model has been trained for each data partition, predictions are generated for each fold and repetition, and the results are averaged.
For the examples presented here, we employed a grid-search cross-validation approach to train an MLPRegressor, Ridge, and NPE model using the parameter configurations listed in Table 7. To ensure stable results, we set up RepeatedKFold cross-validation with 20 repeats and 10 splits, resulting in a total of 200 folds. The cortical circuit model used in our study is a ‘general-purpose’ model that captures some important properties of cortical dynamics, such as asynchronous-irregular spiking activity23, but it is not intended to reproduce all properties of the neural circuit. For this reason, it is necessary to train the neural network in a manner that prevents overfitting of the simulation data, but such that it can still learn the general trends of the simulated cortical activity. To achieve this, we constrained the hyperparameter search to explore values within a narrow range (e.g., number of hidden layers ≤ 2 and number of neurons per layer ≤ 50 for the MLPRegressor), which would return regression models characterized by low complexity. After the training process was finished, we defined the metric E/I = (JEE/JEI)/(JIE/JII) as the ratio between the predicted E/I of excitatory and inhibitory populations, respectively. This summary measure quantifies the net effect of excitatory and inhibitory processes in the circuit model and may be seen as a global E/I ratio from model simulations.
Empirical data
We did not generate new data for this study; instead, the data were obtained from previous studies38,83. All methods and experiments involving live vertebrates were conducted in accordance with relevant ethical guidelines and regulations, as detailed in the cited publication. Please refer to the corresponding publications for further details.
Details of the dataset on developmental activity in mice are as described in ref. 38. Briefly, experiments were carried out on C57BL/6J mice of both sexes. In vivo extracellular recordings were performed from the prelimbic subdivision of the PFC in over 100 non-anesthetized mice of ages from 2nd to 12th postnatal days. We selected mice of age ≥ 4 postnatal days, as the neural activity of younger mice was so sparse that it was insufficient to compute an accurate estimate of parameter predictions. Extracellular signals were acquired and digitized at a 32 kHz sampling rate and band-pass filtered (0.1–9000 Hz) using an extracellular amplifier (Digital Lynx SX; Neuralynx, Bozeman, MO, Cheetah, Neuralynx, Bozeman, MO). To obtain the LFP data, the extracellular signals recorded during baseline activity were downsampled to 100 Hz, summed across channels, and segmented into 5-second epochs. Epochs were z-score normalized.
The EEG dataset consists of 201 eyes-closed resting-state recordings of human neural activity acquired with a 19-channel EEG system (Nihon Kohden Neurofax JE-921A) using a sampling rate of 500 Hz83. The EEG recordings were acquired from 51 healthy controls (HCs), 50 AD patients at a mild severity stage (ADMIL), 50 moderate AD patients (ADMOD), and 50 AD patients at a severe stage (ADSEV). AD patients were diagnosed according to the criteria of the NIA-AA (National Institute on Aging and Alzheimer’s Association)100. Signals were preprocessed to reduce the presence of noise and segmented into 5-second epochs; deeper insights on the preprocessing pipeline can be found in ref. 83. Epochs were z-score normalized.
Statistical tests
We used LME models to test whether the inferred parameters from the empirical databases were statistically different between groups, by also considering inter-subject variability. LME models were implemented using the lme4 package from R. For the ‘mice-LFP’ data, we fitted an LME model with the inferred parameter as a dependent variable, the group variable (i.e., age) as a fixed effect, and the mouse identifier as a random effect. To verify the significance of the mouse’s random effect, the Bayesian Information Criterion (BIC) was compared between the LME model including this variable (Eq. 4) and the model without it (Eq. 5), and the best fit was selected.
For ‘AD-EEG’ analysis, we incorporated electrode differences in the statistical test. For each inferred parameter, different models were fitted to examine the fixed effects of group and electrode, and their interaction, taking into account patient-specific fluctuations. The BIC was calculated for two LME models: with (Eq. 6) and without patient random effect (Eq. 7). The group variable values were HC, ADMIL, ADMOD, and ADSEV.
After choosing the best-performing model according to the BIC value, a likelihood-ratio test (LRT) was conducted between it and the respective model excluding the interaction between electrode and group, i.e., Eq. 8 was compared with Eqs. 6 and 9 with Eq. 7.
The resulting optimal model was used for post-hoc analyses. Specifically, we statistically compared marginal means of parameter values estimated from the model using the emmeans package. All p-values were Holm-Bonferroni adjusted for the number of comparisons made.
