Abstract
De novo pyrimidine biosynthesis is achieved by cytosolic carbamoyl-phosphate synthetase II, aspartate transcarbamylase and dihydroorotase (CAD) and uridine 5′-monophosphate synthase (UMPS), and mitochondrial dihydroorotate dehydrogenase (DHODH). However, how these enzymes are orchestrated remains enigmatical. Here we show that cytosolic glutamate oxaloacetate transaminase 1 clusters with CAD and UMPS, and this complex then connects with DHODH, which is mediated by the mitochondrial outer membrane protein voltage-dependent anion-selective channel protein 3. Therefore, these proteins form a multi-enzyme complex, named ‘pyrimidinosome’, involving AMP-activated protein kinase (AMPK) as a regulator. Activated AMPK dissociates from the complex to enhance pyrimidinosome assembly but inactivated UMPS, which promotes DHODH-mediated ferroptosis defence. Meanwhile, cancer cells with lower expression of AMPK are more reliant on pyrimidinosome-mediated UMP biosynthesis and more vulnerable to its inhibition. Our findings reveal the role of pyrimidinosome in regulating pyrimidine flux and ferroptosis, and suggest a pharmaceutical strategy of targeting pyrimidinosome in cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository48,49 with the dataset identifier PXD041017. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Further information on the research design is available in the Nature Research Reporting Summary linked to this article.
References
Yamaoka, T. et al. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. J. Biol. Chem. 272, 17719–17725 (1997).
Evans, D. R. & Guy, H. I. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J. Biol. Chem. 279, 33035–33038 (2004).
Moffatt, B. A. & Ashihara, H. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book 1, e0018 (2002).
An, S., Kumar, R., Sheets, E. D. & Benkovic, S. J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106 (2008).
French, J. B. et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science 351, 733–737 (2016).
Kyoung, M., Russell, S. J., Kohnhorst, C. L., Esemoto, N. N. & An, S. Dynamic architecture of the purinosome involved in human de novo purine biosynthesis. Biochemistry 54, 870–880 (2015).
Robitaille, A. M. et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339, 1320–1323 (2013).
Ben-Sahra, I., Howell, J. J., Asara, J. M. & Manning, B. D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328 (2013).
Lane, A. N. & Fan, T. W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 43, 2466–2485 (2015).
Jones, M. E. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 49, 253–279 (1980).
Zhu, J. & Thompson, C. B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 20, 436–450 (2019).
Mayers, J. R. & Vander Heiden, M. G. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem. Sci. 40, 130–140 (2015).
Melendez-Rodriguez, F. et al. HIF1α suppresses tumor cell proliferation through inhibition of aspartate biosynthesis. Cell Rep. 26, 2257–2265.e4 (2019).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Sullivan, L. B. et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015).
Garcia-Bermudez, J. et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 20, 775–781 (2018).
Sullivan, L. B. et al. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 20, 782–788 (2018).
Alkan, H. F. et al. Cytosolic aspartate availability determines cell survival when glutamine is limiting. Cell Metab. 28, 706–720.e6 (2018).
Kremer, D. M. et al. GOT1 inhibition promotes pancreatic cancer cell death by ferroptosis. Nat. Commun. 12, 4860 (2021).
Liu, M. et al. Inhibiting both proline biosynthesis and lipogenesis synergistically suppresses tumor growth. J. Exp. Med. 217, e20191226 (2020).
Bajzikova, M. et al. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 29, 399–416.e10 (2019).
Wang, Y. et al. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat. Commun. 10, 201 (2019).
Sullivan, J. E. et al. Inhibition of lipolysis and lipogenesis in isolated rat adipocytes with AICAR, a cell-permeable activator of AMP-activated protein kinase. FEBS Lett. 353, 33–36 (1994).
Cool, B. et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 3, 403–416 (2006).
Myers, R. W. et al. Systemic pan-AMPK activator MK-8722 improves glucose homeostasis but induces cardiac hypertrophy. Science 357, 507–511 (2017).
