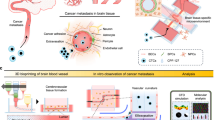
Abstract
In brain metastasis, cancer cells remain in close contact with the existing vasculature and can use vessels as migratory paths—a process known as vessel co-option. However, the mechanisms regulating this form of migration are poorly understood. Here we use ex vivo brain slices and an organotypic in vitro model for vessel co-option to show that cancer cell invasion along brain vasculature is driven by the difference in stiffness between vessels and the brain parenchyma. Imaging analysis indicated that cells move along the basal surface of vessels by adhering to the basement membrane extracellular matrix. We further show that vessel co-option is enhanced by both the stiffness of brain vasculature, which reinforces focal adhesions through a talin-dependent mechanism, and the softness of the surrounding environment that permits cellular movement. Our work reveals a mechanosensing mechanism that guides cell migration in response to the tissue’s intrinsic mechanical heterogeneity, with implications in cancer invasion and metastasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
Code availability
Code use in this article can be made available upon request to the corresponding author.
References
Paul, C. D., Mistriotis, P. & Konstantopoulos, K. Cancer cell motility: lessons from migration in confined spaces. Nat. Rev. Cancer 17, 131–140 (2017).
Riching, K. M. et al. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys. J. 107, 2546–2558 (2014).
Fraley, S. I. et al. Three-dimensional matrix fiber alignment modulates cell migration and MT1-MMP utility by spatially and temporally directing protrusions. Sci. Rep. 5, 14580 (2015).
Provenzano, P. P. et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med 4, 38 (2006).
Alexander, S., Koehl, G. E., Hirschberg, M., Geissler, E. K. & Friedl, P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem. Cell Biol. 130, 1147–1154 (2008).
Marcadis, A. R. et al. Rapid cancer cell perineural invasion utilizes amoeboid migration. Proc. Natl Acad. Sci. USA 120, e2210735120 (2023).
Weigelin, B., Bakker, G.-J. & Friedl, P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. Intravital 1, 32–43 (2012).
Beunk, L., Brown, K., Nagtegaal, I., Friedl, P. & Wolf, K. Cancer invasion into musculature: mechanics, molecules and implications. Semin. Cell Dev. Biol. 93, 36–45 (2019).
Bentolila, L. A. et al. Imaging of angiotropism/vascular co-option in a murine model of brain melanoma: implications for melanoma progression along extravascular pathways. Sci. Rep. 6, 23834 (2016).
Carbonell, W. S., Ansorge, O., Sibson, N. & Muschel, R. The vascular basement membrane as ‘soil’ in brain metastasis. PLoS ONE 4, e5857 (2009).
Er, E. E. et al. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat. Cell Biol. 20, 966–978 (2018).
Farin, A. et al. Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia 53, 799–808 (2006).
Fornabaio, G. et al. Angiotropism and extravascular migratory metastasis in cutaneous and uveal melanoma progression in a zebrafish model. Sci. Rep. 8, 10448 (2018).
Gritsenko, P. G. & Friedl, P. Adaptive adhesion systems mediate glioma cell invasion in complex environments. J. Cell Sci. 131, jcs216382 (2018).
Hirata, E. et al. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J. Cell Sci. 125, 858–868 (2012).
Kienast, Y. et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122 (2010).
Winkler, F. et al. Imaging glioma cell invasion in vivo reveals mechanisms of dissemination and peritumoral angiogenesis. Glia 57, 1306–1315 (2009).
Valiente, M. et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014).
Charras, G. & Sahai, E. Physical influences of the extracellular environment on cell migration. Nat. Rev. Mol. Cell Biol. 15, 813–824 (2014).
Mekhdjian, A. H. et al. Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol. Biol. Cell 28, 1467–1488 (2017).
Kechagia, J. Z., Ivaska, J. & Roca-Cusachs, P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20, 457–473 (2019).
Zimmermann, D. R. & Dours-Zimmermann, M. T. Extracellular matrix of the central nervous system: from neglect to challenge. Histochem. Cell Biol. 130, 635–653 (2008).
Barnes, J. M., Przybyla, L. & Weaver, V. M. Tissue mechanics regulate brain development, homeostasis and disease. J. Cell Sci. 130, 71–82 (2017).
Kim, Y. & Kumar, S. CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol. Cancer Res. 12, 1416–1429 (2014).
Mooney, K. L. et al. The role of CD44 in glioblastoma multiforme. J. Clin. Neurosci. 34, 1–5 (2016).