We employed ANOVA models to analyze the distribution of features computed from the simulated data, aiming to test for statistical differences across parameter bins. Effect sizes for the ANOVA models were quantified using η2. We computed the Hellinger distance (DH) between the parameter error distributions of multi-feature and single-feature models to assess their similarity.
Computational resources and processing time
Simulations of neural activity and field potentials, feature extraction methods, training of machine-learning algorithms, and circuit parameter predictions were parallelized on: (i) a high-performance computing server equipped with a 64-core CPU and 256 GB of RAM, (ii) a high-performance cluster consisting of 6 nodes, with each node equipped with 24 cores and 64 GB of RAM, and (iii) the supercomputer Albaicin (Huawei FusionServer Pro series) of University of Granada. Simulation time varied depending on the model parameters and the computing system. On average, each multithreaded simulation of the LIF network model, including field potential computations, took approximately 10 seconds. Distributing these simulations across multiple computing resources reduced the total computation time to about three months. Other computationally intensive tasks included feature extraction and training the learning models. For instance, processing 2 million samples, each with 10,000 sampling points, using 64 threads on the 64-core CPU took around 2 hours. Additionally, training the MLP model on all samples, with a single RepeatedKFold cross-validation round (20 repeats, 10 splits), took approximately 30 minutes using the 64-core CPU.
Data availability
Code of the ncpi library, as well as code supporting the findings of this study, can be found at https://github.com/necolab-ugr/ncpi. The simulation datasets generated with the LIF network model and the trained machine learning models are publicly available in a Zenodo repository: https://doi.org/10.5281/zenodo.15351117. The LFP dataset is available for download from the following repository: https://gin.g-node.org/mchini/development_EI_decorrelation. EEG data will be made available by the authors upon reasonable request.
Code availability
Code of the ncpi library, as well as code supporting the findings of this study, can be found at https://github.com/necolab-ugr/ncpi. The simulation datasets generated with the LIF network model and the trained machine learning models will be made available in Zenodo repositories. The LFP dataset is available for download from the following repository: https://gin.g-node.org/mchini/development_EI_decorrelation. EEG data will be made available by the authors upon reasonable request.
References
D’Angelo, E. & Jirsa, V. The quest for multiscale brain modeling. Trends Neurosci. 45, 777–790 (2022).
Einevoll, G. T. et al. The scientific case for brain simulations. Neuron 102, 735–744 (2019).
Amunts, K. et al. The Human Brain Project-Synergy between neuroscience, computing, informatics, and brain-inspired technologies. PLoS Biol. 17, e3000344 (2019).
Madadi Asl, M., Valizadeh, A. & Tass, P. A. Decoupling of interacting neuronal populations by time-shifted stimulation through spike-timing-dependent plasticity. PLoS Comput. Biol. 19, e1010853 (2023).
Martínez-Cañada, P. et al. Combining aperiodic 1/f slopes and brain simulation: An EEG/MEG proxy marker of excitation/inhibition imbalance in Alzheimer’s disease. Alzheimer’s. Dement. Diagn. Assess. Dis. Monit. 15, e12477 (2023).
Sanzeni, A., Histed, M. H. & Brunel, N. Emergence of irregular activity in networks of strongly coupled conductance-based neurons. Phys Rev. X 12, 011044 (2022).
Zerlaut, Y., Zucca, S., Panzeri, S. & Fellin, T. The spectrum of asynchronous dynamics in spiking networks as a model for the diversity of non-rhythmic waking states in the Neocortex. Cell Rep. 27, 1119–1132.e1117 (2019).
Cavallari, S., Panzeri, S. & Mazzoni, A. Comparison of the dynamics of neural interactions between current-based and conductance-based integrate-and-fire recurrent networks. Front. Neural Circuits 8, 12 (2014).
Ostojic, S. Two types of asynchronous activity in networks of excitatory and inhibitory spiking neurons. Nat. Neurosci. 17, 594–600 (2014).
Mazzoni, A., Brunel, N., Cavallari, S., Logothetis, N. K. & Panzeri, S. Cortical dynamics during naturalistic sensory stimulations: experiments and models. J. Physiol. Paris 105, 2–15 (2011).
Ostojic, S., Brunel, N. & Hakim, V. Synchronization properties of networks of electrically coupled neurons in the presence of noise and heterogeneities. J. Comput. Neurosci. 26, 369–392 (2009).
