Abstract
Engineered circular RNAs (circRNAs) are emerging as promising platforms for RNA-based vaccines in cancer treatment. We summarize the recent advances of design, synthesis, and delivery of circRNA-based cancer vaccines, and highlight the applications and challenges of circRNA vaccines in cancer therapy. Further enhancements are required in areas such as antigen selection, targeted delivery, multidimensional crosstalks, and clinical trial assessments to advance the efficacy and safety of circRNA vaccines in cancer.
Similar content being viewed by others
Introduction
Circular RNAs (circRNAs) represent a unique type of RNA characterized by their covalently closed circular structure1, and they play crucial roles in cancer development through various mechanisms. Cancer is a systemic disease marked by tumor-promoting inflammation and immune evasion, leading to changes in the immune landscape. Immunotherapy has revolutionized cancer treatment by targeting the immune system2, with vaccines being critical in public health and cancer management. Despite advancements in therapeutic cancer vaccines, challenges remain in candidate identification, immune response evaluation, and overcoming tumor immune microenvironment suppression3. RNA-based vaccine are emerging as a promising approach in cancer immunotherapy, particularly with the success of messenger RNA (mRNA) vaccines4. Recent developments in synthetic circRNAs suggest their potential as a new category of RNA therapeutics and vaccines for cancer treatment5. This review discusses the current state of circRNA-based vaccines in cancer, including design, synthesis, purification, delivery, application, and the challenges and future perspectives in precision oncology.
Discovery of circRNA vaccines
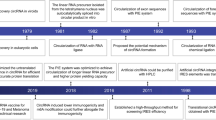
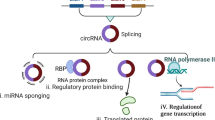
CircRNAs were initially identified in plant pathogenic viroids6 and in eukaryotic cells7. Subsequently, circRNAs were yielded by exon splicing during in vivo maturation of eukaryotic nuclear mRNAs8. Natural circRNAs have been found to function as efficient microRNA (miRNA) sponges9,10. The advent of artificial in vitro circRNA synthesis has enabled the expression of cancer antigens in the development of circRNA vaccines11.
CircRNAs as therapeutic targets in cancer
The circular structure of naturally occurring circRNAs confers resistance to exonuclease activity, enhancing their stability compared to linear RNAs, suggesting their roles as promising biomarkers for liquid biopsy in cancer detection and potential therapeutic targets for cancer treatment12. Dysregulated circRNA expression can contribute to cancer initiation, underscoring their potential as targets for therapeutic intervention. Noteworthy examples include circNUP50, which promotes cisplatin resistance in ovarian cancer (OC)13, and various other circRNAs such as hsa_circ_000791914, circRBM3315, circPLPP416, circPVT117 implicated in drug resistance across different cancer types. In addition, circRNAs like circHERC118, and circITGB619 have been associated with tumor progression and show promise as targets for cancer therapy. Interestingly, the corresponding linear transcripts of circRNAs are not considered essential. Although the levels of the corresponding linear mRNA are usually verified to be unaffected, off-target effects beyond their linear counterparts are less predictable. Importantly, ongoing clinical trials worldwide are investigating the therapeutic targeting of circRNAs (Table 1), although many are in early phases and have garnered significant interest in exploring the roles of circRNAs in cancer therapy.
CircRNAs as promising vaccine platforms
The potential of artificial circRNAs as a vaccine platform was initially recognized following the discovery of translated circRNAs in human and Drosophila cells20. In contrast to the short half-lives of less than 20 h for linear transcripts, circRNA isoforms exhibit high stability and sustained expression for up to 168 h21,22. The key step in synthesizing in vitro-transcribed (IVT) circRNA involves circularizing linear RNA molecules through diverse methods. Advances in artificial RNA circularization techniques have paved the way for circRNAs to be considered as promising vaccine platforms. A recent milestone in circRNA vaccine development involved the creation of a circRNA vaccine targeting the severe acute respiratory syndrome coronavirus (SARS-CoV-2). This circRNA vaccine demonstrated superior and more sustained antigen production compared to linear mRNA vaccines, eliciting a higher proportion of neutralizing antibodies and distinct Th1-skewed immune responses23. Furthermore, Li et al. explored the potential of a circRNA platform for protein expression and compared its duration with linear RNA, as well as its anti-tumor efficacy in challenging malignancies24. Together, circRNA cancer vaccines hold the potential to boost immune responses against cancer cells.
Design of circRNA-based cancer vaccines
To improve the stability and efficacy, diverse strategies should be considered during the development of circRNA vaccines25.
Optimization of circRNA vaccine backbone to promote translation efficiency
The efficacy of circRNA vaccines relies significantly on the translation machinery of their host organisms to generate antigens that can subsequently trigger an immune response. As such, the open reading frame (ORF) responsible for encoding the antigen and the elements facilitating its translation are crucial considerations in the design of linear precursors of circRNAs (pre-circRNAs). Some elements that promote circularization and reduce immunogenicity are also important for the design of artificial circRNAs. Given that internal ribosome entry sites (IRES) and continuous ORFs can initiate translation on artificial circRNA26, IRES-ORF cassettes serve as core components in pre-circRNAs20,27. The translation of desired proteins commences when 40S ribosomal subunits engage with the start coding codons of ORF and terminate at the stop codons of ORF in both prokaryotic and eukaryotic cells28,29. Although IRES-mediated translation can occur via an intron splicing scar, it is less efficient than having the IRES immediately upstream of a gene. Furthermore, incorporating spacers between the IRES and the gene of interest, as well as optimizing the 5’ and 3’ untranslated regions (UTRs), can improve circRNA translation30. These findings highlight the relevance of vector topology and non-coding RNA element tuning for enhanced circRNA translation.
