Abstract
Candida auris is an emerging fungal pathogen notable for its intrinsically high resistance to fluconazole, the most prescribed antifungal drug. However, the genetic regulators underlying fluconazole susceptibility in C. auris remain unclear. Here we performed a pooled screen of piggyBac (PB) transposition mutants and identified significant enrichment of mitochondrial genes whose inactivation reduces fluconazole susceptibility. A genome-wide genetic interaction analysis of a mitochondrial gene deletion mutant, pet309Δ, suggests that the vacuolar calcium pump homologue CDT1 (Calcium and Drug Transporter 1) is responsible for its reduced fluconazole susceptibility. Fluconazole induces significant upregulation of CDT1 through the calcineurin signalling pathway. Cdt1, beyond its canonical calcium-pumping function, has evolved another function in mediating fluconazole efflux through its fluconazole-induced, calcineurin- and ATP hydrolysis-dependent plasma membrane localization. In addition, Cdt1 accelerates the evolution of fluconazole resistance or tolerance, and its transcript levels are substantially elevated across resistant clinical isolates. Our findings reveal a neofunctionalized role for Cdt1 in mediating fluconazole efflux in C. auris.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its supplementary files. The PB library sequencing data are available under BioProject accession no. PRJNA1273510. The RNA-seq data are deposited under Gene Expression Omnibus accession no. GSE300783. The whole-genome sequence data are available under BioProject accession no. PRJNA1278929. Source data are provided with this paper.
References
Calvo, B. et al. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J. Infect. 73, 369–374 (2016).
Lockhart, S. R. et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64, 134–140 (2017).
Satoh, K. et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53, 41–44 (2009).
Magobo, R. E., Corcoran, C., Seetharam, S. & Govender, N. P. Candida auris-associated candidemia, South Africa. Emerg. Infect. Dis. 20, 1250 (2014).
Chowdhary, A. et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur. J. Clin. Microbiol. Infect. Dis. 33, 919–926 (2014).
Ben-Ami, R. et al. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis. 23, 195 (2017).
Gaitán, A. C. R. et al. Nosocomial fungemia by Candida auris: first four reported cases in continental Europe. Rev. Iberoam. Micol. 34, 23–27 (2017).
Schelenz, S. et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 5, 35 (2016).
Morales-López, S. E. et al. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg. Infect. Dis. 23, 162 (2017).
Li, C. et al. Genetic mutations across multiple pathways drive filamentous growth and virulence in the emerging fungal pathogen Candida auris. Sci. China Life Sci. 68, 2484–2486 (2025).
Alanio, A. et al. First patient-to-patient intrahospital transmission of clade I Candida auris in France revealed after a two-month incubation period. Microbiol. Spectr. 10, e0183322 (2022).
Huang, X. et al. Murine model of colonization with fungal pathogen Candida auris to explore skin tropism, host risk factors and therapeutic strategies. Cell Host Microbe 29, 210–221.e6 (2021).
Antifungal Susceptibility Testing for C. auris (Centers for Disease Control and Prevention (CDC), 2024).
Ostrowsky, B. Candida auris isolates resistant to three classes of antifungal medications – New York, 2019. MMWR Morb. Mortal. Wkly Rep. 69, 6–9 (2020).
Lyman, M. Notes from the field: transmission of pan-resistant and echinocandin-resistant Candida auris in health care facilities ― Texas and the District of Columbia, January–April 2021. MMWR Morb. Mortal. Wkly Rep. 70, 1022–1023 (2021).
Antibiotic Resistance Threats in the United States (Centers for Disease Control and Prevention (CDC), 2019).
WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action (World Health Organization, 2022).
Vanden Bossche, H., Koymans, L. & Moereels, H. P450 inhibitors of use in medical treatment: focus on mechanisms of action. Pharmacol. Ther. 67, 79–100 (1995).
Perlin, D. S., Rautemaa-Richardson, R. & Alastruey-Izquierdo, A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect. Dis. 17, e383–e392 (2017).
Muñoz, J. F. et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 9, 5346 (2018).
