Abstract
Multicellular cytokine networks drive intestinal inflammation and colitis-associated cancer (CAC). Interleukin-22 (IL-22) exerts both protective and pathogenic effects in the intestine, but the mechanisms that regulate this balance remain unclear. Here, we identify that IL-22 directly induces responsiveness to the IL-6 family cytokine oncostatin M (OSM) in intestinal epithelial cells (IECs) by activating STAT3 and upregulating the OSM receptor. In turn, OSM synergizes with IL-22 to sustain STAT3 activation in IECs and promote proinflammatory epithelial adaptation and immune cell chemotaxis to the inflamed intestine. Conditional deletion of the OSM receptor in IECs protects mice from both colitis and CAC, and pharmacological blockade of OSM attenuates established CAC. Thus, IL-22 and OSM form a pathogenic circuit that drives inflammation and tumorigenesis. Our findings reveal a previously unknown mechanism by which OSM supports intestinal pathology and highlight the IL-22–OSM axis as a promising therapeutic target for inflammatory bowel disease and CAC.
Similar content being viewed by others
Main
Multicellular cytokine circuits play crucial roles in promoting intestinal inflammation and colorectal cancer (CRC)1,2. Deciphering the dependencies within these circuits could offer new insights into inflammatory bowel disease (IBD) pathogenesis and prioritization of therapeutic targets. Interestingly, cytokines implicated in gut pathophysiology can exhibit dual functionality by facilitating epithelial repair and barrier function following acute injury, while promoting inflammation, epithelial transformation and tumorigenesis in the context of chronic colitis2,3. Separating the distinct mechanisms driving beneficial versus harmful cellular responses could provide means to exploit these pathways for therapeutic benefit.
Oncostatin M (OSM) can drive pathological responses in several human diseases, including psoriasis, rheumatoid arthritis, infections and IBD4,5,6. The expression of OSM is highly increased in the inflamed mucosa of individuals with IBD, including both ulcerative colitis (UC) and Crohn’s disease (CD), and associates closely with treatment-refractory disease7. OSM is produced by hematopoietic cells and acts on cells bearing the OSM receptor (OSMR), which is expressed primarily in tissues of mesenchymal origin (especially fibroblasts and endothelial cells) and to a lesser extent in epithelial cells4,5,6. OSM signaling involves several pathways, including STAT3, the MAPK cascade and PI3K, with occasional involvement of STAT1, STAT5 and potentially STAT6 (refs. 6,8). Although OSM is implicated in promoting intestinal inflammation, the underlying mechanisms remain undefined.
Here, we comprehensively characterized the OSM–OSMR axis in intestinal inflammation by applying state-of-the-art single-cell sequencing and conditional deletion of Osmr in specific tissue-resident cell populations in two independent mouse models of chronic intestinal inflammation. Our investigations reveal substantial upregulation of OSMR on epithelial cells during intestinal inflammation, both in mouse models and in tissues from individuals with IBD. Importantly, we identify OSMR as a key mediator of intestinal inflammation via its impact on epithelial cells. Notably, group 3 innate lymphoid cell (ILC3)-derived interleukin-22 (IL-22) is essential for both induction and sustained expression of OSMR expression in epithelial cells during colitis. Similarly, IL-22 drives epithelial OSMR expression in a mouse model of colitis-associated cancer (CAC), in which disruption of epithelial OSMR signaling curtails tumor development. In summary, we provide new evidence for a pivotal role of the IL-22–OSM axis in intestinal pathophysiology.
Results
Epithelial OSMR induction in intestinal inflammation
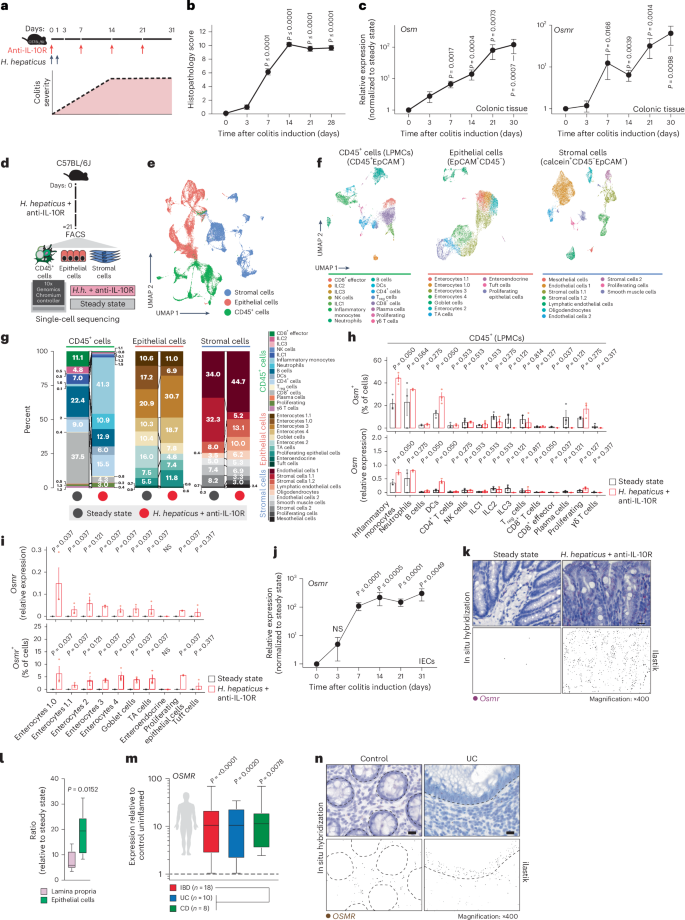
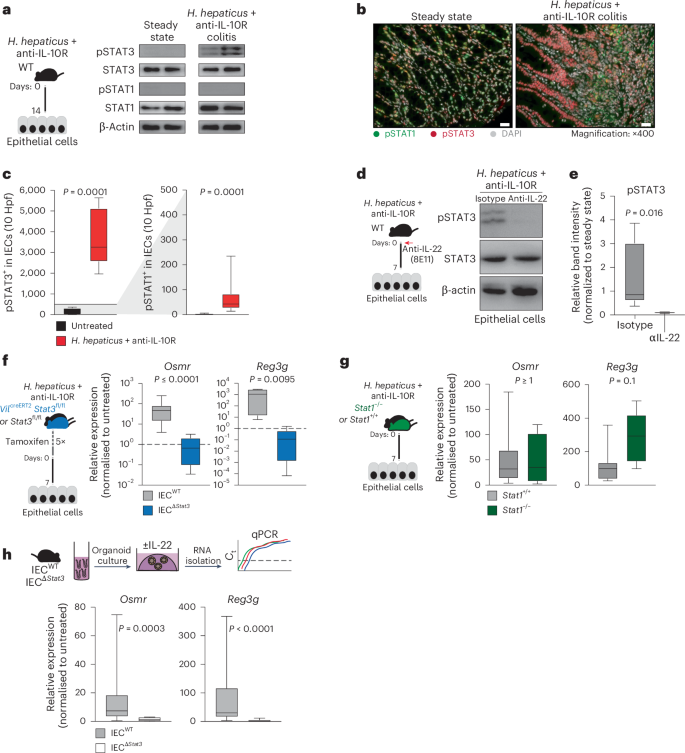
To investigate the cellular components of the OSM–OSMR axis in intestinal inflammation, we used the Helicobacter hepaticus + anti-IL-10R-induced colitis model9,10, which is characterized by prolonged chronic inflammation and microbial dysbiosis, reproducing key pathogenic features of human IBD11. In this model, inflammation becomes evident 3–7 days following initiation, and colitis persists for up to 30 days (Fig. 1a,b). The expression of Osm and Osmr is upregulated early in the course of intestinal inflammation and remains elevated for up to 30 days (Fig. 1c).
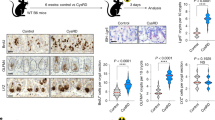
a, Schematic of the H. hepaticus + anti-IL-10R-induced colitis time course study. b, Histopathology scores of colitis severity at days 3 (n = 6), 7 (n = 41), 14 (n = 23), 21 (n = 16) and >28 (n = 6), pooled from two to four experiments. Data are shown as mean ± s.e.m. and were analyzed by a Kruskal–Wallis test with a Dunn’s post hoc test. c, qPCR analysis of Osm and Osmr expression in colon tissue at days 3 (Osm/Osmr, n = 4/4), 7 (Osm/Osmr, n = 14/20), 14 (Osm/Osmr, n = 9/14), 21 (Osm/Osmr, n = 20/21) and >28 (Osm/Osmr, n = 10/13), normalized to steady-state controls. Data were pooled from two to four experiments and are shown as mean ± s.e.m. P values were calculated by (two-tailed) one-sample t-tests and Wilcoxon rank tests. d–i, Single-cell workflow: stromal (calcein⁺CD45⁻EpCAM⁻), immune (CD45⁺) and epithelial (EpCAM⁺) cells were sorted, pooled (three mice per pool, three pools per group) and processed with 10x Genomics (18 mice total). d, Experimental approach for single-cell workflow. e, Uniform manifold approximation and projection (UMAP) showing CD45⁺, epithelial and stromal cell clusters. f,g, UMAP (f) and quantification (g) of major cell populations within CD45+, epithelial and stromal clusters; DCs, dendritic cells; LPMCs, lamina propria mononuclear cells; NK, natural killer; Treg, regulatory T cells; TA cells, transit-amplifying cells. h, Relative Osm expression and percentage of Osm⁺ cells within the CD45⁺ cluster. i, Relative Osmr expression and Osmr⁺ cell frequency within epithelial clusters. Data are shown as mean ± s.e.m. and were analyzed by Wilcoxon rank-sum test; NS, not significant. j, qPCR analysis of epithelial Osmr expression at days 3, 7, 14, 21 and >28 (n = 4, 23, 12, 15 and 11, respectively), normalized to steady-state controls and pooled from two to four experiments. Data are shown as mean ± s.e.m. P values were calculated by (two-tailed) one-sample t-tests and Wilcoxon tests. k, ISH of Osmr in colon tissue from colitic (day 10, n = 3) and steady-state mice (n = 6). Purple signal indicates Osmr expression. Data were analyzed by ilastik-based quantification; scale bar, 20 μm. l, Quantification of Osmr⁺ cells by ISH in the lamina propria (pink) and epithelium (green) of colitic (day 7) versus steady-state mice (n = 6). Data were analyzed by Mann–Whitney U-test. m, qPCR of epithelial OSMR expression in healthy individuals and individuals with IBD (UC, n = 10; CD, n = 8). Data are shown as mean ± s.e.m.; statistical tests used were the same as in j. n, ISH of OSMR in mucosal biopsies from healthy individuals (n = 5) and individuals with UC (n = 12); brown dots indicate OSMR⁺ cells. Dashed lines demarcate the epithelium; scale bar, 20 μm.
We next acquired single-cell transcriptomes from distinct cell populations, including colonic epithelial (EpCAM+CD45–), stromal (calcein+CD45–EpCAM–) and CD45+ immune cells (Fig. 1d). Three independent samples were profiled per condition and cell type (Extended Data Fig. 1a,b) to capture all relevant cell populations at steady-state and during peak colitis (day 22; Fig. 1e,f, Supplementary Fig. 1a–c and Supplementary Table 1). Various inflammatory genes were expressed in CD45+, epithelial and stromal cells (Supplementary Fig. 2a). Colitis was characterized by a significant increase in inflammatory monocytes and neutrophils, accompanied by prominent alterations in epithelial and stromal cells (Fig. 1g and Extended Data Fig. 1c).
In healthy mice, Osm expression was detected exclusively in hematopoietic cell types, including neutrophils, monocytes, B cells, ILCs and dendritic cells (Fig. 1h and Extended Data Fig. 1d,e). During colitis, the number of Osm-expressing cells in the colon increased, with inflammatory monocytes, neutrophils and dendritic cells being the primary sources of OSM (Fig. 1h and Extended Data Fig. 1f). Conversely, Osmr was expressed broadly among mesenchymal cell types at steady state (Extended Data Fig. 1g), whereas it was notably absent in immune and epithelial cells (Fig. 1i and Extended Data Fig. 1g,h). By contrast, Osmr expression in colonic epithelial cells was clearly increased during colitis (Fig. 1i and Supplementary Fig. 2b). This expression occurred predominantly in enterocytes (Fig. 1i and Supplementary Fig. 2b). Using quantitative PCR (qPCR), we confirmed that the expression of Osmr was upregulated in intestinal epithelial cells (IECs) as early as day 3 following colitis induction and persisted for at least 28 days (Fig. 1j and Extended Data Fig. 2a–c). Notably, epithelial Osmr expression was upregulated in H. hepaticus + anti-IL-10R-induced colitis but not in the absence of inflammation following H. hepaticus colonization or anti-IL-10R treatment alone (Extended Data Fig. 2c)12,13. The induction of OSMR protein expression in epithelial cells was confirmed by flow cytometry (Extended Data Fig. 2d,e). Exploring putative receptor–ligand interactions in our single-cell RNA-sequencing (scRNA-seq) dataset revealed strong putative interactions between enterocytes and Osm+ immune cells, particularly inflammatory monocytes and neutrophils (Extended Data Fig. 2f). Visualizing Osmr expression in inflamed colonic tissue by in situ hybridization (ISH) confirmed strong Osmr expression in epithelial cells during H. hepaticus + anti-IL-10R-induced colitis and its absence under steady-state conditions (Fig. 1k and Extended Data Fig. 3a,b). Notably, Osmr expression was more prominently upregulated in epithelial cells following colitis induction than in the lamina propria (Fig. 1k,l and Extended Data Fig. 3c–e). Specifically, the upregulation of Osmr expression in epithelial cells correlated significantly with histopathology score, whereas this association was less clear in the lamina propria (Extended Data Fig. 3f,g).
In mucosal samples from individuals with IBD, the expression of both OSM and OSMR is increased7. OSM is primarily enriched in inflammatory monocytes and type 2 dendritic cells, whereas OSMR is most highly expressed by inflammation-associated fibroblasts14, closely resembling the pattern seen in H. hepaticus + anti-IL-10R-induced colitis. Next, we examined the expression of OSMR in epithelial cells in human IBD using publicly available single-cell sequencing data14,15. Remarkably, OSMR was expressed by enterocytes in both individuals with UC (Supplementary Fig. 3a) and individuals with CD (Supplementary Fig. 3b). To validate this finding, we quantified OSMR expression in epithelial cells isolated from colonic mucosal biopsies from healthy individuals and individuals with IBD by using qPCR (n = 10 UC and n = 8 CD) and ISH (n = 10 UC, n = 2 CD and n = 5 healthy individuals). Our analysis revealed significantly elevated epithelial OSMR expression in IBD samples (Fig. 1m,n and Extended Data Fig. 4a,b). Additionally, we confirmed OSMR expression at the protein level and observed that it correlated with histopathology score (Extended Data Fig. 4c).
