Abstract
The emergence of SARS-CoV-2 variants with defined mutations that enhance pathogenicity or facilitate immune evasion has resulted in a continual decline in the protective efficacy of existing vaccines. Therefore, there is a pressing need for a vaccine capable of combating future variants. In this study, we designed new mRNA vaccines, BSCoV05 and BSCoV06, and generated point mutations in the receptor-binding domain (RBD) of the original Wuhan strain to increase their broad-spectrum antiviral activity. Additionally, we used the BA.1 RBD as a control. Both vaccines elicited a robust immune response in BALB/c and K18-hACE2 mice, generating high levels of specific binding antibodies against the BA.2 RBD. Moreover, all three vaccines induced neutralizing antibodies against the prototype viral strain and relevant variants, including the Alpha and Beta strains and the Omicron variants BA.1, BA.2, BA.5, XBB.1.5, XBB.1.16, EG.5.1, and EG.5.1.1, with BSCoV06 demonstrating broader neutralizing antibody activity. Both BSCoV05 and BSCoV06 also elicited a cellular immune response. After the challenge, both BSCoV05 and BSCOV06 provided protection against the EG.5.1 strain in both mouse strains. Therefore, these two vaccines merit further evaluation in nonhuman primates, and this vaccine design strategy should be explored for its potential application in combating future SARS-CoV-2 variants, offering valuable insights into broad-spectrum vaccine development.
Similar content being viewed by others
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in a significant number of deaths and immense economic losses worldwide since the first outbreak in 2019, becoming a serious public health emergency. As of August 2024, the World Health Organization (WHO) reported a cumulative infection count of 775 million people, with over 7 million fatalities1.
Effective vaccination is the primary method for preventing SARS-CoV-2 infection. Several SARS-CoV-2 vaccines, including the BioNtech and Moderna mRNA vaccines2,3, inactivated vaccines4, and Novavax subunit vaccines5, have been approved for intramuscular injection in humans. These vaccines have significantly reduced the number of severe cases and mortality rate, playing a crucial role in controlling the pandemic6.
The spike (S) protein is a key target for vaccine development. The receptor-binding domain (RBD) of the S protein, which binds to host receptors to facilitate viral entry into cells7, is the primary epitope for neutralizing antibodies8. Many vaccines, such as mRNA-1273, have been developed using the S protein as the antigen. However, research has shown that the majority of neutralizing antibodies in the serum of patients bind to the RBD, making it another important antigen for vaccine development9.
As time has progressed, the highly mutable nature of the SARS-CoV-2 S protein has led to the emergence of numerous variants and subvariants, many of which have acquired mutations in the RBD that influence immune escape. This has resulted in a diminishing efficacy of previously approved vaccines. Following the Wuhan strain, several dominant variants emerged, including Alpha, Beta, Gamma, and Delta. In November 2021, the Omicron variant (B.1.529) appeared, followed by various Omicron subvariants, such as BA.2, BA.4, BA.5, BA.2.75, BQ.1.1, XBB, XBB.1.5, XBB.1.16, EG.5.1, JN.1, KP.2, and KP.3, which have increasingly enhanced immune evasion10,11. To address the emergence of new variants, bivalent vaccines containing the Wuhan-1 spike protein and BA.4/5 have been approved. However, studies indicate that the neutralizing activity of sera from vaccinated individuals against BQ.1, BQ.1.1, XBB, and XBB.1 has significantly diminished, with antibody titers against BQ variants decreasing by 13–81 times and antibody titers against XBB variants decreasing by 66–155 times12. This highlights the need for a broad-spectrum vaccine to combat the continuously emerging variants.
Mutations in the S protein can facilitate immune escape, with certain mutations in the RBD significantly altering the surface structure of the RBD that corresponds to antibody epitopes7,13. The E484 mutation has been shown to reduce the neutralizing activity of convalescent sera by more than tenfold14. The B.1.1.529 variant, due to mutations K417N, G446S, E484A, and Q493R, has evaded most previously generated antibodies. Plasma samples from individuals who received three doses of CoronaVac and were not infected indicated that the neutralization titers (NT50) decreased by 1.5–1.7 times against the BA.4 or BA.5 sublineages harboring mutations R346I (BA.5.9), R346T (BA.4.6), and R346S (BA.4.7). A similar reduction in neutralization titers was observed in the plasma of individuals recovering from breakthrough infections with BA.1 or BA.2 variants. Importantly, the BA.4 or BA.5 subvariants with R346I, R346T, or R346S mutations can significantly evade antibody neutralization by plasma samples from individuals with BA.5 breakthrough infections, resulting in a 2.4–2.6-fold decrease in the 50% neutralization titer (NT50)15. Therefore, considering these immune escape sites is crucial in the design of future vaccines.
The development of broad-spectrum vaccines often relies on multivalent vaccines to enhance the immune response. However, the production of multivalent vaccines is complex and costly and also risks significantly reducing protective efficacy as new variants arise. In this study, we based our approach on the RBD sequence of the Wuhan-1 strain. We generated variant strains by retaining conserved antibody binding sites and incorporating characteristic critical mutations from various variants. Moreover, we developed novel mRNA vaccines on the basis of these strains to evaluate their immunogenicity and effectiveness, with the aim of achieving broad-spectrum efficacy.
