Abstract
During DNA replication, the replisome must remove barriers and roadblocks including the transcription machinery1,2. Transcription–replication conflicts (TRCs) occur when there are collisions between the replisome and transcription machinery, and are increasingly recognized as an important source of mammalian genome instability3. How cells facilitate replisome bypass at sites of TRCs is incompletely understood. Here we show that the CUL3–KCTD10 E3 ligase senses TRCs and promotes remodelling of the RNA polymerase complex to allow replisome bypass. We found that the substrate adaptor KCTD10 interacts with the replisome and the transcription machinery and regulates both in unstressed conditions. These bivalent interactions allow KCTD10 to detect co-directional TRCs and facilitate higher-order assembly of KCTD10 complexes that recruit CUL3 to induce the ubiquitination and removal of the RNA polymerase factor TCEA2. In the absence of KCTD10, there is increased retention of TCEA2 and the RNA polymerase complex, causing an accumulation of TRCs and increased DNA damage. Our results demonstrate how replication can proceed through transcriptionally active regions, utilizing a unique bridging function of the CUL3–KCTD10 complex. These findings provide a framework for how the coordination between transcription and replication may contribute to the maintenance of genome stability.
Similar content being viewed by others
Main
To ensure accurate propagation of genetic information, DNA replication must be able to move through proteins and RNA that coat the template strand1. Paradoxically, replication in mammalian cells preferentially starts at actively transcribed regions, with euchromatic regions being the first to replicate in mammalian genomes and replication origins enriched around transcription start sites4,5. This creates situations in which DNA replication must move through chromatin domains that are enriched for RNA polymerase (RNAP) complexes and nascent RNA, increasing the potential for direct conflicts between the replisome and transcription machinery. These TRCs can cause DNA damage and lead to genome instability, particularly in situations where replication or transcription are uncoupled from their normal regulatory processes, such as in cancer2,3,6,7. However, the degree of DNA damage is not uniform for all TRCs. Head-on TRCs are generally thought to promote DNA damage, whereas co-directional TRCs are thought to be less deleterious to genome stability3,8. This is not a general phenomenon for all co-directional TRCs, as there are specific contexts in which co-directional TRCs have been described as deleterious9,10,11; however, it is unclear what factors promote the resolution of co-directional TRCs to limit genome instability.
KCTD10 controls replication and transcription
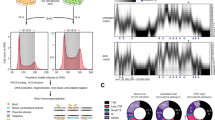
The substrate-specific CUL3 adaptor KCTD10 has a key role in regulating genome stability. In the absence of KCTD10, cells display increased γH2AX staining during interphase12 (Fig. 1a,b and Extended Data Fig. 1a). In addition, cells are markedly sensitized to cytotoxic effects of the catalytic TOP2 inhibitors ICRF193 and ICRF18713 (Fig. 1c and Extended Data Fig. 1b,c). These inhibitors bind to the ATPase domain of TOP2, inhibiting the second ATP hydrolysis step of the catalytic cycle and stabilizing the complex in a closed clamp conformation after G strand passage and re-ligation14. However, KCTD10 loss does not sensitize cancer cells to killing with canonical TOP2 poisons (such as etoposide) or ionizing radiation (Extended Data Fig. 1e,f). As a member of the Broad-complex, Tramtrack and Bric-à-brac (BTB) protein family, KCTD10 is likely to mediate the recruitment of CUL3 to ubiquitin substrates15,16 (Extended Data Fig. 1d). The importance of this CUL3-dependent pathway in genome stability is supported by the additional identification of CUL3 and E2 enzyme UBE2M as sensitizers to ICRF187 (Extended Data Fig. 1b,d). Finally, KCTD10 loss also increases the basal rate of micronuclei in cells (Extended Data Fig. 1g). Taken together, these data support a role for KCTD10 in regulating genome stability and repairing lesions caused by catalytic TOP2 inhibitors.
a, γH2AX foci from KCTD10-deficient U2OS cells. Left, representative images. Right, quantification of foci. Data are mean ± s.d. (n = 3 biologically independent experiments). shKCTD10, shRNA targeting KCTD10; shScr, scramble shRNA. Scale bars, 10 μm. b, Western blot from KCTD10-deficient and control (Scr) U2OS cells. c, MTS assay in KCTD10-deficient or control U2OS cells with the TOP2 inhibitor ICRF193. d, Nascent DNA imaging in KCTD10-deficient or control U2OS cells pulsed with EdU for 30 min. Each dot represents the ratio of cells with EdU foci to cells with pan-nuclear EdU staining. Data are mean ± s.d. (n = 3 biologically independent experiments). Scale bars, 10 μm. e, DNA fibre assays in KCTD10-deficient or control U2OS cells pulsed with CldU and IdU as shown. Data are mean ± s.d. (n = 150 fibres examined over 3 independent experiments). f, Nascent transcription imaging in KCTD10-deficient or control U2OS cells pulsed with EU for 1 h. Data are mean ± s.d. (n = 3 biologically independent experiments). a.u., arbitrary units; max, maximum; min, minimum. Scale bars, 10 μm. g,h, DNA fibre assays in KCTD10-deficient or control U2OS cells, treated with the transcription inhibitor PG490 or PARPi (Extended Data Fig. 2f). g, Schema for fibre experiments (top) and representative fibre tracts (bottom). h, Quantification of fork speed in KCTD10-deficient or control U2OS cells, treated with PG490 (1 μM) as indicated. Data are mean ± s.d. (n = 150 fibres examined over 3 independent experiments). i,j, PLA for POLR2A–PCNA in KCTD10-deficient or control U2OS cells. i, Representative images. Scale bars, 10 μm. j, Quantification of foci. Data are mean ± s.d. (n = 200 cells examined over 3 independent experiments). P values were calculated with a two-tailed extra sum of squares F-test (c) or with one-way ANOVA with Bonferroni’s multiple comparisons test.
Besides an N-terminal BTB domain, the only other region of KCTD10 that is characterized is a C-terminal PCNA-interacting protein (PIP) box. PIP boxes are widespread short linear motifs that mediate protein–protein interactions with the key DNA replication and repair protein PCNA17,18,19. Based on the lack of sensitivity of KCTD10-deficient cells to ionizing radiation (Extended Data Fig. 1f), we hypothesized that KCTD10 may have a role in DNA replication. Consistently, we found that cell lines deficient in KCTD10 had a higher proportion of cells with EdU foci compared with pan-nuclear EdU staining than in control cells (Fig. 1d). Additionally, DNA fibre assays revealed that KCTD10-deficient cells had a slower rate of fork progression (Fig. 1e). These results suggested that KCTD10 may facilitate proper fork progression in cells. Accordingly, we detected a modest increase in the S phase population in KCTD10-deficient cells that was only statistically significant with one short hairpin RNA (shRNA) (Extended Data Fig. 1h). In the nucleus, KCTD10 was primarily localized to chromatin and its expression was relatively unchanged throughout the cell cycle (Extended Data Fig. 1i,j). Despite having a clear effect on DNA replication, Gene Ontology (GO) analysis of genes that clustered together with KCTD10 based on shared phenotypes in a CRISPR-based imaging screen12 revealed a strong enrichment for terms associated with transcription-associated pathways (GO:0045899 and GO:0043922) and ubiquitin pathways (GO:0016567 and GO:0043161) (Extended Data Fig. 1k). Thus, in addition to regulating genome stability, these data raised the possibility that KCTD10 may have a role in transcription.
To investigate whether KCTD10 regulates transcription, we used an immunofluorescence-based assay to image nascent RNA synthesis in cells. These experiments showed that KCTD10 knockdown increased the rate of 5-ethynyl-uridine (EU) incorporation in KCTD10-deficient U2OS cells compared with controls (Fig. 1f and Extended Data Fig. 2a,b). We further confirmed that the observed changes in EU incorporation reflected alterations in transcription by treating cells with the transcription inhibitor PG490 for 4 h before RNA labelling and found that this abolished EU incorporation (Extended Data Fig. 2a,b). This 4-h treatment with PG490 induced RNAPII degradation in our cells (Extended Data Fig. 2c), as previously reported20,21.
In these experiments, there was notably higher EU staining in the nucleolus relative to the nucleoplasm. To determine whether KCTD10 selectively affects transcription in either nuclear compartment, we co-stained cells with the nucleolar marker fibrillarin to quantify the EU intensity in both regions. These experiments showed that KCTD10 knockdown increased the EU intensity in both the nucleolus and the nucleoplasm (Extended Data Fig. 2d,e).
These data suggested that KCTD10 may function as an endogenous limiter for transcription and raised the question of whether its role in DNA replication depends on limiting transcription. To test this, we performed DNA fibre assays in cells that were pre-treated with PG490 before labelling with 5-chloro-2′-deoxyuridine (CldU) and 5-iodo-2′-deoxyuridine (IdU) (Fig. 1g,h). In these experiments, transcriptional inhibition fully rescued the fork speed defect observed in KCTD10-deficient cells. By contrast, direct acceleration of fork progression with a PARP inhibitor did not rescue the fork speed defect (Fig. 1g and Extended Data Fig. 2f). These results indicate that KCTD10 coordinates DNA replication and transcription in human cells and may function to resolve TRCs. To directly assess this, we performed proximity ligation assay (PLA) experiments using probes for PCNA and POLR2A to quantify potential TRCs in KCTD10-deficient cells (Fig. 1i,j and Extended Data Fig. 2g,h). POLR2A–PCNA proximity pairs were significantly enriched in KCTD10-deficient cells (Fig. 1i,j), supporting the idea that KCTD10 has a role in preventing or resolving TRCs.
KCTD10 responds to co-directional TRCs
Conflicts between transcription and DNA replication can occur from head-on collisions or co-directional collisions of both complexes. To investigate this further, we utilized an episomal plasmid system designed to study head-on and co-directional TRCs in cells8. In this system, doxycycline-inducible transcription can be used to study co-directional or head-on TRCs in both an R-loop-forming gene (the mouse gene Airn) and a gene that does not form R loops (enhanced cyan fluorescent protein (ECFP)) (Extended Data Fig. 2i). In these experiments, KCTD10 depletion led to a significant increase in the number of POLR2A–PCNA proximity pairs resulting from co-directional TRCs (Fig. 2a). No such effects were observed during head-on TRCs. This suggested that CUL3–KCTD10 may work specifically to resolve co-directional TRCs. Furthermore, KCTD10 depletion had a much greater effect on TRCs generated by transcription of ECFP compared with Airn, indicating that the TRC to which KCTD10 responds is not associated with strong R-loop formation. In support of this, KCTD10 knockdown did not significantly alter R-loop levels as measured by dot blot assays (Extended Data Fig. 2j–m). This is consistent with previous data showing that head-on TRCs lead to increased R-loop stability compared with co-directional TRCs8.
a, Quantification of POLR2A–PCNA PLA foci from KCTD10-deficient (sh2 and sh3) or control HEK293T cells expressing the indicated reporters8 as shown in Extended Data Fig. 2i (top), with schemas showing the orientation of replication and transcription (bottom). Cells were treated with 1 mg ml−1 doxycycline (Dox) for 24 h to induce TRCs. Data are mean ± s.d. (n = 3 biologically independent experiments). Data are normalized to the number of foci in control cells treated with doxycycline. CD, co-directional TRCs; HO, head-on TRCs. b, Quantification of co-directional and head-on TRCs among KCTD10-associated peaks from CUT&RUN sequencing in HeLa cells expressing Flag-tagged KCTD10 treated with DMSO control or ICRF193 (100 nM for 4 h). c, Western blot analysis of KCTD10-deficient or control U2OS cells. d, Colony-formation assays in KCTD10-deficient or control U2OS cells treated with ATMi (KU55933, top) or CHK2i (BML277, bottom). Data are mean ± s.d. (n = 3 biologically independent experiments). P values were calculated with one-way ANOVA with Bonferroni’s multiple comparisons test.