Voss, C. M. et al. AMPK activation affects glutamate metabolism in astrocytes. Neurochem. Res. 40, 2431–2442 (2015).
Schaffer, B. E. et al. Identification of AMPK phosphorylation sites reveals a network of proteins involved in cell invasion and facilitates large-scale substrate prediction. Cell Metab. 22, 907–921 (2015).
Hardie, D. G., Schaffer, B. E. & Brunet, A. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 26, 190–201 (2016).
Steinberg, G. R. & Hardie, D. G. New insights into activation and function of the AMPK. Nat. Rev. Mol. Cell Biol. 24, 255–272 (2022).
Mao, C. et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 593, 586–590 (2021).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Peters, G. J. et al. In vivo inhibition of the pyrimidine de novo enzyme dihydroorotic acid dehydrogenase by brequinar sodium (DUP-785; NSC 368390) in mice and patients. Cancer Res. 50, 4644–4649 (1990).
Natale, R. et al. Multicenter phase II trial of brequinar sodium in patients with advanced melanoma. Ann. Oncol. 3, 659–660 (1992).
Urba, S. et al. Multicenter phase II trial of brequinar sodium in patients with advanced squamous-cell carcinoma of the head and neck. Cancer Chemother. Pharmacol. 31, 167–169 (1992).
Maroun, J. et al. Multicenter phase II study of brequinar sodium in patients with advanced lung cancer. Cancer Chemother. Pharmacol. 32, 64–66 (1993).
Degterev, A. et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1, 112–119 (2005).
Thornberry, N. A. et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356, 768–774 (1992).
Monteverde, T., Muthalagu, N., Port, J. & Murphy, D. J. Evidence of cancer-promoting roles for AMPK and related kinases. FEBS J. 282, 4658–4671 (2015).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Lee, H. et al. Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat. Cell Biol. 22, 225–234 (2020).
Cantor, J. R. et al. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell 169, 258–272.e17 (2017).
Zhao, H. et al. AMPK-mediated activation of MCU stimulates mitochondrial Ca2+ entry to promote mitotic progression. Nat. Cell Biol. 21, 476–486 (2019).
Wang, S. et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194 (2015).
Kory, N. et al. SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science https://doi.org/10.1126/science.aat9528 (2018).
Li, L. et al. ZNF516 suppresses EGFR by targeting the CtBP/LSD1/CoREST complex to chromatin. Nat. Commun. 8, 691 (2017).
Peng, M., Yin, N. & Li, M. O. SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437 (2017).
Yang, R. et al. Identification of purine biosynthesis as an NADH-sensing pathway to mediate energy stress. Nat. Commun. 13, 7031 (2022).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 50, D1522–D1527 (2022).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Acknowledgements
We thank Xiaohui Liu, X. Wang, Y. Wang and L. Xu (Metabolomics Facility at Tsinghua University Branch of China National Center for Protein Sciences, China) for technical help with metabolomics analysis, and Xueqing Liu from National Center for Protein Sciences (Beijing) for technical help with SEC. We thank L. Zou, L. Li and J. Deng from the Core Facilities of Basic Medical Sciences of Capital Medical University for their assistance with cell flow cytometry, MS and microscopic imaging. This work was supported by the National Natural Science Foundation of China (82030093 and 81972567 to B.L.; 82002958 to C.Y.) and Beijing Natural Science Foundation (5194023 to C.Y.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: B.L. and C.Y.; methodology: B.L., C.Y., Y.W., X.L, J.N. and Q.C.; investigation: C.Y., Y.Z. and L.W.; visualization: C.Y., Y.Z., L.W., L.M., Z.G. and R.Y.; funding acquisition: B.L. and C.Y.; project administration: B.L., Y.F. and C.Y.; supervision: B.L. and C.Y.; writing—original draft: B.L. and C.Y.; writing—review and editing: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks David Hardie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The contribution of GOT1 and GOT2 to pyrimidine biosynthesis.