Safarians, G. et al. Glioblastoma spheroid invasion through soft, brain-like matrices depends on hyaluronic acid–CD44 interactions. Adv. Health. Mater. 12, 2203143 (2023).
Wolf, K. J. et al. A mode of cell adhesion and migration facilitated by CD44-dependent microtentacles. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1914294117 (2020).
Anderson, S. M., Kelly, M. & Odde, D. J. Glioblastoma cells use an integrin- and CD44-mediated motor-clutch mode of migration in brain tissue. Cell. Mol. Bioeng. 17, 121–135 (2024).
Bergonzini, C., Kroese, K., Zweemer, A. J. M. & Danen, E. H. J. Targeting integrins for cancer therapy—disappointments and opportunities. Front. Cell Dev. Biol. 10, 863850 (2022).
Raab-Westphal, S., Marshall, J. F. & Goodman, S. L. Integrins as therapeutic targets: successes and cancers. Cancers 9, 110 (2017).
Haeger, A. et al. Collective cancer invasion forms an integrin-dependent radioresistant niche. J. Exp. Med. 217, e20181184 (2019).
Bui, T. et al. Functional redundancy between β1 and β3 integrin in activating the IR/Akt/mTORC1 signaling axis to promote ErbB2-driven breast cancer. Cell Rep. 29, 589–602.e6 (2019).
Polacheck, W. J., Kutys, M. L., Tefft, J. B. & Chen, C. S. Microfabricated blood vessels for modeling the vascular transport barrier. Nat. Protoc. 14, 1425–1454 (2019).
Di Stasi, A. et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 365, 1673–1683 (2011).
Goult, B. T., Yan, J. & Schwartz, M. A. Talin as a mechanosensitive signaling hub. J. Cell Biol. 217, 3776–3784 (2018).
Elosegui-Artola, A. et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016).
Isomursu, A. et al. Directed cell migration towards softer environments. Nat. Mater. 21, 1081–1090 (2022).
Austen, K. et al. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 (2015).
Folkman, J. Tumor angiogenesis: therapeutic implications. N. Engl. J. Med. 285, 1182–1186 (1971).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Weis, S. M. & Cheresh, D. A. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat. Med. 17, 1359–1370 (2011).
Kuczynski, E. A., Vermeulen, P. B., Pezzella, F., Kerbel, R. S. & Reynolds, A. R. Vessel co-option in cancer. Nat. Rev. Clin. Oncol. 16, 469–493 (2019).
Dai, J. et al. Astrocytic laminin-211 drives disseminated breast tumor cell dormancy in brain. Nat. Cancer 3, 25–42 (2022).
Griveau, A. et al. A glial signature and Wnt7 signaling regulate glioma-vascular interactions and tumor microenvironment. Cancer Cell 33, 874–889.e7 (2018).
Voutouri, C. et al. Experimental and computational analyses reveal dynamics of tumor vessel cooption and optimal treatment strategies. Proc. Natl Acad. Sci. USA 116, 2662–2671 (2019).
Stoletov, K. et al. Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci. 126, 904–913 (2013).
Slack, R. J., Macdonald, S. J. F., Roper, J. A., Jenkins, R. G. & Hatley, R. J. D. Emerging therapeutic opportunities for integrin inhibitors. Nat. Rev. Drug Discov. 21, 60–78 (2022).
Barbazán, J. et al. Liver metastasis is facilitated by the adherence of circulating tumor cells to vascular fibronectin deposits. Cancer Res. 77, 3431–3441 (2017).
Beaty, B. T. et al. Talin regulates moesin–NHE-1 recruitment to invadopodia and promotes mammary tumor metastasis. J. Cell Biol. 205, 737–751 (2014).
Sakamoto, S., McCann, R. O., Dhir, R. & Kyprianou, N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 70, 1885–1895 (2010).
Budday, S., Ovaert, T. C., Holzapfel, G. A., Steinmann, P. & Kuhl, E. Fifty shades of brain: a review on the mechanical testing and modeling of brain tissue. Arch. Comput. Methods Eng. 27, 1187–1230 (2020).
Reuten, R. et al. Basement membrane stiffness determines metastases formation. Nat. Mater. 20, 892–903 (2021).
Bangasser, B. L. et al. Shifting the optimal stiffness for cell migration. Nat. Commun. 8, 15313 (2017).
Osmani, N. et al. Metastatic tumor cells exploit their adhesion repertoire to counteract shear forces during intravascular arrest. Cell Rep. 28, 2491–2500.e5 (2019).