To better understand the genomic sites that recruit KCTD10, we performed CUT&RUN to capture KCTD10 binding sites in cells expressing a Flag-tagged KCTD10 with or without ICRF193 treatment. In total, 2,515 KCTD10 binding peaks associated with 868 genes were identified for untreated samples while there were 15,603 peaks associated with 8,297 genes in ICRF193-treated cells. In both samples, KCTD10 peaks predominantly associated within 1 kb from promoters (Extended Data Fig. 3a,b). This supports previous data suggesting that co-directional TRCs are enriched near the transcription start sites3,22,23. To understand whether these KCTD10 binding sites may occur at sites of co-directional TRCs, we utilized previously developed methods to classify head-on or co-directional TRCs based on replication fork direction (RFD) from Okazaki fragment sequencing (OK-seq) data24,25. RFD was calculated using prior OK-seq data26 and integrated with transcription orientation at KCTD10 peaks to identify potential head-on versus co-directional TRCs (Extended Data Fig. 3c). In these analyses, 213 KCTD10-associated peaks exhibited TRCs, with 165 (77%) being co-directional (Fig. 2b). After treatment with ICRF193, we identified 1,932 KCTD10-associated peaks as potential co-directional TRCs (Fig. 2b). These peaks mapped to 1,646 genes, with 140 of these genes being common to control and ICRF193-treated cells (Extended Data Fig. 3d,e). These genes represent targets for several well-known transcription factors including E2F, KMT2D (also known as MLL) and YY1, as well as several unknown regulator motifs (Extended Data Fig. 3f). Together, these results support the idea that KCTD10 binds to co-directional TRCs in cells.
It has previously been observed that co-directional TRCs activate a distinct DNA damage response compared with head-on TRCs—that is, the ATM–CHK2 pathway8. We tested for evidence of ATM activation in our samples and found that KCTD10-deficient cells showed higher levels of phosphorylated CHK2 kinase but not phosphorylated CHK1, which is primarily targeted by ATR (Fig. 2c and Extended Data Fig. 3g). Additionally, KCTD10-deficient cells showed increased sensitivity to inhibition of both ATM (KU55933) and CHK2 (BML277) in colony-formation assays (Fig. 2d). However, KCTD10 loss did not affect the response to inhibition of ATR (VX970) or DNA-PK (NU7441) (Extended Data Fig. 3h,i). Overall, KCTD10 depletion had the largest effect on sensitivity to CHK2 inhibition with a more than 50-fold decrease in the half-maximal inhibitory concentration (IC50) (Extended Data Fig. 3j). PLA experiments for PCNA and POLR2A showed that CHK2i increased the number of TRCs in control cells but did not further increase the number of TRCs in KCTD10-deficient cells (Extended Data Fig. 3k,l), suggesting that CHK2 and KCTD10 function within the same pathway. Altogether, these data further support a role for KCTD10 in resolving co-directional TRCs and show that these TRCs activate a DNA damage response in cells.
Catalytic TOP2 inhibitors cause TRCs
The data described above indicate that ICRF193 may create TRCs; we therefore sought to clarify the consequences of catalytic TOP2 inhibition on replication and transcription. Short-term treatment with ICRF193 induced the formation of γH2AX and 53BP1 foci, and, to a lesser extent, RAD51 foci (Extended Data Fig. 4a). This damage can be exacerbated by depleting KCTD10 and occurs to the greatest degree in S phase cells (Extended Data Fig. 4b,c). Compared with etoposide, ICRF193 induced less DNA damage that accumulates more gradually in the nuclei of cells (Extended Data Fig. 4d–f).
On the basis of previous work showing that catalytic TOP2 inhibition leads to TOP2B degradation27, we hypothesized that the damage observed may depend on the presence of TOP2B. Indeed, TOP2B depletion led to a reduction in the number of DNA damage foci in cells following treatment with ICRF193 (Extended Data Fig. 4g,h). Additionally, co-treating cells with ICRF193 and an RNA polymerase inhibitor or a DNA polymerase inhibitor reduced the amount of DNA damage (Extended Data Fig. 4i). Specifically, we found that co-treating cells with ICRF193 and the transcription inhibitor PG490 had the largest effect. In PLA experiments, treatment with ICRF193 increased the number of POLR2A–TOP2B and POLR2A–PCNA pairs, but not the number of TOP2B–PCNA pairs (Extended Data Fig. 4j–l). These data suggest that catalytic TOP2 inhibition traps TOP2B on DNA strands, promoting the formation of TRCs (Extended Data Fig. 4m).
Prior work has suggested that etoposide can also induce transcription-associated damage in cells28. Of note, etoposide primarily induces TRCs in which the front of the RNAP complex (POLR2E)29 is near PCNA, whereas ICRF193 induces more TRCs in which the back of the polymerase (POLR2J) is near PCNA (Extended Data Fig. 4n). This could influence the difference in sensitivities seen between ICRF193 and etoposide in KCTD10-deficient cells (Fig. 1c and Extended Data Fig. 1a–c,e,f), and suggests they induce TRCs differently.
Our initial hypothesis was that KCTD10 may facilitate ubiquitin-dependent removal of TOP2B from DNA. However, there were no differences in the total protein level or chromatin-bound fraction of TOP2B in untreated cells or in cells treated with ICRF193 with KCTD10 knockdown versus control shRNA (Extended Data Fig. 5a). Additionally, we tested other TOP2-dependent processes that were previously ascribed to catalytic TOP2 inhibitors. To test the ability of KCTD10-deficient cells to progress through the cell cycle, cells were synchronized in late G2 using a CDK1 inhibitor (RO3306) and released for 2 h to allow most cells to enter and complete mitosis. There were no alterations in the ratio of cells in interphase, prophase, metaphase, or anaphase between control and KCTD10-deficient cells (Extended Data Fig. 5b). In addition to this, we tested whether KCTD10 might interfere with a previously described ICRF193-dependent induction of the G2/M checkpoint30. In these experiments, asynchronous cells were incubated with ICRF193 for 2 h, stained for a mitotic-specific histone (phospho-histone H3 (Ser10)) and imaged to quantify the mitotic index (Extended Data Fig. 5c). These experiments showed that there were no differences between control or KCTD10-deficient cells in the mitotic index of control or drug-treated cells, indicating proper entry into mitosis for unstressed cells and proper induction of the TOP2α-dependent G2/M checkpoint in ICRF193-treated cells. Together, these data argue against a role for KCTD10 in directly regulating TOP2 dynamics in cells.
Bivalent recognition of TRCs by KCTD10
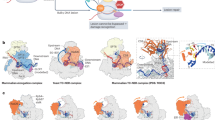
We next tested whether KCTD10 localized to the replication or transcription machinery. In iPOND assays31, KCTD10 localized at nascent DNA in unstressed and stressed cells (Fig. 3a). This was further supported by PLAs showing proximity between KCTD10 and the replication factor and putative KCTD10 binding partner PCNA (Fig. 3b and Extended Data Figs. 4l and 6a). Of note, KCTD10–PCNA proximity pairs were increased by treating cells with the TOP2 inhibitor ICRF193, corroborating the idea that KCTD10 resolves replication-associated lesions created by this drug. Immunoprecipitation of Flag-tagged KCTD10 showed strong co-enrichment of PCNA and the large subunit of RNAPII (POLR2A), which was enhanced by ICRF193 treatment (Fig. 3c). In cells, KCTD10–POLR2A proximity pairs were detected predominantly after treating cells with drugs that affect DNA replication and/or transcription (Fig. 3d,e and Extended Data Figs. 4l and 6a), with the most marked induction occurring after treating cells with ICRF193, PARP inhibition (PARPi) or the splicing inhibitor pladienolide B. Mapping these interactions using co-immunoprecipitation assays showed that the KCTD10 PIP box was essential both for its interaction with PCNA and with POLR2A (Fig. 3f,g). One possible interpretation of this data was that we were detecting an indirect interaction mediated by PCNA or DNA. However, co-immunoprecipitation of PCNA or POLR2A was unaffected by treating lysates with the promiscuous nuclease benzonase (Extended Data Fig. 6b). Furthermore, treating cells with an inhibitor that prevents PCNA loading onto chromatin did not affect ICRF193-induced proximity pairs between POLR2A and KCTD10 (Extended Data Fig. 6c,d). These data suggested that PCNA and RNAPII interact with KCTD10 near the same binding site, thus these interactions are unlikely to co-occur on the same KCTD10 molecule.
a, iPOND assay in U2OS cells treated with EdU for 15 min and DMSO or 1 μM ICRF193 for 4 h prior to EdU. b, PLA for KCTD10–PCNA in U2OS cells treated with 100 nM ICRF193 or DMSO for 4 h. Left, representative images. Right, quantification of foci. Data are mean ± s.d. (n = 200 cells examined over 3 independent experiments). Scale bars, 10 μm. c, Co-immunoprecipitation (IP) assay in HEK293T cells transiently expressing Flag–KCTD10 and treated with 1 μM ICRF193 or DMSO for 4 h. d,e, PLA for KCTD10–POLR2A in U2OS cells treated with indicated drugs for 4 h. d, Quantification of foci. Data are mean ± s.d. (n = 200 cells examined over 3 independent experiments). e, Representative images. PARPi used at 10 μM; PlaB, pladienolide B used at 200 nM; HU, hydroxyurea used at 250 μM; Aph, aphidicolin used at 4 μM. Scale bars, 10 μm. f, schema of KCTD10 truncations. IDR1 and IDR2, intrinsically disordered regions 1 and 2. g, Co-immunoprecipitation assay in HEK293T cells expressing the indicated constructs as depicted in f. EV, empty vector control. h, Co-immunoprecipitation assay in HEK293T cells co-expressing Flag–KCTD10 with the indicated point mutations and Halo–KCTD10 and treated with 1 μM ICRF193 for 4 h. WT, wild type. i, Quantification of PLA foci for POLR2A–PCNA in KCTD10-deficient and control cells rescued with wild-type KCTD10 or KCTD10(R167A) and treated with ICRF193 (100 nM) or DMSO for 4 h. Data are mean ± s.d. (n = 200 cells examined over 3 independent experiments). j, Schema depicting how KCTD10 might bind to PCNA and RNAPII. P values were calculated with a two-tailed unpaired t-test (b) or one-way ANOVA with Bonferroni’s multiple comparisons test (d,i).
Structural and biophysical work has suggested that KCTD10 and other KCTD family members may function as dimers or tetramers through interactions between their BTB domains32,33. To determine whether KCTD10 can self-associate, we performed co-immunoprecipitation experiments using Flag- and Halo-tagged constructs. These assays confirmed that KCTD10 interacts with itself (Extended Data Fig. 6e). Unexpectedly, deletion of the BTB domain did not disrupt binding between Flag–KCTD10 and Halo-tagged KCTD10. By contrast, removal of amino acids 100–238, which span the region between the BTB domain and the PIP box, completely abolished the binding (Extended Data Fig. 6f). In cells, KCTD10 self-association was similarly detected by proximity between Flag- and Halo-tagged KCTD10 (Extended Data Fig. 6g,h). Treatment with ICRF193 enhanced self-association of KCTD10, which was abolished by deletion of residues 100–238, reflecting that KCTD10 undergoes self-association after induction of CD-TRCs.