In summary, although expression of OSMR is primarily confined to mesenchymal cells in healthy intestinal tissue, there is a significant induction of OSMR expression in IECs under inflamed conditions in both mice and humans.
Epithelial OSMR expression drives intestinal inflammation
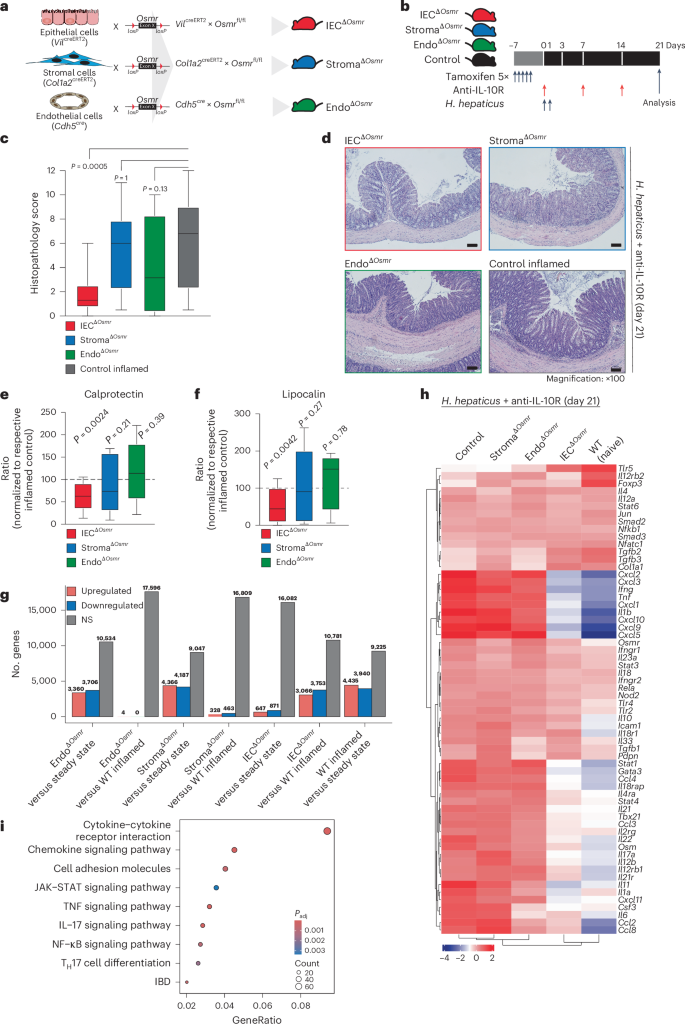
To assess the functional impact of OSMR expression in distinct nonhematopoietic cell types during intestinal inflammation, we implemented conditional deletion strategies by crossing Osmrfl/fl mice with VilcreERT2 (ΙECΔOsmr), Col1a2creERT2 (StromaΔOsmr) and Cdh5cre (EndoΔOsmr) mice to delete Osmr expression in epithelial, stromal and endothelial cells, respectively (Fig. 2a). Cell-type-specific targeting was validated by exploring expression of the deleter genes in our scRNA-seq dataset and by crossing Cre-deleter lines with a fluorescent reporter mouse (Rosa-CAG-LSL-tdTomato (Ai9); Supplementary Fig. 4a,b). OSMR acts as a receptor for IL-31; however, unlike OSM, we did not observe an upregulation of IL-31 in either mouse or human intestinal inflammation (Supplementary Fig. 4c)7. IECΔOsmr and StromaΔOsmr mice were treated with tamoxifen to induce deletion (Fig. 2b), after which colitis was induced in ΙECΔOsmr, StromaΔOsmr and EndoΔOsmr mice using the H. hepaticus + anti-IL-10R approach (Fig. 2b). StromaΔOsmr and EndoΔOsmr mice showed no discernible difference in intestinal pathology compared to controls (Fig. 2c,d). By contrast, histological evidence of colitis and fecal inflammatory markers, such as calprotectin and lipocalin, were significantly reduced in ΙECΔOsmr mice (Fig. 2c–f).
a, Schematic illustrating the Osmr deletion strategy in different cell types. Cell-type-specific deleter mouse lines (VilcreERT2, Cdh5cre and Col1a2creERT2) were crossed with Osmrfl/fl mice to generate the indicated cell-type-specific Osmr knockouts. b, Timeline of tamoxifen administration and subsequent colitis induction. Respective littermate control mice (VilcreERT2–, Cdh5cre– or Col1a2creERT2– crossed with Osmrfl/fl or VilcreERT+, Cdh5cre+ or Col1a2creERT2+ crossed with Osmrfl/wt) were used for each genotype. c, Histopathological scoring 21 days after colitis induction, based on two to three independent experiments (control inflamed, n = 31; IECΔOsmr, n = 13; StromaΔOsmr, n = 21; EndoΔOsmr, n = 12). Adjusted P values calculated from a Kruskal–Wallis test with Dunn’s post hoc comparisons indicate differences between OSMR-deficient and wild-type mice. d, Representative hematoxylin and eosin-stained colon sections from steady-state and inflamed mice of the indicated genotypes at day 21; scale bar, 100 μm. Sample sizes are as indicated in c. e,f, Calprotectin (e) and lipocalin (f) levels in fecal samples from the indicated genotypes, measured by enzyme-linked immunosorbent assay (ELISA) and normalized to their respective inflamed control mice. Data were pooled from two independent experiments (IECΔOsmr, n = 12; StromaΔOsmr, n = 12; EndoΔOsmr, n = 14). P values (two-tailed) were calculated by one-sample t-tests and Wilcoxon tests. g, Differentially expressed genes in colonic tissues (RNA-seq) of the indicated mouse lines compared to either steady-state mice or respective inflamed controls; WT, wild-type. h, Heat map displaying the expression of curated IBD pathway genes in colon tissue of the different mouse genotypes (n = 4–5 per group). i, Top biological processes enriched among differentially expressed genes in colon tissue from IECΔOsmr mice compared to respective inflamed controls (n = 4–5 per group); Padj, adjusted P value.
Using RNA-seq analysis of total colonic tissue, StromaΔOsmr and EndoΔOsmr mice showed an inflamed transcriptional signature compared to steady-state mice but minimal differential gene expression compared to control inflamed mice (Fig. 2g,h, Supplementary Fig. 4d–f and Supplementary Table 2). However, the inflammatory signature was reduced in ΙECΔOsmr mice compared to in wild-type control inflamed mice (Fig. 2g,h, Supplementary Fig. 4g and Supplementary Table 2). KEGG pathway analysis revealed significant alterations in pathways related to cytokine–cytokine receptor interaction, JAK–STAT signaling, IL-17 signaling and IBD pathways (Fig. 2i and Supplementary Table 4).
To confirm the relevance of epithelial Osmr deletion in intestinal inflammation in an independent colitis model, we evaluated OSM and OSMR upregulation in the Citrobacter rodentium infection model. We observed a significant increase in Osm and Osmr expression in colonic tissue following C. rodentium infection (Extended Data Fig. 5a–c). Similarly, Osmr upregulation in epithelial cells was detected as early as day 3 after infection and persisted up to day 21 (Extended Data Fig. 5d,e). Consistent with the H. hepaticus + anti-IL-10R model, C. rodentium infection of IECΔOsmr mice after tamoxifen treatment resulted in reduced histopathology compared to wild-type controls (Extended Data Fig. 5g). Despite this reduction in inflammation, bacterial clearance was unaffected (Extended Data Fig. 5h), indicating that Osmr deletion in epithelial cells mitigates colitis severity without impairing host defence mechanisms.
Overall, these findings reveal a nonredundant pathogenic role for OSM–OSMR signaling in epithelial cells during intestinal inflammation.
Epithelial OSMR expression during colitis requires IL-22
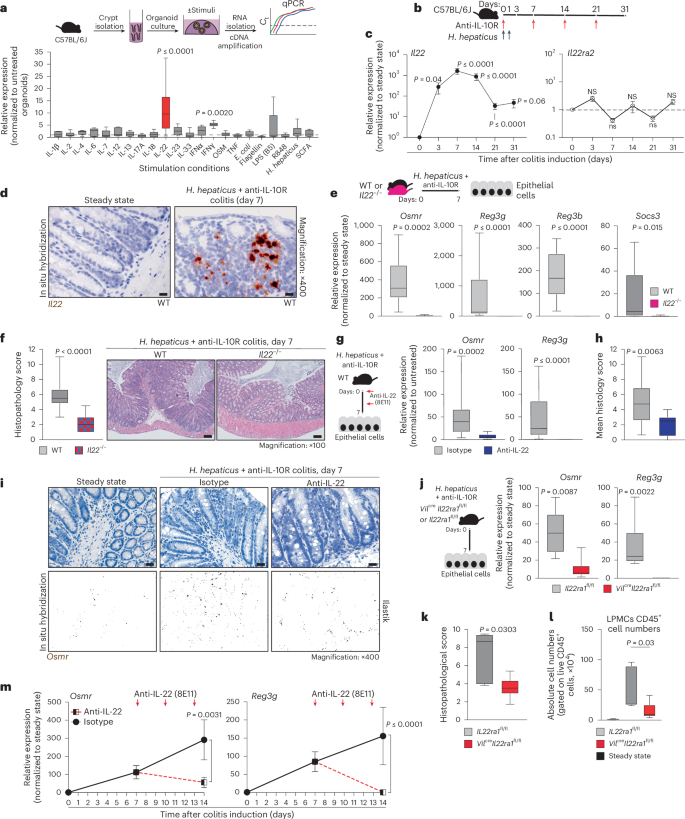
Osmr expression is absent in IECs under healthy conditions but is rapidly induced during colitis (Fig. 1i–k). To identify putative drivers of Osmr expression in IECs, we subjected mouse colon epithelial organoids to various inflammatory stimuli, including cytokines, Toll-like receptor ligands and H. hepaticus bacterial lysates, and analyzed gene expression changes by qPCR (Fig. 3a and Extended Data Fig. 6a). This revealed IL-22 as the most potent inducer of Osmr expression in colon organoids (Fig. 3a). Osmr was induced within 3 h of stimulation with IL-22, suggesting that Osmr is likely a direct target gene of IL-22 signaling (Extended Data Fig. 6b). Consistent with mouse cells, IL-22 also induced OSMR expression in human colon epithelial organoids (Extended Data Fig. 6c).
a, Schematic of colon organoids from female C57BL6/J mice exposed to stimuli for 24 h. Osmr expression was assessed by qPCR (three independent experiments with three biological replicates each); Ct, cycling threshold; IFN, interferon; E. coli, Escherichia coli; LPS, lipopolysaccharide; SCFA, short-chain fatty acid. b, Experimental setup for colitis induction. c, Il22 and Il22ra2 expression in colonic tissues from colitic mice at the following days: day 3 (Il22, n = 8; Il22ra2, n = 9), day 7 (Il22, n = 29; Il22ra2, n = 10), day 14 (Il22, n = 17; Il22ra2, n = 8), day 21 (Il22, n = 14; Il22ra2, n = 11) and >day 28 (Il22, n = 5; Il22ra2, n = 8). Data were pooled from two to five independent experiments and are shown as mean ± s.e.m. P values (two-tailed) were calculated by one-sample t-tests and Wilcoxon rank tests. d, Il22 expression detected by ISH in colon mucosa from steady-state and colitic mice (n = 2); scale bar, 20 μm. e,f, Il22–/– and cohoused wild-type mice analyzed 7 days after colitis induction. qPCR analysis (e; Osmr, Reg3g, Reg3b and Socs3) in epithelial cells (data normalized to steady-state controls; n = 10 per genotype, two experiments) and histopathological scoring (f; n = 10 per genotype, two experiments) are shown; scale bar, 100 μm. g–i, Wild-type mice with H. hepaticus + anti-IL-10R colitis treated with anti-IL-22 (n = 9) or isotype control (n = 15) on days 0 and 3 and analyzed on day 7. g, qPCR of epithelial gene expression. h, Histopathological scoring (two experiments). i, Osmr expression detected by ISH (brown punctae); top, ISH with hematoxylin staining; bottom, ilastik-processed image (n = 3–6 per group); scale bar, 20 μm. j–l, H. hepaticus + anti-IL-10R colitis induced in Vilcre+ × Il22ra1fl/fl or Vilcre– × Il22ra1fl/fl mice, analyzed on day 7 (n = 6 per genotype). j, qPCR for Osmr and Reg3g expression in epithelial cells normalized to steady-state controls. k,l, Histopathological scoring (k) and CD45+ cell abundance (l). m, Wild-type mice with H. hepaticus + anti-IL-10R colitis treated with anti-IL-22 (n = 9) or isotype control (n = 11) on days 7 and 10; data were analyzed on day 14. qPCR was performed on Osmr and Reg3g in epithelial cells (two independent experiments). Data are shown as mean ± s.e.m. P values (two-tailed) were calculated using a Mann–Whitney test for e–h, j, k and m.
IL-22 plays a complex and pleiotropic role in intestinal inflammation, not only exacerbating intestinal inflammation but also promoting epithelial cell survival and regeneration, regulating mucin and antibacterial peptide expression and providing protection in certain colitis models16,17,18. In H. hepaticus + anti-IL-10R-induced colitis, Il22 expression was detected in intestinal tissue as early as day 3 and persisted into the chronic phase of colitis (Fig. 3b,c). Notably, Il22 was predominantly localized near the epithelial barrier in colitis, adjacent to Osmr-expressing IECs, as confirmed by ISH (Fig. 3d and Extended Data Fig. 6d,e). By contrast, IL-22-binding protein (IL-22BP; encoded by Il22ra2), which binds and neutralizes IL-22 (ref. 19), was not induced in H. hepaticus + anti-IL-10R-induced colitis (Fig. 3c).
To evaluate the functional importance of IL-22 in epithelial OSMR expression, we induced colitis in Il22–/– mice (Fig. 3e). Indeed, Il22–/– mice showed reduced intestinal inflammation and failed to upregulate Osmr expression in IECs (Fig. 3e,f). Of note, the intestinal microbiota is altered in Il22–/– mice, potentially impacting colitis development20. To address this potential caveat, we blocked IL-22 using a neutralizing antibody in wild-type mice with H. hepaticus + anti-IL-10R colitis and observed similarly attenuated Osmr expression in IECs, in addition to reduced colitis severity (Fig. 3g–i). Osmr induction required direct IL-22 signaling in IECs, because mice with IEC-restricted IL-22 receptor deficiency (Vilcre+Il22ra1fl/fl)21 also failed to upregulate Osmr in the H. hepaticus + anti-IL-10R model, which correlated with reduced immune cell infiltration in colonic tissue and a concomitant reduction in histopathological features of colitis (Fig. 3j–l and Supplementary Fig. 5a–d). Consistent with the role of IL-22 in upregulating the expression of OSMR on epithelial cells in H. hepaticus + anti-IL-10R colitis, we observed that increased expression of Osmr on IECs in C. rodentium-induced colitis was also IL-22 dependent (Extended Data Fig. 5i,j). This finding confirms the relevance of IL-22 in promoting Osmr expression in IECs across two independent models of intestinal inflammation.