Follain, G. et al. Hemodynamic forces tune the arrest, adhesion, and extravasation of circulating tumor cells. Dev. Cell 45, 33–52.e12 (2018).
Shellard, A. & Mayor, R. Durotaxis: the hard path from in vitro to in vivo. Dev. Cell 56, 227–239 (2021).
Chang, J., Pang, E. M., Adebowale, K., Wisdom, K. M. & Chaudhuri, O. Increased stiffness inhibits invadopodia formation and cell migration in 3D. Biophys. J. 119, 726–736 (2020).
Wang, C., Tong, X. & Yang, F. Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using PEG-based hydrogels. Mol. Pharm. 11, 2115–2125 (2014).
Singh, S. P., Schwartz, M. P., Lee, J. Y., Fairbanks, B. D. & Anseth, K. S. A peptide functionalized poly (ethylene glycol) (PEG) hydrogel for investigating the influence of biochemical and biophysical matrix properties on tumor cell migration. Biomater. Sci. 2, 1024–1034 (2014).
Debnath, J. et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111, 29–40 (2002).
Mishra, A. et al. Imaging pericytes and capillary diameter in brain slices and isolated retinae. Nat. Protoc. 9, 323–336 (2014).
Segel, M. et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature 573, 130–134 (2019).
Qi, L. et al. Talin2-mediated traction force drives matrix degradation and cell invasion. J. Cell Sci. 129, 3661–3674 (2016).
Ershov, D. et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022).
Miyawaki, T. et al. Visualization and molecular characterization of whole-brain vascular networks with capillary resolution. Nat. Commun. 11, 1104 (2020).
Elosegui-Artola, A. et al. Matrix viscoelasticity controls spatiotemporal tissue organization. Nat. Mater. 22, 117–127 (2023).
Acknowledgements
We thank L. Kelleher for technical assistance; R. Alani for providing 1205Lu, B16-F10 and WM983B cells; H. T. Nia for providing E0771 cells; the Memorial Sloan Kettering Cancer Center for providing the MDA cell lines; J. L. Teo, H.-S. Li and W. W. Wong for providing T cells; C. Stashko for his assistance during the AFM measurements; and J. A. White for giving us access to their vibratome. We also thank the Bio-Interface and Technology core facility at Boston University (used for nano-indentation) and the Koch Institute histology core (used for tissue cryosectioning). This work was supported by grants from the National Institutes of Health (NIH) (EB033821) and the National Science Foundation (NSF) Center for Engineering MechanoBiology (CMMI-1548571). M.U. was supported by an European Molecular Biology Organization long-term fellowship (EMBO ALTF811-2018), the Center for Multiscale and Translational Mechanobiology at Boston University and an American Heart Association postdoctoral fellowship (828475). A.E.S. was supported by an NSF Graduate Research Fellowship (1745302 and 2141064) and a Boston University Multicellular Design Program Kilachand Fellowship. B.P.S. was supported by the NIH National Institute of Biomedical Imaging and Bioengineering (no. T32 EB016652). R.O. was supported by an NIH R35 (CA242447-01). C.R.R. was supported by an NSF Graduate Research Fellowship (2021324226). J.B.T. was supported by the NIH through the Translational Research in Biomaterials Training Program (no. T32 EB006359). M.T.N. was supported by an NIH NRSA F32 postdoctoral fellowship (NIH NHLBI 1F32HL165691). J.E. acknowledges support from the NIH (R21 EB28491). A.E.A. acknowledges the support from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 851055). A.E.A. is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (CC2214), the UK Medical Research Council (CC2214) and the Wellcome Trust (CC2214).
Author information
Authors and Affiliations
Contributions
M.U. and C.S.C. conceived the study and designed the experiments. M.U., A.E.S., B.P.S., O.C., R.O., C.R.R., L.L, M.T.N., J.B.T. and J.Y. performed the experiments. M.U. and O.C. analysed the data. R.O. and V.M.W. oversaw the AFM data collection. O.C. and A.E.A. designed alginate gel experiments. C.R.R. and X.H. performed and oversaw brain injections. J.E. contributed to data interpretation. All authors discussed results. M.U., J.E. and C.S.C. wrote the manuscript with feedback from all authors. C.S.C. oversaw the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Steven D. Chang, Gregory Longmore and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
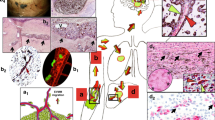
Extended Data Fig. 1 Characterization of cell shape and invasion depth in ex vivo brain cultures.