A customized ORF is critical for enhancing the efficiency of circRNA translation. The reduction in yields due to nicking of longer circRNAs, potentially facilitated by magnesium-catalyzed autohydrolysis, represents a notable drawback that requires enhancement22,31. A growing focus has been directed towards utilizing shorter ORFs (sORFs) to express immunoglobulin neoantigens, which may trigger the desired immune responses32. New methodologies and datasets have also developed for identifying sORFs. Through an integrated workflow for sORF discovery33, a repository of sORFs identified via ribosome profiling (RIBO-seq), and a technique known as ProTInseq have been developed to characterize unnotated sORFs34,35. Recent advances include the use of various technologies and methods to identify unannotated translated sORFs and previously unknown noncanonical peptides in human cancer proteomes36,37.
m6A modification of encoding antigens to reduce circRNA immunogenicity
The N6-methyladenosine (m6A) modification is widely observed in various types of RNA molecules, including mRNA38, long non-coding RNA39, small nuclear RNA40, and circRNA41. The presence of m6A may contribute to circRNA translation and immunoregulation42. To mitigate innate immunogenicity, circRNAs can be engineered as endogenous nucleic acids to evade immune surveillance and modification with m6A. In comparison to unmodified circRNAs that encode detectable reporter proteins, circRNAs with 5% m6A incorporation exhibit similar translation levels and demonstrate increased resistance to nucleases30. This suggests that m6A modification does not hinder circRNA translation and may enhance its stability. Notably, the Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA3C), a latent oncoprotein and tumor antigen, has been found to upregulate the transcription of METTL14, an m6A writer enzyme, and directly interact with METTL14 to enhance its stability43. Moreover, m6A modification of the oncoprotein CUB domain containing protein 1 (CDCP1) in bladder cancer has been shown to synergize with chemical carcinogens in promoting malignant transformation of uroepithelial cells and bladder cancer tumorigenesis44. Further study has revealed that programmable m6A installation on CDCP1 mRNA using RCas9-METTL3 system accelerates bladder cancer progression45. These findings indicate the involvement of m6A modification in tumor antigens in cancer development, suggesting that artificial circRNAs encoding antigens can be modified with m6A to prevent immune responses.
Selection of targeting neoantigens to improve anti-tumor specificity
Personalized vaccines based on neoantigens have shown promise in eliciting broad anti-tumor responses tailored to individual cancer patients. A cancer vaccination strategy can also be designed to induce immunological memory for long-term cancer control46. Veatch et al. utilized genetically modified T cells as a vaccine platform to develop a cancer vaccine consisting of autologous T cells modified with neoantigens and additional adjuvant signals (Tvax). Subsequently, this therapeutic vaccination showed anti-tumor activity in subcutaneous and metastatic preclinical mouse models47. More importantly, concurrent delivery of immune checkpoint blockade might alter T cell dynamics and boost neoantigen vaccine-induced anti-tumor immunity48. Similarly, subcutaneous immunization with a nanovaccine that combined a BCG bacterial cell wall skeleton (BCG-CWS) based nanoscale adjuvant (BCNA) with peptide neoantigens effectively targeted lymph nodes, elicited robust innate and tumor-specific immune responses, enhanced neoantigen immunogenicity49. Conversely, chemotherapy-induced neoantigen nanovaccines comprised multiple neoantigens and damage-associated molecular patterns (DAMPs) exhibited enhanced immune responses in tumor-bearing mice, and increased efficiency of checkpoint blockade cancer immunotherapy50. Clinically, a personalized therapeutic cancer vaccine (PTCV) (GNOS-PV02) encoding up to 40 neoantigens coadministered with plasmid-encoded interleukin-12 plus pembrolizumab in patients with advanced hepatocellular carcinoma (HCC) previously treated with a multityrosine kinase inhibitor, showed clinical responses were associated with the number of neoantigens encoded in the vaccine, and neoantigen-specific T cell responses were confirmed in the vast majority of evaluable patients51. Recently, neoepitopes from circRAPGEF5 and circMYH9 have been found to elicit antigen-specific T cells response and expansion, T cells trained with circMYH9 peptides can specifically target and eliminate tumor-derived organoids in colorectal cancer (CRC)52. In addition, circRNA-based neoantigen vaccine has demonstrated superior tumor immunotherapeutic effects in HCC53. Despite advances in the identification, prioritization, and immunological targeting of personalized neoantigens, challenges such as the limited availability of immunogenic neoantigens, the inadequate efficacy of tumor-specific T cells, and the immune evasion strategies employed by tumor cells continue to hinder the attainment of optimal clinical outcomes in neoantigen-directed immunotherapy.
Synthesis of stable circRNAs
CircRNAs are generally formed through one or multiple precursor linear RNAs, and then circularized to covalently closed loops. This process is often mediated by chemical, enzymatic, or ribozymatic ligation methods.