Results
Antigen design and vaccine preparation
In our preliminary work, we discovered that incorporating characteristic immune escape sites from different variants into the RBD of the original Wuhan strain significantly enhances the broad-spectrum protective efficacy of subunit vaccines against various variants. On the basis of these findings, we introduced the mutations S371F, K417N, N440K, L452R, L455F, F456L, T478K, E484A, F486V, Q493R, Q498R, N501Y, and Y505H into the RBD of the Wuhan-1 strain to generate the candidate vaccine sequence BSCoV05 (Fig. 1a and Supplementary Table 1). Additionally, we introduced the mutations H346T, L368I, S371F, K417N, N440K, V445P, L452R, T478K, E484A, F490S, Q493R, Q498R, N501Y, and Y505H to create the candidate vaccine sequence BSCoV06. Due to the BA.1 strain being the first variant of the Omicron lineage and exhibiting strong immune evasion, it shares a certain degree of homology with subsequent variants and also has a high degree of homology with earlier strains. Therefore, we selected the RBD sequence of BA.1 as a control vaccine sequence, which included the mutations G339D, S371L, S373H, K417N, N440K, G446S, S477N, T478N, E484A, Q493R, N501Y, and Y505H (Fig. 1a).
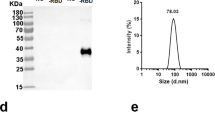
a The vaccine design and mutation schematic, the arrows represent a schematic illustration of mutations on the RBD, and the red mutations are unique to the sequence of the vaccine. b–d The structural predictions for BSCoV05, BSCoV06, and BA.1. e Diagram of the mRNA-LNPs and f the particle size distribution curve of the LNPs.
First, we performed structural predictions of the candidate vaccine protein sequences via AlphaFold3 (Fig. 1b–d). The results indicated that most of these mutations affect the antigen surface structure, potentially inhibiting the recognition of conformational epitopes and the presentation of the antigen. We subsequently incorporated the signal peptide sequence MDWTWILFLVAAATRVHS into the RBD sequence, optimized the codons, and constructed a plasmid containing a 5’ untranslated region (UTR), 3’ UTR, and PolyA tail. This was followed by in vitro transcription to obtain mRNA. We utilized a microfluidic approach to prepare mRNA lipid nanoparticles (mRNA-LNPs) (Fig. 1e) using the lipids SM102, DSPC, PEG2000-DMG, and cholesterol. Dynamic light scattering (DLS) analysis revealed that the size of all vaccine particles was approximately 100 nm (Fig. 1f), and encapsulation efficiency tests revealed that the encapsulation efficiency for all vaccines exceeded 95%.
BSCoV05, BSCoV06, and BA.1 can elicit strong immune responses in BALB/c mice and K18-hACE2 mice
To verify whether BSCoV05 and BSCoV06 activate an immune response postimmunization, BALB/c mice were immunized with the vaccines (Fig. 2a). Serum samples were collected before immunization and at 5 h and 24 h postimmunization to assess changes in cytokine levels and related chemokines. Tris-HCl buffer was used as the placebo control. Compared with those in the placebo group, the levels of most cytokines in the treatment groups significantly increased at 5 h postimmunization and markedly decreased at 24 h (Fig. 2b). However, the MCP-1 and MIP-1b levels remained elevated at 24 h, even surpassing those at 5 h. MCP-1 and MIP-1b are critical chemokines for the activation and migration of antigen-presenting cells16, and their mRNA expression was still high at 24 h. In the placebo group, MIP-1β levels remained elevated after 24 h, with some individuals even demonstrating an increase. However, this increase was not statistically significant, and the cause of this trend may be attributed to testing errors. Additionally, compared to the vaccine group, the increase in levels was more pronounced in the latter. Moreover, IL-6 promotes the differentiation of naive CD4+ T cells into effector and helper cells, bridging innate immunity to adaptive immune responses17, whereas IL-5 is involved in mouse B-cell differentiation16,18. These findings demonstrated that BSCoV05 and BSCoV06 successfully elicited an immune response in BALB/c mice. For K18-hACE2 mice, aside from the less pronounced changes in IL-5 compared to BALB/c mice, the other cytokines showed similar results (Supplementary Fig. 1).
a Schematic diagram of the immunization process in BALB/c mice. b The expression levels of cytokines at 5 h and 24 h postimmunization, with ρ indicating the serum concentration of cytokines (pg/mL) (n = 4). c ELISA results showing the levels of BA.2 RBD-specific binding antibodies in BALB/c mice (n = 5). d Immunization process in K18-hACE2 mice. e ELISA results for BA.2 RBD-specific binding antibodies in K18-hACE2 mice (Placebo: n = 5, BSCoV 05, BSCoV06 and BA.1: n = 4). The detection limit of the ELISA is 200 and the schematic diagram of the immune process was obtained from the BioRender website, “ns” means no significant difference.
To further validate the immunogenicity of the vaccines, BALB/c and K18-hACE2 mice were immunized three times on days 0, 14, and 28. Considering that most prevalent variants have evolved from the BA.2 variant, serum samples were collected at 14, 21, 28, 35, and 42 days postimmunization to measure BA.2 RBD-specific binding antibody levels via ELISA (Fig. 2c). At 14 days postimmunization, RBD-specific antibodies were detected in both BALB/c and K18-hACE2 mice across all three vaccine groups (Fig. 2d, e). Following the three immunizations, the levels of specific antibodies significantly increased, with average antibody titers peaking at 35 days and reaching 105 in both BALB/c and K18-hACE2 mice. By 42 days, a decline was observed, and no significant differences were noted between high and low doses for some vaccines. In K18-hACE2 mice, however, the difference between high and low doses of BSCoV06 was more pronounced. Therefore, all three vaccines demonstrated suitable immunogenicity.