(a) Percentage of 1205Lu cells adhered to vessels after 24h of ex vivo culture (n=7 slices, 4 experiments). Graph shows mean ± standard deviation. (b) Brain slice immunostaining of laminin (magenta), lifeact-GFP (green) and nuclei (hoechst, blue) of a 1205Lu cell spreading over the outer surface of a vessel. 3D view,11µm-thick image stack. Scale bar 10µm. (c) Left: single image plane of the cell in (b), including isolectin-IB4 labelling of endothelial cells (grey). Scale bar 10µm. Right: laminin (magenta), lifeact-GFP (green) and isolectin-IB4 (grey) fluorescence profile along the dashed yellow line. (d) Example of cell invasion depth quantification. Left: immunostaining of laminin (magenta) and lifeact-GFP (green). Yellow dashed lines outline a cell in the bulk (cell 1) and a cell spreading along a vessel (cell 2). Scale bar 50µm. Right: region 2 (centred on the cell of interest) is used to average laminin intensity across a 60µm stack and define the reference point for invasion depth (0µm). Because of the opacity of brain tissue, averaged laminin intensity shows a peak at the top of the slice and decays as imaging deeper into the tissue. The origin of invasion depths is defined as \({z}_{{peak}\_{region}2}-6\mu m\). Region1 (that follows the cell 2D shape), is used to average both cell and laminin fluorescence. The intensity of the GFP signal is used to determine the cell depth within the slice, and local laminin intensity is used to determine if a cell is in the bulk or on a vessel (if observing a local peak of laminin intensity). (e) Violin plots of 1205Lu cell shape circularities when adhered to vessels or to the bulk brain tissue 4 and 24h after seeding. (f) Violin plots of 1205Lu invasion depths within the slice when adhered to vessels or to the bulk brain tissue 4 and 24h after seeding. For e-f, n=562 for Bulk, n=621 for Vessel at 4h, n=337 for Bulk, n=706 for Vessel at 24h, from 6 slices, 3 experiments. Graphs show median ± interquartile range. Dots represent the median values for each slice analysed. Statistical analysis: Kruskal–Wallis test followed by a post hoc Dunn’s test.
Extended Data Fig. 2 Vessel co-option is a common pattern of invasion across many different cell lines.
a-c, Mean invasion depth per slice for cells adhered to vessels or to the bulk brain tissue for 1205Lu (n=7), WM983B (n=4), U2OS (n=4), MCF7 (n=6) and MDA-MB-231 (n=6) (a), A375P (n=8) and A375M2 (n=6) (b), nHDF (n=7), MCF10A (n=5) and HBVPC (n=5) cell lines (c). Full distributions shown in Fig. 1d, j–l. Data corresponds to at least 3 experiments. Statistical analysis: two-sided paired T-test or an ordinary one-way ANOVA test followed by a post hoc Tukey’s test (for b). All graphs show mean ± standard deviation. (d) Violin plots of cell shape circularities for MDA-MB-231 TGL, BrM2-831, BoM-1833 and LM2-4175 cells adhered to the vasculature. (e) Violin plots of bulk invasion depths for MDA-MB-231 TGL, BrM2-831, BoM-1833 and LM2-4174 cells. (f) Violin plots of vessel invasion depths for MDA-MB-231 TGL, BrM2-831, BoM-1833 and LM2-4175 cells. (g) Violin plots of cell shape circularities for cells adhered to vessels or to the bulk brain tissue for B16-F10, E077 and NIH-3T3 mouse-derived cell lines. (h) Violin plots of invasion depths for cells adhered to vessels or to the bulk brain tissue for B16-F10, E0771 and NIH-3T3 mouse-derived cell lines. For MDA-MB-231 TGL (parental cell line): n=394 for Bulk, n=441 for Vessel, from 5 slices, 3 experiments. For BrM2-831 (brain tropic): n=652 for Bulk, n=479 for Vessel, from 6 slices, 3 experiments. For BoM-1833 (bone tropic): n=520 for Bulk, n=496 for Vessel, from 6 slices, 3 experiments. For LM2-4175 (lung-tropic): n=564 for Bulk, n=529 for Vessel, from 6 slices, 3 experiments. For B16-F10 (melanoma): n=239 for Bulk, n=212 for Vessel, from 4 slices, 3 experiments. For E0771 (breast cancer): n=269 for Bulk, n=359 for Vessel, from 6 slices, 3 experiments. For NIH-3T3 (embryonic fibroblast): n=165 for Bulk, n=530 for Vessel, from 6 slices, 3 experiments. (i) Brain slice immunostaining of laminin (magenta) and T-cells labelled with CellTracker-488 (green). Left: z-plane 2µm below the top surface. Right: z-plane 50µm below the top surface. White insert labels zoomed area. Scale bars: 100µm, 10µm (inserts). (j) Violin plots of T-cell shape circularities when adhered to vessels or to the bulk brain tissue. (k) Violin plots of T-cell invasion depths within the slice when adhered to vessels or to the bulk brain tissue. For T-cells: n=380 for Bulk, n=208 for Vessel, from 3 slices, 2 experiments. For all violin plots, dots represent the median value for each slice analysed. Statistical analysis: two-sided Mann–Whitney or a Kruskal–Wallis test followed by a post hoc Dunn’s test (for d-f). Graphs show median ± interquartile range.
