Abstract
Pattern recognition receptors (PRRs) expressed in antigen-presenting cells are thought to shape pathogen-specific immunity by inducing secretion of costimulatory cytokines during T-cell activation, yet data to support this notion in vivo are scarce. Here, we show that the cytosolic PRR Nod-like Receptor CARD 4 (NLRC4) suppresses, rather than facilitates, effector and memory CD4+ T-cell responses against Salmonella in mice. NLRC4 negatively regulates immunological memory by preventing delayed activation of the cytosolic PRR NLR pyrin domain 3 (NLRP3) that would otherwise amplify the production of cytokines important for the generation of Th1 immunity such as intereukin-18. Consistent with a role for NLRC4 in memory immunity, primary challenge with Salmonella expressing flagellin modified to largely evade NLRC4 recognition notably increases protection against lethal rechallenge. This finding suggests flagellin modification to reduce NLRC4 activation enhances protective immunity, which could have important implications for vaccine development against flagellated microbial pathogens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data used to generate the figures presented in this work are available in Cambridge Research Repository Apollo with the identifier https://doi.org/10.17863/CAM.41575. Source data are provided with this paper.
References
Takeda, K. & Akira, S. Toll-like receptors. Curr. Protoc. Immunol. 109, 14.12.1–14.12.10 (2015).
Gross, O., Thomas, C. J., Guarda, G. & Tschopp, J. The inflammasome: an integrated view. Immunol. Rev. 243, 136–151 (2011).
Iwasaki, A. & Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 (2015).
Evavold, C. L. & Kagan, J. C. How inflammasomes inform adaptive immunity. J. Mol. Biol. 430, 217–237 (2018).
McSorley, S. J., Cookson, B. T. & Jenkins, M. K. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 164, 986–993 (2000).
Miao, E. A. et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1 beta via Ipaf. Nat. Immunol. 7, 569–575 (2006).
Franchi, L. et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1 beta in salmonella-infected macrophages. Nat. Immunol. 7, 576–582 (2006).
Miao, E. A. et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107, 3076–3080 (2010).
Lightfield, K. L. et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9, 1171–1178 (2008).
Zhao, Y. et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011).
Kofoed, E. M. & Vance, R. E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 (2002).
Broz, P. & Dixit, V. M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016).
Ding, J. & Shao, F. Growing a gasdermin pore in membranes of pyroptotic cells. EMBO J. 37, e100067 (2018).
Man, S. M. et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl Acad. Sci. USA 111, 7403–7408 (2014).
Qu, Y. et al. NLRP3 recruitment by NLRC4 during Salmonella infection. J. Exp. Med. 213, 877–885 (2016).
Garlanda, C., Dinarello, C. A. & Mantovani, A. The interleukin-1 family: back to the future. Immunity 39, 1003–1018 (2013).
O’donnell, H. et al. Toll-like receptor and inflammasome signals converge to amplify the innate bactericidal capacity of T helper 1 cells. Immunity 40, 213–224 (2014).
Kupz, A. et al. NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8(+) T cells. Nat. Immunol. 13, 162–169 (2012).
Hess, J., Ladel, C., Miko, D. & Kaufmann, S. H. E. Salmonella typhimurium aroA– infection in gene-targeted immunodeficient mice - major role of CD4(+) TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J. Immunol. 156, 3321–3326 (1996).
Nauciel, C. Role of Cd4+ T-cells and T-independent mechanisms in acquired-resistance to Salmonella-Typhimurium infection. J. Immunol. 145, 1265–1269 (1990).
Broz, P. et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207, 1745–1755 (2010).
Lara-Tejero, M. et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J. Exp. Med. 203, 1407–1412 (2006).
Mastroeni, P., Simmons, C., Fowler, R., Hormaeche, C. E. & Dougan, G. Igh-6-/- (B-cell-deficient) mice fail to mount solid acquired resistance to oral challenge with virulent Salmonella enterica serovar Typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect. Immun. 68, 46–53 (2000).
