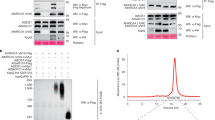
Abstract
Nucleotide-binding leucine-rich repeat (NLR) receptors play crucial roles in plant immunity by sensing pathogen effectors1. In Arabidopsis, certain sensor NLRs function as NADases to catalyse the production of second messengers2,3, which can be recognized by enhanced disease susceptibility 1 (EDS1) with its partner senescence-associated gene 101 (SAG101), to activate helper NLR N requirement gene 1 (NRG1)4. A cryoelectron microscopy structure shows that second-messenger-activated EDS1–SAG101 mainly contacts the leucine-rich repeat domain of NRG1A to mediate the formation of an induced EDS1–SAG101–NRG1A complex. Structural comparisons show that binding of a second messenger induces conformational changes in EDS1–SAG101, which are recognized by NRG1A, leading to its allosteric activation. We further show that an inhibitory NRG1 family member, NRG1C, efficiently outcompetes NRG1A for binding to second-messenger-activated EDS1–SAG101. These findings uncover mechanisms for NRG1A activation through its recognition of a modified host EDS1–SAG101 complex, and NRG1A inhibition by NRG1C through sequestration of the activated EDS1–SAG101, thus shedding light on the activation and constraint of a central plant immune response system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available in the main text, Supplementary Information and the listed Protein Data Bank (PDB) files: Apo-EDS1–SAG101 (PDB 4NFU); EDS1–SAG101 with ADRr-ATP (PDB 7XJP); EDS1–SAG101–NRG1AWHD-LRR (PDB 8YN1); and EDS1–SAG101–NRG1C (PDB 8YN0). Full versions of all gels and blots are provided in Supplementary Fig. 1. Source Data are provided with this paper.
Code availability
No custom code or mathematical algorithms were used in this study.
References
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Ma, S. et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370, eabe3069 (2020).
Martin, R. et al. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Mol. Plant Microbe Interact. 34, eabd9993 (2021).
Jia, A. et al. TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity. Science 377, eabq8180 (2022).
Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 (2006).
Zhou, J. & Zhang, Y. Plant immunity: danger perception and signaling. Cell 181, 978–989 (2020).
Cui, H., Tsuda, K. & Parker, J. E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511 (2015).
Dodds, P. N. & Rathjen, J. P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010).
van der Biezen, E. A. & Jones, J. D. G. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci 23, 454–456 (1998).
Jubic, L. M., Saile, S., Furzer, O. J., El Kasmi, F. & Dangl, J. L. Help wanted: helper NLRs and plant immune responses. Curr. Opin. Cell Biol. 50, 82–94 (2019).
Feehan, J. M., Castel, B., Bentham, A. R. & Jones, J. D. G. Plant NLRs get by with a little help from their friends. Curr. Opin. Cell Biol. 56, 99–108 (2020).
Hu, Z. & Chai, J. Assembly and architecture of NLR resistosomes and inflammasomes. Annu. Rev. Biophys. 52, 207–228 (2023).
Wang, J. et al. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870 (2019).
Förderer, A. et al. A wheat resistosome defines common principles of immune receptor channels. Nature 610, 532–539 (2022).
Zhao, Y. et al. Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism. Sci. Adv. 8, eaav5870 (2022).
Liu, F. et al. Activation of the helper NRC4 immune receptor forms a hexameric resistosome. Cell 187, 4877–4889 (2024).
Jacob, P. et al. Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 373, 420–425 (2021).
Feehan, J. M. et al. Oligomerization of a plant helper NLR requires cell-surface and intracellular immune receptor activation. Proc. Natl Acad. Sci. USA 120, e2210406120 (2023).
Huang, S. et al. Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity. Science 377, eabq3297 (2022).
Dongus, J. A. & Parker, J. E. EDS1 signalling: at the nexus of intracellular and surface receptor immunity. Curr. Opin. Cell Biol. 62, 102039 (2021).
Sun, X. et al. Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat. Commun. 12, 3335 (2021).
Wu, Z., Tian, L., Liu, X., Zhang, Y. & Li, X. TIR signal promotes interactions between lipase-like proteins and ADR1-L1 receptor and ADR1-L1 oligomerization. Plant Physiol. 187, 681–686 (2021).
Lapin, D. et al. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31, 2430–2455 (2019).
Castel, B. et al. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 222, 966–980 (2019).