Extended Data Fig. 3 Depletion of CD44, integrin β1, zyxin or paxillin does not significantly impair invasion into brain slices.
(a) Percentage of scramble control and CD44 KO cells adhered to vessels after 24h of ex vivo culture (n=8 slices, 4 experiments). (b) Left: representative western blot of CD44 expression in scramble control and CD44 KO cells. Right: quantification of all western blots (one per experiment, 4 experiments). (c) Violin plots of invasion depths for scramble control and CD44 KO cells moving through the bulk brain tissue (Scr: n=571, CD44 KO: n=708, 8 slices, 4 experiments). (d) Violin plots of invasion depths for scramble control and CD44 KO cells moving along the vasculature (Scr: n=652, CD44 KO: n=511, 8 slices, 4 experiments). (e) Percentage of scramble control and integrin β1 KO cells adhered to vessels after 24h of ex vivo culture (n=7 slices, 4 experiments). (f) Left: representative western blot of integrin β1 expression in scramble control and integrin β1 KO cells. Right: quantification of all western blots (one per experiment, 4 experiments). (g) Violin plots of invasion depths for scramble control and integrin β1 KO cells moving through the bulk brain tissue (Scr: n=500, Itgβ1 KO: n=607, 7 slices, 4 experiments). (h) Violin plots of invasion depths for scramble control and integrin β1 KO cells moving along the vasculature (Scr: n=720, Itgβ1 KO: n=354, 7 slices, 4 experiments). i-u, Adaptor proteins. The zyxin CRISPR KO cell lines were established as clonal cell lines by single cell sorting after infection with the CRISPR lentivirus, and two cell lines were analysed (data shown in Fig. 2 corresponds to clone 1). (i) Percentage of scramble control, zyxin clone-1 KO and paxillin KO cells adhered to vessels after 24h of ex vivo culture (Scr: n=7 slices, Zyx KO c1: 8 slices, Pxn KO: 8 slices, 3 experiments). (j) Percentage of scramble control and zyxin clone-2 KO cells adhered to vessels after 24h of ex vivo culture (n=6 slices, 3 experiments). (k) Left: representative western blot of zyxin expression in scramble control and zyxin clone-1 KO cells. Right: quantification of all western blots (one per experiment, 4 experiments). (l) Left: representative western blot of paxillin expression in scramble control and paxillin KO cells. Right: quantification of all western blots (one per experiment, 4 experiments). (m) Left: representative western blot of zyxin expression in scramble control and zyxin clone-2 KO cells. Right: quantification of all western blots (one per experiment, 3 experiments). (n) Violin plots of invasion depths for scramble control, zyxin clone-1 KO and paxillin KO cells moving through the bulk brain tissue (Scr: n=467, 7 slices; Zyx KO c1: n=353, 8 slices; Pxn KO: n=660, 8 slices, 4 experiments). (o) Violin plots of invasion depths for scramble control and zyxin clone-2 KO cells moving through the bulk brain tissue (Scr: n=288 for Scr, Zyx KO c2: n=303, 6 slices, 3 experiments). (p) Violin plots of invasion depths for scramble control, zyxin clone-1 KO and paxillin KO cells moving along the vasculature (Scr: n=593, 7 slices; Zyx KO c1: n=704, 8 slices; Pxn KO: n=597, 8 slices, 4 experiments). (q) Violin plots of invasion depths for scramble control and zyxin clone-2 KO cells moving along the vasculature (Scr: n=275, Zyx KO c2: n=481, 6 slices, 3 experiments). r-u, FAK inhibition with FC11. (r) Percentage of cells adhered to vessels in control conditions or under 1µM FC11 after 24h of ex vivo culture (n=6 slices, 3 experiments). (s) Violin plots of cell shape circularities for cells adhered to the vasculature in control conditions or under 1µM FC11 (Control: n=631, FC11 1µM: n=642, 6 slices, 3 experiments). (t) Violin plots of invasion depths for cells moving through the bulk brain tissue in control conditions or under 1µM FC11 (Control: n=466, FC11 1µM: n=289, 6 slices, 3 experiments). (u) Violin plots of invasion depths for cells moving along the vasculature in control conditions or under 1µM FC11 (Control: n=631, FC11 1µM: n=642, 6 slices, 3 experiments). For all violin plots, dots represent the median values per slice analysed. Statistical analysis: two-sided Mann–Whitney or a Kruskal–Wallis test followed by a post hoc Dunn’s test (for n,p). Graphs show median ± interquartile range. For all other graphs, statistical analysis was performed using a two-sided T-test or an ordinary one-way ANOVA test followed by a post hoc Tukey’s test (for i). Graphs show mean ± standard deviation. For western blot quantification, protein expression was normalized to loading control (β-actin), and to the expression on the scramble control condition.
