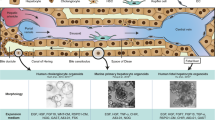
Abstract
The induction of tissue-specific vessels in in vitro living tissue systems remains challenging. Here, we directly differentiated human pluripotent stem cells into CD32b+ putative liver sinusoidal progenitors by dictating developmental pathways. By devising an inverted multilayered air–liquid interface culture, hepatic endoderm, septum mesenchyme, arterial and sinusoidal quadruple progenitors self-organize to generate and sustain hepatocyte-like cells neighboured by divergent endothelial subsets composed of CD32blowCD31high, LYVE1+STAB1+CD32bhighCD31lowTHBD−vWF− and LYVE1−THBD+vWF+ cells. WNT2 mediates sinusoidal-to-hepatic intercellular crosstalk potentiating hepatocyte differentiation and branched endothelial network formation. Intravital imaging reveals the iPS-cell-derived putative liver sinusoidal endothelial progenitor develops fully perfused human vessels with functional sinusoid-like features. Organoid-derived hepatocyte- and sinusoid-derived coagulation factors enable correction of in vitro clotting time with Factor V-, VIII-, IX- and XI-deficient plasma, and rescues the severe bleeding phenotype in haemophilia A mice on transplantation. Advanced organoid vascularization technology allows for interrogating key insights governing organ-specific vessel development, paving the way for coagulation disorder therapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq raw and processed data have been deposited in the GEO under accession numbers GSE217649 (Bulk)85 and GSE270807 (single cell)86 respectively. The relevant data and statistics that support the findings of this study are available within the Supplementary Information. Source data are provided with this paper.
References
Koike, H. et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut–midgut boundary. Nature 574, 112–116 (2019).
Ouchi, R. et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30, 374–384.e6 (2019).
Low, J. H. et al. Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell 25, 373–387.e9 (2019).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Akbari, S., Arslan, N., Senturk, S. & Erdal, E. Next-generation liver medicine using organoid models. Front. Cell Dev. Biol. https://doi.org/10.3389/fcell.2019.00345 (2019).
Garreta, E. et al. Rethinking organoid technology through bioengineering. Nat. Mater. 20, 145–155 (2021).
Takebe, T. & Wells, J. M. Organoids by design. Science 364, 956–959 (2019).
Rafii, S., Butler, J. M. & Ding, B. S. Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325 (2016).
Augustin, H. G. & Koh, G. Y. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science https://doi.org/10.1126/science.aal2379 (2017).
Matsumoto, K., Yoshitomi, H., Rossant, J. & Zaret, K. S. Liver organogenesis promoted by endothelial cells prior to vascular function. Science 294, 559–563 (2001).
Ding, B. Sen et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 468, 310–315 (2010).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Kitamoto, Y., Tokunaga, H. & Tomita, K. Vascular endothelial growth factor is an essential molecule for mouse kidney development: glomerulogenesis and nephrogenesis. J. Clin. Invest. 99, 2351–2357 (1997).
Homan, K. A. et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262 (2019).
Koch, P. S., Lee, K. H., Goerdt, S. & Augustin, H. G. Angiodiversity and organotypic functions of sinusoidal endothelial cells. Angiogenesis 24, 289–310 (2021).
Roberts, H. R. & Cederbaum, A. I. The liver and blood coagulation: physiology and pathology. Gastroenterology 63, 297–320 (1972).
Wisse, E. et al. Structure and function of sinusoidal lining cells in the liver. Toxicol. Pathol. 24, 100–111 (1996).
Braet, F. et al. Structure and dynamics of the fenestrae-associated cytoskeleton of rat liver sinusoidal endothelial cells. Hepatology 21, 180–189 (1995).
Shetty, S., Lalor, P. F. & Adams, D. H. Liver sinusoidal endothelial cells—gatekeepers of hepatic immunity. Nat. Rev. Gastroenterol. Hepatol. 15, 555–567 (2018).
Do, H., Healey, J. F., Waller, E. K. & Lollar, P. Expression of Factor VIII by murine liver sinusoidal endothelial cells. J. Biol. Chem. 274, 19587–19592 (1999).
Fomin, M. E. et al. Production of Factor VIII by human liver sinusoidal endothelial cells transplanted in immunodeficient uPA mice. PLoS ONE 8, e77255 (2013).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Takebe, T. et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep 21, 2661–2670 (2017).
Koui, Y. et al. An in vitro human liver model by iPSC-derived parenchymal and non-parenchymal cells. Stem Cell Rep. 9, 490–498 (2017).
Gage, B. K. et al. Generation of functional liver sinusoidal endothelial cells from human pluripotent stem-cell-derived venous angioblasts. Cell Stem Cell 27, 254–269.e9 (2020).
Tian, S. P. et al. A novel efficient strategy to generate liver sinusoidal endothelial cells from human pluripotent stem cells. Sci. Rep. 14, 13831 (2024).
