Abstract
The naturally occurring channelrhodopsin variant anion channelrhodopsin-1 (ACR1), discovered in the cryptophyte algae Guillardia theta, exhibits large light-gated anion conductance and high anion selectivity when expressed in heterologous settings, properties that support its use as an optogenetic tool to inhibit neuronal firing with light. However, molecular insight into ACR1 is lacking owing to the absence of structural information underlying light-gated anion conductance. Here we present the crystal structure of G. theta ACR1 at 2.9 Å resolution. The structure reveals unusual architectural features that span the extracellular domain, retinal-binding pocket, Schiff-base region, and anion-conduction pathway. Together with electrophysiological and spectroscopic analyses, these findings reveal the fundamental molecular basis of naturally occurring light-gated anion conductance, and provide a framework for designing the next generation of optogenetic tools.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The protein coordinate and atomic structure factor have been deposited in the Protein Data Bank (PDB) under accession number 6CSM. The raw diffraction images have been deposited in the SBGrid Data Bank repository (ID: 569). All other data are available from the corresponding authors upon reasonable request.
References
Zhang, F. et al. The microbial opsin family of optogenetic tools. Cell 147, 1446–1457 (2011).
Ernst, O. P. et al. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014).
Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 (2015).
Deisseroth, K. & Hegemann, P. The form and function of channelrhodopsin. Science 357, eaan5544 (2017).
Nagel, G. et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296, 2395–2398 (2002).
Nagel, G. et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl Acad. Sci. USA 100, 13940–13945 (2003).
Zhang, F. et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 11, 631–633 (2008).
Berndt, A., Yizhar, O., Gunaydin, L. A., Hegemann, P. & Deisseroth, K. Bi-stable neural state switches. Nat. Neurosci. 12, 229–234 (2009).
Gunaydin, L. A. et al. Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392 (2010).
Yizhar, O. et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178 (2011).
Lin, J. Y., Knutsen, P. M., Muller, A., Kleinfeld, D. & Tsien, R. Y. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 16, 1499–1508 (2013).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Kato, H. E. et al. Atomistic design of microbial opsin-based blue-shifted optogenetics tools. Nat. Commun. 6, 7177 (2015).
Rajasethupathy, P. et al. Projections from neocortex mediate top-down control of memory retrieval. Nature 526, 653–659 (2015).
Mattis, J. et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods 9, 159–172 (2011).
Wiegert, J. S., Mahn, M., Prigge, M., Printz, Y. & Yizhar, O. Silencing neurons: tools, applications, and experimental constraints. Neuron 95, 504–529 (2017).
Zhang, F. et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007).
Chow, B. Y. et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102 (2010).
Berndt, A., Lee, S. Y., Ramakrishnan, C. & Deisseroth, K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science 344, 420–424 (2014).
Wietek, J. et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science 344, 409–412 (2014).
Govorunova, E. G., Sineshchekov, O. A., Janz, R., Liu, X. & Spudich, J. L. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 (2015).
Berndt, A. et al. Structural foundations of optogenetics: determinants of channelrhodopsin ion selectivity. Proc. Natl Acad. Sci. USA 113, 822–829 (2016).
Wietek, J. et al. An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo. Sci. Rep. 5, 14807 (2015).
Wietek, J. et al. Anion-conducting channelrhodopsins with tuned spectra and modified kinetics engineered for optogenetic manipulation of behavior. Sci. Rep. 7, 14957 (2017).
Govorunova, E. G., Sineshchekov, O. A. & Spudich, J. L. Proteomonas sulcata ACR1: a fast anion channelrhodopsin. Photochem. Photobiol. 92, 257–263 (2016).
Wietek, J., Broser, M., Krause, B. S. & Hegemann, P. Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. J. Biol. Chem. 291, 4121–4127 (2016).
Govorunova, E. G. et al. The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep. 7, 43358 (2017).
Iyer, S. M. et al. Optogenetic and chemogenetic strategies for sustained inhibition of pain. Sci. Rep. 6, 30570 (2016).
