Abstract
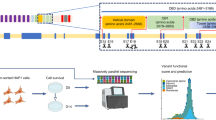
Sequencing-based genetic tests have uncovered a vast array of BRCA2 sequence variants1. Owing to limited clinical, familial and epidemiological data, thousands of variants are considered to be variants of uncertain significance2,3,4 (VUS). Here we have utilized CRISPR–Cas9-based saturation genome editing in a humanized mouse embryonic stem cell line to determine the functional effect of VUS. We have categorized nearly all possible single nucleotide variants (SNVs) in the region that encodes the carboxylate-terminal DNA-binding domain of BRCA2. We have generated function scores for 6,551 SNVs, covering 96.4% of possible SNVs in exons 15–26 spanning BRCA2 residues 2479–3216. These variants include 1,282 SNVs that are categorized as missense VUS in the clinical variant database ClinVar, with 77.2% of these classified as benign and 20.4% classified as pathogenic using our functional score. Our assay provides evidence that 3,384 of the SNVs in the region are benign and 776 are pathogenic. Our classification aligns closely with pathogenicity data from ClinVar, orthogonal functional assays and computational meta predictors. We have integrated our embryonic stem cell-based BRCA2-saturation genome editing dataset with other available evidence and utilized the American College of Medical Genetics and Genomics/Association for Molecular Pathology guidelines for clinical classification of all possible SNVs. This classification is available as a sequence–function map and serves as a valuable resource for interpreting unidentified variants in the population and for physicians and genetic counsellors to assess BRCA2 VUS in patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in the manuscript and in the supplementary file. The raw sequencing reads were deposited to the Gene Expression Omnibus (GEO) under accession GSE248438 and the reads were aligned to GRCh38 human reference genome. The MAVISp dataset for BRCA2 was deposited in the MAVISp database (https://services.healthtech.dtu.dk/services/MAVISp-1.0/). The interactive interface for BRCA2 variant exploration is available at https://appshare.cancer.gov/BRCA2_SGE/.
Code availability
The code used for ES cell-based BRCA2 SGE is available at https://github.com/MelissaGall/AVENGERS.
References
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020).
Dorling, L. et al. Breast cancer risks associated with missense variants in breast cancer susceptibility genes. Genome Med. 14, 51 (2022).
Breast Cancer Association Consortium et al. Breast Cancer Risk Genes—association analysis in more than 113,000 women. N. Engl. J. Med. 384, 428–439 (2021).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Cooper, G. M. & Shendure, J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 12, 628–640 (2011).
Tabet, D., Parikh, V., Mali, P., Roth, F. P. & Claussnitzer, M. Scalable functional assays for the interpretation of human genetic variation. Annu. Rev. Genet. 56, 19.1–19.25 (2022).
Starita, L. M. et al. Variant interpretation: functional assays to the rescue. Am. J. Hum. Genet. 101, 315–325 (2017).
Weile, J. & Roth, F. P. Multiplexed assays of variant effects contribute to a growing genotype-phenotype atlas. Hum. Genet. 137, 665–678 (2018).
Fowler, D. M. & Fields, S. Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801–807 (2014).
Findlay, G. M. et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 (2018).
Findlay, G. M., Boyle, E. A., Hause, R. J., Klein, J. C. & Shendure, J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513, 120–123 (2014).
Li, H. et al. Functional annotation of variants of the BRCA2 gene via locally haploid human pluripotent stem cells. Nat. Biomed. Eng. 8, 165–176 (2024).
Hanna, R. et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e20 (2020).
Cuella-Martin, R. et al. Functional interrogation of DNA damage response variants with base editing screens. Cell 184, 1081–1097.e19 (2021).
Huang, C., Li, G., Wu, J., Liang, J. & Wang, X. Identification of pathogenic variants in cancer genes using base editing screens with editing efficiency correction. Genome Biol. 22, 80 (2021).
Kweon, J. et al. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene 39, 30–35 (2020).
Kim, Y. et al. High-throughput functional evaluation of human cancer-associated mutations using base editors. Nat. Biotechnol. 40, 874–884 (2022).
Erwood, S. et al. Saturation variant interpretation using CRISPR prime editing. Nat. Biotechnol. 40, 885–895 (2022).
Monteiro, A. N. et al. Variants of uncertain clinical significance in hereditary breast and ovarian cancer genes: best practices in functional analysis for clinical annotation. J. Med. Genet. 57, 509–518 (2020).
