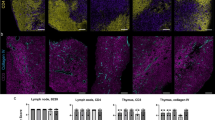
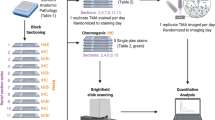
Abstract
High-content imaging is needed to catalog the variety of cellular phenotypes and multicellular ecosystems present in metazoan tissues. We recently developed iterative bleaching extends multiplexity (IBEX), an iterative immunolabeling and chemical bleaching method that enables multiplexed imaging (>65 parameters) in diverse tissues, including human organs relevant for international consortia efforts. IBEX is compatible with >250 commercially available antibodies and 16 unique fluorophores, and can be easily adopted to different imaging platforms using slides and nonproprietary imaging chambers. The overall protocol consists of iterative cycles of antibody labeling, imaging and chemical bleaching that can be completed at relatively low cost in 2–5 d by biologists with basic laboratory skills. To support widespread adoption, we provide extensive details on tissue processing, curated lists of validated antibodies and tissue-specific panels for multiplex imaging. Furthermore, instructions are included on how to automate the method using competitively priced instruments and reagents. Finally, we present a software solution for image alignment that can be executed by individuals without programming experience using open-source software and freeware. In summary, IBEX is a noncommercial method that can be readily implemented by academic laboratories and scaled to achieve high-content mapping of diverse tissues in support of a Human Reference Atlas or other such applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated during the current study are available in the Zenodo repository (https://doi.org/10.5281/zenodo.5244551).
Code availability
All custom code used in this work is freely available as open-source software under the Apache 2.0 license from the National Institute of Allergy and Infectious Diseases (NIAID) GitHub organization. The registration algorithm is available from https://github.com/niaid/sitk-ibex, and the Imaris extension code is available from https://github.com/niaid/imaris_extensions.
References
Regev, A. et al. The Human Cell Atlas. eLife https://doi.org/10.7554/eLife.27041 (2017).
Snyder, M. P. et al. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Börner, K. et al. Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat. Cell Biol. 23, 1117–1128 (2021).
Schubert, W. et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 24, 1270–1278 (2006).
Schubert, W. Topological proteomics, toponomics, MELK-technology. Adv. Biochem. Eng. Biotechnol. 83, 189–209 (2003).
Gerdes, M. J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013).
Lin, J. R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Lin, J. R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife https://doi.org/10.7554/eLife.31657 (2018).
Adams, D. L., Alpaugh, R. K., Tsai, S., Tang, C. M. & Stefansson, S. Multi-Phenotypic subtyping of circulating tumor cells using sequential fluorescent quenching and restaining. Sci. Rep. 6, 33488 (2016).
Tsujikawa, T. et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 19, 203–217 (2017).
Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science https://doi.org/10.1126/science.aar7042 (2018).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 e915 (2018).
Saka, S. K. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Taube, J. M. et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J. Immunother. Cancer https://doi.org/10.1136/jitc-2019-000155 (2020).
Tan, W. C. C. et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. (Lond.) 40, 135–153 (2020).
Bodenmiller, B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst. 2, 225–238 (2016).
Hickey, J. et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods https://doi.org/10.1038/s41592-021-01316-y (2021).
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e1319 (2020).
Kennedy-Darling, J. et al. Highly multiplexed tissue imaging using repeated oligonucleotide exchange reaction. Eur. J. Immunol. 51, 1262–1277 (2021).
Phillips, D. et al. Highly multiplexed phenotyping of immunoregulatory proteins in the tumor microenvironment by CODEX tissue imaging. Front. Immunol. 12, 687673 (2021).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387 e1319 (2018).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Radtke, A. J. et al. IBEX: a versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl Acad. Sci. USA 117, 33455–33465 (2020).
Gola, A. et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136 (2021).
Lowekamp, B. C., Chen, D. T., Ibanez, L. & Blezek, D. The design of SimpleITK. Front. Neuroinform. 7, 45 (2013).
Yaniv, Z., Lowekamp, B. C., Johnson, H. J. & Beare, R. SimpleITK Image-Analysis Notebooks: a Collaborative Environment for Education and Reproducible Research. J. Digit. Imaging 31, 290–303 (2018).
Vaughan, J. C., Jia, S. & Zhuang, X. Ultrabright photoactivatable fluorophores created by reductive caging. Nat. Methods 9, 1181–1184 (2012).
Bolognesi, M. M. et al. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J. Histochem. Cytochem. 65, 431–444 (2017).
Murray, E. et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015).
Baschong, W., Suetterlin, R. & Laeng, R. H. Control of autofluorescence of archival formaldehyde-fixed, paraffin-embedded tissue in confocal laser scanning microscopy (CLSM). J. Histochem. Cytochem. 49, 1565–1572 (2001).
