Abstract
Filoviruses such as Ebola virus (EBOV) give rise to frequent epidemics with high case fatality rates while therapeutic options remain limited. Earlier genetic screens aimed to identify potential drug targets for EBOV relied on systems that may not fully recapitulate the virus life cycle. Here we applied an image-based genome-wide CRISPR screen to identify 998 host regulators of EBOV infection in 39,085,093 cells. A deep learning model associated each host factor with a distinct viral replication step. From this we confirmed UQCRB as a post-entry regulator of EBOV RNA replication and show that small-molecule UQCRB inhibition reduced virus infection in vitro. Using a random forest model, we found that perturbations on STRAP (a spliceosome-associated factor) disrupted the equilibrium between viral RNA and protein. STRAP was associated with VP35, a viral RNA processing protein. This genome-wide screen coupled with 12 secondary screens including validation experiments with Sudan and Marburg virus, presents a rich resource for host regulators of virus replication and potential targets for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw screen images and single-cell features are available in their entirety on Google Cloud Storage at https://console.cloud.google.com/storage/browser/opspublic-east1/EBOVOpticalPooledScreen. A curated subset of raw images from the genome-wide screen in HeLa cells (https://doi.org/10.7910/DVN/YHVWXY), and the targeted screens in HeLa and Huh7 cells at 16 h post infection (https://doi.org/10.7910/DVN/6FQNUA) and at 24 h post infection (https://doi.org/10.7910/DVN/W9WVHG) are available at Harvard’s Dataverse37,38,39. Additional tables are available on Zenodo at https://doi.org/10.5281/zenodo.14741479 (ref. 40). Source data are provided with this paper.
Code availability
Code is available on Zenodo at https://zenodo.org/records/15725070 (ref. 41).
References
Hoenen, T., Groseth, A. & Feldmann, H. Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Microbiol. 17, 593–606 (2019).
Hoenen, T. et al. Inclusion bodies are a site of Ebolavirus replication. J. Virol. 86, 11779–11788 (2012).
Nanbo, A., Watanabe, S., Halfmann, P. & Kawaoka, Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 3, 1206 (2013).
Bruchez, A. et al. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science 370, 241–247 (2020).
Carette, J. E. et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011).
Cheng, H. et al. A parallel genome-wide RNAi screening strategy to identify host proteins important for entry of Marburg virus and H5N1 influenza virus. Virol. J. 12, 194 (2015).
Martin, S. et al. A genome-wide siRNA screen identifies a druggable host pathway essential for the Ebola virus life cycle. Genome Med. 10, 58 (2018).
Feldman, D. et al. Optical pooled screens in human cells. Cell 179, 787–799.e17 (2019).
Carlson, R. J., Leiken, M. D., Guna, A., Hacohen, N. & Blainey, P. C. A genome-wide optical pooled screen reveals regulators of cellular antiviral responses. Proc. Natl Acad. Sci. USA 120, e2210623120 (2023).
Funk, L. et al. The phenotypic landscape of essential human genes. Cell 185, 4634–4653.e22 (2022).
Galão, R. P. et al. TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction. PLoS Pathog. 18, e1010530 (2022).
Hölzer, M. et al. Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells. Sci. Rep. 6, 34589 (2016).
Wynne, J. W. et al. Comparative transcriptomics highlights the role of the activator protein 1 transcription factor in the host response to Ebolavirus. J. Virol. 91, e01174-17 (2017).
Filone, C. M., Dower, K., Cowley, G. S., Hensley, L. E. & Connor, J. H. Probing the virus host interaction in high containment: an approach using pooled short hairpin RNA. Assay Drug Dev. Technol. 13, 34–43 (2015).
Flint, M. et al. A genome-wide CRISPR screen identifies N-acetylglucosamine-1-phosphate transferase as a potential antiviral target for Ebola virus. Nat. Commun. 10, 285 (2019).
Gong, M. et al. Genome-wide CRISPR/Cas9 screen identifies SLC39A9 and PIK3C3 as crucial entry factors for Ebola virus infection. PLoS Pathog. 20, e1012444 (2024).
Fang, J. et al. Functional interactomes of the Ebola virus polymerase identified by proximity proteomics in the context of viral replication. Cell Rep. 38, 110544 (2022).
Bray, M.-A. et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 11, 1757–1774 (2016).