Parsons, M. T. et al. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: an ENIGMA resource to support clinical variant classification. Hum. Mutat. 40, 1557–1578 (2019).
Guidugli, L. et al. Assessment of the clinical relevance of BRCA2 missense variants by functional and computational approaches. Am. J. Hum. Genet. 102, 233–248 (2018).
Richardson, M. E. et al. Strong functional data for pathogenicity or neutrality classify BRCA2 DNA-binding-domain variants of uncertain significance. Am. J. Hum. Genet. 108, 458–468 (2021).
Biswas, K. et al. A computational model for classification of BRCA2 variants using mouse embryonic stem cell-based functional assays. NPJ Genom. Med. 5, 52 (2020).
Ikegami, M. et al. High-throughput functional evaluation of BRCA2 variants of unknown significance. Nat. Commun. 11, 2573 (2020).
Biswas, K. et al. Sequencing-based functional assays for classification of BRCA2 variants in mouse ESCs. Cell Rep. Methods 3, 100628 (2023).
Mishra, A. P. et al. BRCA2–DSS1 interaction is dispensable for RAD51 recruitment at replication-induced and meiotic DNA double strand breaks. Nat. Commun. 13, 1751 (2022).
Mishra, A. P. et al. Characterization of BRCA2 R3052Q variant in mice supports its functional impact as a low-risk variant. Cell Death Dis. 14, 753 (2023).
Hartford, S. A. et al. Interaction with PALB2 is essential for maintenance of genomic integrity by BRCA2. PLoS Genet. 12, e1006236 (2016).
Roy, R., Chun, J. & Powell, S. N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78 (2011).
Kuznetsov, S. G., Liu, P. & Sharan, S. K. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat. Med. 14, 875–881 (2008).
Sharan, S. K. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131, 131–142 (2003).
Sahu, S. et al. Saturation genome editing of 11 codons and exon 13 of BRCA2 coupled with chemotherapeutic drug response accurately determines pathogenicity of variants. PLoS Genet. 19, e1010940 (2023).
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Cline, M. S. et al. BRCA challenge: BRCA exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 14, e1007752 (2018).
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Clark, K. A. et al. Comprehensive evaluation and efficient classification of BRCA1 RING domain missense substitutions. Am. J. Hum. Genet. 109, 1153–1174 (2022).
Pejaver, V. et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am. J. Hum. Genet. 109, 2163–2177 (2022).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Brnich, S. E. et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 12, 3 (2019).
Tavtigian, S. V., Harrison, S. M., Boucher, K. M. & Biesecker, L. G. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum. Mutat. 41, 1734–1737 (2020).
Sahu, S. et al. Protocol for the saturation and multiplexing of genetic variants using CRISPR–Cas9. STAR Protoc. 4, 102702 (2023).
Easton, D. F. et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 81, 873–883 (2007).
Sirisena, N. et al. Functional evaluation of five BRCA2 unclassified variants identified in a Sri Lankan cohort with inherited cancer syndromes using a mouse embryonic stem cell-based assay. Breast Cancer Res. 22, 43 (2020).
Biswas, K. et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood 118, 2430–2442 (2011).
Arnaudi, M. et al. MAVISp: multi-layered assessment of variants by structure for proteins. Preprint at bioRxiv https://doi.org/10.1101/2022.10.22.513328 (2023).
Yang, H. et al. BRCA2 function in DNA binding and recombination from a BRCA2–DSS1–ssDNA structure. Science 297, 1837–1848 (2002).
Li, H. et al. Risks of breast and ovarian cancer for women harboring pathogenic missense variants in BRCA1 and BRCA2 compared with those harboring protein truncating variants. Genet Med. 24, 119–129 (2022).
Spurdle, A. B. et al. Towards controlled terminology for reporting germline cancer susceptibility variants: an ENIGMA report. J. Med. Genet. 56, 347–357 (2019).
Hu, C. et al. Functional analysis and clinical classification of 462 germline BRCA2 missense variants affecting the DNA binding domain. Am. J. Hum. Genet. 111, 584–593 (2024).
Shimelis, H. et al. BRCA2 hypomorphic missense variants confer moderate risks of breast cancer. Cancer Res. 77, 2789–2799 (2017).