To further define the self-association interface, we used ColabFold34 (MMseqs2 plus AlphaFold 2-multimer) to model KCTD10 self-association. The model predicted several candidate residues in the proper position to participate in electrostatic interactions between KCTD10 monomers (Extended Data Fig. 6i–k). Among these, R167 emerged as a key residue since the KCTD10(R167A) was unable to undergo self-association in co-immunoprecipitation and in vitro binding assays (Fig. 3h and Extended Data Fig. 6l). To test the importance of KCTD10 self-association, we reintroduced either wild-type KCTD10 or KCTD10(R167A) into KCTD10-deficient cells and quantified TRCs with POLR2A–PCNA PLA (Fig. 3i). Whereas wild-type KCTD10 reduced POLR2A–PCNA proximity pairs, KCTD10(R167A) did not do so. Furthermore, KCTD10(R167A) mutant did not abolish KCTD10 binding with POLR2A or PCNA in co-immunoprecipitation experiments (Extended Data Fig. 6m), although it did modestly decrease the interaction with POLR2A. These findings support the idea that KCTD10 dimerization or oligomerization is important for resolving TRCs and suggest that bivalent recognition of PCNA and the RNAP complex may facilitate KCTD10 self-association to remove TRCs.
KCTD10 recruits CUL3 to co-directional TRCs
We speculated that the dimerization or oligomerization of KCTD10 might be a signal to recruit the CUL3 E3 ligase to TRCs. This model depends on KCTD10 forming a complex with both PCNA and RNAP complexes at TRCs (Fig. 3j). In lysates from untreated cells, size-exclusion chromatography revealed that KCTD10 and PCNA co-eluted in overlapping fractions, whereas POLR2A was present only in a subset of these fractions (Fig. 4a). Following ICRF193 treatment, these proteins tended to co-elute in the same fractions. In fractions in which all three proteins co-eluted after ICRF193 treatment, KCTD10 co-immunoprecipitated both PCNA and POLR2A (Fig. 4b and Extended Data Fig. 7a). By contrast, KCTD10 coprecipitated with PCNA primarily in untreated fractions (Fig. 4b and Extended Data Fig. 7a). These data suggest that KCTD10 may form complexes with both PCNA and POLR2A in cells after ICRF193 treatment, potentially allowing KCTD10 dimers or oligomers to form at TRCs.
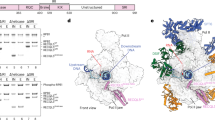
a, Western blot of fractions from size-exclusion chromatography from control (DMSO) and ICRF193-treated (1 μM, 4 h) U2OS cells. b, KCTD10 co-immunoprecipitation assay from the indicated fractions from ICRF193-treated cells as indicated in a. c,d, PLA for CUL3–POLR2A in KCTD10-deficient U2OS cells treated with 100 nM ICRF193 or DMSO for 4 h. c, Quantification of foci. Data are mean ± s.d. (n = 200 cells examined over 3 independent experiments). d, Representative images. e,f, PLA for CUL3–POLR2A in HEK293T cells expressing the co-directional ECFP reporter treated with 1 mg ml−1 doxycycline for 20 h and 100 nM ICRF193 or DMSO for 4 h. Scale bars, 10 μm. e, Quantification of foci. Data are mean ± s.d. (n = 200 cells examined over 3 independent experiments). f, Representative images. Scale bars, 10 μm. g, iPTM scores from AlphaFold 2-multimer predictions with KCTD10. h, PAE plots for the top-ranked AlphaFold 2-multimer predictions for POLR2C–KCTD10 and TCEA2–KCTD10. Right, schematic for the uncharacterized supersecondary structure located in KCTD10 as depicted. i, Predicted structure of KCTD10 bound to POLR2C. j, Predicted structure of KCTD10 bound to TCEA2. k, Co-immunoprecipitation assay in HEK293T cells expressing Flag-tagged KCTD10 and treated with 1 μM ICRF193 or DMSO. P values were calculated with one-way ANOVA with Bonferroni’s multiple comparisons test.
Consistent with a model in which KCTD10 senses TRCs and functions as a platform to recruit CUL3, CUL3–POLR2A proximity pairs were enriched in cells after treatment with ICRF193 (Fig. 4c,d). These pairs were markedly reduced in the absence of KCTD10, indicating that KCTD10 is required for CUL3 recruitment to RNAP complexes. A similar phenotype was seen when testing for ubiquitin–POLR2A proximity pairs in cells (Extended Data Fig. 7b–d). CUL3–POLR2A proximity pairs could also be induced after creating co-directional TRCs in ECFP-expressing reporter cells (Fig. 4e,f and Extended Data Fig. 2i). In these cells, KCTD10–POLR2A pairs were also induced by creating co-directional TRCs (Extended Data Fig. 7e,f). Furthermore, when CUL3 activity was inhibited with a small molecule neddylation inhibitor (MLN4924), we observed an increase in the number of TRCs independent of KCTD10 (Extended Data Fig. 7g,h). These data show that KCTD10 recruits CUL3 to TRCs, and that CUL3 activity is important for the resolution of TRCs in cells. Notably, treatment with etoposide did not induce CUL3–POLR2A proximity (Extended Data Fig. 7i,j), suggesting that this CUL3–KCTD10 axis is specifically activated by lesions that are stabilized by the catalytic TOP2 inhibitor ICRF193, but not by TOP2 inhibitors more broadly.
To test whether dimerization is sufficient to induce the binding with CUL3, we inserted a modified FKBP domain (DmrB) at the C terminus of KCTD10, which enabled us to artificially induce dimerization using a ligand (AP20187) (Extended Data Fig. 8a). Inducing KCTD10 dimerization led to increased co-immunoprecipitation of CUL3 from cell lysates (Extended Data Fig. 8b). CUL3–POLR2A proximity pairs could not be induced by ICRF193 in G1-arrested cells (Extended Data Fig. 8c,d), indicating that ongoing DNA replication is necessary for the response. Inhibition of PCNA loading (Extended Data Fig. 8e–h) and inactivation of the KCTD10 PIP box (Extended Data Fig. 8i,j) also reduced both CUL3–POLR2A and ubiquitin–POLR2A proximity pairs, indicating that PCNA specifically contributes to CUL3 engagement. Furthermore, deletion of the PIP box in KCTD10 increased sensitivity to ICRF193 (Extended Data Fig. 8k) and DNA damage in the presence of ICRF193 (Extended Data Fig. 8l). As KCTD10 is recruited to RNAP complexes independently of PCNA (Extended Data Fig. 6c,d), these data demonstrate that KCTD10 dimerization or oligomerization is critical for CUL3 recruitment.
Finally, we measured CUL3–POLR2A proximity pairs after reintroducing the dimerization-defective KCTD10(R167A) mutant. KCTD10(R167A) did not rescue CUL3 recruitment to RNAP complexes after ICRF193 treatment (Extended Data Fig. 8m). Together, these data support a model in which CUL3–KCTD10 responds to TRCs such as those created by the catalytic TOP2 inhibitor ICRF193. Once recruited, KCTD10 facilitates ubiquitination events at the TRCs.
KCTD10 interacts with POLR2C and TCEA2
The core RNAPII complex consists of 12 proteins. In addition to these core proteins, a number of accessory factors are required to facilitate each step of transcription29. Therefore, despite our work showing potential interactions between KCTD10 and the largest subunit, POLR2A, we aimed to further clarify which protein was responsible for direct binding with KCTD10 within the complex. To accomplish this, we utilized ColabFold34 to screen for potential interaction partners among each of the core RNAPII proteins and all proteins associated with transcription by RNAPII (GO:0006366) (Extended Data Fig. 9a). For each protein, we generated five models, and the interface-predicted TM score (iPTM) was used to score potential hits (Fig. 4g and Extended Data Fig. 9b). As a control, models were built for KCTD10 with a PCNA monomer, dimer or trimer. The best iPTM score between KCTD10 and PCNA was found for models containing a PCNA trimer (iPTM = 0.58) (Fig. 4g), supporting the idea that KCTD10 binds to the functional complex on chromatin. To prioritize hits across our screen of transcriptional proteins, we selected top hits (iPTM > 0.4) and visualized them as networks (Extended Data Fig. 9c). This analysis revealed potential interactions with a broad range of RNAPII components that are involved in transcription initiation and elongation (for example, POLR2C, TBP, CCNT1, TCEA2, TCEA3 and CDK12), chromatin-modifying enzymes (for example, SETD2 and ARID4B), RNAPII termination (for example, PCF11) and several lineage-specific transcription factors (MEN1, GATA3, BCL9, TBX5 and WNT3A).
Among all core RNAPII proteins tested, the best iPTM score was between the third-largest RNAPII subunit POLR2C (iPTM = 0.62), with POLR2A coming in a distant second (iPTM = 0.3) (Fig. 4g). In addition, the TFIIS isoforms TCEA2 and TCEA3 were among our top hits, supporting previous identification of KCTD10 binding with TCEA2 in affinity purification–mass spectrometry experiments35. TCEA2 and TCEA3 are two of the three vertebrate isoforms of TFIIS, an elongation factor that increases RNAP fidelity and speed36,37,38,39. Additionally, TFIIS is responsible for rescuing permanently stalled or arrested RNAPs, thereby promoting their return to active elongation complexes29,40,41.
On the basis of these results, we further explored interactions with POLR2C and TCEA2. The top-ranked structure for each of these proteins in complex with KCTD10 revealed potential binding sites near the PIP box of KCTD10 (Fig. 4h–j). Predicted alignment error (PAE) plots showed that interprotein interactions occurred predominantly across mutually exclusive regions of KCTD10 (Fig. 4h). A C-terminal tail near the PIP box of KCTD10 inserts into a groove formed by POLR2C (Fig. 4h,i). Meanwhile, an uncharacterized mixed secondary structure domain in KCTD10 interacts with domain II and part of the interdomain linker in TCEA2, which have previously been shown to insert into the jaw and funnel, respectively, of RNAPII38 (Fig. 4h,j). This interaction is conserved across all three TFIIS isoforms (Fig. 4h and Extended Data Fig. 9d,e), however the predictions show a much lower confidence for interactions between KCTD10 and TCEA1 (Extended Data Fig. 9d). These predicted interactions between KCTD10 and POLR2C, TCEA2 or TCEA3 were validated by co-immunoprecipitations (Fig. 4k and Extended Data Fig. 9f,g).
These data suggest that KCTD10 engages the RNAPII complex from the rear. To test this spatially, we assessed proximity between KCTD10 and either front-facing (POLR2I and POLR2E) or rear-facing (POLR2C and POLR2J) RNAPII subunits29. In the presence of ICRF193, KCTD10 formed PLA foci with POLR2C and POR2J, but not with POLR2I or POLR2E (Extended Data Fig. 9h–j), confirming its selective association with the rear of RNAPII.
KCTD10 mediates TCEA2 ubiquitination
We hypothesized that KCTD10 may ubiquitinate one of the RNAPII complex members identified in our AlphaFold screen. To test this, we expressed His-tagged ubiquitin in cells and performed denaturing pulldowns to enrich ubiquitin-conjugated proteins. In response to ICRF193 treatment, we detected ubiquitination of core RNAPII subunits (POLR2A, POLR2B and POLR2C) (Extended Data Fig. 9k). However, the ubiquitination of these subunits was unaffected by KCTD10 knockdown, suggesting they are not direct targets of the CUL3–KCTD10 complex.