Chemical synthesis
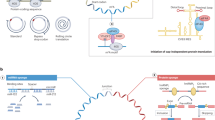
The chemical synthesis involves the use of cyanogen bromide (BrCN) or 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) as coupling agents to activate the chemical ligation of linear RNA precursors (Fig. 1a). Although chemically synthesized circRNAs can serve as translation templates, the presence of unnatural phosphoramidate linkages raises concerns regarding biosafety in protein production and RNA-based therapy54. To address these concerns, the application of in vivo chemistry in bioconjugation has gained significant attention due to its rapid reaction kinetics, high yields, minimal byproducts, and strong chemospecificity and biocompatibility55. Therefore, further study is warranted to enhance translation efficacy in living systems.
a Chemical synthesis by the treatment of T4 polynucleotide kinase (T4 PNK) and calf intestinal alkaline phosphatase (CIAP) followed by the conjugation of 5’-end phosphate with 3’-end hydroxyl catalyzed by the treatment of condensing agents (BrCN or EDC). b Enzymatic synthesis catalyzed by T4 RNA ligases using a DNA splint in a complementary base-paring manner to facilitate site-specific ligation. c Ribozymatic synthesis by Group I intron-based permuted intron‒exon (PIE) system. Permutation of a native group I intron and insertion of a custom sequence (C-S) into the exonic region (E2 and E1). And then this PIE system spontaneously ligates in the presence of free guanosine to form circRNA and release the two half-intron fragments.
Enzymatic synthesis
Enzymatic methods commonly utilize RNA ligases, such as T4 RNA ligase I and T4 RNA ligase II, which facilitate the formation of a covalent 3’-5’ phosphodiester bonds between 5’-phosphate and 3’-hydroxyl end groups of linear RNA precursors in an ATP-dependent manner (Fig. 1b). T4 RNA ligase I is particularly effective in linking single-stranded RNA precursors smaller than 500 nt with unstructured ends56, although it has been observed to produce unexpected products due to partial reversal and the lower reaction specificity. T4 RNA ligase II has been shown to selectively and efficiently produce circRNAs without the need for a splint, through the strategic design of precursor strands57. The ligation of two single-stranded RNA strands by T4 RNA ligase II is facilitated by the formation of a “nick-like intermediate”, which can lead to intermolecular ligation and byproduct formation58. However, RNA circles synthesized by T4 RNA ligases without additional fragments demonstrate minimized immunogenicity59. Improving the circularization efficiency of T4 RNA ligases can be accomplished by integrating unstructured elements and homologous sequences.
Ribozymatic synthesis
Longer RNA circularization is mediated by ribozymatic ligation and it involves consecutive ester exchange reactions facilitated by the permuted intron-exon (PIE) system. Both group I and group II introns, which are autocatalytic ribozymes, catalyze the ligation of longer linear RNA precursors through self-catalyzed splicing reactions in circRNA synthesis (Fig. 1c). However, group I introns are more frequently employed compared to group II introns. In addition, engineered circRNA regulators can enhance the production of endogenous circRNAs by specifically binding to linear precursors60. Furthermore, a specific RNA binding protein (RBP) has been identified to bind to sites within the flanking intron splice sites of linear RNA, promoting back-splicing and playing essential roles as back-splicing factors in regulating circRNA biosynthesis61. Therefore, the design of RBP motifs in linear precursors can significantly enhance in vitro circularization and circRNA production. It is also imperative to explore novel approaches for constructing a PIE system devoid of exogenous fragments to facilitate the efficient circularization of linear RNA precursors62.
Purification of circRNAs
Effective purification of circRNAs is crucial due to the manifestation of immunodeficiency and protein degradation resulting from an imbalanced circRNA composition. The gel electrophoresis has frequently employed for the differentiation and isolation of intact circRNAs from other RNAs63. However, it is important to note that while the electrophoresis system is suitable for quality control of circRNA purification for vaccine development, it may not be ideal for large-scale preparation and/or GMP production from the gels. An improved method has been established for circRNA purification using RNase R to remove linear RNAs64, the efficiency of RNase R can be influenced by the specific recognition sequences and structured 3’ ends present linear RNAs. By tailoring the length and characteristics of interested RNAs, high performance liquid chromatography (HPLC) can achieve high purity and significant quantities of RNAs65. In addition, HPLC is effective in eliminating unwanted byproducts during IVT66. Size-exclusion chromatography (SEC) is capable of separating molecules based on their size, and the combination of SEC with HPLC (SEC-HPLC) has been utilized to enhance the homogeneity of recombinant hepatitis B vaccine and to characterize constituents of influenza virus vaccine67,68. In the context of in vitro engineered circRNAs, complete elimination of nicked circRNAs is challenging due to degradation during processing. Wesselhoeft and colleagues successfully obtained notably pure circRNA (90% circular, 10% nicked) by employing gel extraction for small quantities and SEC-HPLC for larger quantities of splicing reaction starting material22. Despite the currently available methods for purifying circRNAs, it is crucial to either combine these techniques or qualify novel methodologies to improve purification processes for vaccine development.
Delivery of circRNA vaccines
Due to the negatively charged nature and large molecular size of RNA molecules, the passage of circRNAs through the cell membrane is challenging69,70. Various delivery strategies have been devised to address this issue.