BSCoV06 has broad-spectrum antiviral activity
To further validate the broad-spectrum activity of the three vaccines postimmunization, we collected serum from BALB/c mice on day 42 and conducted live virus neutralization assays against the original strain, alpha strain, beta strain, and the Omicron variants BA.1, BA.2, BA.5, XBB.1.5, XBB.1.6, EG.5.1, and EG.5.1.1. The results indicated that all three vaccines elicited relatively high levels of neutralizing antibodies against BA.1, BA.2, and BA.5, with the strongest neutralizing activity observed against BA.2 (Fig. 3a–c). The geometric mean titers (GMTs) for the high and low doses were as shown in Supplementary Table 2. Both the BSCoV05 and BA.1 vaccines exhibited lower neutralization activity against the XBB variants and EG.5 variants (GMT < 100). In contrast, BSCoV06 generated high levels of neutralizing antibodies against the XBB and EG.5 variants, with GMTs as shown in Supplementary Table 2. However, the neutralizing antibody levels elicited by BSCoV06 against the original strain, alpha, beta, and delta strains were slightly lower than those elicited by BSCoV05, with GMTs of 291, 334, 291, and 253, respectively. BSCoV05 vaccination resulted in the highest neutralizing antibody levels against the original strain, alpha, and delta strains, surpassing those against BA.1. The neutralizing activity of antibodies induced by BSCoV05 was comparable to that induced by BA.1. Conversely, there was a significant difference in neutralizing activity between BSCoV06 and BA.1.
a–c The GMTs of neutralizing antibodies against different strains in the serum of BALB/c mice immunized with BSCoV05, BSCoV06, and BA.1. d The titers of neutralizing antibodies against various pseudoviruses in the serum of BALB/c mice immunized with BSCoV06 or BA.1. e The neutralizing antibody titers against various live strains in the serum of K18-hACE2 mice 42 days postimmunization with each vaccine. (Placebo: n = 5, BSCoV 05,BSCoV06 and BA.1: n = 4) The data are expressed as means ± SEM. Statistical analysis was conducted using one-way ANOVA and Tukey’s multiple comparison test. The significance of differences between the experimental group and the unimmunized control group is indicated by asterisks, the significance of no marker indicates no significance. *P < 0.05, **P < 0.01, ***P < 0.005. The detection limit of the authentic virus neutralization experiment is 16 and the pseudovirus neutralization experiment is 20 as indicated by the red dashed line.
We also assessed the neutralizing antibody levels against the pseudoviruses of strains BF.7, BQ.1, CH.1.1, JN.1, and KP.2 (Fig. 3d). The findings revealed that BSCoV06 produced high levels of neutralizing antibodies against the evolutionarily older strains BF.7, BQ.1, and CH.1.1, whereas BSCoV05 demonstrated significantly lower neutralization activity against CH.1.1 than BSCoV06 did. Neutralizing antibodies were also detected in each vaccine group for the newer strains JN.1 and KP.2. K18-ACE2 mice presented similar results. In K18-hACE2 mice (Fig. 3e), neutralizing antibody levels against the original strains, BA.1, BA.5, XBB.1.5, and EG.5.1 were assessed, revealing similar trends but overall lower antibody levels than those in BALB/c mice. Both vaccines achieved seroconversion against various strains in both BALB/c and K18-hACE2 mice; notably, BSCoV06 induced higher neutralizing antibody levels against new strains, such as the XBB series, demonstrating greater broad-spectrum activity.
To further observe the decay of antibodies, we collected serum samples at 56 days post-primary immunization and selected the prototype strain, BA.1, BA.5, XBB.1.15, and EG.5.1 for neutralization assays. Considering that excessive blood collection prior to the challenge might affect the mice’s condition during the challenge, we opted to conduct neutralizing antibody detection seven days before the challenge and at 56 days post-primary immunization. The results indicated a certain degree of decline in neutralizing antibodies against all strains, with a more pronounced decrease observed for the BA.1 vaccine in K18-hACE2 mice (Supplementary Fig. 2). However, overall, the neutralizing antibodies for BSCoV05 and BSCoV06 still maintained a certain level and broad-spectrum efficacy.
BSCoV05 and BSCoV06 can elicit cellular immune responses
To further investigate the cellular immune responses induced by the two vaccines, we collected peripheral blood mononuclear cells (PBMCs) from BALB/c mice 42 days postimmunization and used spike proteins from the BA.1.1, XBB, and EG.5 strains as antigen stimulators to conduct ELISpots assays (Fig. 4 and Supplementary Fig. 3). The results indicated that both BSCoV05 and BSCoV06 generated specific spots for IFN-γ (Fig. 4a–c), IL-2 (Fig. 4d–f), and IL-4 (Fig. 4g–i). In contrast, the number of specific spots targeting the spike protein of the EG.5 strain was lower, with no significant differences observed between the BSCoV05 and BSCoV06 groups compared with the BA.1 group.
a–c The number of IFN-γ-specific spots generated after stimulation in each group (n = 5). d–f The number of IL-2-specific spots produced following stimulation in each group (n = 5). g–i The number of IL-4-specific spots generated after stimulation in each group (n = 5). The data are expressed as the means ± SEMs. Statistical analysis was conducted via one-way ANOVA and Tukey’s multiple comparison test. The significance of differences between the experimental group and the unimmunized control group is indicated by asterisks, and the significance of no marker indicates no significance. *P < 0.05, **P < 0.01, ***P < 0.005.