Huang, H. et al. Functional evaluation and clinical classification of BRCA2 variants. Nature https://doi.org/10.1038/s41586-024-08388-8 (2024).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Walker, L. C. et al. Using the ACMG/AMP framework to capture evidence related to predicted and observed impact on splicing: recommendations from the ClinGen SVI Splicing Subgroup. Am. J. Hum. Genet. 110, 1046–1067 (2023).
Drost, M. et al. A functional assay-based procedure to classify mismatch repair gene variants in Lynch syndrome. Genet. Med. 21, 1486–1496 (2019).
Sorrentino, E. et al. Integration of VarSome API in an existing bioinformatic pipeline for automated ACMG interpretation of clinical variants. Eur. Rev. Med. Pharmacol. Sci. 25, 1–6 (2021).
Kopanos, C. et al. VarSome: the human genomic variant search engine. Bioinformatics 35, 1978–1980 (2019).
Tate, J. G. et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 (2019).
de Bruijn, I. et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 83, 3861–3867 (2023).
Tiberti, M. et al. MutateX: an automated pipeline for in silico saturation mutagenesis of protein structures and structural ensembles. Brief. Bioinformatics 23, bbac074 (2022).
Sora, V. et al. RosettaDDGPrediction for high-throughput mutational scans: from stability to binding. Protein Sci. 32, e4527 (2023).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023).
Frazer, J. et al. Disease variant prediction with deep generative models of evolutionary data. Nature 599, 91–95 (2021).
Rentzsch, P., Schubach, M., Shendure, J. & Kircher, M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 13, 31 (2021).
Feng, B.-J. PERCH: a unified framework for disease gene prioritization. Hum. Mutat. 38, 243–251 (2017).
Ioannidis, N. M. et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885 (2016).
Tavtigian, S. V., Deffenbaugh, A. M., Yin, L., Judkins, T., Scholl, T., Samollow, P. B., de Silva, D., Zharkikh, A. & Thomas, A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 43, 295–305 (2005).
Acknowledgements
The authors thank K. Biswas, R. Sharan, S. Modak, D. Chakraborty, S. Acharya and members of our laboratory for helpful discussions and suggestions on this project; J. Carrel and M. Karwan; and E. Conner. The ACMG/AMP codes were derived from a paid subscription to Varsome (Saphetor). We obtained segregation and frequency data for SNVs in exons 15–26 from the BRCA2 gene variant database of the Global Variome shared LOVD installation. The S.K.S. laboratory is funded by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, US National Institutes of Health. S.S. is supported by a K99 grant from National Cancer Institute, US National Institutes of Health (K99CA279648). R.C. is funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261201500003I. The E.P. laboratory is supported by Danmarks Grundforskningsfond (DNRF125), Novo Nordisk Fonden Bioscience and Basic Biomedicine (NNF20OC0065262) and Hartmanns Fond (R241-A33877). M.A. is supported by a PhD Fellowship from the Danish Data Science Academy (DDSA).
Author information
Authors and Affiliations
Contributions
S.S. and S.K.S. conceived the study and designed the experimental approach. M.G. performed the bioinformatics analysis. S.S., E.S., D.C. and T.S. performed the experiments. J.G. performed cloning of sgRNA. M.A. and E.P. performed MAVISp analyses. R.C. helped in genome editing experiments. K.M. and M.Z. helped in analysing breast cancer datasets. S.S. and S.K.S. wrote the manuscript, and all authors were involved during the discussion and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Yongsub Kim, Sean Tavtigian and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Generation of all possible SNVs using CRISPR-Cas9 based SGE.
a, Heatmap showing the total number of expected and recovered variants across 51 experimental pools to saturate the C-terminal DNA binding domain. The value in the boxes represents the number of variants expected/recovered ranging from 80 SNVs (light blue) to 234 SNVs (dark blue). A total of 96.4% of all possible SNVs were recovered (6551 out of 6796 SNVs). b,c, Bar plot showing the distribution of (b) indel and (c) HDR rates calculated for each experimental pool. HDR rates are calculated based on the percentage of reads for SNVs with fixed PAM modification in each experimental pool. d, Pearson correlation between the read counts of all SNVs recovered after HDR at a frequency of 1 in 105 reads in both replicates across the experimental pools.