Direct injection of circRNAs
Most vaccines are typically administered through injection, making direct injection a more expedient and cost-effective approach. Naked RNA can be selectively absorbed by dendritic cells (DCs) through micropinocytosis, leading to the activation of T-cell responses71,72. Frequently, naked RNA vaccines are formulated in a solution without a carrier to aid RNA uptake (Fig. 2a)73,74. Following direct injection, naked RNAs can induce antigen-specific antibodies and T-cell-specific responses. While in vivo experiments have showed significant expression of circRNA post direct injection75, the efficacy of the vaccine is limited by the degradation of naked circRNA. To address this challenge, continuous long-term injection of circRNA vaccines is necessary to sustain anti-tumor activity. Alternatively, coadministration of circRNA vaccines with liposomes can protect circRNAs against degradation and enhance the cellular uptake of circRNAs.
a CircRNAs are formulated in solution and then directly injected into mouse. b CircRNAs are ex vivo transfected into dendritic cells (DCs) by the nanochannel electro-injection (NEI) system. c CircRNAs are entrapped in lipid with microfluidic mixer to form lipid-based nanoparticle (LNP). Virus-like particle (VLP) (d) and adeno-associated virus (AAV) (e) can be engineered to encapsulate and deliver circRNAs. The icons of tube, dendritic cell, lipid, virus, mouse, and syringe were freely sourced from https://bioicons.com/.
Ex vivo loading of circRNAs into DCs
Owing to DC-mediated MHC-antigen binding and various cytokines for T-cell activation76,77, DC vaccination has been recognized as an active immunotherapy in cancer treatment and shown to be safe in clinical trials, both in terms of short- and long-term side effects. The primary approach for loading DCs with antigens ex vivo involves the introduction of mRNA encoding the desired antigens78. DCs can be transfected with circRNAs encoding tumor antigens or neoantigens, and subsequently administered to the host to trigger immune responses against the antigens. The nanochannel electro-injection (NEI) system has been utilized for the safe and efficient delivery of various nucleic acid molecules into DCs (Fig. 2b). NEI has demonstrated the ability to efficiently deliver circRNA into primary mouse bone marrow DC2.4 cells with 68.3% efficiency rate, enabling the expression of target proteins without significant cytotoxic effects. Furthermore, DCs transfected with circRNA do not remarkably impact cellular viability or induce DC maturation79. These findings suggest that NEI serves as a safe and effective transfection platform for the in vitro transformation of DCs, showing promise for the development of DC vaccines against cancer. While ex vivo DC loading offers precise control over transfection efficiency and cellular targeting, this strategy is costly and labor-intensive.
Lipid-based delivery
Liposomes are frequently used to deliver RNA due to their versatility as nanocarriers capable of transporting both hydrophobic and hydrophilic molecules across cell membranes (Fig. 2c)80. Lipid nanoparticles (LNPs) characterized by favorable biocompatibility, are currently considered the most promising materials for organ-selective nucleic acid drug delivery and have demonstrated success in cancer treatment81. The delivery of self-amplifying mRNA vaccines via cationic LNPs has been shown to elicit robust humoral and cellular-mediated immune responses in mice, indicating high RNA vaccine encapsulation efficiency82. Furthermore, the incorporation of helper polymer can be employed to enhance the efficacy of LNP delivery system83. By encapsulating the antigen-coding circRNA within LNPs, Li and colleagues have established a novel circRNA vaccine platform that effectively stimulates robust innate and adaptive immune responses, showing enhanced anti-tumor efficacy across various mouse tumor models24. In addition, LNP-mediated targeted delivery of mRNA cancer vaccine is predicted to reduce side effects and increase the immune response84. To prevent against hepatic damage, optimizations of LNP structure and organ-targeted system are necessary to improve the delivery of circRNA vaccines to target cells.
Other delivery strategies
RNA-enriched extracellular vesicles show therapeutic potential in RNA delivery85, making them less likely to be rejected when delivering circRNA cargos into cells. Virus-like particle (VLP) is self-assembling spherical nanocarrier that has been designed to encapsulate and deliver mRNA vaccines86. Adeno-associated virus (AAV) has been also employed to deliver DNA molecules encoding circRNA precursors for efficient circRNA expression (Fig. 2d)87. Despite the early phase of clinical delivery of circRNA vaccines, it is essential to focus on improving the efficiency of circRNA delivery.
Current circRNA vaccines in cancer therapy
CircRNA vaccines show promising efficacy and potential superiority over mRNA vaccines in cancer treatment. Nevertheless, the progress of circRNA vaccine development remains in its initial stage, with limited human studies reported thus far.
CircRNAOVA-luc-LNP vaccine
A recently in vivo investigation examined the efficacy of a circRNA vaccine in challenging mouse tumor models24. This vaccine, known as circRNAOVA-luc-LNP, utilized the shared antigen OVA (257-264, SIINFEKL) to trigger an immune response and applied LNP as a delivery system. Both group I intron-mediated PIE system and backbone elements were employed for in vitro synthesis of translatable circRNA. The circRNA products were purified using HPLC as referring to previous study88. The in vitro assay showed that circRNA exhibited greater longevity compared to its modified mRNA counterpart. The circRNAOVA-luc-LNP vaccine was successfully delivered into cells, translated into protein, and elicited an anti-tumor immune response24. These findings indicate that circRNA-LNP platform holds promise as a compelling vaccination strategy for clinical translation.