BSCoV05 and BSCoV06 protected BALB/c mice challenged with EG.5.1
The original intent of this vaccine design was to better address the emergence of new variants. The EG.5 series variants, which circulated in 2023, represent a newer set of mutations within the Omicron lineage. They are also the most recent mutations accessible to us due to regional constraints, hence we have employed the EG.5.1 variant for our challenge experiments. In BALB/c mice, three immunizations were completed, followed by a challenge with the EG.5.1 strain on day 63 postimmunization. Nasopharyngeal swabs were collected daily for five days following the challenge, and changes in body temperature and weight were recorded. On day 5 postchallenge, the mice were euthanized for dissection (Fig. 5a). Weight loss was observed in all challenged mice (Fig. 5b), with no significant differences between the vaccine and control groups, and no notable changes in body temperature were recorded (Supplementary Fig. 4). Analysis of the lung viral RNA loads revealed that all the vaccine groups presented significantly reduced viral RNA levels (Fig. 5c), which was negatively correlated with the levels of neutralizing antibody levels (Supplementary Fig. 8). The high-dose BSCoV06 group presented the lowest viral load. However, the BA.1 vaccine group also presented significantly decreased viral RNA levels. Additionally, pathological examination revealed that lung injuries in infected mice were characterized by pulmonary hemorrhage, inflammatory cell infiltration, vascular thrombosis, bronchial obstruction, and protein exudation (Fig. 5d, e). The pathological scoring results indicated that the degree of pathological damage in the placebo group was significantly greater than that in the vaccine group.
a Schematic of BALB/c mouse viral infection. b Changes in the body weight of the mice after infection. c Viral RNA load in the lungs on day 5 postinfection. d Pathological scoring of the lung tissue. e Histopathological sections demonstrating lung tissue damage. f Viral RNA load detected in nasopharyngeal swabs within five days postinfection. g Viral RNA load detected in throat swabs within five days postinfection. The data are expressed as the means ± SEMs. Statistical analysis was conducted via one-way ANOVA and Tukey’s multiple comparison test. The significance of differences between the experimental group and the unimmunized control group is indicated by asterisks, and the significance of no marker indicates no significance. *P < 0.05, **P < 0.01, ***P < 0.005.
To further confirm the lung viral load, immunofluorescence staining was performed using N protein antibodies (Supplementary Fig. 5). The viral N protein was present in all groups, but a significant reduction in N protein levels was noted in the vaccine groups compared with the placebo group, with the group treated with BSCoV06 showing the most pronounced decrease. Although the viral RNA load of the BA.1 group was significantly reduced, a substantial amount of viral N protein remained detectable. The results from nasopharyngeal swabs postchallenge indicated no differences in the viral load between the vaccine and placebo groups during the first two days (Fig. 5f, g); however, from days 3–5, the vaccine groups showed a marked reduction.
BSCoV05 and BSCoV06 protected K18-hACE2 mice challenged with EG.5.1
To further evaluate the protective efficacy of the vaccines, we conducted a challenge experiment using humanized K18-hACE2 mice. Following the established challenge protocol (Fig. 6a), after three immunizations, the mice were challenged with the EG.5.1 strain on day 63 postimmunization. Nasopharyngeal swabs were collected daily for five days following the challenge, and changes in body temperature and weight were recorded. Postchallenge, the placebo group presented significant weight loss (Fig. 6b). In contrast, the weight loss of the mice in all the vaccine groups, except the low-dose BA.1 group was within 10% of their original body weight. By day 5, only 40% of the placebo group mice survived, whereas all other vaccine group mice remained alive (Fig. 6c). The placebo group mice presented a more significant decrease in body temperature (Supplementary Fig. 4). On days 3–5 post-challenge, the viral RNA loads in nasopharyngeal swabs from the vaccine groups were notably lower than that in the placebo group (Supplementary Fig. 6). Pathological sections and scoring results demonstrated that the degree of pathological damage in most vaccine groups was significantly lower than that in the control group (Fig. 6e, f).
a Schematic of K18-hACE2 mouse viral infection. b Changes in the body weights of the mice after infection. c Survival curve within five days postinfection. d Viral RNA load in the lungs on day 5 postinfection. e Pathological scoring of lung tissue. f Histopathological sections demonstrating lung tissue damage (Placebo: n = 5, BSCoV 05, BSCoV06 and BA.1: n = 4). g Immunofluorescence of lung tissue in K18-hACE2 mice. The data are expressed as the means ± SEMs. Statistical analysis was conducted via one-way ANOVA and Tukey’s multiple comparison test. The significance of differences between the experimental group and the unimmunized control group is indicated by asterisks, and the significance of no marker indicates no significance. *P < 0.05, **P < 0.01, ***P < 0.005.
On day 5 postchallenge, the mice were euthanized, and lung tissues were collected for viral RNA load detection (Fig. 6d). Except for the high-dose BA.1 group and the low-dose BSCoV06 group, all the other vaccine groups presented significantly reduced viral RNA levels. Furthermore, immunofluorescence staining of lung tissues revealed a substantial presence of the SARS-CoV-2 N protein in the placebo group (Fig. 6g), while the low-dose BSCoV06 and BA.1 groups also presented considerable fluorescence signals. Notably, the levels of N protein detected in the high-dose BSCoV05 and BSCoV06 groups were significantly reduced.