Baldi, P. Autoencoders, unsupervised learning, and deep architectures. In Proc. ICML Workshop on Unsupervised and Transfer Learning 37–49 (JMLR, 2012).
Moon, K. R. et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482–1492 (2019).
Luthra, P. et al. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antiviral Res. 158, 288–302 (2018).
Batra, J. et al. Protein interaction mapping identifies RBBP6 as a negative regulator of Ebola virus replication. Cell 175, 1917–1930.e13 (2018).
Woolsey, C. et al. A VP35 mutant Ebola virus lacks virulence but can elicit protective immunity to wild-type virus challenge. Cell Rep. 28, 3032–3046.e6 (2019).
Jung, H. J. et al. Terpestacin inhibits tumor angiogenesis by targeting UQCRB of mitochondrial complex III and suppressing hypoxia-induced reactive oxygen species production and cellular oxygen sensing. J. Biol. Chem. 285, 11584–11595 (2010).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Söderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Replogle, J. M. et al. Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–2575.e28 (2022).
Gentili, M. et al. Classification and functional characterization of regulators of intracellular STING trafficking identified by genome-wide optical pooled screening. Cell Syst. 15, 1264–1277.e8 (2024).
Feldman, D. et al. Pooled genetic perturbation screens with image-based phenotypes. Nat. Protoc. 17, 476–512 (2022).
Ramezani, M. et al. A genome-wide atlas of human cell morphology. Nat. Methods 22, 621–633 (2025).
Köster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012).
Singh, S., Bray, M.‐A., Jones, T. R. & Carpenter, A. E. Pipeline for illumination correction of images for high-throughput microscopy. J. Microsc. 256, 231–236 (2014).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 39, btac757 (2023).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Proc. Part III 18 (pp. 234–241) (Springer International Publishing, 2015).
Carlson, R. Ebola virus optical pooled screen | genome-wide screen images. Harvard Dataverse 10.7910/DVN/YHVWXY (2025).
Carlson, R. Ebola virus optical pooled screen | secondary screen early timepoint (16 hours) images. Harvard Dataverse 10.7910/DVN/6FQNUA (2025).
Carlson, R. Ebola virus optical pooled screen | secondary screen late timepoint (24 hours) images. Harvard Dataverse 10.7910/DVN/W9WVHG (2025).
Carlson, R. Additional data for single-cell image-based screens identify host regulators of Ebola virus infection dynamics [Data set]. Zenodo https://doi.org/10.5281/zenodo.14741479 (2025).
Stefanakis, G. & Carlson R. beccajcarlson/EBOVOpticalPooledScreen: v01. Zenodo https://zenodo.org/records/15725070 (2025).
Carroll, S. A. et al. Molecular evolution of viruses of the family Filoviridae based on 97 whole-genome sequences. J. Virol. 87, 2608–2616 (2013).
Acknowledgements
We thank members of the Blainey, Davey, Uhler and Hacohen labs for critical feedback and discussions; C. Diaz and J. Bauman in the lab of J. T. Neal at the Broad Institute for assistance in developing custom antibody conjugations; and Y. Qin for assistance with data management. The HeLa cell line was used in this research. Henrietta Lacks, and the HeLa cell line that was established from her tumour cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We thank Lacks, now deceased, and the Lacks family for their contributions to biomedical research. This work was supported by the Broad Institute through startup funding (to P.C.B.) and the BN10 programme, and two grants from the National Human Genome Research Institute (HG009283 and RM HG006193). P.C.B. was supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. R.J.C. was supported by a Fannie and John Hertz Foundation Fellowship and an NSF Graduate Research Fellowship. C.F.B. was supported by R01AI148663 and P01AI120943. A.R. was supported by a George F. Carrier Postdoctoral Fellowship. A.R. and C.U. acknowledge support from the Eric and Wendy Schmidt Center at the Broad Institute, NCCIH/NIH (1DP2AT012345), and ONR (N00014-22-1-2116). G.K.A., D.W.L, C.F.B and R.A.D. are supported by NIH P01AI120943.