Heng, T. S. P., Painter, M. W. & Project, I. G. The immunological genome project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Bruchard, M. et al. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat. Immunol. 16, 859–870 (2015).
Mariathasan, S. et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004).
Tsutsui, H., Matsui, K., Okamura, H. & Nakanishi, K. Pathophysiological roles of interleukin-18 in inflammatory liver diseases. Immunol. Rev. 174, 192–209 (2000).
Mastroeni, P., Villarreal-Ramos, B. & Hormaeche, C. E. Role of T cells, TNF alpha and IFN gamma in recall of immunity to oral challenge with virulent salmonellae in mice vaccinated with live attenuated Aro-Salmonella. Vaccines. Micro. Pathogenesis 13, 477–491 (1992).
Kupz, A. et al. Contribution of Thy1+ NK cells to protective IFN-gamma production during Salmonella typhimurium infections. Proc. Natl Acad. Sci. USA 110, 2252–2257 (2013).
Letran, S. E. et al. TLR5-deficient mice lack basal inflammatory and metabolic defects but exhibit impaired CD4 T cell responses to a flagellated pathogen. J. Immunol. 186, 5406–5412 (2011).
Yang, J. et al. Flagellins of Salmonella Typhi and nonpathogenic Escherichia coli are differentially recognized through the NLRC4 pathway in macrophages. J. Innate Immun. 6, 47–57 (2014).
Amer, A. et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281, 35217–35223 (2006).
Franchi, L. et al. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur. J. Immunol. 37, 3030–3039 (2007).
Suzuki, T. et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 3, e111 (2007).
Sauer, J. D. et al. Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc. Natl Acad. Sci. USA 108, 12419–12424 (2011).
Yoshimoto, T. et al. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. J. Immunol. 161, 3400–3407 (1998).
Neighbors, M. et al. A critical role for interleukin 18 in primary and memory effector responses to Listeria monocytogenes that extends beyond its effects on Interferon gamma production. J. Exp. Med. 194, 343–354 (2001).
Maxwell, J. R. et al. IL-18 bridges innate and adaptive immunity through IFN-gamma and the CD134 pathway. J. Immunol. 177, 234–245 (2006).
Pepper, M. & Jenkins, M. K. Origins of CD4+ effector and central memory T cells. Nat. Immunol. 12, 467–471 (2011).
Ben-Sasson, S. Z. et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl Acad. Sci. USA 106, 7119–7124 (2009).
Sokolovska, A. et al. Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nat. Immunol. 14, 543–553 (2013).
Mastroeni, P. et al. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192, 237–247 (2000).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Mo, E., Peters, S. E., Willers, C., Maskell, D. J. & Charles, I. G. Single, double and triple mutants of Salmonella enterica serovar Typhimurium degP (htrA), degQ (hhoA) and degS (hhoB) have diverse phenotypes on exposure to elevated temperature and their growth in vivo is attenuated to different extents. Micro. Pathog. 41, 174–182 (2006).
Man, S. M. et al. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1 beta production. J. Immunol. 191, 5239–5246 (2013).
Harrison, J. A., Villarreal-Ramos, B., Mastroeni, P., DeHormaeche, R. D. & Hormaeche, C. E. Correlates of protection induced by live Aro(-) Salmonella Typhimurium vaccines in the murine typhoid model. Immunology 90, 618–625 (1997).
Dehormaeche, R. D., Jessop, H. & Bundell, C. Antibodies to the C epitope of Neisseria gonorrhoeae are present in patients with gonorrhea and absent in normal sera. J. Gen. Microbiol. 134, 1289–1297 (1988).
Acknowledgements
This work was supported by grants from Biotechnology and Biological Sciences Research Council (BBSRC) (nos. BB/H003916/1 and BB/K006436/1), Zoetis UK (no. BB/K006436/1) and Wellcome Trust (no. 108045/Z/15/Z) to C.E.B. A.S.B was supported by a Wellcome Trust 4-year PhD studentship. We thank K. A. Fitzgerald and D. Golenbock for suppling the gene-deficient mouse strains, S. M. Man for critical review of the manuscript and helpful discussions, and A. Cooke and members of her group, especially P. Zaccone and S. Newland, for sharing their expertise with P.T. and helpful discussions.