Discussion
Owing to the global transmission of SARS-CoV-2, the emergence of variants with mutations that promote immune escape is inevitable19,20,21, necessitating the development of broad-spectrum vaccines to combat new variants. In this study, we designed two mRNA vaccines by designing viral strains using the RBD of the original Wuhan strain and key mutated sites from different strains.
Broad-spectrum vaccines against SARS-CoV-2 are often produced using S proteins from multiple strains to create multivalent vaccines, such as the BNT162b2 BA.4/5 bivalent mRNA vaccine22, which has shown some efficacy. However, with the continual emergence of new mutations, the protective efficacy of these vaccines has diminished. Moreover, compared with bivalent or multivalent mRNA vaccines, monovalent mRNA vaccines reduce production complexity and, to some extent, lower production costs. An important question arises regarding whether there could be antigenic interference among components in bivalent or multivalent mRNA vaccines. Therefore, we utilized the RBD of the prototype to retain essential conserved residues and incorporated key mutations from significant strains to enhance the vaccine’s broad-spectrum efficacy.
We designed two distinct RBD vaccines by introducing various mutations, resulting in notable differences in the antibody levels elicited. Notably, BSCoV05 had unique mutations—L455F, F456L, and F486V—compared with BSCoV06. The F486 mutation has been observed in the BA.4/5 variant and all subsequent variants23, occurring as a valine mutation only in the BA.4/5, BQ.1.1, and BF.7 variants. There was no significant increase in broad-spectrum activity of BSCoV05 compared with that of the BA.1 vaccine, suggesting that these three mutations may not be critical antigenic epitopes. In contrast, the introduction of the unique mutations H346T, L368I, V445P, and F490S in BSCoV06 significantly altered the resulting antibody response. H346T originates from BQ.1.124 and is also present in XBB.1, whereas the L368I mutation is uniquely present in XBB.1. The introduction of these mutations markedly enhanced the neutralizing activity of antibodies against XBB and EG.524. These findings indicate that these four mutations not only affect viral viability but also may represent important antigenic sites. Structural predictions revealed no significant structural changes due to mutations among the three vaccines, with most mutations modifying the antigen surface. While the introduction of these mutations significantly increased the broad-spectrum activity of the vaccines against new strains, it resulted in a slight reduction in neutralizing activity against old strains, such as the original strain and the alpha and beta variants.
Previous mRNA vaccines targeting SARS-CoV-2 typically required two immunizations25,26,27. In BALB/c mice, preliminary results revealed that neutralizing antibody levels remained relatively low after two immunizations (Supplementary Fig. 7). The serum-neutralizing activity induced by the vaccine is directly related to the vaccine’s protective efficacy, prompting a third immunization. Following two booster immunizations, neutralizing antibody levels significantly increased, with GMTs reaching as high as 104, and neutralizing activity against other strains also markedly improved. Additionally, we observed a decline in antibody levels in some groups on days 28 and 42 (Fig. 2c, e), indicating that antibody levels may wane after the initial immunization. This further underscores the importance of a second booster immunization. Thus, the third immunization is crucial for increasing neutralizing antibody production.
Prior studies have indicated that clearance of the virus after SARS-CoV-2 infection plays a key role and that administering T-cell vaccines in the absence of neutralizing antibodies can protect mice from SARS-CoV-2 infection28,29. The cellular immune responses induced by BSCoV05 and BSCoV06 were not particularly strong and were even weaker than those induced by the BA.1 vaccine, potentially due to mutations altering certain T-cell epitopes. In BALB/c mice, although the GMT for neutralizing antibodies against EG.5.1.1 reached 103, the lung RNA load significantly decreased after the challenge but was not completely eliminated, which may be related to insufficient cellular immunity. In K18-hACE2 mice, neutralizing antibody levels were significantly lower than those in BALB/c mice, yet both groups protected the mice from death postinfection, with reduced lung viral loads. This finding suggests that non-neutralizing antibodies may also play a role in antiviral immunity via Fc mechanisms. Previous studies have shown that vaccination and infection can elicit antibodies with Fc effector functions, such as antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP)30. In recovering individuals, antibodies with Fc effector functions appear to be more stable than neutralizing antibodies, and these antibodies provide protection against SARS-CoV-2 across various animal models30. Moreover, although vaccine-associated enhanced respiratory disease has been linked to Th2-biased CD4 + T-cell responses, no such phenomenon was observed among the three vaccines in this study31.
This study has several limitations. First, the experiments were conducted on mice without further evaluation of the vaccine in other animal models, and the number of animals used in each group was limited, leading to considerable individual variability in some groups. Second, neutralization experiments involving new live viruses, such as JN.1 and KP.2, are lacking, and the challenge strains used were somewhat singular. Additionally, the mechanisms underlying the broad-spectrum immunity of the vaccines were not explored in depth. Nevertheless, this study serves as a preliminary exploration of the feasibility of designing a broad-spectrum SARS-CoV-2 vaccine, warranting further investigation.
Methods
Virus and cells
The SARS-CoV-2 variants used in this study were all sourced from the National Kunming High-level Biosafety Primate Research Center. All experiments involving the engineered viruses were conducted in BSL-3/4 laboratories, strictly following relevant regulations. Prior to experimentation, viral titers were confirmed through control assays. The Vero cells and 293T-hACE2 cells used in this study were both obtained from our laboratory’s stock.