Author information
Authors and Affiliations
Contributions
R.J.C. and J.J.P. designed the approach with input from all authors. R.J.C, J.J.P., B.Y.S. and N.T. performed experiments. R.J.C. performed analysis aside from developing the deep learning model. A.R. and G.S. developed the deep learning methodology and designed the architecture for it with input from R.J.C., J.J.P. and C.U., and G.S. trained the model to obtain the single-cell embeddings. A.S. provided critical feedback and performed custom antibody conjugation. D.W.L. and K.C.F.S. developed the VP35 antibody. G.K.A., K.C.F.S., D.W.L., C.F.B., N.H., C.U., R.A.D. and P.C.B. supervised the research. R.J.C. and J.J.P. wrote the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
P.C.B. is a consultant to or holds equity in 10X Genomics, General Automation Lab Technologies/Isolation Bio, Next Gen Diagnostics, Cache DNA, Concerto Biosciences, Stately, Ramona Optics, Bifrost Biosystems, and Amber Bio. His laboratory received research funding from Calico Life Sciences, Merck, and Genentech for work related to genetic screening. N.H. holds equity in and advises Danger Bio/Related Sciences, owns equity in BioNtech and receives research funding from Bristol Myers Squibb. C.U. serves on the Scientific Advisory Board of Immunai, Relation Therapeutics, and Focal Biosciences, and receives research funding from AstraZeneca and Janssen Pharmaceuticals. The Broad Institute and MIT may seek to commercialize aspects of this work, and related applications for intellectual property have been filed. A.S. is an employee at Genentech and R.J.C. is an employee at Flagship Pioneering. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Stefan Bonn, Thomas Hoenen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
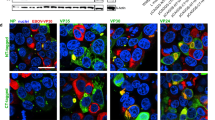
Extended Data Fig. 1 A genome-wide optical pooled screen identifies genes affecting VP35 protein, RNA, and c-Jun.
(a) Integration of optical pooled screening workflow with RNA FISH detection using HCR amplification. (b) Histograms of intensity features in five channels for non-targeting controls cells that were infected or not infected in the genome-wide optical pooled screen at 28 h. (c) Top 40 hits with increased or decreased VP35 protein levels and the number of non-Ebola virus genetic screens or Ebola-specific genetic screens they scored in. Genes not previously associated with Ebola in the literature are marked with an orange asterisk. (d) Gene set enrichment analysis of genes with significantly decreased (purple) or increased (gold) Ebola virus VP35 protein levels. (e) Volcano plot showing genes that scored significantly for changes in VP35 RNA levels by FISH. (f) Enrichr analysis of gene ontology terms significantly enriched in genes that reduced VP35 RNA levels. (g) Volcano plot showing genes that scored significantly for changes in c-Jun levels. (h) Enrichr analysis of gene ontology terms significantly enriched in genes that reduced or enhanced c-Jun levels.
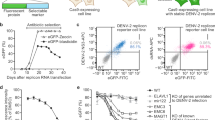
Extended Data Fig. 2 Quality control and filtering information for genome-wide optical pooled screen.
(a) Per-channel and per-plate mean intensities across 2562 fields of view for two out of eight plates in the genome-wide screen. (b) Same as (a) but post illumination-correction. (c) Same as (b) but after averaging across all channels for each plate and removing the bottom 10th percentile of each image. (d) Scatterplots of i and j coordinates for VP35 median intensity per-cell (top) and vimentin median intensity per-cell (bottom) for 10,000 randomly selected cells across the entire genome-wide screen post post-illumination correction. (e) Frequency of i and j coordinates for cells pre-filtering for duplicate phenotyping cells and cells at field of view edges and post-filtering. (f) Pre-normalization mean per-field of view FISH median intensity (plate I, well A1) pre-normalization and post-scaling to mean and unit variance based on features for non-targeting cells within -/+ the width of a field of view of each cell of interest. (g) Performance for the random forest classification of apoptotic, mitotic, and interphase cells trained on manual annotations.
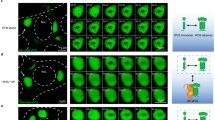
Extended Data Fig. 3 Additional unsupervised and fine-tuned autoencoder metrics.
(a) Fully unsupervised autoencoder reconstruction losses for training and test sets across 25 epochs. (b) Examples of manually labelled faint, punctate, cytoplasmic, and peripheral input cell images with accompanying unsupervised autoencoder reconstructions. (c) Fine-tuned autoencoder trained using negative log likelihood loss with balanced validation accuracy also reported across 50 epochs of training. (d) Best model train and test set accuracies for the VP35 protein localization prediction task using SVMs on latent embeddings from the unsupervised autoencoder, predefined features, a Resnet-50 architecture trained on the prediction task, or the fine-tuned autoencoder. Predefined features include intensity, correlation, and texture morphological features similar to those previously described for Cell Painting18. (e) Confusion matrix of model predictions vs manually labelled classifications on model test set. (f) Proportion of cells in each VP35 localization category for non-targeting controls and the genes with the largest proportion of faint (NPC1), punctate (UQCRB), and peripheral (ITGB1) cells. Error bars indicate SEM across sgRNAs targeting the same gene.