By contrast, TCEA2 was robustly ubiquitinated following ICRF193 treatment, and this ubiquitination was markedly reduced by KCTD10 knockdown (Fig. 5a) or pharmacological inhibition of CUL3 with a neddylation inhibitor (Extended Data Fig. 9l). This appeared specific to TCEA2, as TCEA3 was not detectably ubiquitinated under the same conditions (Extended Data Fig. 9k). Mapping the modification site revealed that K184 is the primary ubiquitination site on TCEA2 following ICRF193 treatment (Fig. 5b and Extended Data Fig. 9m). Together, these results identify TCEA2 as a specific substrate of the CUL3–KCTD10 complex within the RNAPII machinery.
a, Denaturing Ni-NTA pulldown from HEK293T cells expressing Flag-tagged TCEA2 and His-tagged ubiquitin (His–Ub) and treated with 1 μM ICRF193 or DMSO in the presence of 50 μM MG132 for 4 h. b, Denaturing Ni-NTA pulldown from KCTD10-deficient (sh3) and control HEK293T cells expressing wild-type or K184R mutant Flag–TCEA2 and/or His–Ub in the presence of 50 μM MG132. c, Western blot of the chromatin fraction from KCTD10-deficient or control U2OS cells treated with 1 μM ICRF193, 1 μM etoposide (ETP) or 2.5 μM VCP inhibitor (VCPi; NMS-873) for 6 h. d, Western blot of chromatin fractions from TCEA2-deficient HEK293T cells expressing wild-type TCEA2, TCEA2(K184R) or empty vector (−) treated with ICRF193 (1 μM) or DMSO for 4 h. siTCEA2, small interfering RNA (siRNA) targeting TCEA2. e, EdU incorporation in KCTD10-deficient (sh3), TCEA2-deficient (siTCEA2) and control U2OS cells treated with 10 μM EdU and 100 nM ICRF193 or control for 6 h. Data are mean ± s.d. (n = 3 biologically independent experiments). siCtr, control siRNA. f,g, γH2AX foci from KCTD10-deficient U2OS cells with control siRNA (siCtr) or siTCEA2 and treated with 100 nM ICRF193 or DMSO for 6 h. f, Quantification of foci. Data are mean ± s.d. (n = 200 cells examined over 3 independent experiments). g, Representative images. Scale bars, 10 μm. Data are mean ± s.d. P values were calculated with one-way ANOVA with Bonferroni’s multiple comparisons test.
Prolonged ICRF193 treatment (6 h) led to reduced chromatin-bound RNAPII and TCEA2 in a KCTD10-dependent manner (Fig. 5c). This raised the question of whether TCEA2 ubiquitination might contribute to RNAPII removal. Supporting this idea, KCTD10-deficient cells exhibited a marked increase in chromatin-bound TCEA2 and RNAPII (Fig. 5c and Extended Data Fig. 9n) and an enhanced interaction between POLR2A and TCEA2, as shown by co-immunoprecipitation (Extended Data Fig. 9o). To assess whether TCEA2 removal influences RNAPII retention, we depleted TCEA2 and reconstituted cells with wild-type TCEA2 or TCEA2(K184R). We observed a corresponding decrease in chromatin-bound RNAPII after ICRF193 treatment in TCEA2-depleted cells, and cells expressing TCEA2(K184R) showed stronger RNAPII retention (Fig. 5d). Notably, inhibition of the AAA+ ATPase VCP (also known as p97) blocked RNAPII unloading under these conditions (Fig. 5c, lanes 2 and 3), suggesting that RNAPII removal requires active remodelling. However, VCP inhibition had no effect in KCTD10-deficient cells and no effect on TCEA2 unloading from chromatin (Fig. 5c, lanes 7 and 8), placing KCTD10 function upstream of VCP in the pathway controlling RNAPII turnover at TRCs.
Based on our finding that RNAPII degradation rescues the replication defect in KCTD10-deficient cells (Fig. 1h), we hypothesized that active RNAPII remodelling—mediated by TCEA2 ubiquitination and removal—may facilitate replisome bypass at TRCs. Consistent with this hypothesis, co-depletion of TCEA2 restored DNA replication in KCTD10-deficient cells treated with ICRF193 (Fig. 5e and Extended Data Fig. 9p). Furthermore, supporting the idea that persistent TCEA2 retention causes DNA damage at TRCs, TCEA2 depletion suppressed ICRF193-induced DNA damage regardless of KCTD10 status (Fig. 5f,g). These findings support a model in which CUL3–KCTD10 ubiquitinates TCEA2, promoting its removal from chromatin and enabling RNAPII remodelling to facilitate replication bypass at TRCs.
Discussion
These data support a model in which KCTD10 functions as a key component of the mammalian replisome, facilitating replisome bypass at TRCs (Extended Data Fig. 10). In support of this model, previous work has shown that TFIIS is bound to active elongation complexes, and its presence promotes the transformation to higher-energy elongation-competent states with increased processivity41,42,43,44,45,46. Of note, the bacterial TFIIS homologue has been shown to inhibit DNA repair by maintaining elongation-competent RNAP47, raising the possibility that TCEA2 and its other family members may have a conserved role in coordinating transcription with genome stability.
KCTD10 is one of three members in clade C of the KCTD superfamily of BTB-domain proteins32. It has been proposed that KCTD family members form higher-order dimeric and oligomeric states that may provide a suitable platform for CUL3 recruitment despite their lack of a C-terminal 3-box extension of the BTB domain that is important for BTB-CUL332,33,48,49. Our data suggest that the BTB domain itself does not mediate this oligomerization; instead, KCTD10 appears to bridge distinct substrates (the replisome and RNAP complex) prior to CUL3 recruitment. It remains to be seen whether other clade C KCTD family members, which share a similar PIP box32, have analogous functions in replication.
Our findings reveal that perturbing CUL3–KCTD10 exposes several therapeutic vulnerabilities, as KCTD10-deficient cells have increased sensitivity to catalytic inhibitors of TOP2, ATM or CHK2. In the case of ICRF193, replication stress depends on the presence of RNAPII, suggesting that damage arises primarily from collisions with stalled RNAPII, rather than direct inhibition of the replisome. This implies that ICRF193-induced TOP2 lesions present a greater barrier to transcription than to replication, and that alternative replication-associated pathways exist for resolving ICRF193-induced lesions. These lesions do not significantly recruit RAD51, suggesting that they are not repaired by homologous recombination. This contrasts with other reports showing that TRC-associated damage typically activates homologous recombination repair50,51, and suggests that the type of TRC induced by ICRF193 may contrast significantly with other TOP2 inhibitors such as etoposide. Alternatively, ICRF193 may elicit a weaker replication checkpoint response than etoposide owing to slower or less severe induction of damage.
This work invites broader questions about how KCTD10 shapes the interplay between DNA replication and transcription. In mammalian genomes, transcription and replication are preferentially co-oriented, a bias that is thought to minimize head-on TRCs26. The existence of the pathway that we describe here may contribute to this bias, reducing selective pressure for alternative genome configurations. This mechanism could be particularly critical for maintaining the stability of regions with high transcriptional activity, given the relationship between KCTD10 and TOP2 and the importance of TOP2 for the rapid transcription of stimulus-responsive genes28,52,53,54,55. Finally, whether KCTD10 also contributes to the maintenance of epigenetic information, beyond its role in genome stability, remains an open question.
Methods
Cell lines
HEK293T, HeLa and U2OS cells were purchased from ATCC. All cell lines were authenticated by short tandem repeat analysis performed at Mayo Clinic. Cell lines were routinely tested for Mycoplasma contamination. HEK293T and HeLa cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM). U2OS cells were cultured in DMEM:F12 supplemented with 10% FBS. All cells were grown at 37 °C with 5% CO2.
Plasmid transfection and lentiviral infection
Full-length KCTD10 and the IDR2Δ deletion were cloned into pLVX3-CMV-puro (3×Flag at the N terminus) vector using PCR amplification of cDNA with restriction sites added as a 5′ primer overhang. BTBΔ, PIP3A and PIPΔ mutants of KCTD10 were generated by site-directed mutagenesis of the full-length KCTD10-pLVX3 plasmid (Q5 site-directed mutagenesis, New England Biolabs). KCTD10 shRNAs and TOP2B shRNAs were purchased from Sigma-Aldrich. Scramble shRNA was a gift from D. Sabatini56. MYC–CUL3 (pcDNA3-MYC-CUL3) was a gift from Y. Xiong57. TCEA1, TCEA2 and TCEA3 cDNA was synthesized (Genewiz), digested and ligated into pLVX3-CMV-puro.
Cells were transfected with TransIT-X2 (Mirus) according to manufacturer’s instructions. For transient expression, cells were analysed 48 h after transfection. To generate lentivirus, HEK293T cells were transfected with the plasmid of interest, psPAX2 and pMD2G (at a ratio of 5:3.75:1.25). Thirty-six to forty hours after transfection, culture supernatants were collected, centrifuged at 800g for 5 min and passed through a 0.2-μm filter to remove cells and debris. Filtered virus was then split into 1 ml aliquots and stored at −80 °C until use.
For lentiviral transduction, cells were seeded in one well of a 6-well plate 16–18 h prior to infection. At the time of infection, 1 ml of medium was removed and replaced with 1 ml of viral supernatant together with 10 μg ml−1 polybrene. For experiments with transient knockdown or expression, cells were used 48 h later for experiments. For stable cell lines, cells were selected with the appropriate antibiotic for at least one week. Stable cell lines were frozen at this stage and not used for more than 15 passages after this point.
Immunoprecipitation and immunoblotting
Cells were washed three times with PBS and lysed in NETN buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, supplemented with 50 mM β-glycerophosphate, 10 mM NaF and protease inhibitors) at 4 °C with rotation for 30 min. Cell lysates were clarified by sonication with a Bioruptor Plus (Diagenode) for 8 cycles (30 s ON/30 s OFF, at 4 °C) and centrifuged at 12,500g for 15 min. For immunoprecipitations, clarified lysates were incubated with anti-Flag M2 Affinity Gel (Sigma) or with protein A/G magnetic beads (ThermoFisher) for 2 h at 4 °C with rotation. Beads were washed four times with lysis buffer. For SDS–PAGE, clarified lysates were then diluted into Laemmli buffer and boiled for 5–10 min at 95 °C. Samples were migrated through stacking gels at 75 V and resolving gels at 200 V using Tris-glycine-SDS buffer. Wet transfers were performed for 60 min at 300 mA using Tris-glycine buffer. Membranes for phosphoprotein blots were blocked in 5% BSA in TBST, and all other membranes were blocked in 5% fat-free milk in TBST for 30 min at room temperature. Membranes were incubated with primary antibody solutions in TBST overnight at 4 °C and in secondary antibody solutions in TBST for 1 h at room temperature. Membranes were imaged using SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher) on a LI-COR Odyssey Fc.
Size-exclusion chromatography
Size-exclusion chromatography was performed as described58. In brief, cells were treated with ICRF193 or control (DMSO). Cell lysates were run on an AKTA Purifier FPLC system with a Superdex 200 gel filtration column (GE Healthcare) to separate different fractions. Collected fractions were subjected to western blotting and the indicated fractions were subsequently incubated with anti-KCTD10 antibody (Sigma) overnight. The immunocomplexes were then separated by SDS–PAGE and western blotting was done as described above.