a, A schematic to show the incorporation of 15N-amine-glutamine into pyrimidines. b, Immunoblotting analysis of GOT1 and GOT2 expression in HeLa/shScramble, HeLa/shGOT1, HeLa/shGOT2 cells. c-j, Relative levels (c) and fractional labeling of glutamine (d), glutamate (e), aspartate (f), carbamoyl aspartate (g), dihydroorotate (h) and orotate (i) and UMP (j) in HeLa/shScramble, HeLa/shGOT1 and HeLa/shGOT2 cells cultured with 1 mM 15N-amine-glutamine for 8 h. k–m, Labeled fractions of UMP (k) and relative intracellular levels of aspartate, glutamate (l), carbamoyl aspartate, dihydroorotate, orotate and UMP (m) in HeLa/shScramble and HeLa/shGOT1 cells cultured with 10 mM 13C4, 15N-aspartate for 8 h. (c–m) Values are shown as the mean ± SD, n = 3 biologically independent samples. P values were obtained using one-way ANOVA with Tukey test. (b) Data are verified in three replicates with similar results.
Extended Data Fig. 2 The enzymes involved in pyrimidine biosynthesis form the multi-enzyme complex.
a, HEK293T cells expressing Flag-CAD were subjected to IP analysis, followed by immunoblotting. Flag-vector was used as a negative control. b, IP analysis of the interaction between endogenous CAD, DHODH, UMPS and GOT1 in HEK293T cells with anti-UMPS antibody. Rabbit IgG was used as a negative control. c, HEK293T cells expressing Flag-UMPS were subjected to IP analysis, followed by immunoblotting. d, HEK293T cells expressing Flag-tagged truncations of GOT1 were subjected to IP analysis using anti-Flag M2 beads, followed by immunoblotting. N-terminal region, 1-214aa; C-terminal region, 215-413aa. e, HEK293T cells expressing truncations of UMPS were subjected to IP analysis using anti-Flag M2 beads, followed by immunoblotting. f, Schematic showing the interaction between CAD, GOT1 and UMPS to promote the incorporation of newly synthesized aspartate into pyrimidine. (a-e) Data are verified in three replicates with similar results.
Extended Data Fig. 3 DHODH is indispensable for the synthesis of orotate.
a, A schematic to show the metabolism of isotope-labeled glutamine via the reductive and oxidative pathways. b,c, Fractional labeling (b) and relative levels (c) of carbamoyl aspartate, dihydroorotate, orotate and UMP in WT and DHODH−/− Hela cells that were supplemented with 400 μM uridine and cultured with 1 mM 13C5-glutamine for 8 h before harvest. d, In vitro pull-down analysis of the interaction between GST-tagged proteins and His-VDAC3 purified from E. coli with GST protein as a negative control. e, HEK293T cells expressing Flag-tagged truncations of CAD were subjected to IP analysis using anti-Flag M2 beads, followed by immunoblotting. f, HEK293T cells expressing Flag-tagged truncations of DHODH were subjected to IP analysis using anti-Flag M2 beads, followed by immunoblotting. N-terminal region, 1-99aa; C-terminal region, 100-395aa. g, HeLa cells were homogenized and fractionated. The cytosolic and intact mitochondrial fractions were analyzed by immunoblotting. (b,c) Values are shown as the mean ± SD, n = 3 biologically independent samples. P values were obtained using unpaired two-tailed Student’s t-test. (d-g) Data are verified in three replicates with similar results.