Saile, S. C. et al. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol. 18, e3000783 (2020).
Wu, Z. S. et al. Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 222, 938–953 (2019).
Pruitt, R. N. et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598, 495–499 (2021).
Tian, H. et al. Activation of TIR signalling boosts pattern-triggered immunity. Nature 598, 500–503 (2021).
Wu, Z. et al. The N-terminally truncated helper NLR NRG1C antagonizes immunity mediated by its full-length neighbors NRG1A and NRG1B. Plant Cell 34, 1621–1640 (2022).
Wang, J., Song, W. & Chai, J. Structure, biochemical function, and signaling mechanism of plant NLRs. Mol. Plant 16, 75–95 (2023).
Ao, K. & Li, X. Indirect recognition of pathogen effectors by NLRs. Essays Biochem. 66, 485–500 (2022).
Contreras, M. P. et al. The nucleotide binding domain of NRC-dependent disease resistance proteins is sufficient to activate downstream helper NLR oligomerization and immune signaling. New Phytol. 243, 345–361 (2024).
Chai, J. J., Song, W. & Parker, J. E. New biochemical principles for NLR immunity in plants. Mol. Plant Microbe Interact. 36, 468–475 (2023).
Locci, F. & Parker, J. E. Plant NLR immunity activation and execution: a biochemical perspective. Open Biol. 14, 230387 (2024).
Wang, Z. et al. Plasma membrane association and resistosome formation of plant helper immune receptors. Proc. Natl Acad. Sci. USA 120, e2222036120 (2023).
Danev, R., Yanagisawa, H. & Kikkawa, M. Cryo-electron microscopy methodology: current aspects and future directions. Trends Biochem. Sci. 44, 837–848 (2019).
Ariga, H. et al. NLR locus-mediated trade-off between abiotic and biotic stress adaptation in Arabidopsis. Nat. Plants 3, 17072 (2017).
Van der Hoorn, R. A., De Wit, P. J. & Joosten, M. H. Balancing selection favors guarding resistance proteins. Trends Plant Sci. 7, 67–71 (2002).
Gust, A. A., Pruitt, R. & Nurnberger, T. Sensing danger: key to activating plant immunity. Trends Plant Sci. 22, 779–791 (2017).
Bhandari, D. D. et al. An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat. Commun. 10, 772 (2019).
Dongus, J. A. et al. Cavity surface residues of PAD4 and SAG101 contribute to EDS1 dimer signaling specificity in plant immunity. Plant J. 110, 1415–1432 (2022).
Chini, A., Grant, J. J., Seki, M., Shinozaki, K. & Loake, G. J. Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38, 810–822 (2004).
Zhu, Y., Qian, W. & Hua, J. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 6, e1000844 (2010).
Volz, R., Harris, W., Hirt, H. & Lee, Y. H. ROS homeostasis mediated by MPK4 and SUMM2 determines synergid cell death. Nat. Commun. 13, 1746 (2022).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Hough, M. A. & Wilson, K. S. From crystal to structure with CCP4. Acta Crystallogr. D Struct. Biol. 74, 67 (2018).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Struct. Biol. 60, 2126–2132 (2004).
Zheng, S. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Scheres, S. H. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016).
Scheres, S. H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Pettersen, E. F. et al. UCSF chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Struct. Biol. 66, 213–221 (2010).
Acknowledgements
We thank staff at the X-ray crystallography platform, National Protein Science Facility, Tsinghua University, and at Shanghai Synchrotron Radiation Facility, for assistance with X-ray diffraction data collection. We thank J. Wang, J. Zhao and B. Xu at the Institute of Advanced Agricultural Sciences, Peking University for provision of cryo-EM facility support. This project was supported by the Research Center for Industries of the Future at Westlake University, the National Natural Science Foundation of China (no. 32171193 to Z.H.), the National Key Research and Development Program of China (no. 2021YFA1300701 to Z.H.), the Max-Planck Society and Deutsche Forschungsgemeinschaft funding (German Research Foundation, no. SFB-1403–414786233 to J.W. and J.E.P.).