Denaturing Ni-NTA pulldowns
Cells were lysed in 8 M urea, 100 mM NaH2PO4, 300 mM NaCl, 10 mM Tris-HCl pH 8.0. Lysates were then sonicated with a Bioruptor Plus (Diagenode) for 8 cycles (30 s ON/30 s OFF, at 4 °C) and incubated with Ni-NTA agarose beads (Qiagen) for 1–2 h at room temperature. Beads were washed five times with 8 M urea, 100 mM NaH2PO4, 300 mM NaCl, 10 mM Tris-HCl pH 8.0. Samples were then boiled in loading buffer and subjected to SDS–PAGE and immunoblotting as described above.
Isolation of proteins on nascent DNA
iPOND assays were performed as described59. In brief, 3× 150-mm dishes per sample were labelled with 10 μM EdU for 15 min at 37 °C. After this, cells were immediately fixed by adding 1% paraformaldehyde in PBS to cells after decanting the culture supernatant. After 20 min, cross-linking was quenched with 1 ml of 1.25 M glycine and cells were collected by scraping and transferred to a 50-ml tube. Cells were washed and permeabilized with 0.25% Triton X-100 in PBS for 30 min at room temperature. Click reactions were performed with 10 mM sodium ascorbate, 2 mM CuSO4 and 10 μM biotin azide for 2 h at room temperature. After washing, biotin-labelled cells were lysed (50 mM Tris-HCl pH 8.0, 1% SDS) and sonicated. The cell lysates were incubated with streptavidin magnetic beads overnight at 4 °C. Beads were washed with once with lysis buffer, once with 1 M NaCl, and then twice with lysis buffer. Washed beads were incubated with 2× Laemmli buffer (1:1 (v:v) with beads) and were analysed by SDS–PAGE and immunoblotting.
In vitro binding assay
Flag-tagged KCTD10 protein was generated using the TNT system with wild-type KCTD10 per the manufacturers protocol (Promega). To generate glutathione S-transferase (GST)-tagged wild-type and R167A-mutant KCTD10, the full-length protein was cloned into the pGEX-4T-2 and purified from Escherichia coli using GST Resin (Sigma). The purification was verified by SDS–PAGE as shown in Extended Data Fig. 7l. Purified Flag–KCTD10 and GST–KCTD10 were then incubated together at a 1:1 ratio in ice-cold buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, supplemented with 50 mM β-glycerophosphate, 10 mM NaF and protease inhibitors) at 4 °C for 4 h. Following this, GST–KCTD10 was immunoprecipitated and subsequently immunoblotted with an anti-Flag antibody.
Immunofluorescence
Cells were grown on coverslips for 18–24 h before experiments and then treated as indicated for each experiment. As described previously60, nuclei for RAD51 foci were pre-extracted with (20 mM HEPES pH 7.4, 20 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol and protease inhibitor cocktail) for 3 min on ice and fixed in 4% paraformaldehyde for 10 min on ice. After fixation, coverslips were permeabilized with 0.25% Triton X-100 for 10 min at room temperature. Cells were blocked in 3% BSA in PBS for 30 min. Primary and secondary antibodies were diluted in 3% BSA in PBS and incubated on coverslips for 1 h at room temperature. Nuclei were counterstained with Hoechst 33342. Coverslips were mounted onto glass slides using a glycerol-based mounting medium with p-phenylenediamine (10 mg ml−1; Sigma). Coverslips were sealed and imaged on an ImageXpress Micro Confocal system (Molecular Devices).
Nascent DNA imaging
Cells, grown on coverslips, were labelled with EdU (10 μM) for 30 min. Following this, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 20 min at room temperature. After permeabilization, cells were washed twice in 3% BSA in PBS. Click reactions were then performed with 10 mM sodium ascorbate, 2 mM CuSO4 and 10 μM Alexa Fluor 488 azide for 30 min at room temperature, protected from light. For co-staining proteins, primary and secondary antibodies were added at this point in the same manner described for general immunofluorescence. Nuclei were counterstained with Hoechst 33342, mounted on coverslips and imaged as described above.
Nascent transcription imaging
Nascent transcription imaging was performed using an RNA synthesis kit (abcam). Cells were grown on coverslips and labelled with EU for 1 h. Fixation, permeabilization and click reactions were performed according to the manufacturer’s instructions. Nuclei were counterstained with DAPI as detailed in the protocol. Coverslips were imaged on an ImageXpress Micro Confocal system (Molecular Devices). Nuclear EU intensity was measured in ImageJ using the DAPI channel to create image masks of the nuclei. Intensities for each nuclei are plotted as a single point. For representative images, the mpl-inferno lookup table was applied to the EU channel images.
In situ PLA
Cells were grown on coverslips for 18–24 h before experiments. For PLA assays using 293T cells, cells were grown on poly-l-lysine-coated coverslips to enhance attachment. Cells were pre-extracted for 3 min on ice with nuclear extraction buffer60 (20 mM HEPES, pH 7.4, 20 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol and protease inhibitor cocktail). Following this, cells were fixed with 4% paraformaldehyde for 10 min on ice and permeabilized with 0.2% Triton X-100 for 10 min. PLA was performed by a Duo-link in situ PLA kit (Sigma) according to the manufacturer’s instructions. In brief, samples were blocked in blocking solution at 37 °C for 1 h and incubated with primary antibodies (1:500) at room temperature for 1 h. Then, probes were incubated at room temperature for 1 h. Ligation and hybridization were performed at 37 °C for 30 min and 90 min, respectively. Nuclei were counterstained with Hoechst 33342. Coverslips were mounted onto glass slides using a glycerol-based mounting medium with p-phenylenediamine (10 mg ml−1; Sigma) and visualized with an ImageXpress Micro Confocal system (Molecular Devices).
DNA fibre assays
DNA fibre assays were performed as previously described61. In brief, cells were pulsed with 25 μm CldU for 20 min, washed with PBS (37 °C), and pulsed with 250 μm IdU for 20 min. Cells were washed in PBS, counted and resuspended at 1 × 106 per ml. Approximately 2–3 μl of cells were added to the top of a glass slide and dried for approximately 2 min. Cells were then lysed by adding 15 μl lysis buffer (200 mM Tris-HCl pH 7.4, 50 mM EDTA, 0.5% SDS) directly to the cell droplet. The slides were then tilted at an angle by placing them on one end of a tissue culture plate lid allowing DNA fibres to stretch across the slide. Fibres were fixed in 3:1 methanol:acetic acid for 10 min and then air dried. After washing slides with PBS, the fibres were denatured in 2.5 M HCl overnight at 4 °C. Following this, fibres were blocked in 2% BSA in PBS for 30 min and stained with primary antibodies diluted in 2% BSA in PBS for 2 h at room temperature (mouse anti-BrdU (1:25, BD Biosciences) and rat anti-BrdU (1:1,000, abcam)). After washing three times with PBS, fibres were stained with secondary antibodies for 1 h at room temperature (Rhodamine Red anti-rat (1:500) and Alexa Fluor 488 anti-mouse (1:500)). Coverslips were mounted onto slides and images were acquired on a Nikon eclipse 80i fluorescence microscope with a 63× objective. Fibre lengths were measured in ImageJ.
Cell fractionation
Subcellular fraction was performed as described previously with modifications62. In brief, cells were collected, washed in PBS, and lysed for 20 min on ice (10 mM HEPES, pH 7.4, 10 mM KCl, 0.05% NP-40, supplemented with 50 mM β-glycerophosphate, 10 mM NaF and protease inhibitors). After this, nuclei were pelleted by centrifugation in a microcentrifuge (14,000 rpm at 4 °C for 10 min). The nuclei were washed once with lysis buffer and then incubated in low salt buffer for 15 min on ice (10 mM Tris-HCl pH 7.4, 0.2 mM MgCl2, 1% Triton X-100, supplemented with 50 mM β-glycerophosphate, 10 mM NaF, and protease inhibitors). After centrifugation, the soluble fraction was saved as the soluble nuclear proteins. The pellet was resuspended in NETN buffer and sonicated with a Bioruptor Plus (Diagenode) for 8 cycles (30 s ON/30 s OFF, at 4 °C) and centrifuged at 12,500g for 15 min. Lysates were then diluted into Laemmli buffer and analysed by SDS–PAGE with immunoblotting as described above.
RNA/DNA dot blotting
DNA/RNA hybrids were extracted as described63. In brief, cells were lysed in TE buffer with 0.625% SDS and 62.5 μg ml−1 proteinase K for 3 h at 37 °C. DNA was extracted by phenol-chloroform extraction and ethanol precipitated. Precipitated DNA was spooled and washed with 70% ethanol without centrifugation. After air drying DNA, it was resuspended in TE buffer and sonicated. The concentration of DNA was analysed by (Qubit high-sensitivity dsDNA; Invitrogen). Samples were normalized based off the total amount of double stranded DNA (dsDNA) (100 ng μl−1) and were pre-treated with nucleases (RNase H or RNase T1) if indicated. A 2-μl dot of sample was added to a nylon membrane. After the membranes had dried, the nucleic acids were crosslinked by UV irradiation (120 mJ cm−2). Membranes were hydrated in TBS, blocked in 5% milk in TBS, and incubated either S9.6 or dsDNA antibodies (abcam) overnight at 4 °C. Membranes were then washed, stained with secondary antibodies and imaged.
Clonogenic assays
Cells were seeded in triplicate in each well of a 6-well plate (800–1,000 cells per well). After cells attached (16 h later), cells were treated with indicated drugs and left in medium containing the drug for 10–14 days. Colonies were stained with Giemsa solution (Sigma) and counted. Data were normalized to plating efficiencies.
Cell cycle analysis
Cells were trypsinized, collected and filtered through a cell strainer to achieve a single-cell suspension. Ethanol was then added drop-wise with constant mixing to fix and permeabilize cells. Fixed cells were washed three times with PBS, pelleting cells each time with centrifugation at 900g for 5 min at 4 °C. Fixed cells were then resuspended in PI/RNase solution (Thermo Fisher) at room temperature for 30 min. Stained samples were analysed on an AttuneNxT flow cytometer (Thermo Fisher). Cell cycle analysis was performed using FlowJo.
Screening for KCTD10 interaction partners
FASTA sequences belonging the GO term ‘transcription by RNA polymerase II’ (GO:0006366) were retrieved from the European Bioinformatics Institute (EMBL-EBI) Gene Ontology and GO Annotations database (www.ebi.ac.uk/QuickGO/). Using Linux shell commands, the FASTAs were split, concatenated with the full-length FASTA sequence for KCTD10 (UniprotKB ID: Q9H3F6), and then pooled into batches of 100 files. Each batch was run through ColabFold (v1.5.5)34,64,65 using Tesla T4 GPUs as a first pass, then larger complexes were completed using Tesla A100 GPUs. One protein could not be completed due to limitations of AlphaFold 2 (CHD7). iPTM scores were extracted from individual json files and used to create a heat map with rows clustered using Seaborn and matplotlib (Extended Data Fig. 10c). To prioritize top hits, an iPTM score >0.4 was chosen, corresponding to >1 s.d. above the average iPTM scores for the complex formed between KCTD10 and two copies of PCNA. Protein structures were visualized and images for figures were created using ChimeraX-1.6.1. The PAE generated by AlphaFold 2 was plotted as 2D heat maps using ChimeraX-1.6.1.