Zhao, Z. et al. A central catecholaminergic circuit controls blood glucose levels during stress. Neuron 95, 138–152 (2017).
Mohammad, F. et al. Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods 14, 271–274 (2017).
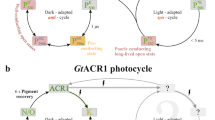
Sineshchekov, O. A., Govorunova, E. G., Li, H. & Spudich, J. L. Gating mechanisms of a natural anion channelrhodopsin. Proc. Natl Acad. Sci. USA 112, 14236–14241 (2015).
Li, H., Sineshchekov, O. A., Wu, G. & Spudich, J. L. In vitro activity of a purified natural anion channelrhodopsin. J. Biol. Chem. 291, 25319–25325 (2016).
Sineshchekov, O. A., Li, H., Govorunova, E. G. & Spudich, J. L. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc. Natl Acad. Sci. USA 113, E1993–E2000 (2016).
Kato, H. E. et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482, 369–374 (2012).
Volkov, O. et al. Structural insights into ion conduction by channelrhodopsin 2. Science 358, eaan8862 (2017).
Krause, N., Engelhard, C., Heberle, J., Schlesinger, R. & Bittl, R. Structural differences between the closed and open states of channelrhodopsin-2 as observed by EPR spectroscopy. FEBS Lett. 587, 3309–3313 (2013).
Sattig, T., Rickert, C., Bamberg, E., Steinhoff, H. J. & Bamann, C. Light-induced movement of the transmembrane helix B in channelrhodopsin-2. Angew. Chem. Int. Edn Engl. 52, 9705–9708 (2013).
Pescitelli, G. et al. Exciton circular dichroism in channelrhodopsin. J. Phys. Chem. B 118, 11873–11885 (2014).
Yi, A., Mamaeva, N., Li, H., Spudich, J. L. & Rothschild, K. J. Resonance raman study of an anion channelrhodopsin: effects of mutations near the retinylidene Schiff base. Biochemistry 55, 2371–2380 (2016).
Hontani, Y. et al. Reaction dynamics of the chimeric channelrhodopsin C1C2. Sci. Rep. 7, 7217 (2017).
Bruun, S. et al. Light–dark adaptation of channelrhodopsin involves photoconversion between the all-trans and 13-cis retinal isomers. Biochemistry 54, 5389–5400 (2015).
Maeda, A., Tomson, F. L., Gennis, R. B., Balashov, S. P. & Ebrey, T. G. Water molecule rearrangements around Leu93 and Trp182 in the formation of the L intermediate in bacteriorhodopsin’s photocycle. Biochemistry 42, 2535–2541 (2003).
Greenhalgh, D. A., Farrens, D. L., Subramaniam, S. & Khorana, H. G. Hydrophobic amino acids in the retinal-binding pocket of bacteriorhodopsin. J. Biol. Chem. 268, 20305–20311 (1993).
Berndt, A. et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl Acad. Sci. USA 108, 7595–7600 (2011).
Yi, A. et al. Structural changes in an anion channelrhodopsin: formation of the K and L intermediates at 80 K. Biochemistry 56, 2197–2208 (2017).
Berndt, A. & Deisseroth, K. Expanding the optogenetics toolkit. Science 349, 590–591 (2015).
Govindjee, R. et al. Effects of substitution of tyrosine 57 with asparagine and phenylalanine on the properties of bacteriorhodopsin. Biochemistry 34, 4828–4838 (1995).
Kolbe, M., Besir, H., Essen, L. O. & Oesterhelt, D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science 288, 1390–1396 (2000).
Betancourt, F. M. & Glaeser, R. M. Chemical and physical evidence for multiple functional steps comprising the M state of the bacteriorhodopsin photocycle. Biochim. Biophys. Acta 1460, 106–118 (2000).
Facciotti, M. T., Rouhani, S. & Glaeser, R. M. Crystal structures of bR(D85S) favor a model of bacteriorhodopsin as a hydroxyl-ion pump. FEBS Lett. 564, 301–306 (2004).
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004).