Yap, K. K. et al. Liver specification of human iPSC-derived endothelial cells transplanted into mouse liver. JHEP Rep. 6, 101023 (2024).
Cast, A. E., Walter, T. J. & Huppert, S. S. Vascular patterning sets the stage for macro and micro hepatic architecture. Dev. Dyn. 244, 497–506 (2015).
Hikspoors, J. P. J. M. et al. The fate of the vitelline and umbilical veins during the development of the human liver. J. Anat. 231, 718–735 (2017).
Zovein, A. C. et al. Vascular remodeling of the vitelline artery initiates extravascular emergence of hematopoietic clusters. Blood 116, 3435–3444 (2010).
DeSesso, J. M. Vascular ontogeny within selected thoracoabdominal organs and the limbs. Reprod. Toxicol. 70, 3–20 (2017).
Marcelo, K. L., Goldie, L. C. & Hirschi, K. K. Regulation of endothelial cell differentiation and specification. Circ. Res. 112, 1272–1287 (2013).
Minn, K. T., Dietmann, S., Waye, S. E., Morris, S. A. & Solnica-Krezel, L. Gene expression dynamics underlying cell fate emergence in 2D micropatterned human embryonic stem cell gastruloids. Stem Cell Rep. 16, 1210–1227 (2021).
Chhabra, S., Liu, L., Goh, R., Kong, X. & Warmflash, A. Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids. PLoS Biol. 17, e3000498 (2019).
Niwa, A. et al. A novel serum-free monolayer culture for orderly hematopoietic differentiation of human pluripotent cells via mesodermal progenitors. PLoS ONE 6, e22261 (2011).
Honda-Ozaki, F. et al. Pluripotent stem cell model of Nakajo-Nishimura syndrome untangles proinflammatory pathways mediated by oxidative stress. Stem Cell Rep. 10, 1835–1850 (2018).
Scarfò, R. et al. CD32 captures committed haemogenic endothelial cells during human embryonic development. Nat. Cell Biol. 26, 719–730 (2024).
Rosa, S. et al. Functional characterization of iPSC-derived arterial- and venous-like endothelial cells. Sci. Rep. 9, 1835–1850 (2019).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Wesley, B. T. et al. Single-cell atlas of human liver development reveals pathways directing hepatic cell fates. Nat. Cell Biol. 24, 1487–1498 (2022).
Popescu, D. M. et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019).
Zhang, H. et al. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat. Genet. 48, 537–543 (2016).
Clarke, P. A., Dickson, J. H., Harris, J. C., Grabowska, A. & Watson, S. A. Gastrin enhances the angiogenic potential of endothelial cells via modulation of heparin-binding epidermal-like growth factor. Cancer Res. 66, 3504–3512 (2006).
Vasse, M. et al. Oncostatin M induces angiogenesis in vitro and in vivo. Arterioscler. Thromb. Vasc. Biol. 19, 1835–1842 (1999).
Alves-Lopes, J. P., Söder, O. & Stukenborg, J. B. Use of a three-layer gradient system of cells for rat testicular organoid generation. Nat. Protoc. 13, 248–259 (2018).
Alves-Lopes, J. P., Söder, O. & Stukenborg, J. B. Testicular organoid generation by a novel in vitro three-layer gradient system. Biomaterials 130, 76–89 (2017).
Malerød, L., Juvet, L., Gjøen, T. & Berg, T. The expression of scavenger receptor class B, type I (SR-BI) and caveolin-1 in parenchymal and nonparenchymal liver cells. Cell Tissue Res. 307, 173–180 (2002).
Strauss, O., Phillips, A., Ruggiero, K., Bartlett, A. & Dunbar, P. R. Immunofluorescence identifies distinct subsets of endothelial cells in the human liver. Sci. Rep. 7, 44356 (2017).
Su, T. et al. Single-cell transcriptomics reveals zone-specific alterations of liver sinusoidal endothelial cells in cirrhosis. Cell Mol. Gatroenterol. Hepatol. 11, 1139–1161 (2021).
Wisse, E. et al. Fat causes necrosis and inflammation in parenchymal cells in human steatotic liver. Histochem. Cell Biol. 157, 27–38 (2022).
Takebe, T. et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat. Protoc. 9, 396–409 (2014).
Itkin, T. et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature 532, 323–328 (2016).
Guan, Y. et al. A human multi-lineage hepatic organoid model for liver fibrosis. Nat. Commun. 12, 6138 (2021).
Harrison, S. P. et al. Scalable production of tissue-like vascularized liver organoids from human PSCs. Exp. Mol. Med. 55, 2005–2024 (2023).
Hess, A. et al. Single‐cell transcriptomics stratifies organoid models of metabolic dysfunction‐associated steatotic liver disease. EMBO J. 42, e113898 (2023).
Shinozawa, T. et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell–derived organoids. Gastroenterology 160, 831–846.e10 (2021).
Velazquez, J. J. et al. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst. 12, 41–55.e11 (2021).