CUT&RUN sequencing
CUT&RUN was performed according to published protocols66. In brief, 5 × 105 fresh cells were washed twice with wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 1× protease inhibitor cocktail) at room temperature, and incubated with Concanavalin-coated (ConA) magnetic beads (Bangs Laboratories, BP531) prewashed with binding buffer (20 mM HEPES pH 7.9, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) at room temperature for 10 min. Then the samples were incubated with primary antibody (mouse anti-Flag, 1:100) in antibody buffer (wash buffer containing 0.05% Digitonin, 2 mM EDTA) on a rotating platform at 4 °C overnight. The beads were then washed with digitonin-containing buffer (wash buffer, 0.05% digitonin), and resuspended with pA/G-MNase (700 ng ml−1 in digitonin buffer) and rotated for 1 h at 4 °C. After two washes with ice-cold digitonin buffer to remove unbound pA/G-MNase, beads were subjected to digestion in pA/G-MNase digestion buffer (digitonin buffer containing 2 mM CaCl2) on ice for 1 h. Solubilized chromatin was then released using CUT&RUN stop buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% digitonin, 50 µg ml−1 glycogen and 100 μg ml−1 RNase A) and incubated at 37 °C for 30 min. CUT&RUN DNA was extracted with phenol-chloroform-isoamyl alcohol (Invitrogen, 15593049) and ethanol precipitation and then dissolved in 12 μl 1 mM Tris-HCl pH 8 with 0.1 mM EDTA. The DNA libraries were prepared by NEBNext Ultra II DNA Library Prep Kit for Illumina (E7645) v.2.
Sequencing analysis
DNA libraries were sequenced on a NovaSeq X Plus sequencer. Reads were mapped to the GRCh38 human reference genome using BWA-MEM with default settings67. Sequencing duplicates were marked and removed using Picard implemented in the Genome Analysis Toolkit (GATK4)68. KCTD10 peaks were called using Macs269. Peaks with false discovery rate (FDR) ≤0.05 were considered significant. Sequencing depth in peaks were normalized as reads per kilobase of transcript per million mapped reads (RPKM) using the bamCoverage function in deepTools. Okazaki sequencing data were retrieved from NCBI BioProject PRJNA30158826. RFD was computed using OKseqHMM70. Co-directional or head-on TRCs were identified as described24,25 using a custom script. Genomic feature enrichment in peaks was classified using ChIPseeker package71. Representative regions showing KCTD10 binding regions with TRCs were visualized in Integrative Genomics Viewer (IGV)72. Gene overlap and transcription factor target enrichment analysis was performed using Metascape73.
Figures and schemas
All individual figure panels were made in GraphPad Prism and assembled into figures using Adobe illustrator. Schema were created in Adobe illustrator.
Statistics and reproducibility
All experiments were repeated at least three times independently unless otherwise stated in the figure legends. No statistical methods were used to determine sample size. Investigators were not blinded to the experiment owing to staffing constraints but data were analysed by multiple investigators, some of whom were blinded to the experiment. Immunofluorescence images were acquired and analysed in a blinded fashion. Samples were randomly allocated into experimental groups before treatment and were performed under the same conditions. For example, plates were randomly assigned to groups for experiments. The statistical tests used and resulting P values are indicated in the figures and corresponding legends. Statistical analyses were performed using GraphPad Prism.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The CUT&RUN data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession GSE295262. Data associated with predicted KCTD10 interactions are available on Dryad (https://doi.org/10.5061/dryad.15dv41p7z). We used the following publicly available datasets: γH2AX interphase scores (interphase_nucleus_gH2AX_mean_score; column AK) and KCTD10 phenotypic clusters (interphase cluster 106, mitotic cluster 15) (Extended Data Fig. 1a,k) are available at https://vesuvius.wi.mit.edu/genes.xlsx: gene–drug interactions for KCTD10 and DNA damaging agents (Extended Data Fig. 1b,c) are available at https://durocher.shinyapps.io/GenotoxicScreens/ and on Mendeley Data (https://data.mendeley.com/datasets/gfcn2wmrpf/1)74. The Homo sapiens h38 genome assembly is available from NCBI (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.40/). Okazaki sequencing data were retrieved from NCBI BioProject PRJNA301588. The GO lists referenced in this study were downloaded on Quick Go (https://www.ebi.ac.uk/QuickGO/annotations). All materials are available upon request. All plasmids and cell lines require a material transfer agreement (MTA). Source data are provided with this paper.
Code availability
ColabFold is available on Github (https://github.com/sokrypton/ColabFold). Metascape is available at https://metascape.org/gp/index.html#/main/step1. Scripts for major genomic data analyses are included in Supplementary Note 1 and available at https://github.com/xiaoguanghuan123/KCTD10_TRC.
References
Stewart-Morgan, K. R., Petryk, N. & Groth, A. Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 22, 361–371 (2020).
Kotsantis, P., Petermann, E. & Boulton, S. J. Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov. 8, 537–555 (2018).
Goehring, L., Huang, T. T. & Smith, D. J. Transcription–replication conflicts as a source of genome instability. Annu. Rev. Genet. 57, 157–179 (2023).
Marchal, C., Sima, J. & Gilbert, D. M. Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell Biol. 20, 721–737 (2019).
Hu, Y. & Stillman, B. Origins of DNA replication in eukaryotes. Mol. Cell 83, 352–372 (2023).
Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017).
Gaillard, H., García-Muse, T. & Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015).
Hamperl, S., Bocek, M. J., Saldivar, J. C., Swigut, T. & Cimprich, K. A. Transcription–replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170, 774–786.e19 (2017).
Wang, J. et al. Persistence of RNA transcription during DNA replication delays duplication of transcription start sites until G2/M. Cell Rep. 34, 108759 (2021).
Brüning, J.-G. & Marians, K. J. Replisome bypass of transcription complexes and R-loops. Nucleic Acids Res. 48, 10353–10367 (2020).
Dutta, D., Shatalin, K., Epshtein, V., Gottesman, M. E. & Nudler, E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543 (2011).
Funk, L. et al. The phenotypic landscape of essential human genes. Cell 185, 4634–4653.e22 (2022).
Olivieri, M. et al. A genetic map of the response to DNA damage in human cells. Cell 182, 481–496.e21 (2020).
Ling, E. M. et al. A comprehensive structural analysis of the ATPase domain of human DNA topoisomerase II beta bound to AMPPNP, ADP, and the bisdioxopiperazine, ICRF193. Structure 30, 1129–1145.e3 (2022).
Pintard, L., Willems, A. & Peter, M. Cullin‐based ubiquitin ligases: Cul3–BTB complexes join the family. EMBO J. 23, 1681–1687 (2004).
Maekawa, M. & Higashiyama, S. KCTD10 biology: an adaptor for the ubiquitin E3 complex meets multiple substrates: emerging divergent roles of the cullin-3/KCTD10 E3 ubiquitin ligase complex in various cell lines. Bioessays 42, e1900256 (2020).
Moldovan, G.-L., Pfander, B. & Jentsch, S. PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007).
Wang, Y. et al. KCTD10 interacts with proliferating cell nuclear antigen and its down-regulation could inhibit cell proliferation. J. Cell. Biochem. 106, 409–413 (2009).
He, H., Tan, C.-K., Downey, K. M. & So, A. G. A tumor necrosis factor α- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase δ and proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA 98, 11979–11984 (2001).
Leuenroth, S. J. & Crews, C. M. Triptolide-induced transcriptional arrest is associated with changes in nuclear substructure. Cancer Res. 68, 5257–5266 (2008).
Vispé, S. et al. Triptolide is an inhibitor of RNA polymerase I and II–dependent transcription leading predominantly to down-regulation of short-lived mRNA. Mol. Cancer Ther. 8, 2780–2790 (2009).
St Germain, C. P. et al. Genomic patterns of transcription–replication interactions in mouse primary B cells. Nucleic Acids Res. 50, 2051–2073 (2022).
Sankar, T. S., Wastuwidyaningtyas, B. D., Dong, Y., Lewis, S. A. & Wang, J. D. The nature of mutations induced by replication–transcription collisions. Nature 535, 178–181 (2016).
Bayona-Feliu, A. et al. The chromatin network helps prevent cancer-associated mutagenesis at transcription-replication conflicts. Nat. Commun. 14, 6890 (2023).
Bayona-Feliu, A., Barroso, S., Muñoz, S. & Aguilera, A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription–replication conflicts. Nat. Genet. 53, 1050–1063 (2021).
Petryk, N. et al. Replication landscape of the human genome. Nat. Commun. 7, 10208 (2016).
Isik, S. et al. The SUMO pathway is required for selective degradation of DNA topoisomerase IIβ induced by a catalytic inhibitor ICRF-1931. FEBS Lett. 546, 374–378 (2003).
Uusküla-Reimand, L. & Wilson, M. D. Untangling the roles of TOP2A and TOP2B in transcription and cancer. Sci. Adv. 8, eadd4920 (2022).
Schier, A. C. & Taatjes, D. J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34, 465–488 (2020).
Luo, K., Yuan, J., Chen, J. & Lou, Z. Topoisomerase IIα controls the decatenation checkpoint. Nat. Cell Biol. 11, 204–210 (2009).
Sirbu, B. M. et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25, 1320–1327 (2011).
Pinkas, D. M. et al. Structural complexity in the KCTD family of cullin3-dependent E3 ubiquitin ligases. Biochem. J 474, 3747–3761 (2017).
Esposito, L., Balasco, N. & Vitagliano, L. Alphafold predictions provide insights into the structural features of the functional oligomers of all members of the KCTD family. Int. J. Mol. Sci. 23, 13346 (2022).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Luck, K. et al. A reference map of the human binary protein interactome. Nature 580, 402–408 (2020).
Weaver, Z. A. & Kane, C. M. Genomic characterization of a testis-specific TFIIS (TCEA2) gene. Genomics 46, 516–519 (1997).
Conaway, R. C., Kong, S. E. & Conaway, J. W. TFIIS and GreB: two like-minded transcription elongation factors with sticky fingers. Cell 114, 272–274 (2003).
Kettenberger, H., Armache, K.-J. & Cramer, P. Architecture of the RNA polymerase II–TFIIS complex and implications for mRNA cleavage. Cell 114, 347–357 (2003).
Labhart, P. & Morgan, G. T. Identification of novel genes encoding transcription elongation factor TFIIS (TCEA) in vertebrates: conservation of three distinct TFIIS isoforms in frog, mouse, and human. Genomics 52, 278–288 (1998).
Noe Gonzalez, M., Blears, D. & Svejstrup, J. Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 22, 3–21 (2021).
Erie, D. A. The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim. Biophys. Acta 1577, 224–239 (2002).
Pokholok, D. K., Hannett, N. M. & Young, R. A. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809 (2002).
Sekimizu, K., Kobayashi, N., Mizuno, D. & Natori, S. Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry 15, 5064–5070 (1976).
Erie, D. A., Hajiseyedjavadi, O., Young, M. C. & von Hippel, P. H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science 262, 867–873 (1993).
Kettenberger, H., Armache, K.-J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004).
Zhang, C. & Burton, Z. F. Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J. Mol. Biol. 342, 1085–1099 (2004).
Sivaramakrishnan, P. et al. The transcription fidelity factor GreA impedes DNA break repair. Nature 550, 214–218 (2017).
Zhuang, M. et al. Structures of SPOP–substrate complexes: insights into molecular architectures of BTB–Cul3 ubiquitin ligases. Mol. Cell 36, 39–50 (2009).
Canning, P. et al. Structural basis for Cul3 protein assembly with the BTB–Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 288, 7803–7814 (2013).
Groelly, F. J. et al. Mitotic DNA synthesis is caused by transcription–replication conflicts in BRCA2-deficient cells. Mol. Cell 82, 3382–3397.e7 (2022).