Using a recently identified IL-22-induced gene signature in human IBD tissue22, we investigated whether expression of the IL-22 signature and OSMR are correlated in mucosal samples from individuals with IBD. Indeed, the IL-22 gene signature correlated strongly with OSMR expression across four independent IBD cohorts, comprising more than 868 mucosal samples in total (Extended Data Fig. 6f,g).
To determine whether IL-22 is required to sustain OSMR expression in IECs, colon epithelial organoids were subjected to IL-22 treatment, followed by IL-22 removal and subsequent assessment of Osmr expression. After removing IL-22, Osmr expression significantly decreased (Supplementary Fig. 5e). Similarly, a 7-day regimen of IL-22 blockade in mice with established H. hepaticus + anti-IL-10R colitis caused significantly reduced Osmr expression in IECs (Fig. 3m), demonstrating that IL-22 is required for both induction and maintenance of epithelial OSMR expression during colitis.
IL-22-induced OSMR expression requires STAT3
IL-22 predominantly activates STAT3 and, to a lesser extent, STAT1 (refs. 17,23). Examination of the Eukaryotic Promoter Database revealed predicted binding sites for STAT1 and STAT3 upstream of the Osmr transcription start site. We therefore explored STAT1 and STAT3 phosphorylation in IECs from healthy and colitic mice by western blotting and immunofluorescence and observed clear activation of STAT3, but not STAT1, under inflamed conditions (Fig. 4a–c). By contrast, STAT1 activation was observed mainly in the lamina propria (Extended Data Fig. 7a). As expected, STAT3 phosphorylation in IECs required IL-22 (Fig. 4d,e). Corroborating these results, stimulation of colonic epithelial organoids with IL-22 resulted in clear phosphorylation of STAT3 but not STAT1 (Extended Data Fig. 7b–d).
a, H. hepaticus + anti-IL-10R colitis was induced in wild-type mice for 14 days. Protein analysis of IECs from steady-state (n = 4) and H. hepaticus + anti-IL-10R (n = 5) colitic mice was conducted. The western blot depicts phosphorylated STAT1 (pSTAT1), STAT1, pSTAT3, STAT3 and β-actin for representative samples from two experiments. b,c, Immunofluorescence staining for pSTAT1 and pSTAT3 in steady-state (n = 7) and inflamed (n = 10) mouse samples (b) with subsequent quantification (c) in the epithelium; scale bar, 20 μm. Hpf, high-power field. Data were pooled from two experiments. d,e, H. hepaticus + anti-IL-10R colitis was induced in wild-type mice for 7 days, and mice were treated with anti-IL-22 (clone 8e11, n = 4) or isotype control (mouse IgG1, GP120:9709, n = 5) on days 0 and 3 of colitis induction. Protein analysis of IECs from steady-state and H. hepaticus + anti-IL-10R colitic mice and anti-IL-22 or isotype-treated inflamed mice was performed. d, Western blot depicting STAT3 phosphorylation in epithelial cells from anti-IL-22- or isotype-treated colitic mice. Data are representative of two independent experiments. e, Quantification of STAT3 phosphorylation using ImageJ. The relative band intensity of blots from inflamed mice was normalized to that of steady-state untreated mice. f,g, Colitis was induced in VilcreERT2Stat3fl/fl (IECΔStat3) or VilcreERT2Stat3fl/wt (IECWT; f) and Stat1–/– or Stat1+/+ (g) mice using the H. hepaticus + anti-IL-10R model for 7 days. qPCR analysis was performed to assess the expression of Osmr and Reg3g in epithelial cells from inflamed mice normalized to epithelial cells isolated from untreated respective control mice. Data are representative of one experiment; n = 5–7 mice per genotype. h, Mouse colon organoids generated from Vilcre+Stat3fl/fl (IECΔStat3) or Vilcre-Stat3fl/fl (IECWT) mice and stimulated with 10 ng ml–1 IL-22. Relative expression of Osmr was analyzed by qPCR. Data are representative of three independent experiments from three independent biological replicates. P values (two-tailed) were calculated using a Mann–Whitney test for c and e–h.
To determine the relative contribution of STAT1 and STAT3 to the induction of OSMR expression in IECs, mice deficient in STAT1 or STAT3 were subjected to H. hepaticus + anti-IL-10R-induced colitis. IECs deficient in STAT3 failed to upregulate Osmr expression during colitis, whereas STAT1 deficiency had no discernible impact (Fig. 4f,g). Consistent with these findings, genetic STAT3 deficiency or pharmacologic blockade of JAK1/JAK2 or STAT3 inhibited upregulation of Osmr expression in colonic epithelial organoids treated with IL-22 (Fig. 4h and Extended Data Fig. 7e).
In summary, IL-22 plays a critical role in driving Osmr expression in IECs both in vivo and in vitro, and this process is contingent on STAT3 activation.
ILCs drive early OSMR induction in IECs in acute colitis
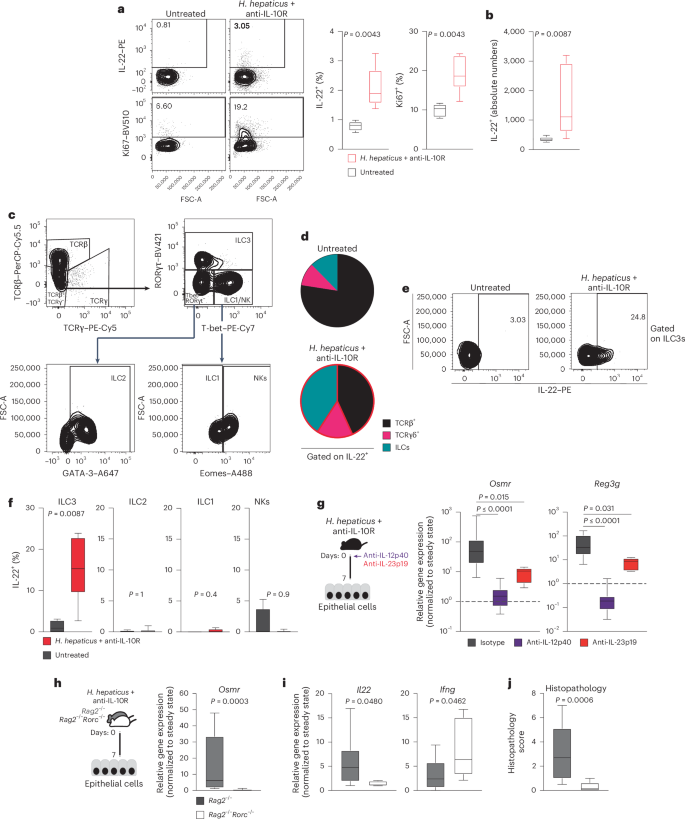
IL-22 is expressed predominantly by hematopoietic cells, particularly ILCs and IL-17-producing helper T (TH17) cells17,24,25. To identify the cellular source of IL-22 in H. hepaticus + anti-IL-10R-induced colitis, we evaluated lamina propria T cells and ILCs (live CD45+CD19–CD11c–Ly6G/C–F4/80–FcεRIα–TCRγ– TCRβ–) 7 days after colitis induction using flow cytometry and observed an increased frequency of IL-22+ and proliferating cells (Fig. 5a and Extended Data Fig. 8a,b), consistent with increased absolute numbers of IL-22-producing cells in colitic mice (Fig. 5b). IL-22 production was significantly elevated in ILCs during colitis (Fig. 5c,d and Extended Data Fig. 8c). Specifically, we observed increased abundance of IL-22+ ILC3s after colitis induction (Fig. 5e,f and Extended Data Fig. 8d). IL-22 expression by intestinal ILCs is known to be controlled by the cytokine IL-23, which is comprised of two subunits, IL-12p40 (also shared with IL-12) and IL-23p19 (refs. 17,26,27). We therefore investigated whether inhibition of IL-23 signaling affects OSMR expression in IECs during colitis. Indeed, antibody blockade of either IL-12p40 or IL-23p19 resulted in decreased expression of Osmr and Reg3g in IECs, along with reduced Il22 expression in colon tissue and alleviated intestinal pathology (Fig. 5g and Extended Data Fig. 8e–h). Similarly, IL-23-dependent induction of IL-22 production by ILC3s was observed during colitis in the C. rodentium model (Extended Data Fig. 5k,l).
a,b, H. hepaticus + anti-IL-10R colitis was induced in wild-type mice for 7 days. Percentage (a) and absolute cell numbers (b), respectively, of IL-22- or Ki67-expressing CD45+Lin– (that is, live CD45+CD19–CD11c–Ly6G/C–F4/80–FcεRIα–) LPMCs in naive mice and colitic mice were assessed. Flow cytometry-assessed cytokine expression after stimulation with phorbol 12-myristate 13-acetate (PMA)/ionomycin, IL-23 and IL-1β is shown. Data are representative of two experiments; n = 12. c, Representative gating strategy for identifying αβ and γδ T cells and different ILC subsets in colitic mice. ILC subsets were identified by flow cytometry of mouse LPMCs. The ILC population was characterized as CD45+Lin– (that is, CD19–CD11c–Ly6G/C–F4/80–FcERIα–TCRα–TCRγ–) and live cells. Subsets were further defined based on transcription factor expression: ILC3 (RORγt+Lin–TCRα–TCRγ–), ILC2 (GATA3+RORγt–T-bet–Eomes–Lin–TCRα–TCRγ–), ILC1 (T-bet+Eomes–RORγt–Lin–TCRα–TCRγ–) and natural killer cells (T-bet+Eomes+RORγt–Lin–TCRα–TCRγ–). d, Pie chart showing the indicated cell populations within total IL-22+CD45+CD19–CD11c–Ly6G/C–F4/80–FcεRIα– cells in steady-state and inflamed mice on day 7 of colitis. e,f, IL-22 production by different ILC populations identified as described in c. Data are representative of two experiments; n = 12. P values are derived from Mann–Whitney U-tests. g, H. hepaticus + anti-IL-10R colitis was induced in wild-type mice. Mice were treated with anti-IL-12p40, anti-IL-23p19 or isotype control on day 0 and analyzed on day 7. Osmr (left) and Reg3g (right) expression was quantified in epithelial cells by qPCR. Data are representative of one to two independent experiments; n = 6–29 per time point. Adjusted P values were obtained using a Kruskal–Wallis test followed by a Dunn’s multiple comparisons test. h–j, H. hepaticus + anti-IL-10R colitis was induced in Rag2–/–Rorc–/– and Rag2–/– mice for 7 days. h,i, qPCR gene expression analysis of Osmr (h) and Il22 and Ifng (i) in colonic IECs from inflamed Rag2–/–Rorc–/– and Rag2–/– mice normalized to steady-state mice. j, Histopathological scoring of colitis in Rag2–/–Rorc–/– and Rag2–/– mice on day 7 of colitis. Data are from one to two independent experiments; n ≥ 6. P values (two-tailed) were calculated using a Mann–Whitney test in a, b, f and h–j.
To further delineate the contributions of innate immune cells in driving early Osmr expression in IECs, we compared Rag2–/– mice (which lack T and B cells) and wild-type mice in the H. hepaticus + anti-IL-10R model. Colitis severity in Rag2–/– mice was significantly less than that of wild-type mice, consistent with a lack of adaptive immunity (Extended Data Fig. 8i), but Osmr was nevertheless induced in IECs from colitic Rag2–/– mice at levels approaching those of wild-type animals for at least 14 days (Extended Data Fig. 8j). Thus, innate immune cell-derived IL-22 is sufficient to induce sustained Osmr expression in IECs. ILC3 development requires the transcription factor RΟRγt (encoded by Rorc). Therefore, we next evaluated the functional relevance of ILC3 by inducing colitis in Rorc–/–Rag2–/– mice (which lack ILC3) and observed reduced Il22 expression in the colon, reduced epithelial Osmr expression and attenuated colon histopathology compared to Rorc-sufficient Rag2–/– mice (Fig. 5h–j).
These findings highlight the role of innate immune cells in driving IL-22-induced upregulation of OSMR expression in IECs. Additionally, these findings demonstrate that IL-22 can sustain OSMR expression in IECs independently of T cells, thus initiating the IL-22–OSM inflammation circuit.
OSMR in IECs drives STAT3 and leukocyte recruitment
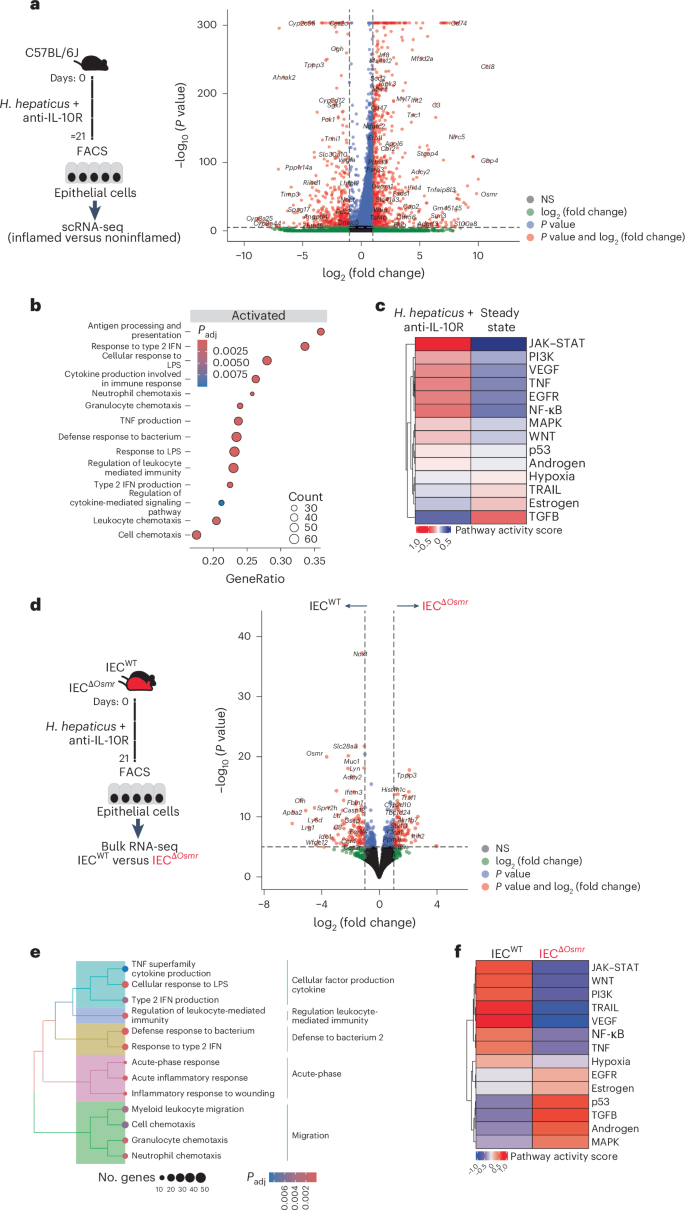
To better characterize the biological impact of OSM on IECs during colitis, we first evaluated our scRNA-seq data from epithelial cells collected at steady-state or at the peak of colitis (day 21). IECs from colitic mice showed upregulation of inflammation-associated genes, such as Ccl8, S100a8, Saa3 and Reg3b, in addition to several inflammatory pathways, including cellular response to lipopolysaccharides, response to type 2 interferons and antigen processing and presentation (Fig. 6a,b and Supplementary Tables 3 and 4). Prediction of potential upstream pathways identified JAK–STAT signaling as a highly probable driver of the colitis-associated transcriptional response in IECs (Fig. 6c), consistent with a potential role for cytokines such as IL-22 and OSM.
