Abstract
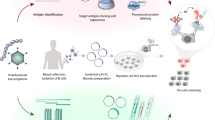
Antibody-based research applications are critical for biological discovery. Yet there are no industry standards for comparing the performance of antibodies in various applications. We describe a knockout cell line-based antibody characterization platform, developed and approved jointly by industry and academic researchers, that enables the systematic comparison of antibody performance in western blot, immunoprecipitation and immunofluorescence. The scalable protocols, which require minimal technological resources, consist of (1) the identification of appropriate cell lines for antibody characterization studies, (2) development/contribution of isogenic knockout controls, and (3) a series of antibody characterization procedures focused on the most common applications of antibodies in research. We provide examples of expected outcomes to guide antibody users in evaluating antibody performance. Central to our approach is advocating for transparent and open data sharing, enabling a community effort to identify specific antibodies for all human proteins. Mid-level graduate students with training in biochemistry and prior experience in cell culture and microscopy can complete the protocols for a specific protein within 1 month while working part-time on this effort. Antibody characterization is needed to meet standards for resource validation and data reproducibility, which are increasingly required by journals and funding agencies.
Key points
-
YCharOS is a knockout-based consensus platform for antibody characterization developed through a collaboration between academia and industry. The platform enables direct comparisons among research antibodies that target a specific protein in three common applications: western blot, immunoprecipitation and immunofluorescence.
-
The use of the YCharOS consensus protocols and open data dissemination facilitates a community-driven approach to identifying one, or ideally two, renewable and specific antibodies for each human protein.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
Underlying data can be found at Zenodo via the following links: https://doi.org/10.5281/zenodo.8356134 (ref. 49), https://doi.org/10.5281/zenodo.10844512 (ref. 50), https://doi.org/10.5281/zenodo.10108291 (ref. 51), https://doi.org/10.5281/zenodo.10149969 (ref. 52), https://doi.org/10.5281/zenodo.12666747 (ref. 53), https://doi.org/10.5281/zenodo.12636746 (ref. 54), https://doi.org/10.5281/zenodo.7459248 (ref. 55), https://doi.org/10.5281/zenodo.10835290 (ref. 56), https://doi.org/10.5281/zenodo.7671286 (ref. 57), https://doi.org/10.5281/zenodo.10835327 (ref. 58), https://doi.org/10.5281/zenodo.7459387 (ref. 59), https://doi.org/10.5281/zenodo.10838677 (ref. 60), https://doi.org/10.5281/zenodo.4724176 (ref. 61), https://doi.org/10.5281/zenodo.10845536 (ref. 62), https://doi.org/10.5281/zenodo.13151151 (ref. 63), https://doi.org/10.5281/zenodo.7971951 (ref. 64), https://doi.org/10.5281/zenodo.10839338 (ref. 65), https://doi.org/10.5281/zenodo.7459541 (ref. 66), https://doi.org/10.5281/zenodo.7671135 (ref. 67), https://doi.org/10.5281/zenodo.10819348 (ref. 68), https://doi.org/10.5281/zenodo.10927535 (ref. 69), https://doi.org/10.5281/zenodo.10819189 (ref. 70) and https://doi.org/10.5281/zenodo.10839647 (ref. 71).
Code availability
YCharOS imaging analysis scripts are available on GitHub via https://github.com/ABIF-McGill/YCharOS_IF_characterization/.
References
Baker, M. When antibodies mislead: the quest for validation. Nature 585, 313–314 (2020).
Voskuil, J. L. A. et al. The Antibody Society’s antibody validation webinar series. MAbs 12, 1794421 (2020).
Ellis, M. J. et al. Identification of high-performing antibodies for the reliable detection of Tau proteoforms by Western blotting and immunohistochemistry. Acta Neuropathol. 147, 87 (2024).
Andersson, S. et al. Insufficient antibody validation challenges oestrogen receptor beta research. Nat. Commun. 8, 15840 (2017).
Laflamme, C. et al. Implementation of an antibody characterization procedure and application to the major ALS/FTD disease gene C9ORF72. eLife https://doi.org/10.7554/eLife.48363 (2019).
Rot, A. et al. Murine bone marrow macrophages and human monocytes do not express atypical chemokine receptor 1. Cell Stem Cell 29, 1013–1015 (2022).
Virk, H. S. et al. Validation of antibodies for the specific detection of human TRPA1. Sci. Rep. 9, 18500 (2019).
Elliott, S. et al. Anti-Epo receptor antibodies do not predict Epo receptor expression. Blood 107, 1892–1895 (2006).
Schrohl, A. S., Pedersen, H. C., Jensen, S. S., Nielsen, S. L. & Brunner, N. Human epidermal growth factor receptor 2 (HER2) immunoreactivity: specificity of three pharmacodiagnostic antibodies. Histopathology 59, 975–983 (2011).