Extended Data Fig. 4 Depletion of talin significantly impairs invasion into brain slices.
a-e, Talin 1 KO. The talin 1 CRISPR KO cell lines were established as clonal cell lines by single cell sorting after infection with the CRISPR lentivirus, and two cell lines were analysed (data shown in Fig. 2 corresponds to clone 1). (a) Percentage of scramble control, talin 1 clone-1 KO and talin 1 clone-2 KO cells adhered to vessels after 24h of ex vivo culture (Scr: n=7, Tln1 KO: n=8, 4 experiments). (b) Left: representative western blot of talin 1 and talin 2 expression in scramble control, talin 1 clone-1 KO and talin 1 clone-2 KO cells. Right: quantification of all western blots (one per experiment, 4 experiments). (c) Violin plots of cell shape circularities for scramble control, talin 1 clone-1 KO and talin 1 clone-2 KO cells adhered to the vasculature (Scr: n=826, 7 slices; Tln1 KO c1: n=725, 8 slices; Tln1 KO c2: n=741, 8 slices; 4 experiments). (d) Violin plots of invasion depths for scramble control, talin 1 clone-1 KO and talin 1 clone-2 KO cells moving through the bulk brain tissue (Scr: n=425, 7 slices; Tln1 KO c1: n=521, 8 slices; Tln1 KO c2: n=721, 8 slices; 4 experiments). (e) Violin plots of invasion depths for scramble control, talin 1 clone-1 KO and talin 1 clone-2 KO cells moving along the vasculature (Scr: n=826, 7 slices; Tln1 KO c1: n=725, 8 slices; Tln1 KO c2: n=741, 8 slices; 4 experiments). f-i, Talin1+2 KO. (f) Percentage of scramble control, talin 2 KO and talin 1 and 2 KO cells adhered to vessels after 24h of ex vivo culture (n=8 slices, 4 experiments). (g) Left: representative western blot of talin 1 and talin 2 expression in scramble control, talin 2 KO and talin 1 and 2 KO cells. Right: quantification of all western blots (one per experiment, 4 experiments). (h) Violin plots of invasion depths for scramble control, talin 2 KO and talin 1 and 2 KO cells moving through the bulk brain tissue (Scr: n=560, Tln2 KO: n=509, Tln1+Tln2 KO: n=515, 8 slices, 4 experiments). (i) Violin plots of invasion depths for scramble control, talin 2 KO and talin 1 and 2 KO cells moving along the vasculature (Scr: n=717, Tln2 KO: n=640, Tln1+Tln2 KO: n=544, 8 slices, 4 experiments). For all violin plots, dots represent the median values per slice analysed. Statistical analysis: Kruskal–Wallis test followed by a post hoc Dunn’s test. Graphs show median ± interquartile range. For all other graphs, statistical analysis was performed using a one-way ANOVA test followed by a post hoc Tukey’s test. Graphs show mean ± standard deviation. For western blot quantification, protein expression was normalized to loading control (β-actin), and to the expression on the scramble control condition.
Extended Data Fig. 5 Roughness ratio calculation.
(a) 1205Lu lifeact-GFP image (grey). From left to right, representative images of a scramble control and a talin1 and 2 KO tumour one week after brain injection. Overlayed, the tumour edge (green) and boundary (magenta) that were used to calculate the roughness ratio as (boundary length)/(edge length). Scalebar 500µm.
Extended Data Fig. 6 Modelling vessel co-option in vitro.