Bhowmick, R. et al. RAD51 protects human cells from transcription–replication conflicts. Mol. Cell 82, 3366–3381.e9 (2022).
Das, S. K. et al. MYC assembles and stimulates topoisomerases 1 and 2 in a “topoisome”. Mol. Cell 82, 140–158.e12 (2022).
Ju, B.-G. et al. A Topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006).
Ma, J., Bai, L. & Wang, M. D. Transcription under torsion. Science 340, 1580–1583 (2013).
Bunch, H. et al. Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 6, 10191 (2015).
Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. Phosphorylation and regulation of Akt/PKB by the Rictor–mTOR complex. Science 307, 1098–1101 (2005).
Ohta, T., Michel, J. J., Schottelius, A. J. & Xiong, Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541 (1999).
Huang, J. et al. SLFN5-mediated chromatin dynamics sculpt higher-order DNA repair topology. Mol. Cell 83, 1043–1060.e10 (2023).
Sirbu, B. M., Couch, F. B. & Cortez, D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7, 594–605 (2012).
Zhao, F. et al. ASTE1 promotes shieldin-complex-mediated DNA repair by attenuating end resection. Nat. Cell Biol. 23, 894–904 (2021).
Quinet, A., Carvajal-Maldonado, D., Lemacon, D. & Vindigni, A. in Methods in Enzymology, Vol. 591 (ed. Eichman, B. F.) 55–82 (Academic Press, 2017).
Huang, J. et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11, 592–603 (2009).
Crossley, M. P., Bocek, M. J., Hamperl, S., Swigut, T. & Cimprich, K. A. qDRIP: a method to quantitatively assess RNA–DNA hybrid formation genome-wide. Nucleic Acids Res. 48, e84 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2022).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arXiv.1303.3997 (2013).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP–seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012).
Liu, Y., Wu, X., d’Aubenton-Carafa, Y., Thermes, C. & Chen, C.-L. OKseqHMM: a genome-wide replication fork directionality analysis toolkit. Nucleic Acids Res. 51, e22 (2023).
Yu, G., Wang, L.-G. & He, Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics 14, 178–192 (2013).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Kloeber, J. A. et al. Alphafold2 modeling of KCTD10 interactions with RNAPII machinery. Dryad https://doi.org/10.5061/dryad.15dv41p7z (2025).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Palangat, M. & Landick, R. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J. Mol. Biol. 311, 265–282 (2001).
Artsimovitch, I. & Landick, R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl Acad. Sci. USA 97, 7090–7095 (2000).
Foster, J. E., Holmes, S. F. & Erie, D. A. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell 106, 243–252 (2001).
Acknowledgements
The authors thank L. Karnitz, L. Wang, H. Dong, A. Kanakkanthara, S. Kaufmann and all members of the Lou laboratory for helpful comments and discussions throughout the duration of this project, and the Cimprich laboratory for providing the episomal replication–transcription reporters. Molecular graphics and analyses were performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. This work was supported by the National Institutes of Health (T32GM145408 (J.A.K.)), (CA264600 and CA285345 (Z.L.)), the Dr. Gerald J. Robbins, MD, Career Development Award in Cancer Research (J.H.), the Whitney and Beth MacMillan Fund in Lung Cancer Research Honoring Julian R. Molina, MD, PhD (J.H.) and the Mayo Foundation (Z.L.).
Author information
Authors and Affiliations
Contributions
J.A.K., B.C., J.H. and Z.L. conceived of the project, designed experiments and prepared the manuscript. J.A.K. and B.C. performed most of the experiments and data analysis. Z. Wang provided helpful input on analysis and provided reagents for the CUT&RUN experiments. G.S. performed bioinformatic analyses on the CUT&RUN experiments. C.S.K. contributed to experiments focused on ICRF193-induced DNA damage. S.Z., Y.O., H.X., K.L., F.Z., Y.J., J.L., L.W. and X.W. provided valuable advice and expertise on experimental design. R.W.M., F.Z., H.Q., H.X., Y.O., L.W., S.Z., Z. Wu, X.T., K.L., P.Y., Y.J. and X.W. provided helpful input on data analysis and provided reagents. All authors reviewed the manuscript and provided constructive comments. J.H. and Z.L. supervised the entire project and secured funding for the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Initial characterization of KCTD10.
a, normalized γH2AX scores in interphase cells for all gRNAs tested from a CRISPR imaging screen12. b-c, a CRISPR drug screen identifies CUL3-KCTD10 as a key survival factor in the presence of catalytic TOP2 inhibition13. b, NormZ scores for all gRNAs in the presence of ICRF187. c, NormZ scores for KCTD10 gRNAs amongst all DNA damaging agents tested. d, schema depicting a CUL3-KCTD10 ubiquitin ligase complex. e-f, colony formation assays in KCTD10-deficient or control U2OS cells treated with etoposide (e) or ionizing radiation (f). Data are presented as mean values ± SD (n = 3 biologically independent experiments). g, micronucleus (MN) quantification in KCTD10-deficient or control U2OS cells. Data are presented as mean values ± SD (n = 4 biologically independent experiments). h, cell cycle analysis of KCTD10-deficient or control U2OS cells. Left, representative cell cycle profiles, Right, quantification of the percent of S-phase cells. Data are presented as mean values ± SD (n = 3 biologically independent experiments). i, western blot of nuclear soluble and chromatin fractions from U2OS cells treated with ICRF193 (1 μM), PARPi (olaparib, 10 μM), pladienolide B (PladB, 200 nM), or DMSO for 4 h. j, western blot from U2OS cells released from a double thymidine block for the indicated times. Right, representative FACS profiles from cells released from a double thymidine block for the indicated times. k, gene ontology analysis of genes that cluster together with KCTD10 from a CRISPR imaging screen12. p-values were calculated with a one-way ANOVA with Bonferroni’s multiple comparisons test.
Extended Data Fig. 2 KCTD10 negatively regulates transcription.
a-b, nascent transcription assay in KCTD10-deficient or control U2OS cells pre-treated with PG490 (1 μM) or DMSO for 4 h prior to EU labeling. a, quantification of EU nuclear intensity. Data are presented as mean values ± SD (n = 3 biologically independent experiments). b, representative images depicting EU intensity with color scaled to the EU fluorescence intensity. c, western blot from KCTD10-deficient or control U2OS cells treated with PG490 (1 μM) or DMSO for 4 h. d-e, nascent transcription assay in KCTD10-deficient or control U2OS cells co-stained with fibrillarin. d, quantification of EU nuclear intensity. Data are presented as mean values ± SD (n = 3 biologically independent experiments). e, representative images and analysis strategy to quantify EU intensity in the nucleolus and nucleoplasm. Each dot represents the mean EU intensity per nucleus for >50 nuclei per independent replicate; n = 3. f, quantification of fork speed in KCTD10-deficient or control U2OS cells, treated with PARPi (olaparib, 10 μM) as indicated. Data are presented as mean values ± SD (n = 150 fibers examined over 3 independent experiments). g, schema depicting how PLA assays quantify TRCs. h, single antibody controls for PLAs in Fig. 1i,j. i, schema for head-on and co-directional TRC reporters8. j-m, DNA/RNA hybrid dot blot assay from KCTD10-deficient or control U2OS cells treated with ICRF193 (100 nM) or DMSO for 4 h. Purified DNA/RNA samples were treated with the indicated nucleases prior to blotting. j, representative blot. k, quantification of undigested blots. l, quantification of samples treated with RNase H. m, quantification of samples after treatment with RNase T1. Data are presented as mean values ± SD (n = 3 biologically independent experiments). Scalebars = 10 μm. p-values were calculated with a one-way ANOVA with Bonferroni’s multiple comparisons test.
Extended Data Fig. 3 Identification and characterization of KCTD10 binding genome wide.
a, genomic features associated with KCTD10 peaks from cells transfected with empty vector (EV) or FLAG-tagged KCTD10 (FL-KCTD10) and treated with control (DMSO) or ICRF193 (100 nM) for 4 h. b, mean coverage of FL-KCTD10 in control or ICRF193-treated cells along the gene body (± 3 kb). c, representative window showing FL-KCTD10 binding in control and ICRF193-treated cells and the association with gene bodies and replication fork direction (RFD). d, venn diagram showing genome-wide co-occurrence of KCTD10-bound genes with co-directional conflicts in control and ICRF193-treated cells. e, circos plot showing overlap between KCTD10-bound genes in control and ICRF193-treated cells. Magenta curves link exact gene matches. Teal curves link genes that belong to the same enriched ontology term. f, enrichment analysis of transcription factor targets from KCTD10-bound genes in control and ICRF193 treated cells. p-values reported from a hypergeometric test with a Benjamini-Hochberg correction. g, quantification of relative pCHK2 and pCHK1 levels to total protein controls (CHK2 and CHK1, respectively. Data are presented as mean values ± SD (n = 3 biologically independent experiments). h-i, colony formation assays with KCTD10-deficient or control U2OS cells in the presence of ATRi (VX970, h) and DNA-PKi (NU7441, i). Data are presented as mean values ± SD (n = 3 biologically independent experiments). j, IC50 values derived from experiments shown in panels b-c and Fig. 2d. k-l, PLA for POLR2A-PCNA in KCT10-deficient U2OS cells treated with CHK2i (BML277, 1 μM) or DMSO for 24 h. k, quantification of PLA foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). l, representative images. Scalebars = 10 μm. p-values were calculated with a one-way ANOVA with Bonferroni’s multiple comparisons test unless otherwise stated.
Extended Data Fig. 4 Catalytic TOP2 inhibitors induce TRCs at trapped TOP2B.
a, γH2AX, 53BP1, and RAD51 foci in U2OS cells after ICRF193 (100 nM) or DMSO treatment (4 h). Left, representative images. Right, quantification foci. Data are presented as mean values ± SD (n = 3 biologically independent experiments). b, 53BP1 foci in KCTD10-deficient or control U2OS cells treated with ICRF193 (100 nM) for 4 h. Data are presented as mean values ± SD (n = 3 biologically independent experiments). c, 53BP1 foci in U2OS cells, treated with EdU and DMSO or ICRF193 for 1 h. Left, quantification of foci in EdU + or – nuclei. Data are presented as mean values ± SD (n = 3 biologically independent experiments). Right, representative images. d-f, co-staining for γH2AX and 53BP1 in U2OS cells treated with etoposide (ETP, 1 μM) or ICRF193 (100 nM) for the indicated times. d, representative images. e, quantification of 53BP1 foci. f, quantification of γH2AX foci. Data are presented as mean values ± SD (n = 3 biologically independent experiments). g, Western blot of TOP2B-deficient or control U2OS cells. h, 53BP1 foci in TOP2B-depleted U2OS cells treated with ICRF193 (100 nM) or DMSO for 4 h. Left, quantification of foci. Data are presented as mean values ± SD (n = 3 biologically independent experiments). Right, representative images. i, Quantification of 53BP1 foci from U2OS cells co-treated with ICRF193 (100 nM), PG490 (1 μM), aphidicolin (Aph, 4 μM), or ATRi (VX-970 500 nM). Data are presented as mean values ± SD (n = 3 biologically independent experiments). j-k, proximity ligation assays (PLA) testing proximity between POLR2A-TOP2B, POLR2A-PCNA, and PCNA-TOP2B in U2OS cells treated with ICRF193 or DMSO for 4 h. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). l, representative PLA controls for POLR2A, PCNA, and TOP2B antibodies used throughout this manuscript. (m)PCNA = mouse anti-PCNA; used for PLAs with TOP2B. (r)PCNA = rabbit anti-PCNA; used with PLAs for KCTD10 and POLR2A. m, schema depicting the proposed TOP2B-dependent TRC that occurs in the presence of catalytic TOP2 inhibition. n, PLA for POLR2E-PCNA and POLR2J-PCNA in U2OS cells treated with ICRF193 (100 nM) or etoposide (ETP, 1 μM). Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). Error bars indicate the mean ± SD. Scalebars = 10 μm. p-values were calculated with a two-tailed, unpaired t-test (a,j) or a one-way ANOVA with Bonferroni’s multiple comparisons test.