Extended Data Fig. 2 Function scores based on cell fitness and drug response enhances classification accuracy of ESC-based BRCA2 SGE.
a, The Pearson correlation of Function Scores (FS) for 6551SNVs between cell fitness (DMSO treated) and response to drugs (cisplatin treated). Additionally, we assessed the correlation between cell fitness and olaparib treatment, as well as the correlation between cisplatin and olaparib responses. b, Schematic showing our filtering strategy to integrate classification scores based on cell fitness and drug response data to accurately categorize SNVs. 6551 SNVs were categorized using Mixture-modelling based on their survival at day 14, and integrated FS values were computed by combining cell fitness and drug response scores. c, By employing GMM, we categorized variants based on the integrated values, revealing that 6209 (94.7%) SNVs exhibited strong concordance, while only 353 (5.4%) SNVs showed an opposite classification with cell fitness data. d, Heatmap represents the percentage of variant recovered and then finally classified by integrating the scores and excluding the uncertain variants. The value in the boxes represent the percentages ranging from 82% (light blue) to 100% (dark blue).
Extended Data Fig. 3 SGE experiments reveal temporal depletion of Cas9-induced indels.
Histogram showing the distribution of indels identified in each experimental pool, along with their corresponding function scores. In this analysis, we considered indels that met specific criteria: they had to align with the reference genome, displaying a single insertion or deletion within 50 base pairs of the anticipated Cas9 cleavage site defined by the sgRNAs used in each experimental pool. These indels were only counted if their frequency was at least 1 in 106 reads in both replicates, and in-frame indels were marked when their size was divisible by 3. Function scores were determined by comparing the frequency of indels at day 14 relative to day 3. We observed depletion of frameshifting indels in the pool, while a few in-frame indels were occasionally tolerated.
Extended Data Fig. 4 Mixture modelling of function scores to classify BRCA2 SNVs.
a, Correlation of function scores of all SNVs between individual replicates demonstrate the distribution of functional and nonfunctional thresholds. b, Correlation of function scores of all SNVs between individual replicates for individual exons. The dashed line represents the thresholds for functional and nonfunctional thresholds derived from GMM. c–e, Swarm plots demonstrating the distribution of (c) synonymous and nonsense SNVs (n = 1648), (d) ClinVar-reported SNVs (n = 783) and (e) all classified variants (n = 6198). The dashed line represents the median of the nonsense SNVs (red) and synonymous SNVs (blue).
Extended Data Fig. 5 ESC-based BRCA2 SGE outperforms computational meta predictors.
a, ROC curves indicate the performance of computational models at categorizing ClinVar-reported missense variants in the BRCA2 CTDB domain. ROC shows an AUC value of 0.96 in classifying ClinVar variants and AUC value of 0.96 in accurately categorizing nonsense and synonymous SNVs. We observed a moderate concordance to other computational predictors like CADD, BayesDel, REVEL, AlignGV-GD grade, PRIOR scores, EVE and AlphaMissense. b, Correlation between SGE-derived function scores and computational metrics in determining ClinVar-reported BRCA2 SNVs. The colour code represents the benign, likely benign, pathogenic, and likely pathogenic class reported in ClinVar.
Extended Data Fig. 6 ESC-based BRCA2 SGE map for key functional domains reveal strong concordance to AlphaMissense prediction.
a–d, 3D structural plots derived from BRCA2 CTDB domain from Alphafold depicting the helical domain, OB1, OB2 and OB3. The color code of the 3D structure plot is derived from AlphaFold and correlated with the pLDDT score, a score to provide information about the confidence of the prediction of a specific residue, in particular regions with a pLDDT score <50 are coloured in orange, regions with a pLDDT score between 50 and 70 are coloured in yellow, regions with a pLDDT score between 70 and 90 are coloured in light blue, regions with a pLDDT score > 90 are coloured in blue. In MAVISP we used structures or portion of structures with a pLDDT score > 70. Sequence-function map displaying the concordance of functional and non-functional categorization of individual amino acid changes across (a) helical domain residues 2479-2668, (b) OB1 residues from 2682-2794, (c) OB2 residues from 2804-3054 and (d) OB3 domain residues from 3073-3167 of the BRCA2 CTDB domain. The first box represents ESC-based BRCA2 SGE dataset and their concordance with MAVISP saturation scan and Alpha-Missense prediction. The box colour signifies “functional (blue)”, “non-functional (red)”, and “uncertain (turquoise blue)” class. The wildtype amino acid is shown as a circle, while white boxes indicate excluded amino acids. The Y-axis denotes the alternative amino acid changes. The magnified image of the 3D ribbon plot depicts the position of amino acid residues where clusters of functional/non-functional SNVs are reported across individual domains.
