Abstract
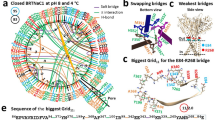
Broad-range thermal receptor 1 (BRTNaC1), activated by heat at low extracellular pH, was recently identified in myriapods. Although the overexpression of BRTNaC1 leads to robust heat-activated current with a cation selectivity profile, the structure of this receptor and how it is gated by proton and heat remain to be investigated. Here we determine cryogenic electron microscopy structures of BRTNaC1 in the apo, proton-induced and heated states. Based on these structures, patch-clamp recordings and molecular dynamic simulations, we found that a ‘twist the wrist’ mechanism is used for proton activation of BRTNaC1, while heat induces broad conformational changes in BRTNaC1, including rotation and shift in the transmembrane helices to open this channel. Moreover, as testosterone inhibited BRTNaC1 activation, we identified four clustered residues important for such inhibition. Therefore, our study has established the structural basis for ligand and temperature gating in the BRTNaC1 ion channel.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The structure coordinates and cryo-EM density maps have been deposited in the PDB under accession numbers 8YMR and EMD-39405 for BRTNaC1pH8, 8YMS and EMD-39406 for BRTNaC1pH4, 8YMU and EMD-39408 for BRTNaC1MβCD_pH4, 8YMT and EMD-39407 for BRTNaC1MβCD_pH4_TEST_bound, 8YMW and EMD-39409 for BRTNaC14°C, 8YMX and EMD-39410 for BRTNaC140°C. The materials and data can be obtained from the corresponding authors upon request. Source data are provided with this paper.
References
Julius, D. & Nathans, J. Signaling by sensory receptors. Cold Spring Harb. Perspect. Biol. 4, a005991 (2012).
Dhaka, A., Viswanath, V. & Patapoutian, A. Trp ion channels and temperature sensation. Annu. Rev. Neurosci 29, 135–161 (2006).
Qu, Z. et al. Millipede genomes reveal unique adaptations during myriapod evolution. PLoS Biol. 18, e3000636 (2020).
Yao, Z. et al. A thermal receptor for nonvisual sunlight detection in myriapods. Proc. Natl Acad. Sci. USA 120, e2218948120 (2023).
Boscardin, E., Alijevic, O., Hummler, E., Frateschi, S. & Kellenberger, S. The function and regulation of acid-sensing ion channels (ASICs) and the epithelial Na+ channel (ENaC): IUPHAR Review 19. Br. J. Pharmacol. 173, 2671–2701 (2016).
Jasti, J., Furukawa, H., Gonzales, E. B. & Gouaux, E. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 (2007).
Yoder, N., Yoshioka, C. & Gouaux, E. Gating mechanisms of acid-sensing ion channels. Nature 555, 397–401 (2018).
Sun, D. et al. Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1. eLife 9, e57096 (2020).
Noreng, S., Bharadwaj, A., Posert, R., Yoshioka, C. & Baconguis, I. Structure of the human epithelial sodium channel by cryo-electron microscopy. eLife 7, e39340 (2018).
Sun, D. et al. Cryo-EM structure of the ASIC1a–mambalgin-1 complex reveals that the peptide toxin mambalgin-1 inhibits acid-sensing ion channels through an unusual allosteric effect. Cell Discov. 4, 27 (2018).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph 14, 354–360 (1996).
Yang, J. et al. PAC, an evolutionarily conserved membrane protein, is a proton-activated chloride channel. Science 364, 395–399 (2019).
Sherwood, T. W., Frey, E. N. & Askwith, C. C. Structure and activity of the acid-sensing ion channels. Am. J. Physiol. Cell Physiol. 303, C699–C710 (2012).
Li, T., Yang, Y. & Canessa, C. M. Outlines of the pore in open and closed conformations describe the gating mechanism of ASIC1. Nat. Commun. 2, 399 (2011).
Yao, Z. et al. A thermal receptor for nonvisual sunlight detection in myriapods. Proc. Natl Acad. Sci. USA, 120, e2218948120 (2023).
Baconguis, I., Bohlen, C. J., Goehring, A., Julius, D. & Gouaux, E. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na+-selective channel. Cell 156, 717–729 (2014).
Cristofori-Armstrong, B. & Rash, L. D. Acid-sensing ion channel (ASIC) structure and function: insights from spider, snake and sea anemone venoms. Neuropharmacology 127, 173–184 (2017).
Olsson, M. H. M., Søndergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Korkosh, V. S. & Tikhonov, D. B. Analysis of residue-residue interactions in the structures of ASIC1a suggests possible gating mechanisms. Eur. Biophys. J. 52, 111–119 (2023).
Zhou, H. X. & Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 118, 1691–1741 (2018).
Su, N. et al. Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands. Nat. Chem. Biol. 19, 72–80 (2023).