(a) Device immunostaining of HMBEC lifeact-Ruby (magenta), 1205Lu lifeact-GFP (green) and nuclei (hoechst, blue). From left to right, representative images of a scramble control, a talin 1 KO, a talin 2 KO and a talin 1 and 2 KO cell adhering to the micro-vessel. Maximum projection, scale bar 20µm. (b) Cell sphericity of scramble control, talin 1 KO, talin 2 KO and talin 1 and 2 KO cells adhered to the micro-vessel (Scr: n=53, 7 devices; Tln1 KO: n=25, 6 devices; Tln2 KO: n=42, 6 devices; Tln1+Tln2 KO: n=29, 8 devices; 3 experiments). Lighter dots represent each cell value and darker dots represent the median value per device. Statistical analysis: Kruskal–Wallis test followed by a post hoc Dunn’s test. Graphs show median ± interquartile range. (c) Cell velocity of scramble control and talin 1 and 2 KO cells moving along the micro-vessel (Scr: n=47, 7 devices; Tln1+Tln2 KO: n=42, 8 devices; 3 experiments). Lighter dots represent each cell value and darker dots represent the mean value per device. Statistical analysis: two-sided T-test. Graph show mean ± standard deviation. d-e, Laminin intensity (d) and cell sphericity (e) of 1205Lu cells spreading on control devices or in devices where the apoptosis of endothelial cells was triggered with the small molecule CID (Control: n=26, 7 devices; CID 5nM: n=118, 18 devices; 4 experiments). Lighter dots represent each cell value and darker dots represent the median value per device. Statistical analysis: two-sided Mann–Whitney test. Graphs show median ± interquartile range.
Extended Data Fig. 7 Depletion of vinculin significantly impairs invasion into brain slices.
The vinculin CRISPR KO cell lines were established as clonal cell lines by single cell sorting after infection with the CRISPR lentivirus, and two cell lines were analysed (data shown in Fig. 5 corresponds to clone 2). (a) Percentage of scramble control, vinculin clone-1 KO and vinculin clone-2 KO cells adhered to vessels after 24h of ex vivo culture (n=5, 3 experiments). Statistical analysis: one-way ANOVA test followed by a post hoc Tukey’s test. Graphs show mean ± standard deviation. (b) Left: representative western blot of vinculin expression in scramble control, vinculin clone-1 KO and vinculin clone-2 KO cells. Right: quantification of all western blots (one per experiment, 3 experiments). (c) Violin plots of cell shape circularities for scramble control, vinculin clone-1 KO and vinculin clone-2 KO cells adhered to the vasculature (Scr: n=411, Vcl KO c1: n=453, Vcl KO c2: n=474, 5 slices, 3 experiments). (d) Violin plots of invasion depths for scramble control, vinculin clone-1 KO and vinculin clone-2 KO cells moving through the bulk brain tissue (Scr: n=242, Vcl KO c1: n=385, Vcl KO c2: n=411, 5 slices, 3 experiments). (e) Violin plots of invasion depths for scramble control, vinculin clone-1 KO and vinculin clone-2 KO cells moving along the vasculature (Scr: n=411, Vcl KO c1: n=453, Vcl KO c2: n=474, 5 slices, 3 experiments). For all violin plots, dots represent the median values per slice analysed. Statistical analysis: Kruskal–Wallis test followed by a post hoc Dunn’s test. Graphs show median ± interquartile range. For western blot quantification, protein expression was normalized to loading control (β-actin), and to the expression on the scramble control condition.
Extended Data Fig. 8 Tuning the stiffness of the vasculature by filling it with a stiff gel or by inhibiting contractility.