Zhang, C. J. et al. A human liver organoid screening platform for DILI risk prediction. J. Hepatol. 78, 998–1006 (2023).
Ton, M. L. N. et al. An atlas of rabbit development as a model for single-cell comparative genomics. Nat. Cell Biol. 25, 1061–1072 (2023).
Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 40, 245–253 (2022).
Géraud, C. et al. GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. J. Clin. Invest. 127, 1099–1114 (2017).
Winkler, M. et al. Endothelial GATA4 controls liver fibrosis and regeneration by preventing a pathogenic switch in angiocrine signaling. J. Hepatol. 74, 380–393 (2021).
Preziosi, M., Okabe, H., Poddar, M., Singh, S. & Monga, S. P. Endothelial Wnts regulate β‐catenin signaling in murine liver zonation and regeneration: a sequel to the Wnt–Wnt situation. Hepatol. Commun. 2, 845–860 (2018).
Rocha, A. S. et al. The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Rep. 13, 1757–1764 (2015).
Weyand, A. C. & Pipe, S. W. New therapies for hemophilia. Blood 133, 389–398 (2019).
Lascaris, B. et al. Production of physiological amounts of hemostatic proteins by human donor livers during ex situ long-term normothermic machine perfusion for up to 7 days. J. Thromb. Haemost. https://doi.org/10.1016/j.jtha.2024.08.004 (2024).
Kalucka, J. et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779.e20 (2020).
Marcu, R. et al. Human organ-specific endothelial cell heterogeneity. iScience 4, 20–35 (2018).
Wong, B. W., Zecchin, A., García-Caballero, M. & Carmeliet, P. Emerging concepts in organ-specific lymphatic vessels and metabolic regulation of lymphatic development. Dev. Cell 45, 289–301 (2018).
Hen, G. et al. Venous-derived angioblasts generate organ-specific vessels during zebrafish embryonic development. Development 142, 4266–4278 (2015).
Cakir, B. et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 16, 1169–1175 (2019).
Wakabayashi, T. et al. CD157 marks tissue-resident endothelial stem cells with homeostatic and regenerative properties. Cell Stem Cell 22, 384–397.e6 (2018).
Lee, L. K. et al. LYVE1 marks the divergence of yolk sac definitive hemogenic endothelium from the primitive erythroid lineage. Cell Rep. 17, 2286–2298 (2016).
Sugiyama, Y. et al. Sinusoid development and morphogenesis may be stimulated by VEGF-Flk-1 signaling during fetal mouse liver development. Dev. Dyn. 239, 386–397 (2010).
Lotto, J. et al. Single-cell transcriptomics reveals early emergence of liver parenchymal and non-parenchymal cell lineages. Cell 183, 702–716.e14 (2020).
Sherief, L. M. et al. Factor VIII inhibitor development in Egyptian hemophilia patients: does intron 22 inversion mutation play a role? Ital. J. Pediatr. 46, 129 (2020).
Lai, J., Hough, C., Tarrant, J. & Lillicrap, D. Biological considerations of plasma-derived and recombinant Factor VIII immunogenicity. Blood 129, 3147–3154 (2017).
Pasi, K. J. et al. Multiyear follow-up of AAV5-hFVIII-SQ gene therapy for hemophilia. New Engl. J. Med. 382, 29–40 (2020).
Gage, B. K., Merlin, S., Olgasi, C., Follenzi, A. & Keller, G. M. Therapeutic correction of hemophilia A by transplantation of hPSC-derived liver sinusoidal endothelial cell progenitors. Cell Rep. 39, 110759 (2022).
Son, J. S. et al. Therapeutic correction of hemophilia A using 2D endothelial cells and multicellular 3D organoids derived from CRISPR/Cas9-engineered patient iPSCs. Biomaterials 283, 121429 (2022).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Yoshimori, T. et al. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11, 747–763 (2000).
Kamikubo, Y. et al. Selective Factor VIII activation by the tissue factor–Factor VIIa–Factor Xa complex. Blood 130, 1661–1670 (2017).
Saiki N., Kono T., Koido M. & Takebe T. Bulk RNA sequencing for gene expression profileing of primary liver sinusoidal endothelial cells and iPSC-derived hepatic endoderm, septum mesenchymal cells, endothelial cells, and liver bud organoids. Gene Expression Omnibus www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217649 (2025).
Saiki N. & Takebe T. Longitudinal scRNA-seq analysis of human liver bud organoid (HLBO) with iPS-derived liver sinusoidal endothelial progenitor (iLSEP). Gene Expression Omnibus www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE270807 (2025).