Yamashita, K., Hirata, K. & Yamamoto, M. KAMO: towards automated data processing for microcrystals. Acta Crystallogr. D. 74, 441–449 (2018).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
DiMaio, F. Advances in Rosetta structure prediction for difficult molecular-replacement problems. Acta Crystallogr. D 69, 2202–2208 (2013).
Tanimoto, T., Furutani, Y. & Kandori, H. Structural changes of water in the Schiff base region of bacteriorhodopsin: proposal of a hydration switch model. Biochemistry 42, 2300–2306 (2008).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. P. & Lanyi, J. K. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 291, 899–911 (1999).
Kato, H. E. et al. Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature 521, 48–53 (2015).
Pei, J., Kim, B. H. & Grishin, N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 42, W320–W324 (2014).
Acknowledgements
We thank C. Lee, M. Lo, K. Geiselhart and M. Lima for technical support; K. K. Kumar, N. R. Latorraca, M. Inoue and K. Katayama for critical comments; and the APS beamline staff at 23ID-B and 23ID-D for assistance in data collection. We acknowledge support by the Stanford Bio-X and the Kwanjeong Foundation (Y.S.K.), JST PRESTO (JPMJPR1782 to H.E.K., JPMJPR15P2 to K.I.), the US Department of Energy, Scientific Discovery through Advanced Computing (SciDAC) program (R.O.D.), MEXT (17H03007 to K.I., 25104009/15H02391 for H.K.), J.S.T. CREST (JPMJCR1753, H.K.) and Mathers Charitable Foundation (B.K.K.). K.D. was supported by a grant for channelrhodopsin crystal structure determination from the NIMH (R01MH075957 to K.D.).
Reviewer information
Nature thanks P. Scheerer, L. Tian and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Y.S.K. and H.E.K. contributed equally and either has the right to list himself first in bibliographic documents. Y.S.K. and H.E.K. expressed, purified and crystallized GtACR1, harvested crystals, and collected diffraction data. H.E.K. and K.Y. processed the diffraction data and solved the structure. Y.S.K. and L.E.F. performed electrophysiology. Y.S.K. measured UV-vis spectra. S.I. performed FTIR experiments under the guidance of K.I. and H.K. J.M.P. and R.O.D. provided input on structural considerations. C.R. and K.E.E. performed cell cultures and molecular cloning for electrophysiology. K.D. initiated and supervised this ChR structure/function project; Y.S.K., H.E.K., B.K.K. and K.D. planned and guided the work, and interpreted the data. Y.S.K., H.E.K. and K.D. prepared the manuscript and wrote the paper with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Crystallography.
a, Size exclusion chromatogram of the purified GtACR1 protein used for crystallography. Similar results were seen in more than 20 independent experiments. b, Electrophysiology of full-length GtACR1 (left) and the final crystallization construct (right); whole-cell voltage-clamp recordings in five cells held at −70 mV, with 513 nm light at 1.0 mW mm−2 irradiance delivered with timing as shown with green-coloured bars, while cells were held at resting potentials from −95 mV (lowest trace) to +5 mV (uppermost trace) in steps of 10 mV. Similar results were seen in 3–5 cells from each group, and no significant difference was seen in resting potential, input resistance, reversal potential or photocurrent magnitude. c, Confocal images of cultured hippocampal neurons expressing full-length GtACR1 (left) and the final crystallization construct (right). Similar results were seen in more than five cells from 3–5 coverslips. Note the markedly reduced aggregation of the truncated construct. d, Crystals of GtACR1. Similar crystals were generated in more than 200 experiments. e, Lattice packing of GtACR1 crystals, viewed parallel to the x axis (left) and the y axis (right). f, Different amino acid configurations at different chains within the asymmetric unit of GtACR1. g, C-terminal interactions among different chains within the asymmetric unit of GtACR1.