Extended Data Fig. 7 Correlation of SNV classification by ESC-based BRCA2 SGE and homology directed repair (HDR) assay.
Robust alignment between our ESC-based BRCA2 SGE-based classification and HDR score, particularly for 440 germline BRCA2 missense variants distributed across crucial functional regions (Helical domain and OB1) within the BRCA2 C-terminal DNA binding domain. The box colour represents light blue as benign, red as pathogenic, pooled from strong, moderate and supporting categories. The light red denotes the uncertain class of SNVs. The HR score for individual SNVs were available from Hu et al.49.
Extended Data Fig. 8 ESC-based BRCA2 SGE-based classification show concordance with orthogonal functional assay.
a, Concordance between HDR score and SGE-based classification for OB2 and OB3 domain SNVs. Notably, SNVs in the Tower domain exhibit a high HDR score, consistent with the functional nature of our SGE-based classification. b, Sankey plot representing the overall distribution of HDR-normal and abnormal SNVS and their concordance with different tiers of pathogenicity and benignity in the functional assay. c, Comparison of SGE-based classification with CRISPR-independent, BAC recombineering-based method. d, Strong concordance of SGE-based classification with CRISPR-based prime editing.
Extended Data Fig. 9 Categorizing 149 uncertain SNVs as potential hypomorphic variants.
a, Comparison of ESC-based BRCA2 SGE-based classification of SNVs previously classified using the MANO-B assay based on their responses to various PARP inhibitors (olaparib, rucaparib, niraparib) and carboplatin. b, Experimental strategy demonstrating the filtering of “uncertain” class SNVs with conflicting classifications between cell fitness and drug response data to identify potential hypomorphs (n = 302 SNVs). The uncertain variants were filtered to identify SNVs that fall in the uncertain and pathogenic categories and further identified if they represent pathogenic class in both cisplatin and olaparib dataset. c, Heatmap showing potential hypomorphic variants that survive in the DMSO pool but are sensitive to DNA damaging agents (n = 158 SNVs).
Extended Data Fig. 10 Comparison of ACMG/AMP classification of BRCA2 SNVs by ESC-based BRCA2 SGE with VarSome and HAP1 cell line-based MAVE classification.
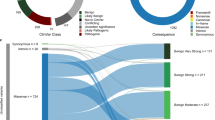
a, Sankey plot representing the concordance between ACMG/AMP classification performed by VarSome and ESC-based BRCA2 SGE. The number of VUSs also reduced by integrating our SGE functional assay data into PS3/BS3 evidence categories. b, Sankey plot represents the concordance between ESC-based BRCA2 SGE and cell viability-based MAVEs developed using human HAP1 cell line.
Supplementary information
Supplementary Table 1
Details on the oligonucleotide sequences used for sgRNA cloning and for amplicon sequencing.
Supplementary Table 2
Details on the oligonucleotide pool used for the ES cell-based BRCA2 SGE.
Supplementary Table 3
Percentage of variants recovered and classified across individual experimental pools.
Supplementary Table 4
Case–control, family history, pathology and segregation dataset on BRCA2 variants used for calibration.
Supplementary Table 5
Function scores for 6,551 SNVs across exons 15 to 26 encoding the CTDB of BRCA2.
Supplementary Table 6
Points-based ACMG/AMP clinical classification of 6,551 BRCA2 SNVs.
Supplementary Table 7
Validation of the ES cell-based BRCA2 SGE and concordance with orthogonal functional assays.
Rights and permissions
About this article
Cite this article
Sahu, S., Galloux, M., Southon, E. et al. Saturation genome editing-based clinical classification of BRCA2 variants. Nature 638, 538–545 (2025). https://doi.org/10.1038/s41586-024-08349-1
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-08349-1
This article is cited by
-
Creating an atlas of variant effects to resolve variants of uncertain significance and guide cardiovascular medicine
Nature Reviews Cardiology (2025)
-
Tracing the evolution of sequencing into the era of genomic medicine
Nature Reviews Genetics (2025)
-
Global research trends in PARP inhibitors for ovarian cancer: a bibliometric analysis
Discover Oncology (2025)
-
Functional evaluation and clinical classification of BRCA2 variants
Nature (2025)
-
Analysis of more than 400,000 women provides case-control evidence for BRCA1 and BRCA2 variant classification
Nature Communications (2025)