Combination of circRNA vaccines with other treatments
Chimeric antigen receptor (CAR) T-cell therapy has significantly revolutionized the management of hematological malignancies89. A recent clinical trial demonstrated that the integration of CAR T-cell therapy with sequential mRNA vaccine administrations resulted in potent anti-neoplastic effects in patients with genitourinary cancers, offering a promising avenue for a novel targeted therapeutic strategy90. In addition, a personalized mRNA vaccine combined with adjuvant pembrolizumab exhibited superior efficacy compared to pembrolizumab alone in patients with resected, high-risk melanoma91. Administration of naked circRNA elicits innate immune responses in mice upon injection, activating DCs and promoting antigen presentation and robust T cell reactions when engineered circRNAs encoding proteins are employed. Immunization with a circOVA vaccine complexed with a charge-altering releasable transporter has demonstrated anti-tumor efficacy92. Recently, a circRNA vaccine encoding cytokines has been shown to modulate intratumoral immune responses and impede tumor growth in colon and melanoma models. Functioning as an adjuvant-type circRNA, this vaccine has the potential to enhance the anti-programmed cell death protein 1 (PD-1) antibody-induced tumor repression93. As an intrinsic adjuvant, CXCL13 can promote broad immune protection induced by circRNA vaccines and offer a higher level of safety94. Such antigen-adjuvant-circRNA vaccine make it adaptable for their applications in cancer therapy.
Conclusions and perspectives
DNA vaccines represent a promising strategy for the prevention and treatment of cancer, as they facilitate the delivery of protein antigens to elicit immune responses in both animals and humans95,96. Despite the lower cost and better stability than those of mRNA or circRNA vaccines, DNA vaccines have not yet been widely adopted in clinical practice. Similarly, mRNA vaccines have shown considerable potential in cancer immunotherapy with lower price than circRNA vaccines. Nevertheless, the low immunogenicity resulting from suboptimal mRNA expression poses a challenge to the efficiency of mRNA vaccines97,98. Tumor-specific cryptic antigenic peptides translated from circRNAs have been found to trigger immune responses, with vaccines containing these tumor-specific circRNAs or the encoded peptides proving effective in mouse cancer models99. Although engineered circRNAs have emerged as a novel platform for developing cancer vaccines, circRNA vaccines in precision oncology are still at the early stage and need more achievements.
First, the selection of tumor-specific antigens is important for the efficacy of cancer vaccines. Conventional tumor-associated antigen (TAA) may trigger both central and peripheral tolerance, leading to suboptimal efficacy of TAA-targeting cancer vaccines. In contrast, neoantigens are truly tumor-specific and highly immunogenic100. Nevertheless, neoantigen-specific cytotoxic type 1 regulatory T (Tr1) cells can inhibit anti-tumor responses and thereby impede immune control of cancer101. Notably, the application of artificial intelligence (AI) has been utilized to predict and identify immunogenic neoantigens, advancing neoantigen-based vaccine development102,103. These discoveries will promote the discovery of novel neoantigens and the development of circRNA-based vaccines.
Next, targeted delivery of circRNA vaccines is crucial for enhanced therapeutic effects. Currently, LNPs are the primary delivery system for circRNAs. The development of targeted LNPs should be further refined to ensure effective delivery performance. Alternatively, novel delivery systems that can replace LNPs may be explored104. Recent study indicates that LNPs modified with mannose maintain their physical properties even after lyophilization, offering long-term lymph node-targeting delivery stability and inducing potent and persistent immune responses105. Therefore, reducing off-target effects through targeted delivery systems is essential to maximize the therapeutic efficacy of circRNA vaccines.
Then, multidimensional crosstalks between exogenous circRNAs and endogenous biomolecules can influence the therapeutic efficacy of circRNA vaccines in cancer treatment. While circRNAs encoding neoantigens do not regulate parental gene transcription in normal cells, exogenous circRNAs can modulate gene expression through miRNA-mediated competitive endogenous RNA (ceRNA) crosstalks106. In addition, exogenous circRNAs can interact with RBPs to regulate their parental genes107. Together, these multidimensional crosstalks may impact the translation of circRNAs encoding neoantigens, potentially leading to undesired side effects in cancer treatment.
Last, many early phase trials of circRNA-related therapeutics are currently ongoing. However, no circRNA-based cancer vaccines have received regulatory approval. To improve personalized therapeutic effects, combinations of circRNA cancer vaccines with immune checkpoint inhibitors (ICIs) or CAR T cells should be considered. Given their promising tolerability, circRNA vaccines in combination with other regimens could be further expanded to broaden immunotherapeutic platforms for enhanced synergy.
In brief, circRNA-based vaccines hold significant promise for cancer therapy. However, it is essential to address challenges related to large-scale purification and biosafety determination of circRNA vaccines. While circRNA-based vaccines have shown success in animal studies, further clinical trials are urgently required to evaluate their effectiveness in human cancer treatment. The comparative advantages of different approaches will become clearer as clinical trials progress. In addition to effectiveness, factors like production costs will also impact the clinical acceptance of circRNA vaccines. It is important to note that a vaccine alone may not be adequate to enable the immune system to overcome a tumor’s defenses, and ongoing trials are combing vaccines with drugs to boost T-cell responses108. With advancements in technologies, the widespread use of circRNA vaccines for preventing infectious diseases and treating tumor malignancies holds promise for the future.
Data availability
As this is a review, there are no original datasets. However, all referenced data sources are cited within the manuscript.
References
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691 (2019).
Hiam-Galvez, K. J., Allen, B. M. & Spitzer, M. H. Systemic immunity in cancer. Nat. Rev. Cancer 21, 345–359 (2021).
Katsikis, P. D., Ishii, K. J. & Schliehe, C. Challenges in developing personalized neoantigen cancer vaccines. Nat. Rev. Immunol. 24, 213–227 (2024).
Liu, C. et al. mRNA-based cancer therapeutics. Nat. Rev. Cancer 23, 526–543 (2023).
Liu, X. et al. Circular RNA: an emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J. Control Release 348, 84–94 (2022).