Extended Data Fig. 2 Structural analysis of GtACR1.
a, 2Fo − Fc maps (blue mesh, contoured at 1σ) for the retinal-binding pockets of chains A–D. b, 2Fo − Fc maps (blue mesh, contoured at 1σ) for the lipid molecules. c, 2Fo − Fc maps (blue mesh, contoured at 1σ) and Fo − Fc maps (green and red meshes, contoured at 3.0σ and −3.0σ, respectively) for the Schiff base region of chains A–D. Water molecules are shown as red spheres. d, Table describing data collection and refinement statistics of GtACR1. Dataset was collected from 80 crystals. Values in parentheses are for the highest-resolution shell.
Extended Data Fig. 3 Structure-based sequence alignment of microbial opsin genes.
The sequences are GtACR1 (GenBank accession AKN63094.1), GtACR2 (GenBank AKN63095.1), ZipACR (GenBank APZ76709.1), PsuACR1 (GenBank ID: KF992074.1), the chimaeric channelrhodopsin between CrChR1 and CrChR2 (C1C2, PDB code 3UG9)34, CrChR1 (GenBank 15811379), CrChR2 (GenBank 158280944), ChR1 from Volvox carteri (VcChR1, UniProtKB B4Y103), ChR1 from V. carteri (VcChR2, UniProtKB ID: B4Y105), Chrimson (GenBank ID: AHH02126.1), ChR from Tetraselmis striata (TsChR, GenBank ID: KF992089.1), HsBR (PDB code 1C3W)59, HsHR (PDB code 1E12)48, and Krokinobacter eikastus rhodopsin 2 (KR2, PDB code 3X3B)60. The sequence alignment was created using PROMALS3D61 and ESPript 362 servers. Secondary structure elements for GtACR1 are shown as coils and arrows. ‘TT’ represents turns. Cysteine residues forming intermolecular and intramolecular disulfide bridges are highlighted in green and yellow, respectively. The residues of retinal-binding pockets are coloured pink. The residues in the Schiff base region are coloured cyan. The residues forming the ECS2 and CCS are coloured orange and blue, respectively.
Extended Data Fig. 4 Structural comparison among GtACR1, HsBR, HsHR, C1C2 and CrChR2.
a, b, Side view and extracellular view of the superimposed transmembrane regions of GtACR1 (blue) and HsBR (cyan) (a), GtACR1 (blue) and HsHR (beige) (b), C1C2 (green) and CrChR2 (yellow) (c). The ATRs are shown as stick models, and are coloured orange (GtACR1), salmon (HsBR), light-yellow (HsHR), green (C1C2) and yellow (CrChR2).
Extended Data Fig. 5 Interactions between N- and C-terminal regions and the 7-TM domain.
a, Interactions between the C-terminal region and the 7-TM domain. Hydrogen bonds are shown by dashed lines. b, Fluorescent size-exclusion chromatography traces of the full-length GtACR1 (1–295), the crystallized construct (1–282), and the C-terminal truncated construct (∆C: 1–253), showing possible importance of the C terminus in proper folding and/or stability. Similar results were observed in three independent experiments. c, Interactions between the N-terminal region and the ECL1. Hydrogen bonds are shown by dashed lines. d, Fluorescent size-exclusion chromatography traces of wild-type and C-to-S mutants of GtACR1. Labels indicate estimated elution positions of the aggregate, GtACR1–eGFP, and free eGFP; C-to-S mutants show decreased (<1/3) expression compared to the wild type. Similar results were observed in three independent experiments. e, Stained SDS–PAGE gel image of wild-type and N-terminal 6-amino-acid-truncated GtACR1 in the presence and absence of reducing reagent (β-mercaptoethanol); the wild type runs as a mixer of monomer and dimer in β-mercaptoethanol,whereas N-terminal-truncated GtACR1 stays monomeric even in the absence of β-mercaptoethanol. This experiment was performed once, but similar experiments with different concentrations of β-mercaptoethanol were performed three times, all with similar results. f–h, Dimer interfaces of GtACR1 (f), C1C2 (g) and CrChR2 (h) viewed at two angles from the side; note reduced interface area (outlined) for GtACR1. For gel source data, see Supplementary Fig. 1.