Hill, S. & Tononi, G. Modeling sleep and wakefulness in the thalamocortical system. J. Neurophysiol. 93, 1671–1698 (2005).
Compte, A., Sanchez-Vives, M. V., McCormick, D. A. & Wang, X. J. Cellular and network mechanisms of slow oscillatory activity (<1 Hz) and wave propagations in a cortical network model. J. Neurophysiol. 89, 2707–2725 (2003).
Brunel, N. & Wang, X. J. What determines the frequency of fast network oscillations with irregular neural discharges? I. Synaptic dynamics and excitation-inhibition balance. J. Neurophysiol. 90, 415–430 (2003).
Pronold, J. et al. Multi-scale spiking network model of human cerebral cortex. Cereb. Cortex 34, bhae409 (2024).
Haufler, D., Ito, S., Koch, C. & Arkhipov, A. Simulations of cortical networks using spatially extended conductance-based neuronal models. J. Physiol. 601, 3123–3139 (2023).
Rimehaug, A. E. et al. Uncovering circuit mechanisms of current sinks and sources with biophysical simulations of primary visual cortex. Elife 12, e87169 (2023).
Billeh, Y. N. et al. Systematic integration of structural and functional data into multi-scale models of Mouse Primary Visual Cortex. Neuron 106, 388–403.e318 (2020).
Schmidt, M. et al. A multi-scale layer-resolved spiking network model of resting-state dynamics in macaque visual cortical areas. PLoS Comput. Biol. 14, e1006359 (2018).
Markram, H. et al. Reconstruction and simulation of neocortical microcircuitry. Cell 163, 456–492 (2015).
Potjans, T. C. & Diesmann, M. The cell-type specific cortical microcircuit: relating structure and activity in a full-scale spiking network model. Cereb. Cortex 24, 785–806 (2014).
Einevoll, G. T., Kayser, C., Logothetis, N. K. & Panzeri, S. Modelling and analysis of local field potentials for studying the function of cortical circuits. Nat. Rev. Neurosci. 14, 770–785 (2013).
Hagen, E. et al. Brain signal predictions from multi-scale networks using a linearized framework. PLoS Comput. Biol. 18, e1010353 (2022).
Martínez-Cañada, P., Ness, T. V., Einevoll, G. T., Fellin, T. & Panzeri, S. Computation of the electroencephalogram (EEG) from network models of point neurons. PLoS Comput. Biol. 17, e1008893 (2021).
Mazzoni, A. et al. Computing the Local Field Potential (LFP) from Integrate-and-Fire Network Models. PLoS Comput. Biol. 11, e1004584 (2015).
Næss, S. et al. Biophysically detailed forward modeling of the neural origin of EEG and MEG signals. Neuroimage 225, 117467 (2021).
Hagen, E. et al. Hybrid scheme for modeling local field potentials from point-neuron networks. Cereb. Cortex 26, 4461–4496 (2016).
Huang, Y., Parra, L. C. & Haufe, S. The New York Head-A precise standardized volume conductor model for EEG source localization and tES targeting. Neuroimage 140, 150–162 (2016).
Mazzoni, A., Whittingstall, K., Brunel, N., Logothetis, N. K. & Panzeri, S. Understanding the relationships between spike rate and delta/gamma frequency bands of LFPs and EEGs using a local cortical network model. Neuroimage 52, 956–972 (2010).
Mazzoni, A., Panzeri, S., Logothetis, N. K. & Brunel, N. Encoding of naturalistic stimuli by local field potential spectra in networks of excitatory and inhibitory neurons. PLoS Comput. Biol. 4, e1000239 (2008).
Bertelsen, N. et al. Electrophysiologically-defined excitation-inhibition autism neurosubtypes. medRxiv, 1–41 (2024).
Trakoshis, S. et al. Intrinsic excitation-inhibition imbalance affects medial prefrontal cortex differently in autistic men versus women. Elife 9, e55684 (2020).
Gao, R., Peterson, E. J. & Voytek, B. Inferring synaptic excitation/inhibition balance from field potentials. Neuroimage 158, 70–78 (2017).
Ahmad, J. et al. From mechanisms to markers: novel noninvasive EEG proxy markers of the neural excitation and inhibition system in humans. Transl. Psychiatry 12, 467 (2022).
Karalunas, S. L. et al. Electroencephalogram aperiodic power spectral slope can be reliably measured and predicts ADHD risk in early development. Dev. Psychobiol. 64, e22228 (2022).