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J. & Kleinschmidt, A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA 73, 3852–3856 (1976).
Hsu, M. T. & Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280, 339–340 (1979).
Bailleul, B. During in vivo maturation of eukaryotic nuclear mRNA, splicing yields excised exon circles. Nucleic Acids Res. 24, 1015–1019 (1996).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Meganck, R. M. et al. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 23, 821–834 (2021).
Li, J. et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am. J. Cancer Res. 5, 472–480 (2015).
Zhu, Y. et al. CircNUP50 is a novel therapeutic target that promotes cisplatin resistance in ovarian cancer by modulating p53 ubiquitination. J. Nanobiotechnol. 22, 35 (2024).
Xu, L. et al. hsa_circ_0007919 induces LIG1 transcription by binding to FOXA1/TET1 to enhance the DNA damage response and promote gemcitabine resistance in pancreatic ductal adenocarcinoma. Mol. Cancer 22, 195 (2023).
Zhong, C. et al. M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int. J. Biol. Sci. 19, 1543–1563 (2023).
Li, H. et al. N6-methyladenosine-modified circPLPP4 sustains cisplatin resistance in ovarian cancer cells via PIK3R1 upregulation. Mol. Cancer 23, 5 (2024).
Yi, J. et al. CircPVT1 promotes ER-positive breast tumorigenesis and drug resistance by targeting ESR1 and MAVS. EMBO J. 42, e112408 (2023).
Cui, Y. et al. CircHERC1 promotes non-small cell lung cancer cell progression by sequestering FOXO1 in the cytoplasm and regulating the miR-142-3p-HMGB1 axis. Mol. Cancer 22, 179 (2023).
Li, K. et al. A circular RNA activated by TGFbeta promotes tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability. Nat. Commun. 14, 6876 (2023).
Wang, Y. & Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 21, 172–179 (2015).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629 (2018).
Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744 e1716 (2022).
Li, H. et al. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics 12, 6422–6436 (2022).
Niu, D., Wu, Y. & Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target Ther. 8, 341 (2023).
Chen, C. Y. & Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417 (1995).
Chen, C. K. et al. Structured elements drive extensive circular RNA translation. Mol. Cell 81, 4300–4318.e4313 (2021).
Abe, N. et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew. Chem. Int. Ed. Engl. 52, 7004–7008 (2013).
Abe, N. et al. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 5, 16435 (2015).
Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 41, 262–272 (2023).
Li, Y. F. & Breaker, R. R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 121, 5364–5372 (1999).
Khodadoust, M. S. et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 543, 723–727 (2017).
Cao, K., Hajy Heydary, Y., Tong, G. & Martinez, T. F. Integrated workflow for discovery of microprotein-coding small open reading frames. STAR Protoc. 4, 102649 (2023).
Miravet-Verde, S. et al. ProTInSeq: transposon insertion tracking by ultra-deep DNA sequencing to identify translated large and small ORFs. Nat. Commun. 15, 2091 (2024).
Olexiouk, V., Van Criekinge, W. & Menschaert, G. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 46, D497–D502 (2018).
Tong, G., Hah, N. & Martinez, T. F. Comparison of software packages for detecting unannotated translated smallopen reading frames by Ribo-seq. Brief Bioinform 5, bbae268 (2024).
Zhu, C. et al. moPepGen: rapid and comprehensive proteoform identification. Preprint at bioRxiv https://doi.org/10.1101/2024.03.28.587261 (2024).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Olazagoitia-Garmendia, A. et al. m(6)A methylated long noncoding RNA LOC339803 regulates intestinal inflammatory response. Adv. Sci. 11, e2307928 (2024).
Xu, X. et al. A positive feedback circuit between RN7SK snRNA and m(6)A readers is essential for tumorigenesis. Mol. Ther. 31, 1615–1635 (2023).
Ye, Y. et al. Genome-wide identification and characterization of circular RNA m(6)A modification in pancreatic cancer. Genome Med. 13, 183 (2021).
Chen, R. X. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 10, 4695 (2019).
Lang, F. et al. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 15, e1007796 (2019).
Yang, F. et al. Dynamic m(6)A mRNA methylation reveals the role of METTL3-m(6)A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene 38, 4755–4772 (2019).
Ying, X. et al. Programmable N6-methyladenosine modification of CDCP1 mRNA by RCas9-methyltransferase like 3 conjugates promotes bladder cancer development. Mol. Cancer 19, 169 (2020).
Fritah, H., Rovelli, R., Chiang, C. L. & Kandalaft, L. E. The current clinical landscape of personalized cancer vaccines. Cancer Treat. Rev. 106, 102383 (2022).
Veatch, J. R. et al. A therapeutic cancer vaccine delivers antigens and adjuvants to lymphoid tissues using genetically modified T cells. J. Clin. Invest. 131, e144195 (2021).
Liu, L. et al. Concurrent delivery of immune checkpoint blockade modulates T cell dynamics to enhance neoantigen vaccine-generated antitumor immunity. Nat. Cancer 3, 437–452 (2022).
Liu, K. et al. Expanding the potential of neoantigen vaccines: harnessing Bacille Calmette-Guerin cell-wall-based nanoscale adjuvants for enhanced cancer immunotherapy. ACS Nano 18, 11910–11920 (2024).
Chen, G. et al. Chemotherapy-induced neoantigen nanovaccines enhance checkpoint blockade cancer immunotherapy. ACS Nano 17, 18818–18831 (2023).