Extended Data Fig. 4 Involvement of VDAC3 in the pyrimidine biosynthetic complex.
a, Fractional labeling of carbamoyl aspartate, dihydroorotate, orotate and UMP in pooled VDAC3 knockout and control HeLa cells cultured in medium containing 1 mM 13C5-glutamine for 8 h. b, Pooled VDAC3 knockout and control HeLa cells were cultured in various concentrations of uridine. Cell viability was measured after 3 days of treatment. The results were normalized to those of cells infected with control sgRNA without uridine supplementation. The expression of VDAC3 was detected by immunoblotting. c, d, Proliferation of control and CAD silencing (c) or control and DHODH silencing (d) HeLa cells cultured in medium containing uridine (200 μM) or vehicle. The expression of CAD or DHODH was detected by immunoblotting. e, Cell viability of control and GOT1 silencing HeLa cells cultured with or without uridine (400 μM) supplementation. The results were normalized to those of cells infected with scrambled shRNA. GOT1 expression was detected by immunoblotting. f, OCR analysis of Pooled VDAC3 knockout and control HeLa cells. VDAC3 expression was detected by immunoblotting. g, DHOA uptake into proteoliposomes containing VDAC3. Empty liposomes with glycerol were used as a control. h, A schematic indicating how the VDAC3 channel acts as a platform to integrate the pyrimidine biosynthetic complex. (a-g) Values are shown as the mean ± SD, n = 3 (a-e,g) or 4 (f) biologically independent samples. P values were obtained using one-way ANOVA with Tukey test (a), two-way ANOVA with Sidak test (b-d) and unpaired two-tailed Student’s t-test (e,g). (b-e,g) Western blots are verified in three replicates with similar results.
Extended Data Fig. 5 Dynamic formation of de novo pyrimidine biosynthetic complexes.
a, The chromatography elution profiles of the fractions were shown. b, Co-fractionation of pyrimidinosome by fast protein liquid chromatography with Superose 6 columns. The size-exclusion chromatography of whole cell lysates from HEK293T/shSramble and HEK293T/shCAD cells was analyzed by immunoblotting. The dashed box indicates pyrimidinosome-containing fractions. The elution positions of a series of protein standards with known molecular masses were indicated. c, Sucrose density gradient centrifugation analysis of the pyrimidinosome using the whole cell lysates from HEK293T cells. d,e, HEK293T cells expressing CAD were cultured under pyrimidine-depleted medium with or without uridine supplementation for 12 h and subjected to IP analysis. f,g, Immunofluorescence analysis of the co-localization of Flag-CAD and GFP-GOT1 (f) or mCherry/mCherry-UMPS and VDAC3-Flag (g) proteins in HeLa cells growing in pyrimidine-rich or pyrimidine-deprived medium. Flag-CAD was labeled in red (f) and VDAC3-Flag (g) was labeled in green. Scale bar, 10 μm. h,i, Co-localization of GFP-fusion proteins and mCherry-fusion proteins in HeLa cells cultured in pyrimidine-deprived medium. GFP, DHODH-GFP, or GFP-GOT1 was co-transfected with mCherry-UMPS, and DHODH-mCherry was co-transfected with GFP-GOT1 (i). Scale bar, 10 μm. Pearson’s correlation coefficients for co-localization of the indicated proteins under pyrimidine-deprived conditions in i (h). The clusters from different cells in each group were analyzed using the co-localization analysis tools in ZEN software. Data are shown as the mean ± SD, n = 41 (GFP/mCherry-UMPS), n = 36 (GOT1/mCherry-UMPS), n = 60 (DHODH/mCherry-UMPS) and n = 45 (GOT1/DHODH-mCherry) biologically independent clusters. P values were obtained using one-way ANOVA with Sidak test. j, Immunofluorescence analysis of the co-localization of DHODH-GFP and MitoTracker Deep Red in HeLa cells growing in pyrimidine-rich or pyrimidine-deprived medium. Scale bar, 10 μm. (b-g, i, j) Data are verified in three replicates with similar results.