Bruining, H. et al. Measurement of excitation-inhibition ratio in autism spectrum disorder using critical brain dynamics. Sci. Rep. 10, 9195 (2020).
Molina, J. L. et al. Memantine effects on electroencephalographic measures of putative excitatory/inhibitory balance in Schizophrenia. Biol. Psychiatry Cogn. Neurosci. Neuroimag. 5, 562–568 (2020).
Chini, M., Pfeffer, T. & Hanganu-Opatz, I. An increase of inhibition drives the developmental decorrelation of neural activity. Elife 11, e78811 (2022).
Schaworonkow, N. & Voytek, B. Longitudinal changes in aperiodic and periodic activity in electrophysiological recordings in the first seven months of life. Dev. Cogn. Neurosci. 47, 100895 (2021).
Voytek, B. et al. Age-related changes in 1/f neural electrophysiological noise. J. Neurosci. 35, 13257–13265 (2015).
Montobbio, N., Maffulli, R., Abrol, A. & Martínez-Cañada, P. Editorial: Computational modeling and machine learning methods in neurodevelopment and neurodegeneration: from basic research to clinical applications. Front Comput. Neurosci. 18, 1514220 (2024).
Peng, G. C. Y. et al. Multiscale modeling meets machine learning: What can we learn? Arch. Comput. Methods Eng. 28, 1017–1037 (2021).
von Rueden, L., Mayer, S., Sifa, R., Bauckhage, C. & Garcke, J. Combining Machine Learning and Simulation to a Hybrid Modelling Approach: Current and Future Directions. in Advances in Intelligent Data Analysis XVIII (ed. M. R. Berthold, A. Feelders & G. Krempl) 548–560 (Springer International Publishing, Cham, 2020).
Baker, R. E., Peña, J. M., Jayamohan, J. & Jérusalem, A. Mechanistic models versus machine learning, a fight worth fighting for the biological community? Biol. Lett. 14, 20170660 (2018).
Procopio, A. et al. Combined mechanistic modeling and machine-learning approaches in systems biology - A systematic literature review. Comput. Methods Prog. Biomed. 240, 107681 (2023).
Willard, J., Jia, X., Xu, S., Steinbach, M. & Kumar, V. Integrating scientific knowledge with machine learning for engineering and environmental systems. ACM Comput. Surv. 55, 1–37 (2022).
García, J. M. et al. A hybrid machine learning and mechanistic modelling approach for probing potential biomarkers of excitation/inhibition imbalance in cortical circuits in dementia. Available at SSRN https://doi.org/10.2139/ssrn.4977918 (2024).
Haghighat, E., Raissi, M., Moure, A., Gomez, H. & Juanes, R. A physics-informed deep learning framework for inversion and surrogate modeling in solid mechanics. Comput. Methods Appl. Mech. Eng. 379, 113741 (2021).
Boelts, J., Lueckmann, J. M., Gao, R. & Macke, J. H. Flexible and efficient simulation-based inference for models of decision-making. Elife 11, e77220 (2022).
Beck, J., Deistler, M., Bernaerts, Y., Macke, J. H. & Berens, P. Efficient identification of informative features in simulation-based inference. Adv. Neural Inf. Process. Syst. 35, 19260–19273 (2022).
Gonçalves, P. J. et al. Training deep neural density estimators to identify mechanistic models of neural dynamics. Elife 9, e56261 (2020).
Cranmer, K., Brehmer, J. & Louppe, G. The frontier of simulation-based inference. Proc. Natl. Acad. Sci. USA 117, 30055–30062 (2020).
Ziaeemehr, A. et al. Virtual Brain Inference (VBI): A flexible and integrative toolkit for efficient probabilistic inference on virtual brain models. bioRxiv, 2025.01.21.633922 (2025).
Hashemi, M. et al. Simulation-based inference on virtual brain models of disorders. Mach. Learn.: Sci. Technol. 5, 035019 (2024).
Hashemi, M. et al. Amortized Bayesian inference on generative dynamical network models of epilepsy using deep neural density estimators. Neural Netw. 163, 178–194 (2023).
Tolley, N., Rodrigues, P. L. C., Gramfort, A. & Jones, S. R. Methods and considerations for estimating parameters in biophysically detailed neural models with simulation based inference. PLoS Comput Biol. 20, e1011108 (2024).
Gewaltig, M.-O. & Diesmann, M. Nest (neural simulation tool). Scholarpedia 2, 1430 (2007).