Author information
Authors and Affiliations
Contributions
P.T. designed, performed and analysed all in vivo, ex vivo and in vitro experiments. P.M., J.A.W., L.J.H., A.S.B. and S.J.W performed part of some in vivo experiments. O.J.B. performed the bacterial motility assays. J.A.W supervised all bacterial mutagenesis work. A.S.B. performed all bacterial mutagenesis work. P.M., D.J.M. and C.E.B. conceived the study and secured the funding. C.E.B. and P.T. supervised the study. P.T., J.A.W. and C.E.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 NLRC4 is required to restrict microbial spread during an oral sub-lethal but not an oral lethal infection with S. Typhimurium.
a, Wild-type and nlrc4−/− mice were challenged orally with 4.4 × 109 CFU S. Typhimurium M525P to establish a sub-lethal infection and microbial burden was determined in the spleen and liver at 5 days (n = 6 for wild-type and Nlrc4−/−) and 14 days (n = 7 for wild-type and n = 6 for Nlrc4−/−) post-challenge. b, Wild type and Nlrc4−/− mice were challenged orally 8 × 108 CFU S. Typhimurium SL1344 to establish a lethal infection and microbial burden was determined in the spleen and liver at 4 days post-challenge. Each symbol represents one mouse and horizontal lines delineate the mean. Statistical significance was calculated by 2-way ANOVA followed by Sidak’s multiple comparisons test for a and two-tailed Mann Whitney test for b. Data are representative of two independent experiments.
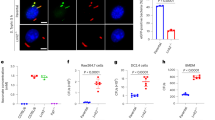
Extended Data Fig. 2 The effect of infection dose on the potency of ex vivo CD4+ T cell-mediated responses.
Wild-type mice were challenged with either 1.58 × 105 CFU, 1.72 × 104 CFU or 1.53 × 103 CFU S. Typhimurium M525P. Mice were euthanised 104-125 days after challenge and their splenic CD4+ T cells were stimulated with medium only (w/o stim.), anti-mouse CD3e (anti-CD3e) or whole bacterial cell extract (extract). a, IFN-γ measured by ELISA in the cell culture supernatant after 72 hours. b, IL-2 measured by ELISA in the cell culture supernatant after 24 hours. Data are shown as mean ± s.e.m. Statistical significance was calculated by a one-way ANOVA. Data were generated from one experiment.
Extended Data Fig. 3 Flow cytometric analysis reveals that CD4+ T cells secreting IFN-γ ex vivo have the profile of effector memory, rather than central memory, CD4+ T cells.
CD4+ T cells isolated from the spleen of wild-type and Nlrc4−/− mice 100 days after primary challenge with 1.24 × 104 CFU S. Typhimurium M525P were stimulated ex vivo with whole bacterial cell extract and phenotypically characterised via flow cytometry. Viable cells were first selected using a fixable viability dye (FVD) and single cells only were further included in the analysis based on their forward scatter area vs. forward scatter height. Single viable cells expressing CD4 were then separated to IFN-γ- and IFN-γ+ with both groups analysed for expression of CD44 and CD62L. For CD4+IFN-γ- cells, the phenotype CD44+CD62−represents effector T cells, while the phenotype CD44−CD62+ represents naïve T cells. For CD4+IFN-γ+ cells, the phenotype CD44+CD62−represents effector memory T cells, while the phenotype CD44+CD62+ represents central memory T cells. Percentages shown in the respective quadrants are mean values ± SD from N = 5 wild-type and N = 6 Nlrc4−/− mice. Data were generated from one experiment.
Extended Data Fig. 4 NLRC4 does not affect protection against lethal re-challenge.