a, Workflow schematic for scRNA-seq sample preparation (left) and volcano plot depicting differentially expressed genes in enterocytes between inflamed mice after colitis induction and steady-state (see Fig. 1d). Red dots represent genes that are expressed at least twofold higher with statistical significance. b, Significantly enriched (adjusted P < 0.05) gene set enrichment analysis (GSEA) terms in enterocytes from scRNA-seq data derived from a; adjusted P values were calculated using the Benjamini–Hochberg test. c, PROGENy pathway activity scores derived from epithelial scRNA-seq data. PROGENy is a computational method that leverages a large compendium of publicly available perturbation experiments to identify a core set of pathway responsive genes, enabling the inference of pathway activity from transcriptomic data. d, Left, experimental workflow of bulk RNA-seq of sorted epithelial cells from IECΔOsmr and control mice. H. hepaticus + anti-IL-10R colitis was induced in IECΔOsmr and control mice (littermates). IECs were sorted from both genotypes on day 21 and subjected to bulk RNA-seq. Right, volcano plot depicting differentially expressed genes in IECs between inflamed IECΔOsmr and control mice (n = 5 per group). Red dots represent genes that are expressed at least twofold higher with statistical significance. e, Significantly enriched (adjusted P < 0.05) GSEA terms in epithelial cells from bulk RNA-seq data derived from d; adjusted P values were calculated using the Benjamini–Hochberg test. f, PROGENy pathway activity scores derived from epithelial bulk RNA-seq data in d.
To precisely delineate the transcriptional alterations in IECs mediated by OSM, we sorted epithelial cells from IECsΔOsmr and control mice after colitis induction and subjected them to RNA-seq analysis (Fig. 6d and Supplementary Table 3). Pathway analysis identified reduced inflammatory gene expression in OSMR-deficient IECs, including response to lipopolysaccharides and type 2 interferons (Fig. 6e and Supplementary Table 4). As expected, JAK–STAT signaling was identified as a key upstream cascade regulating IEC response in control mice but was predicted to have low activity in the absence of OSMR (Fig. 6f). Previous studies have shown that OSM can stimulate STAT3 activation in human epithelial cell lines28. Interestingly, we observed that OSM induced a robust and sustained activation of STAT3 compared to IL-22 in colon epithelial cells (Extended Data Fig. 9a). Furthermore, OSMR-deficient IECs failed to maintain STAT3 phosphorylation following colitis induction (Extended Data Fig. 9b), along with a reduction in the OSM-responsive transcriptional program in these cells (Extended Data Fig. 9c,d and Supplementary Table 6).
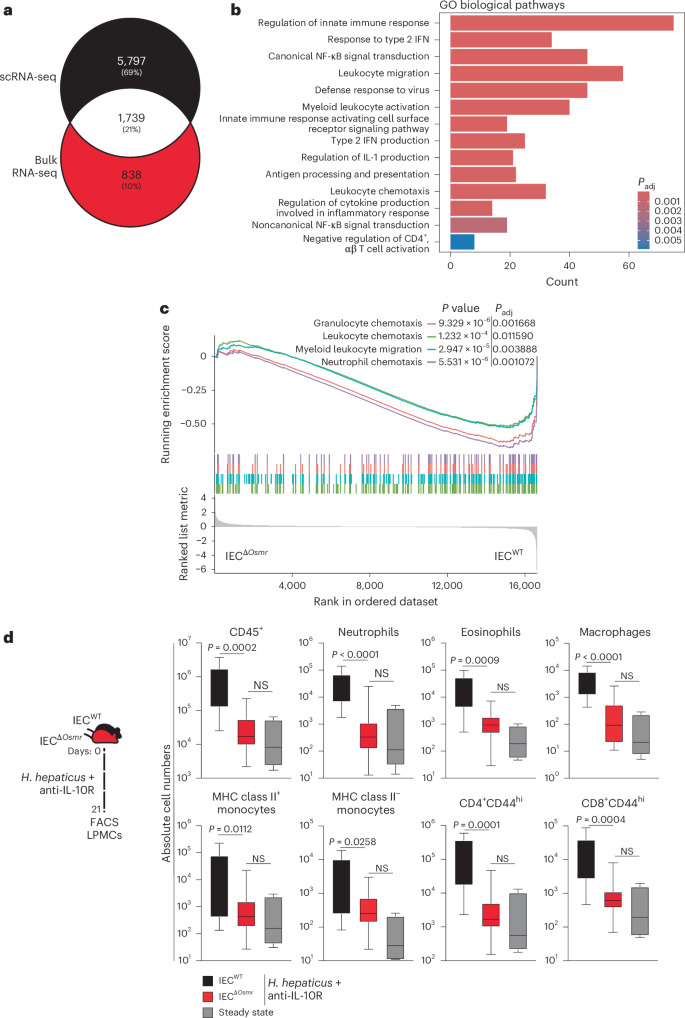
We next compared the colitis-associated changes identified in single-cell analysis of wild-type IECs and bulk RNA-seq analyses of OSMR-deficient IECs and identified >1,700 overlapping genes (Fig. 7a and Supplementary Table 4). Pathway analysis of overlapping genes identified strong enrichment of leukocyte migration and chemotaxis pathways (Fig. 7b,c and Supplementary Table 4), suggesting that OSM may act on IECs to modulate leukocyte recruitment during intestinal inflammation. Indeed, flow cytometry analysis of colonic immune cells from IECΔOsmr and control mice following colitis induction revealed reduced infiltration of granulocytes, monocytes and T cells in IECΔOsmr mice compared to respective controls (Fig. 7d). A similar reduction in leukocyte recruitment was observed in IECΔOsmr mice during C. rodentium-induced colitis (Extended Data Fig. 5m).
a, Venn diagram representing the overlap of differentially expressed genes in epithelial bulk and scRNA-seq datasets from Fig. 6a,d. b, Significantly enriched (adjusted P < 0.05) Gene Ontology terms of the overlapping genes from both comparisons in Fig. 6a,d; adjusted P values were calculated using the Benjamini–Hochberg test. c, GSEA plot depicting the enrichment of differentially expressed genes from pathways related to cell chemotaxis in epithelial cells comparing inflamed IECΔOsmr and control mice, as in Fig. 6d. d, H. hepaticus + anti-IL-10R colitis was induced in IECΔOsmr (n = 13) and control mice (n = 11). The immune cell composition in the lamina propria was analyzed by flow cytometry on day 21 of inflammation. Data are representative of two independent experiments. P values (two-tailed) were calculated using a Mann–Whitney U-test comparing IECΔOsmr and control mice.
Together, OSMR signaling promotes and sustains STAT3 activation and the JAK–STAT transcriptional signature in IECs, thereby promoting intestinal immune cell recruitment during colitis.
Epithelial OSMR is essential for CAC
Chronic mucosal inflammation is a well-established risk factor for CRC, with sustained STAT3 activation being a key driver of its development17,29. Our findings indicate that OSM promotes STAT3 activation and epithelial cell proliferation during colitis (Extended Data Fig. 9b and 10a). Therefore, we investigated the role of OSM in CAC using the dextran sodium sulfate/azoxymethane (DSS/AOM) model, in which epithelial transformation is initiated by administration of the procarcinogen AOM and tumorigenesis is promoted by repeated treatment with the intestinal irritant DSS, causing chronic colitis and consequent tumor growth30,31.
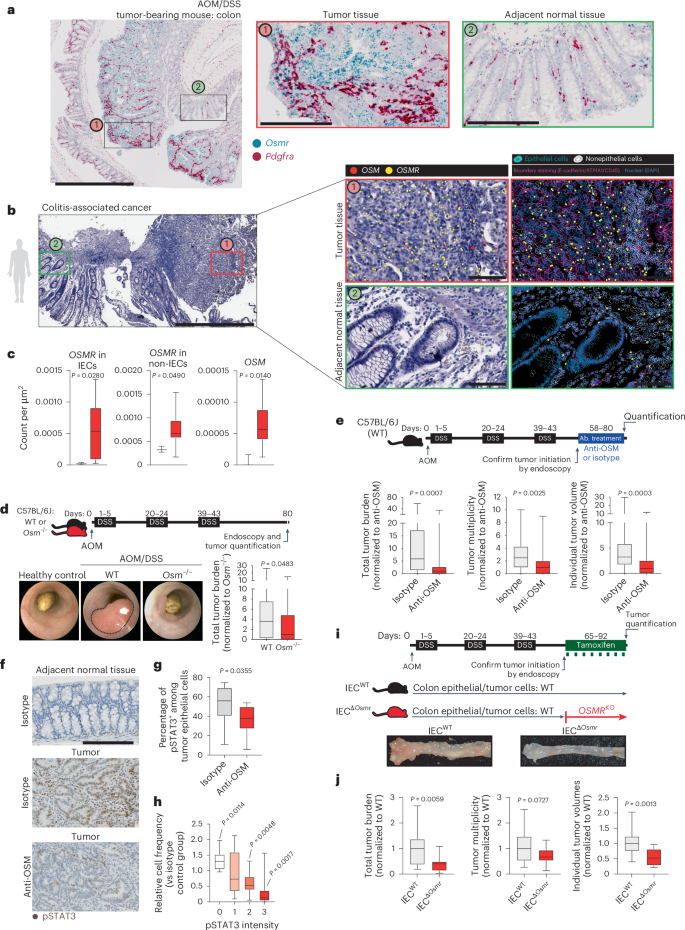
We confirmed increased expression of Osm and Osmr in DSS/AOM-induced colon tumors using a previously published RNA-seq dataset (Extended Data Fig. 10b)32. Osm and Osmr expression in CAC correlated with the expression of Stat3 as well as Socs3, a well-known target gene of STAT3 (Extended Data Fig. 10c,d). We then confirmed that the expression of Osmr was induced in IECs during DSS colitis and in DSS/AOM-induced tumors using qPCR (Extended Data Fig. 10f). Using ISH, we observed strong Osmr expression in tumor epithelial and stromal cells (Pdgfra+; Fig. 8a). However, Osmr was expressed at much lower levels in epithelial cells from adjacent tumor-free and noninflamed tissue (Fig. 8a).
a, Wild-type mice underwent AOM/DSS-induced CAC. Colon tissues from tumor-bearing mice were analyzed by ISH for Osmr (blue) and Pdgfra (red; stromal marker); scale bars, 1,000 μm (left) and 200 μm (two insets on the right). b, Spatial transcriptomics analysis of a biopsy from an individual with CAC, including tumor tissue (red box) and adjacent normal mucosa (green box). OSM and OSMR expression was assessed in epithelial and nonepithelial cells using epithelial (EPCAM, CDH1, KRT20, MUC5B and MKI67) and nonepithelial markers (CD3E, CD4, CD14, CD68, C1QC, CD19, CD79A, PDGFRA, VIM, PECAM1, CD34 and KIT); scale bar, 1,000 μm (left) and 100 μm (insets on the right). c, Quantification of OSMR expression in epithelial and nonepithelial cells in CAC (n = 10, red bars) and healthy control (n = 3, black bars) tissue, along with nonepithelial OSM expression. d, Experimental schematic comparing CAC induction in wild-type versus Osm–/– mice. Representative colonoscopy images and tumor burden on day 80 are shown (n = 25 per genotype). e, Wild-type mice received AOM and DSS, followed by treatment with anti-OSM (n = 28) or isotype antibody (rIgG2a; n = 26). Tumor burden, multiplicity and volume were assessed on day 80. Data are shown normalized to the isotype-treated group. f, Immunostaining for pSTAT3 in colon tumors and adjacent normal tissues from mice treated as in e; scale bar, 200 μm. g, Quantification of pSTAT3+ epithelial cells (n = 10 per group). h, Relative pSTAT3 expression intensity normalized to that observed in control tissue (n = 10 per group). Data are derived from two experiments. P values (two-tailed) were calculated from one-sample t-tests and Wilcoxon rank tests. i,j, VilcreERT2+Osmrfl/fl (IECΔOsmr; n = 15) and VilcreERT2+Osmrwt/wt (IECWT; n = 18) mice were treated with AOM/DSS to induce CAC. Tamoxifen administration induced epithelial-specific Osmr deletion. i, Experimental design and representative colon images. j, Tumor burden, multiplicity and average volumes from two independent experiments. Data are shown normalized to the VilcreERT2+Osmrwt/wt group. P values (two-tailed) were calculated using a Mann–Whitney test in c–e, g and j.
We next investigated the spatial expression and distribution of OSM and OSMR in a cohort of individuals with CAC (n = 10). Consistent with findings in mice, OSMR was highly expressed in tumor epithelial and stromal cells, whereas its expression in epithelial cells was minimal in adjacent tumor-free tissue (Fig. 8b and Extended Data Fig. 10e). Quantitative analysis revealed a significant increase in OSMR expression in tumor epithelial cells compared to in nontumor epithelial cells. Furthermore, OSM expression was elevated in individuals with CAC and remained restricted to nonepithelial cells within the lamina propria (Fig. 8b,c).
To explore the role of OSM in CAC, we next used Osm–/– mice and induced CAC using the DSS/AOM model (Fig. 8d). Interestingly, OSM deficiency significantly reduced tumor burden (Fig. 8d). However, this observation could be due to an impact of OSM on colitis and tumor initiation, an impact of OSM on tumor progression or a combination of the two. To evaluate the impact of OSM on tumor progression, rather than tumor initiation, we induced CAC in wild-type mice and confirmed tumor development by endoscopy, after which mice were treated with anti-OSM or isotype control (Fig. 8e). Anti-OSM treatment reduced tumor size and number, supporting a possible role for OSM in CAC progression (Fig. 8e). Notably, STAT3 phosphorylation was reduced in colonic adenomas after treatment with anti-OSM (Fig. 8f–h).