Acknowledgements
We express our sincere gratitude to S. Yamanaka and S. Izumo, Y. Kajii, and A. Nakanishi for supporting the collaboration research between Takeda Pharmaceutical Company and Center for iPS Cell Research and Application (CiRA), Kyoto University (T-CiRA). We thank T. Shinozawa and H. Anayama (Takeda Pharmaceutical Company) for advice on the research strategy; Y. Noguchi, M. Fukui and A. Nukuda (Orizuru Therapeutics, Inc.) for technical assistance with in vitro experiments; M. Ide and M. Nomura (RABICS, Ltd) for technical assistance with animal experiments. We also thank K. Hashikami, T. Yamamura and M. Takeyama (Axcelead Drug Discovery Partners, Inc.) for generating and providing NSI-FVIII KO mice; T. Sasaki and K. Aoyama (Axcelead Drug Discovery Partners) for liquid chromatography with tandem mass spectrometry analysis; C. Moriya (Science Tokyo) for providing NOD-SCID mice with cranial window; Y. Sakamaki (Science Tokyo) for her expert technical assistance and advice with electron microscopy; A. Niwa (CiRA) for advice and discussion on differentiation of the haemogenic endothelium; Y. Kamikubo (Thrombo Translational Research Lab Inc.) for advice and discussion on coagulation experiment; H. Matsumoto (T-CiRA), M. Maezawa and M. Mori (Science Tokyo) for kind administrative assistance. This work was supported by the T-CiRA Joint Program from Takeda Pharmaceutical Company and by Japan Agency for Medical Research and Development (AMED) under grant nos. JP23bm1223006 and JP24ym0126809. This work was also supported by Cincinnati Children’s Research Foundation grant, CURE award, NIH Director’s New Innovator Award (grant no. DP2 DK128799-01), grant nos. R01DK135478, UG3/UH3 DK119982, PHS grant no. P30 DK078392 (Integrative Morphology Core and Pluripotent Stem Cell and Organoid Core) of the Digestive Disease Research Core Center in Cincinnati, the Falk Transformational Awards Program, JST Moonshot R&D grant no. JPMJMS2033 and JPMJMS2022, Takeda Science Foundation, Mitsubishi Foundation, Cannon Foundation and AMED CREST (grant no. JP22gm1210012), grant nos. JP24bm1223006, JP24ym0126809, JP24gm1210012, JP24fk0210150, JP23gm1610005, JP20fk0210037, JP20bm0704025, JP21fk0210060, JP21bm0404045, JP22fk0210106, JSPS JP18H02800, 19K22416, 21H04822 and World Premier International Research Center Initiative (WPI) PRIMe, MEXT, Japan. Fetal tissue analysis was conducted with the support of the Developmental Origins of Health and Disease Biorepository at Icahn School of Medicine at Mount Sinai. We are grateful to the Biorepository laboratory team and Biorepository participants for their contributions to this research.
Author information
Authors and Affiliations
Contributions
N.S. and T.T. conceived the study. N.S., Y.N. and T.T. designed the experiments. N.S. wrote the original draft of the manuscript. Y.Y., W.L.T. and T.T. reviewed and edited the manuscript. N.S. performed almost all in vitro and animal experiments, bioinformatics and analysed data. Y.N. performed and optimized haemophilia A animal model experiments. Y.Y. performed the scanning electron microscopy experiment. S.K. performed cranial window animal experiments. E.K. and K.A. performed and optimized the fibrin clot experiment. J.F. and T.S. performed the cell culture and biochemical assays. K.I., R.O., P.C.-E. and Y.-W.C. performed sectioning, staining and analysis. T.K. and M.K. obtained and analysed some RNA-seq data. M.F. performed and optimized siRNA transfection experiment.
Corresponding author
Ethics declarations
Competing interests
N.S. and T.T. are the inventors of the patent applications WO2020203713A1 and WO2022230919A1. N.S., E.K. and T.T. are the inventors of the patent application WO2020175594A1. T.T. received research funding from Takeda Pharmaceutical Company, Ltd. Y.N., E.K., T.K. and M.F. are employees of Takeda Pharmaceutical Company, Ltd. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Directed differentiation and characterization of iLSEP.
a) Time-lapse morphological representation of differentiation of iLSEP in micropatterned colony. Red arrows indicate expanding CD34+ endothelial regions (together with Extended Data Fig. 1b). b) Immunofluorescent staining for CD34 and Brachyury at day6. c) Immunofluorescent staining for CD31 and CD32b at day6. d) Scatter dot-plot of the flow cytometry analysis of CD31/CD32 expression in CD34+CD43− iLSEP and iAEC after CD34+ purification at day 8. CD43 is a marker to exclude fractions of the hematopoietic population. e) Heatmap of iPSC, iSTM, conventional EC (iAEC), iLSEP, and primary LSEC showing lineage marker gene set. Mes.: Mesenchyme; Endo.: pan-endothelium; LSEC: liver sinusoidal endothelial cell. f) Scatter dot-plot of the flow cytometry analysis of CD31/CD32 expression in CD34+CD43− VA at day8.
Extended Data Fig. 2 Induction of LSEPs into LSECs by OSM, guided by single-cell analysis of endothelial development.