Kwon, Y. J. et al. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J. Clin. Microbiol. 57, 01624-18 (2019).
Chow, N. A. et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11, 03364-19 (2020).
Rybak, J. M. et al. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio 11, 00365-20 (2020).
Li, J. et al. Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob. Agents Chemother. 65, 02663-20 (2021).
Li, J., Coste, A. T., Bachmann, D., Sanglard, D. & Lamoth, F. Deciphering the Mrr1/Mdr1 pathway in azole resistance of Candida auris. Antimicrob. Agents Chemother. 66, e0006722 (2022).
Chow, E. W. et al. The transcription factor Rpn4 activates its own transcription and induces efflux pump expression to confer fluconazole resistance in Candida auris. mBio 14, e02688-23 (2023).
Gao, J. et al. LncRNA DINOR is a virulence factor and global regulator of stress responses in Candida auris. Nat. Microbiol. 6, 842–851 (2021).
Li, Z. et al. Genome-wide piggyBac transposon-based mutagenesis and quantitative insertion-site analysis in haploid Candida species. Nat. Protoc. 5, 2705–2727 (2020).
Gao, J. et al. Candida albicans gains azole resistance by altering sphingolipid composition. Nat. Commun. 9, 4495 (2018).
Sun, N., Parrish, R. S., Calderone, R. A. & Fonzi, W. A. Unique, diverged, and conserved mitochondrial functions influencing Candida albicans respiration. mBio 10, 00300-19 (2019).
Sun, N. et al. Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob. Agents Chemother. 57, 532–542 (2013).
Gulshan, K., Schmidt, J. A., Shahi, P. & Moye-Rowley, W. S. Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol. Cell. Biol. 28, 5851–5864 (2008).
Hallstrom, T. C. & Moye-Rowley, W. S. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275, 37347–37356 (2000).
Traven, A., Wong, J. M., Xu, D., Sopta, M. & Ingles, C. J. Interorganellar communication: altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem. 276, 4020–4027 (2001).
Tsai, H.-F., Krol, A. A., Sarti, K. E. & Bennett, J. E. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50, 1384–1392 (2006).
Zhang, X. & Moye-Rowley, W. S. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the Fo component of the mitochondrial ATPase. J. Biol. Chem. 276, 47844–47852 (2001).
Moye-Rowley, W. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 354, 15–21 (2005).
Shingu-Vazquez, M. & Traven, A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot. Cell 10, 1376–1383 (2011).
Brun, S. et al. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48, 1788–1796 (2004).
Hossain, S. et al. Mitochondrial perturbation reduces susceptibility to xenobiotics through altered efflux in Candida albicans. Genetics 219, iyab095 (2021).
Cheng, S., Clancy, C. J., Nguyen, K. T., Clapp, W. & Nguyen, M. H. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob. Agents Chemother. 51, 1855–1858 (2007).
Thomas, E. et al. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob. Agents Chemother. 57, 5580–5599 (2013).
Li, J., Brandalise, D., Coste, A. T., Sanglard, D. & Lamoth, F. Exploration of novel mechanisms of azole resistance in Candida auris. Antimicrob. Agents Chemother. 68, e0126524 (2024).
Maesaki, S., Marichal, P., Bossche, H. V., Sanglard, D. & Kohno, S. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 44, 27–31 (1999).
Cunningham, K. W. & Fink, G. R. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124, 351–363 (1994).
Sanglard, D., Ischer, F., Marchetti, O., Entenza, J. & Bille, J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48, 959–976 (2003).
Juvvadi, P. R. et al. Calcium-mediated induction of paradoxical growth following caspofungin treatment is associated with calcineurin activation and phosphorylation in Aspergillus fumigatus. Antimicrob. Agents Chemother. 59, 4946–4955 (2015).
Liu, F.-f. et al. Calcium signaling mediates antifungal activity of triazole drugs in the Aspergilli. Fungal Genet. Biol. 81, 182–190 (2015).
Li, Y. et al. Mitochondrial dysfunctions trigger the calcium signaling-dependent fungal multidrug resistance. Proc. Natl Acad. Sci. USA 117, 1711–1721 (2020).