To address the specific role of OSM in epithelial cells and modulation of tumor progression, we used IECΔOsmr mice to control temporal OSMR expression in IECs in CAC. Tamoxifen was administered after tumor initiation to both IECΔOsmr and control mice, and tumor burden was assessed after 3–4 weeks (Fig. 8i). Remarkably, Osmr deletion in IECs significantly reduced the number and size of tumors, confirming an epithelial-intrinsic role for OSMR signaling in promoting colitis-induced tumorigenesis (Fig. 8i,j).
Osmr upregulation and maintenance in the H. hepaticus + anti-IL-10R colitis model depended on IL-22 and IL-12/IL-23. Notably, DSS treatment also induced epithelial Osmr expression in an IL-22-dependent manner (Extended Data Fig. 10g,h). Next, we evaluated whether IL-12/IL-23 and IL-22 were also required to maintain OSMR expression in established CAC. We observed a correlation between Il12b and Osmr in CAC (Extended Data Fig. 10i). In wild-type mice with established AOM/DSS-induced tumors, blockade of IL-12/IL-23 (using anti-IL-12p40) or IL-22 for 1 week caused reduced expression of Osmr and Socs3 in adenoma epithelial cells (Extended Data Fig. 10j–l). IL-12p40 blockade similarly reduced Il22 expression in adenomas (Extended Data Fig. 10m).
Together, tumor epithelial cells express Osmr in CAC, which promotes STAT3 phosphorylation and tumor growth. Finally, Osmr expression on epithelial cells in CAC is mediated by IL-22.
Discussion
Unchecked inflammation exacerbates intestinal pathology and increases the risk of cancer. Inflammation promotes tumor initiation, in which normal epithelial cells acquire genomic alterations that initiate tumorigenesis and promote sustained proliferation of cancer cells2,33. Various pathways have been implicated in supporting CAC, with STAT3 activation in epithelial cells being one of the most relevant protumorigenic pathways29. Here, we identified close cooperation between two related mucosal cytokine pathways acting on epithelial cells to promote intestinal inflammation and carcinogenesis. Our data demonstrate that OSMR is broadly expressed by intestinal stromal and endothelial cells but not epithelial cells under steady-state conditions, whereas the expression of OSMR is upregulated by epithelial cells during intestinal inflammation and in CAC. Notably, ILC3-derived IL-22 was a key inducer and rheostat of epithelial OSMR expression, as OSMR expression was unstable in the absence of IL-22 both in colitis and CAC. STAT3 was essential for upregulation of Osmr in IECs, supporting the relevance of IL-22 in modulating Osmr expression. Deletion of Il22ra1 or Osmr in IECs ameliorated colitis, highlighting the individual role of both cytokines in modulating inflammation in vivo. Furthermore, deletion of Osmr in IECs after tumor development resulted in tumor reduction.
OSM plays a central role in orchestrating tissue repair and maintaining immune homeostasis through its cell-type-specific effects across diverse organ systems. It promotes epithelial and stromal cell proliferation and migration, drives extracellular matrix remodeling and supports angiogenesis, processes essential for barrier integrity and tissue regeneration4,8. However, beyond its reparative functions, OSM has also been implicated in driving inflammation, particularly in barrier tissues such as the skin, lung and intestine. Notably, in IBD, OSM emerged as the most transcriptionally upregulated cytokine in inflamed intestinal mucosa compared to in healthy individuals, highlighting its potential as both a mediator and biomarker of mucosal inflammation7,34. Increased OSM expression in the intestinal mucosa is mirrored by overexpression of its receptor OSMR, suggesting a specific role for OSM–OSMR interactions in IBD pathogenesis. Although prior studies suggested a key role for stromal cells in OSM-mediated pathology7,35, our conditional deletion experiments, in which Osmr was deleted in Col1a2+ fibroblasts and Cdh5+ endothelial cells, failed to impact acute intestinal pathology. However, due to the difficulty in achieving complete target gene deletion in these cell types, it is possible that small residual populations of OSMR+ stromal cells contributed to inflammation in our experiments. Nevertheless, our data clearly support a previously unappreciated pathogenic role for OSM–OSMR signaling in IECs, based on the protective effect of Osmr deletion in this cell type. OSMR expression by stromal cells may also be relevant for tissue remodeling and tissue repair in intestinal inflammation, which may be more evident in chronic human IBD. Indeed, OSM pathway dysregulation has been shown to play a fibrogenic role in other organs8,36.
Our data highlight a previously underappreciated role for OSMR in epithelial responses and adaptation to tissue inflammation. OSMR signaling in IECs led to sustained STAT3 activation and the expression of chemokines that regulate the recruitment of proinflammatory immune cells. Notably, OSMR expression in IECs was maintained through IL-22-mediated STAT3 activation, both of which are dysregulated in human IBD16,37. Furthermore, IL-23 was required for IL-22 expression and, consequently, for the upregulation of epithelial OSMR expression in inflamed colon tissue. These findings suggest that the IL-22–OSMR axis could be influenced by anti-IL-12p40 and anti-IL-23p19 therapies, which are currently used in IBD management38. However, recent studies indicate that in individuals resistant to anti-IL-12p40 therapy, a strong IL-22 signature persists22. This signature correlates with elevated OSMR expression, suggesting an active IL-22–OSMR–OSM inflammatory circuit in these therapy-refractory individuals. Further investigations into the activity of the IL-22–OSMR–OSM pathway in individuals receiving anti-IL-23p19 therapy could provide critical insights into the clinical relevance of targeting this axis in specific populations of individuals with IBD and CAC.
The role of IL-22 in inflammation and cancer is complex, and both cancer-promoting and inhibitory functions have been described for IL-22 (refs. 16,17). Although IL-22 promotes inflammation in the H. hepaticus + anti-IL-10R model, it plays an essential role in epithelial repair in chemically induced colitis and is thought to be protective in the context of acute epithelial damage16,17,18. IL-22 has also been shown to play a double-edged role in CAC; dysregulated or sustained IL-22 expression (as occurs in the absence of IL-22BP) can promote tumorigenesis in AOM/DSS- and bacteria-induced colon cancer models, but it can also promote protective DNA damage responses in the epithelium19,21,39. IL-22 can promote colon cancer via STAT3 activation in epithelial cells, a known oncogenic signaling pathway40, and we found that Osmr expression in IECs was modulated by STAT3 activation and sustained only in the presence of IL-22. Interestingly, OSMR expression by epithelial cells was required to maintain STAT3 phosphorylation. Thus, the IL-22–OSM axis is necessary to maintain STAT3 and immune cell recruitment to the intestine to maintain the pathogenic impact of IL-22 in inflammation and carcinogenesis. Targeting the OSM–OSMR axis in intestinal inflammation and CAC could separate the beneficial effects of IL-22 on epithelial cell recovery and repair from its ability to sustain harmful STAT3 activation and immune cell recruitment.
The cellular source of IL-22 and its role within this inflammatory circuit are crucial for understanding how IL-22 modulates the OSM–OSMR axis. Our findings indicate that ILC3s serve as an early source of IL-22 in the H. hepaticus + anti-IL-10R colitis model, as well as in C. rodentium infection. However, IL-22 production is not restricted to ILC3s; rather, it is derived from a diverse range of immune cells depending on the inflammatory context25,41,42,43. Although early studies primarily identified IL-17⁺CD4⁺ T cells (TH17 cells) as key producers of IL-22, subsequent research has demonstrated that multiple ILC subsets contribute significantly to IL-22 secretion. In addition to ILC3s, natural killer cell subsets, lymphoid tissue inducer cells and CD11c⁺ colonic cells activated via Toll-like receptor signaling have been shown to produce IL-22. Studies using IL-23R reporter mice further revealed that γδ⁺ T cells within the intestinal lamina propria, along with certain CD11b⁺ myeloid cells, express IL-23R and are capable of producing IL-22 in vivo44. More recently, neutrophils have also been implicated as a source of IL-22, particularly in the setting of acute DSS-induced colitis.
These findings suggest a dynamic and context-dependent regulation of IL-22 production, wherein distinct immune cell populations may contribute to IL-22 secretion at different time points during inflammation. This temporal and cellular heterogeneity is particularly relevant in chronic inflammatory conditions, where the interplay between adaptive and innate immune responses may shape disease progression. Understanding how these diverse IL-22-producing populations integrate with the OSM–OSMR axis will be critical for delineating their roles in tissue homeostasis, repair and inflammation-driven tumorigenesis.
Independent from the impact of IL-22 in inflammation and cancer, OSM has been shown to support tumor progression and metastasis in various settings45,46. OSM produced by infiltrating immune cells can alter the tumor microenvironment and expression of OSMR by tumor cells, and tumor-associated stromal cells may be important for tumor progression47. Downstream of OSMR activation, the MAPK, PI3K and STAT3 pathways can all be activated by OSM engagement and participate in tumor progression. OSM is frequently overexpressed in cancer and associates with poor prognostic features in many tumor types, including colon cancer48. However, the functional role of the OSM–OSMR axis in CAC remains unexplored, and our findings highlight the pathogenic cooperation of IL-22 and OSM in CAC. A phase 2 clinical trial investigating anti-OSMR therapy (vixarelimab) for UC is currently underway, which holds the potential to provide important clinical insights into the role of the OSM–OSMR axis in intestinal inflammation and CAC.
In summary, we show that the IL-22 and OSM pathways collaborate to promote intestinal inflammation and colitis-driven colon cancer. This axis drives persistent activation of STAT3 in epithelial cells, which promotes high expression of chemotactic factors to fuel sustained recruitment of inflammatory leukocytes during colitis. Our data thus support the concept of OSM/OSMR blockade for IBD treatment, particularly in individuals at increased risk of CRC.
Methods
Clinical samples
All human samples were collected following written informed consent and in compliance with protocols approved by the Institutional Review Board (EA1/200/17). Samples were obtained from individuals undergoing intestinal resection at Charité–Universitätsmedizin Berlin. For ISH analysis in Extended Data Fig. 4a, additional samples were sourced from individuals at the Mayo Clinic, enrolled in an observational study approved by the Mayo Clinic Institutional Review Board (10-006628). Informed consent was obtained in accordance with institutional guidelines. Surgical specimens were collected, fixed in formalin and processed for histological analysis. Participant demographics are summarized in Supplementary Table 5. All samples were processed within 2–4 h of collection. Sample collection adhered to the ethical standards of the World Medical Association, including the Declaration of Helsinki and its subsequent amendments.
Mice
All mice used were bred under specific pathogen-free conditions at the animal facilities of the Federal Institute for Risk Assessment, the Research Institute for Experimental Medicine of Charité, Genentech or The Jackson Laboratory. The animals were housed under a 12-h light/12-h dark cycle at a temperature of 22 ± 2 °C and a relative humidity of 55 ± 10%. Animal experiments were performed in compliance with German animal protection laws and were approved by the responsible governmental authority (Landesamt für Gesundheit und Soziales) under applications G 0291/18 and G 0016/18. Female C57BL/6 mice were obtained from Charles River. Osmrfl/fl mice (B6;129-Osmrtm1.1Nat/J; 011081) were obtained from The Jackson Laboratory and genotyped via PCR to confirm the targeted allele deletion. VilcreERT2 (ref. 49), Col1a2creERT2 (ref. 50) or Cdh5cre (ref. 51) were crossed with Osmrfl/fl to establish cell-type-specific OSMR-deficient mouse lines including VilcreERT2Osmrfl/fl (IECΔOsmr), Col1a2creERT2Osmrfl/fl (StromaΔOsmr) or Cdh5cre Osmrfl/fl (EndoΔOsmr) or with B6.Cg-Gt(Rosa)26Sortm9(CAG-tdTomato)Hze/J (Ai9)52 to generate VilcreERT2 Ai9, Col1a2creERT2 Ai9 or Cdh5cre Ai9 and with Stat3fl/fl to generate VilcreERT2Stat3fl/fl. VilcreERT2 mice were crossed with Stat3fl/fl (ref. 53). Rag2–/–Rorc–/–54,55 mice were provided by the laboratory of C.R., VilcreStat3fl/fl mice were obtained from the laboratory of S. Wirtz, and VilcreIl22ra1fl/fl21, Il22[–/–56 and Stat1[–/–57 mouse lines were provided by the laboratories of A.D. and M.L., respectively.
For AOM/DSS studies, Osmrfl/fl mice were generated and bred at Genentech. The Osmr conditional knockout allele was generated using C57BL/6N C2 embryonic stem cells and established methods. A targeting vector consisting of a 5′ homology arm, a loxP site, the exon 3 region to be floxed, an FRT-Neo-FRT-loxP cassette and a 3′ homology arm was used to create the conditional knockout allele in embryonic stem cells. The Neo selection cassette was excised by FlpO treatment, leaving a residual FRT site. The conditional knockout allele was verified by sequencing before microinjection. The floxed Osmr region corresponds to GRCm39 chromosome 15: 6881773–6882254.
Mice (6–20 weeks old) were cohoused with wild-type mice (Charles River) for at least 4 weeks before colitis induction to normalize microbiota. To minimize cage effects, animals were randomly assigned to different cages before experimental procedures. Subsequent to this cohousing period, and before the onset of colitis, Col1a2creERT2 and VilcreERT2Osmrfl/fl mice and VilcreERT2Stat3fl/fl mice were administered tamoxifen (Sigma-Aldrich) via intraperitoneal injection. The dosage was set at 75 mg per kg (body weight) administered daily over 5 days, in alignment with the protocol delineated by The Jackson Laboratory52,58. Control mice for the conditional deletion experiments were matched littermates with cre+, creERT2+Osmrfl/wt or creERT2–Osmrfl/fl and were also treated with tamoxifen.
Colitis induction with H. hepaticus
H. hepaticus (DSMZ, 22909) was revived from glycerol stocks and cultured on blood agar (Fisher Scientific) under microaerobic conditions at 37 °C. After quality confirmation by microscopy, bacteria were transferred to tryptone soy broth (Fisher Scientific) with 10% fetal calf serum (FCS; Sigma), 10 μg ml–1 vancomycin, 5 μg ml–1 trimethoprim and 2.5 IU ml–1 polymyxin B and grown at 37 °C and 180 rpm. Mice were orally inoculated with 1 × 108 colony-forming units (c.f.u.) on 2 consecutive days using a 22-gauge curved blunted needle and received 1 mg of anti-IL-10R (1B1.2, BioXcell) intraperitoneally on day 0 and weekly thereafter7,9.
At designated time points, mice were killed, and tissues (colon, small intestine, cecum, liver, spleen and mesenteric lymph nodes) were collected. Intestinal segments (~0.3 cm) were fixed in 4% formaldehyde (SAV) for histology or stored in RNAlater (Qiagen) for gene expression analysis.
C. rodentium infection
C. rodentium ICC169 was cultured overnight in LB broth (Sigma) with nalidixic acid (50 μg ml–1) at 37 °C with shaking. Bacterial concentration was assessed by optical density at 600 nm, and c.f.u. was determined by serial dilution and plating. Mice were orally infected with 5 × 109 c.f.u. in 200 μl using a 22-gauge curved blunted needle.