Author information
Authors and Affiliations
Contributions
Experimental design was the responsibility of J.C., J.E.P., Z.H., S.H. and J.W. Recombinant protein expression assays, purification, structure determination, modelling and data analysis were performed by S.H., R.S., A.J., Y.X., Y.S. and J.C. Biochemical assays were carried out by S.H., R.S., A.J., L.W., J.W., D.M., Z.W., Z.H., X.L., J.E.P. and J.C. Plant cell death assays and immunoblot analysis were undertaken by J.W., D.M. and J.E.P. All authors carried out data analysis. J.C. wrote the manuscript draft. J.E.P., Z.H., X.L., S.H., J.W. and R.S. edited the manuscript, with contributions from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Tian-Min Fu, John Rathjen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Proteins used for pull-down assays and ion leakage measurement assays.
a, b, e, f, h, Proteins used for pull-down assays in Fig. 1a (a), Fig. 3c (b), Fig. 4a (e), Fig. 4e (f) and Fig. 5b (h) were purified using Strep beads and analysed by SDS-PAGE. c, d, g, Proteins in Fig. 3d (c), Fig. 3e (d), Fig. 4f (g) were detected via western blot. Ponceau S served as loading control. The experiments were independently repeated three (a, b, d, e, f, g, h) or four (c) times with similar results. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Negative staining and cryo-EM micrographs of the purified EDS1-SAG101-NRG1A∆N57 protein.
The EDS1-SAG101-NRG1A protein complex was purified through affinity and size-exclusion chromatography. The peak fractions from gel filtration chromatography were used for negative staining and cryo-EM. The pentamer-like particles are highlighted in red circles (top) in negative staining and orange (bottom) in cryo-EM with a size of approximately 26 nm. Scale bar, 100 nm. Protein purification and corresponding micrograph identification were repeated three times with similar results.
Extended Data Fig. 3 Cryo-EM reconstruction of the EDS1-SAG101-NRG1AWHD-LRR complex.
a, Reconstitution of EDS1-SAG101-NRG1AWHD-LRR. NRG1AWHD-LRR was co-expressed with EDS1, SAG101, RPP1 and ATR1 in insect cells. The protein complex was purified using GS4B beads, following the Strep beads, and analysed by SDS-PAGE. The experiment was repeated three times with similar results. For gel source data, see Supplementary Fig. 1. b, A representative cryo-EM micrograph selected from 6,913 micrographs of the EDS1-SAG101-NRG1AWHD-LRR complex. c, Representative views of 2D class averages of the EDS1-SAG101-NRG1AWHD-LRR complex. d, The cryo-EM image processing workflow for the EDS1-SAG101-NRG1AWHD-LRR complex. e, FSC curves at 0.143 of the final reconstructions of the EDS1-SAG101-NRG1AWHD-LRR complex unmasked (blue) or masked (black). f, FSC curves at 0.5 for model refined against the final masked map (black) and the unmasked map (blue).
Extended Data Fig. 4 Structural comparison of ADPr-ATP-bound EDS1-SAG101 with EDS1-SAG101-NRG1AWHD-LRR and EDS1-SAG101-NRG1C.
a, The cryo-EM structures of ADPr-ATP-bound EDS1-SAG101 (PDB code: 7XJP) and EDS1-SAG101 from EDS1-SAG101-NRG1AWHD-LRR were aligned with PyMOL. Colour codes are shown as indicated. b, ADPr-ATP of the cryo-EM structure of EDS1-SAG101-NRG1AWHD-LRR complex. c, ADPr-ATP of the crystal structure of EDS1-SAG101-NRG1C. ADPr-ATP is shown in stick. The cryo-EM density (b) and the electron density (c) surrounding the small molecule are shown in grey mesh. The map was contoured at 1.2 sigma in PyMOL.
Extended Data Fig. 5 Sequence alignment of Arabidopsis PAD4 with SAG101, NRG1 family proteins with ADR1 family proteins, and NRG1 from different plant species.
a, Amino acid sequence alignment among Arabidopsis PAD4 and SAG101. b, Amino acid sequence alignment among NRG1 and ADR1 family proteins in Arabidopsis. c, Amino acid sequence alignment of NRG1A from different plant species. Putative NRG1A homologues of other species as indicated were obtained from NCBI (National Library of Medicine) using A. thaliana NRG1A sequence as a query. Except Nicotiana benthamiana, other species all belong to Brassicaceae. Clustal Omega was used for the sequence alignment. In a-c, residues of SAG101 and NRG1A involved in interacting are marked with solid purple squares at the bottom. Bulkier residues of NRG1C interacting with EDS1-SAG101 are marked with solid blue squares at the bottom.