Wild-type and Nlrc4−/− mice were challenged with either sterile PBS (naïve) or 1-1.15 × 104 CFU S. Typhimurium M525P and allowed to clear the primary infection for 90-106 days. They were then re-challenged orally with 1.37-7.2 × 107 CFU S. Typhimurium SL1344 to establish a lethal infection and inspected twice daily. They were euthanized upon detection of adverse signs and a humane end point curve was constructed for a, naïve mice and b, mice ‘immunised’ with Salmonella. Data has been pooled from 2 independent experiments. Statistical significance was calculated by a two-sided log-rank test.
Extended Data Fig. 5 Data variation between independent trials comparing CD4+ T cell-mediated responses in wild-type, Nlrc4-/-, Nlrp3-/- and Nlrp3-/-Nlrc4-/- mice is not statistically significant.
Levels of IFN-γ produced by CD4+ T cells stimulated ex vivo with whole bacterial cell extract were compared between wild-type mice used in three independent trials shown in Fig. 1c, Fig. 3a and Fig. 3b. Each symbol represents one mouse and horizontal lines delineate the mean. The data was tested for statistical significance by one-way ANOVA.
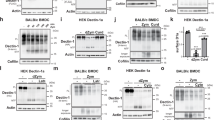
Extended Data Fig. 6 In the absence of NLRC4, Salmonella-infected macrophages do not undergo early cell death enabling them to survive longer and secrete IL−1β via activation of NLRP3.
Unprimed bone marrow derived macrophages (BMDMs) from wild-type, Nlrc4−/−, Nlrp3−/− and Nlrc4−/−Nlrp3−/− mice were infected with S. Typhimurium M525P in logarithmic growth at the MOI=50. a, Cytotoxicity was determined by measuring levels of LDH released in the cell culture supernatant. b, Levels of IL-1β were determined by ELISA in the cell culture supernatant. Data are shown as mean of triplicate wells ± s.e.m. Data are representative of two independent experiments.
Extended Data Fig. 7 Pairwise Sequence Alignment of FliC from E. coli K-12 MG1655 and S. Typhimurium M525P.
Comparison of the predicted FliC proteins from S. Typhimurium M525P and E. coli K-12 MG1655 by Clustal Omega (EMBL-EBI).
Extended Data Fig. 8 Salmonella expressing flagellin from non-pathogenic E. coli (M525P fliCMG1655) is motile but impaired at activating the NLRC4 inflammasome in macrophages.
a, Comparison of the motility diameter between S. Typhimurium M525P, M525P fliCMG1655 and SL1344. Data are expressed as % of the diameter achieved by the SL1344. b–d, Unprimed bone marrow derived macrophages from wild-type and Nlrc4−/− mice were infected for 1 h, 2 h, 6 h and 24 h with S. Typhimurium M525P or M525P fliCMG1655 in logarithmic growth. b, Cytotoxicity was determined by measuring levels of LDH released in the cell culture supernatant at an MOI of 50. c, Levels of IL-1β and d, TNF-α were determined by ELISA in the cell culture supernatant at an MOI of 1. Data has been pooled from 6 independent experiments and shown as mean ± s.e.m. in a. Data are representative of two independent experiments in b–d and shown as mean of triplicate wells ± s.e.m.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Tourlomousis, P., Wright, J.A., Bittante, A.S. et al. Modifying bacterial flagellin to evade Nod-like Receptor CARD 4 recognition enhances protective immunity against Salmonella. Nat Microbiol 5, 1588–1597 (2020). https://doi.org/10.1038/s41564-020-00801-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-020-00801-y
This article is cited by
-
Inflammasomes and pyroptosis in cancer: mechanisms and therapeutic advances
Journal of Hematology & Oncology (2025)
-
Immunology of gut microbiome and liver in non-alcoholic fatty liver disease (NAFLD): mechanisms, bacteria, and novel therapeutic targets
Archives of Microbiology (2024)
-
Inflammasome activation, NLRP3 engagement and macrophage recruitment to tumor microenvironment are all required for Salmonella antitumor effect
Cancer Immunology, Immunotherapy (2022)
-
Insights into inflammasome regulation: cellular, molecular, and pathogenic control of inflammasome activation
Immunologic Research (2022)
-
Inflammasomes and adaptive immune responses
Nature Immunology (2021)