Carnevale, N. T. & Hines, M. L. The NEURON book (Cambridge University Press, 2006).
Hagen, E., Næss, S., Ness, T. V. & Einevoll, G. T. Multimodal Modeling of Neural Network Activity: Computing LFP, ECoG, EEG, and MEG Signals With LFPy 2.0. Front Neuroinform 12, 92 (2018).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Boelts, J. et al. sbi reloaded: a toolkit for simulation-based inference workflows. J. Open Source Softw. 10, 7754 (2025).
McKerns, M. M., Strand, L., Sullivan, T., Fang, A. & Aivazis, M. A. G. Building a framework for predictive science. in Proceedings of the 10th Python in Science Conference (ed. van der Walt, S. & Millman, J.), 76–86 (2011).
Hagen, E. LFPy workspace (https://github.com/LFPy) (2025).
Fulcher, B. D. & Jones, N. S. hctsa: A computational framework for automated time-series phenotyping using massive feature extraction. Cell Syst. 5, 527–531.e523 (2017).
Lubba, C. H. et al. catch22: Canonical time-series characteristics. Data Min. Knowl. Discov. 33, 1821–1852 (2019).
Shafiei, G. et al. Neurophysiological signatures of cortical micro-architecture. Nat. Commun. 14, 6000 (2023).
Hansen, J. Y. et al. Integrating multimodal and multiscale connectivity blueprints of the human cerebral cortex in health and disease. PLoS Biol. 21, e3002314 (2023).
Donoghue, T. et al. Parameterizing neural power spectra into periodic and aperiodic components. Nat. Neurosci. 23, 1655–1665 (2020).
Martínez-Cañada, P., Noei, S. & Panzeri, S. Methods for inferring neural circuit interactions and neuromodulation from local field potential and electroencephalogram measures. Brain Inform. 8, 27 (2021).
Martínez-Cañada, P. & Panzeri, S. Spectral Properties of Local Field Potentials and Electroencephalograms as Indices for Changes in Neural Circuit Parameters. in Brain Informatics (ed. M. Mahmud, M. S. Kaiser, S. Vassanelli, Q. Dai & N. Zhong) 115-123 (Springer International Publishing, Cham, 2021).
Maestú, F., de Haan, W., Busche, M. A. & DeFelipe, J. Neuronal excitation/inhibition imbalance: core element of a translational perspective on Alzheimer pathophysiology. Ageing Res. Rev. 69, 101372 (2021).
Harris, S. S., Wolf, F., De Strooper, B. & Busche, M. A. Tipping the scales: peptide-dependent dysregulation of neural circuit dynamics in Alzheimer’s Disease. Neuron 107, 417–435 (2020).
Busche, M. A. & Hyman, B. T. Synergy between amyloid-β and tau in Alzheimer’s disease. Nat. Neurosci. 23, 1183–1193 (2020).
Sohal, V. S. & Rubenstein, J. L. R. Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol. Psychiatry 24, 1248–1257 (2019).
Rodríguez-González, V. et al. Connectivity-based Meta-Bands: A new approach for automatic frequency band identification in connectivity analyses. Neuroimage 280, 120332 (2023).
Pesaran, B. et al. Investigating large-scale brain dynamics using field potential recordings: analysis and interpretation. Nat. Neurosci. 21, 903–919 (2018).
Cohen, M. X. Where does EEG come from and what does it mean? Trends Neurosci. 40, 208–218 (2017).
Lopes da Silva, F. EEG and MEG: relevance to neuroscience. Neuron 80, 1112–1128 (2013).
Chini, M. & Hanganu-Opatz, I. L. Prefrontal cortex development in health and disease: lessons from rodents and humans. Trends Neurosci. 44, 227–240 (2021).
Zhang, Z., Jiao, Y. Y. & Sun, Q. Q. Developmental maturation of excitation and inhibition balance in principal neurons across four layers of somatosensory cortex. Neuroscience 174, 10–25 (2011).
Kroon, T., van Hugte, E., van Linge, L., Mansvelder, H. D. & Meredith, R. M. Early postnatal development of pyramidal neurons across layers of the mouse medial prefrontal cortex. Sci. Rep. 9, 5037 (2019).
Okaty, B. W., Miller, M. N., Sugino, K., Hempel, C. M. & Nelson, S. B. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. J. Neurosci. 29, 7040–7052 (2009).
Rodríguez-González, V. et al. Consistency of local activation parameters at sensor- and source-level in neural signals. J. Neural Eng. 17, 056020 (2020).