Biddle, M. S. & Virk, H. S. YCharOS open antibody characterisation data: lessons learned and progress made. F1000Res 12, 1344 (2023).
Ayoubi, R. et al. Scaling of an antibody validation procedure enables quantification of antibody performance in major research applications. eLife https://doi.org/10.7554/eLife.91645 (2023).
Biddle, M. et al. Improving the integrity and reproducibility of research that uses antibodies: a technical, data sharing, behavioral and policy challenge. MAbs 16, 2323706 (2024).
Bandrowski, A., Pairish, M., Eckmann, P., Grethe, J. & Martone, M. E. The Antibody Registry: ten years of registering antibodies. Nucleic Acids Res. 51, D358–D367 (2023).
Davies, P. et al. Comprehensive characterization and optimization of anti-LRRK2 (leucine-rich repeat kinase 2) monoclonal antibodies. Biochem. J. 453, 101–113 (2013).
Lis, P. et al. Development of phospho-specific Rab protein antibodies to monitor in vivo activity of the LRRK2 Parkinson’s disease kinase. Biochem. J. 475, 1–22 (2018).
Jong, T., Gehrlein, A., Sidransky, E., Jagasia, R. & Chen, Y. Characterization of novel human β-glucocerebrosidase antibodies for Parkinson’s disease research. J. Parkinsons Dis. 14, 65–78 (2024).
Stadler, C. et al. Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy. J. Proteomics 75, 2236–2251 (2012).
Laflamme, C., Edwards, A. M., Bandrowski, A. E. & McPherson, P. S. Opinion: independent third-party entities as a model for validation of commercial antibodies. N. Biotechnol. 65, 1–8 (2021).
Baker, M. Reproducibility crisis: blame it on the antibodies. Nature 521, 274–276 (2015).
Goodman, S. L. The antibody horror show: an introductory guide for the perplexed. N. Biotechnol. 45, 9–13 (2018).
Bradbury, A. & Pluckthun, A. Reproducibility: standardize antibodies used in research. Nature 518, 27–29 (2015).
Kwon, D. The antibodies don’t work! The race to rid labs of molecules that ruin experiments. Nature 635, 26–28 (2024).
Uhlen, M. et al. A proposal for validation of antibodies. Nat. Methods 13, 823–827 (2016).
Mitchell, K. G. et al. High-volume hybridoma sequencing on the NeuroMabSeq platform enables efficient generation of recombinant monoclonal antibodies and scFvs for neuroscience research. Sci. Rep. 13, 16200 (2023).
Bandrowski, A. et al. The Resource Identification Initiative: a cultural shift in publishing. F1000Res 4, 134 (2015).
Bandrowski, A. E. & Martone, M. E. RRIDs: a simple step toward improving reproducibility through rigor and transparency of experimental methods. Neuron 90, 434–436 (2016).
Bandrowski, A., Pairish, M., Eckmann, P., Grethe, J. & Martone, M. E. The antibody registry: ten years of registering antibodies. Nucleic Acids Res. https://doi.org/10.1093/nar/gkac927 (2022).
Longworth, S. YCharOS antibody characterization data is now linked from the CiteAb antibody search engine! CiteAB https://blog.citeab.com/ycharos-antibody-characterization-data-citeab-search-engine/ (2024).
Marx, V. Change-makers bring on recombinant antibodies. Nat. Methods 17, 763–766 (2020).
Schwende, H., Fitzke, E., Ambs, P. & Dieter, P. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J. Leukoc. Biol. 59, 555–561 (1996).
Bairoch, A. The Cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 29, 25–38 (2018).
Ruiz Moleon, V. et al. A guide to selecting high-performing antibodies for Rab27A (UniProt ID: P51159) for use in western blot, immunoprecipitation, and immunofluorescence. Zenodo https://doi.org/10.5281/zenodo.13685126 (2024).
Ayoubi, R. et al. A guide to selecting high-performing antibodies for CSNK1A1 (UniProt ID: P48729) for use in western blot, immunoprecipitation, and immunofluorescence [version 1; peer review: awaiting peer review]. F1000Res https://doi.org/10.12688/f1000research.155928.1 (2024).
Fotouhi, M. et al. Antibody characterization report for valosin-containing protein VCP. Zenodo https://doi.org/10.5281/zenodo.7971904 (2023).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Uhlen, M. et al. The human secretome. Sci. Signal https://doi.org/10.1126/scisignal.aaz0274 (2019).
Starr, T., Bauler, T. J., Malik-Kale, P. & Steele-Mortimer, O. The phorbol 12-myristate-13-acetate differentiation protocol is critical to the interaction of THP-1 macrophages with Salmonella typhimurium. PLoS ONE 13, e0193601 (2018).