a-b, Brain stiffness characterization. Young’s modulus (a) and force-indentation curves (b) of fresh 700µm-thick brain slices and 20µm-thick cryosections (Fresh: n=120, Cryo: n=120, 6 slices, 3 experiments). Statistical analysis: two-sided Mann–Whitney test. Lighter dots represent each indentation value and darker dots represent the median value per experiment. Graphs show median ± interquartile range. (c) Force-indentation AFM curves for vessel and bulk locations in 20µm-thick cryosections (n=40 pairs of bulk and vessel measurements from 4 brains). Graph shows median ± interquartile range. (d) Left: representative western blot of myosin-IIa expression in scramble control and myosin-IIa KO endothelial cells. Right: quantification of all western blots (one per experiment, 3 experiments). Graph shows mean ± standard deviation. (e) Force-indentation curves for scramble control and myosin-IIa KO endothelial cells (Scr: n=73, Myh9: n=60, 3 experiments). Graph shows median ± interquartile range. (f) Young’s modulus of the collagen-HA gel mixture used in the microfluidic devices (n=81, 9 gels, 3 experiments). Lighter dots represent each indentation value and darker dots represent the median value per experiment. Graph shows median ± interquartile range. g-l, Dextran perfusions. (g) Proton NMR of dextran methacrylate, 70% degree of functionalization (calculated based on a previously published procedure, see methods). (h) Mean Young’s modulus of perfused dextran gels for both soft and stiff gel formulations (n=3 perfusions). For each perfusion, a gel sample was kept for nanoindentation. Graph shows mean ± standard error of the mean. (i) Young’s modulus of vessels in brain cryosections from perfusions in (h) measured with AFM (Soft: n=25, Stiff: n=21, 3 brains). Statistical analysis: two-sided T-test. Graph shows mean ± standard deviation. (j) Percentage of cells adhered to vessels after 24h of ex vivo culture in control, soft and stiff dextran brain slices (Control: n=11, Soft: n=11, Stiff: n=10, 3 experiments). Statistical analysis: one-way ANOVA test followed by a post hoc Tukey’s test. Graphs show mean ± standard deviation. (k) Violin plots of invasion depths for cells moving through the bulk brain tissue after 24h of ex vivo culture in control, soft and stiff dextran brain slices (Control: n=517, 11 slices; Soft: n=727, 11 slices; Stiff: n=741, 10 slices; 3 experiments). (l) Violin plots of invasion depths for cells moving along the vasculature after 24h of ex vivo culture in control, soft and stiff dextran brain slices (Control: n=1349, 11 slices; Soft: n=1278, 11 slices; Stiff: n=1050, 10 slices, 3 experiments). (m) Mean Young’s modulus of the stiff alginate gel formulation in (n) (n=3 experiments). Graph shows mean ± standard error of the mean. (n) Violin plots of cell velocities for cells migrating between alginate gels (Soft-Stiff: n=450, Stiff–Stiff: n=428, 4 experiments). The soft gel formulation was the same as in Fig. 5. For all violin plots, dots represent the median values per slice (k,l) or experiment (n). Statistical analysis: two-sided Mann–Whitney test or a Kruskal–Wallis test followed by a post hoc Dunn’s test for multiple comparisons in k,l. Graphs show median ± interquartile range. For western blot quantification, protein expression was normalized to loading control (β-actin), and to the expression on the scramble control condition.
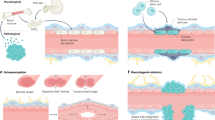
Extended Data Fig. 9 Proposed model of vessel co-option.
The differential stiffness between vessels and parenchyma promotes vessel co-option though a talin-dependent mechanism.
Supplementary information
Supplementary Table 1
CRISPR guides.
Supplementary Video 1
Supplementary Video 1: 1205Lu cell migration in brain slices. The migration of 1205Lu Lifeact–GFP (green) cells on a brain slice is shown. The vessels had been labelled with isolecin-IB4 (magenta) before imaging. The white arrows highlight examples of cells in the bulk tissue. The video is representative of n = 3 independent experiments (maximum projection of a 16 µm stack). Scale bar, 100 µm.
Supplementary Video 2
Supplementary Video 2: control and talin 1- and talin 2-KO tumours. The image stack (42 µm) of a brain slice immunostaining of laminin (magenta) and 1205Lu LifeAct–GFP (green) is shown. The control (left) and talin 1- and talin 2-KO tumours (right) 1 week after brain injection are shown.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1 and Table 1
Statistical source data.
Source Data Extended Data Fig. 2 and Table 2
Statistical source data.
Source Data Extended Data Fig. 3 and Table 3
Statistical source data.
Source Data Extended Data Fig. 3 and Table 3
Unprocessed western blots.
Source Data Extended Data Fig. 4 and Table 4
Statistical source data.
Source Data Extended Data Fig. 4 and Table 4
Unprocessed western blots.
Source Data Extended Data Fig. 6 and Table 6
Statistical source data.
Source Data Extended Data Fig. 7 and Table 7
Statistical source data.
Source Data Extended Data Fig. 7 and Table 7
Unprocessed western blots.
Source Data Extended Data Fig. 8 and Table 8
Statistical source data.
Source Data Extended Data Fig. 8 and Table 8
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Uroz, M., Stoddard, A.E., Sutherland, B.P. et al. Differential stiffness between brain vasculature and parenchyma promotes metastatic infiltration through vessel co-option. Nat Cell Biol 26, 2144–2153 (2024). https://doi.org/10.1038/s41556-024-01532-6
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41556-024-01532-6