Singh, S. D. et al. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 5, e1000532 (2009).
Cunningham, K. W. & Fink, G. R. Calcineurin inhibits VCX 1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2226–2237 (1996).
Rosenberg, A. et al. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat. Commun. 9, 2470 (2018).
Berman, J. & Krysan, D. J. Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 18, 319–331 (2020).
Almagro Armenteros, J. J., Sønderby, C. K., Sønderby, S. K., Nielsen, H. & Winther, O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 33, 3387–3395 (2017).
Allen, G. & Green, N. M. A 31-residue tryptic peptide from the active site of the [Ca++]-transporting adenosine triphosphatase of rabbit sarcoplasmic reticulum. FEBS Lett. 63, 188–192 (1976).
Maruyama, K. & MacLennan, D. H. Mutation of aspartic acid-351, lysine-352, and lysine-515 alters the Ca2+ transport activity of the Ca2+-ATPase expressed in COS-1 cells. Proc. Natl Acad. Sci. USA 85, 3314–3318 (1988).
Kühlbrandt, W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 (2004).
Lutgring, J. D. et al. FDA-CDC Antimicrobial Resistance Isolate Bank: a publicly available resource to support research, development, and regulatory requirements. J. Clin. Microbiol. 56, 01415-17 (2018).
Wang, X. et al. The first isolate of Candida auris in China: clinical and biological aspects. Emerg. Microbes Infect. 7, 93 (2018).
Fan, S. et al. A biological and genomic comparison of a drug-resistant and a drug-susceptible strain of Candida auris isolated from Beijing, China. Virulence 12, 1388–1399 (2021).
Bing, J. et al. A case of Candida auris candidemia in Xiamen, China, and a comparative analysis of clinical isolates in China. Mycology 13, 68–75 (2022).
Simm, C. et al. Disruption of iron homeostasis and mitochondrial metabolism are promising targets to inhibit Candida auris. Microbiol. Spectr. 10, e0010022 (2022).
Paulusma, C. C. et al. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 47, 268–278 (2008).
Davis, J. A. et al. The lipid flippases ALA4 and ALA5 play critical roles in cell expansion and plant growth. Plant Physiol. 182, 2111–2125 (2020).
Hiraizumi, M., Yamashita, K., Nishizawa, T. & Nureki, O. Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 365, 1149–1155 (2019).
Andersen, J. P. et al. P4-ATPases as phospholipid flippases—structure, function, and enigmas. Front. Physiol. 7, 275 (2016).
Axelsen, K. B. & Palmgren, M. G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 (1998).
Sonntag, Y. et al. Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes. Nat. Commun. 2, 304 (2011).
Hemenway, C. S. & Heitman, J. Calcineurin: structure, function, and inhibition. Cell Biochem. Biophys. 30, 115–151 (1999).
Rusnak, F. & Mertz, P. Calcineurin: form and function. Physiol. Rev. 80, 1483–1521 (2000).
Cowen, L. E., Carpenter, A. E., Matangkasombut, O., Fink, G. R. & Lindquist, S. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot. Cell 5, 2184–2188 (2006).
Cowen, L. E. & Lindquist, S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309, 2185–2189 (2005).
Goldman, A. et al. The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity. Mol. Cell 55, 422–435 (2014).
Park, H.-S. et al. Calcineurin targets involved in stress survival and fungal virulence. PLoS Pathog. 12, e1005873 (2016).
Lew-Smith, J., Binkley, J. & Sherlock, G. The Candida Genome Database: annotation and visualization updates. Genetics 229, iyaf001 (2025).
Engel, S. R. et al. Saccharomyces Genome Database: advances in genome annotation, expanded biochemical pathways, and other key enhancements. Genetics 229, iyae185 (2025).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the \({2}^{-\varDelta \varDelta {C}_{{\rm{T}}}}\) method. Methods 25, 402–408 (2001).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Xie, J. L., Singh-Babak, S. D. & Cowen, L. E. Minimum inhibitory concentration (MIC) assay for antifungal drugs. Bio Protoc. 2, e252 (2012).