Subedi, P. et al. Comparison of methods to isolate proteins from extracellular vesicles for mass spectrometry-based proteomic analyses. Anal. Biochem. 584, 113390 (2019).
Shirai, A. et al. Global analysis of gel mobility of proteins and its use in target identification. J. Biol. Chem. 283, 10745–10752 (2008).
Marcon, E. et al. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat. Methods 12, 725–731 (2015).
Sikorski, K. et al. A high-throughput pipeline for validation of antibodies. Nat. Methods 15, 909–912 (2018).
Stadler, C., Skogs, M., Brismar, H., Uhlen, M. & Lundberg, E. A single fixation protocol for proteome-wide immunofluorescence localization studies. J. Proteomics 73, 1067–1078 (2010).
Pillai-Kastoori, L., Schutz-Geschwender, A. R. & Harford, J. A. A systematic approach to quantitative western blot analysis. Anal. Biochem. 593, 113608 (2020).
Wang, M., Herrmann, C. J., Simonovic, M., Szklarczyk, D. & von Mering, C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168 (2015).
Wang, M. et al. PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell Proteomics 11, 492–500 (2012).
Antonin, W. et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19, 6453–6464 (2000).
Bonifacino, J. S., Gershlick, D. C. & Dell’Angelica, E. C. Immunoprecipitation. Curr. Protoc. Cell Biol. 71, 7.2.1–7.2.24 (2016).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Ayoubi, R. et al. Antibody characterization report for calponin-3. Zenodo https://doi.org/10.5281/zenodo.8356134 (2023).
Ayoubi, R. & Laflamme, C. Dataset for the calponin-3 antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10844512 (2024).
Ruiz Moleon, V. et al. Antibody characterization report for PLC-γ-2 (PLCG2). Zenodo https://doi.org/10.5281/zenodo.10108291 (2023).
Southern K. Dataset for the PLC-γ-2 antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10149969 (2023).
Biddle, M. S. et al. A guide to selecting high-performing antibodies for synaptotagmin-1 (Uniprot ID P21579) for use in western blot, immunoprecipitation, immunofluorescence and flow cytometry. Zenodo https://doi.org/10.5281/zenodo.12666747 (2024).
Ayoubi, R. & Laflamme, C. Dataset for the synaptotagmin-1 antibody screening study. Zenodo https://doi.org/10.5281/zenodo.12636746 (2024).
Ayoubi, R. et al. Antibody characterization report for endothelin-converting enzyme 1. Zenodo https://doi.org/10.5281/zenodo.7459248 (2022).
Ayoubi, R. & Laflamme, C. Dataset for the endothelin-converting enzyme 1 antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10835290 (2024).
Ayoubi, R., Worrall, D., McPherson, P. S. & Laflamme, C. Antibody characterization report for angiogenin. Zenodo https://doi.org/10.5281/zenodo.7671286 (2023).
Ayoubi, R. & Laflamme, C. Dataset for the angiogenin antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10835327 (2024).
Ayoubi, R. et al. Antibody characterization report for QPRTase (nicotinate–nucleotide pyrophosphorylase (carboxylating)). Zenodo https://doi.org/10.5281/zenodo.7459387 (2022).
Ayoubi, R. & Laflamme, C. Dataset for the QPRTase (nicotinate–nucleotide pyrophosphorylase (carboxylating)) antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10838677 (2024).
Ayoubi, R. et al. Antibody characterization report for plectin. Zenodo https://doi.org/10.5281/zenodo.4724176 (2021).
Ayoubi. R, Gonzalez Bolivar, S., McPherson, P. S. & Laflamme, C. Antibody characterization report for Fc receptor gamma-chain (FCER1G). Zenodo https://doi.org/10.5281/zenodo.10845536 (2024).
Ayoubi, R. & Laflamme, C. Dataset for the Fc receptor γ-chain (FCER1G) antibody screening study. Zenodo https://doi.org/10.5281/zenodo.13151151 (2024).
Ayoubi, R. et al. Antibody characterization report for Prolow-density lipoprotein receptor-related protein1 (LRP-1). Zenodo https://doi.org/10.5281/zenodo.7971951 (2023).
Ayoubi, R. & Laflamme, C. Dataset for the Prolow-density lipoprotein receptor-related protein1 (LRP-1) antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10839338 (2024).
McDowell, I. et al. Antibody characterization report for ubiquilin-2. Zenodo https://doi.org/10.5281/zenodo.7459541 (2022).
Laflamme, C. Dataset for the ubiquilin-2 antibody screening study. Zenodo https://doi.org/10.5281/zenodo.7671135 (2023).