Extended Data Fig. 5 KCTD10 does not directly regulate TOP2 activity.
a, western blot from whole cell lysates (WCL) and chromatin fractions from KCTD10-deficient or control U2OS cells treated with ICRF193 (1 μM) or DMSO for the indicated. b, mitotic progression assay in KCTD10-deficient or control U2OS cells. Left, experimental schema to test mitotic progression. Right, quantification of cells at the indicated cell cycle stages, 2 h after release from G2/M block. Data are presented as mean values ± SD (n = 3 biologically independent experiments). c, G2/M checkpoint assay in KCTD10-deficient or control U2OS cells. Left, representative images of interphase (pHH3-) and mitotic (pHH3 + ) cells. Top right, experimental schema for G2/M checkpoint assay. Bottom right, quantification of pHH3+ cells. Data are presented as mean values ± SD (n = 3 biologically independent experiments). Scalebars = 10 μm. p-values reported from a two-way ANOVA with Dunnett’s multiple comparisons test (b) and a one-way ANOVA with Bonferroni’s multiple comparisons test (c).
Extended Data Fig. 6 Characterization of KCTD10 self-association.
a, representative PLA control for KCTD10 antibody used throughout this manuscript. b, co-immunoprecipitation assay in HEK293T cells expressing FLAG-KCTD10. Prior to immunoprecipitation with anti-FLAG beads, whole cell lysates were digested with benzonase or mock digested. c-d, PLA for POLR2A-KCTD10 in U2OS cells treated as with PCNAi1 (2 μM) and/or ICRF193 (100 nM). Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). e, co-immunoprecipitation assay in HEK293T cells with the indicated FL-KCTD10 constructs and HALO-KCTD10 treated with ICRF193 (1 μM) for 4 h. f, co-immunoprecipitation assay in HEK293T cells with the indicated FL-KCTD10 constructs and HALO-KCTD10 treated with ICRF193 (1 μM) for 4 h. g-h, PLA for HALO-FLAG in U2OS cells expressing HALO-tagged and FLAG-tagged KCTD10 with the indicated constructs as depicted in g and Fig. 3f. g, quantification of foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). h, representative images. i-k, predicted structure of a KCTD10 dimer from Alphafold2-multimer. i, full view of the dimer. j, zoomed-in and rotated view of the dimer interface with residues of interest noted. k, alternative view of the dimer interface. l, in vitro binding assay between GST-tagged wild-type (WT) and R167A-mutant KCTD10 and FLAG-tagged KCTD10. Left, experimental schema. Right, immunoblot after GST pulldown and Coomassie stain of purified GST-KCTD10. m, co-immunoprecipitation assay in HEK293T cells transiently expressing WT and R167A-mutant FLAG-KCTD10 and treated ICRF193 (1 μM) or DMSO (4 h). Error bars indicate the mean ± SD. Scalebars = 10 μm. p-values reported from a one-way ANOVA with Bonferroni’s multiple comparisons test.
Extended Data Fig. 7 KCTD10 recruits CUL3 to co-directional TRCs.
a, co-immunoprecipitation assay for KCTD10 from the indicated fractions shown in Fig. 4a. b-c, PLA for POLR2A and Ubiquitin (Ub) in KCTD10-deficient U2OS cells treated with ICRF193 (100 nM) or DMSO for 4 h. b, representative images. c, quantification of foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). d, representative images from PLA control for the indicated antibodies used throughout the manuscript. e-f, PLA for KCTD10-POLR2A in HEK293T cells expressing the codirectional-ECFP reporter (Extended Data Fig. 2i) treated with doxycycline (1 mg/mL) for 20 h and ICRF193 (100 nM) or DMSO for 4 h. e, quantification of foci. Data are presented as mean values ± SD (n = 150 cells examined over 3 independent experiments). f, representative images. g-h, PLA for POLR2A-PCNA in U2OS cells treated with ICRF193 (100 nM) and MLN4924 (1 μM) for 4 h. g, quantification of foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). h, representative images. i-j, PLA for CUL3-POLR2A in U2OS cells treated with ICRF193 (100 nM) or etoposide (ETP, 1 μM) for 4 h. i, quantification of foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). j, representative images. Error bars indicate the mean ± SD. Scalebars = 10 μm. p-values were calculated with a one-way ANOVA with Bonferroni’s multiple comparisons test.
Extended Data Fig. 8 CUL3 recruitment depends on bivalent recognition of PCNA and RNA polymerase by KCTD10.
a, schema depicting experiment to test dimer-dependent binding of KCTD10 and CUL3. b, co-immunoprecipitation assay in HEK293T cells with MYC-CUL3 and the indicated KCTD10 constructs conjugated to a dimerization domain (DmrB) that is inducible with ligand (AP20187, 100 nM, 4 h). Left, representative blot. Right, quantification of CUL3 pulldown efficiency relative to input. Data are presented as mean values ± SD (n = 3 biologically independent experiments). c-d, PLA for CUL3-POLR2A pairs in U2OS cells after a double thymidine block and treated with ICRF193 (100 nM) or DMSO for 4 h. c, quantification of foci. Each dot represents the mean number of foci per nucleus for >50 nuclei per independent replicate; n = 3. d, representative images. e-f, PLA for CUL3-POLR2A pairs in U2OS cells treated with ICRF193 (100 nM) and PCNAi1 (2 μM) for 4 h. e, quantification of foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). f, representative images. g-h, PLA for POLR2A-Ub pairs in U2OS cells treated with ICRF193 (100 nM) and PCNAi1 (2 μM) for 4 h. g, quantification of foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). h, representative images. i-j, PLA for CUL3-POLR2A (i) and POLR2A-Ub (j) in KCTD10 knockdown U2OS cells rescued with wild-type (WT) or PIP3A KCTD10 treated with ICRF193 (100 nM) or DMSO. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). k, MTS assay in control cells or KCTD10-deficient cells rescued with WT or PIP3A KCTD10. Data are presented as mean values ± SD (n = 3 biologically independent experiments). p-values were calculated with a two-tailed extra sum of squares F-test. l, γH2AX foci from KCTD10-deficient U2OS cells rescued with WT or PIP3A KCTD10 treated with ICRF193 (100 nM) or DMSO. Data are presented as mean values ± SD (n = 3 biologically independent experiments). m, quantification of PLA foci for CUL3-POLR2A in KCTD10-deficient or control U2OS cells with wild-type or R167A KCTD10 reintroduced and treated with ICRF193 (100 nM) or DMSO for 4 h. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). Scalebars = 10 μm. p-values were calculated with a one-way ANOVA with Bonferroni’s multiple comparisons test unless otherwise stated.
Extended Data Fig. 9 Characterizing KCTD10’s function on RNAPII.
a, schematic depicting the method of screening for KCTD10 interacting partners using ColabFold (MMseqs2 + Alphafold2-multimer). b, heatmap depicting the interface predicted TM score (iPTM) score for all proteins screen for interactions with KCTD10. c, network diagram for proteins with an iPTM score > 0.4, generated by MCL clustering with an inflation parameter of 4 using Cytoscape75. d-e, predicted aligned error (PAE) plots for the top ranked Alphafold2-multimer predictions for TCEA1-KCTD10 (d) and TCEA3-KCTD10 (e). f, co-immunoprecipitation assay in HEK293T cells transiently expressing FLAG-tagged POLR2C and treated with ICRF193 (1 μM) for 4 h. g, co-immunoprecipitation assay in HEK293T cells transiently expressing FLAG-tagged TCEA3 treated with ICRF193 (1 μM) for 4 h. h-j, PLA for KCTD10 and POLR2C, POLR2J, POLR2I, and POLR2E in U2OS cells treated with ICRF193 (100 nM) or DMSO for 4 h. h, quantification of foci. Data are presented as mean values ± SD (n = 200 cells examined over 3 independent experiments). i, representative images. j, representative single antibody PLA control for POLR2C, POLR2J, POLR2I, and POLR2E antibodies. Error bars indicate the mean ± SD. p-values were calculated with a one-way ANOVA with Bonferroni’s multiple comparisons test. k, denaturing Ni-NTA pulldown from KCTD10-deficient and control HEK293T cells expressing His-tagged ubiquitin (His-Ub) and treated with ICRF193 (1 μM) or DMSO for 4 h in the presence of MG132 (50 μM). l, denaturing Ni-NTA pulldown from HEK293T cells expressing FL-TCEA2 and His-Ub, treated with ICRF193 (1 μM) and/or MLN4924 (1 μM) in the presence of MG132 (50 μM) for 4 h. m, denaturing Ni-NTA pulldown from HEK293T cells expressing wild-type (WT) and mutant FL-TCEA2 with the indicated mutations and His-Ub, treated with ICRF193 (1 μM) for 4 h in the presence of MG132 (50 μM). n, western blot of chromatin and whole cell lysates (WCL) from KCTD10-deficient or control U2OS cells. Right, quantification of relative TCEA2 chromatin compared to WCL. Data are presented as mean values ± SD (n = 3 biologically independent experiments). o, co-immunoprecipitation assays for endogenous POLR2A in KCTD10-deficient or control U2OS cells. Right, quantification of relative TCEA2 pulldown efficiency. Data are presented as mean values ± SD (n = 3 biologically independent experiments). p, western blot from TCEA2-deficient (siTCEA2) and control U2OS cells. Scalebars = 10 μm. p-values were calculated with a one-way ANOVA with Bonferroni’s multiple comparisons test.
Extended Data Fig. 10 Overall working model.
Upon encountering transcriptional roadblocks such as supercoiled DNA or trapped TOP2 (Extended Data Fig. 4m), the RNA polymerase complex attempts to backtrack and restart. At actively replicating regions, KCTD10 functions together with replication forks to sense the transcription roadblock by self-assembly which facilitates formation of a CUL3-RBX1-KCTD10 complex. This complex ubiquitinates TCEA2, which promotes TCEA2 removal and conversion of the RNAP complex back to a lower energy state41,45,46,76,77,78. This state is amenable to p97/VCP-mediated extraction, allowing the replisome to bypass the site. Whether additional factors work in parallel or downstream of KCTD10 to resolve roadblocks such as trapped TOP2 remains unclear. In the absence of KCTD10, the RNA polymerase retains TCEA2 and remains in an active elongation-competent complex that becomes permanently arrested. This disfavors replisome bypass and can lead to the generation of a persistent TRC. If this is unable to be resolved, this will eventually convert into a DSB11 and activate a DNA damage response mediated by ATM and CHK2.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 (the raw blots and gating strategy), Supplementary Note 1 containing the scripts used for CUT&RUN analysis and Supplementary Table 1, a list of reagents and resources used in the study.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kloeber, J.A., Chen, B., Sun, G. et al. KCTD10 is a sensor for co-directional transcription–replication conflicts. Nature 648, 210–219 (2025). https://doi.org/10.1038/s41586-025-09585-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09585-9