Busche, M. A. et al. Tau impairs neural circuits, dominating amyloid-β effects, in Alzheimer models in vivo. Nat. Neurosci. 22, 57–64 (2019).
Griffiths, J. & Grant, S. G. N. Synapse pathology in Alzheimer’s disease. Semin Cell Dev. Biol. 139, 13–23 (2023).
Delbeuck, X., Van der Linden, M. & Collette, F. Alzheimer’disease as a disconnection syndrome? Neuropsychol. Rev. 13, 79–92 (2003).
Forner, S., Baglietto-Vargas, D., Martini, A. C., Trujillo-Estrada, L. & LaFerla, F. M. Synaptic impairment in Alzheimer’s disease: a dysregulated symphony. Trends Neurosci. 40, 347–357 (2017).
Wang, J. et al. Tau pathology is associated with synaptic density and longitudinal synaptic loss in Alzheimer’s disease. Mol. Psychiatry 29, 2799–2809 (2024).
Lan, G. et al. Association of presynaptic loss with Alzheimer’s disease and cognitive decline. Ann. Neurol. 92, 1001–1015 (2022).
Ranasinghe, K. G. et al. Altered excitatory and inhibitory neuronal subpopulation parameters are distinctly associated with tau and amyloid in Alzheimer’s disease. Elife 11, e77850 (2022).
Donner, T. H. & Siegel, M. A framework for local cortical oscillation patterns. Trends Cogn. Sci. 15, 191–199 (2011).
Sanz-Leon, P., Knock, S. A., Spiegler, A. & Jirsa, V. K. Mathematical framework for large-scale brain network modeling in The Virtual Brain. Neuroimage 111, 385–430 (2015).
Sanz Leon, P. et al. The Virtual Brain: a simulator of primate brain network dynamics. Front Neuroinform. 7, 10 (2013).
Deco, G., Senden, M. & Jirsa, V. How anatomy shapes dynamics: a semi-analytical study of the brain at rest by a simple spin model. Front. Comput. Neurosci. 6, 68 (2012).
Pronold, J. et al. Multi-Scale Spiking Network Model of Human Cerebral Cortex. Cerebral Cortex 34, bhae409 (2024).
Donoghue, T. et al. Evaluating and Comparing Measures of Aperiodic Neural Activity. bioRxiv 2024.09.15.613114 (2024).
Plesser, H. E., Eppler, J. M., Morrison, A., Diesmann, M. & Gewaltig, M.-O. Efficient parallel simulation of large-scale neuronal networks on clusters of multiprocessor computers. 672–681 (Springer).
Hay, E., Hill, S., Schürmann, F., Markram, H. & Segev, I. Models of neocortical layer 5b pyramidal cells capturing a wide range of dendritic and perisomatic active properties. PLoS Comput Biol. 7, e1002107 (2011).
Betancourt, M. Calibrating model-based inferences and decisions. arXiv preprint arXiv:1803.08393 (2018).
McKhann, G. M. et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7, 263–269 (2011).
Acknowledgements
This study was supported by grants PID2022-139055OA-I00, PID2022-137461NB-C31, and PID2022-138286NB-I00, funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”; by “Junta de Andalucía” - Postdoctoral Fellowship Program PAIDI 2021; and by “CIBER en Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Spain” through “Instituto de Salud Carlos III” co-funded with ERDF funds. We would like to thank Roberto Maffulli and Mariangela Panniello for their useful feedback on this work.
Author information
Authors and Affiliations
Contributions
Conceptualization: P.M.C.Data Curation: A.O.V., V.R.G., J.P., C.G. & P.M.C.Formal Analysis: A.O.V., V.R.G, J.P., C.G., N.M., F.P., C.M. & P.M.C.Funding Acquisition: C.M., J.P., C.G. & P.M.C.Investigation: A.O.V. & P.M.C.Methodology: All authors. Resources: V.R.G., J.P., C.G., C.M. & P.M.C.Software: A.O.V. & P.M.C.Supervision: P.M.C.Validation: A.O.V. & P.M.C.Visualization: A.O.V. & P.M.C.Writing – Original Draft Preparation: P.M.C.Writing – Review & Editing: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Orozco Valero, A., Rodríguez-González, V., Montobbio, N. et al. A Python toolbox for neural circuit parameter inference. npj Syst Biol Appl 11, 45 (2025). https://doi.org/10.1038/s41540-025-00527-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41540-025-00527-9