To quantify bacterial burden, 4- to 5-mm segments from proximal, mid and distal colon, spleen and liver were collected, rinsed in PBS/bovine serum albumin (BSA) and stored in 500 μl of PBS/BSA. Tissues were weighed with sterile tweezers between genotypes. Homogenization was performed in bead-containing tubes (Qiagen) at 6.5 m s–1 for ≥60 s in 500 μl of sterile PBS. Homogenates were serially diluted, plated on LB agar with nalidixic acid and incubated at 37 °C for 18–24 h. c.f.u. was counted from the final three dilutions to calculate bacterial loads.
DSS/AOM treatment and tumor induction
To initiate colon tumorigenesis, mice were administered an intraperitoneal injection of AOM (Sigma) at a dosage of 10 mg per kg (body weight). Five days after injection, the mice began a regimen of 3% DSS (MP Biomedicals, molecular mass 36,000–50,000 Da) in drinking water, which was sustained for a duration of 5 days, followed by a 16-day period on regular water. Miniendoscopy was performed at the indicated time points to confirm tumor development, following previously described protocols59. Mice received a total of three DSS cycles, after which they were either killed for tumor evaluation or assigned to additional treatments (as described in the figure legends). In some experiments, mice were treated only with DSS to model colitis in the absence of tumor formation. Cumulative tumor burden in this model rarely exceeded 100 mm3, and no mice were killed due to large tumors or bowel obstruction. However, some mice were killed for humane reasons if body weight decreased by >20% or if they developed rectal prolapse. Tumor induction experiments were performed at Genentech, approved by Genentech’s Institutional Animal Care and Use Committee, and adhere to the NRC Guidelines for the Care and Use of Laboratory Animals.
In vivo antibody neutralization
IL-22 was neutralized via intraperitoneal injection of 450 μg of anti-IL-22 (clone 8E11, Genentech) on days 0 and 3 (7-day model) or days 7 and 10 (14-day model). IL-12p40 (C17.8) or IL-23p19 (G23-8, BioXcell) was neutralized by intraperitoneal injection of 1 mg on day 0. Control mice received isotype-matched mouse IgG1 (GP120:9709, Genentech). In the DSS/AOM model, OSM was blocked using 0.5 mg of anti-OSM (clone 24A8, Genentech)60 or IgG2a isotype control (anti-gp120, Genentech), administered twice weekly.
Histological scoring
Histological assessment of mouse colitis was performed as previously described61. Briefly, proximal, middle and distal colon segments were formalin fixed, paraffin embedded and stained with hematoxylin and eosin. Sections were scored (0–3) across four criteria: leukocyte infiltration, affected area, epithelial hyperplasia and goblet cell depletion. Severe disease features (crypt abscesses, edema and submucosal leukocyte infiltration) were also considered. Scores (0–12 per segment) were averaged across segments to yield a mean score per mouse. Blinded scoring was confirmed by a second blinded observer.
Histomorphological evaluation of human ileal and colonic tissues was performed according to modified scoring systems based on Naini and Cortina62 for CD and Riley et al.63 for UC. To assess the correlation between normalized OSMRβ expression and histopathology scores, histological scores were first normalized across diseases (Riley score for UC and Naini and Cortina score for CD). The normalized histopathology scores were then correlated with normalized OSMRβ expression values.
Culture and stimulation of cell lines
The Caco-2 and HCA-7 human epithelial colorectal adenocarcinoma cell lines were cultured in DMEM (Invitrogen) enriched with 10% FCS (Sigma), 2 mM l-glutamine and 10,000 U ml–1 penicillin/streptomycin (Gibco). The HCA-7 cell line was obtained from the European Collection of Authenticated Cell Cultures and stored by common cell repository at Genentech. Cells used in this study were negative for mycoplasma and authenticated by RNA-seq analysis. Two methods of mycoplasma detection were used to avoid false-positive/false-negative results: Lonza Mycoalert and Stratagene Mycosensor. Caco-2 cells were initially purchased from ATCC and were continuously tested negative for mycoplasma using a Lonza MycoAlert Mycoplasma Detection kit. Both cell lines are not listed as misidentified cell lines by the International Cell Line Authentication Committee.
Cells were maintained at 37 °C in 5% CO2. Before cytokine treatment, they were starved for 24 h. Stimulation was performed with 100 ng ml–1 recombinant human OSM, interferon-γ or IL-22 (PeproTech) for 15 min. Lysates were prepared using RIPA buffer with phosphatase and protease inhibitors (EDTA-free; Thermo Fisher). HCA-7 cells were also stimulated with 100 ng ml–1 recombinant human IL-22 or recombinant human OSM for 30 min, 24 h or 48 h, followed by analysis via MSD pSTAT3 assay (Meso Scale Diagnostics).
To assess the transcriptional program induced by OSM in epithelial cells, bulk RNA-seq analysis was performed using the HCA-7 cell line. HCA-7 cells were stimulated with recombinant human OSM for 24 h, after which RNA was isolated for bulk RNA-seq (see detailed methods below; Supplementary Table 6). Sequencing data related to HCA-7 stimulation are deposited in NCBI’s GEO and can be accessed through GEO Series accession number GSE269578.
Organoid isolation
Colon organoids were generated and cultured, as described previously, with some modifications64. Mouse intestines were opened, cut into 2- to 5-mm pieces and washed with cold PBS (pH 7.4). Tissues were incubated in 10 mM EDTA, and crypts were released by pipetting and centrifugation, filtered (80 μm), pelleted and resuspended in ice-cold Matrigel GFR (Corning). Crypts (500 per 50 μl) were seeded per well in 24-well plates. After Matrigel polymerization (15 min, 37 °C), 500 μl of advanced DMEM/F12-based medium (with HEPES, GlutaMAX, N2, B27, penicillin/streptomycin, epidermal growth factor and N-acetylcysteine) was added, supplemented with colonic crypt-specific growth factors, including gastrin, nicotinamide, A83-01, PGDE2, 20% R-spondin, 10% noggin and 50% WNT-3A conditioned medium. Y-27632 (Abmole) was included for 2 days to prevent anoikis. Medium was changed every 2 days; organoids were passaged 1:2 every 1–2 weeks by mechanical shearing using a pipette tip.
Organoids were stimulated with 10 ng ml–1 cytokines for downstream assays (Supplementary Table 7). For western blot-based STAT phosphorylation, 100 ng ml–1 cytokines was used. For inhibitor experiments, organoids were pretreated for 6 h with 50 μM fludarabine, 25 μM STA-21, 2.5 μM ruxolitinib or 2.5 μM tofacitinib before cytokine stimulation.
Colonic cell isolation
Mice were killed by cervical dislocation, and colons were collected in ice-cold PBS, opened longitudinally, cleaned and cut into 2-mm pieces. Tissues underwent sequential EDTA + DTT washes in RPMI (pH 7.4) with 10% FCS at 37 °C to detach epithelial cells. Remaining tissue was washed (RPMI + 10% FCS + 15 mM HEPES), vortexed and digested with collagenase VIII (Sigma-Aldrich). Digests were filtered, washed and separated on a 30%/40%/70% Percoll gradient (Thermo Scientific). Cells from the 30%/40% and 40%/70% interfaces were collected as stroma- and lamina propria leukocyte-enriched fractions.
Epithelial cells were isolated from pooled supernatants of two sequential EDTA incubations, washed twice in PBS and centrifuged at 300g for 5 min. Lysates were prepared using Qiazol (RNA, Qiagen) or RIPA buffer (protein, Thermo Scientific), depending on downstream applications.
Western blotting
Protein concentration was measured using a bicinchoninic acid assay (Serva) on lysates prepared in RIPA buffer with phosphatase and protease inhibitors (Thermo Fisher). Samples and BSA standards were incubated with BCA reagent at 37 °C, and absorbance was read at 550 nm. Protein concentrations were calculated via linear regression. SDS–PAGE (10%) and transfer to PVDF membranes (Carl Roth) were performed at 200 mA for 1–2 h. Membranes were blocked in 5% milk in Tris-buffered saline with Tween 20 detergent (TBST) and incubated overnight at 4 °C with primary antibodies to STAT1, STAT3, pSTAT1 and pSTAT3 (Cell Signaling; 1:1,000 in 5% BSA-TBST). After washing, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling) for 1 h at room temperature. Bands were visualized using chemiluminescent substrate (Thermo Fisher) and imaged with a LAS-4000 minianalyzer. Densitometry was performed using ImageJ (v1.48V). Antibodies are listed in Supplementary Table 7.
Calprotectin and lipocalin ELISA
Fecal samples stored at –20 °C were homogenized in PBS with protease inhibitors (Serva). ELISAs were performed using capture antibodies (for example, anti-mouse NGAL) and blocking with 1% BSA, followed by biotinylated detection antibodies, horseradish peroxidase-conjugated Avidin (Biolegend) and TMB substrate. Absorbance was measured at 450 nm. Fecal calprotectin was measured using a Mouse S100A8/S100A9 Heterodimer ELISA kit (RD), per the manufacturer’s instructions. Antibodies are listed in Supplementary Table 7.
Flow cytometry
For surface staining, freshly isolated LPMCs and epithelial cells were prepared and allocated for both surface and intracellular staining procedures. Cells were resuspended in a master mix containing surface antibodies and incubated at 4 °C for 30 min. All antibodies used in this study are listed in Supplementary Table 7. After staining, cells were fixed using BD lysis buffer (BD Biosciences) and subsequently washed. Precision Count Beads (Biolegend) were added before acquisition. For Ki67 staining, epithelial cells were first subjected to surface staining, fixed with Fixation/Permeabilization buffer using the FoxP3 Staining Buffer Set (eBioscience) and stained intracellularly with Ki67.
For ILC analysis, various stimulation conditions were initially tested. We selected stimulation with IL-2 (25 ng ml–1; PeproTech), IL-7 (25 ng ml–1; PeproTech), IL-23 (25 ng ml–1; PeproTech) or a combination of IL-23, IL-1β (10 ng ml–1; PeproTech), PMA (5 ng ml–1; Sigma) and ionomycin (500 ng ml–1; Sigma). An unstimulated condition was included as a control. Cells were incubated at 37 °C under these conditions, followed by the addition of brefeldin A (5 µg ml–1; Cayman, Cay11861-25) for 4 h. Surface marker staining was performed, followed by washing and staining with fluorochrome-conjugated streptavidin for 10 min at 4 °C (Supplementary Fig. 6a). Cells were then fixed, permeabilized and stained using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience; Supplementary Fig. 6b). We observed that ILCs from inflamed mice already produced significant amounts of cytokines without stimulation, in contrast to ILCs from steady-state mice. Stimulation with the tested conditions did not alter the extent or profile of cytokine expression (Supplementary Fig. 6c). However, only the combination of PMA/ionomycin, IL-1β and IL-23 enabled the evaluation of cytokine expression in both innate cells and T cells. Therefore, this combination was used for cytokine expression analysis in the main figures of the manuscript.
For OSMRβ staining, cells were treated with Fc Block (15 min and 4 °C) and stained in FACS buffer (PBS + 0.05% BSA, 0.01% NaN3 and 2 mM EDTA, pH 7.4) with either biotin-conjugated goat anti-OSMRβ (R&D, BAF662) for mouse or PE-conjugated anti-OSMRβ (Invitrogen, clone AN-V2) for human (30 min, 4 °C, dark). Amplification was performed using biotin–anti-PE (BioLegend, 408104) and Streptavidin–PE (Miltenyi, 130-106-789) for three to four rounds (10 min each, 4 °C). Specificity was assessed by omitting the primary antibody but including amplification. Cells were counterstained with anti-CD45 (AF700, BioLegend, clone 30-F11 mouse, HI30 human), anti-CD31 (PE-Cy7, clone 390, mouse only), anti-EpCAM (BV421, clone G8.8 mouse; APC, clone 9C4 human) and viability dyes (Calcein AM for mouse, eFluor 780 for human). OSMR expression was normalized by dividing the geometric mean fluorescence of full staining by that of the amplification-only control.
Data acquisition was performed using a BD FACSymphony or BD LSR Fortessa flow cytometer, and FlowJo software (version 10.6.01) was used for data analysis.
Cell sorting
Freshly isolated LPMCs, stromal cells and epithelial cells from colonic tissue were stained and hash tagged using anti-mouse hashtags (Biolegend; IDs 1, 2, 4, 6, 7 and 8) per the 10x Genomics protocol. Following surface staining, cells were washed with PBS + 1% BSA (pH 7.4), centrifuged (300g), resuspended in MEM + 5 mM EDTA, filtered and stained with DAPI or propidium iodide for viability. Viable cells were sorted into MEM + 50% FCS, washed and quantified for scRNA-seq. LPMCs were sorted on a FACS Aria I (BD Biosciences; 70-µm nozzle) and stromal/epithelial cells on an SH800S sorter (Sony; 100-µm nozzle). Gating strategy and purity controls are shown in Supplementary Fig. 7.
Statistical analysis
Sample sizes for each experiment are provided in the corresponding figure captions, along with the specific statistical tests used. Data collection was not performed blind to experimental conditions, and randomization was not applied. However, histopathological scoring was conducted in a blinded manner by two independent observers. No statistical methods were used to predetermine sample sizes, but the numbers used are consistent with those in previous studies7. Except in cases of major experimental error, all data were included in the analyses.
For comparisons between two groups, a two-tailed Mann–Whitney U-test was used, either due to non-normal data distribution or as a conservative nonparametric choice. For comparisons involving more than two groups, a one-way analysis of variance followed by a Tukey’s post hoc test (for paired samples) or a Kruskal–Wallis test followed by a Dunn’s post hoc test (for unpaired samples) was applied. Correlations between gene expression values were evaluated using Spearman’s rank correlation coefficient.
Statistical significance was defined as P ≤ 0.05; nonsignificant results are reported as NS. Error bars represent the standard error of the mean. Box-and-whisker plots show the full data range (whiskers), interquartile range (box) and median (central line). Intragroup variation was not assessed. All statistical analyses, except for RNA-seq (bulk and single cell) data, were performed using GraphPad Prism versions 8 and 10.