Agyare-Tabbi, M. R. et al. The putative error prone polymerase REV1 mediates DNA damage and drug resistance in Candida albicans. npj Antimicrob. Resist. 2, 42 (2024).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019).
Geiser, J. R., van Tuinen, D., Brockerhoff, S. E., Neff, M. M. & Davis, T. N. Can calmodulin function without binding calcium? Cell 65, 949–959 (1991).
Acknowledgements
We thank G. Huang and C. Liu for contributing clinical C. auris isolates to this study; the past and present members of Y.W.’s lab and J.G.’s lab for valuable discussions. This work was supported by the National Key Research and Development Program of China (2025YFA1310100), the National Natural Science Foundation (32370961), the Chinese Academy of Sciences Project for Young Scientists in Basic Research (YSBR-111), the Fundamental Research Funds for the Central Universities (22120250158 and 22120250374), the Open Research Fund of Basic Medicine College (JCKFKT-ZD-004), the National Medical Research Council of Singapore (OFIRG23Jul-0077), and the TRIDENT Program of Singapore (TP_23P2).
Author information
Authors and Affiliations
Contributions
Y.S. and J.C. performed most experiments, conducted bioinformatics analyses and interpreted data. J.W. performed the genetic interaction screens. J.Z. performed the experimental evolution assays. F.S. performed LC–MS experiments and interpreted data. All other authors aided in constructing plasmids and strains used in this study. K.C., Y.W. and J.G. conceptualized the study, designed the experiments and wrote the manuscript. All authors provided comments in editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Yong-Sun Bahn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification of a mitochondrial gene, PET309, involved in C. auris fluconazole susceptibility using the PB-based mutagenesis system.
a Heatmap of the Pearson correlation coefficients (R) calculated across biological triplicates within and between screening conditions. b Volcano plots of the FSI values for PB transposition mutants grown in YPD containing 30, 60, or 120 μg/mL fluconazole compared with the same mutants grown in drug-free YPD. The PB-mutated genes represented by mutants that show statistically significant fold increases (FSI ≥ 1 and p < 0.05) or decreases (FSI ≤ 1 and p < 0.05) after drug treatment are indicated in red or blue, respectively. Three genes known to be associated with fluconazole susceptibility are highlighted in purple. The number of mutants that meet our selection criteria ( | FSI | ≥ 1, p < 0.05) is indicated on the top of each volcano plot. Statistical significance was tested using the Wald test with the Benjamini-Hochberg correction. c Spotting of the indicated strains on YNB plates supplemented with 2% glucose or 2% glycerol. Cells were grown overnight in YPD, washed twice with PBS, and resuspended in PBS at a final concentration of 1 × 107 cells/mL. Subsequently, 3 μL was spotted in serial 10-fold dilutions. Images were taken after 48 h of growth at 30 °C. The experiment was repeated twice with similar results. d Dose-response assays for the indicated strains were performed as described in Fig. 1f. Fluconazole was applied as 2-fold serail dilutions ranging from 0.25 to 256 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Averaged technical triplicates are represented by the color scale. The experiment was repeated twice with similar results.
Extended Data Fig. 2 Characterization of the screening strain, CauW156.
a Genotype description of CauW156. Arrows indicate primers used for genotyping. b PCR-based genotyping of CauW156 using primers shown in (a). WT and CauW08 cells were included as controls. The experiment was repeated twice with similar results. c Dose-response assays for the indicated strains were performed as described in Fig. 1f. Fluconazole was applied as 2-fold serial dilutions ranging from 0.25 to 256 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Averaged technical triplicates are represented by the color scale. The experiment was repeated twice with similar results. d Dose-response assays for the indicated strains were performed as described in Fig. 1f. Fluconazole was applied as 2-fold serial dilutions ranging from 0.25 to 256 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Averaged technical triplicates are represented by the color scale. The experiment was repeated twice with similar results.