Extended Data Fig. 6 Hypoxia promotes the accumulation of pyrimidine precursors.
a, Fractional labeling of carbamoyl aspartate, dihydroorotate, orotate, and UMP in Hela cells cultured in medium containing 1 mM 13C5-glutamine with or without uridine (400 μM) supplementation under normoxia or hypoxia for 8 h. b-d, Labeled fractions of carbamoyl aspartate, dihydroorotate, orotate and UMP (b), relative intracellular levels (c) and fractional labeling of newly synthesized carbamoyl aspartate, dihydroorotate, orotate and UMP (d) in HEK293T cells cultured under normoxia or hypoxia for 8 h with 1 mM 13C5-glutamine. e, Volcano plot of the differential metabolites between hypoxia group and normoxia group. HeLa cells were cultured under normoxia or hypoxia for 8 h. (a-d) Values are shown as the mean ± SD, n = 3 biologically independent samples. P values were obtained using one-way ANOVA with Tukey test (a) and unpaired two-tailed Student’s t-test (b-e).
Extended Data Fig. 7 AMPK associates with the multi-enzyme complex and regulates its formation.
a, Interaction between AMPK and ectopically expressed CAD. HEK293T cells were transfected with Flag-CAD or Flag-vector as a negative control. IP analysis was performed and followed by immunoblotting. b,c, HEK293T cells expressing Flag-tagged AMPKα1 (b) or AMPKα2 (c) were subjected to IP analysis, followed by immunoblotting. d, WT and AMPKα1/2−/− Hela cells expressing Flag-CAD or Flag-vector were subjected to IP analysis with anti-Flag antibody, followed by immunoblotting. e, Interaction between endogenous AMPKα and Flag-tagged truncations of CAD. f,g, In vitro pull-down analysis of the interaction between GST-tagged AMPKα1 and His-tagged proteins. GST protein was used as a negative control. h, Interaction between Flag-tagged UMPS immunoprecipitated from HEK293T cells and GST-tagged AMPKα1 protein purified from E. coli with GST protein as a negative control. GST-pull down analysis was performed, followed by immunoblotting. i, Interaction between Flag-tagged CAD immunoprecipitated from HEK293T cells and His-tagged VDAC3 protein purified from E. coli with Flag-tagged AMPKα1 as a negative control. Ni-pull down analysis was performed, followed by immunoblotting. j, HEK293T cells were cultured under normoxia or hypoxia for 8 h. The expression of indicated proteins was detected by immunoblotting. k, WT and AMPKα1/2−/− Hela cells expressing Flag-GOT1 or Flag-vector were subjected to IP analysis with anti-Flag antibody, followed by immunoblotting. l, A schematic depicting how AMPKα interacts and regulates the pyrimidinosome. (a-k) Data are verified in three replicates with similar results.
Extended Data Fig. 8 AMPKα regulates pyrimidine biosynthesis.
a, Fractional labeling of glutamine, glutamate, aspartate, carbamoyl aspartate, dihydroorotate, orotate and UMP in Hela cells cultured in medium containing 1 mM 13C5-glutamine for the indicated time periods with or without supplementation of A-769662 (200 μM) for 8 h. Values are shown as the mean ± SD, n = 3 biologically independent samples. P values were obtained using unpaired two-tailed Student’s t-test. b, The mass spectrometry results of phosphorylated peptides of Flag-UMPS. HEK293T cells expressing Flag-UMPS were treated with A-769662 (200 μM) for 8 h before immunoprecipitation. Mass spectrometry analysis of Flag-UMPS was performed to identify the phosphorylation sites.
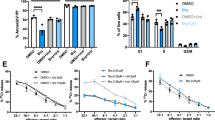
Extended Data Fig. 9 AMPK activation promotes the reliance of cancer cells on DHODH-mediated ferroptosis defence.