Extended Data Fig. 4 Clustering and dimensionality reduction identify information.
(a) Adjusted Rand score for Leiden clustering at different resolutions for 50 folds of 90% of the input data. Box plots here and in (d) indicate median (middle line), 25th, 75th percentile (box) and 1.5 times the IQR (whiskers) as well as outliers (single points). (b) Additional single-cell images of select genetic knockouts from the genome-wide optical pooled screen. (c) Correlation between the PHATE potential distance from the clustering using the fine-tuned model and the adjusted FDR p-value from the Kotliar study, noting genes whose expression significantly increased or decreased along with infection. (d) Correlation between the PHATE potential distance from the supervised clustering and the number of mass spectrometry studies that identified the genes as an interactor with an Ebola virus protein. (e) Venn diagram showing overlap between top optical pooled screen hits, genes that were present in at least one mass spectrometry study, and differentially expressed genes from Kotliar et al’s single-cell RNA sequencing study. (f) Correlation between the PHATE potential distance from the supervised clustering and the 95th percentile z-score for each gene in other virus genetic screens.
Extended Data Fig. 5 Summary of analytical approaches used in the manuscript.
(a) Workflow of analytical approaches used in this manuscript.
Extended Data Fig. 6. Additional secondary screen metrics.
(a) Fraction of non-significant secondary screen hits (p >=0.05) at varying thresholds of the genome-wide FDR-adjusted p-value for the HeLa late infection timepoint VP35 protein intensity. (b) Correlation between genome-wide c-Jun median nuclear delta AUC scores and secondary screen delta AUC scores; black lines indicate standard deviation for non-targeting control sgRNAs in each screen centred around the mean value for non-targeting sgRNAs in the screen. (c) Secondary screen mean viral protein (VP35 for EBOV and SUDV or VP40 for MARV) and RNA intensities in non-targeting control sgRNAs relative to HeLa cells infected with EBOV. (d) Volcano plots for VP35 (EBOV, SUDV) or VP40 (MARV) protein expression in each of the twelve screening conditions. (e) Volcano plot for viral VP35 RNA levels in HeLa cells at the late timepoint condition. (f) Heatmaps showing the difference between HeLa cell and Huh7 cell z-scored delta AUCs for members of the GARP, retromer, and the Sec61 complex. Hierarchical clustering performed using Euclidean distance. (g) Heatmap showing z-scored delta AUC values for genes identified as enriched for a punctate phenotype in the genome-wide screen and also included in secondary screens (white cells indicate conditions where p > 0.05). z-scored dAUC values for VP35 or VP40 protein were calculated on delta AUC values for all genes in each screen condition relative to means and standard deviations for non-targeting sgRNAs. White cells indicate conditions where p > 0.05 relative to non-targeting controls in the same condition. Hierarchical clustering performed using Pearson correlations.
Supplementary information
Supplementary Tables 1, 3–6
Supplementary Table 1. Per-gene mean cumulative ΔAUC scores and P values for VP35 protein, VP35 RNA FISH and c-Jun channels in genome-wide screen. Table 3. Per-gene mean deep learning predictions of VP35 subcellular protein localization and associated ordinal chi-square statistics and P values. Table 4. Summary of PHATE coordinates and gene cluster membership for supervised or unsupervised clustering, as well as the top 15 significant terms for each cluster. Table 5. Per-gene mean ΔAUC scores and P values for VP35 protein and VP35 RNA FISH channels in all secondary screen conditions. Table 6. Summary of key results across all multiple screens and analyses for all genes in the study.
Supplementary Table 2
Per-gene mean random forest regression results for VP35 RNA FISH and c-Jun predictions in genome-wide screen.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Carlson, R.J., Patten, J.J., Stefanakis, G. et al. Single-cell image-based screens identify host regulators of Ebola virus infection dynamics. Nat Microbiol 10, 1989–2002 (2025). https://doi.org/10.1038/s41564-025-02034-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-025-02034-3