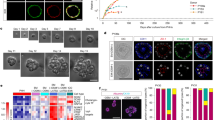
a) Integrated uniform manifold approximation and projection (UMAP) plot of ECs in fetal and adult organs showing yolk sac EC (4 post conception weeks (PCW)), endocardium in fetal heart (4 PCW), fetal liver EC (Early; 7 PCW, Mid; 8-12 PCW and Late; 13−17 PCW), and LSEC zone 2 and 3 in adult liver. The black arrow curve represents the direction of the trajectory based on pseudo-temporal analysis (together with Extended Data Fig. 2b). b) The pseudo-time trajectory of endothelial development to LSEC in adult liver. c) Pseudo-kinetic plots of the expression of markers in different organ groups. d) The aggregate expression level of all genes in gene module, a cluster of genes showing shared expression dynamics among the populations in trajectory, of fetal (early, mid, and late) liver and late fetal + adult liver projected on UMAP. e) Top significantly enriched pathways on dynamics of fetal liver module and late fetal + adult liver module. f) The schema of hiPS cell-derived LSEC differentiation via LSEP. g) Scatter dot-plot of the flow cytometry analysis of CD31/CD32 expression in CD34+CD43− iAEC and iLSEP at day14 under +VEGF and +VEGF + OSM conditions. h) Scatter dot-plots of the flow cytometry analysis of LYVE1/CD32 expression (left). Quantification of CD32+LYVE1+ cell fractions in CD34+CD43− cells (right). Data represent the mean ± SEM (n = 6, iAEC; n = 12; iLSEP; all n are biological replicates; ****, p < 0.0001; ns, not significant; one-way ANOVA with Tukey’s multiple comparisons test). i) The feature plots showing the expression of arterial (CXCR4) and venous (NT5E coding CD73 protein) EC markers in integrated single-cell gene expression data. Gray color indicates no expression. j) Scatter dot-plots of the flow cytometry analysis of CD73/CXCR4 expression. k) Immunofluorescent staining for CD32b/CD31 (upper) and CD32b/FVIII (lower) at day 12 (iSTM, iAEC and iLSEP) and day 16 (iLSEC). l) Scatter dot-plot of the flow cytometry analysis of LYVE1/CD32 expression in VA-derived LSEC (VA-LSEC) at day14. m) Comparison of FVIII protein expression quantified from images of immunofluorescent staining for FVIII/CD32b. Data represent the mean ± SEM (n = 6, VA-LSEC and iLSEC; n = 3, Primary LSEC; all n are biological replicates; **, p < 0.01; ****, p < 0.0001; one-way ANOVA with Dunnett’s multiple comparisons test compared to iLSEC, day14). n) Comparison of secreted FVIII antigen concentration in culture supernatants among iPS-derived STM, endothelial cells, and primary LSEC. Data represent the mean ± SEM (n = 12, iLSEC; n = 8, iAEC and iSTM; n = 9, iLSEP; n = 3, Primary LSEC; n = 4, VA-LSEC; all n are biological replicates; ****, p < 0.0001; one-way ANOVA with Dunnett’s multiple comparisons test compared to iLSEC, day15). o) Scanning electron microscopy (SEM) of iLSEC ( + VEGF and +VEGF, +OSM) and iAEC. Arrowheads indicate the clusters of fenestrae on the cell surface. The right panel is an enlarged image of the dashed square region of iLSEC ( + VEGF, +OSM).
Extended Data Fig. 3 Characterization of endothelial lineages in self-organizing liver bud organoids.
a) Bright-field image of organoids at different time points after mixing cells (day 1, 5, and 11). Scale bar indicates 500 µm. b) Whole-mount single fluorescent channel images of HLBOs including GFP+ iLSEP and mCherry+ iAEC with immunofluorescent staining for FVIII at day 11. Scale bar indicates 200 µm. c) High-magnification whole mount image of HLBO including GFP+ iLSEP with immunofluorescent staining for CD32b and FVIII at day 11. Scale bar indicates 30 µm. d) Scatter dot-plots of the flow cytometry analysis of CD34+ endothelial cells and EPCAM+ (CD326+) epithelial cells in HLBOs at day11. The right panels display CD32 expression in endothelial cells gated by CD34. e) Scatter dot-plots of the flow cytometry analysis of CD31, CD32, and CD36 expression of HLBOs at day11. The panel of CD32 and CD36 was gated by CD31high for both HLBOs, and CD32highCD31low and CD32lowCD31low only for +iLSEP HLBO. f) Cross-sectioning image of HLBOs with immunofluorescent staining for LYVE1/CD32b/STAB1 at day 11. Right panels are enlarged single fluorescent channel images of the dashed square region. g) Transmission immunoelectron microscopy of ultra-thin frozen sections of +iLSEP HLBO labeled with anti-LYVE1 antibody. Right panel is enlarged image of the red dashed square region of left panel. Yellow asterisks indicate pore structures between LYVE1+ endothelial cells. Scale bar indicates 5 µm (left) and 1 µm (right). h) Intravital fluorescence microscopy imaging of the +iLSEP HLBO transplanted mice. The organoid-derived human vessels were observed by GFP+ iLSEP. Host mice vessels and blood flow are visualized using a mice-specific anti-CD31 antibody (mCD31) and 2,000 kDa tetramethylrhodamine-dextran, respectively. Right panels are enlarged image of the dashed square region. White arrow marks the point of connection between the human and mouse blood vessels. i) Alexa Fluor 647 (AF647)-conjugated AcLDL uptake in human blood vessels with blood flow (2,000 kDa tetramethylrhodamine-dextran). Right panels are single fluorescent channel images. Yellow arrowheads indicate regions of AF647-AcLDL merged human blood vessels. Scale bar indicates 50 µm.