Ayoubi, R. et al. Antibody characterization report for TGM2 (protein–glutamine γ-glutamyltransferase 2). Zenodo https://doi.org/10.5281/zenodo.10819348 (2024).
Ayoubi, R. & Laflamme, C. Dataset for the TGM2 (protein–glutamine γ-glutamyltransferase 2) antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10927535 (2024).
Ayoubi, R. et al. Antibody characterization report for sphingosine 1-phosphate receptor 1 (S1PR1). Zenodo https://doi.org/10.5281/zenodo.10819189 (2024).
Ayoubi, R. & Laflamme, C. Dataset for the sphingosine 1-phosphate receptor 1 (S1PR1) antibody screening study. Zenodo https://doi.org/10.5281/zenodo.10839647 (2024).
Acknowledgements
This work was supported by the Emory-Sage-SGC TREAT-AD center established by the National Institute on Aging grant U54AG065187 and additional support by RF1AG057443, by a grant from the Michael J. Fox Foundation for Parkinson’s Research (no. 18331), by a grant from the Motor Neurone Disease Association, the ALS Association and ALS Canada to develop the ALS-Reproducibility Antibody Platform, by the Bill and Melinda Gates Foundation and by the Government of Canada through Genome Canada, Genome Quebec and Ontario Genomics (OGI-210). The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute (grant no. OGI-196), the European Union (EU) and European Federation of Pharmaceutical Industries and Associations through the Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant no. 875510), Janssen, Merck (also known as EMD in Canada and the USA), Pfizer and Takeda. R.A. is supported by a Mitacs postdoctoral fellowship. Image processing workflows for this manuscript were developed with the McGill University Advanced BioImaging Facility (RRID: SCR_017697). CMB is supported by grant number 2020-225398 and 2023-329682 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation. A.B. is the cofounder and serves as the CEO of SciCrunch Inc, a company that works with publishers to improve the rigor and transparency of scientific manuscripts. We thank A. Bairoch at the University of Geneva and manager of the Cellosaurus database, who helped us extract from Cellosaurus the number of KO cell lines and human genes covered by KO lines.
Author information
Authors and Affiliations
Contributions
R.A., A.M.E., P.S.M. and C.L. conceptualized the workflow. R.A., C.L., J.R. and W.A. established the protocols. R.A., A.R.B. Jr., D.C., J.A.C., K.C., K.J.H., D.W.H., K.R, M.R., C.S., H.W., M.S.B, C.M.B., R.A.K., A.B. and H.S.V. contributed to the final version of the protocols. R.A., S.G.B., C.A., V.R.M., M.F. and K.S. carried out the experiments shown here. R.A. and C.L. wrote the manuscript. All authors were involved in editing of the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Fridtjof Lund-Johansen and Cecilia Williams for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Laflamme, C. et al. eLife 8 (2019): https://doi.org/10.7554/eLife.48363
Ayoubi, R. et al. eLife 12 (2023): https://doi.org/10.7554/eLife.91645
Ayoubi, R. et al. F1000Res 12, 810 (2023): https://doi.org/10.12688/f1000research.133899.3
Biddle, M. S. & Virk, H. S. F1000Res 12, 1344 (2023): https://doi.org/10.12688/f1000research.141719.1
Moleon, V.R. et al. F1000Res 12, 1578 (2023): https://doi.org/10.12688/f1000research.143928.2
Extended data
Extended Data Fig. 1 Order of sample loading for antibody screening in WB and IP-WB.
a) A scanned Ponceau S-stained membrane used for antibody screening in WB. Master mixes of MWM, WT and KO lysates were prepared, and samples were loaded in the following order on a 12-well SDS–PAGE gel: MWM, WT and KO lysates. Commercial 12-well gels have two side wells. The right side well was used here for MWM (+). Up to 4 antibodies can be tested in WB on a single 12-well gel. b) Scanned Ponceau S-stained membrane used for antibody screening in IP. Samples are loaded in the following order on a 12-wells SDS–PAGE gel. Up to 3 antibodies can be tested in IP on a single 12-well gel. Transfers from 4-20% TG gels are shown as examples in a) and b). MWM=molecular weight marker, SM=starting material, UB=unbound fraction, IP=immunoprecipitate.
Supplementary information
Supplementary Tables 1–6
This spreadsheet contains six individual sheets, each corresponding to a supplementary table. Each sheet is accompanied by a caption, as outlined in the ‘Description’ tab.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ayoubi, R., Ryan, J., Gonzalez Bolivar, S. et al. A consensus platform for antibody characterization. Nat Protoc 20, 1509–1545 (2025). https://doi.org/10.1038/s41596-024-01095-8
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41596-024-01095-8
This article is cited by
-
YCharOS protocol for antibody validation
Nature Protocols (2025)