Hounsell, E. F., Pickering, N. J., Stoll, M. S., Lawson, A. M. & Feizi, T. The effect of mild alkali and alkaline borohydride on the carbohydrate and peptide moieties of fetuin. Biochem. Soc. Trans. 12, 607–610 (1984).
Yakulis, V., Schmale, J., Costea, N. & Hellerp Production of Fc fragments of IgM. J. Immunol. 100, 525–529 (1968).
Corrodi, H., Hillarp, N. A. & Jonsson, G. Fluorescence methods for the histochemical demonstration of monoamines. 3. Sodium borohydride reduction of the fluorescent compounds as a specificity test. J. Histochem. Cytochem. 12, 582–586 (1964).
Nystrom, R. F., Chaikin, S. W. & Brown, W. G. Lithium borohydride as a reducing agent. J. Am. Chem. Soc. 71, 3245–3246 (1949).
Black, S. et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021).
Westra, W. H. Surgical Pathology Dissection: An Illustrated Guide (Springer, 2003).
Rood, J. E. et al. Toward a common coordinate framework for the human body. Cell 179, 1455–1467 (2019).
Lester, S. C. Manual of Surgical Pathology 3rd edn (Saunders, 2010).
Jonigk, D., Modde, F., Bockmeyer, C. L., Becker, J. U. & Lehmann, U. Optimized RNA extraction from non-deparaffinized, laser-microdissected material. Methods Mol. Biol. 755, 67–75 (2011).
Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J. & Germain, R. N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012).
Kastenmuller, W., Torabi-Parizi, P., Subramanian, N., Lammermann, T. & Germain, R. N. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell 150, 1235–1248 (2012).
Mao, K. et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018).
Baptista, A. P. et al. The chemoattractant receptor Ebi2 drives intranodal naive CD4+ T cell peripheralization to promote effective adaptive immunity. Immunity 50, 1188–1201 e1186 (2019).
Uderhardt, S., Martins, A. J., Tsang, J. S., Lammermann, T. & Germain, R. N. Resident macrophages cloak tissue microlesions to prevent neutrophil-driven inflammatory damage. Cell 177, 541–555 e517 (2019).
Petrovas, C. et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci. Transl. Med. 9, eaag2285 (2017).
Sayin, I. et al. Spatial distribution and function of T follicular regulatory cells in human lymph nodes. J. Exp. Med. 215, 1531–1542 (2018).
Radtke, A. J. et al. Lymph-node resident CD8alpha+ dendritic cells capture antigens from migratory malaria sporozoites and induce CD8+ T cell responses. PLoS Pathog. 11, e1004637 (2015).
Srivastava, S., Ghosh, S., Kagan, J., Mazurchuk, R. & National Cancer Institute’s HTAN Implementation. The making of a PreCancer Atlas: promises, challenges, and opportunities. Trends Cancer 4, 523–536 (2018).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Du, Z. et al. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat. Protoc. 14, 2900–2930 (2019).
Thevenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Guizar-Sicairos, M., Thurman, S. T. & Fienup, J. R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
McRae, T. D., Oleksyn, D., Miller, J. & Gao, Y. R. Robust blind spectral unmixing for fluorescence microscopy using unsupervised learning. PLoS One 14, e0225410 (2019).
Neher, R. A. et al. Blind source separation techniques for the decomposition of multiply labeled fluorescence images. Biophys. J. 96, 3791–3800 (2009).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), NIAID and National Cancer Institute (NCI). This research was also partially supported by a Research Collaboration Agreement (RCA) between NIAID and BioLegend, Inc. (RCA# 2020-0333) and the Chan Zuckerberg Initiative Human Cell Atlas Thymus Seed Network. C.J.C is supported as a UK-US Fulbright Scholar and Fight for Sight Research Scholar. Z.Y. and B.C.L. are supported by the Bioinformatics and Computational Biosciences Branch (BCBB) Support Services Contract HHSN316201300006W/HHSN27200002 to Medical Science & Computing, LLC. D.J. is supported by the grant of the European Research Council (ERC); European Consolidator Grant, XHale (reference #771883). We would like to thank R. Pelletier and M. Aruda from Fluigent for their sterling assistance with the ARIA fluidics device. We are grateful for the technical support provided by G. Portugal, E. Cox and E. Buck from Harvard Apparatus. We thank Dr. S. Pittaluga for her assistance with tissue grossing and orientation. We are appreciative of Drs. G. Cattoretti and M. Bolognesi for sharing their insights on fluorophore inactivation with sodium borohydride.