Additional methods
Detailed descriptions of the methods, including RNA extraction, cDNA synthesis, qPCR (including reverse transcription from low RNA quantities), ISH, histological staining for pSTAT1 and pSTAT3, single-cell library preparation and sequencing, bulk RNA-seq, data processing and analysis (including clustering, annotation and integration of public scRNA-seq datasets), RNA isolation from IBDome cohort samples and Xenium in situ analysis, are provided in the Supplementary Methods section.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data necessary to support the conclusions of this paper are included in the article and the Supplementary Methods. The sequencing data generated or used in this publication are deposited in NCBI’s GEO and can be accessed through the following GEO series accession numbers: GSE269507 (data related to Fig. 1e–i, Extended Data Figs. 1 and 2f and Supplementary Figs. 1, 2 and 4a), GSE269506 (data related to Fig. 2g–i and Supplementary Fig. 4d–g), GSE269505 (data related to Fig. 6d–f), GSE269578 (data related to Extended Data Fig. 9c,d), GSE206285 (data related to Extended Data Fig. 6f), GSE193677 (data related to Extended Data Fig. 6f,g) and GSE57945 (data related to Extended Data Fig. 6g). Additionally, some datasets were obtained from the Broad Institute’s Single Cell Portal and can be accessed through accession numbers SCP259 (data related to Supplementary Fig. 3a,b) and SCP1884 (data related to Supplementary Data Fig. 3a,b). Source data are provided with this paper.
Code availability
The code used for the analysis of the sequencing data in this manuscript is available at https://github.com/msaliutina/OSMR_RNA-Seq.
References
Friedrich, M., Pohin, M. & Powrie, F. Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity 50, 992–1006 (2019).
Francescone, R., Hou, V. & Grivennikov, S. I. Cytokines, IBD and colitis-associated cancer. Inflamm. Bowel Dis. 21, 409–418 (2015).
Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014).
West, N. R., Owens, B. M. J. & Hegazy, A. N. The oncostatin M–stromal cell axis in health and disease. Scand. J. Immunol. 176, e12694 (2018).
Richards, C. D. & Richards, C. D. The enigmatic cytokine oncostatin M and roles in disease. ISRN Inflamm. 2013, 512103 (2013).
Hermanns, H. M. Oncostatin M and interleukin-31: cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev. 26, 545–558 (2015).
West, N. R. et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 23, 579–589 (2017).
Richards, C. D. & Botelho, F. Oncostatin M in the regulation of connective tissue cells and macrophages in pulmonary disease. Biomedicines 7, 95 (2019).
Kullberg, M. C. et al. Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect. Immun. 69, 4232–4241 (2001).
Fox, J. G., Ge, Z., Whary, M. T., Erdman, S. E. & Horwitz, B. H. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 4, 22–30 (2011).
Kaser, A., Zeissig, S. & Blumberg, R. S. Inflammatory bowel disease. Annu. Rev. Immunol. 28, 573–621 (2010).
Kullberg, M. C. et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and γ interferon-dependent mechanism. Infect. Immun. 66, 5157–5166 (1998).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014).
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730 (2019).
Kong, L. et al. The landscape of immune dysregulation in Crohn’s disease revealed through single-cell transcriptomic profiling in the ileum and colon. Immunity 56, 444–458 (2023).
Keir, M. E., Yi, T., Lu, T. T. & Ghilardi, N. The role of IL-22 in intestinal health and disease. J. Exp. Med. 217, e20192195 (2020).
Hernandez, P., Gronke, K. & Diefenbach, A. A catch‐22: interleukin‐22 and cancer. Eur. J. Immunol. 48, 15–31 (2018).
Morrison, P. J. et al. Differential requirements for IL-17A and IL-22 in cecal versus colonic inflammation induced by Helicobacter hepaticus. Am. J. Pathol. 185, 3290–3303 (2015).
Huber, S. et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263 (2012).
Zenewicz, L. A. et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 190, 5306–5312 (2013).
Gronke, K. et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253 (2019).
Pavlidis, P. et al. Interleukin-22 regulates neutrophil recruitment in ulcerative colitis and is associated with resistance to ustekinumab therapy. Nat. Commun. 13, 5820 (2022).
Hernández, P. P. et al. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 16, 698–707 (2015).
Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature 517, 293–301 (2015).
Sonnenberg, G. F., Fouser, L. A. & Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011).
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375 (2010).
Geremia, A. et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 208, 1127–1133 (2011).
Beigel, F. et al. Oncostatin M mediates STAT3-dependent intestinal epithelial restitution via increased cell proliferation, decreased apoptosis and upregulation of SERPIN family members. PLoS ONE 9, e93498 (2014).
Bollrath, J. et al. gp130-mediated STAT3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 15, 91–102 (2009).
Tanaka, T. et al. A novel inflammation‐related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 94, 965–973 (2003).
Becker, C. et al. TGF-β suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity 21, 491–501 (2004).
Itakura, H. et al. Tumor-suppressive role of the musculoaponeurotic fibrosarcoma gene in colorectal cancer. iScience 26, 106478 (2023).
Grivennikov, S. I., Greten, F. R. & Karin, M. Immunity, inflammation, and cancer. Cell 140, 883–899 (2010).
Verstockt, S., Verstockt, B. & Vermeire, S. Oncostatin M as a new diagnostic, prognostic and therapeutic target in inflammatory bowel disease (IBD). Expert Opin. Ther. Targets 23, 943–954 (2019).
Kim, W. M., Kaser, A. & Blumberg, R. S. A role for oncostatin M in inflammatory bowel disease. Nat. Med. 23, 535–536 (2017).
Patel, P., Rai, V. & Agrawal, D. K. Role of oncostatin-M in ECM remodeling and plaque vulnerability. Mol. Cell. Biochem. 478, 2451–2460 (2023).
Li, Y., de Haar, C., Peppelenbosch, M. P. & van der Woude, C. J. New insights into the role of STAT3 in IBD. Inflamm. Bowel Dis. 18, 1177–1183 (2012).
Verstockt, B. et al. IL-12 and IL-23 pathway inhibition in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 20, 433–446 (2023).
Kirchberger, S. et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J. Exp. Med. 210, 917–931 (2013).
Bollrath, J. & Greten, F. R. IKK/NF‐κB and STAT3 pathways: central signalling hubs in inflammation‐mediated tumour promotion and metastasis. EMBO Rep. 10, 1314–1319 (2009).
Spits, H. & Santo, J. P. D. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2010).
Liang, S. C. et al. Interleukin (IL)-22 and IL-17 are coexpressed by TH17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203, 2271–2279 (2006).
Zindl, C. L. et al. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc. Natl Acad. Sci. USA 110, 12768–12773 (2013).
Awasthi, A. et al. Cutting edge: IL-23 receptor GFP reporter mice reveal distinct populations of IL-17-producing cells. J. Immunol. 182, 5904–5908 (2009).
Junk, D. J. et al. Oncostatin M promotes cancer cell plasticity through cooperative STAT3–SMAD3 signaling. Oncogene 36, 4001–4013 (2017).
Soler, M. F., Abaurrea, A., Azcoaga, P., Araujo, A. M. & Caffarel, M. M. New perspectives in cancer immunotherapy: targeting IL-6 cytokine family. J. Immunother. Cancer 11, e007530 (2023).
West, N. R. Coordination of immune–stroma crosstalk by IL-6 family cytokines. Front. Immunol. 10, 1093 (2019).
Chen, M. et al. Exploring the oncostatin M (OSM) feed-forward signaling of glioblastoma via STAT3 in pan-cancer analysis. Cancer Cell Int. 21, 565 (2021).
Marjou, F. E. et al. Tissue‐specific and inducible Cre‐mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Zheng, B., Zhang, Z., Black, C. M., Crombrugghe, Bde & Denton, C. P. Ligand-dependent genetic recombination in fibroblasts a potentially powerful technique for investigating gene function in fibrosis. Am. J. Pathol. 160, 1609–1617 (2002).
Alva, J. A. et al. VE‐cadherin‐Cre‐recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev. Dyn. 235, 759–767 (2006).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Moh, A. et al. Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab. Investig. 87, 1018–1028 (2007).
Hao, Z. & Rajewsky, K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 194, 1151–1164 (2001).
Choi, G. B. et al. The maternal interleukin-17A pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016).
Zheng, Y. et al. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007).
Durbin, J. E., Hackenmiller, R., Simon, M. C. & Levy, D. E. Targeted disruption of the mouse STAT1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450 (1996).
Sohal, D. S. et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 89, 20–25 (2001).
Neurath, M. F. et al. Assessment of tumor development and wound healing using endoscopic techniques in mice. Gastroenterology 139, 1837–1843 (2010).
Headland, S. E. et al. Oncostatin M expression induced by bacterial triggers drives airway inflammatory and mucus secretion in severe asthma. Sci. Transl. Med. 14, eabf8188 (2022).
Izcue, A. et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity 28, 559–570 (2008).
Naini, B. V. & Cortina, G. A histopathologic scoring system as a tool for standardized reporting of chronic (ileo)colitis and independent risk assessment for inflammatory bowel disease. Hum. Pathol. 43, 2187–2196 (2012).
Riley, S. A., Mani, V., Goodman, M. J., Dutt, S. & Herd, M. E. Microscopic activity in ulcerative colitis: what does it mean? Gut 32, 174 (1991).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Acknowledgements
We thank N. Sommer for support with sample collection within the IBDome consortium; J. Kirsch and T. Kaiser (Flow Cytometry Core Facility, DRFZ, Berlin) for technical assistance with cell sorting and C. Heldt and I. Plumbom (Berlin Institute of Health at Charité–Universitätsmedizin Berlin) for support with Xenium sample acquisition and analysis. We would like to express our gratitude to all members of the laboratory of A.N.H. for their assistance as well as to our clinician and nurse colleagues in both the IBD inpatient and outpatient clinics. We also thank our Genentech colleagues in the Genetically Engineered Mouse Models, Microinjection and Embryo Technology labs for allele design and creation and our colleagues in the Genetic Analysis Lab and Animal Resources for technical assistance. This work was supported by grants from the German Research Foundation (DFG-375876048-TRR 241-A05, INST 335/597-1), the ERC Starting Grant ‘iMOTIONS’ (101078069), the BIH Clinician Scientist Program, a Lichtenberg Professorship and the ‘Corona Crisis and Beyond’ grant from the Volkswagen Foundation to A.N.H.; the German Research Foundation (DFG-375876048-TRR 241-B01, Z02; CRU 5023 (50474582); CRC 1449-B04, Z02 (431232613); CRC 1340-B06 (372486779); INST 335/597-1) to B.S.; the German Research Foundation (LO 1542/5-1, LO 1542/4-1), the Willy Robert Pitzer Foundation (21-033) and the Dr. Rolf M. Schwiete Foundation (2021-035) to M.L.; the German Federal Ministry of Education and Research (BMBF, TReAT and CONAN projects), the state of Berlin and EFRE (PersMedLab), the Leibniz Association (TargArt, ImpACt) and the DZKJ (01GL2401A) to M.-F.M.; the BIH Clinical Fellow Program and the German Research Foundation (TRR 241-C03 (375876048), GRK 2318-B02 (318905415)) to M. Schumann; the German Research Foundation (TRR 241-A03 (375876048)) to S. Writz; the German Research Foundation (TRR 241-A01 (375876048), SFB1444/P11 (427826188), SPP1937-DI764/9-2), the ERC Advanced Grant ‘ILCAdapt’ (101055309), the EU Horizon Europe project GlycanTrigger (101093997) and the Einstein Foundation Berlin (Einstein Professorship) to A.D. and the ERC Advanced Grant ‘MEM-CLONK’ (101055157), the German Research Foundation (TRR 241-C03 B02 (375876048), RO 3565/7-1) and the Leibniz ScienceCampus Chronic Inflammation to C.R.
Author information
Authors and Affiliations
Consortia
Contributions
Study design and conceptualization: A.N.H. and N.R.W. Writing, original draft preparation: A.N.H. and R.C. Commenting and editing: N.R.W., Y.L., C.A.C., A.A.K., A.D., C.R., M.L. and B.S. Visualization: A.N.H., R.C. and N.R.W. Experimental data generation: R.C., Y.L., D.B., S.H., L.W., C.A.C., L.V.B., A.K., A.N.H., N.R.W., J.E.K., B.B., L.G., J.H., and J.Z. ISH and immunofluorescence/immunohistochemistry: A.A.K., A.A., D.B. and A.S. scRNA-seq library preparation and sequencing: G.M.G. and M.-F.M. scRNA-seq bioinformatic analysis: M. Saliutina, P.D. and F.H. Bulk RNA-seq analysis: M. Saliutina, J.R. and G.K. FACS staining, acquisition and analysis: R.C., D.B., Y.L., C.A.C., L.V.B. and A.N.H. Xenium acquisition and analysis: S.M. and T.C. Cell sorting: A.N.H., D.B. and R.C. Organoid cultures and stimulations: R.C., S.H. and L.G. Fluidigm qPCR: E.C. and A.N.H. Procurement of consumables, mice and experimental advice: C.R., A.D., M.L., N.M., B.S., S.W., M.-F.M., M.K., N.D. and M. Schumann. All authors revised the manuscript and approved the final version submitted for publication. M. Saliutina, S.M. and C.A.C. contributed equally as second authors. R.C. and Y.L. are shared first authors.
Corresponding author
Ethics declarations
Competing interests
A.S., B.B., J.E.K., J.H., J.Z., M.K., J.R., L.W., M.E.K. and N.R.W. are current or former members of Genentech. N.R.W. and A.N.H. are inventors of patents relating to OSM as a therapeutic target for IBD (WO 2016/120625 A1). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Jochem Bernink, Samuel Huber and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in collaboration with the Nature Immunology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Single-cell RNA sequencing of lamina propria CD45+ cells, epithelial cells, and stromal cells.
(a) UMAP visualization of all sequenced single-cell transcriptomes derived from stromal cells, epithelial cells, and CD45+ cells, labeled by cell hashtags. (b) UMAP visualization of all sequenced single-cell transcriptomes derived from stromal cells, epithelial cells, and CD45+ cells, labeled by experimental condition. (c) Cell proportion changes (y-axis) for non-inflamed (black) and inflamed (red) samples (3 mice/pool, 3 pools/group, Mean ± SEM; Wilcoxon rank-sum test). (d, e) Relative Osm gene expression in stromal (d) (calcein+, CD45-, EpCAM-) and epithelial (e) (EpCAM+, CD45-) cells (3 mice/pool, 3 pools/group,Mean ± SEM; Wilcoxon rank-sum test). (f) Absolute counts (upper panel) and proportions (lower panel) of immune cell populations among Osm-expressing cells in steady-state mice and after H. hepaticus + αIL-10R colitis induction. (g) Osmr relative expression and percentage of Osmr-expressing stromal cells (3 mice/pool, 3 pools/group, Mean ± SEM; Wilcoxon rank-sum test). (h) Osmr relative gene expression in CD45+ cells (3 mice/pool, 3 pools/group, Mean ± SEM; Wilcoxon rank-sum test).
Extended Data Fig. 2 Regulation of Osmr expression in mouse colon during steady-state and colitis.