Yarchoan, M. et al. Personalized neoantigen vaccine and pembrolizumab in advanced hepatocellular carcinoma: a phase 1/2 trial. Nat. Med. 30, 1044–1053 (2024).
Ren, Y. et al. Circular RNA as a source of neoantigens for cancer vaccines. J. Immunother. Cancer 12, e008402 (2024).
Wang, F. et al. Circular RNA-based neoantigen vaccine for hepatocellular carcinoma immunotherapy. MedComm 5, e667 (2024).
Nakamoto, K. et al. Chemically synthesized circular RNAs with phosphoramidate linkages enable rolling circle translation. Chem. Commun. 56, 6217–6220 (2020).
Schauenburg, D. & Weil, T. Chemical reactions in living systems. Adv. Sci. 11, e2303396 (2024).
Costello, A., Lao, N. T., Barron, N. & Clynes, M. Reinventing the wheel: synthetic circular RNAs for mammalian cell engineering. Trends Biotechnol. 38, 217–230 (2020).
Chen, H. et al. Preferential production of RNA rings by T4 RNA ligase 2 without any splint through rational design of precursor strand. Nucleic Acids Res. 48, e54 (2020).
Cheng, K. et al. RNA ligation of very small pseudo nick structures by T4 RNA ligase 2, leading to efficient production of versatile RNA rings. RSC Adv. 9, 8620–8627 (2019).
Liu, C. X. et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 82, 420–434.e426 (2022).
Qi, Y. et al. Engineering circular RNA regulators to specifically promote circular RNA production. Theranostics 11, 7322–7336 (2021).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 (2014).
Rausch, J. W. et al. Characterizing and circumventing sequence restrictions for synthesis of circular RNA in vitro. Nucleic Acids Res. 49, e35 (2021).
Abe, B. T., Wesselhoeft, R. A., Chen, R., Anderson, D. G. & Chang, H. Y. Circular RNA migration in agarose gel electrophoresis. Mol. Cell 82, 1768–1777.e1763 (2022).
Xiao, M. S. & Wilusz, J. E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3’ ends. Nucleic Acids Res. 47, 8755–8769 (2019).
Baronti, L., Karlsson, H., Marusic, M. & Petzold, K. A guide to large-scale RNA sample preparation. Anal. Bioanal. Chem. 410, 3239–3252 (2018).
Vaidyanathan, S. et al. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. Nucleic Acids 12, 530–542 (2018).
Garcia-Canas, V. et al. Approach to the profiling and characterization of influenza vaccine constituents by the combined use of size-exclusion chromatography, gel electrophoresis and mass spectrometry. Biologicals 38, 294–302 (2010).
Kimia, Z. et al. A novel application of ion exchange chromatography in recombinant hepatitis B vaccine downstream processing: Improving recombinant HBsAg homogeneity by removing associated aggregates. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1113, 20–29 (2019).
Feng, Z. et al. An in vitro-transcribed circular RNA targets the mitochondrial inner membrane cardiolipin to ablate EIF4G2(+)/PTBP1(+) pan-adenocarcinoma. Nat. Cancer 5, 30–46 (2024).
Loan Young, T., Chang Wang, K., James Varley, A. & Li, B. Clinical delivery of circular RNA: lessons learned from RNA drug development. Adv. Drug Deliv. Rev. 197, 114826 (2023).
Diken, M. et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 18, 702–708 (2011).
Selmi, A. et al. Uptake of synthetic naked RNA by skin-resident dendritic cells via macropinocytosis allows antigen expression and induction of T-cell responses in mice. Cancer Immunol. Immunother. 65, 1075–1083 (2016).
Probst, J. et al. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 14, 1175–1180 (2007).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Li, F. et al. A novel circular RNA circITGa9 predominantly generated in human heart disease induces cardiac remodeling and fibrosis. Research 7, 0303 (2024).
Ding, Y. et al. The lectin Siglec-G inhibits dendritic cell cross-presentation by impairing MHC class I-peptide complex formation. Nat. Immunol. 17, 1167–1175 (2016).
Qiao, J. et al. Targeting tumors with IL-10 prevents dendritic cell-mediated CD8(+) T cell apoptosis. Cancer Cell 35, 901–915.e904 (2019).
Dorrie, J., Schaft, N., Schuler, G. & Schuler-Thurner, B. Therapeutic cancer vaccination with ex vivo RNA-transfected dendritic cells-an update. Pharmaceutics 12, 92 (2020).
Liu, J. et al. Nanochannel electro-injection as a versatile platform for efficient RNA/DNA programming on dendritic cells. Small 19, e2303088 (2023).
Nele, V., Campani, V., Alia Moosavian, S. & De Rosa, G. Lipid nanoparticles for RNA delivery: self-assembling vs driven-assembling strategies. Adv. Drug Deliv. Rev. 208, 115291 (2024).
Hu, M., Li, X., You, Z., Cai, R. & Chen, C. Physiological barriers and strategies of lipid-based nanoparticles for nucleic acid drug delivery. Adv. Mater. 36, e2303266 (2024).
Lou, G. et al. Delivery of self-amplifying mRNA vaccines by cationic lipid nanoparticles: the impact of cationic lipid selection. J. Control Release 325, 370–379 (2020).
Ball, R. L., Hajj, K. A., Vizelman, J., Bajaj, P. & Whitehead, K. A. Lipid nanoparticle formulations for enhanced co-delivery of siRNA and mRNA. Nano Lett. 18, 3814–3822 (2018).