Extended Data Fig. 3 Examination of the role of 000175 and its homologs in calcium and fluconazole susceptibility in C. auris, C. albicans, and S. cerevisiae.
a Spotting of the indicated strains on YPD plates supplemented with 0, 0.4, 0.6, or 0.8 M CaCl2. Cells were grown overnight in YPD and then diluted to 1 × 107 cells/mL. Subsequently, 3 μL was spotted in 10-fold serial dilutions. Images were taken after 48 h of growth at 30 °C. The experiment was repeated twice with similar results. b Spotting assays for the indicated strains were performed on YPD plates supplemented with 0, 0.4, 0.6, or 0.8 M CaCl2 as described in panel (a). Images were taken after 48 h of growth at 30 °C. The experiment was repeated twice with similar results. c Spotting assays for the indicated strains were performed on YPD plates supplemented with 0, 0.4, 0.6, or 0.8 M CaCl2 as described in panel (a). Images were taken after 48 h of growth at 30 °C. The experiment was repeated twice with similar results. d Spotting assays for the indicated strains were performed on YPD plates supplemented with 0, 0.4, 0.6, or 0.8 M CaCl2 or containing 5, 10, or 20 µg/mL fluconazole as described in panel b Images were taken after 48 h of growth at 30 °C. The experiment was repeated twice with similar results.
Extended Data Fig. 4 Calcineurin promotes Cdt1’s calcium pump-independent function through a post-transcriptional mechanism.
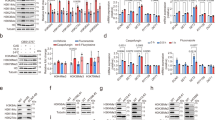
a Calcium-binding loops in C. auris calmodulin, CMD1. Four potential calcium-binding loops were predicted based on the structure of S. cerevisiae calmodulin85. The residues highlighted in red were mutated (D to A and E to V) to abolish its calcium-binding activity, yielding CMD1-4. b Spotting assays for the indicated strains were performed on YPD plates supplemented with 0, 0.4, 0.6, or 0.8 M CaCl2 as described in Extended Data Fig. 3a. Images were taken after 48 h of growth at 30 °C. The experiment was repeated twice with similar results. c Domain organization of the catalytic subunit of C. auris calcineurin, calcineurin A (CNA1). Catalytic, the phosphatase domain; CNB-binding, calcineurin B subunit domain; CAM-binding, calmodulin-binding domain; AI, the auto-inhibitory domain. The boundaries of these domains are defined according to the structure of the rat calcineurin A α-isoform70. Two stop codons were introduced at L453 to generate a constitutively activated calcineurin A, CNA1tr. d Dose-response assays for the indicated strains were performed as described in Fig. 3a. Fluconazole was applied as 2-fold serial dilution ranging from 0.25 to 256 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Data are presented as mean ± SD of technical triplicates. Statistical differences were determined by two-way ANOVA multiple comparison test with the Geisser-Greenhouse correction. The experiment was repeated twice with similar results. e Corresponding MIC50 and SMG levels calculated from data in panel (d). MIC50 and SMG were calculated as described in Fig. 3b. f qPCR analysis of CDT1 transcript levels. The indicated strains were grown overnight in YPD and then subcultured at a 1:100 dilution into YPD containing 0 or 30 µg/mL fluconazole. Cells were collected for RNA extraction after 4 h of growth at 30 °C. CDT1 expression levels were normalized to ACT1, with the level of the untreated WT strain set to 1. Data are presented as mean ± SD of technical triplicates. Statistical differences were determined by two-tailed, unpaired Student’s t-test. The experiment was repeated twice with similar results. g Dose-response assays for the indicated strains were performed as described in Fig. 3a. Fluconazole was applied as 2-fold serial dilution ranging from 0.25 to 256 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Data are presented as mean ± SD of technical triplicates. Statistical differences were determined by two-way ANOVA multiple comparison test with the Geisser-Greenhouse correction. The experiment was repeated twice with similar results. h Corresponding MIC50 and SMG levels calculated from data in panel (g). MIC50 and SMG were calculated as described in Fig. 3b. i The calcium-calcineurin signaling pathway drives Cdt1’s plasma membrane localization post-transcriptionally. The indicated strains expressing GFP-Cdt1 under the ADH1 promoter were grown overnight in YPD and then subcultured at a 1:100 dilution into YPD with 0 or 30 µg/mL fluconazole. Images were taken after 4 h of growth at 30 °C. Scale bar, 5 μm. The experiment was repeated twice with similar results.