Animals and ethics statements
The 6- to 8-week-old female BALB/c mice used in the experiments were obtained from the Institute of Medical Biology, Chinese Academy of Medical Sciences (Production License SCXK (DIAN) 2022-0002). and The 6- to 8-week-old female K18-hACE2 were obtained from Jiangsu Jitui Kang Biotechnology Co., Ltd. (the animal production license SCXK (SU) 2023-0009). Animal experiments were approved by the Animal Experiment Ethics Committee of the Institute of Medical Biology (license DWSP202308005) and conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals established by the National Institutes of Health.
In the mouse experiment, the total number of mice used was 197. Specifically, for each immunogenicity experiment, 5 mice were utilized, for Elispots: 5 mice, for the true virus and pseudo virus neutralizing antibody assays 5 mice, for the challenge experiment 5 mice, and for cytokine detection 4 mice, totaling 24 mice. Across 7 groups, a total of 168 BALB/c mice were employed. In the K18-hACE2 mouse model, the placebo group consisted of 5 mice, while the remaining 6 groups each contained 4 mice, summing up to 29 mice.
During animal experiments, anesthesia is required prior to the procedures. Mice are anesthetized in a sealed chamber using isoflurane gas until they are completely unconscious. The dosage varies due to individual differences and is determined by observing whether the mice have lost consciousness. Subsequently, blood collection and other experimental operations are performed. When the endpoint of the experiment is reached, euthanasia is conducted: the mice are anesthetized with isoflurane and then subjected to cervical dislocation, followed by dissection.
mRNA vaccines
The designed RBD sequence was subsequently cloned and inserted into a plasmid containing a T7 promoter, 5’ UTR, 3’ UTR, and PolyA tail. In vitro, transcription reactions were performed using a commercial kit (NEB, Cat# 2080S) to produce the mRNA, which was then encapsulated in LNPs composed of ionizable lipids, DSPC, cholesterol, and PEG2000-DMG.
The preparation of lipid nanoparticles and testing of encapsulation efficiency
The prepared mRNA is dissolved in a citrate buffer solution (50 mM, pH = 4) and adjusted to a concentration of 108 ng/μL to prepare the buffer phase. The lipid SM102, DMG-PEG2000, DSPC, and cholesterol are dissolved in ethanol at a molar ratio of 50:1.5:10:38.5 to prepare a lipid mixture solution with a total lipid concentration of 16 mM, which is prepared as the ethanol phase. Subsequently, a microfluidic method is used to mix the buffer phase with the ethanol phase in a volume ratio of 3:1 to obtain LNPs. The resulting LNPs are processed with a 100 KD ultrafiltration tube, and the ethanol is replaced and diluted using a 20 mM Tris-HCl buffer to obtain mRNA-LNPs. Encapsulation efficiency testing is performed using a commercial assay kit (Thermo Fisher, R11490) following the manufacturer’s instructions for preparing a standard curve. Free RNA is measured using the standard curve, and total RNA is measured after disrupting the LNPs with Triton X-100. The encapsulation efficiency is calculated as follows:
Animal immunizations and SARS-CoV-2 challenge
Six- to eight-week-old female and healthy BALB/c mice and K18-hACE2 mice were immunized on days 0, 14, and 28. Serum samples were collected on days 14, 21, 28, 35, and 42 to measure S protein-binding antibodies, and serum from day 42 was used for live virus neutralization assays. Spleen lymphocytes from BALB/c mice on day 42 were collected for ELISpot assays. At 63 days postimmunization, both BALB/c mice and K18-ACE2 mice were intranasally administered 100 µL (5 × 105 TCID50 for K18-hACE2; 1 × 105 TCID50 for BALB/c) of the EG.5.1 strain virus, followed by continuous observation for 5 days, during which changes in body temperature and weight were recorded and nasopharyngeal swabs were collected. On the fifth day, the mice were euthanized to assess the lung viral load and pathological damage, and Tris-HCl was used as the placebo. During the experimental process, any deaths of animals that occur under abnormal conditions, such as those resulting from fighting, are excluded from the experimental results. The study proceeds normally, and all individuals are included in the final results.
Cytokine and chemokine analysis
The mouse cytokine 23-Plex immunoassay was conducted using a Bio-Plex Pro assay kit following the manufacturer’s protocol for the Bio-Plex system (Bio-Rad). The Tris-HCl group served as the control group.. The inflammatory cytokines analyzed included eotaxin, G-CSF, GM-CSF, IFN-γ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12, IL-12 (p70), IL-13, IL-17A, KC, MCP-1, MIP-1α, MIP-1b, RANTES, and TNF-α.
ELISA for RBD-specific binding antibodies
A 96-well plate (442404, Thermo Fisher Scientific) was coated overnight at 4 °C with 1 μg/mL BA.2 The RBD protein (440592-V08H123, Sino Biological) was used. After being washed three times with PBS containing 0.05% Tween-20 (PBST), the plate was blocked for 1 h at 37 °C with PBS containing 2% BSA. Serum samples were diluted 1:200 in dilution buffer (0.5% BSA in PBST), and 100 μL of each dilution was added to the wells. The plate was incubated for 1 h at 37 °C, followed by three washes with PBST. HRP-conjugated goat anti-mouse IgG (A-10668, Invitrogen) was diluted 1:25,000 in sample dilution buffer, and 100 μL was added to each well. The plate was incubated again for 1 h at 37 °C and washed three times with PBST. Subsequently, 100 μL of substrate solution (34028, Thermo Fisher Scientific) was added to the wells, which were subsequently incubated at room temperature for 15 min. The reaction was terminated with a stop solution (C1058, Solarbio). The absorbance was measured at 450 nm and 630 nm. Wells without serum samples served as blank controls, with the threshold set at 2.1 times the control value for positive results. The titer of RBD-specific IgG was determined as the reciprocal of the highest serum dilution at which the OD450–630 nm was equal to or greater than the cutoff value (OD450–630 nm > 0.1).