Extended Data Fig. 4 scRNA-seq profiling of HLBOs and primary human liver.
a) Heatmap showing marker gene expression for cell subtype layer group in the HLBO+iLSEP dataset. b) Proportion of cell cycle phase per cell class in each time series. c) Sankey diagram for hierarchical overview of cell composition in HLBO+iLSEP. The hierarchical structure consists of three annotation levels: cell class (Layer 1), which is the upper domain grouping cell types, cell type (Layer 2), and subtype (Layer 3), and the numbers in parentheses indicate the total number of annotations in each layer. d) UMAP of integrated primary liver dataset including fetal and adult stage colored by cell type. e) UMAPs showing common cell type marker expression between primary liver and HLBO+iLSEP.
Extended Data Fig. 5 Comparative analysis of HLBO with published human organoid datasets.
a) UMAPs for human pluripotent stem cell-derived liver organoid cell atlas colored by cell type and organoid source. b) UMAPs display the maximum spearman correlation of (left) adult liver and (right) fetal liver dataset. The right UMAP highlights HLBO+iLSEP cells. c-f) Ridge plots displaying spearman correlations for each organoid dataset against c) adult or fetal hepatocytes, d) adult or fetal cholangiocytes, e) both adult and fetal stellate cells, and f) adult or fetal endothelial cells. Organoid groups that did not contain enough cells to show data distribution are not plotted.
Extended Data Fig. 6 Single-cell characterization of endothelial cell in liver organoids.
a) UMAPs showing LSEC (zone 2 and 3) marker gene expression (together with Extended Data Fig. 2c), whereas organoids are color-coded by data resource. b) Violin plots showing differentiation signature expression by endothelial subset. c) UMAP showing extracted subpopulation of arterial-like endothelial cell (EC1) (left) and Scatter plots showing latent time versus spliced gene expression of arterial markers (right). Black line on scatter plot is linear regression fit line to data points to represent trends of change.
Extended Data Fig. 7 Analysis of hepatic functionalization and architecture in HLBOs.
a) qRT-PCR analysis of AFP and ALB. Data represent the mean ± SEM (n = 3 biological replicates; **, p < 0.01; ****, p < 0.0001; multiple two-sided t-test with Holm-Sidak correction). b) Measurement of albumin in organoid supernatants at day 11. Data represent the mean ± SEM (n = 3 biological replicates; **, p < 0.01; ****, p < 0.0001; one-way ANOVA with Dunnett’s multiple comparisons test compared to +iLSEP HLBO). c) Cross-sectioning image of HLBO without EC (-EC) with H&E staining (left). Distribution of hepatocytes with different zonal marker protein identified by immunofluorescent staining for GS (peri-central) and GLS2 (pan-lobular) in HLBOs (right). d) Cross-sectioning images showing distribution of hepatocytes identified by immunofluorescent staining for ASGR1 and GFP+ iAECs or iLSEPs embedded in a second cell layer. Lower panels are enlarged images of the white dashed square region. The yellow dashed lines indicate the boundaries of the gel layer in organoid generation as illustrated in Fig. 1a. Each section is a serial specimen of the stained images in Fig. 3a, b. e) Localization of ASGR1+ hepatocyte and CD32b+ LSEC determined by whole-mount immunofluorescent staining of HLBOs. f) Measurement of glutamine / glutamate ratio in HLBO supernatants at day11. Data represent the mean ± SEM (n = 3 biological replicates; *, p < 0.05; two-sided Welch’s t-test). g) Determination of ammonia uptake from the medium in HLBOs at day 11. Ammonia change in 24 h was quantified by the difference between the HLBO supernatant and the culture medium. Data represent the mean ± SEM (n = 7 biological replicates; **, p < 0.01; two-sided Welch’s t-test). h-i) Whole-mount immunostaining images of HLBO with/without CYP inducers stained for CYP3A4/PXR/LYVE1 and CYP1A2/GS/LYVE1. Right panels are fluorescence channel split images of LYVE1 combined with each hepatocyte markers in +iLSEP HLBO. j) Representative enrichment plots in scRNA-seq analysis compared between hepatocyte in +iLSEP HLBO and those in other liver organoids (9 publicly available organoids) of integrated dataset. Shown pathways are significantly enriched in +iLSEP HLBO (NES > 0; FDR-adjusted p < 0.05; one-sided permutation test).