Yoder, N. & Gouaux, E. The His–Gly motif of acid-sensing ion channels resides in a reentrant ‘loop’ implicated in gating and ion selectivity. eLife 9, e56527 (2020).
Mahammad, S. & Parmryd, I. in Methods in Membrane Lipids (ed. Owen, D. M.) 91–102 (Springer, 2015).
Gonzales, E. B., Kawate, T. & Gouaux, E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460, 599–604 (2009).
Rook, M. L., Williamson, A., Lueck, J. D., Musgaard, M. & Maclean, D. M. β11-12 linker isomerization governs acid-sensing ion channel desensitization and recovery. eLife 9, e51111 (2020).
Askwith, C. C., Benson, C. J., Welsh, M. J. & Snyder, P. M. DEG/ENaC ion channels involved in sensory transduction are modulated by cold temperature. Proc. Natl Acad. Sci. USA 98, 6459–6463 (2001).
Cao, E., Liao, M., Cheng, Y. & Julius, D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013).
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013).
Tominaga, M. et al. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543 (1998).
Zheng, J. Molecular mechanism of TRP channels. Compr. Physiol. 3, 221–242 (2013).
Yang, F., Cui, Y., Wang, K. & Zheng, J. Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proc. Natl Acad. Sci. USA 107, 7083–7088 (2010).
Kwon, D. H. et al. Heat-dependent opening of TRPV1 in the presence of capsaicin. Nat. Struct. Mol. Biol. 28, 554–563 (2021).
Nadezhdin, K. D. et al. Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel. Nat. Struct. Mol. Biol. 28, 564–572 (2021).
Kim, J., Won, J., Chung, D. K. & Lee, H. H. FRET analysis of the temperature-induced structural changes in human TRPV3. Sci. Rep. 13, 10108 (2023).
Clapham, D. E. & Miller, C. A thermodynamic framework for understanding temperature sensing by transient receptor potential (TRP) channels. Proc. Natl Acad. Sci. USA 108, 19492–19497 (2011).
Liu, R. et al. Choline kinase alpha 2 acts as a protein kinase to promote lipolysis of lipid droplets. Mol. Cell 81, 2722–2735.e9 (2021).
Ma, Q. et al. Coupling HDAC4 with transcriptional factor MEF2D abrogates SPRY4-mediated suppression of ERK activation and elicits hepatocellular carcinoma drug resistance. Cancer Lett. 520, 243–254 (2021).
Caterina, M. J. et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824 (1997).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Li, Z. et al. Uni-Fold: an open source platform for developing protein folding models beyond AlphaFold. Preprint at bioRxiv https://doi.org/10.1101/2022.08.04.502811 (2022).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
PyMOL, v. 1.8. (Schrödinger, 2015).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Laguerre, M., Saux, M., Dubost, J. P. & Carpy, A. MLPP: a program for the calculation of molecular lipophilicity potential in proteins. Pharm. Pharmacol. Commun. 3, 217–222 (1997).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Páll, S. et al. Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J. Chem. Phys. 153, 134110 (2020).
Huang, J. & MacKerell, A. D. Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Acknowledgements
Single-particle cryo-EM data were collected at the Center of Cryo-Electron Microscopy at Zhejiang University. We thank X. Zhang and C. Ma for their support in facility access and data acquisition. We thank S. Chang and L. Wu in the Center of Cryo-Electron Microscopy, Zhejiang University for their technical assistance on cryo-EM. We thank J. Guo for the constructive suggestions. This work was supported by the National Key R&D Program of China (2023YFF1304903 to S.Y.), the National Natural Science Foundation of China (32122040, 31971040 and 32421001 to F.Y.; 32170486 and 32430012 to S.Y.; 81571916, 81372079 and 81201478 to Y.X.), National Forestry and Grassland Administration (2020132610 to S.Y.), Heilongjiang Province (JQ2021C001 to S.Y.), China Postdoctoral Science Foundation (2021M692818 to N.S.; 2023M743097 to X.C.), the Ministry of Science and Technology of China (2022YFF1201901 to H.W.), STI2030-Major Project (2022ZD0212700 to H.W.) and Zhejiang Provincial Natural Science Foundation (LR20C050002 to F.Y.). F.Y. was supported by MOE Frontier Science Center for Brain Science and Brain-Machine Integration, Zhejiang University. This work was supported by the Fundamental Research Funds for the Central Universities. This work was also supported by the Core Facilities in Zhejiang University School of Medicine, including the Zeiss LSM800 confocal fluorescence imaging microscope.