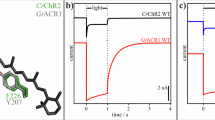
Extended Data Fig. 6 Conductances, reversal potentials, absorption spectra and kinetics of wild-type GtACR1 and mutants.
a–c, Photocurrents (a), reversal potentials (b) and absorption spectra (c) of wild-type GtACR1 and ten mutants of the retinal-binding pocket. λmax values are listed in the table (c, bottom). Photocurrents are measured in whole-cell voltage-clamp recordings held at −70 mV, with 513 nm light at 1.0 mW mm−2 irradiance. Data are mean and s.e.m.; n = 9 for WT, 6 for E163Q, 5 for C102A, M105A, C133A, C133R, C153A, E163A and C237A, and 4 for the rest. *P < 0.05, **P < 0.01, one-way ANOVA followed by Dunnett’s test. Reversal potentials are measured with identical light stimulation while cells were held at resting potentials from −95 mV to +15 mV in steps of 10 mV. Data are mean and s.e.m. n = 10 for WT and C237A, 6 for E163A and E163Q, 5 for C102A, M105A, C133A and C153A, and 4 for the rest. **P = 0.0022, one-way ANOVA followed by Dunnett’s test. Spectra measurement was performed in two independent trials, with wild type as a positive control. d, Comparison of fast closing (left) and slow closing (right) coefficients of wild-type and Y72F mutant GtACR1. Data are mean and s.e.m. n = 10 for WT and 5 for Y72F. P = 0.7 for both graphs, two-tailed t-test.
Extended Data Fig. 7 Current–voltage (I–V) relationships of wild-type GtACR1 and mutants.
The I–V relationship between −95 mV and +15 mV was determined from the single current amplitude at the indicated potentials. Each measurement is normalized to the current amplitude measured at −25 mV. Data are mean and s.e.m. n = 10 for WT and C237A, 8 for E223A, 6 for Q46C, E163A and E163Q, 4 for E68S, E68T, C102S and M105I, and 5 for the rest.
Extended Data Fig. 8 Representative traces of the I–V measurement of wild-type GtACR1 and mutants.
Voltage clamp traces corresponding to the I–V relationships in Extended Data Fig. 7 between −95 mV and +15 mV.
Extended Data Fig. 9 Spectroscopic characterization of wild-type GtACR1 and the D234N mutant.
a, Absorption spectra of wild-type GtACR1 (top left) and the D234N mutant (top right) measured from pH 3.0 to 10.0. The λmax value at each pH is listed in the table (bottom). b, Difference FTIR spectra of wild-type GtACR1 and the D234N mutant measured at 77 K, 170 K and 200 K. c, Difference FTIR spectra of wild-type GtACR1 in the 1,690–1,770 cm−1 region measured at pH 5.0, 7.0 and 9.0. Forty identical recordings at 77 K and seven identical recordings at 170 K and 200 K were averaged.
Extended Data Fig. 10 Comparison of surface electrostatic potential of GtACR1 and C1C2.
a, b, Electrostatic potential surfaces of GtACR1 (a) and C1C2 (b) viewed from four angles. The surface is coloured on the basis of the electrostatic potential contoured from −15 kT (red) to +15 kT (blue). c, d, Representation of positively charged amino acids (lysine and arginine residues) in GtACR1 (c), and negatively charged amino acids (aspartate and glutamate residues) in C1C2 (d).
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion.
Rights and permissions
About this article
Cite this article
Kim, Y.S., Kato, H.E., Yamashita, K. et al. Crystal structure of the natural anion-conducting channelrhodopsin GtACR1. Nature 561, 343–348 (2018). https://doi.org/10.1038/s41586-018-0511-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-018-0511-6
Keywords
This article is cited by
-
A second photoactivatable state of the anion-conducting channelrhodopsin GtACR1 empowers persistent activity
Communications Biology (2025)
-
Cryo-EM structure of a blue-shifted channelrhodopsin from Klebsormidium nitens
Nature Communications (2025)
-
Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states
Nature Structural & Molecular Biology (2025)
-
Channelrhodopsins with distinct chromophores and binding patterns
Nature Communications (2024)
-
Kalium channelrhodopsins effectively inhibit neurons
Nature Communications (2024)