Authentic virus neutralization experiment
Mouse serum was initially diluted 1:16 and then subjected to twofold serial dilutions, with a volume of 50 μL per well. The SARS-CoV-2 virus was diluted in a maintenance medium to a titer of 100 TCID50/50 μL. Equal volumes of the diluted virus mixture were added to the serum at different dilutions. The mixture was gently mixed with a pipette and incubated for 1 h at 37 °C with 5% CO2. Vero cells were digested, counted, and resuspended at a concentration of 100,000 cells/mL. Subsequently, 100 μL of Vero cells (10,000 cells/well) was added to the virus-serum mixture in a 96-well plate, which was incubated at 37 °C with 5% CO2. Cytopathic effects (CPEs) were observed daily, and the final results were determined on day 7. The reciprocal of the serum dilution that could protect 50% of the cells from CPE was defined as the antibody titer.
Pseudovirus neutralization experiment
The serum to be tested was heat-inactivated at 56 °C for 30 min. Starting with an initial dilution of 1:20, the serum was serially diluted threefold in a 96-well plate, resulting in a final volume of 100 μL of medium per well. The pseudovirus titer was diluted to 2 × 106 TCID50/mL in complete medium, and after mixing with serum, 50 μL was added to each well and incubated at 37 °C for 1 h. Additionally, 100 μL of medium mixed with the pseudovirus was used as a positive control. hACE2-293T cells were digested and suspended at a density of 5 × 105 cells/mL, with 100 μL added to each well, and incubated in a 5% CO2 incubator for 48 h. The medium was discarded, and 100 μL of DPBS was added to each well, followed by the addition of 100 μL of luciferase detection reagent (6066769, PerkinElmer). After 3 min, the relative light units were measured to determine the luciferase activity. Neutralizing antibodies were determined as the serum dilution that inhibited 50% of the luciferase activity, indicating neutralization of 50% of the viral infection.
ELISPots assay
ELISPots assays were performed according to the manufacturer’s instructions (Mabtech). In brief, after the mice were euthanized, the spleens were harvested, and a single-cell suspension was obtained through mechanical disruption using a cell strainer. Lymphocytes were then isolated following the protocol provided with the isolation kit (P8860, Solarbio). ELISPOT plates (3321-4APT-10, 3441-4APW-10, 3311-4APW-2, Mabtech) were precoated and equilibrated according to the instructions. The isolated lymphocytes were counted and plated, followed by the addition of stimuli (1.5 µg/well), with wells without stimuli serving as negative controls and PHA-stimulated wells serving as positive controls. After 32 h of stimulation, subsequent processing was carried out according to the kit instructions.
Tissue immunofluorescence
Tissues collected 5 days postviral infection were fixed in 4% paraformaldehyde solution for 3 days, embedded in paraffin, and sectioned at a thickness of 5 μm. Deparaffinization was performed by three washes in xylene followed by rehydration in an ethanol series (100%, 95%, 85%, and 75%) and distilled water/PBS washes. The sections were subjected to antigen retrieval by heating in an antigen retrieval solution at 100 °C, followed by cooling for 20 min after three cycles of heating for 3 min each. After being washed three times with PBS, the sections were permeabilized with 0.5% Triton X-100 in PBS at room temperature for 30 min, blocked with goat serum for 1 h, and then incubated overnight at 4 °C with a primary antibody against nucleocapsid protein. Following three washes with PBST, the sections were incubated in the dark with fluorescent secondary antibodies for 1 h. After being washed with PBST, the sections were mounted with a mounting medium containing DAPI and scanned using 3DHISTECH for analysis.
Histopathology
The tissue was fixed in 10% formalin for three days. After being embedded in paraffin, the tissue was sectioned into 5 μm-thick slices and subjected to hematoxylin and eosin (HE) staining. The sections were scanned and analyzed using 3DHISTECH. An experienced pathologist evaluated the HE-stained sections via the CaseViewer software provided by the manufacturer for scoring purposes. The scoring criteria included inflammatory cell infiltration, pulmonary hemorrhage, vascular thrombosis, bronchial obstruction, protein exudation, thickening of the alveolar walls, and consolidation. The scores for each criterion were summed to obtain a total pulmonary pathology score.
Statistical analysis
Statistical analysis and graphing of the data were conducted using Excel, GraphPad Prism 8.0.2, and Origin 2024. The values are presented as the means ± standard errors of the means (SEMs). One-way analysis of variance (ANOVA) combined with Tukey’s post hoc test was employed to determine the most significant differences between groups, and P < 0.05 was considered statistically significant.
Data availability
No datasets were generated or analyzed during the current study.
Code availability
No unreported custom computer code or algorithm has been used to generate or process the data presented in the manuscript.
References
WHO. WHO COVID-19 Dashboard. https://covid19.who.int/ (2024).