Author information
Authors and Affiliations
Contributions
A.J.R, C.J.C. and R.N.G. wrote the manuscript. A.J.R., C.J.C., H.I. and R.T.B. designed and executed the experiments. Z.R.Y. and B.L. developed image analysis software. L.Y. designed Figs. 1 and 2, and A.J.R. designed Figs. 3 and 4. A.G., A.J.R. and J.K. prepared supplementary videos. J.M. integrated the Leica microscope with the fluidics device. E.S., N.T., J.C., D.J., J.L.D. and J.M.H. provided technical insight, reagents and tissues. All authors offered guidance for the development and optimization of the workflows.
Corresponding authors
Ethics declarations
Competing interests
J.C. is an employee of Biolegend, Inc., and J.M. is an employee of Leica Microsystems, Inc.
Peer review
Peer review information
Nature Protocols thanks Fan Zhang, Yongxin Zhao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Radtke, A. J. et al. Proc. Natl Acad. Sci. USA 117, 33455–33465 (2020): https://doi.org/10.1073/pnas.2018488117
Gola, A. et al. Nature 589, 131–136 (2021): https://doi.org/10.1038/s41586-020-2977-2
Speranza, E. et al. Preprint at bioRxiv (2021): https://doi.org/10.1101/2021.09.08.459430
Extended data
Extended Data Fig. 1 Critical steps in the manual IBEX protocol.
a, Photo depicting central placement of tissue within a two-well chambered coverglass. Glass surface is coated with chrome gelatin alum (invisible when fully dry). b, Picture of small bubbles that form during successful LiBH4 treatment. c, Visual instructions on how to match unique nuclear shapes (Hoechst in yellow, blue box) across the imaging volumes. The left image corresponds to a live image in the Leica LAS X Navigator software. The right image corresponds to the image captured from the previous IBEX cycle. Red circles indicate that the described alignment procedure is being done at the first z-slice (‘Begin’) of the z-stack.
Extended Data Fig. 2 Equipment and assembly of imaging chamber for automated IBEX protocol.
a, Tissues are sectioned onto coated 22 × 22 mm square coverslips and assembled into the RC-21B Large Closed Bath Imaging Chamber. b,c, Top view of imaging chamber placed into PM-2 Platform for Series 20 chambers without (b) and with (c) magnetic platform clamp. d, Equipment footprint of automated IBEX setup; FLPG. e, Complete assembly of PM-2 Platform with RC-21B chamber onto SA-20PL Series 20 stage adapter. Fluid inlet and vacuum outlet highlight the fluid path. Heating electrodes are attached to the top and bottom of the platform using metal prongs that must be bent by user to allow placement into the stage. Temperature probe is inserted into small hole at top of platform to maintain 37 °C for the duration of the protocol.
Supplementary information
Supplementary Information
Tables 1–4 and legends for Supplementary Videos 1–7.
Supplementary Video 1
High dimensional imaging of human lymph node using manual IBEX method. Confocal images of human mesenteric lymph node from a 9 cycle 38 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 2
High dimensional imaging of human spleen using manual IBEX method. Confocal images of human spleen from a 4 cycle 25 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 3
High dimensional imaging of human liver using manual IBEX method. Confocal images of human liver from a 4 cycle 22 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 4
High dimensional imaging of human lymph node using automated IBEX method. THUNDER widefield images of human mesenteric lymph node from a 6 cycle 24 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 5
High dimensional imaging of human jejunum using automated IBEX method. THUNDER widefield images of human jejunum from a 6 cycle 24 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 6
High dimensional imaging of human skin using automated IBEX method. THUNDER widefield images of human skin from a 5 cycle 19 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 7
High dimensional imaging of human kidney (FFPE) using automated IBEX method. THUNDER widefield images of human FFPE kidney sections from a 5 cycle 16 parameter IBEX experiment with Hoechst serving as a fiducial.
Rights and permissions
About this article
Cite this article
Radtke, A.J., Chu, C.J., Yaniv, Z. et al. IBEX: an iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues. Nat Protoc 17, 378–401 (2022). https://doi.org/10.1038/s41596-021-00644-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-021-00644-9
This article is cited by
-
Inhibitory PD-1 axis maintains high-avidity stem-like CD8+ T cells
Nature (2026)
-
Distinct spatial organization governs oral mucosal immunity
Nature Immunology (2026)
-
Human liver and pancreas innervation: resolving 3D neurohistological challenges and advancing insights
Journal of Biomedical Science (2025)
-
Kupffer cell and recruited macrophage heterogeneity orchestrate granuloma maturation and hepatic immunity in visceral leishmaniasis
Nature Communications (2025)
-
Highly multiplexed 3D profiling of cell states and immune niches in human tumors
Nature Methods (2025)