Extended Data Fig. 6 Structure comparisons of the AlphaFold2-predicted structure of inactive NRG1ANOD-LRR with experimental structures.
a, The AlphaFold2-predicted structure of inactive NRG1ANOD-LRR resembles that of the corresponding segment of the inactive ZAR1NOD-LRR. The cryo-EM structure of inactive ZAR1-RKS1 (PDB code: 6J5W) and Alphafold2-predicted structure of inactive NRG1ANOD-LRR were aligned with PyMOL. b, The AlphaFold2-predicted structure of inactive NRG1ANOD-LRR and NRG1AWHD-LRR of EDS1-SAG101-NRG1AWHD-LRR were aligned with PyMOL. Structures were shown in cartoon representations. Colour codes are shown as indicated.
Extended Data Fig. 7 Competition between NRG1A and NRG1C for the interaction with the EDS1-SAG101 complex.
a, EDS1-SAG101 and NRG1C form a monomeric ternary complex in gel filtration. Shown in the left are gel filtration profiles (Superdex 200 Increase 10/300) of EDS1-SAG101 (purple), NRG1C (red), and EDS1-SAG101-NRG1C (green) proteins. A280, absorbance at 280 nm; mAU, milli-absorbance units. Peak fractions in the left were visualized by SDS-PAGE followed by Coomassie blue staining and are shown on the right. b, Purification of NRG1A∆CC protein. N terminal GST tagged NRG1A∆CC (residues 160-809) was expressed in insect cells. GST beads were used to purify proteins, which were further purified by Superose 6 Increase 10/300 GL. Left: a gel filtration profile of NRG1A∆CC. Right: SDS-PAGE analysis of peak fractions. c, NRG1C fails to efficiently outcompete NRG1AWHD-LRR in EDS1-SAG101- NRG1AWHD-LRR. 0.08 μmol the EDS1-SAG101-NRG1AWHD-LRR (with a GST tag) complex protein was incubated with a varying amount of NRG1C (0, 0.04, 0.08, 0.16, 0.32, 0.64 μmol). The mixture was flowed through the Strep beads. After extensive washing, the beads were analysed by SDS-PAGE. In a-c, the experiments were repeated three times with similar results. For gel source data, see Supplementary Fig. 1. d, Structural comparison of EDS1-SAG101-NRG1C with AlphaFold2-predicted EDS1-SAG101-NRG1ANOD-LRR. The cryo-EM structure of EDS1-SAG101-NRG1C (PDB code: 8YN0) was aligned with AlphaFold2-predicted EDS1-SAG101-NRG1ANOD-LRR in PyMOL. The NBD of NRG1A is shown in surface and others in cartoon representation. The NBD of NRG1A clashes with EP domains of EDS1-SAG101.
Extended Data Fig. 8 Working model on activation and regulation of the EDS1-SAG101-NRG1 signaling branch in Arabidopsis.
TNLs form resistosomes upon recognizing their corresponding pathogen effectors. These resistosomes catalyse the production of the nucleotide-derived small molecules ADPr-ATP and di-ADPR, which bind to the apo-EDS1-SAG101 complex. Binding of these small molecules induces conformational changes in EDS1-SAG101, which are recognized by NRG1 family proteins, including NRG1A, NRG1B, and NRG1C. This results in the oligomerization of EDS1-SAG101-NRG1A/B and the assembly of NRG1A/B resistosomes in plasma membrane (and/or other endo-membranes), activating their Ca2+-permeable channel activity to initiate ETI signalling. NRG1C, which lacks the NOD module, forms monomeric complex with EDS1-SAG101. Because of its higher affinity than inactive NRG1A/B, NRG1C efficiently outcompetes NRG1A/B for binding to the ADPr-ATP/di-ADPR-bound EDS1-SAG101 complex, sequestering the activated EDS1-SAG101 from inactive NRG1A/B and negatively regulating NRG1A/B immune signalling.
Supplementary information
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, S., Wang, J., Song, R. et al. Balanced plant helper NLR activation by a modified host protein complex. Nature 639, 447–455 (2025). https://doi.org/10.1038/s41586-024-08521-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-08521-7
This article is cited by
-
Manipulation of the nucleotide pool in human, bacterial and plant immunity
Nature Reviews Immunology (2026)
-
Unraveling plant immunity: from pathogen perception to resistance engineering
Science China Life Sciences (2025)