Extended Data Fig. 8 Inter-cellular lineage interaction by ligand-receptor analysis.
a) The heatmap visualizing the relative importance of each cell type of late fetal liver (13-17 PCW) based on the centrality score of WNT signaling pathway. b) The chord diagram describing significant ligand-receptor pairs involving WNT signaling within fetal liver (p < 0.05; one-sided permutation test without adjustment). c) The heatmap and bar plot summarizing outgoing or incoming signaling strength across cell types. Color scale of the heatmap represents the relative signaling strength of each pathway. Top bar plot shows total strength of all pathways displayed in the heatmap for each cell type. d) The chord diagram describing significant ligand-receptor pairs involved all signals sending to iLSEP (p < 0.05; one-sided permutation test without adjustment). The ligand and receptor names that overlap with intercellular communication in fetal liver are highlighted. e) The chord diagram describing significant ligand-receptor pairs involved all signals sending to fetal liver endothelial cell (p < 0.05; one-sided permutation test without adjustment).
Extended Data Fig. 9 Evaluation of iLSEP-specific siRNA knockdown.
a) qRT-PCR analysis of WNT2, VEGFA, RSPO3, and F8. Data represent the mean ± SEM (n = 4 biological replicates; *, p < 0.05; multiple two-sided t-test with controlling FDR). In the “+Control,” siRNAs of non-targeted negative controls were used. b-c) Scatter dot-plot of the flow cytometry analysis of CD34/CD32 and CD73/CXCR4 expression in CD34+CD43- iLSEC with siRNA treatment at day14. d) Observation of fluorescent reporters with bright-field image to track GFP+ iLSEP and AlexaFlour555 (AF555)-tagged siRNA embedded in the same layer of multilayered gel. Scale bar indicates 500 µm. For at least 9 days, AF555-tagged siRNA remained in the packed gel or inside the organoids. e) Scatter dot-plot of the flow cytometry analysis of whole HLBO cells for separation of GFP−tagged iLSEP in second layer and GFP−negative cells in first layer in HLBO by FACSAria Fusion cell sorter. f) qRT-PCR analysis of WNT2 to validate GFP-tagged iLSEP specific knockdown efficiency. WNT2 mRNA expression levels were determined relative to the negative control siRNA treated for each GFP+ and GFP- fraction isolated. Data represent the mean ± SEM (n = 8, GFP+ groups; n = 7, GFP- in +Control; n = 6, GFP- in +siWNT2; all n are biological replicates; ***, p < 0.001; multiple two-sided t-test with Holm-Sidak correction). g) Cross-sectional H&E staining image of HLBOs with siRNA treatment. h) Measurement of VEGF-A in siRNA-treated HLBO supernatants at day3. Data represent the mean ± SEM (n = 4 biological replicates; ND, not detected). i) Whole-mount image of siRNA-treated HLBOs with immunofluorescent staining for CD31, STAB1 and LYVE1. j) Quantification of the percentage of cells expressing CD31 or LYVE1 alone or co-expressing CD31 and LYVE1. Data represent the mean ± SEM (n = 4 biological replicates; *, p < 0.05; **, p < 0.01; two-way ANOVA with Sidak’s multiple comparison test). k) Quantification of the percentage of STAB1 co-expressing endothelial cells (CD31+ or LYVE1+). Data represent the mean ± SEM (n = 4 biological replicates; ns, not significant; two-sided Welch’s t-test).
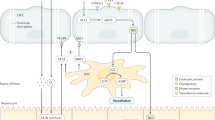
Extended Data Fig. 10 Assessment of coagulation factors in HLBOs and transplanted HLBO into FVIII knockout mice.