Author information
Authors and Affiliations
Contributions
F.Y., N.S. and S.Y. conceived and supervised the project. X.C. and L.Y. performed sample preparation, data acquisition and structure determination. X.C., L.Y., Q.M. and Z.Y. did functional studies. H.W. and Z.D. performed the MD simulation. F.Y., N.S., S.Y., Y.W. and Y.X. performed data analysis. All authors participated in paper preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Sebastian Brauchi, Thomas Voets and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation of PAC-KO cells.
a, Western blot of the expression of PAC in HEK293T cells and PAC-KO HEK293T cells. b, Representative whole-cell recording of PAC current induced by extracellular acidification in HEK293T cells. c, PAC current induced by extracellular acidification was abolished in PAC-KO cells.
Extended Data Fig. 2 Representative whole-cell current recordings of BRTNaC1 mutants.
High concentration of proton (pH3.5) were applied to activate the mutants in the patch-clamp recordings. Whole-cell mode recordings were conducted at ±80 mV for 450 ms, respectively. A dashed line in black indicated zero current level.
Extended Data Fig. 3 The confocal imaging of BRTNaC1 mutant and cell membrane.
The expression pattern of BRTNaC1 and its mutants with cell membranes. Representative bright-field (first row), fluorescent of green fluorescent protein (GFP)-tagged WT BRTNaC1 and mutants (second row), fluorescent of cell membrane labeled by DiD dye (third row) and merged images of GFP tag and DiD signals (forth row) were showed. The fluorescence intensity profile from a line scan (indicated by a dashed line) is showed on the right side, which indicated WT and mutant BRTNaC1 exhibited similar expression patterns.
Extended Data Fig. 4 G392R mutant is anion permeable.
a, Voltage ramps (800 ms) ranging from −80 to +80 mV were used to record current in different extracellular solutions of G392R mutant transfected cells. Extracellular solution (140 mM NaCl, 5 mM MES, 5 mM EDTA, pH 3.5) was substituted with sucrose-supplemented extracellular solution (14 mM NaCl, 5 mM MES, 5 mM EDTA, pH 3.5, osmolarity constant is adjust by sucrose), respectively in a. b, The reversal potentials tested in hPAC-KO cells expressing G392R (n = 9 biological replicates). Data points and bars represent mean ± sem.
Extended Data Fig. 5 Inhibition of BRTNaC1 by amiloride.
a, Representative whole-cell current recording showing that amiloride inhibited BRTNaC1 channel in a concentration-dependent manner. b, Concentration dependent curve of amiloride inhibition of BRTNaC1 in whole-cell patch recordings (IC50: 168.4 ± 7.96 µM, n = 3 biological replicates). Data points and bars represent mean ± sem.
Extended Data Fig. 6 Inhibition of BRTNaC1 by cholesterol.
a, Representative whole-cell current recording of BRTNaC1, where cholesterol up to 300 µM exhibited little inhibitory effect. b, Concentration dependent curve of cholesterol inhibition of BRTNaC1 in whole-cell patch recordings (n = 3 biological replicates). Data points and bars represent mean ± sem.
Extended Data Fig. 7 Cation selectivity pattern of I400A and C401A.
a and c, Voltage ramps (500 ms) ranging from −60 to +60 mV were used to record current in different extracellular solutions of I400A or C401A transfected cells, respectively. Bath solution (140 mM NaCl, 5 mM MES, pH 3.5) was substituted with different cationic compositions (140 mM KCl, 140 mM CsCl, 140 mM LiCl, 110 mM CaCl2, 110 mM MgCl2, or 110 mM BaCl2). b, Cation selectivity in I400A transfected cells (n = 6 biological replicates; Mg2+, P = 0.3487; Ba2+, P = 0.6575; Ca2+, P = 0.6855; Cs+, P = 0.9; Li+, P = 0.0001; K+, P = 0.0002). d, Cation selectivity in C401A transfected cells (n = 9 biological replicates; Mg2+, P = 0.8969; Ba2+, P = 0.6443; Ca2+, P = 0.1272; Cs+, P = 0.0992; Li+, P = 0.0015; K+, P = 0.114). Data points and bars represent mean ± sem (two-tailed t-test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; N.S, not statistically significant).
Supplementary information
Supplementary Information
Supplementary Figs. 1–7 and corresponding figure legends.
Supplementary Video 1
Proton-induced closed-to-open conformational changes in BRTNaC1 channel simulated by MDs. Residue G392 is highlighted in green.
Source Data for Supplementary figures
The source data and unprocessed gels for supplementary figures.
Source Data for Extended Data Fig. 3
Unprocessed confocal images for Extended Data Fig. 3.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Table 2
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, X., Yuan, L., Wen, H. et al. Structure and function of a broad-range thermal receptor in myriapods. Nat Struct Mol Biol 32, 1081–1090 (2025). https://doi.org/10.1038/s41594-025-01495-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01495-8
This article is cited by
-
Thermoring basis for the proton-driven heat activation of a cation-selective channel in myriapods
Scientific Reports (2025)
-
A proton-gated channel identified in the centipede antenna
EMBO Reports (2025)