Uraki, R. et al. An mRNA vaccine encoding the SARS-CoV-2 receptor-binding domain protects mice from various Omicron variants. npj Vaccines 9, 4 (2024).
Islam, N., Sheils, N. E., Jarvis, M. S. & Cohen, K. Comparative effectiveness over time of the mRNA-1273 (Moderna) vaccine and the BNT162b2 (Pfizer-BioNTech) vaccine. Nat. Commun. 13, 2377 (2022).
Xia, S. et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA 324, 951–960 (2020).
Heidary, M. et al. A comprehensive review of the protein subunit vaccines against COVID-19. Front. Microbiol. 13, 927306 (2022).
Suthar, A. B. et al. Public health impact of COVID-19 vaccines in the US: observational study. BMJ 377, e069317 (2022).
Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
Yang, Y. & Du, L. SARS-CoV-2 spike protein: a key target for eliciting persistent neutralizing antibodies. Signal Transduct. Target. Ther. 6, 95 (2021).
Premkumar, L. et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 5, eabc8413 (2020).
Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 21, 361–379 (2023).
Branda, F., Ciccozzi, M. & Scarpa, F. Features of the SARS-CoV-2 KP. 3 variant mutations. Infect. Dis. 56, 1–3 (2024).
Wang, Q. et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286. e278 (2023).
Li, L. et al. Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA. 2.86, JN. 1, EG. 5, EG. 5.1, and HV. 1 sub-variants. Structure 32, 1055–1067. e1056 (2024).
Gupta, D. L. et al. RBD mutations at the residues K417, E484, N501 reduced immunoreactivity with antisera from vaccinated and COVID-19 recovered patients. Drug Target Insights 18, 20 (2024).
Jian, F. et al. Further humoral immunity evasion of emerging SARS-CoV-2 BA. 4 and BA. 5 subvariants. Lancet Infect. Dis. 22, 1535–1537 (2022).
Vazquez, M. I., Catalan-Dibene, J. & Zlotnik, A. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine 74, 318–326 (2015).
Hu, B., Huang, S. & Yin, L. The cytokine storm and COVID‐19. J. Med. Virol. 93, 250–256 (2021).
He, B. et al. Comparative global B cell receptor repertoire difference induced by SARS-CoV-2 infection or vaccination via single-cell V (D) J sequencing. Emerg. Microbes Infect. 11, 2007–2020 (2022).
Ma, W., Fu, H., Jian, F., Cao, Y. & Li, M. Immune evasion and ACE2 binding affinity contribute to SARS-CoV-2 evolution. Nat. Ecol. Evol. 7, 1457–1466 (2023).
Cao, Y. et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663 (2022).
Dejnirattisai, W. et al. SARS-CoV-2 Omicron-B. 1.1. 529 leads to widespread escape from neutralizing antibody responses. Cell 185, 467–484. e415 (2022).
Rudolph, A. E. et al. Effectiveness of BNT162b2 BA. 4/5 bivalent mRNA vaccine against symptomatic COVID-19 among immunocompetent individuals testing at a large US retail pharmacy. J. Infect. Dis. 229, 648–659 (2024).
Cao, Y. et al. BA. 2.12. 1, BA. 4 and BA. 5 escape antibodies elicited by Omicron infection. Nature 608, 593–602 (2022).
Ao, D., He, X., Hong, W. & Wei, X. The rapid rise of SARS‐CoV‐2 Omicron subvariants with immune evasion properties: XBB. 1.5 and BQ. 1.1 subvariants. MedComm 4, e239 (2023).
Turner, J. S. et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 596, 109–113 (2021).
Fang, E. et al. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 7, 94 (2022).
Payne, R. P. et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell 184, 5699–5714. e5611 (2021).
Israelow, B. et al. Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 6, eabl4509 (2021).
Bange, E. M. et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 27, 1280–1289 (2021).
Beaudoin-Bussières, G. & Finzi, A. Deciphering Fc-effector functions against SARS-CoV-2. Trends Microbiol. 32, 756–768 (2024).
Ruckwardt, T. J., Morabito, K. M. & Graham, B. S. Immunological lessons from respiratory syncytial virus vaccine development. Immunity 51, 429–442 (2019).
Acknowledgements
This study was supported by the Key Project of Applied Basic Research in Yunnan Province (202401AS070049), the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-038, 2022-I2M-CoV19-002), the National Key R & D Program of China (2021YFC230170402), and the Yunnan Key R & D Project (202103AQ100001).
Author information
Authors and Affiliations
Contributions
S.L., X.L., and Y.Y. conceived this project and designed experiments. S.L. supervised this study. X.L. wrote the paper. X.L., Y.Y, J.W., and T.C performed the majority of the experiments and analyzed the data. S.L., J.W., C.T., Y.Y., Y.Z., H.Y., Q.H., W.Y., and H.W. performed the experiments with the ABSL3 facility. Y.Y., D. L., Y. L., X. D., L. Y., W.Q., and D.W. contributed specific experiments and data analysis. S.L. reviewed the manuscript. X.L, X.Y, S.L, J.W, and C.T are aware of the grouping details in all experimental procedures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liang, X., Yuan, Y., Wang, J. et al. mRNA vaccines with RBD mutations have broad-spectrum activity against SARS-CoV-2 variants in mice. npj Vaccines 10, 7 (2025). https://doi.org/10.1038/s41541-025-01066-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41541-025-01066-4