a) Bright-field image of organoids at day11. Scale bar indicates 500 µm. b) Cross-sectioning image of HLBOs with immunofluorescent staining for FVIII and GFP-tagged VA or iLSEP embedded in a second cell layer, and LYVE1 at day 11. c) FVIII antigen concentration in organoid supernatants at day 11. Data represent the mean ± SEM (n = 6, +VA HLBO; n = 5, +iLSEP HLBO; both n are biological replicates; ***, p < 0.001; two-sided Welch’s t-test). d) FVIII activity of organoid supernatants at day 11 determined based on APTT mixing with FVIII deficient plasma. To assess quality of the secreted proteins, each relative activity was normalized by FVIII antigen level. Data represent the mean ± SEM (n = 5 biological replicates; **, p < 0.01; two-sided Welch’s t-test). e) Relative activity of coagulation factors (FV, FVIII, FIX, and FXI) in HLBO supernatants at day 11, determined based on APTT mixing with corresponding factor deficient plasma. Data represent the mean ± SEM (n = 3 biological replicates; *, p < 0.05; ***, p < 0.001; multiple two-sided t-test with controlling FDR). f) Time-series specific activity of FVIII in HLBO supernatants at day 4 and 11. Data represent the mean ± SEM (n = 3 biological replicates; *, p < 0.05; **, p < 0.01; ***, p < 0.001; two-way ANOVA with uncorrected Fisher’s LSD test. g) Specific activity of coagulation factors (FV, FIX, and FXI) in HLBO supernatants at day 11 and NPP, calculated by activity (%) / protein level (%) (both parameters relative to NPP). Data represent the mean ± SEM (n = 3 biological replicates; ns, not significant; multiple two-sided t-test with controlling FDR q-value). h) The schema of FVIII gene editing to establish the immunodeficient NS-IL2rg KO/ FVIII KO hemi (NSI-FVIII KO) mice. i) Cross-sectioning image of HLBO graft and mice liver tissue with immunofluorescent staining for human FVIII, LYVE1, CD32b, and human cell marker (Ku80 or human nuclear antigen). Right figure for each group is enlarged single channel images of the orange dashed square region in left figure. j) Measurement of HLBO derived human coagulation factors circulating in mouse blood 8 weeks after liver transplantation (LTx) (n = 5 biological replicates; ****, q < 0.0001; multiple two-sided t-test with controlling FDR (q)). k) Actual APTTs reflecting the activity of FVIII secreted from transplanted HLBO 2 months after transplantation compared with NPP and PBS. Eash sample was diluted to the same FVIII antigen level with inhibitor (+) or (-) FVIII deficient plasma. Data represent the mean ± SEM (n = 3 biological replicates; ***, p < 0.001; ****, p < 0.0001; one-way ANOVA with Sidak’s multiple comparisons test between inhibitor (+) and (-)). l) FVIII antigen concentration in culture supernatants on day 7 after re-seeding (iLSEC) or self-organization (iLSEC in HLBO). Values were normalized by the number of LSEC. Data represent the mean ± SEM (n = 6, iLSEC; n = 8, iLSEC in HLBO; both n are biological replicates; ****, p < 0.0001; two-sided Welch’s t-test). m) The blood trickled from tail-tip in saline solution during 20 min bleeding test. n) Total blood loss during 20 min bleeding evaluated by hemoglobin concentration. Data represent the mean ± SEM (n = 3 biological replicates; **, p < 0.01; two-sided Welch’s t-test).
Supplementary information
Supplementary Information
Supplementary Video legend, Table legend, Methods and Table 8.
Supplementary Table 1
Supplementary Table 1: Significant marker statistics for each group in integrated single-cell transcriptome of endothelial cells in adult liver, fetal liver, yolk sac and fetal heart (q < 0.05), related to Extended Data Fig. 2a–c.
Supplementary Table 2
Supplementary Table 2: Top 100 marker gene list of all endothelial groups in integrated single-cell transcriptome, related to Extended Data Fig. 2a–c.
Supplementary Table 3
Supplementary Table 3: The list of genes in modules that changed in trajectory to LSEC zones 2 and 3, related to Extended Data Fig. 2d,e.
Supplementary Table 4
Supplementary Table 4: The detailed list of significantly enriched pathways on dynamics of gene module along the trajectory to LSEC zones 2 and 3, related to Extended Data Fig. 2e.
Supplementary Table 5
Supplementary Table 5: Significant marker statistics for each group in integrated single-cell transcriptome of +iLSEP HLBO days 3, 7 and 11 (qval_adj < 0.05 and logfold_change > 2), related to Fig. 2a.
Supplementary Table 6
Supplementary Table 6: Description of collected data for construction of a dataset integrated with published human liver organoids, related to Extended Data Fig. 5.
Supplementary Table 7
Supplementary Table 7: Significant marker statistics for each endothelial subpopulation in HLBO (Padj < 0.05), related to Fig. 2b.
Supplementary Video 1
Supplementary Video 1: Whole-mount 3D reconstructed video of +iLSEP HLBO with immunofluorescent staining for CD32b (green) and ASGR1 (red) with DAPI (blue) at day 11, related to Fig. 1c.
Supplementary Video 2
Supplementary Video 2: Time-lapse video of 70 or 2,000 kDa tetramethylrhodamine-dextran visualized-blood flow in GFP+ organoid-derived human vessels for 60 min, related to Fig. 1i.
Supplementary Video 3
Supplementary Video 3: In vivo haemophilia A correction by the HLBO supernatant injection, related to Fig. 5i,j.
Supplementary Video 4
Supplementary Video 4: In vivo haemophilia A correction 8 weeks after orthotopic transplantation of HLBO, related to Fig. 6d–f.
Source data
Source Data Figs. 1 and 3–6 and Extended Data Figs 2, 7, 9 and 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saiki, N., Nio, Y., Yoneyama, Y. et al. Self-organization of sinusoidal vessels in pluripotent stem cell-derived human liver bud organoids. Nat. Biomed. Eng (2025). https://doi.org/10.1038/s41551-025-01416-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41551-025-01416-6