(a) To confirm that epithelial cells are the primary source of OSMR expression in epithelial washes, we enriched epithelial cells using MACS from epithelial washes of inflamed colitic mice. FACS analysis of the epithelial washes is shown before and after the MACS enrichment. Subsequently, Osmr was quantified by qPCR (right bar graph). Data representative of one experiment (n = 6). (b) qPCR analysis of the indicated genes within epithelial washes was conducted to confirm the absence of cellular contamination. Data representative of one experiment (n = 6). (c) Quantification of Osmr expression in epithelial cells by qPCR (Mean ± SEM; aIL-10R, n = 3, H. hepaticus, n = 4; H.h. + αIL-10R, n = 17)). Epithelial cells were isolated from the indicated groups. (d) OSMR expression was assessed by flow cytometry in epithelial cells isolated from the indicated groups (left, histogram). Normalized OSMR expression is shown at the indicated time points after colitis induction (right, bar graph), calculated relative to the staining control for each individual mouse (SS, n = 15: day 7, n = 6, day 14, n = 4). (c, d) P-values were calculated using the Kruskal-Wallis test followed by Dunn’s multiple comparisons test. (e) Representative flow cytometry gating strategy for the identification of colon epithelial (EpCAM⁺) and endothelial (CD31⁺) cells in steady-state conditions. Histograms display OSMR expression levels in EpCAM⁺ epithelial cells (bottom) and CD31⁺ endothelial cells (top) from Cdh5Cre+ × Osmrfl/fl and Cdh5Cre- × Osmrfl/fl mice. The geometric mean fluorescence intensity (Geo mean) of OSMR-PE is indicated for each group, including a control stained only with the secondary antibody. (f) Cell-cell interaction analysis (CellPhoneDB) between Osm+ immune cells and Osmr+ epithelial cells. Analysis derived from scRNAseq analysis (Fig. 1d).
Extended Data Fig. 3 Temporal dynamics of Osmr expression in colonic epithelium and lamina propria during colitis progression.
(a-g) ISH was performed to detect Osmr expression in colon tissue from steady-state and H.h. + αIL-10R-induced colitic mice, with hematoxylin counterstaining. Image processing was conducted using Ilastik, and Osmr expression was quantified with ImageJ. At various time points after colitis induction, the following numbers of mice and images were made and analyzed: Day 0 (steady state), 6 mice with 130 images; Day 3, 2 mice with 50 images; Day 7, 8 mice with 100 images; Day 14, 3 mice with 80 images; and Day 21, 4 mice with 100 images. (a) Representative images of colon sections at different time points (day 0, 3, 7, 14, and 21) after colitis induction. Top row: original image with Osmr ISH and hematoxylin staining. Middle row: Segmentation analysis using Ilastik showing Osmr localization in epithelial cells (blue), lamina propria (orange), and submucosa/muscularis (black). Bottom row: Osmr mRNA expression segregated according to location. Scale bar: 20 μm. (b) Representative images showing peptide/prolyl isomerase B (Ppib) expression as a positive control for ISH and a duplex negative control probe. Scale bar: 20 μm. (c-d) Quantification of Osmr+ counts (c) and Osmr⁺ stain area in epithelial (blue) and lamina propria (red) compartments (d) over time. Statistical analysis was performed using the Kruskal-Wallis test with multiple comparisons relative to Day 0 (steady state). (e) To better visualize relative fold changes in Osmr expression in epithelial vs lamina propria compartments, Osmr counts from (c) were normalized to total tissue area in epithelial cells and lamina propria. Statistical analysis was performed using two-way ANOVA with multiple comparisons. The overall significance of time and expression is depicted in bold. (f, g) Correlation analyses between Osmr expression in different tissue compartments and histopathology scores. Scatter plots illustrate the relationship between Osmr⁺ stain area or Osmr⁺ counts and the severity of colitis, as assessed by histopathological scoring. Pearson correlation coefficients (r) and p-values are provided for each analysis.
Extended Data Fig. 4 Increased OSMR expression in ulcerative colitis correlates with histopathological severity.
(a) Representative images of OSMR mRNA expression detected by ISH in colon tissue from non-IBD controls (n = 3) and UC (n = 3) patients. Epithelial cell regions are identified by dashed lines. Scale bar: 50 μm. (b) Exemplary positive and negative control probes for human RNAScope in ISH. Scale bar: 20 μm. (c) Correlation between OSMR protein expression in colon epithelial cells and histopathology score in patients with IBD (UC, n = 4; CD, n = 5) and healthy controls (n = 2), with each patient having 2–4 sampled locations. Scatter plot showing the relationship between normalized epithelial OSMR protein expression assessed by flow cytometry and histopathology score in IBD patients. Pearson correlation and significance depicted.
Extended Data Fig. 5 Epithelial Osmr expression is induced during C. rodentium infection and regulated by IL-22 and IL-23.
(a) Experimental timeline of C. rodentium infection and colitis over 21 days. (b) Colon histopathology scores in wild-type mice at various time points post-infection (n = 12/time point, 2 experiments; Kruskal-Wallis with Dunn’s test). (c, d) Time-resolved expression of Osm and Osmr mRNA in colonic tissue (n = 12/time point; one-sample Wilcoxon test). (e) OSMR protein levels in epithelial cells on day 7 post-infection vs. uninfected controls (n = 7/group; Mann-Whitney U test). (f) Schematic of tamoxifen-induced IECΔOsmr model for epithelial-specific Osmr deletion. (g) Histopathology scores comparing IECΔOsmr and control mice after infection (n = 12/group; Mann-Whitney U test). (h) Bacterial burden (CFU) in colon, spleen, and liver of IECΔOsmr vs. control mice (n = 12/group; Mann-Whitney U test). (i) Il22 mRNA expression at different time points post-infection (n = 12/time point; one-sample Wilcoxon test). (j) Osmr and Reg3g expression in epithelial cells following IL-22 blockade (n = 6–8/group; Mann-Whitney U test). (k) Frequency of IL-22⁺ ILC3 cells in uninfected (n = 6) and infected mice (n = 8/group; Mann-Whitney U test). (l) Osmr and Il22 expression after IL-23 blockade during infection (uninfected, n = 6; C. rodentium, n = 7; Mann-Whitney U test). (m) Immune profiling in IECΔOsmr vs. wild-type mice, highlighting altered frequencies of CD45⁺ cells, neutrophils, monocytes, eosinophils, and CD4⁺CD44⁺ T cells (n = 12/group; Kruskal-Wallis with Dunn’s test).
Extended Data Fig. 6 Analysis of Osmr expression and response to IL-22 in mouse and human epithelial organoids.
(a) High-throughput q-PCR analysis performed on the Fluidigm Biomark platform using the depicted primers. Data are from pooled from three independent biological replicates. Expression levels were normalized to the median expression and log2 transformed. (b) Colon epithelial organoids derived from C57BL/6 J wild-type mice treated with 10 ng/ml IL-22 over a time course of 3-48 h. Gene expression of Osmr, Reg3g, and Socs3 at each time point was analyzed by q-PCR and normalized to the unstimulated control. Two independent replicates are shown. (c) Human colon epithelial organoids established from specimens of healthy individuals and patients with IBD stimulated with indicated stimuli for 24 h. Statistical analysis was one-way ANOVA with Dunn’s multiple comparisons test; n = 4. (d) ISH detection of Il22 mRNA in colonic mucosal samples from inflamed Il22-/- mice (punctate brown signal) confirming the specificity of the RNAscope Il22 probe. Scale bar: 20 μm. (e) ISH detection of Osmr (left, red punctate) and Il22 (right, brown punctate) mRNA expression in colonic tissue of sequential slides following 14 days of colitis induction in wild-type mice. Scale bar: 20 μm. (f-g) Correlation between human OSMR expression and IL22 enrichment scores in three independent datasets. (f) Correlation analyses in UC colon samples from three independent cohorts: UNIFI trial (GSE206285; n = 532), Mount Sinai cohort (GSE193677; n = 293), and IBDome cohort (n = 64).(g) Correlation analyses in CD ileum samples from three independent cohorts: RISC cohort (GSE57945; n = 191), Mount Sinai cohort (GSE193677; n = 162), and IBDome cohort (n = 81). Spearman’s correlation coefficient (r) and corresponding P-values are indicated in each panel.
Extended Data Fig. 7 STAT signaling in mouse colon organoids and epithelial cell line following cytokine stimulation.
(a) Immunofluorescence staining for pSTAT1 and pSTAT3 in steady-state (n = 7) and inflamed (n = 10) mouse samples with subsequent quantification in colon lamina propria. Data pooled from two experiments. P-values are from Mann-Whitney U tests. (b) Western blot analysis of pSTAT3, total STAT3, and β-actin in mouse colon organoids treated with the indicated cytokines. Data are representative of two independent experiments. (c) Western blot analysis of pSTAT1, total STAT1, and β-actin in mouse colon organoids treated with indicated cytokines. Data are representative of two independent experiments. (d) Western blot analysis of pSTAT1, total STAT1, and β-actin in the human Caco2 cell line (left panel) was performed to confirm functionality of pSTAT1 antibody. Epithelial cells from C57BL/6 J mice with induced inflammation at days 7 and 14 (n = 5) were also analyzed (right panel). pSTAT1, total STAT1, and β-actin in steady state epithelial cells from C57BL/6 J mice are shown in Fig. 4a. (e) Colon epithelial organoids isolated from C57BL/6 mice were treated with JAK/STAT pathway inhibitors (50 μM fludarabine, 25 μM STA-21, 2.5 μM ruxolitinib, or 2.5 μM tofacitinib) for 6 h, and 10 ng/mL of IL-22 was added for another 6 h. 0.1% DMSO was used as inhibitor control. Relative expression of Osmr was analyzed by qPCR. Data are shown for n = 2 biological replicates with n = 3 technical replicates. Graph displays mean ± SEM. P-values are derived from Kruskal-Wallis test.
Extended Data Fig. 8 Flow cytometry analysis of IL-22 production by ILC subsets in naive and colitic mice.
(a) Gating strategy used to identify CD45⁺ cells for analysis in Fig. 5c. (b) Ki67 expression in total ILCs (Lineage⁻, TCRα⁻, TCRγ−, CD90⁺) from colitic mice. n = 12 per condition from two independent experiments. Mann-Whitney U test. (c) IL-22 frequency in ILCs and T cells from colitic wild-type mice (H.h. + αIL-10R, day 7), measured by flow cytometry following PMA/ionomycin, IL-23, and IL-1β stimulation. n = 12; two experiments. (d) Cytokine profiles of ILC subsets: ILC3s (RORγt⁺), ILC1s (T-bet⁺, Eomes⁻), ILC2s (GATA3⁺), and NK cells (T-bet⁺, Eomes⁺); all Lineage⁻, TCRα⁻, TCRγ⁻. Assessed post-stimulation as in (c). n = 12; two experiments. (e, f) H.h. + αIL-10R colitis with anti-IL-12p40 or isotype control treatment (day 0, analysis on day 7). (e) Il22 expression in colon tissue by qPCR. (f) Histopathology scores. Data pooled from two experiments, n = 8 mice/group. (g, h) H.h. + αIL-10R colitis with anti-IL-23p19 or isotype control treatment (day 0, analysis on day 7). (g) Il22 expression in colon tissue by qPCR. (h) Histopathology scores. Data from one experiment, n = 6 mice/group. (i, j) Colitis induced in Rag2⁻/⁻ and Rag2⁺/⁺ mice for two weeks. (i) Colon histopathology scores. (j) Osmr expression in IECs normalized to steady-state controls. n = 6–8 mice/group from two independent experiments. All graphs show mean ± SEM; P-values determined by Mann-Whitney U test.
Extended Data Fig. 9 OSM impact on intestinal epithelial cells.
(a) Human HCA-7 colon epithelial cells were stimulated with 100 ng/ml of rhIL-22 or rhOSM for 30 min, 24 or 48 h. Subsequently, pSTAT3 was quantified in cell lysates using an electrochemiluminescence assay (MSD). Data are representative of two independent experiments, each with three technical replicates. P-values derived from a 2-way comparison using Tukey’s multiple comparison test. (b) Analysis of STAT3 phosphorylation after colitis induction in IECs from IECΔOsmr and control mice (values normalized to those from steady-state mice). P-values are derived from Mann–Whitney U test. Data are representative of two independent experiments; n = 6 mice per group. (c) Volcano plot depicting differentially expressed genes in the epithelial cell line, HCA-7, after treatment with rhOSM for 24 h as described in (a). (d) GSEA plot depicting enrichment of OSM-activated genes in epithelial cells (defined in analysis shown in panel c) comparing inflamed IECΔOsmr and control mice, as in Fig. 6d.
Extended Data Fig. 10 OSM and OSMR expression in colitis-associated cancer.
(a) Percentage of Ki67⁺ cells within Epcam⁺ epithelial cells from IECΔOsmr and control mice at day 21 post-colitis. Data represent two experiments (n = 12); Mann–Whitney U test. (b) Osm and Osmr expression in normal colon (n = 10) vs. CAC (n = 14) tissue from the GSE210970 dataset (Mann–Whitney U test). (c, d) Spearman correlation of Osm (c) and Osmr (d) with Socs3 or Stat3 in GSE210970 dataset. (e) Representative healthy colon (n = 3) biopsy analyzed by spatial transcriptomics shows OSM and OSMR expression in epithelial and non-epithelial cells. Cell types identified via canonical epithelial (EPCAM, CDH1, KRT20, MUC5B, MKI67) and non-epithelial markers (CD3E, CD4, CD14, CD68, C1QC, CD19, CD79A, PDGFRA, VIM, PECAM1, CD34, KIT). (f) Osmr expression in colon epithelial washes from wild-type mice following either DSS-induced colitis (1 cycle, n = 10) or CAC induction via AOM + DSS (3 cycles, n = 18). Mann–Whitney U test with direct comparison with steady state (n = 5). (g, h) DSS colitis mice treated with anti-IL-22 (clone 8e11, n = 10) or isotype control (n = 10). Osmr and Socs3 expression in epithelial cells assessed by qPCR; one-way Anova with Tukey’s multiple comparisons test. (i) Spearman correlation of Osmr with Il12b in GSE210970 dataset. (j–m) CAC induced via AOM + DSS. Tumor development confirmed by endoscopy, followed by treatment with anti-IL-22 (8e11, n = 4), anti-IL-12p40 (C17.8, n = 6), or isotype controls (n = 6) for one week. (k, l) Osmr and Socs3 expression in tumor epithelial cells; (m) Il22 expression in total tumor tissue. One-way ANOVA with Holm-Sidak’s test.
Supplementary information
Supplementary Information
Supplementary Methods and Figs. 1–7.
Supplementary Tables
Supplementary Tables 1–8.
Source data
Source Data Fig. 4
Western blots for Fig. 4a,d.
Source Data Extended Data Fig. 7
Western blots for Extended Data Fig. 7b–d.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cineus, R., Luo, Y., Saliutina, M. et al. The IL-22–oncostatin M axis promotes intestinal inflammation and tumorigenesis. Nat Immunol 26, 837–853 (2025). https://doi.org/10.1038/s41590-025-02149-z
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41590-025-02149-z