Chen, J. et al. Lipid nanoparticle-mediated lymph node-targeting delivery of mRNA cancer vaccine elicits robust CD8(+) T cell response. Proc. Natl Acad. Sci. USA 119, e2207841119 (2022).
Muskan, M. et al. Therapeutic potential of RNA-enriched extracellular vesicles: the next generation in RNA delivery via biogenic nanoparticles. Mol. Ther. 32, 2939–2949 (2024).
Zhang, P. et al. A multiclade env-gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques. Nat. Med. 27, 2234–2245 (2021).
Meganck, R. M. et al. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol. Ther. Nucleic Acids 13, 89–98 (2018).
Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520.e504 (2019).
June, C. H. & Sadelain, M. Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018).
Mackensen, A. et al. CLDN6-specific CAR-T cells plus amplifying RNA vaccine in relapsed or refractory solid tumors: the phase 1 BNT211-01 trial. Nat. Med. 29, 2844–2853 (2023).
Carvalho, T. Personalized anti-cancer vaccine combining mRNA and immunotherapy tested in melanoma trial. Nat. Med. 29, 2379–2380 (2023).
Amaya, L. et al. Circular RNA vaccine induces potent T cell responses. Proc. Natl Acad. Sci. USA 120, e2302191120 (2023).
Yang, J. et al. Intratumoral delivered novel circular mRNA encoding cytokines for immune modulation and cancer therapy. Mol. Ther. Nucleic Acids 30, 184–197 (2022).
Wan, J. et al. CXCL13 promotes broad immune responses induced by circular RNA vaccines. Proc. Natl Acad. Sci. USA 121, e2406434121 (2024).
Disis, M. L. N. et al. Safety and outcomes of a plasmid DNA vaccine encoding the ERBB2 intracellular domain in patients with advanced-stage ERBB2-positive breast cancer: a phase 1 nonrandomized clinical trial. JAMA Oncol. 9, 71–78 (2023).
Neeli, P. et al. DNA vaccines against GPRC5D synergize with PD-1 blockade to treat multiple myeloma. NPJ Vaccines 9, 180 (2024).
Guo, X. et al. mRNA compartmentalization via multimodule DNA nanostructure assembly augments the immunogenicity and efficacy of cancer mRNA vaccine. Sci. Adv. 10, eadp3680 (2024).
Li, Y. et al. mRNA vaccine in cancer therapy: current advance and future outlook. Clin. Transl. Med. 13, e1384 (2023).
Huang, D. et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature 625, 593–602 (2024).
Schumacher, T. N., Scheper, W. & Kvistborg, P. Cancer neoantigens. Annu. Rev. Immunol. 37, 173–200 (2019).
Sultan, H. et al. Neoantigen-specific cytotoxic Tr1 CD4 T cells suppress cancer immunotherapy. Nature 632, 182–191 (2024).
Chu, Y. et al. A transformer-based model to predict peptide–HLA class I binding and optimize mutated peptides for vaccine design. Nat. Mach. Intell. 4, 300–311 (2022).
Chuwdhury, G. S. et al. ImmuneMirror: a machine learning-based integrative pipeline and web server for neoantigen prediction. Brief. Bioinform 25, bbae024 (2024).
Packer, M., Gyawali, D., Yerabolu, R., Schariter, J. & White, P. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat. Commun. 12, 6777 (2021).
Wan, J. et al. Circular RNA vaccines with long-term lymph node-targeting delivery stability after lyophilization induce potent and persistent immune responses. mBio 15, e0177523 (2024).
Tay, Y., Rinn, J. & Pandolfi, P. P. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014).
Sun, X. et al. Potential therapeutic strategy for cancer: multi-dimensional cross-talk between circRNAs and parental genes. Cancer Lett. 588, 216794 (2024).
Drew, L. Cancer-vaccine trials give reasons for optimism. Nature 627, S33 (2024).
Acknowledgements
The authors thank their respective laboratory members and collaborators for critical review of this article. The authors apologize that space constraints prevent them from citing all relevant publications. The language of the manuscript was improved by using an online intelligence tool (WORDVICE.AI). This work was supported in part by research grants from Zhejiang Provincial Natural Science Foundation (LMS25C060001), Ningbo Natural Science Foundation (2024J330), Ningbo Clinical Research Center for Respiratory Diseases (2022L004), Ningbo University Student Scientific Research Innovation Project (2024SRIP1908), and K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Contributions
Z.G. conceived and conceptualized the idea of the review, obtained the funding, wrote the original draft, reviewed, and revised the manuscript. W.H., C.Z., and J.G. provided minor input to writing the original draft and reviewed the manuscript. L.Y. and B.W. reviewed the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gong, Z., Hu, W., Zhou, C. et al. Recent advances and perspectives on the development of circular RNA cancer vaccines. npj Vaccines 10, 41 (2025). https://doi.org/10.1038/s41541-025-01097-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41541-025-01097-x
This article is cited by
-
Progress in cancer vaccines enabled by nanotechnology
Nature Nanotechnology (2025)
-
Circular RNA, A Molecule with Potential Chemistry and Applications in RNA-based Cancer Therapeutics: An Insight into Recent Advances
Topics in Current Chemistry (2025)
-
RNA-based cancer vaccines: mechanisms, clinical progress, and translational challenges
Immunologic Research (2025)
-
Decoding the functional roles of circular RNAs linked to NF-κB pathway in carcinogenesis
Discover Oncology (2025)