a, HeLa cells were treated with A-769662 (200 μM), AICAR (200 μM) or MK-8722 (5 μM) for 12 h. The expression of indicated proteins was detected by immunoblotting. b, HeLa cells were treated with brequinar (0.3 μM) or/and AICAR (40 μM)/A-769662 (200 μM)/MK-8722 (1 μM). Survival of the cells was measured after 3 days of treatment. c, Cell viability of HeLa cells treated with brequinar (0.3 μM) or/and AICAR (40 μM)/A-769662 (200 μM)/MK-8722 (1 μM) for 72 h following pretreatment with vehicle or cell death inhibitors for 24 h. Ferr-1 (10 μM), Z-VAD-FMK (10 μM), necrosulfonamide (2 μM). d,e, Cell viability of H1299 (d) and KYSE-150 (e) cells treated with brequinar (0.3 μM) or/and AICAR (40 μM)/A769662 (200 μM)/MK-8722 (1 μM) for 72 h following pretreatment with vehicle or Ferr-1 (10 μM) for 24 h. f, The body weights of mice with the indicated treatments at the endpoint of experiments. n = 5 (vehicle), n = 5 (AICAR), n = 5 (brequinar), n = 5 (AICAR and brequinar) or n = 6 (vehicle), n = 6 (MK-8722), n = 6 (brequinar), n = 6 (MK-8722 and brequinar). g, HeLa cells were treated with the indicated concentrations of brequinar(BQR) or vidofludimus (Vido). Survival of the cells was measured after 2 days of treatment. h, WT and AMPKα1/2−/− HeLa cells were treated with 2.5 μM brequinar in combination with 0 or 400 μM uridine. Cell survival was measured after 3 days of treatment. (b-h) Values are shown as the mean ± SD, n = 3 (b-e,g,h) biologically independent samples or n = 5 or 6 biologically independent mice (f). P values were obtained using one-way ANOVA with Tukey test (b) and unpaired two-tailed Student’s t-test (c-e, g,h). (a) Data are verified in three replicates with similar results.
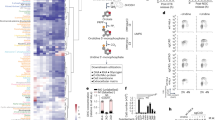
Extended Data Fig. 10 AMPKα1 expression levels in tissue microarrays.
a, b, Tissue microarray staining for AMPKα1 (a) and quantified data of IHC (b). I, liver cancer; II, lung cancer; III, cervical cancer; IV, esophageal cancer; V, colon cancer; VI, gastric cancer; VII, pancreatic cancer. Scale bar, 40 μm. Data are verified in two replicates with similar results(a). Error bars represent mean ± SD, n = 7 biologically independent samples (b). c, A model for the pyrimidinosome under different physiological conditions. Under normal conditions, GOT1, CAD, UMPS, VDAC3 and DHODH form the pyrimidinosome. AMPK interacts with the pyrimidinosome and restricts the association between the components of the complex and de novo synthesis of pyrimidine. When AMPK expression is low, the compactness of the pyrimidine biosynthetic complex is enhanced and de novo synthesis of pyrimidine is promoted. Upon energy stress, AMPK is activated and dissociates from the complex, and thus promoting the association between the components of the complex. Under such conditions, the pyrimidinosome enhances the biosynthesis of OA but not UMP, due to the suppression of UMPS by AMPK, which renders cells more resistant to ferroptosis.
Supplementary information
Supplementary Information
Supplementary Fig. 1. Figure exemplifying the gating strategy.
Supplementary Table 1
MS analysis of DHODH-interacting proteins in HEK293T cells expressing DHODH-Flag. HEK293T expressing Flag-Vector was included as a control.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, C., Zhao, Y., Wang, L. et al. De novo pyrimidine biosynthetic complexes support cancer cell proliferation and ferroptosis defence. Nat Cell Biol 25, 836–847 (2023). https://doi.org/10.1038/s41556-023-01146-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-023-01146-4
This article is cited by
-
AMPK at the interface of nutrient sensing, metabolic flux and energy homeostasis
Nature Metabolism (2026)
-
Regulation of apoptosis, ferroptosis, and pyroptosis mediated by acetylation
Cell Death Discovery (2026)
-
Integrated analysis reveals an immune evasion prognostic signature for predicting the overall survival in patients with hepatocellular carcinoma
Cancer Cell International (2025)
-
Uridine as a hub in cancer metabolism and RNA biology
Experimental & Molecular Medicine (2025)
-
Glycine decarboxylase advances IgA nephropathy by boosting mesangial cell proliferation through the pyrimidine pathway
EMBO Molecular Medicine (2025)