Extended Data Fig. 5 Cdt1’s calcium pump-independent function in fluconazole tolerance depends on its localization to the plasma membrane.
a Spotting assays for the indicated strains were performed on YPD plates supplemented with 0 or 0.4 M CaCl2 as described in Extended Data Fig. 3a. Images were taken after 48 h of growth at 30 °C. The experiment was repeated twice with similar results. b Effect of the K603E substitution in Cdt1 on its plasma membrane localization. The indicated strains expressing GFP-Cdt1 or GFP-Cdt1K603E were grown overnight in YPD and then subcultured at a 1:100 dilution into YPD with 0 or 30 µg/mL fluconazole. Images were taken after 4 h of growth at 30 °C. Scale bar, 5 μm. The experiment was repeated twice with similar results. c Quantification of the fluorescence intensity of GFP-Cdt1 at the plasma membrane observed in panel (b). Box plot format is the same as described in Fig. 3g. n = 50 for both WT and cdt1Δ:PCDT1-CDT1K603E cells. Statistical differences were determined by two-tailed, unpaired Student’s t-test. d Dose-response assays for the indicated strains were performed as in Fig. 3a. Fluconazole was applied as 2-fold serial dilutions ranging from 0.25 to 256 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Data are presented as mean ± SD of technical triplicates. Statistical differences were determined by two-way ANOVA multiple comparison test with the Geisser-Greenhouse correction. The experiment was repeated twice with similar results. e Corresponding MIC50 and SMG levels calculated from data in panel (d). MIC50 and SMG were calculated as described in Fig. 3b.
Extended Data Fig. 6 Sequence alignment of Cdt1 and its homologs using Clustal Omega (v.1.2.4).
From the top, the sequences of S. cerevisiae Pmc1, C. albicans Pmc1, and C. auris Cdt1. The conserved motif, DKTGTLT, around the phosphorylatable D429 (highlighted in red) is marked within a blue square.
Extended Data Fig. 7 Characterization of C. auris clinical isolates utilized in this study.
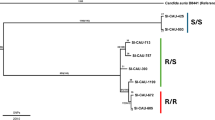
a Phylogenic analyses of 17 clinical strains isolated from hospitals in Beijing, China (marked in red), and four strains obtained from other labs are shown in blue. Fourteen representative isolates from all six C. auris clades are shown in black. The maximum-likelihood phylogenetic tree was generated based on WGS data. The scale bar represents the mean number of nucleotide substitutions per site. b Dose-response assays for the indicated strains were performed as described in Fig. 1f. Fluconazole was applied as 2-fold serial dilutions ranging from 0.5 to 512 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Averaged technical triplicates are represented by the color scale. The experiment was repeated twice with similar results. c Dose-response assays for the indicated strains were performed as described in Fig. 1f. Fluconazole was applied as 2-fold serial dilutions ranging from 1 to 1,024 µg/mL. After incubation at 30 °C for 48 h, growth was measured and normalized to the no-drug control. Data are presented as mean ± SD of technical triplicates. The experiment was repeated twice with similar results.
Supplementary information
Supplementary Information
Extended Data figure legends, Supplementary Result, Fig. 1 and figure legend, and Tables 4–6.
Supplementary Table 1
Genome-wide profiling of genes that affect C. auris’s fluconazole susceptibility.
Supplementary Table 2
GO term enrichment analysis of cellular components represented in the PB-inserted genes associated with 58 concentration-independent mutants.
Supplementary Table 3
Differentially expressed genes (DEGs) in pet309Δ cells.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data and unprocessed gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Song, Y., Chen, J., Wan, J. et al. Candida auris vacuolar calcium pump mediates fluconazole efflux and resistance evolution. Nat Microbiol (2026). https://doi.org/10.1038/s41564-026-02270-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41564-026-02270-1